Abstract
Colicins are toxins that mediate interference competition in microbial ecosystems. They serve as a “common good” for the entire producer population but are synthesized by only few members which pay the costs of colicin production. We have previously shown that production of colicin Ib (cib), a group B colicin, confers a competitive advantage to Salmonella enterica serovar Typhimurium (S. Tm) over commensal E. coli strains. Here, we studied regulation of S. Tm cib expression at the single cell level. Comparative analysis of a single- and a multicopy gfp-reporter for the colicin Ib promoter (Pcib) revealed that the latter yielded optimal signal intensity for a diverse range of applications. We further validated this reporter and showed that gfp expression correlated well with colicin Ib (ColIb) protein levels in individual cells. Pcib is negatively controlled by two repressors, LexA and Fur. Only a small fraction of S. Tm expressed cib under non-inducing conditions. We studied Pcib activity in response to mitomycin C mediated DNA damage and iron limitation. Both conditions, if applied individually, lead to an increase in the fraction of GFP+ S. Tm, albeit an overall low fluorescence intensity. When both conditions were applied simultaneously, the majority of S. Tm turned GFP+ and displayed high fluorescence intensity. Thus, both repressors individually confine cib expression to a subset of the population. Taken together, we provide the first thorough characterization of a conventional gfp-reporter to study regulation of a group B colicin at the single cell level. This reporter will be useful to further investigate the costs and benefits of ColIb production in human pathogenic S. Tm and analyze cib expression under environmental conditions encountered in the mammalian gut.
Introduction
Colicins are narrow-spectrum antimicrobials produced by members of the Enterobacteriaceae family (e. g. E. coli, Salmonella spp. and Shigella) and used for interference competition among close relatives. They serve as “public good” for the population of producers but are only synthesized by a small fraction of the population which lyse (and eventually die) and release colicins. This “division of labor” is supposed to increase the overall fitness of the producer population in competition against colicin-sensitive strains [1, 2]. Colicins bind specific outer-membrane receptors in order to kill susceptible target bacteria. Common killing mechanisms of colicins include pore formation (e.g. colicins A, B, E1, Ia, Ib, K, N and U), nuclease activity (e.g. colicins E2-E9) and the interference with peptidoglycan synthesis (colicin M) [3]. Colicins are classified in two groups (A and B) according to the mode of entry into target bacteria [3]. Group A colicins (A, E1 to E9, K, L, N, S4, U, and Y) translocate via the Tol system while group B colicins (B, D, H, Ia, Ib, M) use the TonB import pathway. Group A colicin-loci are encoded on small, high-copy plasmids and comprise a cluster of three genes: an activity gene encoding the colicin, an immunity gene to protect the producer population against self-killing and a lysis gene, required for cell lysis and concomitant colicin release.
We have recently shown that Salmonella-induced gut inflammation leads to parallel blooms of Salmonella and commensal Enterobacteriaceae in the mouse intestine [4]. Salmonella enterica serovar Typhimurium (S. Tm) strain SL1344 produces Colicin Ib (ColIb), a group B colicin. ColIb binds to the E. coli outer-membrane receptor CirA, translocates to the periplasm in a TonB-dependent fashion and kills by pore formation in the inner membrane [5–7]. The locus encoding the colicin activity gene (cib) and the gene for its corresponding immunity protein (imm) is located on the plasmid pColIB9 (86.9 kB; further termed p2) [8, 9]. In inflammation-inflicted blooms, ColIb confers a significant fitness benefit to S. Tm over competing commensal E. coli [9, 10].
Cib expression is tightly repressed to ensure, that the fraction of producers is kept at low rates under conditions, when colicin is not required. Most group A colicin promoters harbour two overlapping binding sites for the LexA repressor downstream of the −10 box. In the course of DNA damage and the consequent SOS-response, LexA is cleaved by the activated RecA protease and colicin expression is triggered. DNA damage was identified as main stimulus of colicin expression while other tested stressors alone (osmolarity, heat/cold shock, starvation) had a minor influence [11].
Rather, negative regulation by environmental signals seems to be an additional strategy for various colicins to ensure tight repression. This is termed “double-locking” [12]. Cka expression is growth phase dependent and induced by nutrient depletion and positively affected by ppGpp [13]. Further, the iron–sulphur cluster regulator IscR was shown to stabilize LexA at the promoter and is de-repressed upon nutrient starvation [12]. The lysis genes of ColE7 and ColE2 are controlled by CsrA in response to different carbon sources [14, 15]. Besides nutrient conditions, other environmental cues can also affect colicin expression. For example, comparison of ColE7 expression in biofilm and planktonic environments revealed a two to three-fold upregulation in biofilms [16]. Thus, signals derived from DNA damage, growth conditions and the nutritional status of the bacteria converge to regulate colicin expression.
The colicin Ib promoter (Pcib) harbors one LexA binding site [17] (Fig 1A). Furthermore, a putative binding site for Fur is located upstream of the -35 region which mediates iron-dependent repression of Pcib {Nedialkova, 2014 #2}. Thus, Fur takes over a “double-locking” function for Pcib. Accordingly, cib expression in S. Tm is maximally de-repressed under iron limitation and upon exposure to DNA damaging agents, such as the drug mitomycin C (MitC) and conditions prevailing in the inflamed intestine [10].
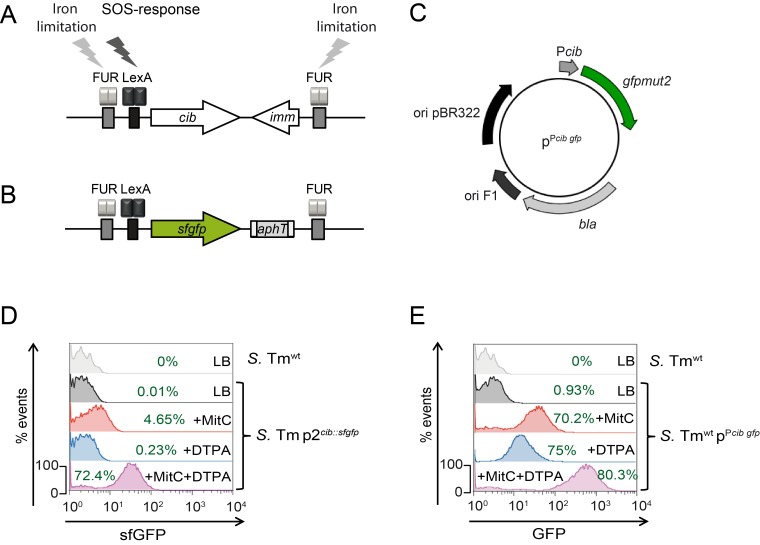
Fig 1. Characterization of the single- and multicopy gfp-reporter.
(A) Genetic organization of the ColIb locus in S. Tmwt and (B) the singlecopy reporter S. Tm p2cib::sfgfp. (C) Schematic view of the multicopy reporter pPcib gfp. (D) S. Tm p2cib::sfgfp and (E) S. Tm pPcib gfp were grown in LB (grey), or in LB supplemented with 0.25μg/ml MitC (red), 100μM DTPA (blue) or with both supplements (purple). Bacteria were analyzed for sfGFP or GFP -signal intensities by flow cytometry, respectively. S. Tmwt lacking the reporter was used as negative control and for calculating the fraction (%) of GFP+ bacteria (green).
