Summary
Phenotypic heterogeneity is commonly observed between isolates of a given pathogen. Epidemiological analyses have identified that some serotypes of the group A Streptococcus (GAS) are non-randomly associated with particular disease manifestations. Here, we present evidence that a contributing factor to the association of serotype M3 GAS isolates with severe invasive infections is the presence of a null mutant allele for the orphan kinase RocA. Through use of RNAseq analysis, we identified that the natural rocA mutation present within M3 isolates leads to the enhanced expression of more than a dozen immunomodulatory virulence factors, enhancing phenotypes such as hemolysis and NAD+ hydrolysis. Consequently, an M3 GAS isolate survived human phagocytic killing at a level 13-fold higher than a rocA complemented derivative, and was significantly more virulent in a murine bacteremia model of infection. Finally, we identified that RocA functions through the CovR/S two-component system as levels of phosphorylated CovR increase in the presence of functional RocA, and RocA has no regulatory activity following covR or covS mutation. Our data are consistent with RocA interfacing with the CovR/S two-component system, and that the absence of this activity in M3 GAS potentiates the severity of invasive infections caused by isolates of this serotype.
Keywords: Phenotypic variation, regulation, virulence factors, cross-talk, Streptococcus pyogenes
Introduction
Bacterial pathogens commonly exhibit variation in virulence factor expression and disease potential between isolates (Willems et al., 2011, Fierer & Guiney, 2001, Carroll et al., 2011, Heithoff et al., 2012, Suerbaum & Josenhans, 2007). For example, Staphylococcus aureus isolates from the USA300 lineage have emerged as the predominant cause of community-acquired cutaneous infections in the United States (Uhlemann et al., 2014), and these isolates show enhanced virulence factor expression and virulence relative to those from the USA200 lineage (Cheung et al., 2011, Uhlemann et al., 2014, Li et al., 2009). Despite pathogen variation being a potential public health concern, due to the possibility of treatment failures and escape from preventative regimes (Croucher et al., 2011, Nicol & Wilkinson, 2008, Aracil et al., 2006), in most cases there is a dearth of information regarding the molecular mechanisms that drive this variation. Possible mechanisms behind pathogen variation include gene acquisition through horizontal gene transfer, gene loss through genome reduction, or the modulation of regulatory systems that lead to altered patterns of expression.
The group A Streptococcus (GAS, Streptococcus pyogenes) is a human-specific, Gram-positive pathogen that causes diseases ranging from the mild (e.g. GAS pharyngitis, a.k.a. strep throat) to the severe (e.g. necrotizing fasciitis, a.k.a. the flesh-eating infection) (Cunningham, 2000). GAS strains are divided into serotypes based upon the sequence of the 5′ end of the emm (M protein-encoding) gene (Steer et al., 2009). Some GAS serotypes show non-random associations with particular disease manifestations. For example, serotype M3 GAS strains are associated with unusually severe invasive infections and a high mortality rate (Beres et al., 2006), serotype M18 strains are associated with outbreaks of acute rheumatic fever (Smoot et al., 2002), and M28 strains are associated with cases of puerperal sepsis (Green et al., 2005). Despite more than five decades passing since these associations were identified, the molecular basis for them remains to be fully elucidated (Mitchell, 1962, Wilmers et al., 1954). While GAS strains show variation in gene content, primarily the result of different integrated bacteriophage, no bacteriophage are unique to one serotype (Beres & Musser, 2007, Maripuu et al., 2008, Beres et al., 2006). As the core genomes of all GAS strains are highly conserved and harbor an extensive arsenal of common virulence factors, we hypothesize that it is the differential regulation of these common virulence factors, rather than the presence of serotype-specific genes, that is the crux of most GAS-serotype disease-phenotype associations.
We and others have described strain-specific variation in GAS regulatory networks (McIver, 2009, Perez et al., 2009, Sumby et al., 2006, Kreikemeyer et al., 2003). For example, the genes encoding the negative regulatory two-component system CovR/S (also known as CsrR/S) contain higher numbers of genetic alterations than the genome average, indicative of selective pressure against a functional regulatory system (Ikebe et al., 2010, Beres et al., 2010). Disruption of the CovR/S system is selected for during invasive infections as this protects GAS from the host immune response through the enhanced expression of multiple immunomodulatory virulence factors (Kappeler et al., 2009, Sumby et al., 2006, Hollands et al., 2010, Li et al., 2014). While covR/S mutation enhances GAS virulence during invasive infections, it is hypothesized that covR/S mutant strains do not persist in the population, explaining their relatively low level of isolation, due to a reduced ability to cause non-invasive (e.g. pharyngeal) infections which represent the vast majority of GAS infections. Consistent with this hypothesis are the findings that covR/S mutant strains are significantly outcompeted by wild-type strains during growth in human saliva (Trevino et al., 2009), and are attenuated in their ability to colonize the murine upper respiratory tract (Alam et al., 2013). Recently, we identified that CovS activates CovR through phosphorylation (Horstmann et al., 2015). While the site of phosphorylation was not identified, it is hypothesized that this occurs at aspartate-53 (Asp-53), a residue that is phosphorylated in vitro in the presence of the phospho-donor acetyl phosphate (Horstmann et al., 2015). Phosphorylation of Asp-53 promotes CovR dimerization and DNA-binding activities (Gusa et al., 2006). In addition to CovS, we have also identified that the eukaryotic-like serine/threonine protein kinase (Stk) also phosphorylates CovR, with threonine-65 (Thr-65) being the site of phosphorylation (Horstmann et al., 2014). Phosphorylation of CovR Thr-65 by Stk inhibits CovR Asp-53 phosphorylation and therefore CovR activity (Horstmann et al., 2014). Thus, CovS and Stk have dueling roles with regard to CovR activation.
