Abstract
Neuronal cell death, in neurodegenerative disorders, is mediated through a spectrum of biological processes. Excessive amounts of free radicals, such as reactive oxygen species (ROS), has detrimental effects on neurons leading to cell damage via peroxidation of unsaturated fatty acids in the cell membrane. Edaravone (3-methyl-1-phenyl-2-pyrazolin-5-one) has been used for neurological recovery in several countries, including Japan and China, and it has been suggested that Edaravone may have cytoprotective effects in neurodegeneration. Edaravone protects nerve cells in the brain by reducing ROS and inhibiting apoptosis. To gain further insight into the cytoprotective effects of Edaravone against oxidative stress condition we have performed comparative two-dimensional gel electrophoresis (2DE)-based proteomic analyses on SH-SY5Y neuroblastoma cells exposed to oxidative stress and in combination with Edaravone. We showed that Edaravone can reverse the cytotoxic effects of H2O2 through its specific mechanism. We observed that oxidative stress changes metabolic pathways and cytoskeletal integrity. Edaravone seems to reverse the H2O2-mediated effects at both the cellular and protein level via induction of Peroxiredoxin-2.
Keywords: Parkinson’s disease, Edaravone, Peroxiredoxin-2, L-Dopa, oxidative stress, proteomics
1. Introduction
Neurodegeneration is characterized by the progressive death of neurons. Neurodegenerative processes cause many neurodegenerative disorders including Parkinson’s disease (PD), Amyotrophic lateral sclerosis (ALS), Huntington’s disease (HD), and Alzheimer’s diseases (AD). For instance, PD is the most common neurodegenerative movement disorder, which affects approximately 1% of individuals above 60 years of age, and is characterized by cumulative neuronal loss (de Lau and Breteler, 2006). Due to the increase in the aging population the prevalence of PD is very likely to double within the next two decades (Dorsey et al., 2007). The preferential dopaminergic neuronal cell death in the substantia nigra results in reduced dopamine levels in the striatum, which causes the motor symptoms associated with PD. At clinical diagnosis this region of the brain has irreversibly lost 50–70% of its neurons compared to unaffected individuals (Davie, 2008).
Free radicals, like reactive oxygen species (ROS), play a crucial role in neurodegeneration and brain injury by exacerbating neuronal cell membrane damage via peroxidation of unsaturated fatty acids. Since the substantia nigra of PD patients exhibit higher levels of oxidized lipids (Bosco et al., 2006), proteins and DNA (Nakabeppu et al., 2007), and lower levels of reduced glutathione (GSH) (Zeevalk et al., n.d.), oxidative stress and mitochondrial dysfunction are known to be potential integrated causes of neurodegeneration in PD (Dawson et al., 2010).
Although neuroprotection and recanalization are main neurodegenerative disease treatment targets, the only current symptomatic approach for treating PD is based on a dopamine replacement strategy, using the dopamine precursor L-3,4-dihydroxyphenylalanine (L-Dopa), which at best only alleviates the motor symptoms. It is known that L-Dopa can also damage dopaminergic (DA) neurons (Blessing et al., 2003; Mena et al., 2009) as chronic treatment is associated with the development of complications such as dyskinesia, motor fluctuation, and dementia (Liu et al., 2014).
Recently a number of studies have focused on developing alternative neuroprotective therapeutic approaches with the ultimate aim of potentially slowing down neurodegeneration. In several countries, including Japan and China, Edaravone (3-methyl-1-phenyl-2-pyrazolin-5-one) has been widely used during the last decade for neurological recovery following acute brain ischemia and subsequent cerebral infarction (Lapchak, 2010). This neuroprotective drug acts as a potent antioxidant protecting against oxidative stress and neuronal apoptosis (Kikuchi et al., 2011: Yoshida et al., 2006) and its beneficial effects has been studied in several clinical trials (Kikuchi et al., 2014). Indeed it has been shown that oxidative cell damage in rat brain is reduced by repeated Edaravone treatment (Yamamoto et al., 2009). Moreover, other studies on HT22 cells demonstrated that Edaravone protects against H2O2-induced injury by inhibiting the production of ROS and activating the MAPK signaling pathway (Zhao et al., 2013). Edaravone has lipophilic groups, exhibiting good cell membrane permeability and the ability to cross the blood–brain barrier. Recent studies show that Edaravone protects brain nerve cells by reducing ROS, inhibiting apoptosis, blocking non-enzymatic peroxidation and lipoxygenase activity, and preventing vascular endothelial cell injury (Shinohara and Inoue, 2013). These findings suggest that Edaravone may have value in the treatment of neurodegenerative disorders such as PD.
In our previous study we performed a comparative two-dimensional gel electrophoresis (2DE)-based proteomic analysis to gain further insight into the effects of L-DOPA in response to H2O2-mediated oxidative stress in SH-SY5Y neuronal cells. We showed that exposure to L-DOPA may aid hypoxia condition in cells and therefore induction of ORP150 (hypoxia up-regulated protein 1) with its concomitant cytoprotective effects (Jami et al., 2014b). Here we have performed a similar study to analyze the cytoprotective effects of Edaravone. We have shown that similar to L-DOPA, Edaravone can reverse the cytotoxic effects of H2O2 but through a different mechanism.
2. Material and Methods
2.1 Reagents and cell culture
Fetal bovine serum (FBS), phosphate buffered saline (PBS), Dulbecco’s minimum essential medium plus F12 (DMEM/F12), penicillin and streptomycin were purchased from Invitrogen (Gaithersburg, MD). Acrylamide/ bis-acrylamide, tris base, glycine, ammonium persulfate, PVDF membrane, TEMED, DTT, SDS, urea, thiourea, glycerol, ammonium bicarbonate, DMSO, ECL reagent, bromoplenol blue were purchased from Fisher Scientific (Pittsburgh, PA). Trypsin and trypan blue were obtained from Sigma-Aldrich (St. Louis, MO).
SH-SY5Y cells were cultured in DMEM/F12 containing 10% FBS, 100 U/ml penicillin and 100 mg/ml streptomycin at 37°C in an atmosphere containing 5% CO2. In order to perform treatments the media were supplemented with 2 mM H2O2 (Sigma-Aldrich, MO), 25 μM Edaravone (Abcam, MA) or a combination of H2O2/Edaravone for 8 h (Kaste et al., 2013).
2.2. Lactate dehydrogenase (LDH) Cytotoxicity assay
Cellular damage following each treatment condition was assessed by measurement of LDH released into the medium. At the end (8h) of each treatment, 50 μl of medium was collected and transferred to a 96-well plate. The measurement of LDH reaction level was performed using the Pierce LDH Cytotoxicity Assay Kit (Thermo Scientific, USA) following the manufacturer’s instructions. The experiment was performed in three biological and two technical replicates. The average LDH release values obtained from the control condition was set to 100% and the relative LDH release levels for each treatment condition was compared to the control condition (p < 0.05).