Quantitative analyses of regulation and expression dynamics at the single cell level can yield further insights into the evolutionary stabilization of lethal phenotypes such as production of colicins in bacterial populations. At the single cell level, expression has mainly been studied in case of group A colicins, including ColK, E7 and E2 [11, 17–19]. In this work, we studied ColIb production by S. Tm at the single cell level. We generated singlecopy and multicopy gfp-reporters for the colicin Ib promoter Pcib. The multicopy reporter yielded optimal signal-intensity for a diverse range of applications and was therefore characterized and validated in detail.
Materials and Methods
Generation of bacterial mutant strains and plasmids
Bacterial strains and plasmids used in this study are listed in Table 1. S. Tm p2 cib::sfgfp (SJB15-2) and S. Tm p2cib-HA (M1400) were generated by λ-Red recombination. To this end, plasmid pWRG7 or pSU315 [20] were used as templates for a sfgfp gene or HA (Hemagglutinin) -epitope tag flanked by a kanamycin-cassette (sfgfp aphT and HA aphT), respectively. The gene for superfolder GFP (sfgfp, [21]) was synthesized codon-optimized for expression in Salmonella enterica (Life Technologies, Regensburg, Germany). After amplification of the sfgfp gene using primers NotI-SFGFP-for and XhoI-SFGFP-rev (S1 Table), the resulting product was digested NotI/XhoI and subsequently cloned in the similarly-digested vector p2795 [22], yielding pWRG7. To construct S. Tm p2cib-HA, sfgfp aphT was amplified from pWRG7 by PCR using primers SFGFP_cib_fwd/SFGFP_cib_rev (S1 Table) and transformed into SB300 pKD46. Correct insertion was validated by PCR and by sequencing using primers Check up_SFGFP_fwd/ Check up_SFGFP /RFP _rev (S1 Table). The cib::sfgfp aphT allele was then transduced into a clean S. Tmwt (SB300 pWKS30) background by P22-transduction [23] to generate S. Tm p2cib::sfgfp. To construct S. Tm p2cib-HA (M1400), the HA aphT epitope was amplified by PCR from pSU315 using primers Colicin-HA-fwd/Colicin-HA-rev (S1 Table). Correct insertion was validated by PCR and by sequencing using primers Col-ÜE-XbaI/ Col-ÜE-XhoI (S1 Table). The cib-HA aphT allele was then transduced into a fresh S. Tmwt (SB300 pWKS30) background by P22-transduction [23]. Expression of the HA-tagged ColIb in S. Tmwt p2cib-HA was verified by Western Blot using HA-specific antiserum (Santa Cruz). ColIb-HA shows similar bactericidal activity against sensitive strains as untagged ColIb and can therefore be considered as functional (not shown). To construct the plasmid pSJB16 (pPcib), pM1437 (pPcib gfp) was hydrolyzed by EcoRI to remove the gfpmut2 gene and then re-ligated. A similar approach was employed to construct the control plasmid pSJB17 (pcontrol): pM968 (pgfp) was hydrolyzed by EcoRI, relegated and the gfpmut2 gene removed.
Table 1. Bacteria and plasmids used in this study.
| S. Tm strains | Designation | Description/genotype | Reference |
|---|---|---|---|
| SB300 | S. Tmwt | S. Tm SL1344, SmR | [24] |
| M1400 | S. Tm p2cib-HA | S. Tm SL1344 cib-HA-aphT, SmR, KanR | This study |
| SJB15-2 | S. Tm p2cib::sfgfp | S. Tm SL1344 cib imm::sfgfp-aphT SmR, KanR | This study |
| E. coli strains | |||
| DH5α | Ec DH5α | F- Φ80lacZΔM15 Δ(lacZYA-argF) U169 recA1 endA1 hsdR17(rk -, mk +) phoA supE44 thi-1 gyrA96 relA1 λ- | Invitrogen |
| MG1655 | Ec MG1655 | E. coli K-12 strain MG1655, F- lambda- ilvG- rfb-50 rph-1; RifR, SmR | [25, 26] |
| Plasmids | |||
| pWKS30 | Low-copy vector; pSC101-based replicon; ampicillin-resistance marker | [27] | |
| pKD46 | Temperature sensitive replication (repA101ts); encodes lambda Red genes (exo, bet, gam); native terminator (tL3) after exo gene; arabinose-inducible promoter for expression (ParaB); encodes araC for repression of ParaB promoter; ampicillin resistance | [28] | |
| pWRG7 | High-copy vector, colE1-replicon; carries sfgfp aphT flanked by FRT sequences; ampicillin resistance | This study | |
| pSU315 | Low-copy vector; R6K gamma replicon | [20] | |
| pSJB17 | pcontrol | pBR322-derivative, ampicillin resistance | This study |
| pSJB16 | pPcib | pBR322-derivative, Pcib, ampicillin resistance | This study |
| pM968 | pgfp | pBR322-derivative, promoterless, gfpmut2; ampicillin resistance | [29] |
| pM1437 | pPcib gfp | pBR322-derivative, Pcib, gfpmut2; ampicillin resistance | [10] |
| pM979 | pPrpsm gfp | pBR322-derivative, PrpsM, (ribosomal rpsM promoter, constitutive), gfpmut2, ampicillin resistance | [29] |
| pM974 | pPsicA gfp | pBR322-derivative, PsicA (locus in Salmonella pathogenicity island 1), gfpmut2, ampicillin resistance | [29, 30] |
Bacterial growth conditions
If not otherwise stated, S. Tm and E. coli strains were grown in 3ml LB medium for 12h under mild aeration at 37°C in test tubes in a rotor wheel. Antibiotic concentrations used were ampicillin (100μg/ml), streptomycin (50μg/ml) and kanamycin (30μg/ml). The (o.n.) culture was then used for inoculation (1:20) of 3ml LB subcultures. The subcultures were supplemented with antibiotics and with either 0.25μg/ml mitomycin C (MitC; Roth) or with 100μM of diethylene triamine pentaacetic acid (DTPA; Sigma) or both as described [10], and grown for 4h at 37°C under mild aeration.
Determination of plasmid stability
The S. Tmwt pPcib gfp strain was inoculated on LB (Sm50, Kan30) from -80°C cryostock and incubated o.n. at 37°C. The following day, a single colony was used to inoculate 10ml LB liquid medium (no antibiotics). This culture was incubated o.n. at 37°C with 180rpm shaking. Subsequently, a sample of 1ml for an OD600 of 0.4 was taken and used to set up a subculture in 10ml LB liquid media (no antibiotics). This subculture was incubated until OD600 = 2–3. Four more passages were carried out in a similar fashion. The experiment was done in triplicates.
From each passage (1–5), samples were taken (OD600 of 1), diluted and plated on LB agar plates to obtain single colonies. A minimum of 100 colonies were screened for ampicillin resistance as a measure for pPcib gfp plasmid stability.
