Induced pluripotent stem cell (iPSC) technology provides a great interface to bring the cells of hyptertensive patients with their genomic data into the laboratory and to study hypertensive responses. As an initial step, the present study established an iPSC bank from patients with primary hypertension and demonstrated an effective and reproducible method of generating functional vascular smooth muscle cells.
Keywords: Hypertension, Induced pluripotent stem cells, Personalized medicine, Vascular smooth muscle cells
Abstract
Studies in hypertension (HTN) pharmacogenomics seek to identify genetic sources of variable antihypertensive drug response. Genetic association studies have detected single-nucleotide polymorphisms (SNPs) that link to drug responses; however, to understand mechanisms underlying how genetic traits alter drug responses, a biological interface is needed. Patient-derived induced pluripotent stem cells (iPSCs) provide a potential source for studying otherwise inaccessible tissues that may be important to antihypertensive drug response. The present study established multiple iPSC lines from an HTN pharmacogenomics cohort. We demonstrated that established HTN iPSCs can robustly and reproducibly differentiate into functional vascular smooth muscle cells (VSMCs), a cell type most relevant to vasculature tone control. Moreover, a sensitive traction force microscopy assay demonstrated that iPSC-derived VSMCs show a quantitative contractile response on physiological stimulus of endothelin-1. Furthermore, the inflammatory chemokine tumor necrosis factor α induced a typical VSMC response in iPSC-derived VSMCs. These studies pave the way for a large research initiative to decode biological significance of identified SNPs in hypertension pharmacogenomics.
Significance
Treatment of hypertension remains suboptimal, and a pharmacogenomics approach seeks to identify genetic biomarkers that could be used to guide treatment decisions; however, it is important to understand the biological underpinnings of genetic associations. Mouse models do not accurately recapitulate individual patient responses based on their genetics, and hypertension-relevant cells are difficult to obtain from patients. Induced pluripotent stem cell (iPSC) technology provides a great interface to bring patient cells with their genomic data into the laboratory and to study hypertensive responses. As an initial step, the present study established an iPSC bank from patients with primary hypertension and demonstrated an effective and reproducible method of generating functional vascular smooth muscle cells.
Introduction
Hypertension (HTN) is a major health burden in the U.S., affecting approximately 75 million people [1]. Although numerous antihypertensive agents are currently available, blood pressure (BP) control remains suboptimal with, about half of hypertensive patients experiencing uncontrolled BP [2]. In addition, a portion of these patients continue to have high BP despite the use of three or more medications, a condition referred to as resistant hypertension (RHTN). Poorly controlled BP can lead to serious adverse cardiovascular outcomes including coronary artery disease, myocardial infarction, heart failure, stroke, and renal failure [3]. Together, this presents an impetus for better recognition and management of BP to improve cardiovascular outcomes of hypertensive patients.
Variability in BP response to nearly all prescribed antihypertensive medications exists; genetic variants in the genes regulating BP or in the drug’s pharmacological pathway may contribute [4]. Through the identification of genetic predictors of BP response and adverse effects to the prescribed medications, pharmacogenomics has the potential to lead to individualized treatment or precision medicine. Several genetic polymorphisms have been replicated for their association with antihypertensive response, including functional variants in ADRB1 and NEDD4L that were studied in candidate gene studies [5] and discoveries arising from genomewide association studies (GWASs) [6]. The effect sizes of these pharmacogenetic associations are approximately 5- to 10-fold greater (i.e., 3–5 mm Hg per allele) than the effect sizes observed in hypertension GWASs (approximately 0.5 mm Hg per allele) [7–9], suggesting the potential of a panel of genetic variants that could be used to guide treatment decisions. The Pharmacogenomic Evaluation of Antihypertensive Responses studies (PEAR and PEAR2), conducted as part of the NIH Pharmacogenomics Research Network, have contributed some of these findings [9–11].
In precision medicine, the ultimate goal is to identify clinically actionable genetic variants that can guide selection of medications. Clinical use of genetic information to guide decisions requires only that the genetic association is sufficiently robust to be predictive in the clinical setting; the biological or functional consequences of the genetic variation need not be known. In most of the examples of disease genetics or pharmacogenetics for which there is clinical application, the functional mechanism of the genetic association is understood. In addition, such information is particularly important for understanding mechanisms underlying differential drug responses and ultimately may facilitate identification of new drug targets, both of which are additional goals of pharmacogenomics studies. To fully exploit available pharmacogenomics findings, it is imperative to perform molecular studies of the associated variants in the appropriate tissues of relevance for the phenotype of interest. Animal models have been used intensively for studying systemic diseases like hypertension; however, they are not useful for understanding the biological impact of human genetic variants. An approach for studying human cells and tissues is needed. To this end, recent advances in human induced pluripotent stem cell (iPSC) technology offer an attractive alternative approach.
iPSC technology has been widely pursued since its initial introduction in 2006 by Takahashi and Yamanaka [12] and provides a unique opportunity to evaluate diseases in a “dish” [13–15]. The technology allows somatic cells, collected from peripheral blood, to be reprogrammed to a stem cell state. With the appropriate differentiation protocols, these cells can then be used to generate any cell type of interest. Importantly, patient genomic information is maintained during the reprogramming and differentiation processes. Since its introduction, the technology has been used to better understand a variety of diseases including neurologic, hepatic, diabetic, and cardiovascular diseases [14, 15].
Coupling of pharmacogenomics and iPSC technology is still in its infancy, but in recent years, the technology used to understand cardiovascular diseases has improved, mainly through the use of patient iPSC-derived cardiomyocytes (iPSC-CMs) [16, 17]. Moretti et al. [18] used iPSC-CMs to elucidate mutations related to long QT syndrome; Zhi et al. [19] combined whole-exome sequencing and iPSC-CMs to study left ventricular hypertrophy, a heritable predictor for cardiovascular disease in African Americans; and Lan et al. [20] demonstrated pharmacological restoration of function in iPSC-CMs from patients with inherited cardiomyopathies.
The current study presents the beginning of a major collaborative effort between the PEAR group and the University of Florida Center for Cellular Reprogramming to combine clinical GWAS data with iPSC technology to decode the biological significance of GWAS-identified SNPs. iPSCs from PEAR patients were generated and differentiated to vascular smooth muscle cells (VSMCs). These cells showed functionality in both contractile and inflammatory responses to physiologically relevant signals. To our knowledge, this study is the first to establish multiple iPSC lines from an HTN pharmacogenomics cohort and to drive these iPSCs toward a functional vascular phenotype. This study is a crucial initial step for advancing findings from GWASs to understand biological significance, which will increase the potential to advance patient care in HTN and RHTN.
