Abstract
Objective
Phosphoinositide 3-kinase γ (PI3Kγ) is a G-protein-coupled receptor-activated lipid kinase mainly expressed in leukocytes and cells of the cardiovascular system. PI3Kγ plays an important signaling role in inflammatory processes. Since subclinical inflammation is a hallmark of atherosclerosis, obesity-related insulin resistance, and pancreatic β-cell failure, we asked whether common genetic variation in the PI3Kγ gene (PIK3CG) contributes to body fat content/distribution, serum adipokine/cytokine concentrations, alterations in plasma lipid profiles, insulin sensitivity, insulin release, and glucose homeostasis.
Study Design
Using a tagging single nucleotide polymorphism (SNP) approach, we analyzed genotype-phenotype associations in 2,068 German subjects genotyped for 10 PIK3CG SNPs and characterized by oral glucose tolerance tests. In subgroups, data from hyperinsulinaemic-euglycaemic clamps, magnetic resonance spectroscopy of the liver, whole-body magnetic resonance imaging, and intravenous glucose tolerance tests were available, and peripheral blood mononuclear cells (PBMCs) were used for gene expression analysis.
Results
After appropriate adjustment, none of the PIK3CG tagging SNPs was significantly associated with body fat content/distribution, adipokine/cytokine concentrations, insulin sensitivity, insulin secretion, or blood glucose concentrations (p>0.0127, all; Bonferroni-corrected α-level: 0.0051). However, six non-linked SNPs displayed at least nominal associations with plasma HDL-cholesterol concentrations, two of them (rs4288294 and rs116697954) reaching the level of study-wide significance (p = 0.0003 and p = 0.0004, respectively). More precisely, rs4288294 and rs116697954 influenced HDL2-, but not HDL3-, cholesterol. With respect to the SNPs’ in vivo functionality, rs4288294 was significantly associated with PIK3CG mRNA expression in PBMCs.
Conclusions
We could demonstrate that common genetic variation in the PIK3CG locus, possibly via altered PIK3CG gene expression, determines plasma HDL-cholesterol concentrations. Since HDL2-, but not HDL3-, cholesterol is influenced by PIK3CG variants, PI3Kγ may play a role in HDL clearance rather than in HDL biogenesis. Even though the molecular pathways connecting PI3Kγ and HDL metabolism remain to be further elucidated, this finding could add a novel aspect to the pathophysiological role of PI3Kγ in atherogenesis.
Introduction
Phosphoinositide 3-kinases (PI3Ks) are a family of enzymes which catalyze the phosphorylation of intracellular phosphoinositides, an important step in many signaling pathways mediating cell growth, proliferation, and differentiation [1]. PI3Ks are divided into three different classes based on their structure, function, and substrate specificity [2]. Class-I kinases are the best characterized PI3Ks catalyzing the phosphorylation of phosphatidylinositol 4,5-bisphosphate to phosphatidylinositol 3,4,5-trisphosphate. All class-I PI3Ks are heterodimers consisting of two subunits: one for the catalytic function, the other one acting as an adapter or regulatory protein. Class-I catalytic subunits have a molecular mass of about 110 kDa (referred to as p110 subunits). Different genes encode for the four different p110 subunits, i.e., PIK3CA, PIK3CB, PIK3CG, and PIK3CD, that give rise to the four class-I PI3K isoforms termed PI3Kα, -β, -γ, and -δ. With respect to their function, class-I PI3Ks are subdivided into class-IA (PI3Kα, -β, and -δ) and class-IB (PI3Kγ) kinases [3]. Class-IA kinases are associated with a regulatory subunit termed p85 and are activated by receptor tyrosine kinases and the GTPase Ras (e.g., PI3Kα) [4] or by G-protein-coupled receptors via the Gβγ subunit of heterotrimeric G-proteins and Rho-family GTPases (e.g., PI3Kβ) [5,6]. Prominent signaling pathways including class-IA kinases are the insulin and insulin-like growth factor signaling pathways mediating activation of Akt/protein kinase B and other downstream mediators which are important for the regulation of glucose metabolism and cell proliferation.
PI3Kγ, as the only class-IB kinase, heterodimerizes with a regulatory p87 or p101 subunit and is activated by G-protein-coupled receptors via Gβγ and Ras [7–9]. PI3Kγ is mainly expressed in leukocytes and cells of the cardiovascular system and plays an important signaling role in inflammatory responses. Chronic low-grade, so-called subclinical, inflammation in turn is a major driver of metabolic diseases and their comorbidities, such as cardiovascular disease, fatty liver disease, and type-2 diabetes [10–12]. Notably, blockade of PI3Kγ either by genetic means (knockout) or by pharmacological inhibition revealed beneficial effects on disease outcome in mouse models of inflammatory disorders like diet-induced obesity, hepatic steatosis, lupus erythematodes, and atherosclerosis [13–17].
Human in vivo studies addressing the role of PI3Kγ in inflammation and inflammation-related metabolic diseases are still lacking. Therefore, we asked whether common genetic variation (minor allele frequency [MAF] ≥0.05) in the PI3Kγ gene PIK3CG exists and whether it affects body fat content and/or distribution, serum cytokine and adipokine concentrations, plasma lipid profiles, insulin sensitivity, insulin release, and glucose homeostasis. To this end, we applied a tagging single nucleotide polymorphism (SNP) approach in a total of 2,068 metabolically characterised subjects at increased risk for type-2 diabetes from the Tübingen Family (TÜF) study for type-2 diabetes.
Material and Methods
Study participants
The TÜF study currently comprises more than 2,500 non-related German Caucasians at increased risk for type-2 diabetes, i.e., non-diabetic subjects with a family history of type-2 diabetes, a body mass index (BMI) ≥27 kg/m2, impaired fasting glycaemia, and/or previous gestational diabetes [18]. All participants underwent physical examination, routine blood tests, and oral glucose tolerance tests (OGTTs). Furthermore, we assessed the medical history, smoking status, and alcohol consumption habits. From the TÜF study, 2,068 subjects with complete anthropometric data sets and documented absence of medication known to influence glucose tolerance, insulin sensitivity, or insulin secretion were genotyped. In the overall study population, 2,066 complete OGTT data sets, 1,243 adiponectin and leptin measurements, and 383 interleukin 6 (IL-6), tumour necrosis factor α (TNF-α), and monocyte chemoattractant protein 1 (MCP-1) measurements were available. Furthermore, data from hyperinsulinaemic-euglycaemic clamps (HECs), magnetic resonance spectroscopy (MRS) of the liver, whole-body magnetic resonance imaging (MRI), and intravenous glucose tolerance tests (IVGTTs) derived from partially overlapping subgroups of 499, 481, 361, and 306 individuals, respectively, were analysed. The clinical characteristics of the overall study population and the major subgroups are given in Table 1. In a very small subgroup of the overall study population (N = 34), high-density lipoprotein (HDL)-cholesterol fractionation data were available from an earlier investigation [19]. This subgroup was not analysed in the earlier investigation [19]. From 29 randomly selected participants, peripheral blood mononuclear cells (PBMCs) were prepared und subjected to gene expression analysis.