Determination of plasmid copy number
To determine plasmid copy number of pPcib gfp , total DNA was extracted from bacterial cultures of S. Tmwt pPcib gfp strain (passage 1–5). Bacteria were harvested by centrifugation and resuspended in 567μl TE-buffer (10 mM Tris-Cl, 1 mM EDTA, pH 8.0), 30μl 10% SDS and 3μl proteinase K (20 mg/ml) and incubated for 1h at 55°C. Thereafter, 100μl of 5M NaCl and 80μl CTAB/NaCl (4.1g NaCl and 10g CTAB [Cetyltrimethylammoniumbromid] dissolved in 100ml ddH2O) were added and incubated at 65°C for 10min. Subsequently, DNA was extracted with an equal volume of phenol/chloroform/isoamylalcohol (25:24:1) and EtOH precipitated. DNA was suspended in 60μl TE-buffer and the DNA concentration was determined using a NanoDrop Spectrophotometer ND-1000 (PEQLAB Biotechnology) and used for qPCR.
qPCR was performed using FastStart Essential DNA Green Master reaction mix for SYBR Green I-based real-time PCR (Roche) on a LightCycler® 96 Instrument. Each reaction was done in triplicates in a reaction mix of 20μl containing: 10μl of 2 x FastStart Essential DNA Green Master (Roche), 0.2μl forward primer (0.3μM), 0.2μl reverse primer (0.3μM), 2.5μl of total DNA (5-10ng/ μl) and 7.1μl RNase-free water. The following oligonucleotides were used: Ampli1_Fwd/Ampli1_Rev for pPcib gfp (amplicon: 73bps) and PsicA_2_fwd/PsicA_2_rev, specific for the sicA promoter which is present at one copy /S. Tm genome (amplicon: 79bps). The reaction was started with 1 cycle 95°C for 10min, following a 3-step amplification for 45 cycles at 95°C for 10s, 50°C (pPcib gfp) or 54°C (16S rRNA) and elongation at 72°C for 10s. To determine the linear dynamic range and reaction efficiency, a standard curve was generated using a 1:10 dilution series of the linearized plasmid pPcib gfp or linearized plasmid pM974 (harboring the psicA) from 2.5 x 107 to 1.0 x 102 copies for pPcib gfp, (efficiency 98%) and 2.5 x 107 to 1.0 x 10−1 copies in case of pM974 (efficiency 93%). Plasmids were linearized, purified and DNA was quantified using Quant-IT Picogreen dsDNA assay kit (Life Technologies). The standard curves were used for absolute quantification of genome and plasmid copy numbers. From this data, the relative copy number of pPcib gfp per S. Tm genome was calculated.
Generation of samples for immunoblotting
Total bacterial protein was harvested from subcultures (250μl of an OD600 = 1). Cells were collected by centrifugation (4°C, 10min, 10.621xg) and pellets were suspended in 250μl 1x loading buffer (50mM Tris-HCl pH 6.8, 100mM DTT, 2% SDS, 0.1% bromphenole blue, 10% glycerol) and lysed by incubation at 95°C for 10min. For supernatant samples, 500μl of subculture were pelleted and 400μl culture supernatant were added to 5x loading buffer and boiled for 10min at 95°C.
SDS-polyacrylamide gel electrophoresis (PAGE) and immunoblotting
SDS-PAGE was used to separate proteins [31]. Subsequently, proteins were transferred using a semidry blot onto a nitrocellulose membrane (GE Healthcare). The membrane was blocked (1x PBS, 0.1% Tween, 5% milk powder) and developed with polyclonal rabbit-anti HA (Y11, Santa Cruz) antiserum and goat-anti-rabbit-HRP (GE Healthcare) as secondary antibody followed by detection with the ECL detection system (GE Healthcare).
Colicin killing-assay
Colicin production and sensitivity was assayed as described [10]. Briefly, the colicin producing strain was grown o.n. as small spot (Ø 5mm) on LB agar containing 0.25μg/ml MitC (Roth). The plate was overlaid with the tester strain (EcMG1655) in top-agar (0.75% agar). Growth of tester strain was analyzed after 24h. Formation of an inhibition zone (halo) around the producer indicated production of colicin and sensitivity of the tester strain.
Immunofluorescence staining of intrabacterial proteins and confocal microscopy
Immunofluorescence staining of intrabacterial proteins was performed as described [32]. Bacteria were grown as described above in sterile polystyrene test tubes (Cultube, Simport). 250μl (OD600 1) culture were spun down for 5min, 4°C, 8000 rpm. Pellets were resuspended with 250μl ice cold 1x phosphate buffer saline (PBS) and 750μl of ice cold 4% paraformaldehyde (PFA) in PBS and incubated for 1h on ice. Subsequently, bacteria were washed three times with ice-cold PBS. Fixed bacteria were immobilized on poly L-lysine coated glass slides (Superfrost Plus, Thermo Scientific) by drying. Immobilized bacteria were additionally fixed for 5min with 4% PFA in PBS and washed 3 times with PBS. Bacteria were treated for 5min with permeabilization buffer A (20mM Tris, 0.1% TritonX-100, 50mM EDTA, 1.8% Glucose; pH 8.0), washed 3 times in ice cold permeabilization buffer B (25mM Tris, 10mM EDTA and 1.8% Glucose; pH 8.0), and incubated for 30min with permeabilization buffer B supplemented with 5mg/ml lysozyme at 4°C. Afterwards, bacteria were washed and blocked with blocking solution (10% normal goat serum diluted in PBS) for 1h. Bacteria were stained with primary antibody (rabbit-anti HA Y11 [1:200] (Santa Cruz); mouse-anti DnaK [1:200] (Enzo Life Sciences) or rabbit-anti GFP [1:200] (antibodies-online.com) diluted in blocking solution and washed 3 times with PBS. Thereafter, bacteria were stained with secondary antibodies (anti-rabbit-Dylight549-conjugate [1:400] (Jackson) or anti-mouse-RhodamineRed-X-conjugate [1:200] (Invitrogen) diluted in blocking solution. Bacterial DNA was stained with 4’6-Diamidine-2-phenylindol (DAPI [1μg/ml], Roth) or Hoechst 33342 ([10μg/ml], Thermo Scientific). Subsequently, bacteria were washed, dried in the dark and mounted with Vectashield (Vector) and sealed with nail varnish.
Confocal microscopy and image analysis
Using 63x oil objective and a magnification of 1 or 2.4 a minimum of 3 images were taken with a Leica SP5 confocal microscope. Image analysis was done using the ImageJ software, version 1.48v (Wayne Rasband, National Institute of Health, USA) [33]. Bacteria were detected in the DAPI channel. A mask was created to define objects (bacteria). This mask was superimposed on the green (GFP-signal) and red (ColIb-HA- or anti-GFP-signal) channels. The cell size, the integrated density and the mean fluorescence intensity (MFI) were calculated for each object. To correlate intrinsic GFP-fluorescence with signals of fluorescently labeled ColIb-HA or GFP, respectively, the corrected total cell fluorescence (CTCF) of the objects was calculated according to the formula: Integrated Density of the selected object–(Area of the selected object X Mean fluorescence of background signal). The detection limit for both MFI and CTCF values is given as the maximum value of MFI or CTCF signal determined for a control strain.
Flow Cytometry
Bacteria were grown as described and diluted in filtered PBS to a concentration of 107cfu/ml. Data were recorded by a FACS Canto II running the FACSDiva software (Aria Becton Dickinson). Data was analyzed using the FlowJo software (Tree Star, Inc.).
Statistical analysis
Statistical analyses were performed with Graph Pad Prism Version 5.01. To calculate statistical significance, the Kruskal-Wallis test with Dunn’s Multiple Comparison test was performed. P-values less than 0.05 were considered as significant. For correlation the Spearman-rank coefficient [ρ] was calculated.
Results and Discussion
Comparison of single-and multicopy gfp-reporters for ColIb gene expression at the single cell level
In order to analyze the expression of the ColIb gene (cib) at the single cell level, we generated single and multicopy Pcib gfp-reporters. For the singlecopy reporter (S. Tm p2cib::sfgfp) the entire ColIb locus on p2 (cib and imm) was exchanged against superfolder gfp (sfgfp) [21], which encodes a bright and photostable GFP-variant (Fig 1B). The singlecopy reporter strain was characterized by flow cytometry under different environmental conditions. S. Tm p2cib::sfgfp was grown for 4h to late logarithmic growth phase either in LB without supplements or in LB supplemented with 0.25μg/ml MitC (to induce DNA damage) or 100μM DTPA (to induce Fe2+-limitation) or both supplements. Flow cytometric analysis showed that the single copy reporter is induced under iron limiting and SOS-inducing conditions and that cib expression was maximal if both supplements were added (Fig 1D). In conclusion, signal intensity of the singlecopy reporter was rather low, which is in line with the notion, that pColIB9 is only present in one copy per cell.