Materials and Methods
Preparation of Peripheral Blood Mononuclear Cells
PEAR [10] and PEAR2 (ClinicalTrials.gov identifier NCT01203852) study participants from the University of Florida site were invited to participate in this study, which led to generation of iPSCs from their samples. The study was approved by the University of Florida institutional review board and registered at ClinicalTrials.gov (identifier NCT01943383). Whole blood (approximately 5 ml) was collected in a collection tube with EDTA. Peripheral blood mononuclear cells (PBMCs) were isolated and used for reprogramming (supplemental online data).
Reprogramming PBMCs to iPSCs
Patient PBMCs were infected with Sendai viral vector SeVdp(KOSM)302L encoding four reprogramming factors (Oct4, Sox2, Klf4, and c-Myc) [21] for 2 hours at 37°C and then plated onto mitotically inactivated mouse embryonic fibroblasts in 6-well plates at 2 × 105 cells per well. Cells were cultured in RPMI 1640 media for the first 3 days and then in Primate ES Medium (ReproCELL, Yokohama, Japan, https://www.reprocell.com) changed every other day. Individual iPSC clones were isolated approximately 2–3 weeks after virus infection. These were transferred to vitronectin-coated dishes and cultured in Essential 8 Medium (StemCell Technologies, Vancouver, Canada, http://www.stemcell.com) following the manufacturer’s protocols (supplemental online data).
Differentiation of iPSCs to Vascular Smooth Muscle Cells
Several previously reported differentiation protocols [22–24] were adapted and modified to identify the following protocol, which yielded the highest percentage of VSMC differentiation in a reproducible manner. In brief, following a 5-minute collagenase IV treatment, iPSC colonies were gently scraped and placed in embryoid body (EB) media (αMEM, 20% fetal bovine serum, 0.05 mM 2-mercaptoethanol, 1% nonessential amino acids, 50 ng/ml BMP4, 50 ng/ml VEGFA and 10 μM ROCK inhibitor) [25]. iPSCs were transferred to Ultra-Low Attachment dishes (Corning, Corning, NY, https://www.corning.com). EBs formed for 4 days, and then EBs were transferred to collagen-coated dishes in VSMC differentiation media composed of Media 231 (Invitrogen, Life Technologies; Thermo Fisher Scientific, Waltham, MA, http://www.thermofisher.com), VSMC differentiation serum (Invitrogen, Life Technologies; Thermo Fisher Scientific), and 10 μM ROCK inhibitor. Cells differentiated for 10 days (supplemental online data).
Flow Cytometry Cell Sorting
On day 10 of VSMC differentiation, cells were stained with CD140b (platelet-derived growth factor receptor β [PDGFR-β]) and CD91 (LRP1) and sorted by double-positive staining (supplemental online data). Sorting was done with a FACSAria II (BD Biosciences, San Jose, CA, http://www.bdbiosciences.com). Sorted cells were plated on collagen type IV-coated dishes and maintained in vascular smooth muscle cell growth media composed of Media 231 (Invitrogen, Life Technologies; Thermo Fisher Scientific) and smooth muscle cell growth supplement (Invitrogen, Life Technologies; Thermo Fisher Scientific).
Gene Expression
RNA was extracted with RNAqueous Total RNA Isolation Kit and DNase treated using Turbo DNA-free (Ambion; Thermo Fisher Scientific). cDNA was made with 4 μl of RNA and the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Life Technologies; Thermo Fisher Scientific) with a C1000 Thermal Cycler (Bio-Rad, Hercules, CA, http://www.bio-rad.com). Real-time polymerase chain reaction on 5 ng/μl of cDNA with 10 nM primer mix was run using the StepOnePlus system (Applied Biosystems, Life Technologies; Thermo Fisher Scientific) with SYBER Green. All genes were normalized to the housekeeping gene GAPDH and were compared for relative quantitation using the ΔΔCT calculation.
Immunocytochemistry
Cells were fixed in 4% paraformaldehyde for 10 minutes, permeabilized for 5 minutes in 0.1% Triton-X with 1× phosphate-buffered saline (PBS) solution, then blocked in 10% serum in 0.05% Triton-X with 1× PBS solution for 1 hour in a humid chamber. Cells were incubated in primary antibody at 4°C overnight. Secondary antibody was added to the cells, away from direct light, for 1 hour at room temperature. Coverslips were dried overnight, mounted with a hardset mounting media containing 4′,6-diamidino-2-phenylindole, and allowed to set for 15 minutes (supplemental online data).
VSMC Contraction Using Cell Tracker Probes
Differentiated VSMCs were plated in 96-well plates coated with collagen IV. The following day, cells were tracked with Cell Trace Oregon Green (Molecular Probes; Thermo Fisher Scientific) at 1:1,000 dilution in serum-free media for 30 minutes at 37°C and then changed to full-serum media. Images of the cells before and after phorbol 12-myristate 13-acetate (PMA; 100 μg/ml for 30 minutes, then 60 minutes with no PMA recovery) were taken. Difference in cell surface area and length were calculated between before and after stimulation using ImageJ (NIH, Bethesda, MD, http://imagej.nih.gov/ij/).
Quantification of Single-Cell Contractility
VSMCs were sparsely plated onto collagen-coated hydrogels with fluorescent beads embedded at the surface, as described previously [26], and allowed to attach overnight. Individual cells were imaged three times at ×200 in fluorescence and phase contrast (a) while attached to the surface of the gel in normal media (basal), (b) while attached to the surface after 30 minutes of submersion in 10 ng/ml of endothelin-1 (ET-1) in media, and (c) while trypsinized to capture the relaxed gel (null).
Each set of three images was aligned and cropped to 272 × 272 µm using ImageJ, and bead movement between basal/null and ET-1/null was calculated using PIVLab in Matlab (MathWorks, Natick, MA, http://www.mathworks.com) [27]. The strain energy generated by each cell contraction was calculated using a Fourier Transform Traction Cytometry algorithm [28, 29]. The resulting strain energy values were compared using analysis of variance with a multicomparison tool in Matlab.