Table 1. Clinical data of the overall study population and the major subgroups.
| Parameter | Overall (N = 2,068) | Adipokines (N = 1,243) | Cytokines (N = 383) | HEC (N = 499) | MRS (N = 481) | MRI (N = 361) | IVGTT (N = 306) |
|---|---|---|---|---|---|---|---|
| N (women/men) | 1,334/734 | 814/429 | 248/135 | 268/231 | 306/175 | 222/139 | 175/131 |
| Age (y) | 39.6 ±12.6 | 39.0 ±12.3 | 40.4 ±12.6 | 39.8 ±12.0 | 44.2 12.0 | 45.4 ±11.6 | 44.6 ±11.3 |
| BMI (kg/m²) | 30.8 ±9.5 | 29.4 ±8.5 | 28.4 ±6.0 | 27.2 ±5.5 | 30.3 ±5.0 | 29.9 ±5.3 | 29.2 ±5.3 |
| Body fat content (%) | 33.5 ±12.8 | 31.7 ±11.6 | 30.9 ±9.5 | 28.3 ±9.6 | 34.6 ±9.6 | 33.0 ±8.9 | 31.8 ±8.8 |
| NGT/IFG/IGT/IFG+IGT | 1,443/258/200/167 | 890/135/118/100 | 271/37/44/31 | 382/39/46/32 | 310/68/55/48 | 230/43/48/40 | 204/31/43/28 |
| Glucose, fasting (mmol/L) | 5.16 ±0.54 | 5.13 ±0.54 | 5.10 ±0.52 | 5.02 ±0.54 | 5.27 ±0.49 | 5.24 ±0.50 | 5.17 ±0.49 |
| 2-h Glucose (mmol/L) | 6.36 ±1.61 | 6.31 ±1.63 | 6.44 ±1.70 | 6.18 ±1.70 | 6.82 ±1.52 | 6.90 ±1.58 | 6.79 ±1.66 |
| Leptin (ng/mL) | - | 28.0 ±31.5 | - | - | - | - | - |
| Adiponectin (μg/mL) | - | 14.4 ±7.4 | - | - | - | - | - |
| IL-6 (pg/mL) | - | - | 0.882 ±0.840 | - | - | - | - |
| TNF-α (pg/mL) | - | - | 2.75 ±5.41 | - | - | - | - |
| MCP-1 (pg/mL) | - | - | 181.7 ±91.6 | - | - | - | - |
| ISI, HEC (*106 L*kg-1*min-1) | - | - | - | 0.084 ±0.052 | - | - | - |
| IHL (% signal) | - | - | - | - | 1.27 ±1.09 | - | - |
| TAT (% body weight) | - | - | - | - | - | 30.5 ±9.2 | - |
| VAT (% body weight) | - | - | - | - | - | 3.33 ±1.71 | - |
| AIR (pmol/L) | - | - | - | - | - | - | 934 ±617 |
Data are given as counts or means ±SD. AIR–acute insulin response; AUC–area under the curve; BMI–body mass index; C-Pep–C-peptide; CRP–C-reactive protein; Glc–glucose; HEC–hyperinsulinaemic-euglycaemic clamp; IFG–impaired fasting glycaemia; IGT–impaired glucose tolerance; IHL–intrahepatic lipids; IL-6—interleukin-6; Ins—insulin; ISI–insulin sensitivity index; IVGTT–intravenous glucose tolerance test; MCP-1 –monocyte chemoattractant protein 1; MRI–magnetic resonance imaging; MRS–magnetic resonance spectroscopy; NGT–normal glucose tolerance; OGTT–oral glucose tolerance test; TAT–total adipose tissue; TNF-α –tumor necrosis factor α; VAT–visceral adipose tissue
Ethics statement
All participants gave informed written consent to the study which adhered to the Declaration of Helsinki. The study protocol was approved by the Ethics Committee of the Medical Faculty of the Eberhard Karls University Tübingen.
Determination of body fat content/distribution
Waist circumference (in cm) as a crude proxy of abdominal fat mass was measured at the midpoint between the lateral iliac crest and the lowest rib in the supine position. BMI as a crude measure of whole-body adiposity was calculated as weight divided by squared height (in kg/m2). The percentage of body fat was measured by bioelectrical impedance (BIA-101, RJL systems, Detroit, MI, USA). To precisely quantify total adipose tissue (TAT) and visceral adipose tissue (VAT) contents, whole-body MRI was performed [20]. The intrahepatic lipid (IHL) content was quantified by localized stimulated echo acquisition mode 1H-MRS [21].
OGTT
After an overnight fasting period of 10 h, a standardised 75-g OGTT was performed. For the determination of plasma glucose, serum insulin and C-peptide levels, venous blood samples were drawn at baseline and at time-points 30, 60, 90, and 120 min of the OGTT.
IVGTT and HEC
In study participants who agreed to undergo both the IVGTT and the HEC, the IVGTT was performed after a 10-h overnight fast and prior to the HEC according to the Botnia regimen [22]. After having collected baseline samples at time-points -10 and -5 min, a glucose bolus of 0.3 g/kg body weight was given intravenously at time-point 0. Blood samples for the measurement of plasma glucose and serum insulin were obtained at time-points 2, 4, 6, 8, 10, 20, 30, 40, 50, and 60 min of the IVGTT. The HEC was started after a 10-h overnight fast or, if combined with an IVGTT, at time-point 60 min after the IVGTT glucose bolus by giving a primed infusion of short-acting human insulin (40 mU/m2/min) for 120 min. Concomitantly, variable infusion of glucose (20%) was started to clamp the plasma glucose concentration at fasting levels. Blood samples for the measurement of plasma glucose were obtained at 5-min intervals. Serum insulin levels were measured at baseline (or prior to the glucose bolus of the IVGTT) and at the steady state (the last 30 min) of the clamp.
Laboratory measurements
Plasma glucose levels (in mmol/L) were measured with a bedside glucose analyser (glucose oxidase method, Yellow Springs Instruments, Yellow Springs, OH, USA). Serum insulin and C-peptide concentrations (in pmol/L, both) were determined by commercial chemiluminescence assays for ADVIA Centaur (Siemens Medical Solutions Diagnostics, Fernwald, Germany). Plasma triglycerides, total cholesterol, HDL and low-density lipoprotein (LDL) cholesterol, and wide-range C-reactive protein (in mg/dL, all) were measured using the ADVIA 1800 clinical chemical analyser. HDL2- and HDL3-cholesterol concentrations (in mg/dL) were determined after ultracentrifugation, as described earlier [23]. Apolipoprotein A1 (apoA1; in mg/dL) was measured by immunonephelometry. Blood cell counts were realized on the ADVIA 2120 haematology analyser (Siemens Healthcare Diagnostics, Eschborn, Germany). Serum adiponectin (in μg/mL) and leptin concentrations (in ng/mL) as well as TNF-α, IL-6, and MCP-1 concentrations (in pg/mL, all) were determined by enzyme-linked immunosorbent assays (adiponectin and leptin–Linco Research, St. Charles, MO, USA; TNF-α and IL-6 –R&D Systems, Wiesbaden-Nordenstadt, Germany; MCP-1 –Bender MedSystems, Vienna, Austria). Plasma free fatty acid (FFA) levels (in μmol/L) were determined enzymatically (NEFAC kit, WAKO Chemicals, Neuss, Germany).
Calculations
Insulin sensitivity was calculated from fasting data, the OGTT, and the HEC. Based on fasting data, homoeostasis model assessment of insulin resistance (HOMA-IR) was calculated as c(Glc0[mmol/L])*c(Ins0[mU/L])/22.5 with c = concentration, Glc = glucose, and Ins = insulin [24]. The OGTT-derived insulin sensitivity index (ISI-OGTT) was calculated as 10,000/{c(Glc0[mmol/L])*c(Ins0[pmol/L])*c(Glcmean[mmol/L])*c(Insmean[pmol/L])}½ [25]. The HEC-derived ISI (ISI-HEC) was calculated as glucose infusion rate necessary to maintain euglycaemia during the last 20 min (steady state) of the clamp (in μmol/kg/min) divided by the steady-state insulin concentration (in pmol/L). Insulin secretion was calculated from the OGTT and the IVGTT. From the OGTT, one insulin-based and one C-peptide-based index were calculated as area under the curve (AUC)0-30 min Insulin/AUC0-30 min Glucose and AUC0-120 min C-Peptide/AUC0-120 min Glucose, respectively [26]. AUC0-30 min Insulin/AUC0-30 min Glucose was calculated as {c(Ins0[pmol/L])+c(Ins30[pmol/L])}/{c(Glc0[mmol/L])+c(Glc30[mmol/L])}. AUC0-120 min C-Peptide/AUC0-120 min Glucose was calculated according to the trapezoid method as ½{½c(C-Pep0[pmol/L])+c(C-Pep30[pmol/L])+c(C-Pep60[pmol/L])+c(C-Pep90[pmol/L])+½c(C-Pep120[pmol/L])}/½{½c(Glc0[mmol/L])+c(Glc30[mmol/L])+c(Glc60[mmol/L])+c(Glc90[mmol/L])+½c(Glc120[mmol/L])} with C-Pep = C-peptide. From the IVGTT, insulin secretion was calculated as acute insulin response (AIR) according to the trapezoid method as ½{½c(Ins0[pmol/L])+c(Ins2[pmol/L])+c(Ins4[pmol/L])+c(Ins6[pmol/L])+c(Ins8[pmol/L])+½c(Ins10[pmol/L])}. The gender-dependent Framingham risk score for cardiovascular disease risk was calculated and converted to 10-year risk categories according to the instructions of the National Institutes of Health (www.nhlbi.nih.gov/health-pro/guidelines/current/cholesterol-guidelines/quick-desk-reference-html/10-year-risk-framingham-table).