2.3. Apoptosis
The rate of apoptosis in each treatment condition was measured using the TUNEL assay (Dead End Fluorimetric Kit, Promega, USA) following the manufacturer’s instruction. The TUNEL positive cells (FITC) were counted in the growth plate zones as a percentage of all cells (FITC and DAPI) in each zone. The average percentage of TUNEL positive cells in each condition was obtained from 10 different zones under fluorescent microscope and data were analyzed using One Way ANOVA statistical test.
2.4. Protein sample preparation and 2-DE gel electrophoresis
Protein sample preparation and 2-DE gel electrophoresis were performed as described before (Jami et al., 2014b). Triplicate batches of SH-SY5Y cells were seeded at 20–30% confluence and harvested when cell density reached 90%. A total amount of 850 μg of soluble proteins in the sample buffer was loaded onto 24-cm IPG strips (GE Healthcare), with non-linear (NL) pH 3-11 gradient. Focusing of proteins, equilibration and the 12.5% SDS-PAGE for the second dimension were performed as previously described (Jami et al., 2010b). Briefly, Proteins were focused at 20 °C according to the following program: 1 h, 0 V and 12 h, 30 V (rehydration); 30-min gradient to 10,000 V; up to 9 h, 10,000 V until 85 kV-h. Gels were stained with Colloidal Coomassie (CC) following the “blue silver” staining method (Jami et al., 2010a), (Candiano et al., 2004).
2.5. Analysis of Differential Protein Abundance
Two-dimensional images were captured by scanning, digitalizing and analysing stained gels as described previously (García-Estrada et al., 2013). Briefly, images were captured using an Image Scanner II (GE Healthcare) previously calibrated by using a gray scale marker (Eastman Kodak Co.), digitalized with Labscan 5.00 (v1.0.8) software (GE Healthcare), and analyzed with the Image Master TM 2D Platinum V7.0 software (GE Healthcare). Three gels for each condition, obtained from three independent cultures (biological replicates), were analysed to guarantee representative results. Differentially expressed proteins between two conditions were considered when the ratio of the relative volume average for one specific spot (present in the three biological replicates) was higher than 1.5 and the p-value was <0.05. False discovery rate (FDR) correction for multiple hypothesis testing was performed according to Storey’s method (q-value <0.05) (Storey, 2002).
2.6. Protein Identification by Mass Spectrometry
The protein spots of interest were manually excised from the gel and washed three times with ammonium bicarbonate/acetonitrile 1:1 (vol/vol) solution. Polyacrylamide fragments were dehydrated in acetonitrile and dried with a vacuum concentrator. Protein reduction and alkylation were performed by reswelling polyacrylamide fragments in 25 mM ammonium carbonate containing 0.15 mg/ml DTT at 56°C. This solution was replaced by 25 mM ammonium bicarbonate containing 10 mg/ml 2-iodoacetamide (Bio-Rad) for 45 min at room temperature and in the dark. Fragments were dried as described above and tryptic cleavage was initiated by reswelling the gel in 25 mM ammonium bicarbonate containing 0.2 μg trypsin per gel slice (Promega, WI, USA) for 20 min on ice. The solution was then replaced by 25 mM ammonium carbonate and digestion carried out overnight at 37°C. Tryptic peptides were extracted at 37°C for 15 min with a 50% (vol/vol) acetonitrile and 10% (vol/vol) acetic acid solution. A second extraction was performed under the same conditions. A third extraction was performed at room temperature for 15 min with 100% acetonitrile. The pooled solution was dried under vacuum and resuspended in 10 μl 0.1% (vol/vol) formic acid as descriebed previously (Zhang et al., 2011) with some modifications.
Five microliters of each sample were loaded onto a 75 μm × 15 cm column self-packed with 3 μmReproSil-Pur C18-AQ beads (Dr. Maisch GmbH, Germany). Peptides were eluted with a gradient of 3–30% acetonitrile in 0.1% formic acid over 60 min at a flow rate of 250 nL/min at 45 °C using a Thermo Scientific EASY-nLC 1000 coupled to a Q Exactive mass spectrometer (Thermo Fisher Scientific). The Q Exactive was operated in data-dependent analysis mode with survey scans acquired at a resolution of 70 000. Up to the top 10 most abundant precursors from the survey scan were selected with an isolation window of 1.6 Thompsons and fragmented by higher-energy collisional dissociation with normalized collision energies of 27. The maximum ion injection times for the survey scan and the MS/MS scans were 20 ms and 60 ms, respectively, and the ion target value for the survey scan and the MS/MS scans was set to 3 000 000 and 1 000 000, respectively. The raw files were processed using the MaxQuant (Cox and Mann, 2008) computational proteomics platform (version 1.2.7.0) for peptide identification and quantitation. The fragmentation spectra were searched against the UniProt human protein database (downloaded June 27, 2014) containing 87 938 protein sequences and allowing up to two missed tryptic cleavages. Carbamidomethylation of cysteine was set as a fixed modification and oxidation of methionine and protein N-terminal acetylation were used as variable modifications for database searching. The precursor and fragment mass tolerances were set to 7 and 20 ppm, respectively. Both peptide and protein identifications were filtered at 1% false discovery rate (FDR) and thus were not dependent on the peptide score.
3. Results and discussion
3.1. Treatment conditions
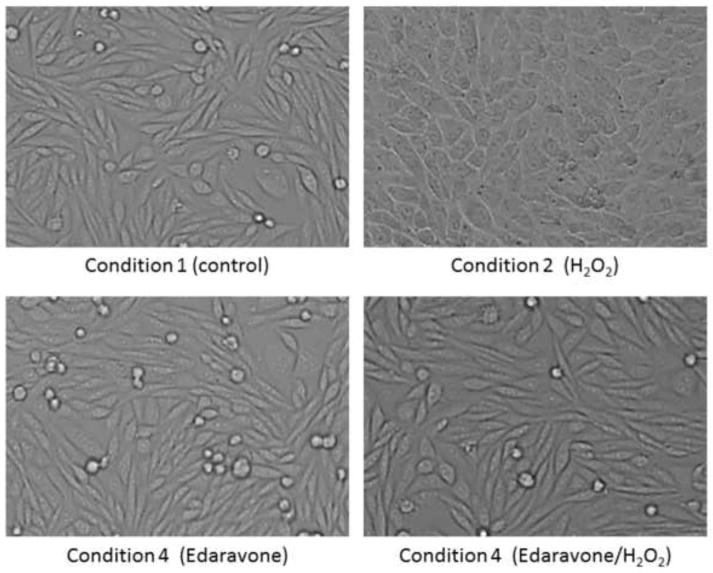
To gain insight into the mechanism by which Edaravone influences the toxic effects of H2O2 in neuronal cells we performed three simultaneous treatments (each one in three biological replicates) on dopaminergic SH-SY5Y neuroblastoma cells and analyzed their proteomic profiles under four conditions: Condition1 (control condition) contained cells grown in standard culture medium without treatment, condition 2 contained cells exposed to a toxic level (2 mM) of H2O2, condition 3 contained cells supplemented with 25 μM Edaravone, and condition 4 contained cells exposed to 25 μM Edaravone and 2 mM H2O2 simultaneously. Several studies have demonstrated that Edaravone concentrations, ranging from 1–100 μM, has no effect on cell viability (Lee et al., 2010) and because of this we chose a concentration of 25 μM for our experiments as this concentration resulted in a reversal of H2O2-induced cytotoxicity (Figure 1).