Little is known with respect to serotype-specific variation in GAS regulatory networks, although recent studies have begun to shed light on this topic. Firstly, it was identified that serotype M18 GAS isolates have a mutant rocA gene which is responsible, through undetermined mechanisms, for the hyper-encapsulation of isolates of this serotype (Lynskey et al., 2013). As RocA has homology to sensor kinases, it is hypothesized that this protein regulates by altering the phosphorylation status of one or more target proteins (Biswas & Scott, 2003). Secondly, we identified that serotype M3 isolates, since at least the 1920’s, harbor mutations in the rivR and fasC regulatory genes, resulting in an increase in protein-G-related α2-macroglobulin binding protein (GRAB) expression and a decrease in streptokinase expression, relative to complemented derivatives (Cao et al., 2014). The increase in GRAB expression, a virulence factor that binds the human protease inhibitor α2-macroglobulin to regulate proteolysis at the GAS cell surface (Rasmussen et al., 1999), is a consequence of the inability of RivR protein to repress GRAB transcription (Trevino et al., 2013). The decrease in streptokinase expression, a virulence factor that promotes dissemination by promoting conversion of human plasminogen into the protease plasmin (McArthur et al., 2012, Syrovets et al., 2012), is a consequence of the fasC mutation drastically reducing the production of the small regulatory RNA (sRNA) FasX which positively regulates streptokinase expression through a post-transcriptional mechanism (Ramirez-Pena et al., 2010, Danger et al., 2015a). Thirdly, we and others recently identified that serotype M3 GAS isolates also harbor a mutant rocA allele and, similar to M18 GAS, that this is responsible for the greater capsule expression seen by M3 isolates (Lynskey et al., 2015, Miller et al., 2015).
Here, as part of our investigation into why serotype M3 GAS isolates are associated with unusually severe invasive infections, we identified that the natural mutant rocA allele of M3 GAS isolates not only enhances capsule expression but also the expression of more than a dozen other immunomodulatory virulence factors. Crucially, the up-regulation of these virulence factors leads to an enhanced ability of M3 isolates to survive in whole human blood, and also to cause disease in a murine model of bacteremia infection. Regarding a mechanism, we discovered that the rocA mutation results in a reduced level of active (phosphorylated) CovR protein. Thus, the inability of the M3 RocA protein to activate CovR leads to the enhanced expression of the virulence factors identified. We propose a model in which rewiring of regulatory systems, as the cumulative consequence of the rivR, fasC, and rocA mutations, is the driving force behind the hyper-virulence of M3 GAS isolates during invasive infections.
Results
A representative serotype M3 GAS isolate is hyper-virulent in a mouse model of soft tissue infection
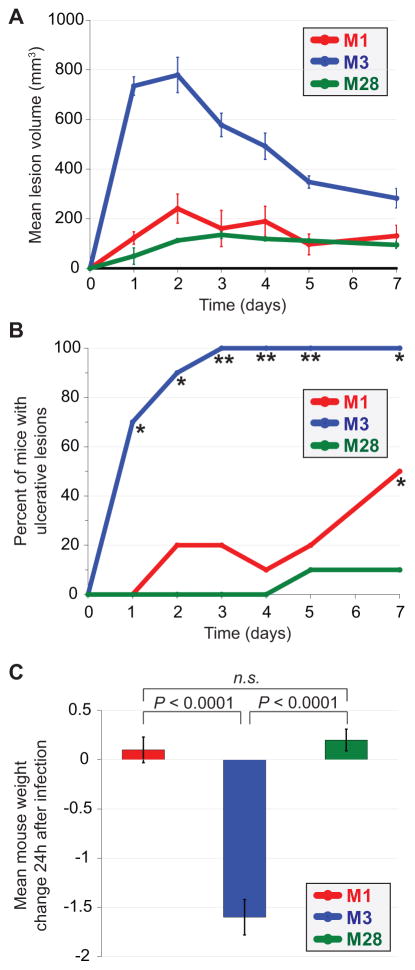
Serotype M3 GAS strains are associated with unusually severe invasive infections in humans and a high mortality rate (Beres et al., 2006). However, it has not been shown that the same is also observed in mouse models of invasive infection, important information given the common use of these animal models in GAS research. To test this we compared representative isolates from three different GAS serotypes, MGAS2221 (M1), MGAS10870 (M3), and MGAS6180 (M28), in a mouse model of soft tissue infection. The M3 isolate was significantly more virulent than the M1 and M28 isolates as exemplified by the formation of significantly larger skin lesions (Figure 1A), enhanced lesion ulceration (Figure 1B), and weight loss during the first 24h of infection (Figure 1C). The data are consistent with M3 GAS isolates showing a similar hyper-virulent phenotype during murine invasive infections as is observed during human invasive infections (Beres et al., 2006).
Figure 1. Enhanced virulence of a serotype M3 GAS isolate in a murine soft tissue infection model.
Clinical isolates representative of M1, M3, and M28 GAS serotypes were each used to infect mice via subcutaneous injection between the shoulder blades. (A) Mean volume of mouse skin lesions that formed at the site of GAS injection. Error bars represent ± the standard error of the mean. The lesion volume for the M3 GAS infected animals was statistically significantly different from the other infected animals (Repeated measures ANOVA; P < 0.0001). (B) Percent of infected mice in which the skin lesion ulcerated. The asterisks highlight data points that are statistically significantly different relative to the other infected animals (Fishers exact test; *P < 0.01, **P < 0.001). (C) Mean animal weight change between days 0 and 1. Error bars represent ± the standard error of the mean. The weight loss observed for the M3 GAS-infected animals was statistically significantly greater than that of the other groups of infected animals (T-test, P < 0.0001).