Selection of tagging SNPs
To identify tagging SNPs, a genomic area of 43.8 kb on human chromosome 7q22 including the complete PIK3CG gene (11 exons, 10 introns) and 2 kb of its 5’-flanking (promoter) region was analysed in silico. Based on genomic data from the Central European (CEU) population of the 1000 Genomes Project (http://browser.1000genomes.org/index.html), we identified 10 representative SNPs that tag all the other common SNPs (MAF ≥0.05) in this region with an r2 ≥0.8 (100% coverage) using the tagger analysis tool of Haploview (http://www.broadinstitute.org/scientific-community/science/programs/medical-and-population-genetics/haploview/haploview). These tagging SNPs were rs4727666 (A/G) in the 5’-flanking region, rs3823963 (T/A) in intron 1, rs1129293 (C/T) in exon 3 (Ser675Ser), rs17401277 (C/T) in intron 6, rs59813697 (A/C), rs4288294 (C/T), rs849405 (A/G), rs116697954 (C/T), and rs2037718 (C/G) in intron 10, and rs10216210 (G/C) in exon 11 (3’-untranslated region). Importantly, the PIK3CG gene is flanked 68 kb upstream by a long intergenic non-coding RNA gene (RNA5SP236) and 137 kb downstream by the PRKAR2B gene, and no obvious linkage blocks were observed that span the PIK3CG locus and one of these adjacent genes.
Genotyping
DNA was isolated from whole blood using a commercial kit (NucleoSpin, Macherey & Nagel, Düren, Germany). Genotyping of the tagging SNPs was performed by mass spectrometry using the massARRAY genotyping platform (Sequenom, Hamburg, Germany) and the manufacturer’s iPLEX software. SNPs rs849405, rs2037718, and rs10216210 resisted massARRAY multiplex assay design and were, therefore, genotyped by allelic discrimination using commercial TaqMan assays (Applied Biosystems, Foster City, CA, USA). The call rates were ≥96%. The genotyping results were validated in 50 randomly selected subjects by bidirectional sequencing, and no deviations were observed.
PBMC isolation and quantitative real-time polymerase chain reaction (qPCR)
PBMCs were isolated from whole blood by density gradient centrifugation using the Ficoll-based Lymphocyte Separation Medium 1077 from PAA Laboratories (Cölbe, Germany). Cells were washed with phosphate-buffered saline, lysed with RLT buffer (Qiagen, Hilden, Germany), and homogenized using QIAshredder (Qiagen). Total-RNA was isolated with RNeasy columns (Qiagen), treated with RNase-free DNase I, and reverse transcribed into cDNA using the Transcriptor First Strand cDNA Synthesis kit from Roche Diagnostics (Mannheim, Germany). QPCR of PIK3CG, CETP (encoding cholesteryl ester transfer protein), PLTP (encoding phospholipid transfer protein), LCAT (encoding lecithin-cholesterol acyltransferase), SCARB1 (encoding scavenger receptor B1), and RPS13 (encoding ribosomal protein S13) mRNA expression was performed in technical duplicates on a LightCycler 480 (Roche Diagnostics) using Probes Master and fluorescent reporter probes from the Universal Probe Library (Roche Diagnostics). Primers were designed using the Roche Probe Design 2.0 software (Roche Diagnostics) and purchased from TIB MOLBIOL (Berlin, Germany). Primer sequences, reporter probes, and qPCR conditions are given in S1 Table. All gene expression data were normalized to the housekeeping gene RPS13 using the ΔCt method.
Statistical analyses
Hardy-Weinberg equilibrium was tested using χ² test with one degree of freedom. Continuous variables with skewed distributions were loge-transformed prior to statistical analysis. Multiple linear regression analyses were performed using the least-squares method. For the analyses in the overall study population, the trait of interest was chosen as outcome variable and the SNP genotype (in the additive inheritance model) as independent variable. Gender, age, BMI, OGTT-derived insulin sensitivity, and lipid-lowering medication (dummy variable for drug classes) were considered as confounding variables and included wherever appropriate. To account for multiple testing (10 SNPs tested in parallel), we corrected the α-level of significance according to Bonferroni and considered a p-value <0.0051 as statistically significant. SNP associations with p-values ≥0.0051 and <0.05 were termed nominal. Associations of SNPs rs4288294 and rs116697954 (in the additive inheritance model) with the gender-dependent Framingham risk score (10-year risk categories as nominal outcome variable) were tested in 694 women and 364 men with BMI and lipid-lowering medication as confounding variables using nominal logistic regression analysis. The effects of the two SNPs (i) on HDL2- and HDL3-cholesterol in 34 subjects and (ii) on gene expression in PBMCs from 29 donors were assessed by multiple linear regression analysis. Due to very low numbers of homozygous minor allele carriers in these latter analyses, the SNPs were tested in the dominant inheritance model only (CC vs. CT+TT). Based on the strictly focused and hypothesis-driven nature of these follow-up investigations (focused on two SNPs only; all other SNPs were not followed up because they failed significance for association with HDL-cholesterol and/or apoA1 in the overall study population), we here abstained from correcting the α-level for multiple testing and considered a p-value <0.05 as statistically significant. All analyses were performed with the JMP 10.0 software (SAS Institute, Cary, NC, USA).
Results
In this study, an overall study population of 2,068 subjects was subjected to genotyping. Of these, nearly two thirds were female (64.5%). The mean age was 39.6 years, the mean BMI 30.8 kg/m², and 69.8% of the participants displayed normal glucose tolerance whereas 12.5% had isolated impaired fasting glycaemia, 9.7% isolated impaired glucose tolerance, and 8.1% impaired fasting glycaemia and impaired glucose tolerance. The clinical characteristics of the overall study population and the major subgroups are given in Table 1.
The 10 non-linked tagging SNPs were representative for all other common genetic variants (MAF ≥0.05) in the PIK3CG gene locus (100% coverage) and were all found to be in Hardy-Weinberg equilibrium (p>0.09). The MAFs observed in our study population were similar to those reported for the CEU population by the 1000 Genomes Project (all differences between MAFTÜF and MAFCEU ≤5%).
In the association analyses, parameters of body fat content and body fat distribution were adjusted for gender and age. Blood glucose levels, insulin sensitivity measures, and adipokine and cytokine levels were adjusted for gender, age, and BMI. Insulin secretion indices were adjusted for gender, age, BMI, and insulin sensitivity, and finally, plasma lipid concentrations were adjusted for gender, age, BMI, and anti-hyperlipidaemic medication (among the 2,016 subjects analyzed for plasma lipids, 97.3% did not receive anti-hyperlipidaemic medication, 2.4% were on statins, 0.1% on fibrates, 0.1% on ezetimibe, and <0.05% on combination therapy).