As fluorescence-intensity of the singlecopy reporter was overall low, we also generated a multicopy reporter by fusing the promoter region Pcib to gfp in a derivative of the pBAD24 ori pBR322 [34] (Fig 1C). pBR322 is a derivative the ColE1-type plasmid pMB1 [35] and shares the replication control mechanism with ColE1 and relatives. Intriguingly, pPcib gfp is therefore the descendant of a group A colicin plasmid.
As expected, the mean signal intensity was higher compared to the single copy reporter under all conditions (Fig 1E). Consequently, the fraction of GFP+ S. Tm was consistently higher for the multicopy reporter, while the overall pattern of the GFP+ S. Tm population under different conditions strongly resembled the singlecopy reporter. The data is in accordance with previous results obtained by bulk assays (Western blot and luciferase reporter) [10].
Intriguingly, different studies on colicin-expression rates in bacterial populations reported rather similar results although media composition, growth conditions and the detection limit of the fluorescent protein (FP) likely varies between different laboratories and analysis methods. Using gfp-reporters based on a low copy number pSC101 plasmid, a rate of ~0.5% was determined for ColA, ColN and ColE1 in the early stationary phase in rich media [36]. For ColE7, the same study determined 1.5% GFP+ cells in the early stationary phase. In a different study using the same gfp-reporter, comparable levels were determined in the stationary phase and slight increases were observed at post-exponential (1.8±0.2%) and exponential phase (2.3±1.2) [16]. For ColK, 3% of the E. coli population was GFP+ in stationary cultures using a reporter based on the natural colicin K-encoding plasmid [19].
Our FACS results using the multicopy gfp-reporter pPcib gfp showed that 0,93% of the Salmonella population were GFP+ in LB during late exponential phase. Measurements using the singlecopy reporter yielded much lower rates (0,01%). This demonstrates that plasmid copy number can introduce a major bias to quantification of gfp expression rates in bacterial populations.
Effect of inducer titration on cib expression in S. Tm
As previously shown for other colicins [19, 36], a small fraction of the S. Tm population carrying the reporters was GFP+ in the absence of supplements (DTPA, MitC). Colicin induction in this subpopulation could be due to spontaneous induction of the SOS-response [37]. Similarly, the noise observed in prophage activation in Corynebacterium glutamicum was also attributed to a spontaneous SOS-response in about 0.2% of cells [38].
When either MitC or DTPA (or both inducers) were added, the entire S. Tm population responded with gfp expression (heterogeneous expression pattern) as shown with both reporters (Fig 1D and 1E). In fact, similar observations were made for ColK. Under MitC or when LexA was defective nearly all cells of the population expressed the cka-gfp fusion and turned GFP+ [19]. Similar observations were made when the LexA binding site of the cka promoter was modified [37]. On the basis of the “division of labor” hypothesis for colicin production, we expected to observe a bimodal expression pattern for a colicin under inducing conditions. In our experiments we used rather non-physiological supplement concentrations optimized for bulk assays to maximally induce Pcib. To test, if lower supplement concentrations would trigger a bimodal expression pattern (i.e. two distinct populations), we generated dose-response curves for the two different supplements MitC and DTPA using the singlecopy reporter S. Tm p2cib::sfgfp (S1 Fig). Curiously, bimodal expression was never observed, not even at low supplement concentrations. We reasoned that due to the presence of two different repressor-binding sites (FUR- and LexA-box) in Pcib, bimodal expression might require both inducing agents at the same time. Therefore, one supplement was kept at a constant concentration (ranging from low to high) while the other supplement was titrated. However, under all possible combinations, fluorescence intensities were log-normally distributed and bimodality (e.g. two peaks of different fluorescence intensity) was not observed (S1 Fig). The experiments were repeated using S. Tm pPcib gfp (multicopy gfp-reporter strain) and similar results were obtained (S1 Fig). In conclusion, cib expression in S. Tm under the tested in vitro conditions does not appear to be bimodal. As the mechanism of ColIb release is still unknown, the actual costs of ColIb production cannot be determined (e.g. release by lysis or secretion). Thus, it is unclear to which extend the principle of “division of labor” applies for cib expression. Further work will be needed to clarify this issue.
The multicopy gfp-reporter is stable and does not influence intrinsic cib expression in S. Tm
The major advantage of the multicopy-gfp-reporter is its high sensitivity. However, the reporter plasmid might be lost due to plasmid instability and thereby bias results. To address this, we determined stability of pPcib gfp in S. Tmwt in the absence of antibiotics. Plasmid stability was >99% after 24 generations during 5 successive passages in LB without ampicillin. Similarly, the copy numbers of pPcib gfp in S. Tmwt was stable at the different rounds of passaging (S2 Fig). From this we concluded, that pPcib gfp stability is sufficient for the planned applications.
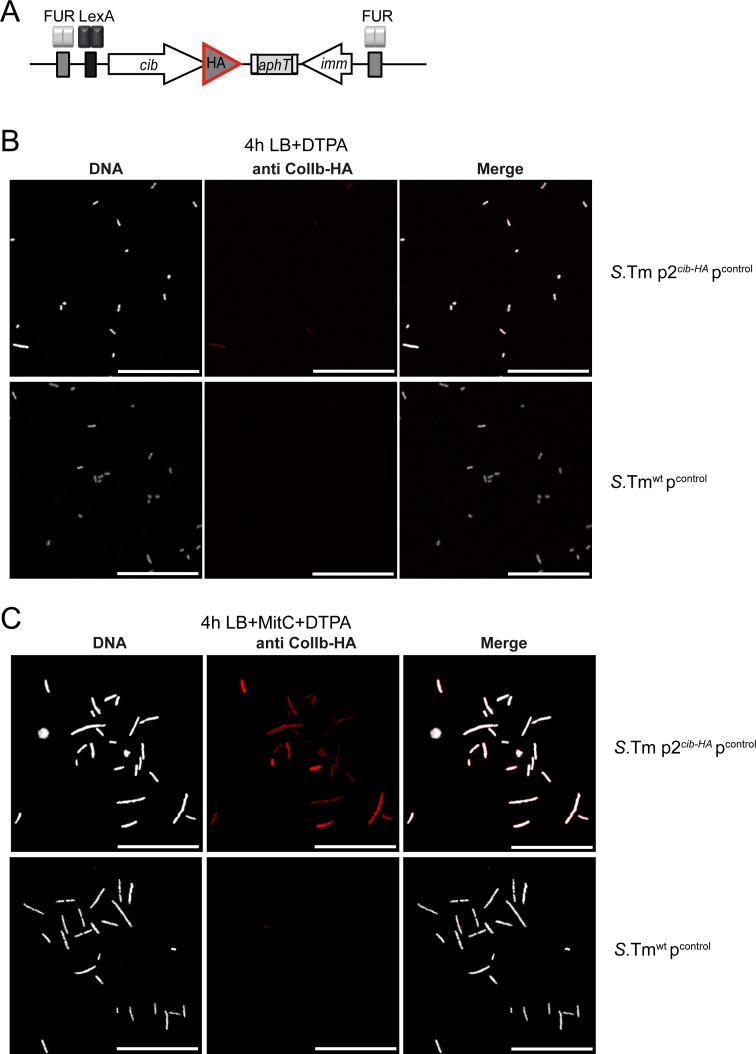
S. Tmwt carrying the multicopy reporter retains its native, functional ColIb locus in addition to the reporter. To test if gfp expression of the multicopy reporter (pPcib gfp) correlates with intrinsic cib expression, we generated a chromosomal ColIb-HA fusion construct in S. Tm wt which could be detected by immunofluorescence (S. Tm p2cib-HA; Fig 2A). ColIb-HA still retains full bactericidal activity against a ColIb-sensitive E. coli strain as determined by halo-assay (not shown). ColIb-HA could be detected within lysozyme-permeabilized bacteria by intrabacterial immunofluorescent staining (Fig 2B and 2C). Efficiency of lysozyme permeabilization was optimized and antibody-specificity confirmed for this assay (S3 Fig; Fig 2B and 2C).
