Protein-induced cardiac progenitor cells (CPCs) that are directly reprogrammed in vitro might be applicable for replacing dead heart muscle after myocardial infarction. The use of undifferentiated CPCs as building blocks to grow specific tissue types in vivo is of great interesting for regenerating the myocardium.
Keywords: Protein, Cardiac differentiation, Cardiac progenitor cells, Cardiac transcription factor, Cell transplantation
Abstract
The reprogramming of fibroblasts to induced pluripotent stem cells raises the possibility that somatic cells could be directly reprogrammed to cardiac progenitor cells (CPCs). The present study aimed to assess highly efficient protein-based approaches to reduce or eliminate the genetic manipulations to generate CPCs for cardiac regeneration therapy. A combination of QQ-reagent-modified Gata4, Hand2, Mef2c, and Tbx5 and three cytokines rapidly and efficiently reprogrammed human dermal fibroblasts (HDFs) into CPCs. This reprogramming process enriched trimethylated histone H3 lysine 4, monoacetylated histone H3 lysine 9, and Baf60c at the Nkx2.5 cardiac enhancer region by the chromatin immunoprecipitation quantitative polymerase chain reaction assay. Protein-induced CPCs transplanted into rat hearts after myocardial infarction improved cardiac function, and this was related to differentiation into cardiomyocyte-like cells. These findings demonstrate that the highly efficient protein-transduction method can directly reprogram HDFs into CPCs. This protein reprogramming strategy lays the foundation for future refinements both in vitro and in vivo and might provide a source of CPCs for regenerative approaches.
Significance
The findings from the present study have demonstrated an efficient protein-transduction method of directly reprogramming fibroblasts into cardiac progenitor cells. These results have great potential in cell-based therapy for cardiovascular diseases.
Introduction
Despite major developments in cardiac infarction therapy, repopulation of the injured heart remains a severe challenge. Because stem cells possess the capacity to produce a large number of functional specific cell types, either in situ or ex vivo, for transplantation, stem cells can be used to replace lost or damaged tissue [1–3]. Although embryonic stem cells and induced pluripotent stem cells (iPSCs) possess cardiogenic potential, the efficiency of cardiac differentiation, the risk of tumor formation, and issues of cellular rejection are barriers to clinical application. Transplantation of cardiac progenitor/stem cells to improve cardiac function holds clinical potential [4–6]. However, autologous cardiac progenitor cells (CPCs) exist in very low numbers within the heart. Also, whether they have great heart regenerative capacity remains controversial. Thus, a larger source of cells for cardiac regeneration is needed [7, 8].
The discovery of iPSCs toppled the view that development can only proceed unidirectionally [9] and suggests that the pluripotent state can be bypassed [10]; that is, noncardiomyocytes can be directly converted into cardiomyocyte-like cells. One group reported on a defined small-molecule cocktail (SB431542, CHIR99021, parnate, and forskolin) with bone morphogenetic protein 4 (BMP4) and only one transcription factor, Oct4, which enabled highly efficient conversion of mouse fibroblasts into cardiac cells without entering the pluripotent state [11]. Takeuchi and Bruneau showed that two cardiac transcription factors, Gata4 and Tbx5, and a cardiac-specific subunit of BRG/Brm-associated factor (BAF) chromatin-remodeling complexes, Baf60c, could direct ectopic differentiation of mouse mesoderm into beating cardiomyocytes [12]. Gata4, Mef2c, and Tbx5 (GMT) could directly reprogram fibroblasts to cardiomyocyte-like cells both in vitro and in vivo [13, 14]. However, it was also reported that GMT overexpression in tail tip fibroblasts and cardiac fibroblasts was inefficient at inducing molecular and electrophysiological phenotypes of mature cardiomyocytes [15]. Another study reprogrammed noncardiomyocytes into functional cardiac-like myocytes by forced expression of Gata4, Hand2, Mef2c, and Tbx5 (GHMT) [16]. Moreover, the addition of ESRRG, MESP1, myocardin, and ZFPM2 to GMT could further enhance reprogramming [17]. Even myocardin, together with Tbx5 and Mef2c, also successfully reprogrammed cells in vitro [18]. MicroRNAs, which repress gene expression, can mediate direct conversion of fibroblasts to cardiomyocytes with or without other reprogramming factors [19–21]. Despite certain successes, some problems remain unresolved. These include safety issues stemming from the integration of foreign genes into the host genome and from the use of viral vectors. A promising alternative strategy for reprogramming is to not use viruses for CPC clinical translation.
Proteins, especially transcription factors, can modulate the gene expression of the host cells, leading to complete transformation of the parental phenotype. Therefore, researchers have sought to reprogram cells by directly delivering proteins of defined transcription factors. Delivering reprogramming proteins to cells is free of virus and no DNA integration method is needed. Because proteins with a high molecular weight cannot freely enter cells, several protein delivery methods are available for the reprogramming strategy. Proteins can be transferred using streptolysin O-mediated reversible permeabilization [22, 23]. 11R-tagged recombinant transcription factors can effectively generate iPSCs [24]. Nanotube-mediated protein delivery systems also have been shown to activate genes for pluripotency in somatic cells [25]. Similarly, delivered proteins can also transdifferentiate fibroblast to cardiomyocytes. It has been demonstrated that the purified 11R-tagged recombinant transcription factors TAT-ETS2 and TAT-MESP1 are capable of reprogramming human dermal fibroblasts (HDFs) into immature cardiomyocytes [26]. However, the reprogramming efficiency is very low, and it is still unclear which transduction system would be the most suitable for protein-based cell reprogramming. The QQ-reagent is a recently developed protein transduction reagent (US patent 2009/0298111 A1). It can deliver proteins into mammalian cells with high efficiency and low cytotoxicity [27]. In the present study, we report the effect of this de novo protein delivery method on reprogramming HDFs to CPCs. Our study lays the foundation for developing methods for protein reprogramming and our results hold promise for the regeneration of cardiomyocytes after myocardial infarction (Ml).
Materials and Methods
A detailed methods section is provided in the supplemental online data.
