Abstract
It is now widely appreciated that drug metabolites, in addition to the parent drugs themselves, can mediate the serious adverse effects of new therapeutic agents, as a result of which there has been heightened interest in the field of drug metabolism from researchers in academia, the pharmaceutical industry, and regulatory agencies. Much progress has been made in recent years in understanding mechanisms of toxicities caused by drug metabolites, and the numerous factors that influence individual exposure to products of drug biotransformation. This review addresses some of these factors, including the role of drug-drug interactions, reactive metabolite formation, individual susceptibility, and species differences in drug disposition caused by genetic polymorphisms in drug metabolizing enzymes. Examples are provided of adverse reactions that are linked to drug metabolism, and the mechanisms underlying variability in toxic response are discussed. Finally, some future directions for research in this field are highlighted in the context of the discovery and development of new therapeutic agents.
Keywords: drug toxicity, reactive metabolites, drug interactions, species differences, pharmacogenomics, safety evaluation
Studies on the metabolic fate of new drugs have been an integral component of the drug discovery and development process for many years, largely in recognition of the potential of circulating metabolites to serve as mediators of both the efficacy and toxicity of their respective parent compounds (1,2). While initial interest in drug metabolites focused primarily on their contribution to the pharmacology of the parent (and led to the successful development of several metabolites as drugs in their own right), the role of metabolites in adverse drug reactions has become a topic of considerable interest in recent years. Given the remarkable advances over the past two decades in analytical technologies (notably liquid chromatography-tandem mass spectrometry, LC-MS/MS) for the detection, identification, and quantitative analysis of drug metabolites in biological fluids, it has now become relatively straightforward to characterize the metabolic profile of a new chemical entity in animals and human subjects. However, the translation of chemical structure to toxicological potential remains challenging, and the effectiveness of preclinical safety assessment programs in predicting drug toxicity in man continues to be debated (3–5). Inter-species differences in drug metabolism are the rule, rather than the exception, and frequently complicate extrapolation of animal safety data to the human situation. In addition, genetic polymorphisms in drug metabolizing enzymes and transporters, both in animals and human subjects, can result in pronounced intra-species differences in drug disposition, further compounding the problem. Nevertheless, much progress has been made in gaining a scientific understanding of these issues, which has led to a deeper appreciation of the role of biotransformation in drug-induced toxicities. This review attempts to summarize current knowledge on the topic, and draws upon selected examples from the literature where metabolism of therapeutic agents is believed to have played a central role in drug-induced toxicity.
Classification of Drug-Induced Toxicities
Based upon an analysis of the primary causes of withdrawal of approved drugs and the attrition of new chemical entities during the drug development process, Park and co-workers proposed a useful classification scheme that describes broad mechanisms of drug-induced toxicities (6). More recently, Smith and Obach (7) added to this classification, such that four fundamental mechanisms of toxicity (designated ‘A’ though ‘D’) may be discerned, as shown in Table 1. Type ‘A’ toxicities result from untoward pharmacology (either on- or off-target), while Types ‘B’ through ‘D’ are believed to be triggered by a discrete chemical insult which results in a cascade of events that ultimately manifest as one of a wide variety of drug-mediated toxic effects. It is noteworthy that products of drug biotransformation are highlighted as playing a causative role in mechanisms ‘B’, ‘C’ and ‘D’, underscoring the important role that drug metabolism may play in mediating the adverse effects of therapeutic agents. It also should be noted that some Type ‘A’ toxicities result indirectly from metabolic events, particularly in cases where pharmacokinetic drug-drug interactions elevate circulating levels of one interactant to the point that an exaggerated pharmacological response, either at the intended or an unrelated biological target, leads to the expression of clinical toxicity. Some examples of these mechanisms are discussed in the following sections.
Table 1.
Mechanisms of drug-induced toxicities (adapted from Park et al, 1998, and Smith & Obach, 2009)
Type A – Normally reversible, involving a defined target leading to a predictable pharmacodynamic outcome. Two sub-types are distinguished:
|
Type B – “Idiosyncratic” drug toxicities (not predictable)
|
Type C – May occur after a single high dose
|
Type D – Occur only after prolonged dosing (carcinogenicity, teratology)
|
Role of Drug Metabolism in Drug-Mediated Organ Damage
From an anatomical and functional perspective, the liver plays a critical role in the disposition of therapeutic agents administered by the oral route, serving as a portal to the tissues, and represents the major site of drug metabolism. As pointed out above, products of drug metabolism have been implicated as causative agents in several types of toxicity and it is not surprising, therefore, that the liver represents a major site of drug-induced toxicity (8–10). Indeed, a wide range of therapeutic agents have been withdrawn from clinical use in the U. S. and elsewhere due to an unacceptably high incidence of hepatotoxicity, including aclofenac, alpidem, amadioquine, benoxaprofen, bromfenac, ibufenac, iproniazid, nefazodone, sudoxicam, tienilic acid, tolresat, troglitazone, trovafloxacin, zileuton, and zomepirac (11). Many other marketed drugs have warnings for a risk of liver toxicity, or severe restrictions in their use, and it is noteworthy that, for most hepatotoxic drugs, bioactivation to chemically reactive metabolites has been demonstrated to occur either in vitro (e.g. in human liver tissue) or in vivo (through characterization of downstream stable metabolites). It is also striking that high dose drugs, administered to patients at doses of 100 mg per day or greater, tend to be the ones which most frequently cause liver injury, while low dose drugs (given at 10 mg per day or less) rarely are problematic in this regard (11,12). For this reason, optimization of lead compounds in drug discovery programs typically focus on improving pharmacokinetics and intrinsic potency as a means of decreasing the projected efficacious clinical dose and the associated “body burden” (of both parent drug and metabolites) as a strategy for attenuating risk of toxicity.
With regard to the biochemical mechanisms by which drugs and other xenobiotics undergo conversion to chemically reactive intermediates, much of our current understanding derives from the pioneering work of Brodie, Mitchell, Gillette and their colleagues at the National Institutes of Health on the popular analgesic and antipyretic agent acetaminophen (APAP) (13). A simplified scheme depicting the metabolic fate of APAP is shown in Fig. 1, which indicates that hepatic conjugation of the phenolic -OH moiety occurs to afford the (inactive) sulfate or glucuronide derivatives (the major routes of clearance), while CYP-mediated oxidation of the drug generates the highly reactive electrophile, N-acetyl-p-benzoquinone imine (NAPQI). Under normal conditions, NAPQI is captured effectively by glutathione (GSH) to afford the corresponding S-linked adduct which, in turn, is cleared via biliary elimination. However, following ingestion of an overdose of APAP, the above detoxification pathways are overwhelmed, and liver tissue is exposed to relatively high levels of NAPQI which binds covalently to hepatic proteins, including the Keap1-Nrf2 cell defense system (14) and also serves as an intracellular oxidizing agent. A number of hypotheses have been advanced to account for the hepatotoxic properties of APAP, and while there remains lack of clarity on the detailed molecular events, it appears that the metabolic formation of NAPQI “upstream” induces cellular stress and triggers a complex series of immune-mediated responses “downstream”. These changes, in turn, perturb the balance of pro- and anti-inflammatory cytokines, ultimately bringing about the centrilobular hepatic necrosis that is characteristic of APAP overdose (9,15). While liver injury has been recognized as a serious consequence of APAP overdose (accidental or otherwise) for the past 40 years, it is remarkable that APAP-mediated hepatotoxicity is claimed to be the most common cause of acute liver failure in the United States today.
Figure 1.
Pathways of metabolism of acetaminophen (APAP), indicating the proposed role of metabolic activation to NAPQI in APAP-mediated liver injury.