Stimulation of VSMC With Tumor Necrosis Factor α
Cells were plated on collagen type IV-coated 6-well dishes at 1–2 × 105 cells per well. Once cells reached 80%–90% confluence, the old media was removed and cells were washed with PBS. Fresh media (1 ml) with or without 30 ng/ml tumor necrosis factor α (TNF-α; R&D Systems, Minneapolis, MN, https://www.rndsystems.com) was added to the cells for 6 hours; RNA was then collected.
Results
Reprogramming PEAR HTN Patient PBMCs to iPSCs
All patient iPSCs were generated at the University of Florida Center for Cellular Reprogramming, as described previously [15, 30]. To date, our HTN iPSC library consists of iPSC clones derived from 17 patients with varying age, sex, and ethnicity (Table 1). All of the patients are registered in PEAR pharmacogenomics studies, thus data are available for genomewide SNP variations as well as clinical responses to antihypertensive drugs. Three different patient iPSC samples—HTN-9, HTN-11, and HTN-21—were used in this study. All three iPSCs demonstrated expression of pluripotent-specific nuclear genes OCT4 and Nanog at comparable levels to human embryonic stem cells (hESCs) (Fig. 1A, 1B). Furthermore, immunofluorescence analysis confirmed expression of the pluripotent-specific cell surface marker SSEA4 (Fig. 2C–2E). We demonstrated previously that the A allele at the rs16960228 SNP on the PRKCA gene correlated with higher expression of the gene and greater BP response to hydrochlorothiazide [9]. Patient HTN-11_iPSC has G/G at rs16960228, whereas patient HTN-9_iPSC has A/A. When we investigated three independent iPSC clones each from patients HTN-11_iPSC and HTN-9_iPSC, they demonstrated that PRKCA expression was higher in HTN-9_iPSC clones at both mRNA and protein levels, consistent with our previously published data [11] (supplemental online Fig. 1). The retention of clinical gene expression data within iPSCs encouraged us to use the cells for further biological analyses.
Table 1.
Currently established iPSCs derived from hypertensive patients at the University of Florida Center for Cellular Reprogramming, with a variety of patient sex, age, and ethnicity characteristics paired with genomewide association study data

Figure 1.
iPSCs derived from peripheral blood mononuclear cells of hypertensive patients express pluripotent-specific nuclear genes OCT4 (A) and Nanog (B); relative mRNA expression compared with control embryonic stem cells, data from three independent experiments, normalized to housekeeping gene GAPDH. Immunocytochemistry demonstrate iPSC expression of pluripotent cell surface protein SSEA4 (C–E). Scale bar = 100 μm. Abbreviations: DAPI, 4′,6-diamidino-2-phenylindole; ES, embryonic stem cells; iPSC, induced pluripotent stem cells.
Figure 2.
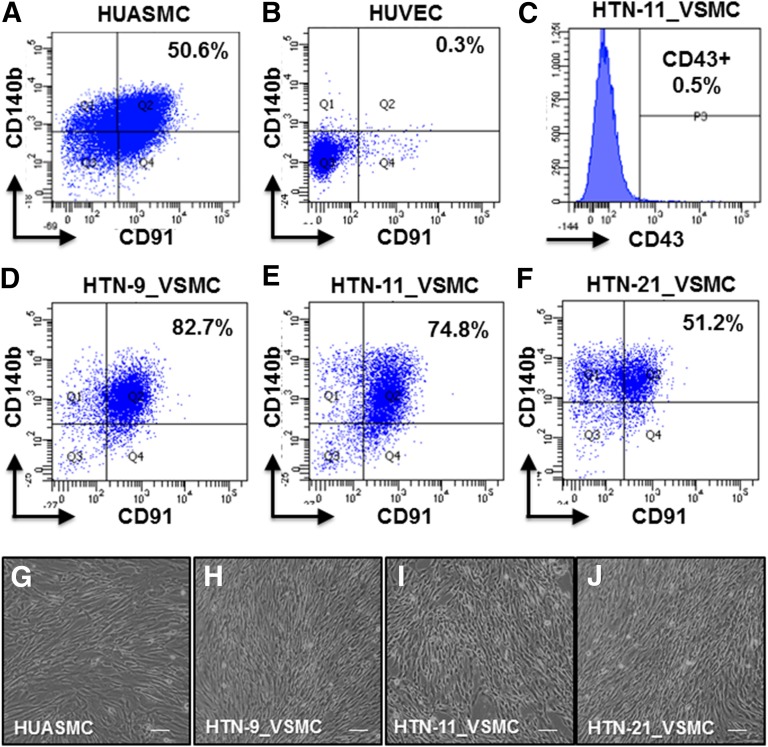
iPSC-derived VSMCs (iPSC-VSMCs) were sorted by flow cytometry for CD91-positive (CD91+)/CD140b+ markers for VSMC populations. Cells were analyzed between passages 2 and 4. CD91+/CD140b+ cells are specific markers for VSMCs (A) and not ECs (B), as demonstrated with primary control cells HUASMC and HUVEC, respectively. The iPSC-VSMC differentiation protocol excluded CD43+ lymphocyte populations, as demonstrated with HTN-11_VSMC sorting (C) and produced a high percentage of the VSMC population in all three iPSC lines (D–F). Differentiated, sorted, and cultured iPSC-VSMCs showed VSMC morphology compared with control cells (G–J). Scale bar = 100 μm. Data are from three independent experiments. Abbreviations: HUASMC, human umbilical arterial smooth muscle cells; HUVEC, human umbilical vein endothelial cells; iPSC, induced pluripotent stem cell; VSMC, vascular smooth muscle cell.
Differentiation of HTN Patient iPSCs to VSMCs
iPSCs were differentiated to VSMCs (supplemental online Fig. 2). Cells were sorted using two cell surface markers, PDRGR-β (CD140b) and LRP1 (CD91), that are present in both progenitor and mature VSMCs (Fig. 2A) but not in endothelial (Fig. 2B) or peripheral blood mononuclear cells [31]. Furthermore, the differentiation protocol demonstrated 99.5% CD43 cells (Fig. 2C), largely excluding lymphocyte lineages when evaluated at day 10 of differentiation. From the CD43-negative cell population at day 10 of differentiation, the protocol yielded 82.7%, 74.8%, and 51.2% CD140b-positive (CD140b+)/CD91+ cells for HTN-9_iPSC_1 (HTN-9_VSMC), HTN-11_iPSC (HTN-11_VSMC), and HTN-21_iPSC (HTN-21_VSMC), respectively (Fig. 2D–2F). The same protocol with the addition of human recombinant proteins PDGF-β (10 ng/ml) and transforming growth factor β (10 ng/ml) yielded similar results, shown with HTN-11_VSMC (supplemental online Fig. 3); therefore, additional growth factors were not necessary to obtain good differentiation yield.