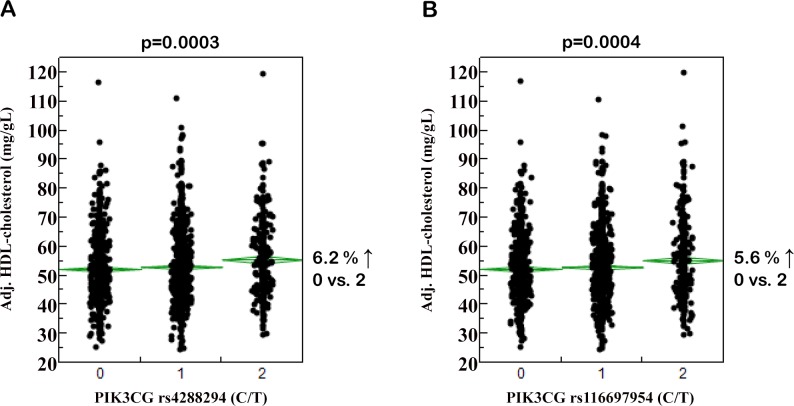
After appropriate adjustment, none of the tested SNPs showed significant association with body fat content and/or distribution, adipokine or cytokine concentrations, insulin sensitivity, insulin secretion, or blood glucose concentrations (p>0.0127, all; S2–S6 Tables). However, six SNPs displayed at least nominal associations with HDL-cholesterol levels without affecting total or LDL-cholesterol, and two of them, i.e., rs4288294 and rs116697954, reached the level of study-wide significance (p = 0.0003 and p = 0.0004, respectively; unadjusted data in Table 2; adjusted data and effect sizes in Fig 1). Both SNPs are located in intron 10, separated from each other by 747 bp. Their minor T-alleles were associated with increased HDL-cholesterol levels. Notably, the minor alleles of the four nominally associated SNPs (rs3823963, rs1129293, rs2037718, and rs10216210) were associated with decreased HDL-cholesterol (Table 2). To further strengthen these results, we tested whether the six SNPs associated with HDL-cholesterol also showed association with the major apolipoprotein of HDL, i.e., apoA1. Among the two SNPs significantly associated with HDL-cholesterol, rs4288294 was nominally (p = 0.0100) and rs116697954 significantly (p = 0.0017) associated with apoA1 levels after adjustment for gender, age, BMI, and anti-hyperlipidaemic medication, and both SNPs’ minor T-alleles were associated with elevated apoA1 levels. Among the four SNPs nominally associated with HDL-cholesterol, rs2037718 and rs10216210 were also nominally associated with apoA1 concentrations (p = 0.0242 and p = 0.0127, respectively; minor alleles associated with reduced apoA1 levels); rs3823963 and rs1129293 did neither reveal significant nor nominal associations with apoA1 (p≥0.1, both). These results reflect our SNP results with HDL-cholesterol.
Table 2. Associations of PIK3CG tagging SNPs with plasma lipid concentrations (NOGTT = 2,016).
| Genotype | NOGTT | FFA, fasting (μmol/L) | Triglycerides, fasting (mg/dL) | Total cholesterol, fasting (mg/dL) | LDL-cholesterol, fasting (mg/dL) | HDL-cholesterol, fasting (mg/dL) | |
|---|---|---|---|---|---|---|---|
| rs4727666 | AA | 1,241 | 593 ±242 | 117 ±78 | 191 ±37 | 119 ±33 | 54.1 ±14.5 |
| AG | 604 | 597 ±261 | 121 ±71 | 192 ±36 | 120 ±33 | 53.1 ±13.5 | |
| GG | 92 | 577 ±205 | 138 ±107 | 200 ±35 | 124 ±30 | 52.6 ±14.2 | |
| p | - | - | 0.6 | 0.0255 | 0.08 | 0.0418 | 0.1 |
| rs3823963 | TT | 671 | 601 ±283 | 121 ±78 | 193 ±37 | 121 ±33 | 54.4 ±14.3 |
| TA | 952 | 590 ±221 | 118 ±73 | 191 ±36 | 119 ±33 | 53.7 ±14.2 | |
| AA | 310 | 595 ±240 | 123 ±93 | 191 ±35 | 119 ±32 | 52.7 ±14.0 | |
| p | - | - | 0.8 | 0.8 | 0.2 | 0.5 | 0.0081 |
| rs1129293 | CC | 927 | 600 ±265 | 121 ±77 | 193 ±37 | 120 ±34 | 54.1 ±14.2 |
| CT | 831 | 589 ±228 | 118 ±74 | 191 ±36 | 119 ±32 | 53.6 ±14.4 | |
| TT | 176 | 589 ±228 | 122 ±100 | 189 ±36 | 118 ±33 | 52.4 ±13.1 | |
| p | - | - | 0.9 | 0.6 | 0.2 | 0.6 | 0.0291 |
| rs17401277 | CC | 1,765 | 596 ±251 | 120 ±77 | 192 ±36 | 120 ±33 | 53.6 ±14.1 |
| CT | 193 | 574 ±213 | 113 ±66 | 189 ±35 | 116 ±31 | 54.3 ±14.3 | |
| TT | 8 | 514 ±210 | 202 ±267 | 238 ±58 | 142 ±40 | 57.0 ±16.6 | |
| p | - | - | 0.1 | 1.0 | 0.9 | 0.5 | 0.8 |
| rs59813697 | AA | 1,574 | 593 ±250 | 119 ±79 | 192 ±37 | 120 ±33 | 53.8 ±14.1 |
| AC | 354 | 599 ±236 | 120 ±73 | 191 ±35 | 118 ±31 | 53.3 ±14.4 | |
| CC | 21 | 599 ±221 | 142 ±109 | 205 ±40 | 134 ±42 | 52.8 ±13.3 | |
| p | - | - | 0.5 | 0.8 | 0.9 | 0.8 | 0.3 |
| rs4288294 | CC | 730 | 588 ±226 | 124 ±85 | 194 ±36 | 121 ±32 | 52.9 ±13.7 |
| CT | 965 | 598 ±251 | 117 ±73 | 190 ±36 | 118 ±33 | 53.6 ±14.2 | |
| TT | 300 | 590 ±280 | 116 ±71 | 195 ±40 | 122 ±36 | 55.9 ±15.0 | |
| p | - | - | 0.7 | 0.09 | 0.9 | 0.4 | 0.0003 |
| rs849405 | AA | 1,607 | 592 ±237 | 119 ±76 | 191 ±37 | 119 ±33 | 54.0 ±14.2 |
| AG | 380 | 608 ±287 | 122 ±83 | 195 ±38 | 123 ±34 | 52.5 ±13.9 | |
| GG | 29 | 551 ±204 | 119 ±51 | 195 ±34 | 122 ±27 | 53.0 ±15.9 | |
| p | - | - | 1.0 | 0.9 | 0.1 | 0.0469 | 0.2 |
| rs116697954 | CC | 650 | 586 ±224 | 126 ±90 | 194 ±36 | 121 ±33 | 52.8 ±13.9 |
| CT | 926 | 603 ±254 | 118 ±72 | 190 ±36 | 118 ±33 | 53.5 ±13.8 | |
| TT | 371 | 588 ±266 | 115 ±70 | 194 ±38 | 121 ±34 | 55.9 ±15.3 | |
| p | - | - | 0.9 | 0.2 | 0.8 | 0.4 | 0.0004 |
| rs2037718 | CC | 710 | 601 ±282 | 117 ±73 | 193 ±37 | 120 ±34 | 54.6 ±14.7 |
| CG | 971 | 592 ±225 | 119 ±76 | 192 ±37 | 119 ±32 | 53.3 ±13.7 | |
| GG | 333 | 585 ±229 | 125 ±91 | 192 ±36 | 121 ±34 | 52.8 ±14.0 | |
| p | - | - | 0.7 | 0.5 | 0.5 | 0.9 | 0.0055 |
| rs10216210 | GG | 1,114 | 595 ±258 | 120 ±75 | 193 ±37 | 120 ±33 | 54.1 ±14.3 |
| GC | 756 | 596 ±233 | 117 ±74 | 191 ±37 | 120 ±33 | 53.3 ±14.1 | |
| CC | 144 | 573 ±220 | 125 ±107 | 190 ±36 | 117 ±33 | 52.4 ±13.2 | |
| p | - | - | 0.8 | 0.5 | 0.3 | 0.6 | 0.0280 |
Metabolic data are shown as unadjusted raw data (means ±SD). Associations between SNP genotypes (additive inheritance model) and plasma lipid concentrations were tested by multiple linear regression analyses (standard least squares method) with gender, age, BMI, and anti-hyperlipidaemic medication as covariates. Nominal associations (p<0.05) are marked by using bold fonts, significant associations (p<0.0051 after Bonferroni correction for 10 SNPs) by using bold fonts and underlining. AUC–area under the curve; BMI–body mass index; FFA–free fatty acids; HDL–high-density lipoproptein; LDL–low-density lipoprotein; OGTT- oral glucose tolerance test; SNP–single nucleotide polymorphism
Fig 1. Associations of PIK3CG SNPs rs4288294 (A) and rs116697954 (B) with plasma HDL-cholesterol concentrations.