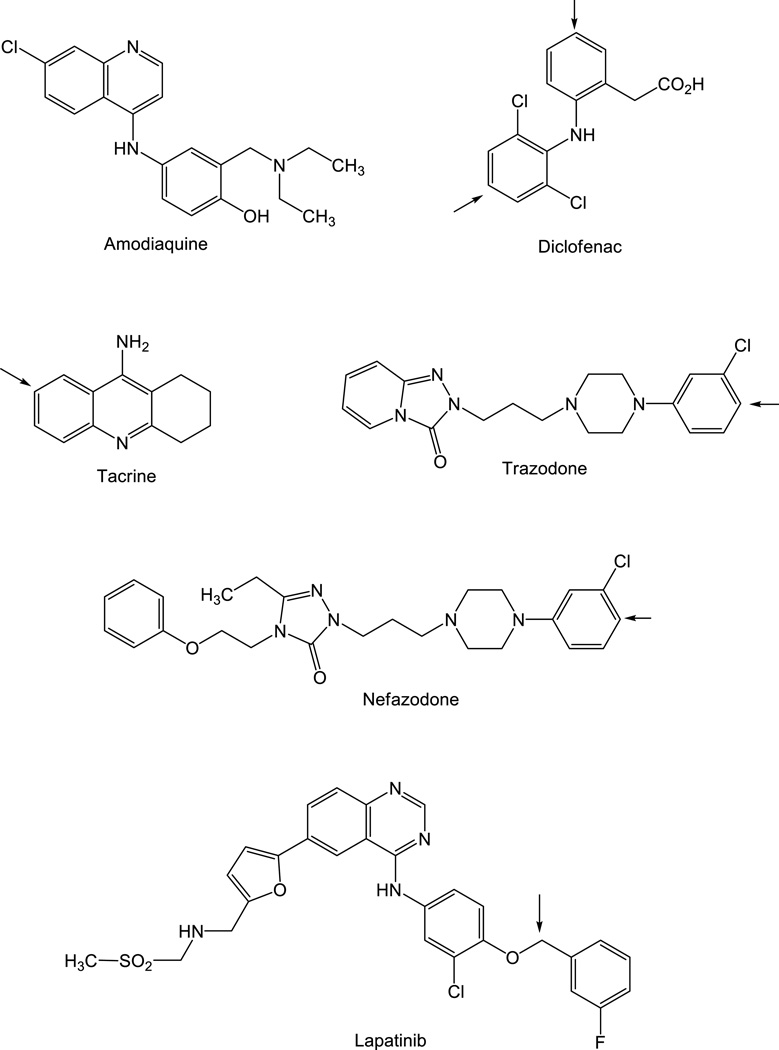
Gaining an understanding of the role of drug metabolism in the liver injury caused by APAP has been important to the field of drug-induced hepatotoxicity from several perspectives. First, it led to the development of intravenous N-acetylcysteine as an antidote for APAP poisoning, which was believed initially to function as a thiol-containing surrogate for GSH, but which was shown subsequently to act as a precursor for GSH biosynthesis and thereby to replenish GSH pools used for detoxification of NAPQI (16). Second, the p-aminophenol sub-structure contained in the APAP molecule is imbedded in many other therapeutic agents, or can be introduced (or unmasked) via metabolic reactions. Awareness of the potential for further metabolic activation of this element to yield an electrophilic quinone imine species has been invoked retrospectively to account for the hepatotoxic properties of drugs such as diclofenac (17), nefazodone (18), trazadone (19), tacrine (20), amodiaquine (21), and lapatinib (22) (Fig. 2), and also may be employed in a prospective sense in screening new chemical entities for possible bioactivation liabilities based on the detection of GSH adducts in vitro or in vivo (23). Indeed, it is now appreciated that a wide variety of compounds with heteroatom-substituted benzene rings can undergo metabolic activation (normally catalyzed by CYP enzymes) to generate electrophilic “quinoid” products (quinones, quinone imines, quinone methides, etc) that bind covalently to cellular macromolecules and, in some cases, oxidative stress via reactive oxygen species; both of these mechanisms can lead to liver toxicity (Fig. 3).
Figure 2.
Drugs which serve as precursors of quinone imine formation. In one case (amodiaquine), the p-aminophenol moiety is present in the parent compound, whereas in the other examples, metabolic oxidation (at the indicated sites) is required to introduce this structural feature.
Figure 3.
Metabolic activation of heteroatom-substituted benzene derivatives to reactive “quinoid” intermediates that can covalently modify proteins and/or cause oxidative stress. For the pathway leading to reactive oxygen species, X = oxygen or nitrogen, and Y = oxygen.
Structure-metabolism relationships such as those discussed above have led to the development of “structural alerts”, or “toxicophores” (15,24), i.e. lists of functional groups that may undergo metabolism to generate chemically reactive, potentially toxic species. Examples include thiophenes and other sulfur-containing heterocycles (via S-oxidation), furans (via epoxidation), anilines (via N- or C-oxidation), nitrobenzenes (via nitro reduction), hydrazines (via oxidation to free radical species), and some carboxylic acid derivatives (via acyl glucuronide or acyl-coenzyme A thioester formation). It should be emphasized, however, that in the context of drug discovery and lead optimization efforts, it is imperative to determine experimentally whether one of these functional groups, if present in a candidate of interest, actually is subject to metabolic activation. In many cases, a thiophene or furan substituent in a particular molecule may not be accessible to drug metabolizing enzymes, and therefore be “silent” with respect to bioactivation. In addition, acyl glucuronide conjugates differ widely in terms of their intrinsic reactivity and propensity to rearrange through acyl migration, and the mere observation of such a conjugate being formed as a metabolite of a carboxylic acid-containing drug should not necessarily be taken as a portent of liver toxicity (25).
The foregoing discussion has focused on the role of reactive, electrophilic metabolites in drug-induced liver injury where the underlying mechanism is believed to involve cellular damage through either covalent modification of critical cellular macromolecules or oxidative stress, or both. However, an additional mechanism of hepatotoxicity that has attracted considerable interest in the area of idiosyncratic drug reactions involves activation of the immune system through haptenization of proteins by reactive drug metabolites (26). Classic examples of this phenomenon are the inhalation anesthetic agent halothane, which undergoes CYP-mediated oxidation to yield trifluoroacetyl chloride, and the anti-inflammatory drug tienilic acid, which is metabolized to a thiophene-S-oxide intermediate. These reactive metabolites covalently modify hepatocellular proteins which, in turn, are recognized as foreign by the immune system and induce an antibody response. Trifluoroacetyl chloride selectively acylates free amino groups on proteins, and antibodies have been detected in the sera of patients exposed to halothane that recognize liver neoantigens containing the trifluoroacetyl moiety (27). In the case of tienilic acid, the reactive metabolite covalently modifies CYP2C9, the P450 isoform responsible for S-oxidation of the drug, resulting in the appearance of “anti-LKM2” autoantibodies in patients who developed hepatitis from tienilic acid therapy (28). Drug-induced toxicities that are mediated by the immune system are especially difficult to predict from preclinical toxicology studies due to the lack of generally applicable animal models for the human immune system. While the protein targets of some reactive metabolites are now known, much remains to be learned on this subject before a knowledge of the chemistry of metabolic activation can be linked to the toxicological outcome. Consequently, it would seem prudent, during the lead optimization phase of early drug development, to evaluate candidate molecules of interest for their propensity to generate chemically reactive metabolites, and to attempt (through appropriate structural modification) to minimize this potential liability in order to reduce the risk of downstream metabolism-dependent toxicities (29).
Whereas drug-induced liver injury has been, and remains, an area of significant concern during both drug development and post-marketing surveillance, other metabolism-based iatrogenic organ toxicities can also occur. Indeed, extra-hepatic tissue-selective toxicities caused by a variety of environmental chemicals are well documented. 3-Methylindole and 4-ipomeanol are two of the most intensively studied lung-selective toxicants, both of which require P450-mediated bioactivation to elicit their toxicity (30,31). The expression of lung-selective CYP2F and CYP4B enzymes has been associated with the metabolic activation of 3-methylindole to 3-methyleneindolenine (32) and 4-ipomeanol to its reactive ene-dial (33), respectively. These two agents are well recognized pulmonary toxins in several experimental animal species. However, extrapolation to humans of the lung-specific toxicities displayed by such model compounds is complicated by the much lower concentrations of cytochrome P450 isoforms that are present in human lung compared to rodents and other susceptible species (34). The organ-specific nephrotoxic potential of halogenated alkenes, such as trichloroethylene, has been long recognized (35). Bioactivation of haloalkenes generally involves the GSH pathway, with a major mechanism for toxicity resulting from the further processing of cysteine conjugates to reactive intermediates by renal cysteine conjugate β-lyase (36). Cytochrome P450-dependent metabolism also is implicated in the renal damage observed with numerous environmental chemicals (37). For example, chloroform can be bioactivated to the reactive metabolite, phosgene, by CYP2E1. Good evidence exists that the kidney toxicity of chloroform in mice is attributable to renal Cyp2e1 because Cyp2e1-null mice were resistant to chloroform-induced kidney toxicity (38), whereas mice harboring a liver-specific P450 reductase knockout still displayed renal damage (39). Kidney damage can also result from the physical deposition of poorly soluble metabolites, such as those derived from ethylene glycol. Case examples illustrating species differences in metabolism-based renal toxicity are provided in a later section.
Metabolic Drug-Drug Interactions and Product Withdrawals
In addition to the drug metabolism-dependent organ toxicities described above, where cellular injury is caused by one or more metabolites of a single parent compound, drug-drug interactions in polytherapy may lead to a serious adverse event. In most cases, the underlying mechanism involves inhibition of cytochrome P450 activity by one of the interactants, resulting in decreased clearance of the second agent and expression of a toxicity that normally is observed only in overdose situations. Although a number of organ systems may be affected by this mechanism, much attention in recent years has focused on the cardiovascular system.
Cardiovascular adverse events most frequently have been a consequence of delayed ventricular polarization (QTc interval prolongation) resulting from drug binding to (and inhibition of) hERG, the α-subunit of the cardiac IKr channel. Numerous approved drugs, including terodiline, sertindole, terfenadine, astemizole, grepafloxacin, cisapride, droperidol, levacetylmethadol, thioridazine, and dofetilide were withdrawn from the market as a result of adverse QTc effects, in many cases precipitated by drug-drug interactions. Perhaps the best documented example of such an interaction was with terfenadine (Seldane), a potent histamine H1 antagonist (IC50 = 1nM) introduced in the U.S. in 1985 for the treatment of seasonal allergies. While terfenadine also inhibits hERG (IC50 = 56 nM), circulating levels of the drug in humans following a therapeutic dose rarely approach toxic concentrations due to extensive first-pass metabolism to yield the corresponding active carboxylic acid metabolite, fexofenadine (IC50 at H1 = 15 nM), and so QTc prolongation with terfenadine was believed only to be an issue in cases of terfenadine overdose. However, the biotransformation of terfenadine to fexofenadine (which binds weakly to hERG) is catalyzed primarily by hepatic and intestinal CYP3A4, and can be blocked by potent inhibitors of this enzyme, e.g. erythromycin and ketoconazole (Fig. 4). Under such conditions, blood levels of terfenadine rise to values that may exceed its IC50 for hERG, with resulting prolongation of the QTc interval and, in severe cases, precipitation of life-threatening cardiotoxicity in the form of Torsades de Pointes (40). Recognition of the mechanism of this toxic drug-drug interaction led to the effective replacement of terfenadine by its active metabolite, fexofenadine (Allegra) in 1996, and terfenadine was withdrawn from the U.S. market the following year. In light of this experience, screening of new chemical entities for hERG binding has been implemented throughout the pharmaceutical industry (41), as have assays for cytochrome P450 inhibition (42) that are designed to eliminate potent P450 inhibitors as development candidates, thereby minimizing the risk of failure due to an underlying drug interaction liability (43). It should be pointed out that one potent mechanism-based P450 inhibitor, ritonavir, has found utility in anti-HIV therapy where it is used primarily to “boost” the systemic exposure of other antiretroviral drugs (44). However, the general utility of such “therapeutic enhancers” likely will be limited to life-threatening diseases due to the incidence of unintended interactions with co-administered drugs.