Furthermore, our protocol has been shown to be highly reproducible by up to three separate researchers, demonstrated with HTN-11_iPSC (HTN-11_VSMC), yielding 74.8%, 67.6%, and 52.6% CD140b+/CD91+ cells for authors N.M.B., B.B.D., and N.E.R., respectively (supplemental online Fig. 4). Differentiated, sorted, and cultured cells exhibited VSMC-like morphology compared with control primary VSMCs (Fig. 2G–2J) and a demonstrated comparable growth profile (supplemental online Fig. 5A). Finally, differentiated cells demonstrated the loss of the pluripotent marker OCT4 and the gain of VSMC receptor PDGFR-β (supplemental online Fig. 5B, 5C). iPSC-derived VSMCs were able to be cryopreserved and subsequently cultured, allowing for flexibility and eliminating a large concern about the use of these cells in future experiments.
Characterization of VSMC Phenotype
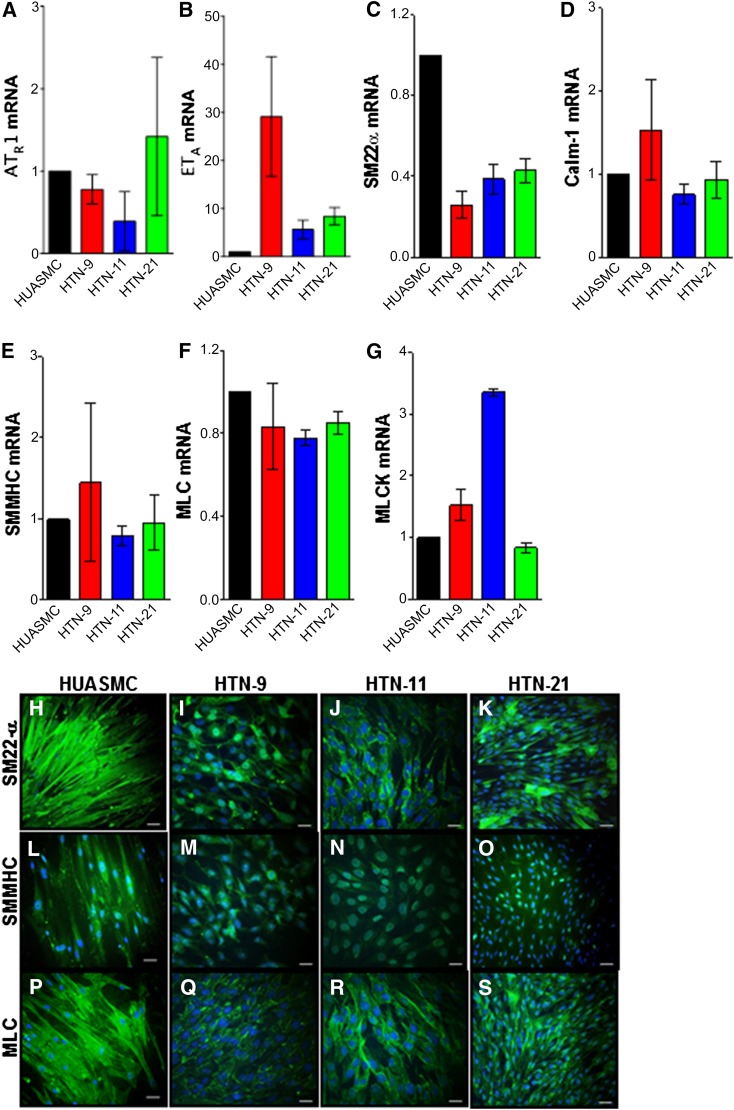
iPSC-derived VSMCs (iPSC-VSMCs) express receptor genes involved in vasotone regulation such as the angiotensin II receptor and endothelin receptor (Fig. 3A, 3B). Furthermore, the cells express genes related to Ca+2 signaling during vasoconstriction, SM22-α, and calmodulin-1 (Fig. 3C, 3D). Finally, vasoconstriction-related genes smooth muscle myosin heavy chain (SMMHC), myosin light chain (MLC), and myosin light chain kinase (MLCK) are also expressed (Fig. 3E–3G). Immunofluorescence demonstrated protein expression of SM22-α, SMMHC, and MLC (Fig. 3H–3S). These data indicate that iPSC-VSMCs express mRNAs and proteins that are highly specific and relevant to VSMC functions.
Figure 3.
iPSC-derived VSMCs expressed vasoconstriction-relevant receptor genes angiotensin II receptor (A), and endothelin receptor type A (B) as well as Ca+2 binding genes SM22α (C) and calmodulin-1 (D). Furthermore, the cells expressed genes involved in contraction; SMMHC (E), MLC (F), and MLCK (G); data from three independent experiments were normalized to housekeeping gene GAPDH. Immunofluorescence demonstrates protein expression of SM22-α (H–K), SMMHC (L–O), and MLC (P–S). Scale bar = 46 μm. Abbreviations: ATR1, angiotensin II receptor; Calm-1, calmodulin-1; ETA, endothelin receptor type A; HUASMC, human umbilical arterial smooth muscle cells; MLC, myosin light chain; MLCK, myosin light chain kinase; SMMHC, smooth muscle myosin heavy chain.
VSMC Contractile Function
In order to further demonstrate the function of iPSC-VSMCs, we next examined their contractility responses. This study demonstrates two different methods of evaluating VSMC contraction to a chemical stimulus, PMA, or to a physiological stimulus, ET-1. PMA (100 ng/ml) stimulation resulted in contraction of cells as measured by a change in cell surface area before and after PMA (Fig. 4A, 4B). Compared with basal conditions before PMA stimulation, human umbilical arterial smooth muscle cell (HUASMC) area decreased by 11% (n = 10) (Fig. 4C), HTN-9_VSMC decreased by 23.3% (n = 16) (Fig. 4D), HTN-11_VSMC decreased by 23.2% (n = 16) (Fig. 4E), and HTN-21_VSMC decreased by 29.8% (n = 12) (Fig. 4F).
Figure 4.