Figure 1. Morphology of SH-SY5Y cells during treatments.
SH-SY5Y cells have normal morphology in both condition 1 (without treatment) and condition 3 (treatment with 25 μM Edaravone for 8 hours). Cellular morphology is altered in response to treatment 2 (2 mM H2O2 for 8 hours). Normal cellular morphology is maintained in condition 4 (2 mM H2O2 and 25 μM Edaravone for 8 hours).
3.2. Oxidative conditions modify cell morphology, cell integrity, and apoptosis rate
As shown in Figure 1 the cell viability and morphology was clearly altered in response to toxic level of H2O2 in condition 2, however co-treatment of the culture media with Edaravone and H2O2 (in condition 4) prevented the viability and morphological changes (Figure 1).
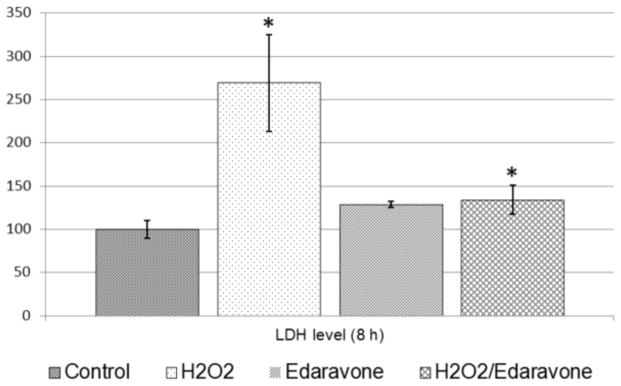
Similar results were observed by the Lactate dehydrogenase (LDH) Cytotoxicity assay that showed a significant increase (p < 0.05) in LDH levels released in the culture media in condition 2 (2.69 fold) and condition 4 (1.34 fold) compared to the control condition (Figure 2). This finding demonstrated a cytotoxic role of H2O2 and a cytoprotective role for Edaravone as it decreased the cellular damage rate from 2.69 fold to 1.34.
Figure 2. Lactate dehydrogenase (LDH) Cytotoxicity assay.
Cellular damage following each treatment condition was assessed by measurement of LDH released into the medium solution. The average LDH release values obtained from the control condition was set to 100% and the relative LDH release levels for each treatment condition was compared to the control condition. LDH levels released in the culture media significantly increased (p < 0.05) in condition 2 (2.69 fold) and condition 4 (1.34 fold).
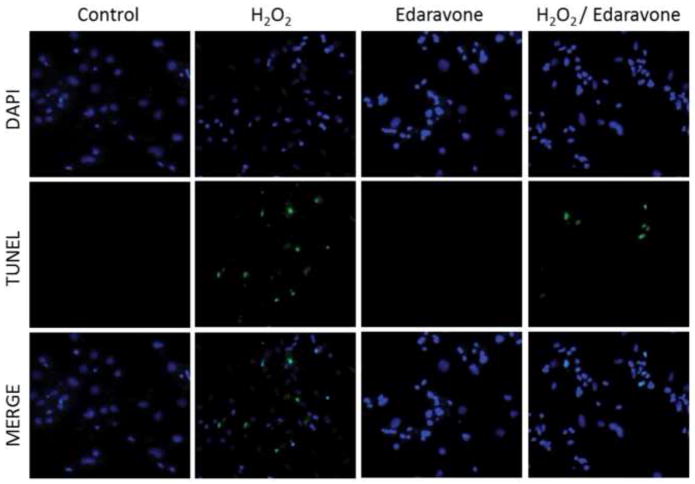
In addition, TUNEL assays were performed to estimate the impact of different treatment condition on cell viability and apoptotic characteristics. These experiments showed averages of 0.84%, 19.38%, 1.32% and 4.83% of apoptotic cells in conditions 1, 2, 3 and 4 respectively (Figure 3). The results of these experiment indicate significantly increased apoptosis in the presence of H2O2 (conditions 2 and 4) and further that Edaravone causes a significant decrease in apoptosis from 19.38% in condition 2 to 4.83% in condition 4.
Figure 3. Evaluation of apoptosis using the TUNEL assay.
The different images illustrate representative zones from each condition stained by DAPI, TUNEL. The merged images are also shown. The average percentage of TUNEL positive cells from 10 different zones in each treatment condition was 0.84%, 19.38%, 1.32% and 4.83% in conditions 1, 2, 3 and 4 respectively.
3.3. Oxidative conditions affect the proteomic profile
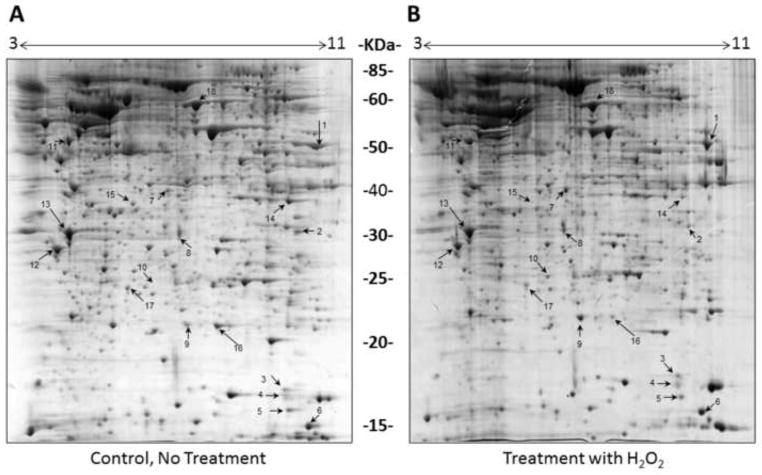
The first set of proteomic analyses (condition 1 vs. condition 2) focused on the effects of H2O2, wherein we compared the proteome profile of SH-SY5Y cells under control condition (Figure 4A) to cells treated with H2O2 (Figure 4B). In the second set (condition1 vs. condition 3) we analyzed the effects of Edaravone exposure on SH-SY5Y cells (Figure 5A) and in the third set (condition1 vs. condition 4) we analyzed the effects of simultaneous exposure to H2O2 and Edaravone (Figure 5B). We evaluated the effects of H2O2 treatment on the cells in three conditions (conditions 1, 2 and 4, Figures 4A, 4B and 5B) and observed an up-regulation of 10 proteins (spots1–6 and 10–13) and down-regulation of 6 proteins (7–9 and 14–16) compared to control condition (Table 1), the same result as we observed in our previous study (Jami et al., 2014b).