Figure 4.
Oxidation of terfenadine to its active carboxylic acid metabolite fexofenadine, and inhibition of this pathway by inhibitors of CYP3A4.
Inter-Species and Intra-Species Differences in Drug Metabolism
Animal studies during preclinical drug development are used to evaluate the absorption, distribution, metabolism and excretion (ADME) profile and safety of new candidate drugs. Ideally, data obtained from such animal studies can be used to screen out severe toxicities and to estimate a safe, maximum starting dose in human Phase I clinical trials. However, this begs the important question, “To what extent can animal studies predict the ADME properties of new chemical entities and (potentially related) human toxicities?” In terms of predicting human toxicities from animal data, a great deal of this information is, of course, not publicly available. Nonetheless, attempts have been made to to address this question, through compilation of available toxicity data in non-human primates, dogs, rats and mice (4). While this analysis has revealed that human cardiovascular, gastrointestinal, and blood cell toxicities are predicted successfully from animal data in more than 80% of cases, overall, about a third of global human toxicities are not predicted in ANY of the standard animal species employed for drug safety assessment. (Dogs and rats are the commonest non-rodent and rodent species, respectively, that are used for pre-clinical toxicology testing). Further analysis demonstrated that dogs are much more useful than rodents in predicting up to 150 different human toxicities. However, the dog was still not predictive for about one-third of drug toxicities reported ultimately in humans. Perhaps surprisingly, non-human primates performed no better than dogs as a predictive species for drug-related toxicities in humans (4).
Differences in drug response, including toxicity, across species (and individuals) will reflect both pharmacokinetic and pharmacodynamic variables. Drug pharmacodynamics often are highly individualized according to the pharmacological target involved, whereas pharmacokinetics are dictated by relatively conserved ADME processes. However, cross-species ADME comparisons also are complicated by a variety of factors, cogently summarized by Lin (45). For example, when considering the oral absorption of a drug, it is well recognized that dogs are poor acid secretors, whereas humans and rats are good acid secretors. Therefore, when the solubility of a drug is pH-dependent, species differences in drug absorption can be expected. In a similar vein, species differences exist in biliary excretion. Rodents and dogs are considered good biliary excretors compared to humans, and this can be reflected in species differences in the extent of fecal excretion and enterohepatic recycling. However, of more importance than biliary excretion to the overall elimination of drugs are renal and metabolic clearance. Predicting renal excretion across species is one area where allometric scaling of relative glomerular filtration rates works quite well, at least for high extraction ratio drugs. On the other hand, scaling metabolism across species is much more problematic. This is particularly unfortunate because it is estimated that some 70% of drugs are cleared primarily by metabolism (46). Moreover, metabolism often is an important determinant of toxicity through the formation of reactive (electrophilic) intermediates that may react with tissue nucleophiles and DNA (vide infra).
As noted above, metabolism dominates clearance routes, with about two-thirds of all drugs cleared primarily by Phase I and Phase II enzymes. Cytochromes P450 and UGTs dominate Phase I and Phase II processes, respectively, and in humans CYP3A4 and UGT2B7 are the specific enzymes that respectively oxidize and conjugate the majority of drugs (46). In terms of animal-to-human predictions, a sensible question would be, “Are all these same drug-metabolizing enzymes present and active in all animal species?” The short answer is NO – and indeed there are many examples of very large differences in metabolizing enzyme capacity across species. For example, species sensitivity to organophosphates (OPs) has been linked to the widely differing plasma levels of paraoxonase (PON1) which hydrolyzes many OPs. Organophosphates elicit neurotoxicity by irreversibly inactivating acetylcholinesterase in the brain, so PON1 serves a protective function against exposure to these toxins. The particularly high sensitivity of birds to OP poisoning has been attributed to their extremely low of plasma PON1 (47). On a more domestic level, inadvertent facile poisoning of cats with APAP is due to a lack of the feline UGT1A6 enzyme that conjugates and eliminates the drug, thereby permitting its build-up to toxic levels (48). Of high relevance to drug disposition studies in industry is the knowledge that dogs (and other canids) completely lack N-acetyltransferase (NAT) genes (49). Therefore, these species have a greatly reduced capacity to metabolize primary aromatic amines by the N-acetylation pathway.
Of substantial relevance to the drug discovery/development process is the extent to which cytochrome P450 activities are conserved across species, especially those species used in safety assessment, such as the rat and dog. Table 2 lists all the important human liver drug metabolizing P450s and identifies homologs in other species. The term ‘homolog’ in biology refers to a related gene, which can be identified on the basis of sequence similarities, whereas ‘ortholog’ is generally reserved for a related gene that maintains functional similarities. For example, rats possess at least six forms of CYP2D enzymes. However, none of these rat forms have the same substrate preferences as human CYP2D6, and should be described as homologs. Only CYP2E1 (and to a lesser extent the CYP1A enzymes) maintain functional similarities across species (50).
Table 2.
Homologs of the major human liver P450s present in experimental animals
| Subfamily | Human | Rat | Mouse | Dog | Monkey |
|---|---|---|---|---|---|
| CYP1A | 1A1, 1A2 | 1A1, 1A2 | 1A1, 1A2 | 1A1, 1A2 | 1A1, 1A2 |
| CYP2A | 2A6, 2A7, 2A13 | 2A1, 2A2, 2A3 | 2A4, 2A5, 2A12, 2A22 | 2A13, 2A25 | 2A23, 2A24 |
| CYP2B | 2B6 | 2B1, 2B2, 2B3 | 2B9, 2B10 | 2B11 | 2B17 |
| CYP2C | 2C8, 2C9, 2C19 | 2C6, 2C7, 2C11, 2C12 + | 2C29, 2C37, 2C38, 2C39 + | 2C21, 2C41 | 2C20, 2C43 |
| CYP2D | 2D6 | 2D1, 2D2, 2D3, 2D4, 2D5, 2D18 | 2D9, 2D10, 2D11, 2D12 + | 2D15 | 2D17, 2D19, 2D29, 2D30 |
| CYP2E | 2E1 | 2E1 | 2E1 | 2E1 | 2E1 |
| CYP3A | 3A4, 3A5, (3A7) | 3A1/23, 3A2, 3A9, 3A18 + | 3A11, 3A13 3A16 + | 3A12, 3A26 | 3A8 |
Denotes the existence of additional members of the species sub-family.
Another way to highlight inter-species differences in P450 metabolism is to compare the activity of a substrate probe for a given human enzyme across different species. Pelkonen and coworkers compared the rates of metabolism of eleven such P450 probes in liver microsomes from mouse, rat, rabbit, dog, rabbit, mini-pig, monkey and human (51), with their data demonstrating very extensive species differences in liver microsomal probe activities. Extension of this type of study to a comparison of cat, dog, horse and human liver microsomal activities revealed a similarly high species variability (52), again with no single species reflecting human metabolism of the probe substrates employed. Notably, these latter workers found extremely high dextromethorphan O-demethylase activity (a CYP2D6 probe in humans) in the horse compared to other species, and extremely low tolbutamide hydroxylase activity (a CYP2C9 probe in humans) in dogs and cats. These findings could have implications in the development of veterinary medicines.
Although no further studies have been reported on horse ‘CYP2D6’, a significant body of data exists regarding the low level of ‘CYP2C9’ activity in dogs, in part because recombinant forms of many canine P450s have been commercially available for several years. Smith and co-workers have identified well over a dozen substrates for CYP2C9, that are hydroxylated very poorly in dog liver microsomes (7). On the basis of amino acid sequence similarities, it might be suspected that dog CYP2C21 (69%) or CYP2C41 (75%) would be the dog ortholog of human CYP2C9 (53). CYP2C21, but not CYP2C41, forms 4’-hydroxy diclofenac (a CYP2C9 probe reaction in human liver microsomes), although the reaction rate is only about 0.5/min, compared to ~20/min for CYP2C9 (54). Therefore, while CYP2C21 appears to be the canine ortholog of human CYP2C9, species differences in metabolism likely occur due to low intrinsic catalytic activity of canine CYP2C21, although further studies are needed for confirmation.
In the case of non-human primates, it might be anticipated that liver P450s in humans and monkeys would be very closely related - and this does, in fact, turn out to be the case. Whereas sequence similarities between human and dog P450 ‘orthologs’ are only about 70%, monkey orthologs of human drug metabolizing P450s in the CYP1-3 families with sequence homologies as high as 95%, are readily identified (55). There are two points of caution, however. First, human CYP2C9 and CYP2C19 share >90% sequence identity, yet they catalyze quite different metabolic reactions. Second, a new monkey P450, CYP2C76, has been identified recently, but this enzyme has no ortholog in humans (55). These considerations provide a molecular basis for the species differences in P450-dependent drug metabolism that can arise between humans and non-human primates. To summarize, highly variable microsomal P450 probe metabolism across species could be a result of: (a) differences in the concentration and/or intrinsic activity of the orthologous enzyme(s) that catalyze the probe reaction, or (b) a lack of functionally similar enzymes, and metabolism by either a homologous enzyme or a completely different form of P450 in that species.