Results
Modified Proteins Can Be Delivered Into Human Dermal Fibroblasts With High Efficiency
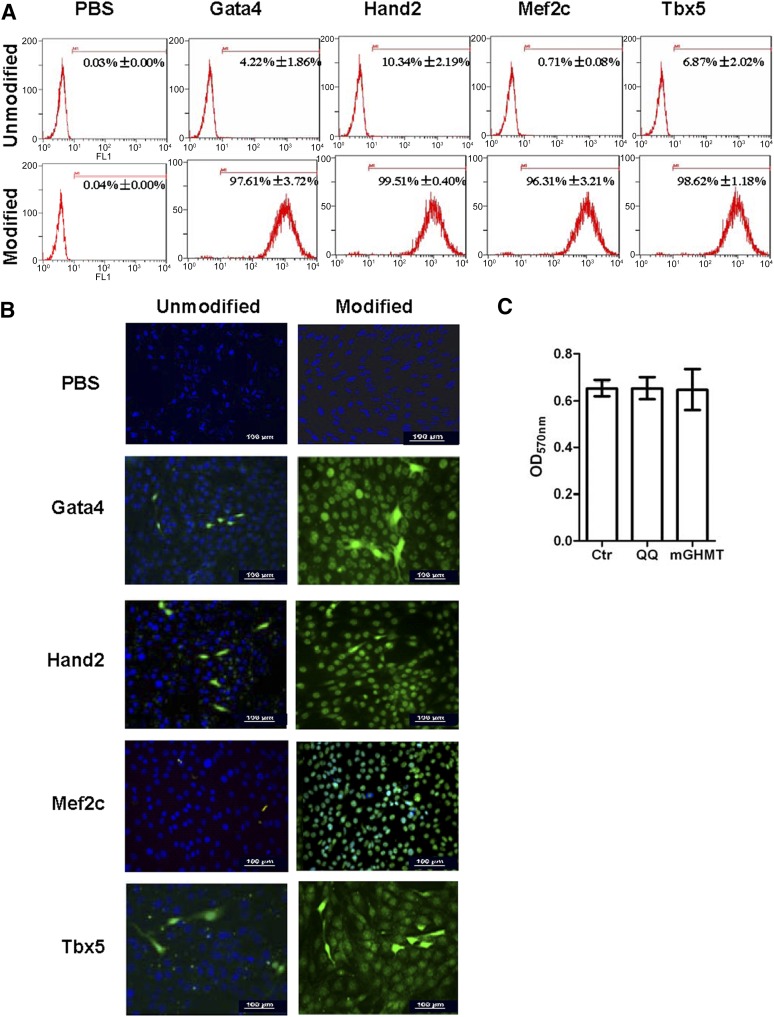
In the present study, we used a simple, nonviral-based protein delivery system, the QQ-reagent, for protein-induced CPCs (piCPCs) from HDFs without genetic modifications. The proteins were individually modified by the QQ-reagent and incubated with fibroblasts for 12 hours. We then tested the uptake of these proteins in HDFs. Flow cytometry analysis demonstrated that >96% of cells were positive for modified proteins after 12 hours of delivery (Fig. 1A). Immunostaining indicated that the proteins were released from the complex and transported to the nucleus of the fibroblasts (Fig. 1B). QQ-modified proteins were more highly transduced into cells compared with unmodified proteins. Moreover, the QQ-reagent and modified proteins had no cytotoxicity on HDFs (Fig. 1C). We also investigated the time-course uptake of modified protein in HDFs. Immunostaining showed that little protein had been delivered into HDFs at 6 hours. However, greater protein uptake was seen at 12 and 24 hours than at 6 hours at the same concentration (supplemental online Fig. 1).
Figure 1.
Cellular uptake of proteins with or without QQ-reagent modification after incubation for 12 hours. Anti-His tag antibody was used to show Gata4/Hand2/Mef2c/Tbx5 proteins. (A): Flow cytometry analysis showed that Gata4 was greatly increased from 4.22% ± 1.86% to 97.61% ± 3.72%; Hand2 was increased from 10.34% ± 2.19% to 99.51% ± 0.40%; Mef2c was increased from 0.71% ± 0.08% to 96.31% ± 3.21%; and Tbx5 was increased from 6.87% ± 2.02% to 98.62% ± 1.18% after QQ-reagent modification. Vehicle was used as a negative control with or without QQ-reagent modification. (B): QQ-reagent-modified or -unmodified proteins were detected by immunostaining in fibroblast after transduction for 12 hours. Green cells were His tag-positive; 4′,6-diamidino-2-phenylindole was used to stain nuclei (blue). All images were merged. Vehicle was used as the negative control with or without QQ-reagent modification. Scale bars = 100 μm. (C): Cell viability was detected by MTT assay. No statistically significant differences were found among these three groups (p > .05; n = 3). Abbreviations: Ctr, untreated cell; mGHMT, modified Gata4/Hand2/Mef2c/Tbx5; OD, optical density; PBS, phosphate-buffered saline; QQ, QQ-reagent treatment.
Gata4, Hand2, Mef2c, and Tbx5 Together Reprogram HDFs Toward a CPC State
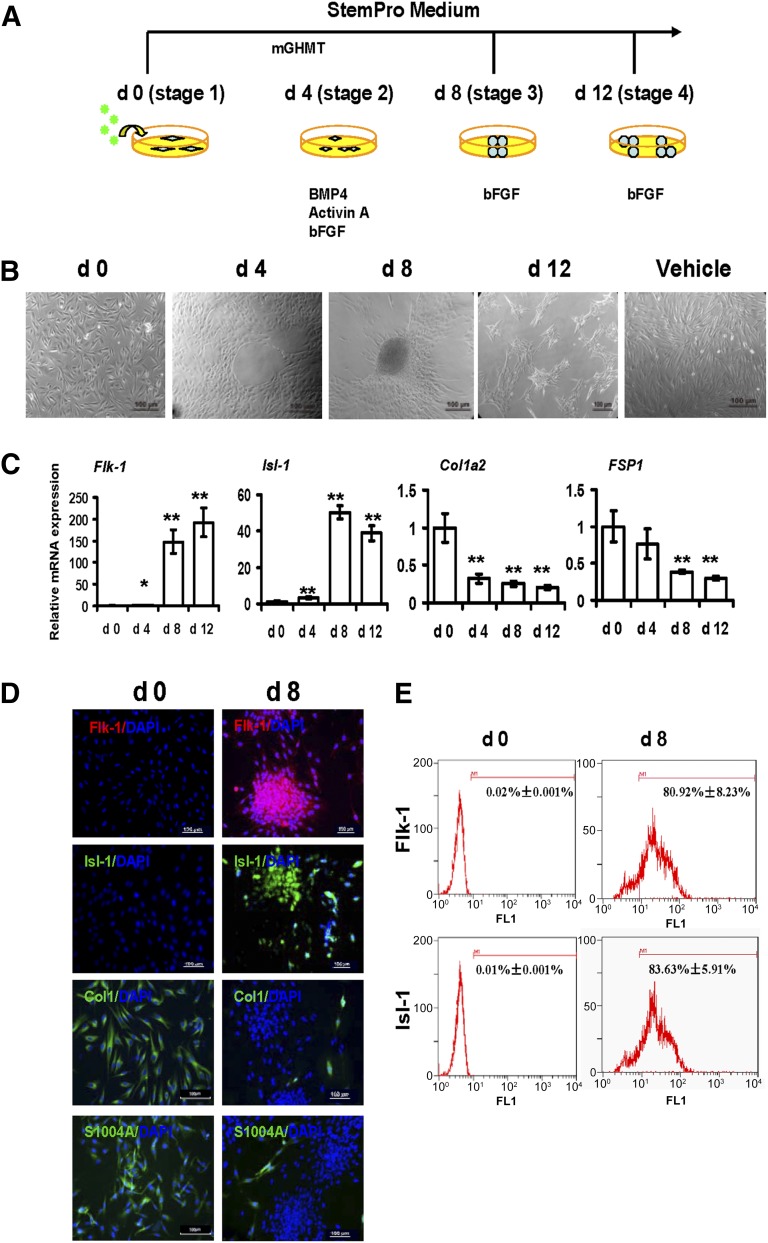
We transduced HDFs using four QQ-reagent-modified cardiac transcription factors (Gata4/Hand2/Mef2c/Tbx5 [mGHMT]). We classified the protein-induced reprogramming step into three stages during piCPC generation by mGHMT (Fig. 2A). We also used BMP4 and activin A as cardioinductive growth factors and basic fibroblast growth factor (bFGF) as a progenitor cell maintenance factor [11]. The Isl-1 gene, a CPC marker, was used to optimize reprogramming efficiency. Isl-1 expression was significantly increased from day 4 to day 32 after mGHMT reprogramming compared with days 0 and 2 (p < .001; supplemental online Fig. 2A). BMP4, activin A, and bFGF were added to the mGHMT reprogramming medium at day 4. mGHMT, plus BMP4 and activin A, greatly upregulated Isl-1 expression compared with the expression in other groups, with or without bFGF at day 