Serotype M3 GAS isolates since at least the 1920s harbor a mutant rocA gene
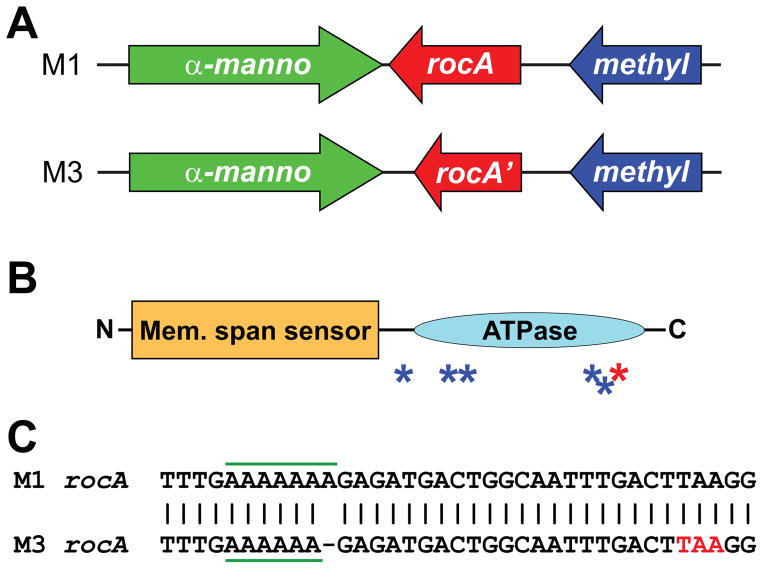
Recently, we and others published notes reporting that serotype M3 isolates harbor a mutant rocA gene, and that this accounts for the increased capsule expression observed for isolates of this serotype (Lynskey et al., 2015, Miller et al., 2015). In comparison to the functional RocA protein from serotype M1 GAS, the M3 GAS RocA protein has a C-terminal truncation (Figures 2A and 2B), as a consequence of a 1 bp deletion in a polynucleotide tract (from 7A’s to 6A’s) that knocks the rocA gene out-of-frame (Figure 2C), and harbors five amino acid substitutions (Figures 2B and S1) as a consequence of non-synonymous single nucleotide polymorphisms (SNPs; Figure S2). In an attempt to identify a timeline for when the observed rocA mutations arose within the M3 GAS population, we sequenced this gene in a collection of M3 GAS isolates. A total of 13 serotype M3 GAS isolates were investigated and these were isolated in a temporally (1920s to 2010) and spatially (Europe, North America, and Japan) diverse manner (Cao et al., 2014). All isolates harbored the same rocA mutations (Table S1), consistent with M3 isolates, since at least the 1920s, harboring a mutant rocA allele.
Figure 2. Serotype M3 GAS isolates harbor a 1 bp deletion in rocA leading to a truncated protein.
(A) Comparison of rocA and neighboring genes between serotype M1 and M3 GAS isolates. Note the truncated rocA gene in M3 GAS. (B) Domain structure of the RocA protein. The N-terminal half of RocA contains multiple putative membrane-spanning regions (Mem. span sensor). The C-terminal half of RocA contains a putative ATPase domain (ATPase). The blue and red asterisks show the relative locations of the amino acid substitutions and the truncation, respectively, in serotype M3 isolates. (C) Schematic of a region of the rocA gene in M1 GAS and also of the corresponding region, which contains a 1 bp deletion, in M3 GAS. The homo-polymeric tract which contains the 1 bp deletion in M3 GAS is highlighted with horizontal green bars. The premature stop codon introduced in the rocA gene of serotype M3 GAS due to the 1 bp deletion is shown in red (TAA).
Regulation of capsule expression by rocA occurs at the RNA level
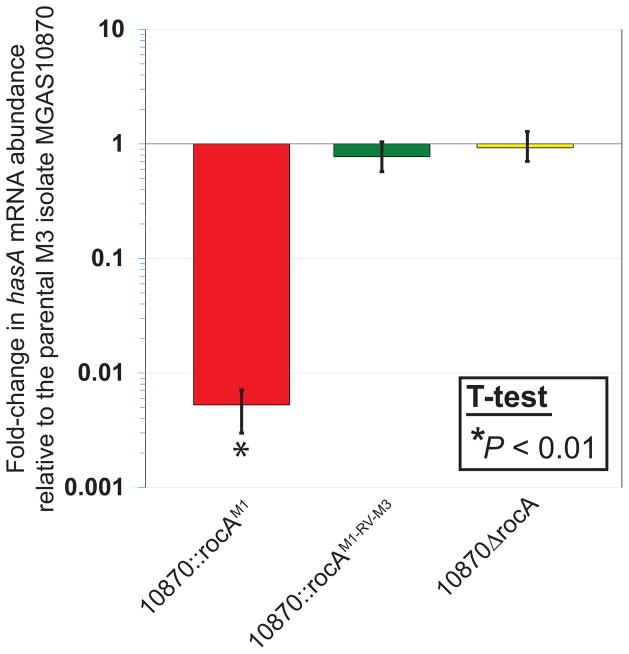
While we and others previously reported that a functional RocA protein can negatively regulate capsule expression in M3 GAS isolates (Lynskey et al., 2015, Miller et al., 2015), it had not been determined at which level regulation occurs. To assess whether the regulation afforded by RocA occurs at the RNA level, we used quantitative RT-PCR to determine the abundance of hasA mRNA, with hasA being the first gene in the capsule biosynthesis operon (Dougherty & van de Rijn, 1994). Our rocA complemented M3 GAS derivative, strain 10870::rocAM1, had an approximately 150-fold decrease in hasA mRNA relative to the parental (MGAS10870), revertant (10870::rocAM1-RV-M3), and deletion mutant (10870ΔrocA) strains (Figure 3). Thus, directly or indirectly, RocA regulates capsule expression at the RNA level.
Figure 3. Complementation of the rocA mutation in serotype M3 GAS reduces transcript abundance from the hyaluronic acid capsule biosynthesis locus.
RNA from exponential phase cultures of the indicated strains was analyzed by Taqman-based quantitative RT-PCR. Experiment was performed in triplicate with mean (± standard deviation) shown. An asterisk (*) highlights the sample that was statistically significantly different, via T-test (P < 0.01), relative to the parental M3 isolate MGAS10870.