Species differences in drug metabolism take on an added significance when they are responsible for species differences in toxicity. The following three examples will illustrate this point for different Phase I and Phase II enzymes. The first example is 4-ipomeanol, a furan-containing compound that was first found to cause lung toxicity after farm animals ingested it unintentionally in their feed (56). 4-Ipomeanol became a model for xenobiotic-induced lung injury and its metabolism has been studied extensively (30). It was established more than 25 years ago that lung CYP4B1 catalyzed the bioactivation of 4-ipomeanol in animals (57), and it has been shown recently that CYP4B1 can convert ipomeanol in vitro to a reactive ene-dial (Fig. 5), presumably following initial epoxidation on the furan ring (33). In the late 1980s, 4-ipomeanol was evaluated in humans by the National Cancer Institute (NCI) as a possible treatment for non-small cell lung carcinoma (58), but the compound was found to be ineffective probably because lung CYP4B1 in humans has little or no catalytic activity (59). In other human studies, a dose-limiting liver toxicity was observed (60). What, then, is the basis for the species difference in organ toxicity? At least two possibilities can be considered; (a) alternative bioactivating liver P450 enzyme(s) in humans, or (b) a lack of detoxifying or conjugating enzyme(s) in human liver. Further studies are needed to discriminate between these possibilities.
Figure 5.
Oxidative bioactivation of 4-ipomeanol to a reactive ene-dial species.
The second example is renal toxicity of the non-nucleoside reverse transcriptase inhibitor, efavirenz. During development, efavirenz was found to cause renal tubule epithelial cell toxicity in rats, but not in monkeys or humans. The drug was metabolized extensively in all three species, but only the cysteinylglycine conjugate M10 (Fig. 6), and its precursor GSH conjugate, M9, were formed exclusively in the rat (61). This observation was critical because the kidney is the major site for processing GSH conjugates through the mercapturic acid pathway, a process that is initiated here by the conversion of M9 to M10 by the enzyme γ-glutamyl-transpeptidase (GGT). It was proposed that metabolite M10 would be subject to further metabolism under the action of a dipeptidase to generate the corresponding cysteine conjugate which, in turn, could be activated, either by S-oxidation or via the cysteine conjugate β-lyase pathway, to yield the ultimate toxic species (Fig. 6). Evidence in support of this hypothesis was obtained from studies in rats pretreated with the glutathione-S-transferase (GST) inhibitor, acivicin, which protected animals from the renal toxicity of efavirenz. It was concluded, therefore, that the basis for the species difference in efavirenz renal toxicity likely is related to species difference in GST activity between rats, monkeys and humans.
Figure 6.
Proposed pathway for the metabolism of efavirenz to a nephrotoxic metabolite(s), which is believed to result from activation of the cysteine conjugate, either through S-oxidation.
Finally, a very recent example of renal toxicity in humans arose with the experimental kinase inhibitor, SGX523 (62). When the drug’s metabolic profile was elucidated, only humans formed substantial quantities of a lactam metabolite, M11, which was determined to be formed by AO. The neutral, AO-derived lactam metabolite was very poorly soluble in patient urine, and so lactam precipitation was suggested to be the cause of the renal crystal deposits. Notably, in vitro studies with liver microsomes would not have revealed this metabolite because AO is a cytosolic enzyme, and in vivo studies in dogs and rats failed to identify M11 as a possible metabolite because AO activity is low or absent in these preclinical animal species. In summary, predicting drug metabolism and drug toxicity in humans from animal data often is problematic. Regarding metabolism, a central issue is that homologous Phase I and Phase II enzymes across species often either: (a) exhibit different substrate specificities, (b) catalyze the same reaction at very different rates, or (c) are present in liver at vastly different concentrations.
Genetic polymorphisms in drug metabolizing enzymes add a further complication in extrapolating metabolic data obtained from a sub-set of human subjects (or even animals) across the larger population. In terms of animal experimentation, this is usually a minor issue because most preclinical animal studies are conducted on inbred strains, which generally results in only a ~2–3 fold inter-animal variation in the metabolic data. Notable exceptions occur in the beagle, which exhibits a pronounced polymorphism in CYP1A2 (63), and in Gunn rats which are deficient in UGT1 activity (64).
In contrast to the typical situation in experimental animals, in humans there can be huge (100-fold) variations in a drug’s metabolic clearance when the pathway involved is under genetic control. Genetic polymorphisms in several Phase I and Phase II enzymes, notably the CYP2D6, CYP2C19, CYP2C9 and UGT1A1 enzymes, grossly affect human drug metabolism (65). The resulting phenotype in carriers of the minor allele is usually one of reduced metabolic function, although rarer polymorphisms in both the CYP2D6 and CYP2C19 genes are known that confer increased metabolic function. The consequences to the patient will also depend on whether metabolism of the parent drug terminates the desired therapeutic activity or, in the case of a prodrugs, confers the pharmacological effect. In this section, genetic polymorphisms in human drug-metabolizing enzymes will be discussed as a complicating factor in assessing drug safety, both from the perspective of drug-induced liver injury and drug-drug interactions - two important reasons for drug failures in the clinic, as noted earlier.
Several drug metabolizing enzyme (and transporter) variants are now recognized as risk factors for drug-induced liver injury. These effects presumably are manifest via an initiating toxic event at the level of reactive metabolite interaction with cellular constituents (66). A major difficulty in this area has been the accumulation of sufficient samples of confirmed pathology for analysis, because the incidence of idiosyncratic hepatotoxicity is very low. However, recently established consortia that address this problem have provided new momentum to this field along with the use of new analytical tools such as genome-wide association studies (GWAS). This latter approach recently enabled identification of the HLA-B*5701 allele as a major risk factor in the development of flucloxacillin-induced liver injury (67).
In terms of drug-metabolizing enzymes, GWAS has yet to be exploited fully, but several candidate gene studies involving isoniazid and diclofenac have appeared. For isoniazid-induced liver injury, the GSTM1 null, NAT2 slow acetylator and CYP2E1 wild-type alleles were found to be significantly associated with the adverse outcome (68,69). These findings are particularly interesting because each of the gene effects may be rationalized in terms of either increasing reactive metabolite formation or decreasing their detoxification. For diclofenac, several possible reactive intermediates have been postulated, including the 2,5- and 2,4’-quinone imines and both the parent and 4’-hydroxy-diclofenac acyl glucuronides. These metabolites may result from combined metabolism involving CYP2C8 and UGT2B7, and in fact the CYP2C8*4 and UGT2B7*2 alleles were found to be associated with diclofenac-induced liver toxicity (70). Again, the pathway analysis used here (Figure C) is compelling because the effect of both variants can be rationalized in terms of increasing toxic metabolite formation. However, these two alleles are at best low-risk genetic variants, and it is clear that interactions between multiple risk factors for idiosyncratic hepatotoxicity are needed to explain the low incidence of this undesirable iatrogenic complication to therapy (66).
Cytochrome P450 genotype is now a well recognized modifying influence on metabolic drug interactions involving CYP2D6 or CYP2C19 poor metabolizers (PMs). As reviewed by Lee et al. (71), the CYP2D6 inhibitors quinidine and fluoxetine had virtually no effect on the pharmacokinetics of the CYP2D6 substrates venlafaxine and risperidone in PMs, but increased the area under the plasma concentration-time curve (AUC) of these two drugs 4–12-fold in extensive metabolizers (EMs). Additionally, the CYP2C19 inhibitors, omeprazole and fluvoxamine, increased the AUC of moclobemide and lansoprazole by 2.2- and 3.8 fold, respectively, in CYP2C19 EMs, but had little or no effect in PMs (72,73). When there is a clear functional P450 deficit, inductive drug interactions also appear to be abrogated in PMs. An early study by the Vanderbilt group demonstrated with mephenytoin that CYP2C19 EMs exhibited a 3–8 fold increase in the ratio of urinary ratio of R:S mephenytoin and a 40–180% increase in the 0–8 hour urinary excretion of the 4’-hydroxy-mephenytoin metabolite. In stark contrast, CYP2C19 poor metabolizers did not respond to either parameter following induction with a course of rifampin (74). More recently, Tracy and coworkers have shown that changes in the oral clearance of flurbiprofen (a probe for CYP2C9) after fluconazole inhibitor co-administration decreased in the order: CYP2C9*1*1 > *1*3 > *3*3 genotypes (75).
Collectively, the foregoing in vivo examples provide the guiding principle: P450 genotype modifies the magnitude of a metabolic drug interaction as a function of the extent to which the polymorphic P450 catalyzes the target reaction. Consequently, one would anticipate that PM status for CYP2D6, CYP2C19 and CYP2C9 would afford protection against certain drug-drug interactions. This would affect approximately 7%, 2% and <1%, respectively, of Caucasian populations, with some pronounced ethnic differences apparent, especially for CYP2C19 PMs which are much more common (1–20%) in Asian populations (76). By extension, graded drug interaction responses would be expected for heterozygotes, or intermediate metabolizer phenotypes, and possibly magnified responses in ultra-rapid CYP2D6 metabolizers, although this remains to be investigated. In contrast to these CYP2 family enzymes, CYP3A4 is not subject to polymorphic variability to the extent that ultrarapid or poor metabolizer phenotype/genotypes are apparent in the general population. Therefore, genotype-dependent modulation of the serious drug-drug interactions that limited the clinical use of terfenadine (and other CYP3A4 substrates) would not be expected.