Adjustment of plasma HDL-cholesterol concentrations (N = 2,016) was achieved by multiple linear regression modelling with gender, age, BMI, and anti-hyperlipidaemic medication as confounding variables. On the x-axes, the number of minor T-alleles is given. The SNPs were tested in the additive inheritance model. HDL–high-density lipoprotein; SNP–single nucleotide polymorphism.
For independent replication of the HDL data, we looked into the meta-analysis data for HDL-cholesterol from genome-wide association studies and Metabochip analyses (N≥94,255) made publicly available by the Global Lipids Genetics Consortium (GLGC, http://www.sph.umich.edu/csg/abecasis/public/lipids2013, [27]). Unfortunately, none of the six SNPs with at least nominal association with HDL-cholesterol in this study was depicted on the arrays used by the GLGC. Using the SNP Annotation and Proxy Search platform of the Broad Institute (http://www.broadinstitute.org/mpg/snap/ldsearch.php), we could identify proxies (with r²>0.8) depicted on the GLGC arrays for four of the six SNPs. However, none of the proxy SNPs reached the level of nominal significance in the GLGC data for association with HDL-cholesterol adjusted for gender and age (p≥0.07, all; subjects on lipid-lowering medication were excluded from the meta-analysis). Association data additionally adjusted for BMI were not available from GLGC. Performing the analyses in our study population without adjustment for BMI raised the p-value of SNP rs4288294 from 0.0003 to 0.0055 (rendering this hit only nominally associated) and of SNP rs116697954 from 0.0004 to 0.0018. Furthermore, BMI exclusion from adjustment abolished the associations of three SNPs described before as nominal hits (rs3823963, rs1129293, and rs10216210; p≥0.08, all).
From 694 female and 364 male study participants, blood pressure and cigarette consumption data were available. Therefore, we tested in these subjects whether the two significantly HDL-associated SNPs, i.e., rs4288294 and rs116697954, associate with the gender-dependent Framingham risk score for cardiovascular disease risk. After adjustment for BMI and anti-hyperlipidaemic medication, SNPs rs4288294 and rs116697954 were significantly associated with the risk score for women (p = 0.0354 and p = 0.0313, respectively; no Bonferroni correction applied in this focused follow-up investigation), with female minor T-allele carriers having a reduced cardiovascular disease risk. Neither SNP was associated with the risk score in the smaller group of men (p = 0.2, both).
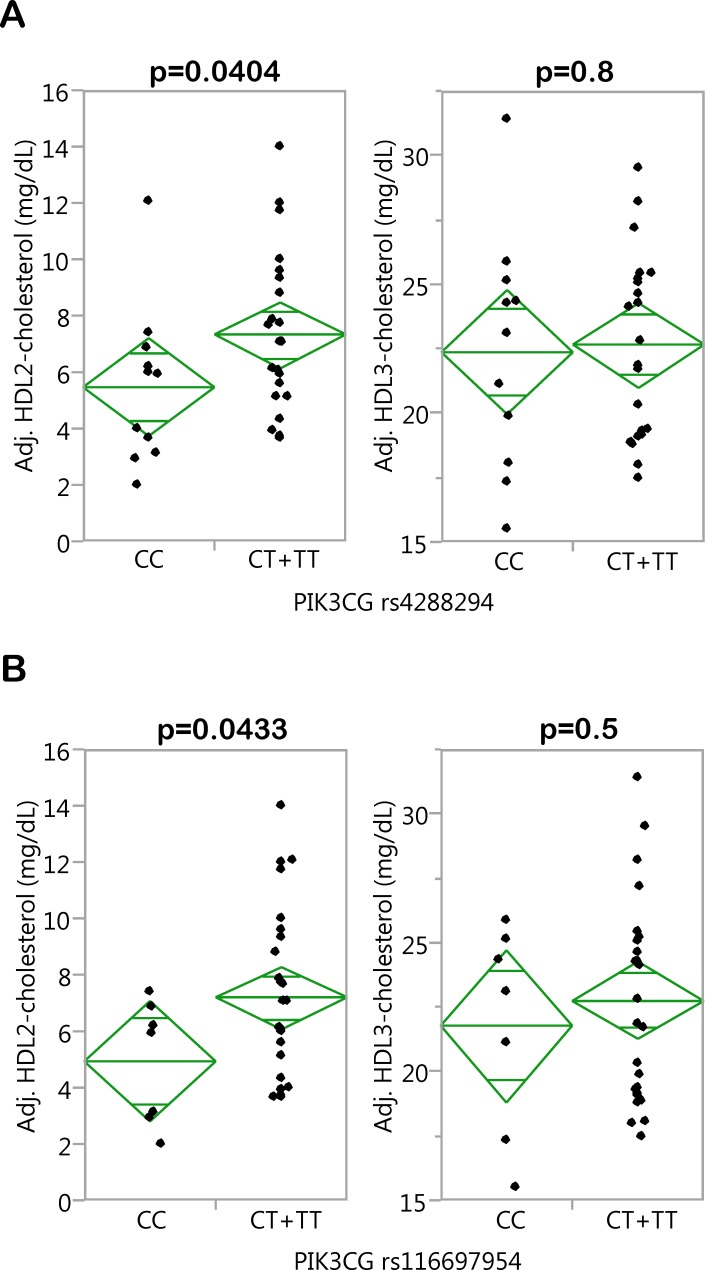
Then, we addressed the question whether the two SNPs affect HDL2- and/or HDL3-cholesterol concentrations. To this end, we looked into HDL fractionation data generated during an earlier investigation [19]. In this subgroup of 34 TÜF participants (clinical characteristics given in S7 Table), the minor T-alleles of SNPs rs4288294 and rs116697954 were significantly associated with higher HDL2- (p = 0.0404 and 0.0433, respectively), but not HDL3- (p = 0.8 and p = 0.5, respectively), cholesterol after adjustment for gender, age, and BMI (Fig 2). In this analysis, CT+TT were jointly analyzed (dominant inheritance model) due to the very low number of TT homozygotes (N = 5 and 7, respectively). Of the 34 participants, 15 had fatty liver (intrahepatic lipid content ≥5.5%) whereas 19 had no fatty liver. Inclusion of the presence/absence of fatty liver as a nominal confounding variable in the multiple linear regression analysis did not affect the association of SNP rs4288294 with HDL2-cholesterol (p = 0.0412; association with HDL3: p = 0.8). The association of SNP rs116697954, the SNP with the smaller effect size on HDL-cholesterol in the overall population (Fig 1B), with HDL2-cholesterol did no longer reach the level of significance (p = 0.06; association with HDL3: p = 0.5).
Fig 2. Associations of PIK3CG SNPs rs4288294 (A) and rs116697954 (B) with HDL2- and HDL3-cholesterol concentrations.