Contraction to the chemical stimulator PMA was measured by the change in cell surface area before (A) and after (B) PMA, analyzed with ImageJ. Scale bar = 100 μm. PMA (100 ng/ml) challenge resulted in contraction of the cells and thus decrease in the cell surface area in HUASMC (C), HTN-9_VSMC (D), HTN-11_VSMC (E), and HTN-21_VSMC (F). Abbreviations: HUASMC, human umbilical arterial smooth muscle cells; PMA, phorbol 12-myristate 13-acetate; VSMC, vascular smooth muscle cells.
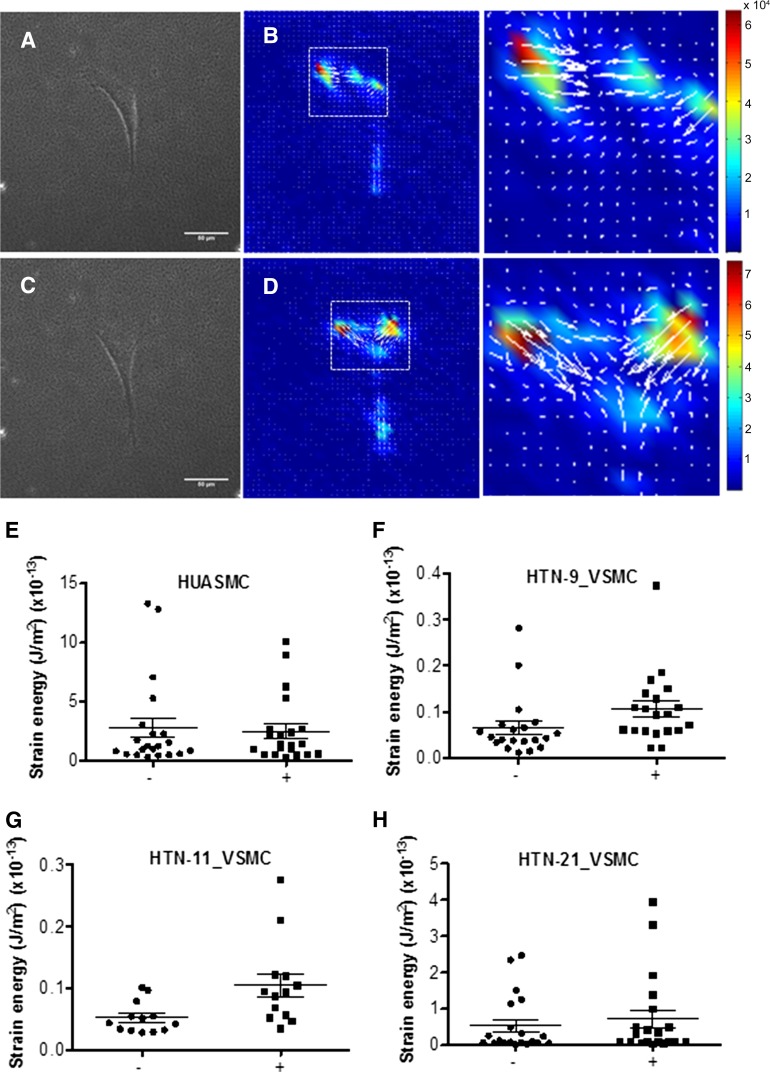
Because chemical activation of VSMC contraction may not demonstrate a physiologically relevant response, we also examined VSMC response to ET-1. To quantify contractile function, we used a sensitive traction force microscopy assay to evaluate the response of VSMCs to ET-1 (10 ng/ml). The strain energy that VSMCs placed on the beaded gel was measured at base line (no stimulation with ET-1) (Fig. 5A, 5B) and in the presence of ET-1 (Fig. 5C, 5D). Results showed that in all cell lines, the strain energy increased, hence the cells contracted in the presence of ET-1 compared with basal. The mean strain energy increased by 1.1-fold in HUASMCs (n = 21) (Fig. 5E), by 1.9-fold in HTN-9_VSMC (n = 20) (Fig. 5F), by 2.0-fold in HTN-11_VSMC (n = 13) (Fig. 5G) and by 1.36-fold in HTN-21_VSMC (n = 20) (Fig. 5H).
Figure 5.
Contraction to the vasoconstrictor ET-1 (10 ng/ml) was measured as a change in strain energy. Cells were plated on soft gel with embedded fluorescent beads (A, C), and the change in cell contractile strain energy generated in basal (A, B) and stimulated (C, D) states was calculated. Scale bar = 50 μm. HUASMC (E), HTN-9_VSMC (F), HTN-11_VSMC (G), and HTN-21_VSMC (H) all showed increased strain energy in the ET-1-stimulated state compared with basal. Abbreviations: ET-1, endothelin-1; HUASMC, human umbilical arterial smooth muscle cells; VSMC, vascular smooth muscle cells.
VSMC Response to Inflammatory Signaling
Hypertension has been linked to inflammation and atherosclerosis [32, 33]. Macrophage-VSMC interaction through fractalkine (CX3CL1)/CX3CR1 in atherosclerosis has been well documented [33–35]. Because such signaling pathways are also likely to be present in HTN, the feasibility of demonstrating responses to such signaling pathways is important.
The three different iPSC-VSMCs were exposed to 6 hours of stimulation with 30 ng/ml TNF-α, which is normally secreted from macrophages. TNF-α stimulation led to a mean increase in the CX3CL1 gene expression by 43.9-fold (HUASMCs), 23.2-fold (HTN-9_VSMC), 3.6-fold (HTN-11_VSMC), and 11.3-fold (HTN-21_VSMC) (Fig. 6A) and a mean increase in MMP9 gene expression by 7.41-fold (HUASMC), 8.5-fold (HTN-9_VSMC), 7.4-fold (HTN-11_VSMC) and 8.7-fold (HTN-21_VSMC) (Fig. 6B) compared with cells that did not receive TNF-α.
Figure 6.
Cells stimulated with TNF-α led to an increase in both gene expression of CX3CL1 (A) and MMP9 (B) compared with cells that did not receive TNF-α. Data from three independent experiments were normalized to housekeeping gene GAPDH. Abbreviations: HUASMC, human umbilical arterial smooth muscle cells; TNF-α, tumor necrosis factor α; VSMC, vascular smooth muscle cells.
Discussion
Hypertension is a major health burden in the U.S. and the world, putting patients at higher risk for cardiovascular and kidney disease if untreated. Treatment of HTN is largely empirical, and the current treatment approaches are suboptimal; therefore, the field needs better ways to select therapy from existing medications and discovery of new therapeutics to advance patient care. Studies in HTN pharmacogenomics seek to uncover genetic determinants of variable response that might be translated into genetically guided clinical care. In addition, hypertension pharmacogenomics studies can provide insights into mechanism of drug effect and lead to identification of novel drug targets; however, accomplishing these latter goals requires a relevant human system for studying biophysiological effects.