Regulatory Guidances on Metabolites in Safety Testing
The growing awareness of the potential role of drug metabolites in the toxicity of their respective parent compounds has led, in recent years, to increased scrutiny from regulatory agencies on the adequacy of safety assessment programs with regard to the toxicological properties of circulating drug metabolites. Following the publication of a “best practices” position paper on this issue in 2002 by the U. S. Pharmaceutical Research and Manufacturers Association (PhRMA) (77), which stimulated much discussion at both scientific meetings and in the peer-reviewed literature, the U. S. Food & Drug Administration (FDA) and the International Conference on Harmonisation (ICH) each published regulatory guidance documents on the safety testing of drug metabolites (78,79). Although the two documents differed in some respects (notably on the definition of what constitutes the exposure threshold above which a metabolite is deemed of interest), the FDA announced recently that they now accept the criteria set out in the ICH document (80). The emphasis of both guidance documents is on biotransformation products that circulate (or are anticipated to circulate) in human plasma, the overall goal of which is to provide assurance that preclinical safety findings are relevant to the human situation in terms of exposure to both parent drug and its circulating metabolites. At the heart of the matter is the question of predicting drug toxicity in humans from results obtained in animals, where the types of species differences in drug metabolism discussed above may confound the interpretation of animal toxicology data in terms of human risk assessment (81).
Key components of the FDA Guidance may be summarized as follows: The document applies only to small molecule non-biologic products, and excludes anti-cancer agents, drug conjugates (other than acyl glucuronides), and chemically reactive metabolites. The primary focus is on stable drug metabolites circulating in human plasma, notably those that are “unique” or “disproportionate” in humans which may require separate testing. Such evaluation (of the preformed metabolite) could include general toxicology studies (3 months duration), assessment of genotoxicity and embryo-fetal developmental toxicity and, in some instances, carcinogenicity studies. An important element of both the FDA and ICH documents relates to the definition of a metabolite of interest. In this regard, the wording of the ICH Guidance (which has now been endorsed by the FDA) states: “Nonclinical characterization of a human metabolite(s) is only warranted when that metabolite(s) is observed at exposures greater than 10 percent of total drug-related exposure and at significantly greater levels in humans than the maximum exposure seen in the toxicity studies.” The document goes on to state, “For drugs for which the daily administered dose is <10 mg, greater fractions of the drug related material might be more appropriate triggers for testing” and “The nonclinical characterization of metabolites with an identified cause for concern (e.g., a unique human metabolite) should be considered on a case-by-case basis.”
These regulatory Guidances, which address an important topic in the safety assessment of new therapeutic agents, raise a number of practical considerations for the pharmaceutical industry. For example, how does one best establish the identity and quantitative importance of individual drug metabolites in human plasma at a sufficiently early stage of clinical development that appropriate toxicological studies, where necessary, can be completed prior to the initiation of large-scale (Phase III) clinical trials? Related to this question, it should be noted that the traditional radiolabeled human ADME study typically is not conducted until after pharmacological proof-of-concept has been obtained (in Phase IIA), which leaves insufficient time to perform toxicology studies on previously unrecognized circulating metabolites with higher exposures in man relative to animals (“disproportionate metabolites”). Also, in the absence of data from a radiolabeled human ADME study, how does one assess whether a given circulating metabolite exceeds 10% of “total drug-related exposure”? One “cold” (i.e. non-radioactive) approach to the problem calls for the use of LC-MS/MS, under conditions of high mass resolution, to screen plasma samples from early (Phase I) clinical trials for components of plasma that exhibit accurate mass values consistent with being structurally related to the parent drug (82). This technique, known as “mass defect filtering” (83), represents a highly sensitive, selective, and relatively unbiased approach to the detection and identification of drug metabolites in biological fluids and, when mass spectrometric responses are calibrated by an external method (e.g. radioactivity measurements from preclinical samples), also can provide quantitative data on metabolite exposure.
Once a circulating human metabolite has been deemed to be “disproportionate” (i.e. exhibits an animal:human exposure margin less than unity), careful consideration needs to be given to the design of appropriate studies to assess its safety profile. While it is sometimes possible to escalate the dose of the parent compound in animal toxicology studies to establish required safety margins for a metabolite, or to employ a different animal species where the metabolite in question is better represented, these approaches are not always successful. In such situations, the FDA Guidance recommends performing toxicological studies with the preformed metabolite, but it is known that the dispositional characteristics of a metabolite that is administered exogenously often differ from those when the metabolite is generated in vivo from the parent drug (84). As such, dosing animals with synthetic metabolites, where feasible, runs the risk of generating data that are not relevant to the safety of the parent drug in man. Clearly, these are complex issues, and it is important that the approach that is adopted for a given development candidate is designed to generate scientifically meaningful data on the safety of circulating human metabolites, while simultaneously taking into consideration the constraints of available resources. In that context, a case-by-case approach to the issue would seem most logical.
Conclusions and Future Prospects
The 1971 Gordon Conference on Drug Metabolism opened with plenary lectures from two of the field’s founders, R. ‘Tec’ Williams and Bernard. R. Brodie, who spoke on the topics: “Factors Involved in Species Differences in Drug Metabolism” and “Biochemical Mechanisms of Drug-Induced Lesions”, respectively (Obach, personal communication). Forty years later, these two subjects continue to attract much attention, not least from scientists in the pharmaceutical industry charged with the development of new therapeutic agents. While the factors involved in species differences in drug metabolism are now reasonably well understood (vide infra), and considerable advances have occurred in our knowledge of both chemical and biochemical mechanisms of iatrogenic organ injury, prediction of the metabolic disposition and safety of new chemical entities in humans from animal studies remains a considerable challenge, although new biochemical and analytical tools continue to evolve to help mitigate this deficit.
For example, in attempts to overcome the lack of conservation of drug metabolizing functions for the major human P450s across different animal species, transgenic (or “humanized”) mice have been created, wherein the gene for a particular human P450 (e.g. CYP2D6) is introduced into their genome. CYP2D6 is a human liver P450 that preferentially metabolizes ‘basic’ compounds and, consequently, this enzyme is responsible for the metabolism of many cardiovascular and CNS-acting drugs. A common probe reaction for CYP2D6 is 4-hydroxylation of debrisoquine, a relatively old antihypertensive drug (85). Mice possess a host of CYP2D genes, but none are efficient at metabolizing debrisoquine. However, when either one or two genes for human CYP2D6 was inserted into wild-type mice, the rate of metabolism of debrisoquine to its 4-hydroxy metabolite increased greatly (86), thereby demonstrating the in vivo function of the transgene. Mice that have been humanized with respect to several other human P450s have been created, but currently remain largely a research tool as opposed to one used extensively in drug development.
Of the various categories of drug-induced toxicity classified in Table 1, the idiosyncratic, Type B, drug toxicities which have an immune basis continue to be the most difficult to rationalize and, therefore, to predict. Drug-induced immunogenicity is an especially serious concern during the development of peptide and protein drugs, and in silico and experimental approaches to preclinical screening for T-cell epitopes derived from normal cellular processing of biotherapeutics have been discussed (87). However, despite recent advances in our knowledge of the role of HLA variants as risk factors for drug-induced hypersensitivity reactions, including those that likely have a bioactivation component (88), devising a rational approach to preclinical screening of small molecules likely to posses this liability remains a daunting task. More promising, perhaps, is the development of lipopolysaccharide-based animal models for studying idiosyncratic toxicities with a strong inflammatory stress component (89) although, again, much remains to be learned about the extrapolation of data of this type to humans.
Finally, the technique of “microdosing”, in which a trace amount (typically 100 µg or less) of a 14C-labeled drug (usually < 1µCi) is administered to a human subject and radioactivity in plasma and excreta is followed by accelerator mass spectrometry, has been proposed as a highly sensitive and effective approach to assessing human drug metabolism during the lead optimization stage of drug development (90). Indeed, by means of this technique, a number of drug candidates can be assessed in humans in terms of their pharmacokinetic characteristics and metabolic profiles before one agent is selected for development. Importantly, this approach allows for metabolic profiles to be compared across species, such that differences in metabolism between humans and the animals employed in safety assessment studies can be taken into consideration in choosing the lead compound. The inclusion of patient cohorts genotyped for the common P450 polymorphisms in tracer dosing studies of this type could extend the utility of this approach to an early assessment of potential pharmacogenetic variability in candidate drug clearance, while minimizing drug exposure.
These, and other, technological developments promise to significantly advance our understanding of the molecular and biochemical basis of species differences in drug metabolism and toxicity, and thereby contribute to the development of safer, more effective therapies for unmet medical needs.
Figure 7.
Candidate gene analaysis of potential pathways of diclofenace bioactivation (70).
Acknowledgments
Development of the section on species differences in drug metabolism and toxicity was supported, in part, by National Institutes of Health grant GM49054.
Abbreviations
- APAP
Acetaminophen
- NAPQI
N-Acetyl-p-benzoquinone imine
- GSH
Glutathione
- GST
Glutathione-S-transferase
- GGT
γ-Glutamyltranspeptidase
- ADME
Adsorption, distribution, metabolism, excretion
- LC-MS/MS
Liquid chromatography – tandem mass spectrometry
- UGT
Uridine dinucleotide phosphate glucuronosyltransferase
- OP
Organophosphate
- PON1
Paraoxonase-1
- AO
Aldehyde oxidase
- NCI
National Cancer Institute
- GWAS
Genome-wide association studies
- NAT
N-Acetyltransferase
- PM
Poor metabolizer phenotype
- EM
Extensive metabolizer phenotype
- PhRMA
Pharmaceutical Research and Manufacturers Association (U.S.A.)