The HDL-cholesterol subfractions HDL2 and HDL3 were obtained by ultracentrifugation. Adjustment of HDL2- and HDL3-cholesterol concentrations (N = 34) was achieved by multiple linear regression modelling with gender, age, and BMI as confounding variables. The SNPs were tested in the dominant inheritance model. HDL–high-density lipoprotein; SNP–single nucleotide polymorphism.
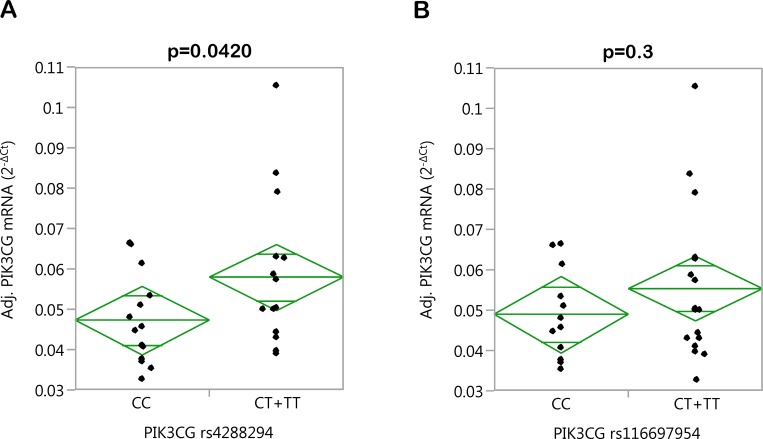
In a first attempt to mechanistically understand the effects of the major HDL-associated SNPs rs4288294 and rs116697954, we measured PIK3CG mRNA expression in PBMCs from 29 randomly selected study participants (22 women, 7 men; age 40.9 ±12.9 years; BMI 32.1 ±4.3 kg/m²; means ±SD) and performed multiple linear regression analysis. Among the anthropometric data available, we identified age (p = 0.0070), body height (p = 0.0209), and waist-hip ratio (p = 0.0275) as significant independent determinants of PIK3CG gene expression explaining about 34% of the variation in PIK3CG mRNA levels and therefore included these parameters as covariates in the multiple linear regression model. Even though not significant, we included gender as well because of the marked gender imbalance in this subgroup (22 women, 7 men). Again, the two SNPs were tested in the dominant inheritance model due to the very low number of TT homozygotes (N = 6 and 7, respectively). Applying this regression model, we observed significantly increased PIK3CG gene expression in minor T-allele carriers of SNP rs4288294 (p = 0.0420; Fig 3A). The effect of SNP rs116697954, which had a smaller effect on HDL-cholesterol in the overall study population, did not pass the significance threshold (p = 0.3; Fig 3B). Even though PBMCs are probably not a major source of cholesteryl ester transfer protein (CETP; mainly expressed in liver and lymph nodes), lecitihin-cholesterol acyltransferase (LCAT; rather ubiquitously expressed), phospholipid transfer protein (PLTP; mainly expressed in thymus, retina, and lung), and scavenger receptor B1 (SRB1; encoded by the SCARB1 gene; mainly expressed in the adrenal cortex), important proteins involved in HDL metabolism/turnover, we reasoned that if a non-coding genetic variant exerts an effect on these genes’ expression at their major expression sites this should also be reflected in cells with more moderate expression levels. Indeed, only CETP mRNA expression ranged at the detection limit (Cp-value >35), whereas the mRNA levels of LCAT (Cp-value 32.7), PLTP (Cp-value 32.0), and SCARB1 (Cp-value 30.5) were reproducibly detectable in PBMCs by qPCR. However, we could not detect any significant association of the two SNPs tested, i.e., rs4288294 and rs116697954, with the expression of these genes (p>0.1, all).
Fig 3. Associations of PIK3CG SNPs rs4288294 (A) and rs116697954 (B) with PIK3CG gene expression in PBMCs.
After isolation of PBMCs from whole blood, the cellular PIK3CG mRNA content was determined by qPCR. Adjustment of PIK3CG mRNA expression (N = 29) was achieved by multiple linear regression modelling with gender, age, body height, and waist-to-hip ratio as confounding variables. The SNPs were tested in the dominant inheritance model. PBMCs–peripheral blood mononuclear cells; qPCR–quantitative real-time polymerase chain reaction; SNP–single nucleotide polymorphism.
Discussion
In this work, we investigated the impact of common genetic variation in the PIK3CG locus, represented by 10 non-linked non-coding tagging SNPs, on inflammatory traits and inflammation-related metabolic traits in individuals at increased risk for type-2 diabetes.
Contrary to our expectations, none of the SNPs directly influenced blood inflammatory markers, such as leukocyte number, plasma CRP levels, or serum IL-6, TNF-α, or MCP-1 concentrations. Moreover, we did not observe any associations with metabolic traits like body fat content, body fat distribution, serum adipokine concentrations, plasma glucose levels, insulin sensitivity, or insulin secretion. However, we identified two SNPs with study-wide significant effects on plasma HDL-cholesterol (rs4288294 and rs116697954). Homozygous carriers of these SNPs’ minor T-alleles had 5–6% higher HDL-cholesterol levels compared to homozygous major allele carriers. Moreover, we found four additional SNPs nominally associated with HDL-cholesterol with their minor alleles being associated with reduced HDL-cholesterol. Even though not significant and with divergent effect directions, these additional findings support our suggestion that the PIK3CG locus affects plasma HDL-cholesterol and argue against a mere chance finding. Opposing effect directions of non-coding SNPs may have multiple reasons: e.g., one SNP in a cis-regulatory element, such as an enhancer structure, may strengthen the binding of a transcription factor to this element whereas another SNP within the same element–and some so-called stretch enhancers can span several kb [28]–may weaken it; or one SNP may enhance/weaken the binding of a transcriptional activator to the locus whereas another SNP may enhance/weaken the binding of a transcriptional repressor.
Based on (i) the observation that the HDL-associated PIK3CG SNPs identified herein were not depicted on the SNP arrays used by the GLGC, (ii) the possibility that the proxies found on the arrays may not perfectly represent our hits, and (iii) the fact that our analyses diverge from those of the GLGC meta-analysis with respect to adjustment for potential confounders, we can add a new candidate locus to the 46 HDL loci identified up to now by meta-analysis of genome-wide association studies [28]. However, this new candidate locus clearly awaits replication in larger study populations applying identical statistical analyses and adjustments of data. In this context, we could show in our study that the lack of adjustment for BMI (as was the case in the GLGC meta-analysis) abolished three of our four nominal HDL-cholesterol hits and raised the p-value of one of our two significant hits to the nominal level. Thus, adjustment for BMI appears crucial to identify associations of PIK3CG SNPs with HDL-cholesterol.
As a first step towards an understanding of the mechanism(s) underlying the SNPs’ association with HDL-cholesterol, we could demonstrate that rs4288294 was significantly associated with PIK3CG gene expression in PBMCs from 29 donors, with minor T-allele carriers displaying higher PIK3CG expression. Larger PBMC collections would be needed to detect a consistent significant effect in minor T-allele carriers of rs116697954 (least significant number estimated from the model: N = 83). Even though we corrected for the gender imbalance observed in our PBMC donors (22 women, 7 men) by including gender as a confounder in our multiple linear regression analyses, this adjustment could have been insufficient to completely exclude any gender bias, and larger PBMC collections with a more balanced gender distribution would allow more stringent conclusions. How the PIK3CG gene product PI3Kγ molecularly influences HDL formation and/or clearance is hitherto unknown, and PIK3CG knockout mice, available for about 15 years now, have not yet been assessed for altered HDL-cholesterol concentrations. There is one very recent interesting report providing evidence for an involvement of PI3Kγ in LDL uptake by macrophages via pinocytosis favouring foam-cell formation [29]. Whether PI3Kγ participates in HDL uptake/clearance by a similar mechanism was unfortunately not investigated, but could be a possibility. Since HDL2 is formed from HDL3, our finding that the minor T-alleles of SNPs rs4288294 and rs116697954 associate with higher HDL2-, but not HDL3-, cholesterol may point to a role of PI3Kγ in HDL clearance rather than in increased HDL biogenesis. With respect to the clinical relevance of these HDL subspecies, low HDL2 concentrations were reported to have a stronger predictive value for coronary heart disease than low HDL3 concentrations in a large prospective study [30].