The rocA mutant allele of M3 GAS isolates leads to the enhanced abundance of multiple immunomodulatory virulence factor-encoding mRNAs
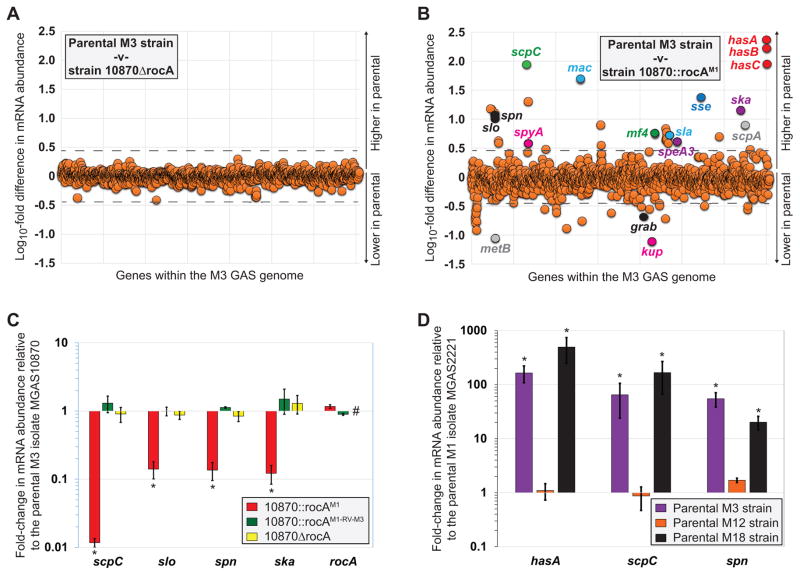
Given the significantly greater virulence of M3 isolates during invasive infections, we hypothesized that the rocA mutation of M3 isolates modifies the abundance of virulence factor-encoding mRNAs in addition to capsule. To test our hypothesis we performed RNAseq analysis to compare the transcriptomes of our parental M3 isolate MGAS10870 and its derivatives 10870ΔrocA and 10870::rocAM1. Comparison of the exponential phase transcriptomes from the parental and rocA deletion mutant strains identified no mRNAs that were statistically significantly different (Kal’s Z-test with false-discovery rate correction (Kal et al., 1999); P ≤ 0.05) between the two isolates at a level 3-fold or greater (Figure 4A). The lack of differentially regulated genes between these two isolates is consistent with the natural rocA mutation of serotype M3 GAS isolates being a null mutation. Comparison of the exponential phase transcriptomes from the parental M3 isolate MGAS10870 and the rocA complemented derivative 10870::rocAM1 identified 56 mRNAs that were statistically significantly different (Kal’s Z-test with false-discovery rate correction (Kal et al., 1999); P ≤ 0.05) between the two isolates at a level 3-fold or greater (Figure 4B). Strikingly, a third of the differentially regulated mRNAs encode for virulence factors, and in almost each case these virulence factor-encoding mRNAs are higher expressed in the parental M3 GAS isolate, consistent with the high virulence of M3 isolates (Table S4). That multiple virulence factor-encoding mRNAs show increased abundance in M3 isolates due to the rocA mutation was verified through quantitative RT-PCR of specific mRNA transcripts (Figure 4C).
Figure 4. The rocA mutation in M3 GAS enhances the abundance of multiple virulence factor encoding mRNAs.
(A) Summary of RNAseq data comparing the parental M3 isolate MGAS10870 with the rocA deletion mutant derivative 10870ΔrocA. Duplicate cultures of each strain were grown to the mid-exponential phase of growth in THY broth. Total RNA was isolated and subjected to RNAseq analysis. The relative expression levels of all genes are graphed, with each represented by a circle. Genes are arranged in the same order as they appear in the chromosome. (B) Summary of RNAseq data comparing the parental M3 isolate MGAS10870 with the rocA complemented derivative 10870::rocAM1. The experiment was performed, and the data are presented, as described in (A) above. (C) Taqman-based quantitative RT-PCR data showing the differential regulation of virulence factor gene expression in clinical M3 GAS due to rocA mutation. The abundance of the indicated mRNAs were determined from triplicate exponential phase GAS cultures. The experiment was performed in triplicate with mean (± standard deviation) shown. The hashtag highlights the lack of rocA transcript in the rocA deletion mutant strain. The asterisks (*) highlight statistical significance relative to the parental M3 isolate MGAS10870 (T-test, P < 0.01). (D) Positive correlation between rocA genotype and virulence factor regulation between isolates of different GAS serotypes. Taqman-based quantitative RT-PCR data showing the differential regulation of virulence factor gene expression between GAS serotypes. The abundance of the indicated mRNAs were determined from triplicate exponential phase GAS cultures. The experiment was performed in triplicate with mean (± standard deviation) shown. The asterisks (*) highlight statistical significance relative to the representative M1 isolate (T-test, P < 0.05).
The expanded RocA regulon is not constrained to M3 GAS isolates
Serotype M18 GAS isolates also harbor a mutant rocA gene (Lynskey et al., 2013), however, it has not been reported that this mutation has the wide regulatory effects as we have observed in M3 GAS. To investigate this issue, and to allow a comparison between four different GAS serotypes (M1, M3, M12, and M18), we performed quantitative RT-PCR analysis of select mRNAs. The representative M3 and M18 GAS isolates had increased (>20-fold) levels of all of the tested (M3 RocA-regulated) mRNAs, relative to the representative M1 and M12 isolates (which harbor a functional rocA; Figure 4D). Thus, the data are consistent with the rocA mutation of M18 GAS isolates also leading to a significant increase in the abundance of multiple virulence factor-encoding mRNAs.