- AUC
Area under the plasma concentration versus time curve
- FDA
U.S. Food and Drug Administration
- ICH
International Conference on Harmonisation
- CNS
Central nervous system
References
- 1.Baillie TA. Metabolism and toxicity of drugs. Two decades of progress in industrial drug metabolism. Chem. Res. Toxicol. 2008;21:129–137. doi: 10.1021/tx7002273. [DOI] [PubMed] [Google Scholar]
- 2.Korfmacher WA. Advances in the integration of drug metabolism into the lead optimization paradigm. Mini-Rev. Med. Chem. 2009;9:703–716. doi: 10.2174/138955709788452694. [DOI] [PubMed] [Google Scholar]
- 3.Olson H, Betton G, Robinson D, Thomas K, Monro A, Kolaja G, Lilly P, Sanders J, Sipes G, Bracken W, Dorato M, Van Deun K, Smith P, Berger B, Heller A. Concordance of the toxicity of pharmaceuticals in humans and in animals. Reg. Toxicol. Pharmacol. 2000;32:56–67. doi: 10.1006/rtph.2000.1399. [DOI] [PubMed] [Google Scholar]
- 4.Greaves P, Williams A, Eve M. First dose of potential new medicines to humans: how animals help. Nature Rev. Drug Discov. 2004;3:226–236. doi: 10.1038/nrd1329. [DOI] [PubMed] [Google Scholar]
- 5.Peters TS. Do preclinical testing strategies help predict human hepatotoxic potentials? Toxicol. Pathol. 2005;33:146–154. doi: 10.1080/01926230590522121. [DOI] [PubMed] [Google Scholar]
- 6.Park BK, Pirmohamed M, Ketteringham N. Role of drug disposition in drug hypersensitivity: a chemical, molecular, and clinical perspective. Chem. Res. Toxicol. 1998;11:969–988. doi: 10.1021/tx980058f. [DOI] [PubMed] [Google Scholar]
- 7.Smith DA, Obach RS. Metabolites in safety testing: considerations of mechanisms of toxicity with dose, abundance, and duration of treatment. Chem. Res. Toxicol. 2009;22:267–279. doi: 10.1021/tx800415j. [DOI] [PubMed] [Google Scholar]
- 8.Lee WM. Drug-induced hepatotoxicity. New Eng. J. Med. 2003;349:474–485. doi: 10.1056/NEJMra021844. [DOI] [PubMed] [Google Scholar]
- 9.Kaplowitz N. Idiosyncratic drug hepatotoxicity. Nature Rev. Drug Discov. 2005;4:489–499. doi: 10.1038/nrd1750. [DOI] [PubMed] [Google Scholar]
- 10.Stevens J, Baker TK. The future of drug safety testing: expanding the view and narrowing the focus. Drug Discov. Today. 2009;14:162–167. doi: 10.1016/j.drudis.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 11.Smith DA, Schmid EF. Drug withdrawals and the lessons within. Curr. Opin. Drug Discov. Develop. 2006;9:38–46. [PubMed] [Google Scholar]
- 12.Uetrecht J. Screening for the potential of a drug candidate to cause idiosyncratic drug reactions. Drug Discov. Today. 2003;8:832–837. doi: 10.1016/s1359-6446(03)02816-2. [DOI] [PubMed] [Google Scholar]
- 13.Mitchell JR, Jollow DJ, Potter WZ, Davis DC, Gillette JR, Brodie BB. Acetaminophen-induced hepatic necrosis. I. Role of drug metabolism. J. Pharmacol. Exp. Ther. 1973;187:185–194. [PubMed] [Google Scholar]
- 14.Copple IM, Goldring CE, Jenkins RE, Chia AJL, Randle LE, Hayes JD, Kitteringham NR, Park BK. The hepatotoxic metabolite of acetaminophen directly activates the Keap1-Nrf2 cell defense system. Hepatology. 2008;48:1292–1301. doi: 10.1002/hep.22472. [DOI] [PubMed] [Google Scholar]
- 15.Williams DP. Toxicophores: investigations in drug safety. Toxicology. 2006;226:1–11. doi: 10.1016/j.tox.2006.05.101. [DOI] [PubMed] [Google Scholar]
- 16.Corcoran GB, Wong BK. Role of glutathione in prevention of acetaminophen-induced hepatotoxicity by N-acetyl-L-cysteine in vivo: studies with N-acetyl-D-cysteine in mice. J. Pharmacol. Exp. Ther. 1986;238:54–61. [PubMed] [Google Scholar]
- 17.Tang W. The metabolism of diclofenac – enzymology and toxicology perspectives. Curr. Drug Metab. 2003;4:319–329. doi: 10.2174/1389200033489398. [DOI] [PubMed] [Google Scholar]
- 18.Kalgutkar AS, Vaz ADN, Lame ME, Henne KR, Soglia J, Zhao SX, Abramov YA, Lombardo F, Collin C, Hendsch ZS, Hop CECA. Bioactivation of the nontricyclic antidepressant nefazodone to a reactive quinone-imine species in human liver microsomes and recombinant cytochrome P450 3A4. Drug Metab. Dispos. 2005a;33:243–253. doi: 10.1124/dmd.104.001735. [DOI] [PubMed] [Google Scholar]
- 19.Kalgutkar AS, Henne KR, Lame ME, Vaz AD, Collin C, Soglia JR, Zhao SX, Hop CE. Metabolic activation of the nontricyclic antidepressant trazodone to electrophilic quinone-imine and epoxide intermediates in human liver microsomes and recombinant P4503A4. Chem.-Biol. Interact. 2005b;155:10–20. doi: 10.1016/j.cbi.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 20.Madden S, Spaldin V, Hayes RN, Woolf TF, Pool WF, Park BK. Species variation in the bioactivation of tacrine by hepatic microsomes. Xenobiotica. 1995;25:103–116. doi: 10.3109/00498259509061837. [DOI] [PubMed] [Google Scholar]
- 21.Jewell H, Maggs JL, Harrison AC, O’Neill PM, Ruscoe JE, Park BK. Role of hepatic metabolism in the bioactivation and detoxication of amodiaquine. Xenobiotica. 1995;25:199–217. doi: 10.3109/00498259509061845. [DOI] [PubMed] [Google Scholar]
- 22.Teng WC, Oh JW, New LS, Wahlin MD, Nelson SD, Ho HK, Chan ECY. Mechanism-based inactivation of cytochrome P450 3A4 by lapatinib. Mol. Pharmacol. 2010 doi: 10.1124/mol.110.065839. [DOI] [PubMed] [Google Scholar]
- 23.Kumar S, Mitra K, Kassahun K, Baillie TA. Approaches for minimizing metabolic activation of new drug candidates in drug discovery. In: Uetrecht J, editor. Adverse Drug Reactions, Handbook of Experimental Pharmacology. Vol. 196. Springer-Verlag; 2010. pp. 511–544. [DOI] [PubMed] [Google Scholar]
- 24.Nelson SD. Structure toxicity relationships – how useful are they in predicting toxicities of new drugs? In: Dansette PM, Snyder R, Delaforge M, Gibson GG, Greim H, Jollow DJ, Monks TJ, Sipes IG, editors. Biological Reactive intermediates VI. Kluwer Academic / Plenum Publishers; 2001. pp. 33–43. [Google Scholar]
- 25.Sawamura R, Okudaira N, Watanabe K, Murai T, Kobayashi Y, Tachibana M, Ohnuki T, Masuda K, Honma H, Kurihara A, Okazaki O. Predictability of idiosyncratic drug toxicity risk for carboxylic acid-containing drugs based on the chemical stability of acyl glucuronide. Drug Metab. Dispos. 2010 doi: 10.1124/dmd.110.034173. [DOI] [PubMed] [Google Scholar]
- 26.Uetrecht J. Idiosyncratic drug reactions: past, present, and future. Chem. Res. Toxicol. 2008;21:84–92. doi: 10.1021/tx700186p. [DOI] [PubMed] [Google Scholar]
- 27.Kenna JG, Satoh H, Christ DD, Pohl LR. Metabolic basis for a drug hypersensitivity: antibodies in sera from patients with halothane hepatitis recognize liver neoantigens that contain the trifluoroacetyl group derived from halothane. J. Pharmacol. Exp. Ther. 1988;245:1103–1109. [PubMed] [Google Scholar]
- 28.Lecoeur S, André C, Beaune PH. Tienilic acid-induced autoimmune hepatitis: anti-liver and -kidney microsomal type 2 autoantibodies recognize a three-site conformational epitope on cytochrome P4502C9. Mol. Pharmacol. 1996;50:326–333. [PubMed] [Google Scholar]
- 29.Kumar S, Kassahun K, Tschirret-Guth RA, Mitra K, Baillie TA. Minimizing metabolic activation during pharmaceutical lead optimization: progress, knowledge gaps and future directions. Curr. Opin. Drug Discov. Develop. 2008;11:43–52. [PubMed] [Google Scholar]
- 30.Gram TE. Chemically reactive intermediates and pulmonary xenobiotic toxicity. Pharmacol. Rev. 1997;49:297–341. [PubMed] [Google Scholar]
- 31.Yost GS. Mechanisms of 3-methylindole pneumotoxicity. Chem. Res. Toxicol. 1989;2:273–279. doi: 10.1021/tx00011a001. [DOI] [PubMed] [Google Scholar]
- 32.Lanza DL, Yost GS. Selective dehydrogenation/oxygenation of 3-methylindole by cytochrome P450 enzymes. Drug Metab. Dispos. 2001;29:950–953. [PubMed] [Google Scholar]