8 (p < .001; supplemental online Fig. 2B). Withdrawing BMP4 and activin A at day 8 maintained Isl-1 expression, but it was downregulated without bFGF at day 12 (supplemental online Fig. 2C). At stage 1, the cells exhibited a long rhombus shape. At stage 2, the rhombus-shaped cells had proliferated and physically touched each other. Also, the cells became more compact and began to form circles. At stage 3, the cells had begun to aggregate and started showing typical colony formation by days 4–8. At stage 4, the cells had also aggregated and had formed many small colonies after digestion and passage (Fig. 2B). No morphology changes were seen in the vehicle control and green fluorescent protein (GFP) control group (Fig. 2B; supplemental online Fig. 3A). Consistent with previous findings [28–31], robust expression of Flk-1 and Isl-1 (cardiac progenitor markers) was detected during the early cardiac reprogramming stage by quantitative polymerase chain reaction (qPCR) (Fig. 2C). Flk-1 and Isl-1 became highly misexpressed by stage 3 after protein induction. Antibodies specific to these markers were increased in the piCPC colonies at day 8 and after cell passage (Fig. 2D). The fibroblast markers, type I collagen a2 (Col1a2) and fibroblast-specific protein 1 (FSP1) [32], were significantly downregulated after the fibroblasts were reprogrammed to piCPCs at the gene (Fig. 2C) and protein (Fig. 2D) level. As a protein delivery control, GFP was highly transduced into fibroblasts (supplemental online Fig. 3B). No change in Flk-1 and Isl-1 expression was detected after GFP transduction (supplemental online Fig. 3B). The percentage of Flk-1- and Isl-1-positive cells had increased approximately 80.92% ± 8.23% and 83.63% ± 5.91% after reprogramming for 8 days compared with those untreated (0.02% ± 0.001% and 0.01% ± 0.001%, respectively; Fig. 2E). These results suggest that the current reprogramming protocol could successfully downregulate fibroblast markers and upregulate cardiac progenitor-specific markers.
Figure 2.
Generation of protein-induced cardiac progenitor cells by modified transcript proteins. (A): Strategy of protein-induced cardiac progenitor cell (piCPC) generation. (B): Cell colonies were initially observed around days 4–8 and could be passaged to many small colonies around day 12. Representative phase contrast images are shown. The control was untreated human dermal fibroblasts in vehicle medium after 8 days. Scale bars = 100 μm. (C): quantitative polymerase chain reaction analysis of cardiac progenitor genes Flk-1 and Isl-1 in piCPCs. Fibroblast markers Col1a2 and FSP1 were also detected (∗, p < .05; ∗∗, p < .01 vs. day 0 control; error bars indicate SD; n = 3). (D): Representative fluorescent images are shown with typical cardiac progenitor markers Flk-1 (red) and Isl-1 (green) and fibroblast markers ColI (green) and FSP-1 (S100A4) (green) before and after reprogramming at day 8. DAPI staining was performed to visualize nuclei (blue) and all images were merged. Scale bars, 100 μm. (E): Flow cytometry analysis demonstrated Flk-1 and Isl-1 expressions were increased from d0 to d8 separately. Abbreviations: bFGF, basic fibroblast growth factor; BMP4, bone morphogenetic protein 4; ColI, collagen I; d, day; DAPI, 4′,6-diamidino-2-phenylindole; FSP1, fibroblast-specific protein 1; mGHMT, modified Gata4/Hand2/Mef2c/Tbx5.