Figure 4. Comparison of the proteomes of SH-SY5Y cells with or without H2O2.
Representative 2-DE gels (n=3 for each treatment) of the proteomes of SH-SY5Y cells grown for 8 hours in the absence (A) or presence (B) of 2 mM H2O2. Protein spots showing differential abundance between the two conditions are numbered and correspond to the proteins listed in Table 1.
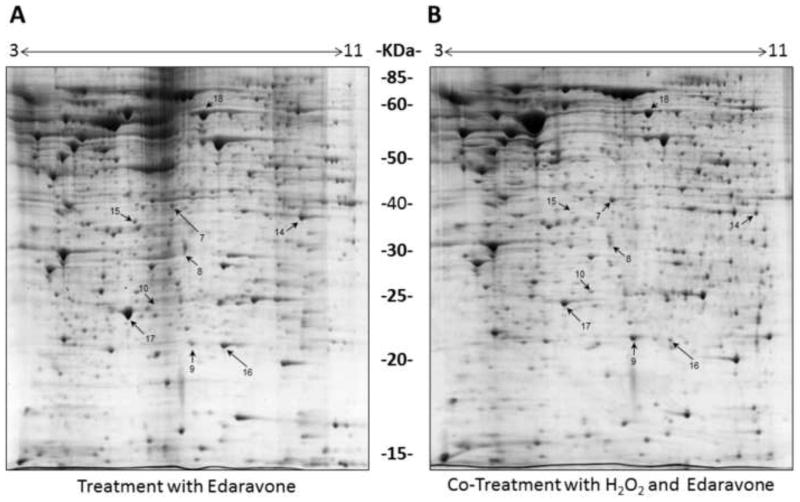
Figure 5. Proteomes of SH-SY5Y cells exposed to Edaravone with or without H2O2.
Representative 2-DE gels (n=3 for each treatment) of the proteomes of SH-SY5Y cells grown in media containing 25 μM Edaravone for 8 hours in the absence (A) or presence (B) of 2 mM H2O2. Protein spots showing differential abundance between the two conditions are numbered and correspond to the proteins listed in Table 1.
Table 1. Detailed information on altered spots during treatment conditions.
Differentially expressed proteins (fold-change higher than 1.5 and p-value <0.05) were classified according to their gene ontology (GO). Several biological processes were affected.
| Spot ID | Protein ID | Description | Mass | PI | Score | Percent protein coverage | AVG Fold change | p-value | q-value | GO (Biological Process) |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | P07954 | Fumarate hydratase, mitochondrial | 54714 | 8.85 | 408 | 27.2 | 7.49 | 3.30E-03 | 1.42E-04 | GO:0009056~catabolic process, GO:0042592~homeostatic process |
| 2 | Q16836 | Hydroxyacyl-coenzyme A dehydrogenase | 34613 | 8.76 | 118 | 21.5 | 5.1 | 4.52E-03 | 1.48E-03 | GO:0055114~oxidation reduction |
| 3 | Q9Y281 | Cofilin-2 | 18764 | 7.66 | 113 | 33.1 | 7.1 | 8.17E-04 | 1.63E-03 | GO:0030042~actin filament depolymerization |
| 4 | P60981 | Destrin | 18852 | 8.14 | 147 | 26.1 | 4.44 | 1.33E-04 | 1.71E-03 | GO:0006928~cell motion, GO:0043243~positive regulation of protein complex disassembly |
| 5 | P23528 | Cofilin-1 | 18776 | 8.22 | 192 | 50.6 | 4.3 | 2.87E-04 | 1.86E-03 | GO:0006928~cell motion, GO:0043243~positive regulation of protein complex disassembly |
| 6 | O00746 | Nucleoside diphosphate kinase | 17469 | 7.77 | 1024 | 41.4 | 2.7 | 3.02E-03 | 2.03E-03 | GO:0009116~nucleoside metabolic process |
| 7 | Q9BYZ2 | L-lactate dehydrogenase A-like 6B | 36817 | 7.62 | 244 | 29.6 | 4.15 | 1.71E-02 | 3.82E-03 | GO:0055114~oxidation reduction, GO:0006096~glycolysis |
| 8 | P04083 | Annexin A1 | 38816 | 6.57 | 142 | 6.62 | 2.07 | 3.78E-03 | 4.26E-03 | GO:00070301~cellular response to hydrogen proxide, GO:0006954~inflammatory response |
| 9 | Q99497 | Protein DJ-1 | 20064 | 6.33 | 501 | 21.2 | 8.52 | 7.77E-04 | 4.15E-03 | GO:0006979~response to oxidative stress, GO:0042592~homeostatic process |
| 10 | P30041 | Peroxiredoxin-6 | 25109 | 5.74 | 348 | 35.7 | 11.3 | 3.10E-04 | 4.95E-03 | GO:0009056~catabolic process, GO:0055114~oxidation reduction |
| 11 | P08670 | Vimentin | 53754 | 5.06 | 1536 | 62.2 | 0.22 | 2.93E-03 | 5.00E-03 | GO:0006928~cell motion |
| 12 | P67936 | Tropomyosin alpha-4 chain | 28620 | 4.65 | 138 | 23.8 | 0.16 | 1.74E-02 | 5.16E-03 | GO:0006928~cell motion |
| 13 | P09493 | Tropomyosin alpha-1 chain | 32746 | 4.69 | 405 | 33.1 | 0.12 | 3.11E-03 | 5.65E-03 | GO:0006979~response to oxidative stress, GO:0006928~cell motion, GO:0046907~intracellular transport |
| 14 | O14556 | Glyceraldehyde-3-phosphate dehydrogenase, | 35953 | 8.49 | 1165 | 48.6 | 0.17 | 6.76E-03 | 7.39E-03 | GO:0055114~oxidation reduction, GO:0006096~glycolysis, GO:0006928~cell motion |
| 15 | Q9UBR2 | Cathepsin Z | 34658 | 6.13 | 308 | 15.7 | 0.15 | 8.00E-03 | 7.92E-03 | GO:0006508~proteolysis |
| 16 | Q08752 | Peptidyl-prolyl cis-trans isomerase D | 20436 | 6.77 | 139 | 8.88 | 0.12 | 3.38E-04 | 8.38E-03 | GO:0043065~positive regulation of apoptotic process |
| 17 | P32119 | Peroxiredoxin-2 | 22049 | 5.66 | 140 | 24 | 12.69 | 9.64E-03 | 8.96E-03 | GO:0032943~mononuclear cell proliferation, GO:0006979~response to oxidative stress, GO:0042592~homeostatic process |
| 18 | P30101 | Protein disulfide-isomerase A3 | 57099 | 6.88 | 1573 | 52.7 | 0.28 | 3.38E-03 | 9.43E-03 | GO:0042592~homeostatic process, GO:0046907~intracellular transport |
When comparing the proteome profiles of cells exposed to Edaravone (either with or without H2O2) to controls, we observed up-regulation of spot 17 (in condition 3) and down-regulation of spot 18 in condition 4. Detailed information on each protein change are summarized in Table 1 where we classify the differentially expressed proteins according to their gene ontology (GO, with focus on biological process) through Uniprot (protein knowledge website: http://www.uniprot.org/) and QuickGO (http://www.ebi.ac.uk/QuickGO/). Upon treatments, several biological processes were affected. For example the majority of protein changes in response to oxidative stress (conditions 2) belong to the “cell motion (GO:0006928)” category of proteins, most of which were prevented by Edaravone co-treatment (condition 4).