- 33.Baer BR, Rettie AE, Henne KR. Bioactivation of 4-ipomeanol by CYP4B1: adduct characterization and evidence for an enedial intermediate. Chem. Res. Toxicol. 2005;18:855–864. doi: 10.1021/tx0496993. [DOI] [PubMed] [Google Scholar]
- 34.Buckpitt A, Boland B, Isbell M, Morin D, Shultz M, Baldwin R, Chan K, Karlsson A, Lin C, Taff A, West J, Fanucchi M, Van Winkle L, Plopper C. Naphthalene-induced respiratory tract toxicity: metabolic mechanisms of toxicity. Drug Metab. Rev. 2002;34:791–820. doi: 10.1081/dmr-120015694. [DOI] [PubMed] [Google Scholar]
- 35.Koren G. The nephrotoxic potential of drugs and chemicals. Pharmacological basis and clinical relevance. Med. Toxicol. Adverse Drug Exp. 1989;4:59–72. doi: 10.1007/BF03259903. [DOI] [PubMed] [Google Scholar]
- 36.Anders MW, Dekant W. Glutathione-dependent bioactivation of haloalkenes. Annu. Rev. Pharmacol. Toxicol. 1998;38:501–537. doi: 10.1146/annurev.pharmtox.38.1.501. [DOI] [PubMed] [Google Scholar]
- 37.Van Vleet TR, Schnellmann RG. Toxic nephropathy: environmental chemicals. Semin. Nephrol. 2003;23:500–508. doi: 10.1016/s0270-9295(03)00094-9. [DOI] [PubMed] [Google Scholar]
- 38.Constan AA, Sprankle CS, Peters JM, Kedderis GL, Everitt JI, Wong BA, Gonzalez FL, Butterworth BE. Metabolism of chloroform by cytochrome P450 2E1 is required for induction of toxicity in the liver, kidney, and nose of male mice. Toxicol. Appl. Pharmacol. 1999;160:120–126. doi: 10.1006/taap.1999.8756. [DOI] [PubMed] [Google Scholar]
- 39.Fang C, Behr M, Xie F, Lu S, Doret M, Luo H, Yang W, Aldous K, Ding X, Gu J. Mechanism of chloroform-induced renal toxicity: non-involvement of hepatic cytochrome P450-dependent metabolism. Toxicol. Appl. Pharmacol. 2008;227:48–55. doi: 10.1016/j.taap.2007.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Monahan BP, Ferguson CL, Killeavy ES, Lloyd BK, Troy J, Cantilena LR. Torsades de pointes occurring in association with terfenadine use. J. Amer. Med. Assoc. 1990;264:2788–2790. [PubMed] [Google Scholar]
- 41.Lagrutta AA, Trepakova ES, Salata J. The hERG channel and risk of drug-acquired cardiac arrhythmia: an overview. Curr. Topics Med. Chem. 2008;8:1102–1112. doi: 10.2174/156802608785700016. [DOI] [PubMed] [Google Scholar]
- 42.Björnsson TD, Callaghan JT, Einolf HJ, Fischer V, Gan L, Grimm S, Kao J, King SP, Miwa G, Ni L, Kumar G, McLeod J, Obach RS, Roberts S, Roe A, Shah A, Snickeris F, Sullivan JT, Tweedie D, Vega JM, Walsh J, Wrighton SA. The conduct of in vitro and in vivo drug-drug interaction studies: a PhRMA perspective. J. Clin. Pharmacol. 2003;43:443–469. [PubMed] [Google Scholar]
- 43.Huang S-M, Temple R, Throckmorton DC, Lesko LJ. Drug interaction studies: study design, data analysis, and implications for dosing and labeling. Clin. Pharm. Ther. 2007;81:298–304. doi: 10.1038/sj.clpt.6100054. [DOI] [PubMed] [Google Scholar]
- 44.Koudriakova T, Iatsimirskaia E, Utkin I, Gangl E, Vouros P, Storozhuk E, Orza D, Marinina J, Gerber N. Metabolism of the human immunodeficiency virus protease inhibitors indinavir and ritonavir by human intestinal microsomes and expressed cytochrome P4503A4/3A5: mechanism-based inactivation of cytochrome P4503A4 by ritonavir. Drug Metab. Dispos. 1998;26:552–561. [PubMed] [Google Scholar]
- 45.Lin JH. Species similarities and differences in pharmacokinetics. Drug Metab. Dispos. 1995;23:1008–1021. [PubMed] [Google Scholar]
- 46.Williams JA, Hyland R, Jones BC, Smith DA, Hurst S, Goosen TC, Peterkin V, Koup JR, Ball SE. Drug-drug interactions for UDP-glucuronosyltransferase substrates: a pharmacokinetic explanation for typically observed low exposure (AUCi/AUC) ratios. Drug Metab. Dispos. 2004;32:1201–1208. doi: 10.1124/dmd.104.000794. [DOI] [PubMed] [Google Scholar]
- 47.Costa LG, Richter RJ, Li WF, Cole T, Guizzetti M, Furlong CE. Paraoxonase (PON 1) as a biomarker of susceptibility for organophosphate toxicity. Biomarkers. 2003;8:1–12. doi: 10.1080/13547500210148315. [DOI] [PubMed] [Google Scholar]
- 48.Court MH, Greenblatt DJ. Molecular genetic basis for deficient acetaminophen glucuronidation by cats: UGT1A6 is a pseudogene, and evidence for reduced diversity of expressed hepatic UGT1A isoforms. Pharmacogenetics. 2000;10:355–369. doi: 10.1097/00008571-200006000-00009. [DOI] [PubMed] [Google Scholar]
- 49.Trepanier LA, Ray K, Winand NJ, Spielberg SP, Cribb AE. Cytosolic arylamine N-acetyltransferase (NAT) deficiency in the dog and other canids due to an absence of NAT genes. Biochem. Pharmacol. 1997;54:73–80. doi: 10.1016/s0006-2952(97)00140-8. [DOI] [PubMed] [Google Scholar]
- 50.Martignoni M, Groothuis GM, de Kanter R. Species differences between mouse, rat, dog, monkey and human CYP-mediated drug metabolism, inhibition and induction. Expert Opin. Drug Metab. Toxicol. 2006;2:875–894. doi: 10.1517/17425255.2.6.875. [DOI] [PubMed] [Google Scholar]
- 51.Turpeinen M, Ghiciuc C, Opritoui M, Tursas L, Pelkonen O, Pasanen M. Predictive value of animal models for human cytochrome P450 (CYP)-mediated metabolism: a comparative study in vitro. Xenobiotica. 2007;37:1367–1377. doi: 10.1080/00498250701658312. [DOI] [PubMed] [Google Scholar]
- 52.Chauret N, Gauthier A, Martin J, Nicoll-Griffith DA. In vitro comparison of cytochrome P450-mediated metabolic activities in human, dog, cat, and horse. Drug Metab. Dispos. 1997;25:1130–1136. [PubMed] [Google Scholar]
- 53.Shou M, Norcross R, Sandig G, Lu P, Li Y, Lin Y, Mei Q, Rodrigues AD, Rushmore TH. Substrate specificity and kinetic properties of seven heterologously expressed dog cytochromes P450. Drug Metab. Dispos. 2003;31:1161–1169. doi: 10.1124/dmd.31.9.1161. [DOI] [PubMed] [Google Scholar]
- 54.Haining RL, Jones JP, Henne KR, Fisher MB, Koop DR, Trager WF, Rettie AE. Enzymatic determinants of the substrate specificity of CYP2C9: role of B'-C loop residues in providing the pi-stacking anchor site for warfarin binding. Biochemistry. 1999;38:3285–3292. doi: 10.1021/bi982161+. [DOI] [PubMed] [Google Scholar]
- 55.Iwasaki K, Uno Y. Cynomolgus monkey CYPs: a comparison with human CYPs. Xenobiotica. 2009;39:578–581. doi: 10.1080/00498250903003135. [DOI] [PubMed] [Google Scholar]
- 56.Boyd MR, Wilson BJ. Isolation and characterization of 4-ipomeanol, a lung-toxic furanoterpenoid produced by sweet potatoes (Ipomoea batatas) J Agric. Food Chem. 1972;20:428–430. doi: 10.1021/jf60180a066. [DOI] [PubMed] [Google Scholar]
- 57.Wolf CR, Statham CN, McMenamin MG, Bend JR, Boyd MR, Philpot RM. The relationship between the catalytic activities of rabbit pulmonary cytochrome P-450 isozymes and the lung-specific toxicity of the furan derivative, 4-ipomeanol. Mol. Pharmacol. 1982;22:738–744. [PubMed] [Google Scholar]
- 58.Christian MC, Wittes RE, Leyland-Jones B, McLemore TL, Smith AC, Grieshaber CK, Chabner BA, Boyd MR. 4-Ipomeanol: a novel investigational new drug for lung cancer. J. Natl. Cancer Inst. 1989;81:1133–1143. doi: 10.1093/jnci/81.15.1133. [DOI] [PubMed] [Google Scholar]
- 59.Zheng YM, Fisher MB, Yokotani N, Fujii-Kuriyama Y, Rettie AE. Identification of a meander region proline residue critical for heme binding to cytochrome P450: implications for the catalytic function of human CYP4B1. Biochemistry. 1998;37:12847–51. doi: 10.1021/bi981280m. [DOI] [PubMed] [Google Scholar]
- 60.Rowinsky EK, Noe DA, Ettinger DS, Christian MC, Lubejko BG, Fishman EK, Sartorius SE, Boyd MR, Donehower RC. Phase I and pharmacological study of the pulmonary cytotoxin 4-ipomeanol on a single dose schedule in lung cancer patients: hepatotoxicity is dose limiting in humans. Cancer Res. 1993;53:1794–1801. [PubMed] [Google Scholar]