For decades, high plasma HDL-cholesterol concentrations were considered as anti-atherosclerotic and cardioprotective based on a large body of epidemiological and experimental data (reviewed in [31,32]). Recently, however, a series of clinical trials aimed at increasing HDL-cholesterol by pharmacological means failed to show improvements in cardiovascular risk [33–38]. Thus, the “HDL hypothesis” that states that decreased plasma HDL concentrations lead to impaired reverse cholesterol transport thereby accelerating atherosclerosis has recently been questioned, and further investigations are needed to clarify the reasons for these treatment studies’ inefficacy. Moreover, it remains to be shown whether the mild but physiological and lifelong elevation in plasma HDL concentrations resulting from naturally occurring genetic variation (as shown here), in contrast to the artificial elevation of plasma HDL by pharmacological intervention, are beneficial for cardiovascular endpoints. Maybe, it is even more HDL functionality, particularly cholesterol efflux capacity, that contributes to the assumed anti-atherosclerotic effects. A recently published work showed a strong association between HDL-cholesterol efflux capacity and cardiovascular events [39]. This capacity, however, is only weekly associated with HDL cholesterol serum levels [39]. Certainly, studies addressing the effects of the PIK3CG SNPs on HDL function and turnover could help defining their contribution to atherosclerosis and cardiovascular disease. We could show here that carriers of the HDL-raising minor T-alleles of rs4288294 and rs116697954 have significantly reduced cardiovascular risk according to the Framingham risk score, at least in the larger, and thus better powered, group of women. This result however has to be interpreted cautiously as HDL-cholesterol is an integral part of the score.
In conclusion, we could demonstrate that common genetic variants in the PIK3CG locus (encoding PI3Kγ), possibly via effects on PIK3CG gene expression, determine plasma HDL-cholesterol concentrations. Since HDL2-, but not HDL3-, cholesterol is influenced by the variants, PI3Kγ may play a role in HDL clearance rather than in HDL biogenesis. Even though the molecular pathways connecting PI3Kγ and HDL metabolism remain to be further elucidated, this finding could add a novel aspect to the pathophysiological role of PI3Kγ in atherogenesis. Our results should encourage genetic studies assessing the effects of PIK3CG SNPs (e.g., rs4288294 and rs116697954) on cardiovascular endpoints that are not available in our study.
Supporting Information
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
We thank all study participants for their cooperation. We gratefully acknowledge the excellent technical assistance of Alke Guirguis, Daniela Thien, and Roman-Georg Werner.
Abbreviations
- AIR
acute insulin response
- apoA1
apolipoprotein A1
- AUC
area under the curve
- BMI
body mass index
- CETP
cholesteryl ester transfer protein
- CEU
Central European
- FFA
free fatty acid
- GLGC
Global Lipids Genetics Consortium
- HDL
high-density lipoprotein
- HEC
hyperinsulinaemic-euglycaemic clamp
- HOMA-IR
homoeostasis model assessment of insulin resistance
- IHL
intrahepatic lipids
- IL-6
interleukin 6
- ISI
insulin sensitivity index
- IVGTT
intravenous glucose tolerance test
- LCAT
lecithin-cholesterol acyltransferase
- LDL
low-density lipoprotein
- MAF
minor allele frequency
- MCP-1
monocyte chemoattractant protein 1
- MRI
magnetic resonance imaging
- MRS
magnetic resonance spectroscopy
- OGTT
oral glucose tolerance test
- PBMCs
peripheral blood mononuclear cells
- PI3K
phosphoinositide 3-kinase
- PLTP
phospholipid transfer protein
- qPCR
quantitative real-time polymerase chain reaction
- SNP
single nucleotide polymorphism
- SRB1
scavenger receptor B1
- TAT
total adipose tissue
- TNF-α
tumour necrosis factor α
- TÜF
TÜbingen Family
- VAT
visceral adipose tissue
Data Availability
Due to ethical restriction, such as patient identifying information, data are held upon request. Interested researchers may contact the principal investigators at www.med.uni-tuebingen.de/Forschung/Kliniken/Medizinische+Klinik/Innere+Medizin+IV.html.
Funding Statement
The study was supported in part by a grant (01GI0925) from the German Federal Ministry of Education and Research (BMBF) to the German Centre for Diabetes Research (DZD e.V.). Norbert Stefan is supported by a Heisenberg professorship from the Deutsche Forschungsgemeinschaft (STE 1096/3-1). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Jean S, Kiger AA. Classes of phosphoinositide 3-kinases at a glance. J Cell Sci. 2014;127: 923–928. 10.1242/jcs.093773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Engelman JA, Luo J, Cantley LC. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet. 2006;7: 606–619. [DOI] [PubMed] [Google Scholar]
- 3. Hirsch E, Costa C, Ciraolo E. Phosphoinositide 3-kinases as a common platform for multi-hormone signaling. J Endocrinol. 2007;194: 243–256. [DOI] [PubMed] [Google Scholar]
- 4. Rodriguez-Viciana P, Warne PH, Dhand R, Vanhaesebroeck B, Gout I, Fry MJ, et al. Phosphatidylinositol-3-OH kinase as a direct target of Ras. Nature 1994;370: 527–532. [DOI] [PubMed] [Google Scholar]
- 5. Fritsch R, de Krijger I, Fritsch K, George R, Reason B, Kumar MS, et al. RAS and RHO families of GTPases directly regulate distinct phosphoinositide 3-kinase isoforms. Cell 2013;153: 1050–1063. 10.1016/j.cell.2013.04.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dbouk HA, Vadas O, Shymanets A, Burke JE, Salamon RS, Khalil BD, et al. G protein-coupled receptor-mediated activation of p110beta by Gbetagamma is required for cellular transformation and invasiveness. Sci Signal. 2012;5: ra89 10.1126/scisignal.2003264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brock C, Schaefer M, Reusch HP, Czupalla C, Michalke M, Spicher K, et al. Roles of G beta gamma in membrane recruitment and activation of p110 gamma/p101 phosphoinositide 3-kinase gamma. J Cell Biol. 2003;160: 89–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kurig B, Shymanets A, Bohnacker T, Prajwal, Brock C, Ahmadian MR, et al. Ras is an indispensable coregulator of the class IB phosphoinositide 3-kinase p87/p110gamma. Proc Natl Acad Sci U S A. 2009;106: 20312–20317. 10.1073/pnas.0905506106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Vadas O, Dbouk HA, Shymanets A, Perisic O, Burke JE, Abi Saab WF, et al. Molecular determinants of PI3Kgamma-mediated activation downstream of G-protein-coupled receptors (GPCRs). Proc Natl Acad Sci U S A. 2013;110: 18862–18867. 10.1073/pnas.1304801110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kaur J. A comprehensive review on metabolic syndrome. Cardiol Res Pract. 2014;2014: 943162 10.1155/2014/943162 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11. Stefan N, Kantartzis K, Haring HU. Causes and metabolic consequences of Fatty liver. Endocr Rev. 2008;29: 939–960. 10.1210/er.2008-0009 [DOI] [PubMed] [Google Scholar]
- 12. van Greevenbroek MM, Schalkwijk CG, Stehouwer CD. Obesity-associated low-grade inflammation in type 2 diabetes mellitus: causes and consequences. Neth J Med. 2013;71: 174–187. [PubMed] [Google Scholar]