The natural rocA mutation of M3 GAS isolates modifies the expression of multiple virulence factors
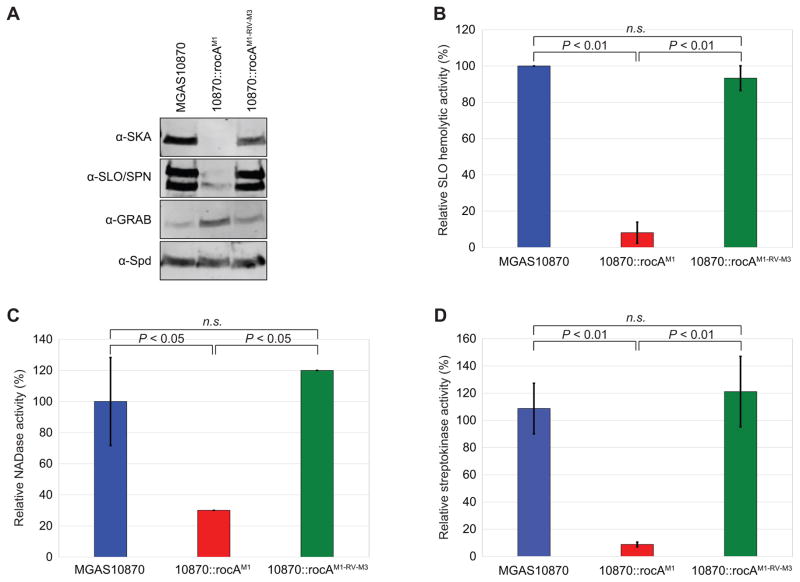
To ensure that differences in the abundance of virulence factor-encoding mRNAs between the parental and rocA-complimented M3 strains results in differences at the protein level, we performed Western blot analyses. Three virulence factors with reduced mRNA levels following rocA complementation (streptokinase, streptolysin O, and NAD-glycohydrolase), one virulence factor with increased mRNA levels following rocA complementation (GRAB), and one virulence factor with unchanged mRNA levels following rocA complementation (Spd, to serve as a loading control) were assayed. The data mirror that seen at the RNA level (Figure 5A), confirming that the natural rocA mutation of M3 GAS isolates modifies the expression of multiple virulence factors by isolates of this hyper-virulent serotype.
Figure 5. Multiple virulence factors show increased expression and activity as a consequence of the rocA mutation in serotype M3 GAS isolates.
(A) Western blot analyses of streptokinase (SKA), streptolysin O (SLO), S. pyogenes NAD-glycohydrolase (SPN), protein-G-related α2-macroglobulin-binding protein (GRAB), and S. pyogenes DNase (Spd). Secreted protein samples from exponential phase cultures of the indicated strains were used. The Spd Western was performed as a loading control as this virulence factor is not RocA-regulated. Note that the upper band in the SLO/SPN Western is SLO, with the lower band being SPN. (B, C, D) Culture supernatants from the indicated strains were isolated and used in multiple assays. (B) Streptolysin O assays. Relative lysis of sheep erythrocytes by SLO present within culture supernatants. (C) NAD-glycohydrolase assays. Relative hydrolysis of NAD+ by SPN present within culture supernatants. (D) Streptokinase assays. Indirect measure of streptokinase activity present within culture supernatants. For each assay statistical significance was tested via T-test.
Increased streptolysin O, NAD-glycohydrolase, and streptokinase activity by M3 isolates as a consequence of the rocA mutation
We performed select functional assays to determine whether there is a decrease in virulence factor activity in a rocA complemented M3 GAS derivative relative to the parental M3 strain. Hemolysis by streptolysin O, hydrolysis of NAD+ by SPN, and conversion of plasminogen to plasmin by streptokinase were all independently measured using filtered GAS culture supernatants. Streptolysin O hemolytic activity was reduced 12-fold (Figure 5B), SPN-mediated NAD+ hydrolytic activity was reduced 3-fold (Figure 5C), and streptokinase-mediated plasminogen activation was reduced 12-fold (Figure 5D), in the rocA complemented M3 GAS derivative 10870rocAM1 relative to the parental and revertant strains. Thus, the changes that occur in the serotype M3 GAS virulence factor expression profile as a consequence of the rocA mutation have direct phenotypic effects.
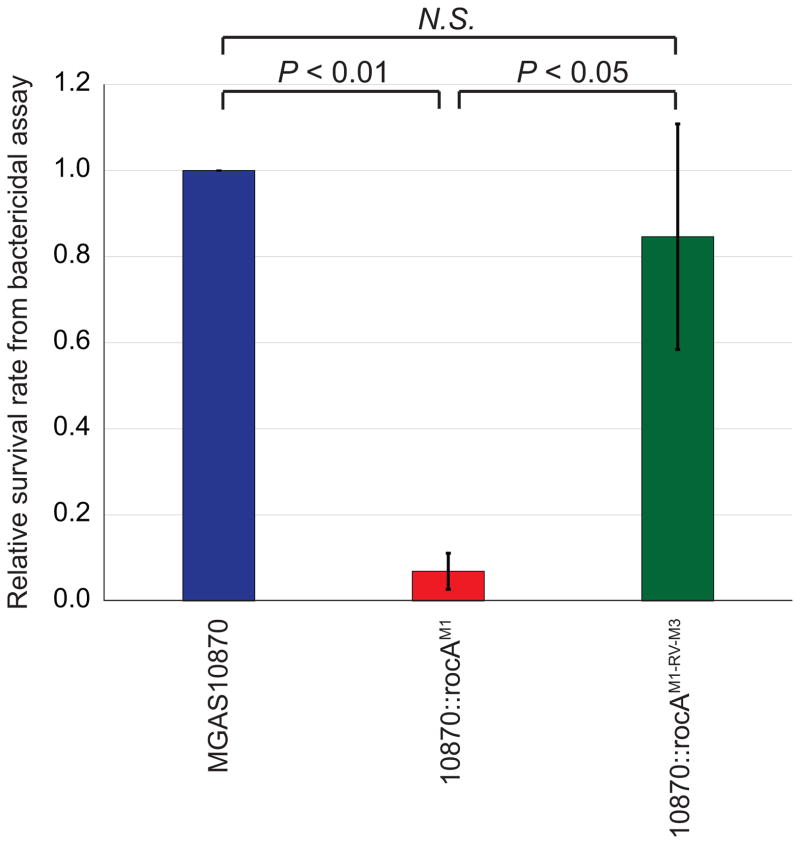
The natural rocA mutation enhances the ability of the parental M3 isolate MGAS10870 to survive incubation in whole human blood
Given the immunomodulatory nature of many of the virulence factors that are expressed in serotype M3 GAS isolates in higher abundance due to the rocA mutation, we tested whether M3 isolates are able to survive human phagocytic killing at a greater rate than rocA complemented derivatives. Bactericidal assays were performed to compare survival rates between the parental M3 isolate MGAS10870, the rocA complemented derivative 10870::rocAM1, and the revertant 10870::rocAM1-RV-M3. The parental and revertant M3 GAS strains survived incubation in human blood at a rate ~13-fold higher than that of the rocA complemented strain (Figure 6).