- 61.Mutlib AE, Gerson RJ, Meunier PC, Haley PJ, Chen H, Gan LS, Davies MH, Gemzik B, Christ DD, Krahn DF, Markwalder JA, Seitz SP, Robertson RT, Miwa GT. The species-dependent metabolism of efavirenz produces a nephrotoxic glutathione conjugate in rats. Toxicol. Appl. Pharmacol. 2000;169:102–113. doi: 10.1006/taap.2000.9055. [DOI] [PubMed] [Google Scholar]
- 62.Diamond S, Boer J, Maduskuie TP, Jr, Falahatpisheh N, Li Y, Yeleswaram S. Species-specific metabolism of SGX523 by aldehyde oxidase and the toxicological implications. Drug Metab. Dispos. 2010;38:1277–1285. doi: 10.1124/dmd.110.032375. [DOI] [PubMed] [Google Scholar]
- 63.Mise M, Hashizume T, Matsumoto S, Terauchi Y, Fujii T. Identification of non-functional allelic variant of CYP1A2 in dogs. Pharmacogenetics. 2004;14:769–773. doi: 10.1097/00008571-200411000-00008. [DOI] [PubMed] [Google Scholar]
- 64.Iyanagi T. Molecular basis of multiple UDP-glucuronosyltransferase isoenzyme deficiencies in the hyperbilirubinemic rat (Gunn rat) J. Biol. Chem. 1991;266:24048–24052. [PubMed] [Google Scholar]
- 65.Evans WE, Relling MV. Pharmacogenomics: translating functional genomics into rational therapeutics. Science. 1999;286:487–491. doi: 10.1126/science.286.5439.487. [DOI] [PubMed] [Google Scholar]
- 66.Russmann S, Jetter A, Kullak-Ublick GA. Pharmacogenetics of drug-induced liver injury. Hepatology. 2010;52:748–761. doi: 10.1002/hep.23720. [DOI] [PubMed] [Google Scholar]
- 67.Daly AK, Donaldson PT, Bhatnagar P, Shen Y, Pe'er I, Floratos A, Daly MJ, Goldstein DB, John S, Nelson MR, Graham J, Park BK, Dillon JF, Bernal W, Cordell HJ, Pirmohamed M, Aithal GP, Day CP. HLA-B*5701 genotype is a major determinant of drug-induced liver injury due to flucloxacillin. Nat. Genet. 2009;41:816–819. doi: 10.1038/ng.379. [DOI] [PubMed] [Google Scholar]
- 68.Huang YS, Chern HD, Su WJ, Wu JC, Chang SC, Chiang CH, Chang FY, Lee SD. Cytochrome P450 2E1 genotype and the susceptibility to antituberculosis drug-induced hepatitis. Hepatology. 2003;37:924–930. doi: 10.1053/jhep.2003.50144. [DOI] [PubMed] [Google Scholar]
- 69.Huang YS. Genetic polymorphisms of drug-metabolizing enzymes and the susceptibility to antituberculosis drug-induced liver injury. Expert Opin. Drug Metab. Toxicol. 2007;3:1–8. doi: 10.1517/17425255.3.1.1. [DOI] [PubMed] [Google Scholar]
- 70.Daly AK, Aithal GP, Leathart JB, Swainsbury RA, Dang TS, Day CP. Genetic susceptibility to diclofenac-induced hepatotoxicity: contribution of UGT2B7, CYP2C8, and ABCC2 genotypes. Gastroenterology. 2007;132:272–281. doi: 10.1053/j.gastro.2006.11.023. [DOI] [PubMed] [Google Scholar]
- 71.Lee LS, Nafziger AN, Bertino JS., Jr Evaluation of inhibitory drug interactions during drug development: genetic polymorphisms must be considered. Clin. Pharmacol. Ther. 2005;78:1–6. doi: 10.1016/j.clpt.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 72.Yu KS, Yim DS, Cho JY, Park SS, Park JY, Lee KH, Jang IJ, Yi SY, Bae KS, Shin SG. Effect of omeprazole on the pharmacokinetics of moclobemide according to the genetic polymorphism of CYP2C19. Clin. Pharmacol. Ther. 2001;69:266–73. doi: 10.1067/mcp.2001.114231. [DOI] [PubMed] [Google Scholar]
- 73.Yasui-Furukori N, Saito M, Uno T, Takahata T, Sugawara K, Tateishi T. Effects of fluvoxamine on lansoprazole pharmacokinetics in relation to CYP2C19 genotypes. J. Clin. Pharmacol. 2004;44:1223–1229. doi: 10.1177/0091270004269015. [DOI] [PubMed] [Google Scholar]
- 74.Zhou HH, Anthony LB, Wood AJ, Wilkinson GR. Induction of polymorphic 4'-hydroxylation of S-mephenytoin by rifampicin. Br. J. Clin. Pharmacol. 1990;30:471–475. doi: 10.1111/j.1365-2125.1990.tb03799.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kumar V, Brundage RC, Oetting WS, Leppik IE, Tracy TS. Differential genotype dependent inhibition of CYP2C9 in humans. Drug Metab. Dispos. 2008;36:1242–1248. doi: 10.1124/dmd.108.020396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mizutani T. PM frequencies of major CYPs in Asians and Caucasians. Drug Metab. Rev. 2003;35:99–106. doi: 10.1081/dmr-120023681. [DOI] [PubMed] [Google Scholar]
- 77.Baillie TA, Cayen MN, Fouda H, Gerson RJ, Grossman SJ, Klunk LJ, LeBlanc B, Perkins DG, Shipley LA. Contemporary issues in toxicology. Drug metabolites in safety testing. Toxicol. Appl. Pharmacol. 2002;182:188–196. doi: 10.1006/taap.2002.9440. [DOI] [PubMed] [Google Scholar]
- 78.U.S. Food and Drug Administration. Guidance for industry. Safety testing of drug metabolites. 2008 http://www.fda.gov/downloads/Drugs/GuidanceComplianceRegulatoryInformation/Guidances/ucm079266.pdf.
- 79.International Conference on Harmonisation. ICH Topic M3 (R2): Non-clinical safety studies for the conduct of human clinical trials and marketing authorization for pharmaceuticals. 2009 http://www.emea.europa.eu/pdfs/human/ich/028695en.pdf. [PubMed] [Google Scholar]
- 80.U.S. Food and Drug Administration. Guidance for industry. M3(R2) nonclinical safety studies for the conduct of human clinical trials and marketing authorization for pharmaceuticals. 2010 http://www.fda.gov/downloads/Drugs/GuidanceComplianceRegulatoryInformation/Guidances/ucm073246.pdf. [PubMed]
- 81.Guengerich FP, MacDonald JS. Applying mechanisms of chemical toxicity to predict drug safety. Chem. Res. Toxicol. 2007;20:344–369. doi: 10.1021/tx600260a. [DOI] [PubMed] [Google Scholar]
- 82.Baillie TA. Approaches to the assessment of stable and chemically reactive drug metabolites in early clinical trials. Chem. Res. Toxicol. 2009;22:263–266. doi: 10.1021/tx800439k. [DOI] [PubMed] [Google Scholar]
- 83.Zhu M, Ma L, Zhang D, Ray K, Zhao W, Humphreys WG, Skiles G, Sanders M, Zhang H. Detection and characterization of metabolites in biological matrices using mass defect filtering of liquid chromatography / high resolution mass spectrometry data. Drug Metab. Dispos. 34:1722–1733. doi: 10.1124/dmd.106.009241. [DOI] [PubMed] [Google Scholar]
- 84.Prueksaritanont T, Lin JH, Baillie TA. Complicating factors in safety testing of drug metabolites: kinetic differences between generated and preformed metabolites. Toxicol. Appl. Pharmacol. 2006;217:143–152. doi: 10.1016/j.taap.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 85.Frank D, Jaehde U, Fuhr U. Evaluation of probe drugs and pharmacokinetic metrics for CYP2D6 phenotyping. Eur. J. Clin. Pharmacol. 2007;63:321–333. doi: 10.1007/s00228-006-0250-8. [DOI] [PubMed] [Google Scholar]
- 86.Corchero J, Granvil CP, Akiyama TE, Hayhurst GP, Pimprale S, Feigenbaum L, Idle JR, Gonzalez FJ. The CYP2D6 humanized mouse: effect of the human CYP2D6 transgene and HNF4alpha on the disposition of debrisoquine in the mouse. Mol Pharmacol. 2001;60:1260–1267. doi: 10.1124/mol.60.6.1260. [DOI] [PubMed] [Google Scholar]
- 87.Kropshofer H, Singer T. Overview of cell-based tools for pre-clinical assessment of immunogenicity of biotherapeutics. J. Immunotoxicol. 2006;3:131–136. doi: 10.1080/15476910600845625. [DOI] [PubMed] [Google Scholar]
- 88.Hung SI, Chung WH, Liu ZS, Chen CH, Hsih MS, Hui RC, Chu CY, Chen YT. Common risk allele in aromatic antiepileptic-drug induced Stevens-Johnson syndrome and toxic epidermal necrolysis in Han Chinese. Pharmacogenomics. 2010;11:349–356. doi: 10.2217/pgs.09.162. [DOI] [PubMed] [Google Scholar]
- 89.Deng X, Luyendyk JP, Ganey PE, Roth RA. Inflammatory stress and idiosyncratic hepatotoxicity: hints from animal models. Pharmacol. Rev. 2009;61:262–282. doi: 10.1124/pr.109.001727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lappin G, Stevens L. Biomedical accelerator mass spectrometry: recent applications in metabolism and pharmacokinetics. Expert Opin. Drug Metab. Toxicol. 4:1021–1033. doi: 10.1517/17425255.4.8.1021. [DOI] [PubMed] [Google Scholar]