piCPCs Differentiate Into Three Cardiac Lineages Under Cardiac Differentiation Conditions
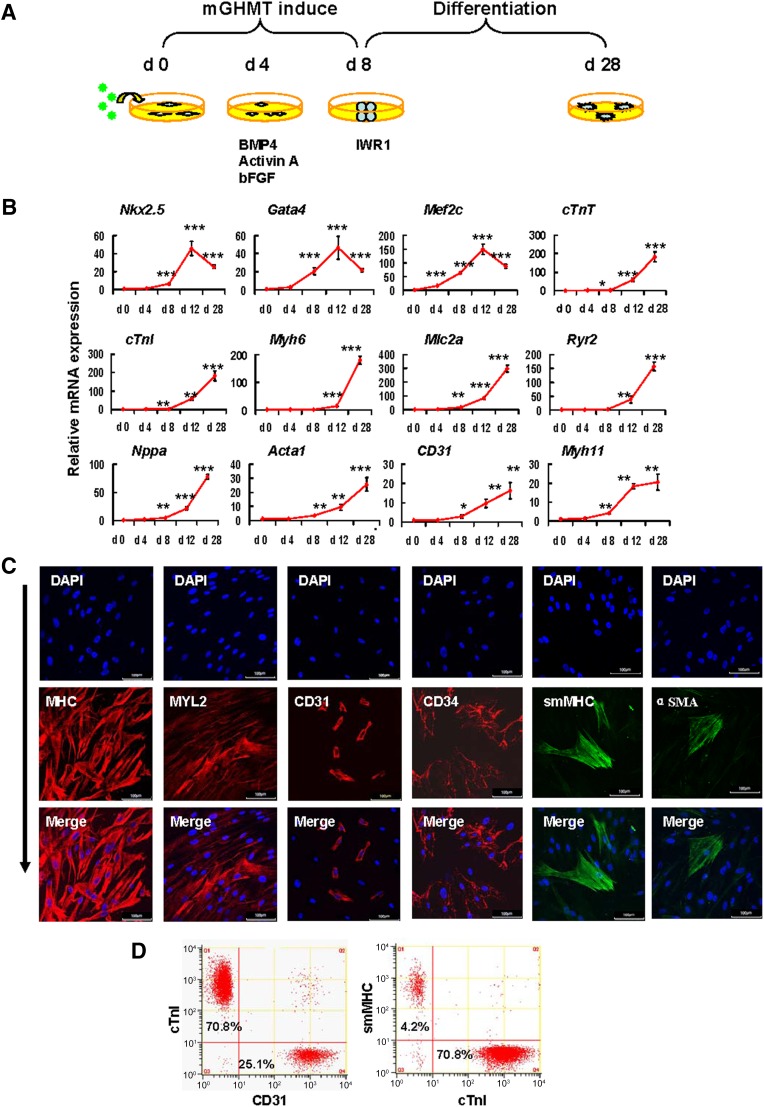
It is inherent for piCPCs to differentiate into three cardiac lineages; however, guiding the progenitor cells to differentiate to a specific lineage is challenging. Moreover, the ability to achieve controlled differentiation toward a specific lineage would further strengthen the clinical application of these cells. To investigate the ability of piCPCs to form the three cardiac lineages, we modified the cardiac differentiation strategy (Fig. 3A) using the findings from a previous report [10]. Wnt inhibition could generate cardiomyocytes from human embryonic stem cell-derived mesoderm cells. The addition of the small molecule IWR-1, an inhibitor of the canonical Wnt pathway, led to the acquisition of terminally differentiated cardiomyocytes [33–35]. However, we showed that piCPCs could differentiate into, not only cardiomyocytes, but also endothelial cells and smooth muscle cells in the presence of IWR1 on gelatin-coated dishes. The gene expression of transcription factors for cardiac myocyte differentiation, including Gata4, Mef2c, and Nkx2.5, were significantly upregulated in differentiated cells. Similarly, the expression levels of cardiomyocyte filament proteins, cardiac troponin I (cTnI) and cardiac troponin T, were also significantly upregulated. CPCs can differentiate into smooth muscle cells and endothelial cells, other than cardiomyocytes, during embryonic development. Consistently, when we switched to differentiation medium, expression of the endothelial marker gene CD31 and the smooth muscle cell maker gene MYH11 was upregulated (Fig. 3B).
Figure 3.
Protein-induced cardiac progenitor cells (piCPCs) differentiated into three cardiac lineages: cardiomyocytes, endothelial cells, and smooth muscle cells. (A): Schematic representation of the strategy to differentiate piCPCs in differentiation medium with IWR1 factor. (B): Quantitative data of mRNA expression of cardiac lineage marker genes (∗, p < .05; ∗∗, p < .01; and ∗∗∗, p < .001 vs. day 0 control; error bars indicate SD; n = 3). (C): Immunofluorescent staining for MHC, MYL2, CD31, CD34, smMHC, and αSMA. The combination of the four factors, GHMT, induces abundant MHC and Myl2, and some expression of CD31 and smMHC 28 days after transduction. Nuclei were counter stained with DAPI. Scale bars = 100 μm. (D): Flow cytometry analysis for cTnI, CD31, and smMHC. mGHMT plus IWR1 significantly enhances cTnI expression, and, to a lesser extent, CD31 and smMHC expression. Abbreviations: αSMA, α-smooth muscle actin; BMP4, bone morphogenetic protein 4; cTnI, cardiac troponin I; cTnT, cardiac troponin T; d, day; DAPI, 4′,6-diamidino-2-phenylindole; GHMT, Gata4/Hand2/Mef2c/Tbx5; mGHMT, modified GHMT; MHC, myosin heavy chain; MYL2, myosin light chain 2; smMHC, smooth muscle myosin heavy chain.
Fluorescent immunostaining showed that cardiac markers (myosin heavy chain [MHC] and myosin light chain 2), endothelial cell markers (CD31 and CD34), smooth muscle cells (smooth muscle MHC [smMHC] and α-smooth muscle actin) were expressed in differentiated cells (Fig. 3C). Of the differentiated cells, 15%–20% expressed CD31 and 2%–5% expressed smMHC, suggesting that few piCPCs had differentiated into endothelial cells and smooth muscle cells. In contrast, most piCPCs (>70%) had differentiated into cTnI-positive cells (Fig. 3D). The results of this comprehensive gene and protein expression analysis suggested that piCPCs could differentiate into the three cardiac lineages, and most of them were cardiomyocyte-like cells.