Figure 6. The naturally occurring rocA mutation of M3 GAS enhances their ability to survive in whole human blood.
Bactericidal assays were performed using heparinized whole human blood and the three indicated GAS strains. The experiment was performed in triplicate with the combined data shown. The data are presented as the GAS survival rate relative to that observed for the parental isolate MGAS10870. Error bars show ± standard deviation. The calculated P-values, determined via T-test, are shown. N.S. = not significant.
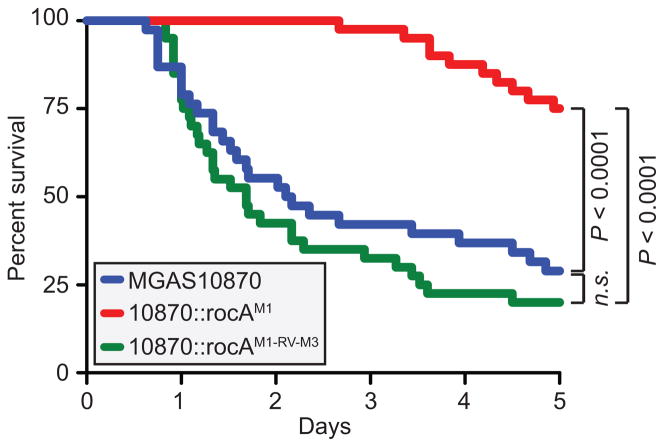
The natural rocA mutation enhances the virulence of serotype M3 GAS isolates
Given their association with causing severe invasive infections, we set out to measure the contribution of the rocA mutation to the ability of M3 GAS isolates to cause invasive infections. To test this, we used a mouse model of bacteremia infection and measured survival over time for animals infected with either the parental, rocA complemented, or rocA revertant strains. Animals infected with the rocA complemented strain survived at a significantly higher rate than the animals infected with the parental or revertant strains (Figure 7). Thus, the mutant rocA gene directly enhances the ability of serotype M3 GAS isolates to cause lethal invasive infections.
Figure 7. The naturally occurring rocA mutation contributes to the invasive disease hyper-virulence of serotype M3 GAS isolates.
Groups of mice were infected intraperitoneally with the parental M3 isolate MGAS10870 (blue), the rocA complemented derivative 10870::rocAM1 (red), or the revertant strain 10870::rocAM1-RV-M3 (green). Survival curves were generated and the data tested for statistical significance by the log-rank test.
RocA functions through CovR
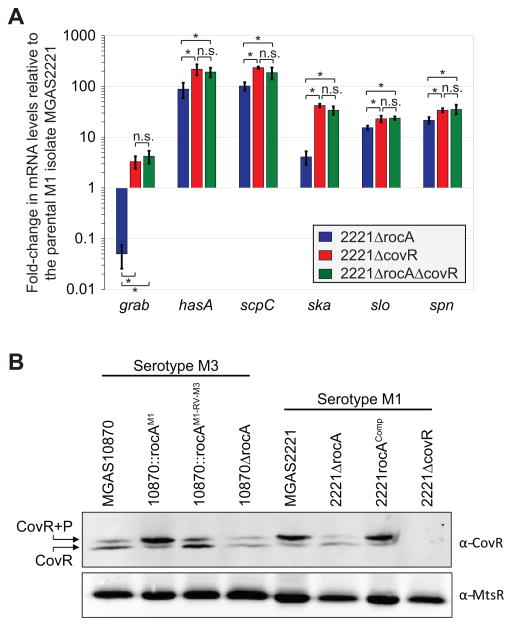
The virulence factors that are differentially regulated between parental and rocA complemented serotype M3 GAS isolates are also regulated by the CovR repressor protein (Cole et al., 2006, Sumby et al., 2006). Thus, we hypothesized that RocA functions through CovR to reduce the expression of the many virulence factors shown in figure 4B. To facilitate testing our hypothesis we created rocA and covR single and double mutant strains in a serotype M1 background. We chose to work in the M1 background to prevent confusion with the fact that M3 isolates are naturally rocA mutants. The parental M1 isolate MGAS2221, the rocA mutant derivative (2221ΔrocA), the covR mutant derivative (2221ΔcovR), and the rocA/covR double mutant derivative (2221ΔrocAΔcovR) were compared via quantitative RT-PCR analysis. All genes tested from MGAS2221 and 2221ΔrocA were statistically significantly different from one another, and both strains were also statistically significantly different from the isolates 2221ΔcovR and 2221ΔrocAΔcovR (Figure 8A). However, 2221ΔcovR and 2221ΔrocAΔcovR were not statistically significantly different from one another. Thus, the rocA mutation does not alter the abundance of the tested mRNAs in the covR mutant background, consistent with RocA functioning through the CovR protein.
Figure 8. RocA functions through CovR to regulate expression of the identified virulence factors by enhancing CovR phosphorylation.