Studying the vasculature, specifically, the contractile vascular smooth muscle cells of the vasculature, could reveal significant biological effects of GWAS-identified SNPs, for example, in patient drug response; however, obtaining vascular cells from individual patients and culturing them in the laboratory is difficult. Induced pluripotent stem cell technology provides a unique opportunity to study individual patient genetics in the laboratory with the use of iPSCs and subsequent differentiation to VSMCs.
Several efforts have been made to establish differentiation protocols for VSMCs from iPSCs. Sone et al. [36] and Taura et al. [37] demonstrated the differentiation of hESCs and iPSCs, respectively, to vascular cells (endothelial and smooth muscle); however, they lacked demonstration of the functionality of these cells. Park et al. showed the differentiation of iPSCs to CD34+ progenitor cells and demonstrated a contractile response of iPSC-VSMCs to carbachol, an agonist for the acetylcholine receptor [38]. Subsequently, Lee et al. showed the ability to differentiate patient artery smooth muscle cells to iPSCs and then differentiate the iPSCs to VSMCs, demonstrating the similarity to the parent cells and contractile function in response to carbachol and angiotensin II [39]. Finally, Xie et al. established feasible iPSC-to-VSMC culturing on nanofibrous scaffolding for tissue engineering [40].
Although patient iPSCs have been proposed to decode biological significance of identified SNPs, no comprehensive approach has been developed to date in the field of hypertension pharmacogenomics. The present study establishes, for the first time, multiple iPSC clones from a hypertensive pharmacogenomics cohort for this purpose. As we have shown in this study with one particular SNP, such a library could be used to determine SNP-gene expression relationships. We envision an iPSC library from 50 HTN patients as a reasonable goal to start. Using such a library, we would be able to identify approximately 5 persons who are carriers of a particular SNP in the heterozygous state of minor allele frequency of 5% (supplemental online Table 1). iPSCs derived from those persons could be compared with controls without the SNP. Moreover, we envision that such a library would also be useful in combination with gene-targeting technologies. Starting from SNP-heterozygous iPSCs, we could establish hemizygous iPSCs by targeting one allele using CRISPR/Cas9. In this manner, we would be able to establish isogenic iPSCs with the wild-type or variant genotype, providing a better chance of analyzing more genuine effects caused by the SNP difference. Altering a single nucleotide at a SNP using gene editing is an option; however, cloning of iPSCs without drug selection is technically challenging and requires more rigorous effort and thus may not be suitable or very practical for a large number of SNP variation studies.
In order to effectively use the iPSC library, especially in such a large research initiative, it is imperative to have a simple yet robust and highly reproducible protocol for HTN-relevant cell types. Furthermore, differentiated cells must not only demonstrate the appropriate phenotype but also function in a disease-relevant manner. Finally, researchers need the ability to culture and expand these cells for substantial periods of time because laboratory experiments are conducted over months and years [15]. Simplicity and reproducibility of the iPSC differentiation protocol is imperative if non-stem cell laboratories are to use this technology for their research. The present study demonstrated in 3 independent iPSC lines, originally from 3 participants with HTN in a PEAR study, that VSMCs can be differentiated with a simple 2-step method using commercially available reagents with high efficiency for >50% of the VSMC population. The method has been shown to be highly reproducible not only by one researcher but by three.
All three iPSC-VSMCs show expression of VSMC-specific and HTN-relevant genes. The cells express both angiotensin II receptor and endothelin-1 receptor type A, receptors for two potent vasoconstrictors essential for future drug-response studies. During vasoconstriction, calcium signaling is observed, and common proteins such as SM22-α and calmodulin-1 are involved [41, 42]. Consequently, the expression of these genes in iPSC-VSMCs would be ideal; the present study showed that all three iPSC-derived cells expressed both of these genes and demonstrated the protein expression of SM22-α via immunofluorescence. Finally, vascular contraction involves the interplay among myosin light chain, myosin light chain kinase, and smooth muscle myosin heavy chain [42, 43]; iPSC-VSMCs showed relevant gene and protein expression of these factors.
In order to truly bridge GWASs and mechanistic understanding using iPSC technology, it is essential to be able to quantitate functions of the differentiated cell types responding to physiological and pharmacological reagents. Vascular smooth muscle cell contraction will be important for drug-response studies, whereas inflammatory responses will play a role in RHTN and discovery of novel therapeutics. VSMC contraction to chemical (PMA) or physiological (ET-1) stimulus was nicely demonstrated in all three iPSC-VSMC lines. Our methods, including traction force microscopy (TFM) used to quantify cell forces, are appropriate for functional assessment of iPSC-VSMCs for many disease models. The greatest weakness of TFM is custom-made polyacrylamide gels, the fabrication and characterization of which will likely vary from laboratory to laboratory. Relative changes in cell response reported for TFM should be consistent, although a 384-well implementation of TFM has been demonstrated [44], suggesting potential for high-throughput screening-compatible substrates in the future.
In addition to contractile function, all three cell lines demonstrated a tremendous response to inflammatory stimulus TNF-α by upregulation of two separate genes, CX3CL1 and MMP9, factors that have been shown to be significant in atherosclerotic settings [33]. Finally, the cells can be cultured, cryopreserved, and recultured from frozen stocks with the maintenance of all genes and functional responses, thereby providing a robust and long-term supply of cells on differentiation.
We are fully aware that the present study is only an initial step for a large research initiative using iPSC technologies to advance HTN pharmacogenomics. Multicellular interaction studies and drug responses therein will be crucial next steps to validate the usefulness of the system. Establishing an isogenic cellular system by gene editing is another critical method to minimize clonal variations. Individual genetic variants may not be sufficient for producing in vitro differences, and studying combined effects of multiple SNPs plus environmental effects may become vital. We strongly believe, however, that it is essential to share ideas, protocols, and materials among research communities, even at early stages, to facilitate this challenging but highly important field of study in the era of personal genomics.