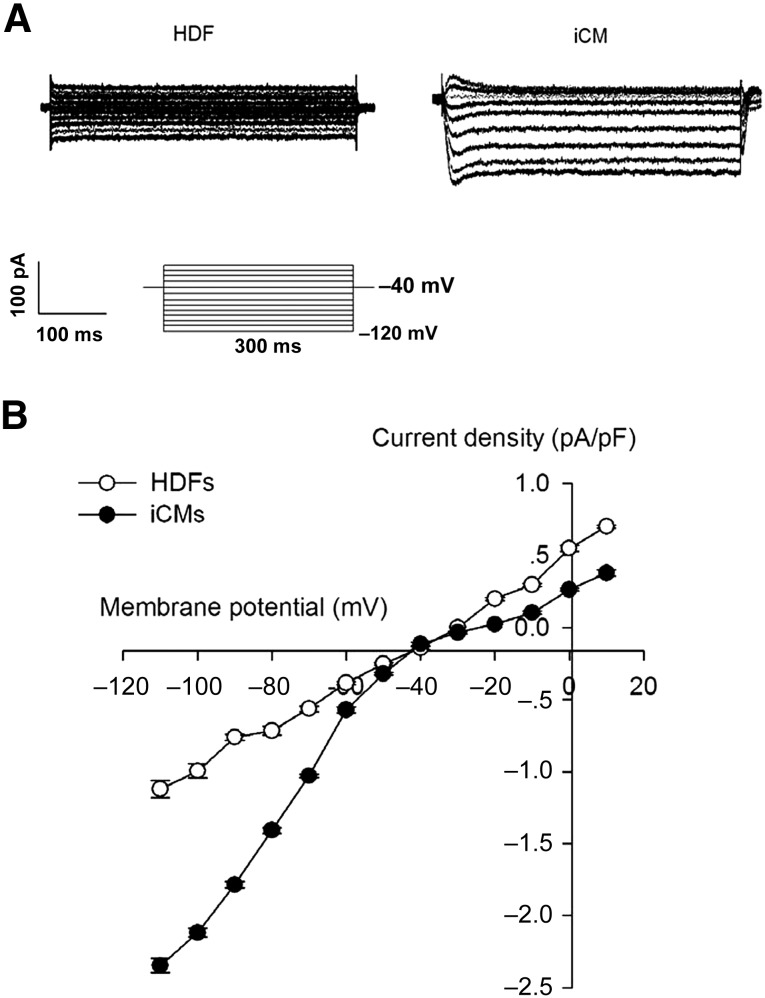
Beating cells were detected after the piCPCs had differentiated for 20 days (supplemental online Movie 1). The current-voltage relationship of IK1 was determined by evoking current-voltage steps to various potentials from −120 mV to 10 mV from a holding potential of −40 mV at 0.2 Hz. No IK1 current was detected in the HDF cells. However, IK1 can be recorded in beating cardiomyocytes after 20 days of differentiation from piCPCs (Fig. 4A). The density of IK1 in the induced-cardiomyocyte group was significantly increased (p < .05) compared with that in the HDF group (Fig. 4B). The proof of cardiomyocyte-like function derived from piCPCs was displayed by calcium signals (supplemental online Movie 2). Those Ik1 current and spontaneous Ca2+ transients revealed that the piCPC-differentiated cells had cardiomyocytes properties.
Figure 4.
Electrophysiology recordings of inward rectifier potassium current (IK1). (A): Typical Ik1 currents in cells. (B): Peak IK1 densities of untreated HDF group and iCM group (p < .05). Data shown as mean ± SD (n = 3). Abbreviations: HDF, human dermal fibroblast; iCM, induced cardiomyocyte.
Epigenetic Chromatin Modifications of Nkx2.5 Enhancer After Protein Reprogramming Reflect a Progenitor Cell State
The cardiomyocyte enhancer recapitulates the expression pattern of the endogenous gene in cardiogenic precursors from the onset of lineage specification to looping of the heart tube [36]. The mammalian homolog of Drosophila tinman, Nkx2.5, plays an early role in regulating cardiac genes and morphogenesis. Our results have demonstrated that Nkx2.5 was upregulated during mGHMT protein reprogramming at both mRNA (Fig. 3A) and protein (Fig. 5A) levels. Immunofluorescence showed that Nkx2.5 was located in the nucleus of piCPCs (Fig. 5B).
Figure 5.
Effect of GHMT on Nkx2.5 chromatin remodeling. (A): Western blotting showed that Nkx2.5 is upregulated by 12 days of treatment with GHMT. Histone H3 was used as an internal control. (B): Immunostaining showed that Nkx2.5 (green) is located in the nucleus of piCPCs after 12 days of reprogramming. DAPI staining was performed to visualize the nucleus (blue), and the images were merged. Scale bar = 100 μm. High-magnification views in insets show the location of Nkx2.5. (C): Diagram of the Nkx2.5 promoter shows the relationship of the −10-kb and −2.8-kb regions that are essential to direct expression in the heart versus stomach, pharynx, and thyroid, respectively. (D): Chromatin immunoprecipitation quantitative polymerase chain reaction assessing Baf60c, H3K4me3, and H3K9ac histone modifications of the Nkx2.5 −10-kb and −2.8-kb enhancers and a −15-kb nonenhancer region in HDFs and in day 12 piCPCs. Fold enrichment was normalized to the IgG control (∗, p < .05; ∗∗, p < .01 vs. relevant HDF control). All data are presented as mean ± SD. Abbreviations: DAPI, 4′,6-diamidino-2-phenylindole; GHMT, Gata4/Hand2/Mef2c/Tbx5; HDFs, human dermal fibroblasts; piCPCs, protein-induced cardiac progenitor cells.
The Nkx2.5 gene is ideally suited for this experiment, because it is activated in CPCs and harbors two well-defined enhancers: an early cardiac-specific enhancer at −9 kb from the start site of transcription and a stomach-, pharynx-, and thyroid-specific enhancer at −2.8 kb [37, 38]. By sequence comparison with human, mouse, rat, and dog genes, evolutionarily conserved regions were found within the 10-kb regulatory sequence [39]. The site of the conserved human cardiac-specific enhancer was at −10 kb (−10,764 base pairs [bp] to −10,464 bp; Fig. 5C). This −10-kb enhancer contains two high-affinity binding sites for Gata and a high-affinity binding site for Mef2. The signals that make the enhancer accessible during lineage commitment are unknown, although, once open, its activity depends on Gata4 [37]. To determine whether mGHMT has any effect on chromatin state, we analyzed the enrichment of histone modifications in the enhancer regions of the cardiac progenitor-specific gene Nkx2.5. We analyzed the enrichment of trimethylated histone H3 lysine 4 (H3K4me3) and monoacetylated histone H3 lysine 9 (H3K9ac), which mark transcriptionally active chromatin [40]. These makers were examined in both HDFs and day-12 piCPCs using chromatin immunoprecipitation (ChIP), followed by qPCR. After reprogramming, H3K4me3 and H3K9ac increased on the −10-kb enhancer region; however, no enrichment was found at the noncardiac enhancer or nonenhancer region of Nkx2.5 in piCPCs compared with HDFs (Fig. 5D).
A BAF chromatin remodeling protein, Baf60c, can initiate cardiogenic differentiation in mouse embryos by recognizing the chromatin of cardiac genes to enhance target selectivity [41]. To further investigate the mechanism of protein reprogramming, we determined whether Baf60c localized to a native cardiac enhancer within chromatin during mGHMT reprogramming. The distribution of Baf60c showed a pattern identical to two histone modifications (H3K4me3 and H3K9ac) associated with active enhancers on the −10-kb cardiac enhancer region by ChIP-qPCR (Fig. 5D). These results suggested that fibroblast-derived piCPCs gained an open chromatin status for at least some cardiac progenitor-specific genes.