Fig 2. Detection of ColIb-HA within individual bacteria.
(A) Schematic of the modified ColIb locus in the strain S. Tm p2cib-HA. S. Tm p2cib-HA pcontrol and S. Tmwt pcontrol were grown in LB supplemented with 100μM DTPA (B) or LB supplemented 0.25μg/ml MitC + 100μM DTPA (C). Bacteria were fixed, permeabilized and intracellular ColIb-HA was detected using HA-specific antiserum (red). DNA was stained with DAPI (greyscale). Scale bar 25μm.
By introducing >100 copies of the promoter Pcib or the gfp gene (S2 Fig), the multicopy reporter may affect regulation of the native Pcib. Therefore, we aimed to address if cib expression is influenced by multicopy effects of pPcib gfp. We generated several derivatives of pPcib gfp (S4 Fig). The plasmid pcontrol lacking the Pcib element and gfp and only retaining the vector backbone served as negative control (S4 Fig). Furthermore, two additional control plasmids were constructed: pPcib lacking the gfp gene (S4 Fig) and pgfp lacking the cib promoter Pcib (S4 Fig). Expression of fluorescent proteins can negatively influence bacterial fitness [39, 40]. To address any possible effects of gfp expression on native Pcib, we also employed a plasmid for constitutive high-level gfp expression pPrpsm gfp (S4 Fig).
S. Tm p2cib-HA was then transformed with pPcib gfp or one the four derivatives, respectively. To quantify cib-HA expression under different environmental conditions, bacteria were grown in LB with or without addition of supplements to late logarithmic growth phase. Additionally, bacteria from a stationary culture (12h grown in LB) were included. Bacterial samples were fixed, permeabilized and DNA and ColIb-HA were stained and analyzed by immunofluorescence microscopy (Fig 2B and 2C). Fluorescence intensity of intrabacterial ColIb-HA levels (Dylight 549 fluorescence) and GFP were quantified using ImageJ (Fig 3).
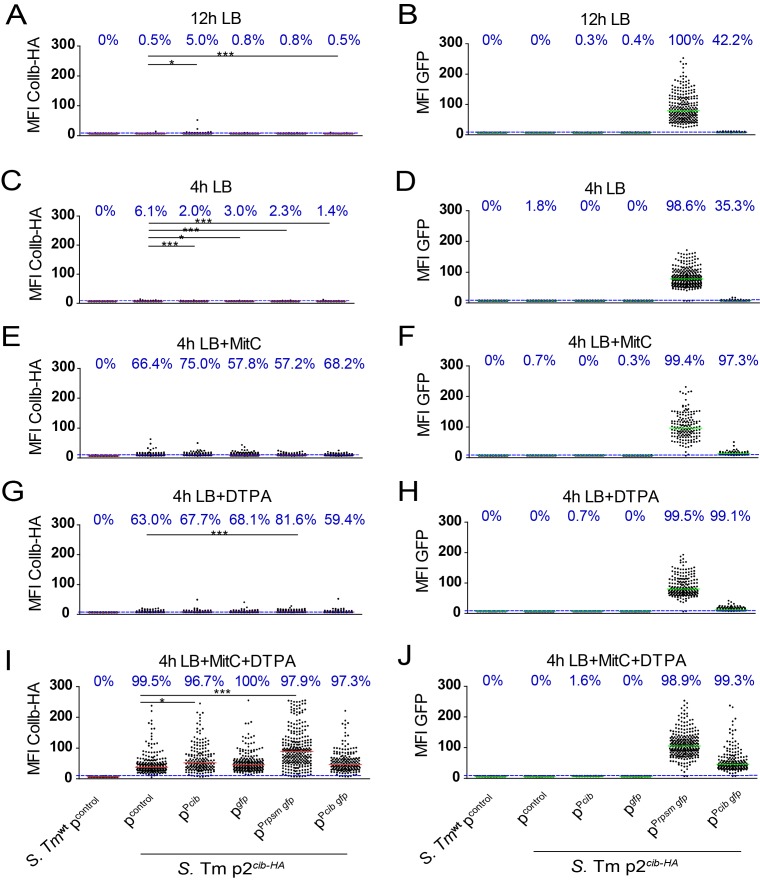
Fig 3. Influence of the multicopy reporter pPcib gfp and its derivatives on expression of native S. Tm p2cib-HA at the single cell level.
S. Tm p2 cib-HA transformed with pPcib gfp or the derivatives (pcontrol, pPcib, pgfp and pPrpsm gfp), respectively, were grown in LB (+ampicillin) overnight (12h; stationary phase) or subcultured for 4h in LB (late logarithmic phase) with or without indicated supplements (0.25μg/ml MitC, 100μM DTPA). Bacteria were fixed, permeabilized and intracellular ColIb-HA was detected using HA-specific antiserum and a DyLight 594-conjugated secondary antibody, DNA was stained with DAPI. Bacteria were imaged by confocal microscopy and fluorescence of DAPI, GFP and DyLight 594-conjugate was recorded. Mean fluorescence intensity (MFI) of ColIb-HA-DyLight594 (A,C,E,G,I) and GFP (B,D,F,H,J) as determined by ImageJ is shown. Dots represent MFI of individual objects (bacteria). Bars represent the median and the dotted line the detection limit (background fluorescence of S. Tmwt pcontrol). Values indicate the fraction (%) of the population above detection limit (blue). Statistical analysis was done using Kruskal-Wallis test with Dunn's post test (* p < 0.05).
In LB without supplements, ColIb-HA and GFP levels were close to the detection limit of this method (Fig 3A–3D). Overall, the relative rates of GFP+ S. Tm were different when compared to the FACSs analysis (Fig 1). This is likely due to different thresholds applied for detection of GFP+ S. Tm cells. Under any of the cib-inducing conditions, no major differences in ColIb-HA levels between S. Tm p2 cib-HA carrying the multicopy reporter pPcib gfp and the derivatives pcontrol, pgfp and pPcib were detected (Fig 3E–3J). Thus, we concluded that the multicopy reporter (pPcib gfp) does not evidently influence expression of native cib under the tested conditions. In contrast, we found that carriage of pPrpsm gfp is accompanied by increased ColIb-HA levels (Fig 3I). This suggests that GFP, or overexpressed proteins in general, may trigger bacterial stress-responses at very high protein concentrations. Alternatively, the requirement for specific tRNAs that become limiting due to the GFP synthesis may account for increased stress.