Conclusion
Based on what we learned through the present study, we present a summary of the envisioned roadmap needed to advance HTN pharmacogenomics using iPSCs: (a) develop and expand an iPSC bank from an HTN pharmacogenomics cohort; (b) validate protocols for efficient differentiation of HTN-relevant cell types from iPSCs; (c) validate sensitive, reproducible, robust, and high-throughput assays using the differentiated cells for pharmacogenomics studies; (d) develop an isogenic cellular system using gene editing; (e) develop cell reference lines that are widely available commercially; and (f) develop a consortia-based model that motivates donation, participation, and membership toward advancing HTN pharmacogenomics.
Supplementary Material
Acknowledgments
We thank the PEAR patients who volunteered to provide their cells for the induced pluripotent stem cell-hypertension initiative and the staff at University of Florida Community Health and Family Medicine who collected the patient samples. We also thank Yan Gong and Caitrin McDonough for analyzing genetic information and Neal Benson at the University of Florida ICBR Flow Core for assistance with live cell sorting. This work was supported in part by National Institutes of Health/National Institute of General Medical Sciences U01 Award GM074492 to J.A.J. and R.M.C.-D., a Clinical and Translational Science Award to the University of Florida UL1 TR000064 to N.M.B. and N.T., and an HHMI Science for Life Award to S.D.C.
Author Contributions
N.M.B.: conception and design, collection and/or assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript; K.E.S, B.B.D., N.E.R., and S.D.C.: collection and/or assembly of data; C.S. and R.M.C.-D.: data analysis and interpretation; M.N.: provision of Sendai virus and experimental protocols; J.A.J.: conception and design, manuscript writing; N.T.: conception and design, data analysis and interpretation, manuscript writing, final approval of manuscript.
Disclosure of Potential Conflicts of Interest
M.N. has uncompensated research funding from Astellas Pharma, Inc. and TAKARA-Bio Inc. and uncompensated stock options in TOKIWA-Bio Inc. The other authors indicated no potential conflicts of interest.
References
- 1.Go AS, Mozaffarian D, Roger VL, et al. Heart disease and stroke statistics—2014 update: A report from the American Heart Association. Circulation. 2014;129:e28–e292. doi: 10.1161/01.cir.0000441139.02102.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vital signs: Awareness and treatment of uncontrolled hypertension among adults--United States, 2003-2010. MMWR Morb Mortal Wkly Rep. 2012;61:703–709. [PubMed] [Google Scholar]
- 3.Rosei EA, Muiesan ML, Rizzoni D. Cardiac and vascular alterations in resistant hypertension. In: Mancia G, ed. Resistant Hypertension: Epidemiology, Pathophysiology, Diagnosis and Treatment. Milan, Italy: Springer-Verlag Italia, 2013:39–50.
- 4.Mellen PB, Herrington DM. Pharmacogenomics of blood pressure response to antihypertensive treatment. J Hypertens. 2005;23:1311–1325. doi: 10.1097/01.hjh.0000173510.52987.68. [DOI] [PubMed] [Google Scholar]
- 5.Gong Y, McDonough CW, Padmanabhan S et al. Hypertension pharmacogenomics. In: Padmanabhan S, ed. Handbook of Pharmacogenomics and Stratified Medicine. London, U.K.: Academic Press, 2014:747–778.
- 6.Johnson JA. Advancing management of hypertension through pharmacogenomics. Ann Med. 2012;44(suppl 1):S17–S22. doi: 10.3109/07853890.2011.653399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Newton-Cheh C, Johnson T, Gateva V, et al. Genome-wide association study identifies eight loci associated with blood pressure. Nat Genet. 2009;41:666–676. doi: 10.1038/ng.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levy D, Ehret GB, Rice K, et al. Genome-wide association study of blood pressure and hypertension. Nat Genet. 2009;41:677–687. doi: 10.1038/ng.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turner ST, Boerwinkle E, O’Connell JR, et al. Genomic association analysis of common variants influencing antihypertensive response to hydrochlorothiazide. Hypertension. 2013;62:391–397. doi: 10.1161/HYPERTENSIONAHA.111.00436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson JA, Boerwinkle E, Zineh I, et al. Pharmacogenomics of antihypertensive drugs: Rationale and design of the Pharmacogenomic Evaluation of Antihypertensive Responses (PEAR) study. Am Heart J. 2009;157:442–449. doi: 10.1016/j.ahj.2008.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turner ST, Bailey KR, Schwartz GL, et al. Genomic association analysis identifies multiple loci influencing antihypertensive response to an angiotensin II receptor blocker. Hypertension. 2012;59:1204–1211. doi: 10.1161/HYP.0b013e31825b30f8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 13.Hankowski KE, Hamazaki T, Umezawa A, et al. Induced pluripotent stem cells as a next-generation biomedical interface. Lab Invest. 2011;91:972–977. doi: 10.1038/labinvest.2011.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu H, Lensch MW, Cahan P, et al. Investigating monogenic and complex diseases with pluripotent stem cells. Nat Rev Genet. 2011;12:266–275. doi: 10.1038/nrg2951. [DOI] [PubMed] [Google Scholar]
- 15.Santostefano KE, Hamazaki T, Biel NM, et al. A practical guide to induced pluripotent stem cell research using patient samples. Lab Invest. 2015;95:4–13. doi: 10.1038/labinvest.2014.104. [DOI] [PubMed] [Google Scholar]
- 16.Adams WJ, García-Cardeña G. Novel stem cell-based drug discovery platforms for cardiovascular disease. J Biomol Screen. 2012;17:1117–1127. doi: 10.1177/1087057112454741. [DOI] [PubMed] [Google Scholar]
- 17.Braam SR, Tertoolen L, van de Stolpe A, et al. Prediction of drug-induced cardiotoxicity using human embryonic stem cell-derived cardiomyocytes. Stem Cell Res (Amst) 2010;4:107–116. doi: 10.1016/j.scr.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 18.Moretti A, Bellin M, Welling A, et al. Patient-specific induced pluripotent stem-cell models for long-QT syndrome. N Engl J Med. 2010;363:1397–1409. doi: 10.1056/NEJMoa0908679. [DOI] [PubMed] [Google Scholar]