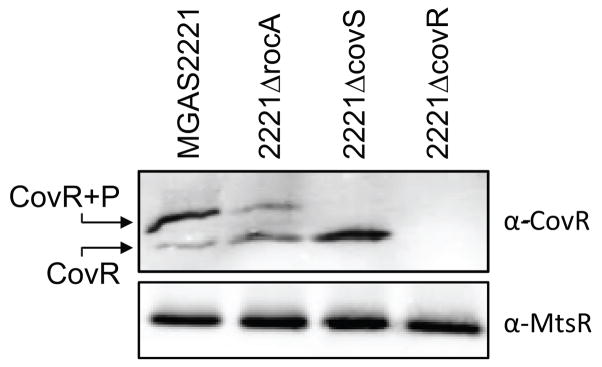
(A) Quantitative RT-PCR data showing that RocA functions through CovR. Parental (MGAS2221), rocA mutant (2221ΔrocA), covR mutant (2221ΔcovR), and rocA/covR double mutant (2221ΔrocAΔcovR) serotype M1 strains were grown to the exponential phase of growth. RNA was isolated, converted to cDNA, and used in Taqman-based quantitative RT-PCR. Note that all three mutant strains differed statistically relative to the parental strain (T-test, P < 0.05). The asterisks highlight additional statistical significance between the datasets (T-test, P < 0.05), and non-significant data are also highlighted (n.s.). (B) Phos-Tag analysis showing that RocA alters the ratio of phosphorylated and non-phosphorylated CovR. Cytoplasmic protein fractions were isolated from the indicated serotype M1 and M3 GAS strains and subjected to Phos-tag gel electrophoresis followed by Western blot. The bands corresponding to non-phosphorylated (lower band) and phosphorylated (upper band) CovR protein are labeled. A Western blot using an anti-MtsR antibody, an unrelated GAS transcriptional regulator, was performed for use as a loading control.
The rocA mutation of serotype M3 isolates lowers the ratio of phosphorylated to non-phosphorylated CovR repressor protein
While the presence of RocA leads to a modest 1.9-fold increase in the abundance of covR mRNA (RNAseq data), we reasoned that the main mechanism by which RocA enhances CovR activity is through phosphorylating, and therefore activating, the CovR repressor. Consequently, given that RocA is not functional in M3 isolates this reduces CovR activity and causes de-repression of virulence factor expression. To test the hypothesis that CovR phosphorylation is altered between the presence and absence of a functional RocA protein, we analyzed GAS cytoplasmic protein fractions via Phos-tag analysis. Phos-tag electrophoresis separates proteins based, in part, on their phosphorylation status. Cytoplasmic protein samples were isolated, separated by Phos-tag electrophoresis, and subjected to Western blot analysis with a highly-specific anti-CovR antibody that we have previously used successfully in Phos-tag analyses (Horstmann et al., 2015). While the majority of CovR recovered from the parental and revertant serotype M3 isolates was non-phosphorylated (lower band in figure 8B), the rocA complemented M3 GAS derivative harbored mostly phosphorylated CovR (upper band in Figure 8B). To further confirm that the RocA protein enhances CovR phosphorylation levels, we utilized our serotype M1 GAS strains and also created a rocA complemented derivative of strain 2221ΔrocA, strain 2221rocAComp. Essentially identical data were gained using our serotype M1 isolates (Figure 8B). Thus, our data are consistent with RocA, directly or indirectly, phosphorylating CovR.
RocA enhances CovR phosphorylation only in the presence of a functional CovS
Several possibilities exist for how RocA could enhance CovR phosphorylation. For example, RocA could directly phosphorylate CovR or could form a heterodimer with CovS, with the CovS/RocA heterodimer having greater kinase activity than a CovS homodimer. To begin to investigate these possibilities we performed Phos-tag analysis using protein samples from strains that included a covS mutant strain. No phosphorylated CovR was observed in the covS mutant strain samples (Figure 9). As RocA is functional in the covS mutant strain the data are consistent with RocA only enhancing the phosphorylation status of CovR in the presence of a functional CovS.
Figure 9. Phos-Tag analysis showing that CovR is not appreciably phosphorylated in a covS mutant strain.
Cytoplasmic protein fractions were isolated from the indicated serotype M1 GAS strains and subjected to Phos-tag gel electrophoresis followed by Western blot. The bands corresponding to non-phosphorylated (lower band) and phosphorylated (upper band) CovR protein are labeled. A Western blot using an anti-MtsR antibody, an unrelated GAS transcriptional regulator, was performed for use as a loading control.
Discussion
Phenotypic heterogeneity at the strain and/or subtype levels is a commonly observed phenomenon within individual bacterial species. For a pathogen, such heterogeneity can impede the efficacy of preventative regimes, for example by resulting in the variable expression of proteins targeted in vaccine formulations. For more than five decades it has been known that some GAS serotypes show non-random associations with particular disease manifestations (Mitchell, 1962, Wilmers et al., 1954). Despite the long timeline of this observation, molecular explanations for GAS-serotype disease-phenotype associations remain to be fully elucidated. Serotype M3 GAS isolates are non-randomly associated with particularly severe invasive disease cases and a high mortality rate (Beres et al., 2006). Here, we have built upon the previous finding that M3 isolates harbor a mutant rocA allele by delineating the regulatory and phenotypic consequences of this mutation, namely the dramatic up-regulation of virulence factor expression and associated activity, as well as an increased ability to resist human phagocytic killing and to cause lethality in a murine invasive infection model. Our findings strongly implicate the acquisition of the rocA mutation as a driving force behind the association of serotype M3 GAS isolates with severe invasive infections.