Correlation of gfp expression of the reporter pPcib gfp with intrinsic cib-HA expression
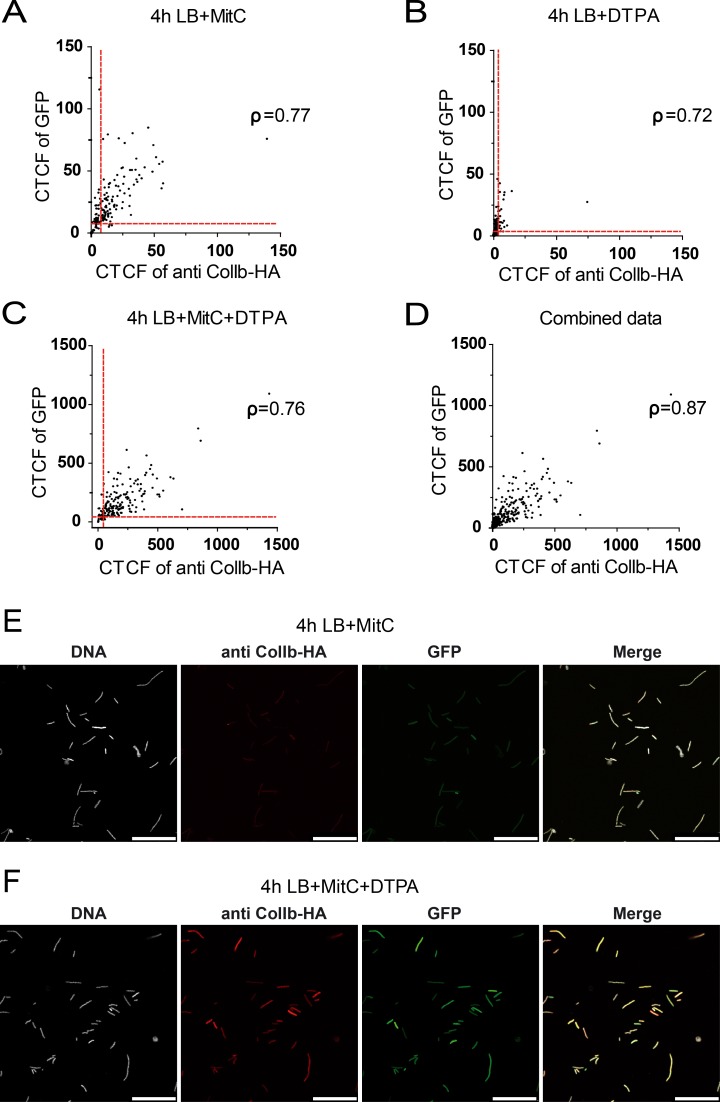
Next, we set out to determine if gfp expression of the multicopy reporter pPcib gfp is a valid proxy for inferring native cib-HA expression in individual bacteria. Formation of the GFP-chromophore depends on the correct folding of the protein. Thus, slow maturation of newly synthesized unfolded GFP-molecules could negatively affect correlation. To address this, we tested if GFP-fluorescence (GFPfl; mature folded GFP) correlated with GFP-protein (total GFP) using a GFP-specific antiserum and intrabacterial immunofluorescence microscopy. This experiment revealed that GFPfl correlates well with GFP protein levels in S. Tm under all conditions tested, indicating fast protein maturation (overall Spearman-rank correlation [ρ] = 0.95; S5 Fig). Next, we correlated gfp expression of the reporter pPcib gfp with cib-HA expression in individual bacteria (Fig 4). To this end, we plotted the GFP-signal against ColIb-HA-DyLight594 signal of individual S. Tm p2cib-HA pPcib gfp from the experiment depicted in Fig 4. GFP-levels correlated well to cib-HA expression under cib-inducing conditions (overall Spearman-rank correlation [ρ] = 0.87; Fig 4D).
Fig 4. Correlation of gfp expression of the reporter pPcib gfp with intrinsic cib-HA expression under different conditions at the single cell level.
S. Tm p2 cib-HA transformed with pPcib gfp was grown in the presence of different supplements as described in Fig 4. Bacteria were fixed, permeabilized and intracellular ColIb-HA was detected using HA-specific antiserum, DNA was stained with DAPI. Bacteria were imaged by confocal microscopy and fluorescence of DAPI, GFP and the DyLight 594-conjugate was recorded. Corrected total cell fluorescence (CTCF) was calculated and GFP-fluorescence of individual bacteria was correlated to Dylight549 fluorescence (A-C). A combination of data from all 3 conditions is shown in (D). Red line: detection limit; [ρ] = Spearman-rank correlation coefficient. Examples are shown for intrabacterial IF for S. Tm supplemented with MitC (E) and MitC+DTPA (F). Scale bar 25μm.
Conclusion
In summary, we provide a comprehensive characterization of reporter tools to study regulation of the group B colicin ColIb at the single cell level in the human enteric pathogen S. Tm. Taken together, the multicopy reporter pPcib gfp yields a high GFP signal and closely reflects expression of native cib. We could rule out that the multicopy-reporter induces substantial changes in regulation of the native ColIb locus at the single cell level, which might have been caused by the introduction of additional copies of the Pcib promoter (including the repressor binding sites). Additionally, toxic effects of GFP might cause bacterial stress responses leading to an erroneous upregulation of the reporter gene [39, 40]. However, we show that intrinsic ColIb-HA levels correlate with the FP-reporter which establishes pPcib gfp as a powerful tool to analyse cib expression at the single cell level. Therefore, this reporter can further be used for time lapse microscopy experiments in microfluidics platforms as well as for studying cib expression in vivo in the murine gut.
Supporting Information
S. Tm p2cib::sfgfp (single copy gfp-reporter) was cultured for 4h in LB with either increasing concentrations of (A) DTPA (6μM, 12μM, 25μM, 50μM, 100μM and 200μM), while MitC concentration was kept constant at 0μg/ml, 0.01μg/ml, 0.1μg/ml or 0.25μg/ml or (B) MitC (0.01μg/ml, 0.05μg/ml, 0.1μg/ml 0.2μg/ml, 0.25μg/ml, 0.5μg/ml and 1μg/ml) while the DTPA concentration was kept constant at 0μM, 6μM, 25μM or 100μM. Bacteria were subsequently analyzed by FACS for GFP-signal intensity. Cultures of S. Tm pPcib (multi-copy gfp-reporter) were grown in LB with either increasing concentrations of (C) DTPA (6μM, 12μM, 25μM, 50μM, 100μM and 200μM), while MitC concentration was kept constant at 0.25μg/ml or (D) MitC (0.01μg/ml, 0.05μg/ml, 0.1μg/ml 0.2μg/ml, 0.25μg/ml, 0.5μg/ml and 1μg/ml) while the DTPA concentration was kept constant at 100μM. Bacteria were analyzed by FACS for GFP-signal intensity. S. Tmwt lacking the reporter was used as negative control and for calculating the fraction (%) of GFP+ bacteria (green).
(TIF)
S. Tmwt harboring pPcib gfp was cultured for five consecutive passages (1–5) in 10ml LB. Briefly, a sample of 1ml for an OD600 of 0.4 was taken and used to set up a subculture in 10ml LB liquid media (no antibiotics). This subculture was incubated until OD600 = 2–3. Four more passages were carried out in a similar fashion. From each passage (1–5), samples were taken (1ml from 1 OD600) and total DNA was extracted for quantitative PCR analysis. (A) Copy number of pPcib gfp per ml culture (OD600 of 1) as determined by absolute quantification. (B) Genome copy number as determined by absolute quantification of PsicA copies per ml culture (OD600 of 1). Data were analyzed by 1-way ANOVA. No significant differences were determined between passages. pPcib gfp copy number in S. Tmwt in the 5 consecutive passages (mean±SD) as calculated from data shown in A and B: passage 1: 169±91; passage 2: 84±18; passage 3: 147±57; passage 4: 198±100; passage 5: 127±67.