These results show the combinatorial contribution of four DNA-binding transcription factors in regulating the cardiac transcriptome and provide evidence that histone modifications and chromatin remodeling proteins are important in this process.
piCPC Transplantation Improves Heart Function After Myocardial Infarction
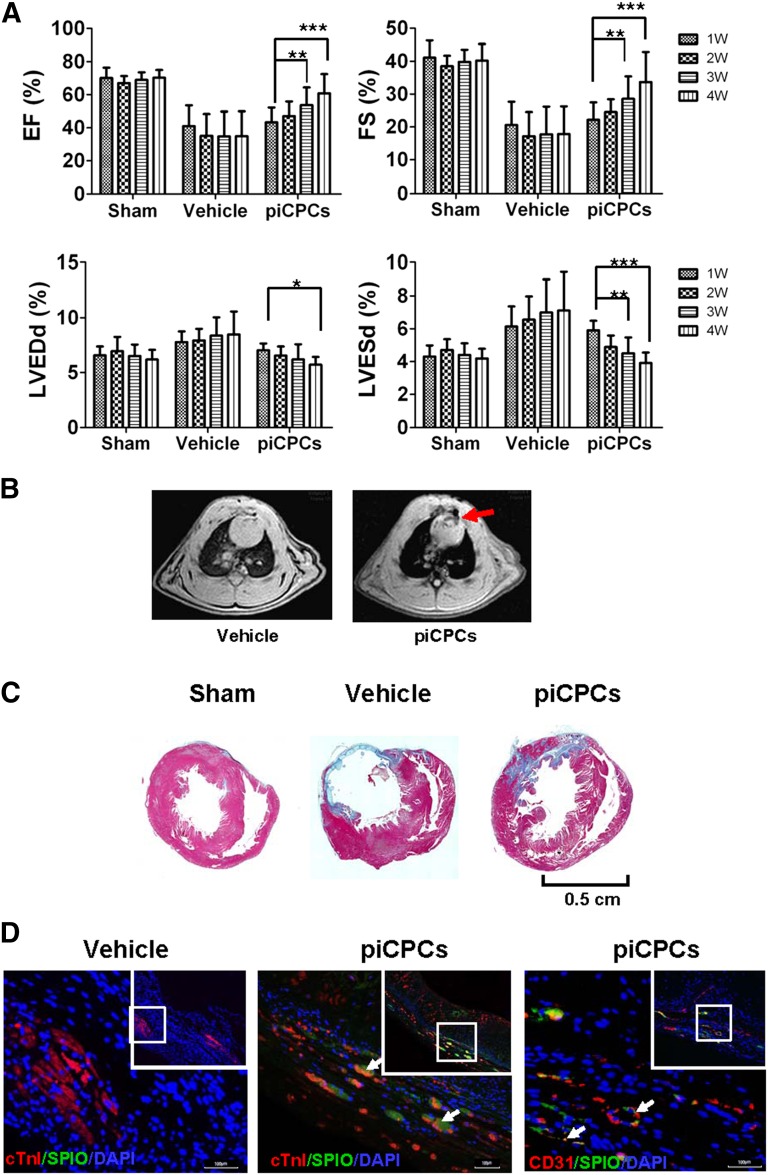
To investigate whether piCPCs can improve cardiac function after myocardial infarction (MI), we injected piCPCs into rat heart tissue immediately after left coronary artery ligation. Echocardiographic analysis showed that both the ejection fraction and fractional shortening were progressively improved at 1–4 weeks in hearts transplanted with piCPCs compared with the vehicle group. The left ventricular internal diameter at end-diastole and left ventricular internal diameter at end-systole were significantly decreased in the hearts receiving piCPCs at 4 weeks after MI compared with the vehicle groups (Fig. 6A; supplemental online Fig. 4).
Figure 6.
In vivo delivery of protein-induced cardiac progenitor cells improves cardiac function after myocardial infarction. (A): EF, FS, LVDd, and LVDs were analyzed by echocardiography (∗, p < .05; ∗∗, p < .01; ∗∗∗, p < .001 vs. relevant 1 week; all data are presented as mean ± SD, n = 8). (B): Transplanted cells were detected by magnetic resonance imaging 4 weeks after myocardial infarction (MI). Red arrow points to the signal loss due to SPIO-labeled cells. (C): Masson trichrome staining on heart sections 4 weeks after MI injection in sham, vehicle, and piCPC groups. Scale bar = 0.5 cm. (D): Immunofluorescent staining for cTnI (red), CD31 (red), and anti-dextran (SPIO, green) of heart sections after piCPCs were transplanted 4 weeks after MI. White arrows point to transplanted cells or colocalization of cTnI or CD31 with SPIO. Scale bars = 100 μm. Abbreviations: cTnI, cardiac troponin I; DAPI, 4′,6-diamidino-2-phenylindole; EF, ejection fraction; FS, fractional shortening; LVDd, left ventricular internal diameter at end-diastole; LVDs, left ventricular internal diameter at end-systole; piCPCs, protein-induced cardiac progenitor cell; SPIO, superparamagnetic iron oxide; W, week.
Rat cardiac axial magnetic resonance T2-weighted imaging showed signal loss in infarcted hearts transplanted with superparamagnetic iron oxide (SPIO)-labeled cells compared with the vehicle control group (Fig. 6B, red arrow). Moreover, fibrosis of infarcted areas was observed in Masson's trichrome-stained myocardial sections after coronary ligation at 4 weeks. Hearts undergoing piCPC transplantation showed decreased fibrosis compared with those treated with vehicle at 4 weeks after MI (Fig. 6C). Immunofluorescence demonstrated that the engrafted piCPCs with the cell tracer SPIO were positive for cTnI and CD31 (Fig. 6D). These results suggest that transplantation of piCPCs played an important role in attenuating left ventricular remodeling after MI. These results further support the observation made in vitro that piCPCs can develop into cardiac lineage cells.
Discussion
It was reported that retroviral expression of GMT resulted in much less efficient cardiomyocyte reprogramming in mouse MI experiments, even in nude mice [42], compared with that of forced overexpression of GHMT [16]. On that basis, we used GHMT with four additional growth factors to directly reprogram CPCs from HDFs. We have demonstrated that the combination of four QQ-modified transcription factors (GHMT) and three growth factors (BMP4, activin A, and bFGF) can rapidly and efficiently induce more than 80% piCPCs from HDFs. The piCPCs were similar to the cardiac progenitors in morphological appearance, colony formation, activation of endogenous cardiac marker genes, and cardiac lineage differentiation potential. The present study has demonstrated that functional cardiac progenitors can be generated from somatic cells via a nonviral method. Although much refinement and characterization of the reprogramming process will be necessary, the findings we have reported provide an improved and potentially safer method of generating CPCs for regenerative purposes.