- 19.Zhi D, Irvin MR, Gu CC, et al. Whole-exome sequencing and an iPSC-derived cardiomyocyte model provides a powerful platform for gene discovery in left ventricular hypertrophy. Front Genet. 2012;3:92. doi: 10.3389/fgene.2012.00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lan F, Lee AS, Liang P, et al. Abnormal calcium handling properties underlie familial hypertrophic cardiomyopathy pathology in patient-specific induced pluripotent stem cells. Cell Stem Cell. 2013;12:101–113. doi: 10.1016/j.stem.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nishimura K, Sano M, Ohtaka M, et al. Development of defective and persistent Sendai virus vector: A unique gene delivery/expression system ideal for cell reprogramming. J Biol Chem. 2011;286:4760–4771. doi: 10.1074/jbc.M110.183780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ross JJ, Hong Z, Willenbring B, et al. Cytokine-induced differentiation of multipotent adult progenitor cells into functional smooth muscle cells. J Clin Invest. 2006;116:3139–3149. doi: 10.1172/JCI28184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marchand M, Anderson EK, Phadnis SM, et al. Concurrent generation of functional smooth muscle and endothelial cells via a vascular progenitor. Stem Cells Translational Medicine. 2014;3:91–97. doi: 10.5966/sctm.2013-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bajpai VK, Mistriotis P, Loh YH, et al. Functional vascular smooth muscle cells derived from human induced pluripotent stem cells via mesenchymal stem cell intermediates. Cardiovasc Res. 2012;96:391–400. doi: 10.1093/cvr/cvs253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rufaihah AJ, Huang NF, Kim J, et al. Human induced pluripotent stem cell-derived endothelial cells exhibit functional heterogeneity. Am J Transl Res. 2013;5:21–35. [PMC free article] [PubMed] [Google Scholar]
- 26.Simmons CS, Ribeiro AJ, Pruitt BL. Formation of composite polyacrylamide and silicone substrates for independent control of stiffness and strain. Lab Chip. 2013;13:646–649. doi: 10.1039/c2lc41110e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thielicke W, Stamhuis EJ. PIVlab – towards user-friendly, affordable and accurate digital particle image velocimetry in MATLAB. J Open Res Softw. 2014;2:e30. [Google Scholar]
- 28.Mierke CT, Kollmannsberger P, Zitterbart DP, et al. Mechano-coupling and regulation of contractility by the vinculin tail domain. Biophys J. 2008;94:661–670. doi: 10.1529/biophysj.107.108472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sabass B, Gardel ML, Waterman CM, et al. High resolution traction force microscopy based on experimental and computational advances. Biophys J. 2008;94:207–220. doi: 10.1529/biophysj.107.113670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xia G, Gao Y, Jin S, et al. Genome modification leads to phenotype reversal in human myotonic dystrophy type 1 iPS-cell derived neural stem cells. Stem Cells. 2015;33:1829–1838. doi: 10.1002/stem.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang CH, Lee YS, Lin SJ, et al. Surface markers of heterogeneous peripheral blood-derived smooth muscle progenitor cells. Arterioscler Thromb Vasc Biol. 2012;32:1875–1883. doi: 10.1161/ATVBAHA.112.245852. [DOI] [PubMed] [Google Scholar]
- 32.Harrison DG, Vinh A, Lob H, et al. Role of the adaptive immune system in hypertension. Curr Opin Pharmacol. 2010;10:203–207. doi: 10.1016/j.coph.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Butoi ED, Gan AM, Manduteanu I, et al. Cross talk between smooth muscle cells and monocytes/activated monocytes via CX3CL1/CX3CR1 axis augments expression of pro-atherogenic molecules. Biochim Biophys Acta. 2011;1813:2026–2035. doi: 10.1016/j.bbamcr.2011.08.009. [DOI] [PubMed] [Google Scholar]
- 34.Liu H, Jiang D. Fractalkine/CX3CR1 and atherosclerosis. Clin Chim Acta. 2011;412:1180–1186. doi: 10.1016/j.cca.2011.03.036. [DOI] [PubMed] [Google Scholar]
- 35.Cybulsky MI, Hegele RA. The fractalkine receptor CX3CR1 is a key mediator of atherogenesis. J Clin Invest. 2003;111:1118–1120. doi: 10.1172/JCI18237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sone M, Itoh H, Yamahara K, et al. Pathway for differentiation of human embryonic stem cells to vascular cell components and their potential for vascular regeneration. Arterioscler Thromb Vasc Biol. 2007;27:2127–2134. doi: 10.1161/ATVBAHA.107.143149. [DOI] [PubMed] [Google Scholar]
- 37.Taura D, Sone M, Homma K, et al. Induction and isolation of vascular cells from human induced pluripotent stem cells--brief report. Arterioscler Thromb Vasc Biol. 2009;29:1100–1103. doi: 10.1161/ATVBAHA.108.182162. [DOI] [PubMed] [Google Scholar]
- 38.Park SW, Jun Koh Y, Jeon J, et al. Efficient differentiation of human pluripotent stem cells into functional CD34+ progenitor cells by combined modulation of the MEK/ERK and BMP4 signaling pathways. Blood. 2010;116:5762–5772. doi: 10.1182/blood-2010-04-280719. [DOI] [PubMed] [Google Scholar]
- 39.Lee TH, Song SH, Kim KL, et al. Functional recapitulation of smooth muscle cells via induced pluripotent stem cells from human aortic smooth muscle cells. Circ Res. 2010;106:120–128. doi: 10.1161/CIRCRESAHA.109.207902. [DOI] [PubMed] [Google Scholar]
- 40.Xie C, Hu J, Ma H, et al. Three-dimensional growth of iPS cell-derived smooth muscle cells on nanofibrous scaffolds. Biomaterials. 2011;32:4369–4375. doi: 10.1016/j.biomaterials.2011.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marchand A, Abi-Gerges A, Saliba Y et al. Calcium signaling in vascular smooth muscle cells: From physiology to pathology. In: Islam S, ed. Calcium Signaling: Advances in Experimental Medicine and Biology. Dordrecht, The Netherlands: Springer Science+Business Media; 2012:795–810. [DOI] [PubMed]
- 42.Salamanca DA, Khalil RA. Protein kinase C isoforms as specific targets for modulation of vascular smooth muscle function in hypertension. Biochem Pharmacol. 2005;70:1537–1547. doi: 10.1016/j.bcp.2005.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kamm KE, Stull JT. The function of myosin and myosin light chain kinase phosphorylation in smooth muscle. Annu Rev Pharmacol Toxicol. 1985;25:593–620. doi: 10.1146/annurev.pa.25.040185.003113. [DOI] [PubMed] [Google Scholar]
- 44.Mih JD, Sharif AS, Liu F, et al. A multiwell platform for studying stiffness-dependent cell biology. PLoS One. 2011;6:e19929. doi: 10.1371/journal.pone.0019929. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.