(TIF)
S. Tm p2cib-HA was grown in LB for 12h and 4h in LB or for 4h supplemented with MitC, DTPA or both. The efficiency of lysozyme permeabilization was validated by staining the cytosolic, constitutively expressed protein DnaK in lysozyme-treated (lower panel) and non-treated samples (upper panel) (A). ColIb-HA was detected within a small fraction of S. Tm grown in LB after lysozyme treatment (arrow) but not in untreated samples (upper panel) (B). ColIb-HA was detected in S. Tm grown in LB MitC+DTPA after lysozyme treatment (lower panel) and also in a small fraction of cells without lysozyme treatment (upper panel) (C). Scale bar 25μm.
(TIF)
Plasmid maps of (A) pPcib gfp (multicopy reporter); (B) pcontrol (only plasmid backbone) (C) pPcib (only promoter of cib); (D) pgfp (only gfp); (E) pPrpsm gfp (constitutive promoter) are shown.
(TIF)
S. Tmwt pPcib gfp was grown for 12h (A) and 4h (B) in LB or for 4h supplemented with MitC (C), DTPA (D) or both (E). Bacteria were fixed, lysozyme permeabilized and stained with a GFP-specific antiserum and a Dylight549-conjugated secondary antibody and analyzed by fluorescence microscopy followed by image analysis. Corrected total cell fluorescence (CTCF) was calculated and GFP-fluorescence of individual bacteria was correlated to Dylight549 fluorescence ([ρ] Spearman-rank correlation coefficient). A combination of all data is shown in (F). Red line: detection limit. Examples for IF-microscopy are shown in (G) and (H). Scale bar 25μm.
(TIF)
(DOCX)
Acknowledgments
We thank Roman Gerlach for kindly providing the plasmid pWRG7, and Ombeline Rossier and members of the Stecher lab for discussions.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The work was supported by grants from the DFG (DFG STE 1971/2-1 and DFG STE 1971/6-1) to BS. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Riley MA, Wertz JE. Bacteriocin diversity: ecological and evolutionary perspectives. Biochimie. 2002;84(5–6):357–64. . [DOI] [PubMed] [Google Scholar]
- 2. Majeed H, Gillor O, Kerr B, Riley MA. Competitive interactions in Escherichia coli populations: the role of bacteriocins. The ISME journal. 2011;5(1):71–81. 10.1038/ismej.2010.90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cascales E, Buchanan SK, Duche D, Kleanthous C, Lloubes R, Postle K, et al. Colicin biology. Microbiology and molecular biology reviews: MMBR. 2007;71(1):158–229. 10.1128/MMBR.00036-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Stecher B, Maier L, Hardt WD. 'Blooming' in the gut: how dysbiosis might contribute to pathogen evolution. Nature reviews Microbiology. 2013;11(4):277–84. 10.1038/nrmicro2989 . [DOI] [PubMed] [Google Scholar]
- 5. Weaver CA, Kagan BL, Finkelstein A, Konisky J. Mode of action of colicin ib: formation of ion-permeable membrane channels. Biochimica et biophysica acta. 1981;645(1):137–42. . [DOI] [PubMed] [Google Scholar]
- 6. Mankovich JA, Hsu CH, Konisky J. DNA and amino acid sequence analysis of structural and immunity genes of colicins Ia and Ib. Journal of bacteriology. 1986;168(1):228–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Davies JK, Reeves P. Genetics of resistance to colicins in Escherichia coli K-12: cross-resistance among colicins of group B. Journal of bacteriology. 1975;123(1):96–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kroger C, Dillon SC, Cameron AD, Papenfort K, Sivasankaran SK, Hokamp K, et al. The transcriptional landscape and small RNAs of Salmonella enterica serovar Typhimurium. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(20):E1277–86. 10.1073/pnas.1201061109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Stecher B, Denzler R, Maier L, Bernet F, Sanders MJ, Pickard DJ, et al. Gut inflammation can boost horizontal gene transfer between pathogenic and commensal Enterobacteriaceae. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(4):1269–74. 10.1073/pnas.1113246109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nedialkova LP, Denzler R, Koeppel MB, Diehl M, Ring D, Wille T, et al. Inflammation fuels colicin Ib-dependent competition of Salmonella serovar Typhimurium and E. coli in enterobacterial blooms. PLoS pathogens. 2014;10(1):e1003844 10.1371/journal.ppat.1003844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ghazaryan L, Tonoyan L, Ashhab AA, Soares MI, Gillor O. The role of stress in colicin regulation. Archives of microbiology. 2014;196(11):753–64. 10.1007/s00203-014-1017-8 . [DOI] [PubMed] [Google Scholar]
- 12. Butala M, Sonjak S, Kamensek S, Hodoscek M, Browning DF, Zgur-Bertok D, et al. Double locking of an Escherichia coli promoter by two repressors prevents premature colicin expression and cell lysis. Mol Microbiol. 2012;86(1):129–39. 10.1111/j.1365-2958.2012.08179.x . [DOI] [PubMed] [Google Scholar]
- 13. Kuhar I, Zgur-Bertok D. Transcription regulation of the colicin K cka gene reveals induction of colicin synthesis by differential responses to environmental signals. Journal of bacteriology. 1999;181(23):7373–80. Epub 1999/11/26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yang TY, Sung YM, Lei GS, Romeo T, Chak KF. Posttranscriptional repression of the cel gene of the ColE7 operon by the RNA-binding protein CsrA of Escherichia coli. Nucleic Acids Res. 2010;38(12):3936–51. Epub 2010/04/10. doi: gkq177 [pii] 10.1093/nar/gkq177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hol FJ, Voges MJ, Dekker C, Keymer JE. Nutrient-responsive regulation determines biodiversity in a colicin-mediated bacterial community. BMC Biol. 2014;12:68. Epub 2014/08/28. doi: s12915-014-0068-2 [pii] 10.1186/s12915-014-0068-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Majeed H, Ghazaryan L, Herzberg M, Gillor O. Bacteriocin expression in sessile and planktonic populations of Escherichia coli. J Antibiot (Tokyo). 2015;68(1):52–5. Epub 2014/07/06. doi: ja201484 [pii] 10.1038/ja.2014.84 . [DOI] [PubMed] [Google Scholar]
- 17. Gillor O, Vriezen JA, Riley MA. The role of SOS boxes in enteric bacteriocin regulation. Microbiology. 2008;154(Pt 6):1783–92. 10.1099/mic.0.2007/016139-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ozeki H, Stocker BA, De Margerie H. Production of colicine by single bacteria. Nature. 1959;184:337–9. . [DOI] [PubMed] [Google Scholar]
- 19. Mulec J, Podlesek Z, Mrak P, Kopitar A, Ihan A, Zgur-Bertok D. A cka-gfp transcriptional fusion reveals that the colicin K activity gene is induced in only 3 percent of the population. Journal of bacteriology. 2003;185(2):654–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Uzzau S, Figueroa-Bossi N, Rubino S, Bossi L. Epitope tagging of chromosomal genes in Salmonella. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(26):15264–9. 10.1073/pnas.261348198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pedelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS. Engineering and characterization of a superfolder green fluorescent protein. Nature biotechnology. 2006;24(1):79–88. 10.1038/nbt1172 . [DOI] [PubMed] [Google Scholar]
- 22. Husseiny MI, Hensel M. Rapid method for the construction of Salmonella enterica Serovar Typhimurium vaccine carrier strains. Infection and immunity. 2005;73(3):1598–605. 10.1128/IAI.73.3.1598-1605.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schmieger H. Phage P22-mutants with increased or decreased transduction abilities. Molecular & general genetics: MGG. 1972;119(1):75–88. . [DOI] [PubMed] [Google Scholar]