3.4. Oxidative stress alters proteins involved in the metabolic routes to generate higher levels of NADPH
Glutathione is one of the main cellular antioxidants that prevents damage caused by ROS (Pompella et al., 2003). During the conversion of reactive H2O2 into H2O the reduced form of glutathione is oxidized by glutathione peroxidase. Then glutathione reductase and NADPH are required to reduce the oxidized glutathione back to its reduced form. Similar to our findings in our previous study, neurons seem to respond to the oxidative stress by generation of higher levels of NADPH (Jami et al., 2014b). One major observation in our work is the down regulation of glyceraldehyde 3-phosphate dehydrogenase (GAPDH). This alteration can result in a metabolic flux change from glycolysis to the pentose phosphate pathway (PPP) (Ralser et al., 2007: Jami et al., 2014a). PPP allows cells to produce higher levels of NADPH to overcome the oxidative conditions and prevent damage caused by ROS (Pompella et al., 2003).
We also observed that lactate dehydrogenase (LDH), involved in oxidation-reduction is up-regulated, a finding that is in concordance with the results of our LDH cytotoxicity assay. As a cytosolic enzyme, LDH catalyzes the conversion of pyruvate to lactate in the absence of oxygen, however it can also perform the reverse reaction in oxidative conditions and generate NADH (Markert, 1984).
We detected a marked up-regulation of mitochondrial nucleoside-diphosphate kinase and fumarate hydratase, both involved in the tricarboxylic acid (TCA) cycle that generates NADH (convertible to NADPH via nicotinomide nucleotide transhydrogenase (NNT)). Our analysis also showed up-regulation of 3-Hydroxyacyl CoA dehydrogenase, an oxidoreductase enzyme involved in beta-oxidation of fatty acids, catalyzing the oxidation of L-3-hydroxyacyl CoA by NAD+ thus generating NADH in response to oxidative stress (Table 1).
3.5. Cell survival mechanisms are activated in response to oxidative conditions
In all of the oxidative stress conditions tested we observed down-regulation of Cathepsin X and Cyclophilin D (both involved in cell death) and up-regulation of three stress sensor proteins (Table 1): Annexin A1, Peroxiredoxin-6 and PARK7/DJ-1. All of these three proteins are involved in cell survival; Annexin A1 can directly improve cell survival by limiting excessive levels of ROS during oxidative stress (Gorecka et al., 2005), Peroxiredoxin-6 can reduce H2O2, short chain fatty acids and phospholipid hydroperoxides (Fisher, 2011) and PARK7/DJ-1 is involved in protecting cells against oxidative stress and cell death (Ariga et al., 2013).
3.6. Oxidative conditions affect proteins responsible for cell integrity
We observed cell morphology modification (Figure 1) and significant cell damage (Figure 2) in condition 2 containing toxic level of H2O2 mostly in the absence of Edaravone. Interestingly, the alteration of several structural proteins indicates a rearrangement of the cytoskeleton due to oxidative stress. For instance, while all three members of the ADF/cofilin family, including Cofilin-1 and Cofilin-2 (which control actin polymerization/depolymerization in a pH-sensitive manner (Bravo-Cordero et al., 2013)) and Destrin (an actin depolymerizing factor) are up-regulated, other actin binding proteins, such as Tropomyosin alpha-4 chain and Tropomyosin alpha-1 chain are down-regulated (Table 1). These proteins bind to actin filaments in muscle and non-muscle cells and play a central role in muscle contraction and in stabilizing cytoskeleton actin filaments in non-muscle cells (Lin et al., 2008).
Down-regulation of Vimentin, that is responsible for maintaining cell shape, cytoplasmic integrity, and stabilization of cytoskeletal interactions, is also interesting (Table 1). This protein is known to play a pivotal role in anchoring organelles in the cytosol (Goldman et al., 1996; Katsumoto et al., 1990). It is possible therefore that the observed neuronal morphological changes in response to H2O2, are due to the attenuation of Vimentin.
3.7. Cytotoxic effects of oxidative stress is reversed by Edaravone
Our proteomic analyses show that the alteration of three mitochondrial enzymes under oxidative stress condition (fumarate hydratase, nucleoside-diphosphate kinase and 3-Hydroxyacyl CoA dehydrogenase) is maintained at baseline in response to Edaravone co-treatments (Table 1). Furthermore, we observed the ability of Edaravone in preventing the toxic effects of H2O2 on cellular integrity (Figures 1 and 2). This is in concert with our proteomic results indicating that Edaravone co-treatments maintained the expression levels of spots 3, 4, 5, 7, 8 and 9 (the H2O2-induced protein alterations affecting the cytoskeleton) at baseline (Figures 4 and 5, Table 1) and is also similar to the results of our previous study where we observed cytoprotective effects of L-Dopa against oxidative stress (Jami et al., 2014b).
3.8. Edaravone protects against oxidative stress through Peroxiredoxin-2
Edaravone treatment resulted in a large increase in the levels of Peroxiredoxin-2 (PRX2, spot 17 with an average of 12.69 fold overexpression) (Table 1). Interestingly, the largest increase in PRX2 is observed in response to Edaravone treatment (conditions 3) whilst the PRX2 induction (5.88 fold), although higher than control conditions, decreases in response to simultaneous Edaravone and H2O2 treatment (condition 4) (Figure 5). This finding suggests that Edaravone scavenges H2O2 via induction of PRX2 and that PRX2 may be degraded during oxidative stress condition.
Peroxiredoxins (PRXs) represent one of the ROS management systems that maintain the intracellular reducing environment via redox reactions at certain cysteine residues. Among them, PRX2 levels are known to be significantly elevated in the substantia nigra of PD patients (Basso et al., 2004). Since PRX2 is the most abundant PRX enzyme in mammalian neurons (Jin et al., 2005), PRX2 is considered a vital PRX in DA neuronal survival protecting cells against oxidative stress (Stresing et al., 2013). It has been documented that PRX2 is inactivated by hyperoxidation during the elimination of peroxides (Peskin et al., 2013).