Heart development is controlled at different molecular levels by cardiac gene regulatory networks, involving transcription factors, signal pathways, epigenetic factors, and microRNAs. It is well known that cardiac transcription factors can bind to DNA in host cells to trigger the specific transcription network. The four reprogramming factors Gata4, Hand2, Mef2c, and Tbx5, are core transcription factors during early heart development [43, 44]. Additionally, cytokines, BMP4, activin A, and bFGF play an important role in promoting piCPC generation in our study. Significant challenges remain in our ability to convert fibroblasts to cardiomyocyte-like cells, and a greater understanding of cardiovascular epigenetics is needed to increase the translational potential of this strategy. In contrast to the relatively advanced knowledge of signaling pathways, the epigenetic alterations that accompany or potentiate cardiogenesis are largely unexplored. The activity of Baf60c is necessary during cardiac commitment but is dispensable for maintenance of the differentiated phenotype. Baf60c can mediate interactions between cardiac transcription factors and the BAF complex adenosine triphosphatase Brg1, thereby potentiating the activation of target genes [45]. In our study, baf60c was enriched at Nkx2.5 cardiac-specific enhancers, which also contain Gata4- and Mef2c-binding sites, suggesting that they might interact during piCPC generation. Similar to a previous report [36], we found that H3K9ac and H3K4me3 are enriched at active promoters of Nkx2.5. They could be potential mechanisms of enhanced cardiac-specific gene expression in our study.
Conclusion
With ischemic cardiomyocyte loss and post-MI fibrosis, the ventricle wall becomes thin. piCPCs that are directly reprogrammed in vitro might be applicable for replacing the dead heart muscle. The use of undifferentiated CPCs as building blocks to grow specific tissue types in vivo is of great interesting for regenerating the myocardium. However, it will be critical to determine whether key physiological properties are faithfully reproduced after reprogramming, such as the electrophysiological properties of action potentials and ionic currents. Further study is also needed to investigate the characteristics of in vivo differentiated cardiomyocytes from piCPCs in their native environment, which might promote survival, maturation, and coupling with neighboring cells.
Supplementary Material
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grants 81120108003 and 81330007), the Guangdong Science and Technology Program of International Cooperation Project (Grant 2014A050503047), and the project of Zhujiang Science and Technology New Star of Guangzhou (Grant 2011J2200049).
Author Contributions
X.-H.L.: conception and design, collection and/or assembly of data, data analysis and interpretation, manuscript writing; Q.L.: conception and design, collection and/or assembly of data; L.J.: collection and/or assembly of data; C.D.: electrophysiology analysis; Z.L.: animal magnetic resonance imaging, data analysis and interpretation; Y.F.: animal model, cell transplantation; M.Z.: echocardiography, collection and/or assembly of data, data analysis and interpretation; H.T.: flow cytometry analysis; Y.F.: collection and/or assembly of data, data analysis and interpretation; Z.S.: construct protein expression vectors; J.W.: conception and design, financial support, final approval of manuscript; X.-Y.Y.: conception and design, data analysis and interpretation, manuscript writing, financial support, final approval of manuscript.
Disclosure of Potential Conflicts of Interest
The authors indicated no potential conflicts of interest.
References
- 1.Zwi L, Caspi O, Arbel G, et al. Cardiomyocyte differentiation of human induced pluripotent stem cells. Circulation. 2009;120:1513–1523. doi: 10.1161/CIRCULATIONAHA.109.868885. [DOI] [PubMed] [Google Scholar]
- 2.Li X, Yu X, Lin Q, et al. Bone marrow mesenchymal stem cells differentiate into functional cardiac phenotypes by cardiac microenvironment. J Mol Cell Cardiol. 2007;42:295–303. doi: 10.1016/j.yjmcc.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 3.Feng Y, Huang W, Meng W, et al. Heat shock improves Sca-1+ stem cell survival and directs ischemic cardiomyocytes toward a prosurvival phenotype via exosomal transfer: a critical role for HSF1/miR-34a/HSP70 pathway. Stem Cells. 2014;32:462–472. doi: 10.1002/stem.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bolli R, Chugh AR, D’Amario D, et al. Cardiac stem cells in patients with ischaemic cardiomyopathy (SCIPIO): Initial results of a randomised phase 1 trial. Lancet. 2011;378:1847–1857. doi: 10.1016/S0140-6736(11)61590-0. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 5.Makkar RR, Smith RR, Cheng K, et al. Intracoronary cardiosphere-derived cells for heart regeneration after myocardial infarction (CADUCEUS): A prospective, randomised phase 1 trial. Lancet. 2012;379:895–904. doi: 10.1016/S0140-6736(12)60195-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Malliaras K, Makkar RR, Smith RR, et al. Intracoronary cardiosphere-derived cells after myocardial infarction: Evidence of therapeutic regeneration in the final 1-year results of the CADUCEUS trial (CArdiosphere-Derived aUtologous stem CElls to reverse ventricUlar dySfunction) J Am Coll Cardiol. 2014;63:110–122. doi: 10.1016/j.jacc.2013.08.724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Laflamme MA, Murry CE. Heart regeneration. Nature. 2011;473:326–335. doi: 10.1038/nature10147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garbern JC, Lee RT. Cardiac stem cell therapy and the promise of heart regeneration. Cell Stem Cell. 2013;12:689–698. doi: 10.1016/j.stem.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 10.Efe JA, Hilcove S, Kim J, et al. Conversion of mouse fibroblasts into cardiomyocytes using a direct reprogramming strategy. Nat Cell Biol. 2011;13:215–222. doi: 10.1038/ncb2164. [DOI] [PubMed] [Google Scholar]
- 11.Wang H, Cao N, Spencer CI, et al. Small molecules enable cardiac reprogramming of mouse fibroblasts with a single factor, Oct4. Cell Rep. 2014;6:951–960. doi: 10.1016/j.celrep.2014.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takeuchi JK, Bruneau BG. Directed transdifferentiation of mouse mesoderm to heart tissue by defined factors. Nature. 2009;459:708–711. doi: 10.1038/nature08039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ieda M, Fu JD, Delgado-Olguin P, et al. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qian L, Huang Y, Spencer CI, et al. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature. 2012;485:593–598. doi: 10.1038/nature11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen JX, Krane M, Deutsch MA, et al. Inefficient reprogramming of fibroblasts into cardiomyocytes using Gata4, Mef2c, and Tbx5. Circ Res. 2012;111:50–55. doi: 10.1161/CIRCRESAHA.112.270264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song K, Nam YJ, Luo X, et al. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature. 2012;485:599–604. doi: 10.1038/nature11139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fu JD, Stone NR, Liu L, et al. Direct reprogramming of human fibroblasts toward a cardiomyocyte-like state. Stem Cell Rep. 2013;1:235–247. doi: 10.1016/j.stemcr.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Protze S, Khattak S, Poulet C, et al. A new approach to transcription factor screening for reprogramming of fibroblasts to cardiomyocyte-like cells. J Mol Cell Cardiol. 2012;53:323–332. doi: 10.1016/j.yjmcc.2012.04.010. [DOI] [PubMed] [Google Scholar]