- 24. Hoiseth SK, Stocker BA. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature. 1981;291(5812):238–9. . [DOI] [PubMed] [Google Scholar]
- 25. Blattner FR, Plunkett G 3rd, Bloch CA, Perna NT, Burland V, Riley M, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277(5331):1453–62. . [DOI] [PubMed] [Google Scholar]
- 26. Moller AK, Leatham MP, Conway T, Nuijten PJ, de Haan LA, Krogfelt KA, et al. An Escherichia coli MG1655 lipopolysaccharide deep-rough core mutant grows and survives in mouse cecal mucus but fails to colonize the mouse large intestine. Infection and immunity. 2003;71(4):2142–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang RF, Kushner SR. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene. 1991;100:195–9. . [PubMed] [Google Scholar]
- 28. Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(12):6640–5. 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Stecher B, Hapfelmeier S, Muller C, Kremer M, Stallmach T, Hardt WD. Flagella and chemotaxis are required for efficient induction of Salmonella enterica serovar Typhimurium colitis in streptomycin-pretreated mice. Infection and immunity. 2004;72(7):4138–50. 10.1128/IAI.72.7.4138-4150.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Stecher B, Robbiani R, Walker AW, Westendorf AM, Barthel M, Kremer M, et al. Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 2007;5(10):2177–89. 10.1371/journal.pbio.0050244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227(5259):680–5. . [DOI] [PubMed] [Google Scholar]
- 32. Schlumberger MC, Muller AJ, Ehrbar K, Winnen B, Duss I, Stecher B, et al. Real-time imaging of type III secretion: Salmonella SipA injection into host cells. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(35):12548–53. 10.1073/pnas.0503407102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nature methods. 2012;9(7):671–5. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. Journal of bacteriology. 1995;177(14):4121–30. Epub 1995/07/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bolivar F, Rodriguez RL, Greene PJ, Betlach MC, Heyneker HL, Boyer HW, et al. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. Epub 1977/01/01. . [PubMed] [Google Scholar]
- 36. Kamensek S, Podlesek Z, Gillor O, Zgur-Bertok D. Genes regulated by the Escherichia coli SOS repressor LexA exhibit heterogeneous expression. BMC microbiology. 2010;10:283 10.1186/1471-2180-10-283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mrak P, Podlesek Z, van Putten JP, Zgur-Bertok D. Heterogeneity in expression of the Escherichia coli colicin K activity gene cka is controlled by the SOS system and stochastic factors. Molecular genetics and genomics: MGG. 2007;277(4):391–401. 10.1007/s00438-006-0185-x . [DOI] [PubMed] [Google Scholar]
- 38. Nanda AM, Heyer A, Kramer C, Grunberger A, Kohlheyer D, Frunzke J. Analysis of SOS-induced spontaneous prophage induction in Corynebacterium glutamicum at the single-cell level. Journal of bacteriology. 2014;196(1):180–8. Epub 2013/10/29. doi: JB.01018-13 [pii] 10.1128/JB.01018-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hebisch E, Knebel J, Landsberg J, Frey E, Leisner M. High variation of fluorescence protein maturation times in closely related Escherichia coli strains. PloS one. 2013;8(10):e75991 10.1371/journal.pone.0075991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liu HS, Jan MS, Chou CK, Chen PH, Ke NJ. Is green fluorescent protein toxic to the living cells? Biochemical and biophysical research communications. 1999;260(3):712–7. 10.1006/bbrc.1999.0954 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
S. Tm p2cib::sfgfp (single copy gfp-reporter) was cultured for 4h in LB with either increasing concentrations of (A) DTPA (6μM, 12μM, 25μM, 50μM, 100μM and 200μM), while MitC concentration was kept constant at 0μg/ml, 0.01μg/ml, 0.1μg/ml or 0.25μg/ml or (B) MitC (0.01μg/ml, 0.05μg/ml, 0.1μg/ml 0.2μg/ml, 0.25μg/ml, 0.5μg/ml and 1μg/ml) while the DTPA concentration was kept constant at 0μM, 6μM, 25μM or 100μM. Bacteria were subsequently analyzed by FACS for GFP-signal intensity. Cultures of S. Tm pPcib (multi-copy gfp-reporter) were grown in LB with either increasing concentrations of (C) DTPA (6μM, 12μM, 25μM, 50μM, 100μM and 200μM), while MitC concentration was kept constant at 0.25μg/ml or (D) MitC (0.01μg/ml, 0.05μg/ml, 0.1μg/ml 0.2μg/ml, 0.25μg/ml, 0.5μg/ml and 1μg/ml) while the DTPA concentration was kept constant at 100μM. Bacteria were analyzed by FACS for GFP-signal intensity. S. Tmwt lacking the reporter was used as negative control and for calculating the fraction (%) of GFP+ bacteria (green).
(TIF)
S. Tmwt harboring pPcib gfp was cultured for five consecutive passages (1–5) in 10ml LB. Briefly, a sample of 1ml for an OD600 of 0.4 was taken and used to set up a subculture in 10ml LB liquid media (no antibiotics). This subculture was incubated until OD600 = 2–3. Four more passages were carried out in a similar fashion. From each passage (1–5), samples were taken (1ml from 1 OD600) and total DNA was extracted for quantitative PCR analysis. (A) Copy number of pPcib gfp per ml culture (OD600 of 1) as determined by absolute quantification. (B) Genome copy number as determined by absolute quantification of PsicA copies per ml culture (OD600 of 1). Data were analyzed by 1-way ANOVA. No significant differences were determined between passages. pPcib gfp copy number in S. Tmwt in the 5 consecutive passages (mean±SD) as calculated from data shown in A and B: passage 1: 169±91; passage 2: 84±18; passage 3: 147±57; passage 4: 198±100; passage 5: 127±67.
(TIF)
S. Tm p2cib-HA was grown in LB for 12h and 4h in LB or for 4h supplemented with MitC, DTPA or both. The efficiency of lysozyme permeabilization was validated by staining the cytosolic, constitutively expressed protein DnaK in lysozyme-treated (lower panel) and non-treated samples (upper panel) (A). ColIb-HA was detected within a small fraction of S. Tm grown in LB after lysozyme treatment (arrow) but not in untreated samples (upper panel) (B). ColIb-HA was detected in S. Tm grown in LB MitC+DTPA after lysozyme treatment (lower panel) and also in a small fraction of cells without lysozyme treatment (upper panel) (C). Scale bar 25μm.
(TIF)
Plasmid maps of (A) pPcib gfp (multicopy reporter); (B) pcontrol (only plasmid backbone) (C) pPcib (only promoter of cib); (D) pgfp (only gfp); (E) pPrpsm gfp (constitutive promoter) are shown.
(TIF)
S. Tmwt pPcib gfp was grown for 12h (A) and 4h (B) in LB or for 4h supplemented with MitC (C), DTPA (D) or both (E). Bacteria were fixed, lysozyme permeabilized and stained with a GFP-specific antiserum and a Dylight549-conjugated secondary antibody and analyzed by fluorescence microscopy followed by image analysis. Corrected total cell fluorescence (CTCF) was calculated and GFP-fluorescence of individual bacteria was correlated to Dylight549 fluorescence ([ρ] Spearman-rank correlation coefficient). A combination of all data is shown in (F). Red line: detection limit. Examples for IF-microscopy are shown in (G) and (H). Scale bar 25μm.
(TIF)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.