PRX2 is also known to protect cells against oxidative dopaminergic neurodegeneration via attenuation of the apoptosis signal-regulating kinase (ASK1) signaling cascade; this protein inhibits ASK1 activity and the subsequent activation of JNK/c-Jun and p38 cell death pathways (Hu et al., 2011).
The catalytic breakdown of H2O2 by typical 2-Cys PRXs is conducted by oxidation of the peroxide-reactive peroxidatic cysteine followed by the formation of a disulfide with the resolving cysteine of a neighboring PRX molecule (Day et al., 2012). Thioredoxin then reduces this disulfide to restore the active peroxidase. In addition to its role in the catalytic cycle of PRX, thioredoxin is a cofactor for many other enzymes and its oxidoreductase activity is important for reduction of protein disulfides under oxidative stress conditions (Day et al., 2012). Interestingly, in our proteomic analyses we observed that the protein disulfide-isomerase A3 or PDIA3 protein is down-regulated (spot 18, 0.28 fold) upon co-treatment of cells with Edaravone and H2O2 (Table 1). The PDIA3 protein belongs to the protein disulfide isomerase family and catalyzes the rearrangement of -S-S- bonds in proteins (Bourdi et al., 1995). Our bioinformatics analysis using BioGraph online software (http://biograph.be/)(Liekens et al., 2011) revealed that this protein contains 2 thioredoxin domains and can therefore participate in redox regulation through the thioredoxin/peroxiredoxin system. Moreover, it has been reported that inhibition of PDIA3, as a pro-apoptotic protein that catalyzes isomerization, reduction, and oxidation of disulfides, can suppress apoptosis suggesting increased neuronal survival (Hoffstrom et al., 2010).
This knowledge combined with our experimental findings suggests that Edaravone protects neuronal cells against oxidative stress either by inhibition of apoptosis or more specifically via direct induction of PRX2. The exact mechanism of Edaravone-mediated neuroprotection warrants further investigation as Edaravone may not only be a potent neuroprotective agent following acute brain ischemia, but also as a component of future combinatorial PD therapeutic strategies. In our previous study we showed that L-Dopa reverses the H2O2-mediated effects at the cell morphology and viability level via indirect induction of the ORP150 protein. Since conversion of L-Dopa to norepinephrine requires molecular oxygen, the exposure to L-Dopa may aid hypoxia condition in cells and therefore induction of ORP150 with its concomitant cytoprotective effects (Jami et al., 2014b).
4. Conclusions
Free radicals are important targets for therapeutic intervention of neurodegenerative diseases as oxidative stress represents an important factor. As a free radical scavenger, Edaravone may offer a novel treatment option for neurodegeneration. Using a 2DE-based proteomic study we have shown that oxidative stress changes neuronal metabolic routes and affects cytoskeletal integrity in neuroblastoma cells. As observed with L-Dopa in our previous study we show here that Edaravone reverses H2O2-mediated effects at the protein level. However, the cytoprotective effects of these two compounds appear to influence different pathways. L-Dopa seems to aid hypoxia condition in cells and therefore induction of ORP150 with its concomitant cytoprotective effects whilst Edaravone protect neuronal cells against oxidative stress via direct induction of PRX2 and inhibition of apoptosis.
Highlights.
Oxidative stress alters proteins involved in neuronal metabolic routes and cytoskeletal integrity.
Edaravone reverses H2O2-mediated effects at the protein level.
Edaravone protects neuronal cells against oxidative stress via direct induction of Peroxiredoxin-2 and inhibition of apoptosis.
Acknowledgments
This work was supported by the Norwegian Research Council (S.G.M.), The Western Norway Regional Health Authority (S.G.M), The Norwegian Centre for Movement Disorders (J.P.L.), NIH NINDS grant P30 NS050276 (T.A.N.), NIH Shared Instrumentation Grant RR027990 (T.A.N.) and Shahrekord University of Medical Sciences grant 246574011394 (M.S.J). We thank Steven Blais for advice on mass spectrometry.
Footnotes
Conflict of interest
The authors declare that they have no conflict of interest.
Contributor Information
Mohammad-Saeid Jami, Email: sjamif@gmail.com.
Zahra Salehi-Najafabadi, Email: Zahra.salehi@live.com.
Fereshteh Ahmadinejad, Email: Fereshte.ahmadi86@ymail.com.
Esthelle Hoedt, Email: Esthelle.Hoedt@med.nyu.edu.
Morteza Hashemzadeh Chaleshtori, Email: mchalesh@yahoo.com.
Thomas A. Neubert, Email: Thomas.Neubert@med.nyu.edu.
Jan Petter Larsen, Email: jpl@sus.no.
Simon Geir Møller, Email: mollers@stjohns.edu.
References
- Ariga H, Takahashi-Niki K, Kato I, Maita H, Niki T, Iguchi-Ariga SMM. Neuroprotective function of DJ-1 in Parkinson’s disease. Oxidative medicine and cellular longevity. 2013;2013:683920. doi: 10.1155/2013/683920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basso M, Giraudo S, Corpillo D, Bergamasco B, Lopiano L, Fasano M. Proteome analysis of human substantia nigra in Parkinson’s disease. Proteomics. 2004;4:3943–52. doi: 10.1002/pmic.200400848. [DOI] [PubMed] [Google Scholar]
- Blessing H, Bareiss M, Zettlmeisl H, Schwarz J, Storch A. Catechol-O-methyltransferase inhibition protects against 3,4-dihydroxyphenylalanine (DOPA) toxicity in primary mesencephalic cultures: new insights into levodopa toxicity. Neurochemistry international. 2003;42:139–51. doi: 10.1016/s0197-0186(02)00075-x. [DOI] [PubMed] [Google Scholar]
- Bosco DA, Fowler DM, Zhang Q, Nieva J, Powers ET, Wentworth P, Lerner RA, Kelly JW. Elevated levels of oxidized cholesterol metabolites in Lewy body disease brains accelerate alpha-synuclein fibrilization. Nature chemical biology. 2006;2:249–53. doi: 10.1038/nchembio782. [DOI] [PubMed] [Google Scholar]
- Bourdi M, Demady D, Martin JL, Jabbour SK, Martin BM, George JW, Pohl LR. cDNA cloning and baculovirus expression of the human liver endoplasmic reticulum P58: characterization as a protein disulfide isomerase isoform, but not as a protease or a carnitine acyltransferase. Archives of biochemistry and biophysics. 1995;323:397–403. doi: 10.1006/abbi.1995.0060. [DOI] [PubMed] [Google Scholar]