- 13. Barber DF, Bartolome A, Hernandez C, Flores JM, Redondo C, Fernandez-Arias C, et al. PI3Kgamma inhibition blocks glomerulonephritis and extends lifespan in a mouse model of systemic lupus. Nat Med. 2005;11: 933–935. [DOI] [PubMed] [Google Scholar]
- 14. Fougerat A, Gayral S, Gourdy P, Schambourg A, Ruckle T, Schwarz MK, et al. Genetic and pharmacological targeting of phosphoinositide 3-kinase-gamma reduces atherosclerosis and favors plaque stability by modulating inflammatory processes. Circulation 2008;117: 1310–1317. 10.1161/CIRCULATIONAHA.107.720466 [DOI] [PubMed] [Google Scholar]
- 15. Chang JD, Sukhova GK, Libby P, Schvartz E, Lichtenstein AH, Field SJ, et al. Deletion of the phosphoinositide 3-kinase p110gamma gene attenuates murine atherosclerosis. Proc Natl Acad Sci U S A. 2007;104: 8077–8082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wymann MP, Solinas G. Inhibition of phosphoinositide 3-kinase gamma attenuates inflammation, obesity, and cardiovascular risk factors. Ann N Y Acad Sci. 2013;1280: 44–47. 10.1111/nyas.12037 [DOI] [PubMed] [Google Scholar]
- 17. Becattini B, Marone R, Zani F, Arsenijevic D, Seydoux J, Montani JP, et al. PI3Kgamma within a nonhematopoietic cell type negatively regulates diet-induced thermogenesis and promotes obesity and insulin resistance. Proc Natl Acad Sci U S A. 2011;108: E854–E863. 10.1073/pnas.1106698108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Stefan N, Machicao F, Staiger H, Machann J, Schick F, Tschritter O, et al. Polymorphisms in the gene encoding adiponectin receptor 1 are associated with insulin resistance and high liver fat. Diabetologia 2005;48: 2282–2291. [DOI] [PubMed] [Google Scholar]
- 19. Kantartzis K, Rittig K, Cegan A, Machann J, Schick F, Balletshofer B, et al. Fatty liver is independently associated with alterations in circulating HDL2 and HDL3 subfractions. Diabetes Care 2008;31: 366–368. [DOI] [PubMed] [Google Scholar]
- 20. Machann J, Thamer C, Schnoedt B, Haap M, Haring HU, Claussen CD, et al. Standardized assessment of whole body adipose tissue topography by MRI. J Magn Reson Imaging 2005;21: 455–462. [DOI] [PubMed] [Google Scholar]
- 21. Machann J, Thamer C, Schnoedt B, Stefan N, Haring HU, Claussen CD, et al. Hepatic lipid accumulation in healthy subjects: a comparative study using spectral fat-selective MRI and volume-localized 1H-MR spectroscopy. Magn Reson Med. 2006;55: 913–917. [DOI] [PubMed] [Google Scholar]
- 22. Tripathy D, Wessman Y, Gullstrom M, Tuomi T, Groop L. Importance of obtaining independent measures of insulin secretion and insulin sensitivity during the same test: results with the Botnia clamp. Diabetes Care 2003;26: 1395–1401. [DOI] [PubMed] [Google Scholar]
- 23. Patsch W, Brown SA, Morrisett JD, Gotto AM Jr, Patsch JR. A dual-precipitation method evaluated for measurement of cholesterol in high-density lipoprotein subfractions HDL2 and HDL3 in human plasma. Clin Chem. 1989;2: 265–270. [PubMed] [Google Scholar]
- 24. Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia 1985;28: 412–419. [DOI] [PubMed] [Google Scholar]
- 25. Matsuda M, DeFronzo RA. Insulin sensitivity indices obtained from oral glucose tolerance testing: comparison with the euglycemic insulin clamp. Diabetes Care 1999;22: 1462–1470. [DOI] [PubMed] [Google Scholar]
- 26. Herzberg-Schafer SA, Staiger H, Heni M, Ketterer C, Guthoff M, Kantartzis K, et al. Evaluation of fasting state-/oral glucose tolerance test-derived measures of insulin release for the detection of genetically impaired beta-cell function. PLoS One 2010;5: e14194 10.1371/journal.pone.0014194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Willer CJ, Schmidt EM, Sengupta S, Peloso GM, Gustafsson S, Kanoni S, et al. Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013;45: 1274–1283. 10.1038/ng.2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Parker SC, Stitzel ML, Taylor DL, Orozco JM, Erdos MR, Akiyama JA, et al. Chromatin stretch enhancer states drive cell-specific gene regulation and harbor human disease risk variants. Proc Natl Acad Sci U S A. 2013;110: 17921–17926. 10.1073/pnas.1317023110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Anzinger JJ, Chang J, Xu Q, Barthwal MK, Bohnacker T, Wymann MP, et al. Murine bone marrow-derived macrophages differentiated with GM-CSF become foam cells by PI3Kgamma-dependent fluid-phase pinocytosis of native LDL. J Lipid Res. 2012;53: 34–42. 10.1194/jlr.M018887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Williams PT, Feldman DE. Prospective study of coronary heart disease vs. HDL2, HDL3, and other lipoproteins in Gofman's Livermore Cohort. Atherosclerosis. 2011;214: 196–202. 10.1016/j.atherosclerosis.2010.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rubenfire M, Brook RD. HDL cholesterol and cardiovascular outcomes: what is the evidence? Curr Cardiol Rep. 2013;15: 349 10.1007/s11886-013-0349-3 [DOI] [PubMed] [Google Scholar]
- 32. Welty FK. How do elevated triglycerides and low HDL-cholesterol affect inflammation and atherothrombosis? Curr Cardiol Rep. 2013;15: 400 10.1007/s11886-013-0400-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Barter PJ, Caulfield M, Eriksson M, Grundy SM, Kastelein JJ, Komajda M, et al. Effects of torcetrapib in patients at high risk for coronary events. N Engl J Med. 2007;357: 2109–2122. [DOI] [PubMed] [Google Scholar]
- 34. Schwartz GG, Olsson AG, Abt M, Ballantyne CM, Barter PJ, Brumm J, et al. Effects of dalcetrapib in patients with a recent acute coronary syndrome. N Engl J Med. 2012;367: 2089–2099. 10.1056/NEJMoa1206797 [DOI] [PubMed] [Google Scholar]
- 35. Boden WE, Probstfield JL, Anderson T, Chaitman BR, Desvignes-Nickens P, Koprowicz K, et al. Niacin in patients with low HDL cholesterol levels receiving intensive statin therapy. N Engl J Med. 2011;365: 2255–2267. [DOI] [PubMed] [Google Scholar]
- 36. Landray MJ, Haynes R, Hopewell JC, Parish S, Aung T, Tomson J, et al. Effects of extended-release niacin with laropiprant in high-risk patients. N Engl J Med. 2014;371: 203–212. 10.1056/NEJMoa1300955 [DOI] [PubMed] [Google Scholar]
- 37. Keene D, Price C, Shun-Shin MJ, Francis DP. Effect on cardiovascular risk of high density lipoprotein targeted drug treatments niacin, fibrates, and CETP inhibitors: meta-analysis of randomised controlled trials including 117,411 patients. BMJ. 2014;349: g4379 10.1136/bmj.g4379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. HPS2-THRIVE Collaborative Group. HPS2-THRIVE randomized placebo-controlled trial in 25 673 high-risk patients of ER niacin/laropiprant: trial design, pre-specified muscle and liver outcomes, and reasons for stopping study treatment. Eur Heart J. 2013;34: 1279–1291. 10.1093/eurheartj/eht055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rohatgi A, Khera A, Berry JD, Givens EG, Ayers CR, Wedin KE, et al. HDL cholesterol efflux capacity and incident cardiovascular events. N Engl J Med. 2014;371: 2383–2393. 10.1056/NEJMoa1409065 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
Due to ethical restriction, such as patient identifying information, data are held upon request. Interested researchers may contact the principal investigators at www.med.uni-tuebingen.de/Forschung/Kliniken/Medizinische+Klinik/Innere+Medizin+IV.html.