- 19.Nam YJ, Song K, Luo X, et al. Reprogramming of human fibroblasts toward a cardiac fate. Proc Natl Acad Sci USA. 2013;110:5588–5593. doi: 10.1073/pnas.1301019110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jayawardena TM, Egemnazarov B, Finch EA, et al. MicroRNA-mediated in vitro and in vivo direct reprogramming of cardiac fibroblasts to cardiomyocytes. Circ Res. 2012;110:1465–1473. doi: 10.1161/CIRCRESAHA.112.269035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muraoka N, Yamakawa H, Miyamoto K, et al. MiR-133 promotes cardiac reprogramming by directly repressing Snai1 and silencing fibroblast signatures. EMBO J. 2014;33:1565–1581. doi: 10.15252/embj.201387605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cho HJ, Lee CS, Kwon YW, et al. Induction of pluripotent stem cells from adult somatic cells by protein-based reprogramming without genetic manipulation. Blood. 2010;116:386–395. doi: 10.1182/blood-2010-02-269589. [DOI] [PubMed] [Google Scholar]
- 23.Bui HT, Kwon DN, Kang MH, et al. Epigenetic reprogramming in somatic cells induced by extract from germinal vesicle stage pig oocytes. Development. 2012;139:4330–4340. doi: 10.1242/dev.086116. [DOI] [PubMed] [Google Scholar]
- 24.Zhou H, Wu S, Joo JY, et al. Generation of induced pluripotent stem cells using recombinant proteins. Cell Stem Cell. 2009;4:381–384. doi: 10.1016/j.stem.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cho SJ, Choi HW, Cho J, et al. Activation of pluripotency genes by a nanotube-mediated protein delivery system. Mol Reprod Dev. 2013;80:1000–1008. doi: 10.1002/mrd.22263. [DOI] [PubMed] [Google Scholar]
- 26.Islas JF, Liu Y, Weng KC, et al. Transcription factors ETS2 and MESP1 transdifferentiate human dermal fibroblasts into cardiac progenitors. Proc Natl Acad Sci USA. 2012;109:13016–13021. doi: 10.1073/pnas.1120299109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Q, Huang Y, Xiao N, et al. Real time investigation of protein folding, structure, and dynamics in living cells. Methods Cell Biol. 2008;90:287–325. doi: 10.1016/S0091-679X(08)00814-5. [DOI] [PubMed] [Google Scholar]
- 28.Kattman SJ, Huber TL, Keller GM. Multipotent flk-1+ cardiovascular progenitor cells give rise to the cardiomyocyte, endothelial, and vascular smooth muscle lineages. Dev Cell. 2006;11:723–732. doi: 10.1016/j.devcel.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 29.Brade T, Gessert S, Kühl M, et al. The amphibian second heart field: Xenopus islet-1 is required for cardiovascular development. Dev Biol. 2007;311:297–310. doi: 10.1016/j.ydbio.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 30.Wu SM, Chien KR, Mummery C. Origins and fates of cardiovascular progenitor cells. Cell. 2008;132:537–543. doi: 10.1016/j.cell.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ishitobi H, Wakamatsu A, Liu F, et al. Molecular basis for Flk1 expression in hemato-cardiovascular progenitors in the mouse. Development. 2011;138:5357–5368. doi: 10.1242/dev.065565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Strutz F, Okada H, Lo CW, et al. Identification and characterization of a fibroblast marker: FSP1. J Cell Biol. 1995;130:393–405. doi: 10.1083/jcb.130.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen B, Dodge ME, Tang W, et al. Small molecule-mediated disruption of Wnt-dependent signaling in tissue regeneration and cancer. Nat Chem Biol. 2009;5:100–107. doi: 10.1038/nchembio.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Willems E, Spiering S, Davidovics H, et al. Small-molecule inhibitors of the Wnt pathway potently promote cardiomyocytes from human embryonic stem cell-derived mesoderm. Circ Res. 2011;109:360–364. doi: 10.1161/CIRCRESAHA.111.249540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hudson J, Titmarsh D, Hidalgo A, et al. Primitive cardiac cells from human embryonic stem cells. Stem Cells Dev. 2012;21:1513–1523. doi: 10.1089/scd.2011.0254. [DOI] [PubMed] [Google Scholar]
- 36.Wamstad JA, Alexander JM, Truty RM, et al. Dynamic and coordinated epigenetic regulation of developmental transitions in the cardiac lineage. Cell. 2012;151:206–220. doi: 10.1016/j.cell.2012.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lien CL, Wu C, Mercer B, et al. Control of early cardiac-specific transcription of Nkx2-5 by a GATA-dependent enhancer. Development. 1999;126:75–84. doi: 10.1242/dev.126.1.75. [DOI] [PubMed] [Google Scholar]
- 38.Reecy JM, Li X, Yamada M, et al. Identification of upstream regulatory regions in the heart-expressed homeobox gene Nkx2-5. Development. 1999;126:839–849. doi: 10.1242/dev.126.4.839. [DOI] [PubMed] [Google Scholar]
- 39.Chen Y, Cao X. NFAT directly regulates Nkx2-5 transcription during cardiac cell differentiation. Biol Cell. 2009;101:335–349. doi: 10.1042/BC20080108. [DOI] [PubMed] [Google Scholar]
- 40.Schlesinger J, Schueler M, Grunert M, et al. The cardiac transcription network modulated by Gata4, Mef2a, Nkx2.5, Srf, histone modifications, and microRNAs. PLoS Genet. 2011;7:e1001313. doi: 10.1371/journal.pgen.1001313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu JI, Lessard J, Crabtree GR. Understanding the words of chromatin regulation. Cell. 2009;136:200–206. doi: 10.1016/j.cell.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Inagawa K, Miyamoto K, Yamakawa H, et al. Induction of cardiomyocyte-like cells in infarct hearts by gene transfer of Gata4, Mef2c, and Tbx5. Circ Res. 2012;111:1147–1156. doi: 10.1161/CIRCRESAHA.112.271148. [DOI] [PubMed] [Google Scholar]
- 43.Olson EN. Gene regulatory networks in the evolution and development of the heart. Science. 2006;313:1922–1927. doi: 10.1126/science.1132292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Srivastava D. Making or breaking the heart: From lineage determination to morphogenesis. Cell. 2006;126:1037–1048. doi: 10.1016/j.cell.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 45.Lickert H, Takeuchi JK, Von Both I, et al. Baf60c is essential for function of BAF chromatin remodelling complexes in heart development. Nature. 2004;432:107–112. doi: 10.1038/nature03071. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.