- Bravo-Cordero JJ, Magalhaes MAO, Eddy RJ, Hodgson L, Condeelis J. Functions of cofilin in cell locomotion and invasion. Nature reviews. Molecular cell biology. 2013;14:405–15. doi: 10.1038/nrm3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candiano G, Bruschi M, Musante L, Santucci L, Ghiggeri GM, Carnemolla B, Orecchia P, Zardi L, Righetti PG. Blue silver: A very sensitive colloidal Coomassie G-250 staining for proteome analysis. Electrophoresis. 2004;25:1327–1333. doi: 10.1002/elps.200305844. [DOI] [PubMed] [Google Scholar]
- Davie CA. A review of Parkinson’s disease. British medical bulletin. 2008;86:109–27. doi: 10.1093/bmb/ldn013. [DOI] [PubMed] [Google Scholar]
- Dawson TM, Ko HS, Dawson VL. Genetic animal models of Parkinson’s disease. Neuron. 2010;66:646–61. doi: 10.1016/j.neuron.2010.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day AM, Brown JD, Taylor SR, Rand JD, Morgan BA, Veal EA. Inactivation of a peroxiredoxin by hydrogen peroxide is critical for thioredoxin-mediated repair of oxidized proteins and cell survival. Molecular cell. 2012;45:398–408. doi: 10.1016/j.molcel.2011.11.027. [DOI] [PubMed] [Google Scholar]
- De Lau LML, Breteler MMB. Epidemiology of Parkinson’s disease. Lancet neurology. 2006;5:525–35. doi: 10.1016/S1474-4422(06)70471-9. [DOI] [PubMed] [Google Scholar]
- Dorsey ER, Constantinescu R, Thompson JP, Biglan KM, Holloway RG, Kieburtz K, Marshall FJ, Ravina BM, Schifitto G, Siderowf A, Tanner CM. Projected number of people with Parkinson disease in the most populous nations, 2005 through 2030. Neurology. 2007;68:384–6. doi: 10.1212/01.wnl.0000247740.47667.03. [DOI] [PubMed] [Google Scholar]
- Effect of a novel free radical scavenger, edaravone (MCI-186), on acute brain infarction. Randomized, placebo-controlled, double-blind study at multicenters. Cerebrovascular diseases (Basel, Switzerland) 2003;15:222–9. doi: 10.1159/000069318. [DOI] [PubMed] [Google Scholar]
- Fisher AB. Peroxiredoxin 6: a bifunctional enzyme with glutathione peroxidase and phospholipase A2 activities. Antioxidants & redox signaling. 2011;15:831–44. doi: 10.1089/ars.2010.3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Estrada C, Barreiro C, Jami MS, Martín-González J, Martín JF. The inducers 1,3-diaminopropane and spermidine cause the reprogramming of metabolism in Penicillium chrysogenum, leading to multiple vesicles and penicillin overproduction. Journal of proteomics. 2013;85:129–59. doi: 10.1016/j.jprot.2013.04.028. [DOI] [PubMed] [Google Scholar]
- Goldman RD, Khuon S, Chou YH, Opal P, Steinert PM. The function of intermediate filaments in cell shape and cytoskeletal integrity. The Journal of cell biology. 1996;134:971–83. doi: 10.1083/jcb.134.4.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gołembiowska K, Dziubina A, Kowalska M, Kamińska K. Paradoxical effects of adenosine receptor ligands on hydroxyl radical generation by L-DOPA in the rat striatum. Pharmacological reports: PR. n.d;60:319–30. [PubMed] [Google Scholar]
- Gorecka KM, Konopka-Postupolska D, Hennig J, Buchet R, Pikula S. Peroxidase activity of annexin 1 from Arabidopsis thaliana. Biochemical and biophysical research communications. 2005;336:868–75. doi: 10.1016/j.bbrc.2005.08.181. [DOI] [PubMed] [Google Scholar]
- Hoffstrom BG, Kaplan A, Letso R, Schmid RS, Turmel GJ, Lo DC, Stockwell BR. Inhibitors of protein disulfide isomerase suppress apoptosis induced by misfolded proteins. Nature chemical biology. 2010;6:900–6. doi: 10.1038/nchembio.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Weng Z, Chu CT, Zhang L, Cao G, Gao Y, Signore A, Zhu J, Hastings T, Greenamyre JT, Chen J. Peroxiredoxin-2 protects against 6-hydroxydopamine-induced dopaminergic neurodegeneration via attenuation of the apoptosis signal-regulating kinase (ASK1) signaling cascade. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2011;31:247–61. doi: 10.1523/JNEUROSCI.4589-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itokawa K, Ohkuma A, Araki N, Tamura N, Shimazu K. Effect of L-DOPA on nitric oxide production in striatum of freely mobile mice. Neuroscience letters. 2006;402:142–4. doi: 10.1016/j.neulet.2006.03.064. [DOI] [PubMed] [Google Scholar]
- Jami MS, Barreiro C, García-Estrada C, Martín JF. Proteome analysis of the penicillin producer Penicillium chrysogenum: characterization of protein changes during the industrial strain improvement. Molecular cellular proteomics MCP. 2010a;9:1182–1198. doi: 10.1074/mcp.M900327-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jami MS, García-Estrada C, Barreiro C, Cuadrado AA, Salehi-Najafabadi Z, Martín JF. The Penicillium chrysogenum extracellular proteome. Conversion from a food-rotting strain to a versatile cell factory for white biotechnology. Molecular cellular proteomics MCP. 2010b;9:2729–2744. doi: 10.1074/mcp.M110.001412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jami MS, Hou J, Liu M, Varney ML, Hassan H, Dong J, Geng L, Wang J, Yu F, Huang X, Peng H, Fu K, Li Y, Singh RK, Ding SJ. Functional proteomic analysis reveals the involvement of KIAA1199 in breast cancer growth, motility and invasiveness. BMC cancer. 2014a;14:194. doi: 10.1186/1471-2407-14-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jami MS, Pal R, Hoedt E, Neubert Ta, Larsen JP, Møller SG. Proteome analysis reveals roles of L-DOPA in response to oxidative stress in neurons. BMC neuroscience. 2014b;15:93. doi: 10.1186/1471-2202-15-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin MH, Lee YH, Kim JM, Sun HN, Moon EY, Shong MH, Kim SU, Lee SH, Lee TH, Yu DY, Lee DS. Characterization of neural cell types expressing peroxiredoxins in mouse brain. Neuroscience letters. 2005;381:252–7. doi: 10.1016/j.neulet.2005.02.048. [DOI] [PubMed] [Google Scholar]
- Kaste M, Murayama S, Ford GA, Dippel DWJ, Walters MR, Tatlisumak T. Safety, tolerability and pharmacokinetics of MCI-186 in patients with acute ischemic stroke: new formulation and dosing regimen. Cerebrovascular diseases (Basel, Switzerland) 2013;36:196–204. doi: 10.1159/000353680. [DOI] [PubMed] [Google Scholar]
- Katsumoto T, Mitsushima A, Kurimura T. The role of the vimentin intermediate filaments in rat 3Y1 cells elucidated by immunoelectron microscopy and computer-graphic reconstruction. Biology of the cell/under the auspices of the European Cell Biology Organization. 1990;68:139–46. doi: 10.1016/0248-4900(90)90299-i. [DOI] [PubMed] [Google Scholar]
- Kikuchi K, Tanaka E, Murai Y, Tancharoen S. Clinical trials in acute ischemic stroke. CNS drugs. 2014;28:929–38. doi: 10.1007/s40263-014-0199-6. [DOI] [PubMed] [Google Scholar]
- Kiyoshi K, Kiyoshi K, Naoki M, Yoko M, Takashi I, Salunya T, Kei M, Chiemi K, Narumi I, Nobuyuki T, Hisaaki U, Naohisa M, Naoto S, Terukazu K, Teruto H, Ikuro M, Motohiro M, Koichi K. Beneficial Effects of the Free Radical Scavenger Edaravone (Radicut) in Neurologic Diseases. Journal of Neurology & Neurophysiology. 2011 doi:org/10.4172/2155-9562.S1-001. [Google Scholar]
- Lapchak PA. A critical assessment of edaravone acute ischemic stroke efficacy trials: is edaravone an effective neuroprotective therapy? Expert opinion on pharmacotherapy. 2010;11:1753–63. doi: 10.1517/14656566.2010.493558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BJ, Egi Y, van Leyen K, Lo EH, Arai K. Edaravone, a free radical scavenger, protects components of the neurovascular unit against oxidative stress in vitro. Brain Research. 2010;1307:22–7. doi: 10.1016/j.brainres.2009.10.026. Epub 2009 Oct 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liekens AML, De Knijf J, Daelemans W, Goethals B, De Rijk P, Del-Favero J. BioGraph: unsupervised biomedical knowledge discovery via automated hypothesis generation. Genome biology. 2011;12:R57. doi: 10.1186/gb-2011-12-6-r57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JJC, Eppinga RD, Warren KS, McCrae KR. Human tropomyosin isoforms in the regulation of cytoskeleton functions. Advances in experimental medicine and biology. 2008;644:201–22. doi: 10.1007/978-0-387-85766-4_16. [DOI] [PubMed] [Google Scholar]
- Liu X, Shao R, Li M, Yang G. Edaravone protects neurons in the rat substantia nigra against 6-hydroxydopamine-induced oxidative stress damage. Cell biochemistry and biophysics. 2014;70:1247–54. doi: 10.1007/s12013-014-0048-8. [DOI] [PubMed] [Google Scholar]
- Markert CL. Lactate dehydrogenase. Biochemistry and function of lactate dehydrogenase. Cell biochemistry and function. 1984;2:131–4. doi: 10.1002/cbf.290020302. [DOI] [PubMed] [Google Scholar]
- Mena MA, Casarejos MJ, Solano RM, de Yébenes JG. Half a century of L-DOPA. Current topics in medicinal chemistry. 2009;9:880–93. [PubMed] [Google Scholar]
- Nakabeppu Y, Tsuchimoto D, Yamaguchi H, Sakumi K. Oxidative damage in nucleic acids and Parkinson’s disease. Journal of neuroscience research. 2007;85:919–34. doi: 10.1002/jnr.21191. [DOI] [PubMed] [Google Scholar]
- Peskin AV, Dickerhof N, Poynton RA, Paton LN, Pace PE, Hampton MB, Winterbourn CC. Hyperoxidation of peroxiredoxins 2 and 3: Rate constants for the reactions of the sulfenic acid of the peroxidatic cysteine. The Journal of biological chemistry. 2013 doi: 10.1074/jbc.M113.460881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pompella A, Visvikis A, Paolicchi A, De Tata V, Casini AF. The changing faces of glutathione, a cellular protagonist. Biochemical pharmacology. 2003;66:1499–503. doi: 10.1016/s0006-2952(03)00504-5. [DOI] [PubMed] [Google Scholar]
- Ralser M, Wamelink MM, Kowald A, Gerisch B, Heeren G, Struys EA, Klipp E, Jakobs C, Breitenbach M, Lehrach H, Krobitsch S. Dynamic rerouting of the carbohydrate flux is key to counteracting oxidative stress. Journal of biology. 2007;6:10. doi: 10.1186/jbiol61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reksidler AB, Lima MMS, Dombrowski PA, Barnabé GF, Andersen ML, Tufik S, Vital MABF. Distinct effects of intranigral L-DOPA infusion in the MPTP rat model of Parkinson’s disease. Journal of neural transmission. Supplementum. 2009:259–68. doi: 10.1007/978-3-211-92660-4_21. [DOI] [PubMed] [Google Scholar]
- Shinohara Y, Inoue S. Cost-effectiveness analysis of the neuroprotective agent edaravone for noncardioembolic cerebral infarction. Journal of stroke and cerebrovascular diseases: the official journal of National Stroke Association. 2013;22:668–74. doi: 10.1016/j.jstrokecerebrovasdis.2012.04.002. [DOI] [PubMed] [Google Scholar]
- Storey JD. A direct approach to false discovery rates. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 2002;64:479–498. doi: 10.1111/1467-9868.00346. [DOI] [Google Scholar]
- Stresing V, Baltziskueta E, Rubio N, Blanco J, Arriba MC, Valls J, Janier M, Clézardin P, Sanz-Pamplona R, Nieva C, Marro M, Petrov D, Dmitri P, Sierra A. Peroxiredoxin 2 specifically regulates the oxidative and metabolic stress response of human metastatic breast cancer cells in lungs. Oncogene. 2013;32:724–35. doi: 10.1038/onc.2012.93. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Yanagisawa M, Tak NW, Watanabe K, Takahashi C, Fujisawa A, Kashiba M, Tanaka M. Repeated edaravone treatment reduces oxidative cell damage in rat brain induced by middle cerebral artery occlusion. Redox report: communications in free radical research. 2009;14:251–8. doi: 10.1179/135100009X12525712409779. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Yanai H, Namiki Y, Fukatsu-Sasaki K, Furutani N, Tada N. Neuroprotective effects of edaravone: a novel free radical scavenger in cerebrovascular injury. CNS drug reviews. 2006;12:9–20. doi: 10.1111/j.1527-3458.2006.00009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeevalk GD, Razmpour R, Bernard LP. Glutathione and Parkinson’s disease: is this the elephant in the room? Biomedicine & pharmacotherapy = Biomédecine & pharmacothérapie. n.d;62:236–49. doi: 10.1016/j.biopha.2008.01.017. [DOI] [PubMed] [Google Scholar]
- Zhao ZY, Luan P, Huang SX, Xiao SH, Zhao J, Zhang B, Gu BB, Pi RB, Liu J. Edaravone protects HT22 neurons from H2O2-induced apoptosis by inhibiting the MAPK signaling pathway. CNS neuroscience & therapeutics. 2013;19:163–9. doi: 10.1111/cns.12044. [DOI] [PMC free article] [PubMed] [Google Scholar]