The RocA protein was first described following a transposon mutagenesis screen looking for factors that negatively regulated the transcription of the covR gene (Biswas & Scott, 2003). A 2.5-fold decrease in covR mRNA levels was identified following rocA disruption, as was an increase in the expression of the CovR-repressed hyaluronic acid capsule (Biswas & Scott, 2003). More recently, it was shown that serotype M18 GAS isolates naturally harbor a mutant rocA gene, and that this is behind the mucoid colony phenotype of M18 isolates (Lynskey et al., 2013). Even more recently, we and others identified that serotype M3 isolates also have increased capsule expression as a consequence of a natural rocA mutation (Lynskey et al., 2015, Miller et al., 2015). By performing the first transcriptome comparison between functional and rocA mutant GAS strains, we have significantly expanded the RocA regulon. We identified that, relative to a rocA complemented derivative, the naturally occurring rocA mutation in serotype M3 isolates has resulted in the dramatic up-regulation of more than a dozen virulence factors (Figure 4B), including streptokinase (SKA) (Sun et al., 2004), streptolysin O (SLO) (Bastiat-Sempe et al., 2014), the C5a peptidase (ScpA) (Ji et al., 1996), the chemokine protease (SpyCEP/ScpC) (Zinkernagel et al., 2008), Mac (Mac/IdeS) (Lei et al., 2001), and the streptococcal esterase (Sse) (Liu et al., 2012a). Given that the same virulence factors are negatively regulated by the CovR/S two-component system (Sumby et al., 2006), that rocA deletion results in a modest decrease in covR mRNA levels (Lynskey et al., 2013, Biswas & Scott, 2003), that rocA deletion reduces the ratio of phosphorylated to non-phosphorylated CovR (Figure 8B), and that RocA has no activity (at least with respect to the regulation of the assayed virulence factor-encoding mRNAs) in a covR mutant strain background (Figure 8A), the data are consistent with RocA functioning through the CovR protein.
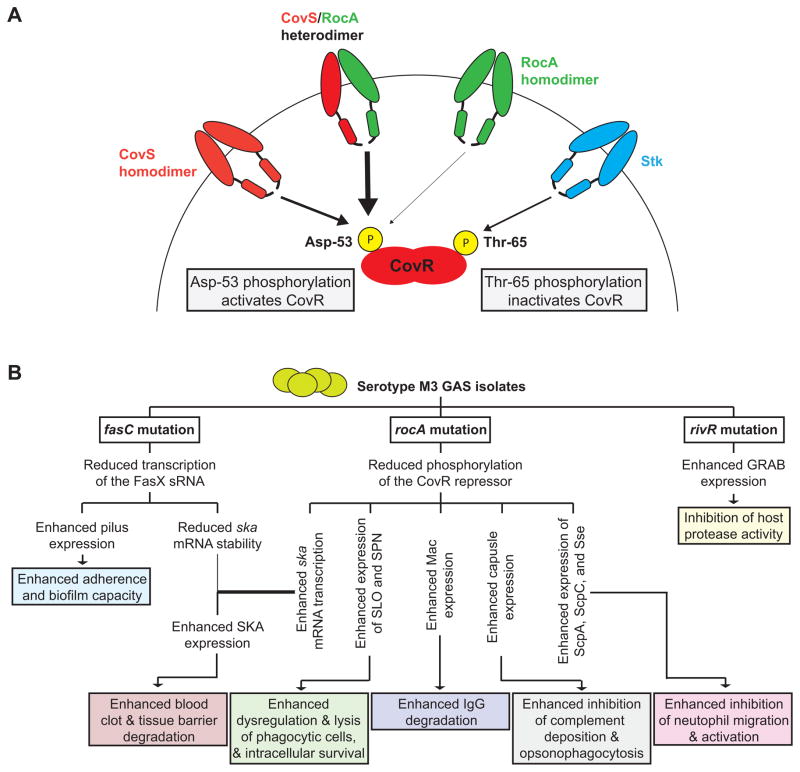
The CovR/S system has been extensively studied since its discovery almost 20 years ago (Federle et al., 1999, Levin & Wessels, 1998). For example, we and others identified that covR/S mutations are selected for during invasive GAS infections, accounting for a large percentage of the strain-specific heterogeneity observed between isolates recovered from invasive and non-invasive infections (Liang et al., 2013, Trevino et al., 2009, Cole et al., 2006, Sumby et al., 2006). Selection for covR disruption results in even higher levels of immunomodulatory virulence factor expression than seen in a rocA mutant strain (Figure 8A), presumably due to the fact that active CovR protein is still present, albeit at a reduced level (Figure 8B), in a rocA mutant. Given that RocA shows structural similarities to sensor kinases (Figure 2B), that the levels of phosphorylated CovR increase in the presence of RocA (Figure 8B), that CovR-repressed genes are transcribed at a higher level following rocA mutation (Figure 8A), and that RocA requires a functioning CovS to promote CovR phosphorylation status (Figure 9), we hypothesize that RocA forms a heterodimer with CovS, and that this heterodimer has greater kinase activity towards CovR at Asp-53 than CovS homodimers (Figure 10A). This model is consistent with our data that a parental M1 GAS isolate has the greatest CovR activity, followed by a rocA mutant, followed by a covS mutant, followed by a covR mutant (Figure 8A and data not shown).
Figure 10. Models of how RocA interacts with the CovR/S system and of how the rocA mutation in M3 isolates contributes to regulatory rewiring that leads to hyper-virulence.
(A) Shown is a model of how CovS, RocA, and Stk feed into the regulation of the CovR repressor. Our data, including the finding that significant CovR phosphorylation is not observed in a covS mutant strain, are consistent with RocA promoting CovR phosphorylation on Asp-53 by forming a heterodimer with CovS, with the heterodimer having greater kinase activity than CovS or RocA homodimers. (B) Three regulatory systems are disrupted in serotype M3 GAS isolates leading to an altered virulence factor profile that, we hypothesize, is the basis behind the association of M3 isolates with severe invasive infections. The consequences of the three regulator gene mutations identified in serotype M3 GAS isolates are shown. Note that while the fasC mutation decreases streptokinase (ska) mRNA levels this is more than offset by the large increase in ska mRNA abundance as a consequence of the rocA mutation, resulting in a net increase in streptokinase expression.
Our data show that serotype M3 GAS isolates since at least the 1920s express multiple immunomodulatory virulence factors at high levels due to the rocA mutation, and we hypothesize that this is a major contributor to the invasive disease hyper-virulence phenotype of M3 isolates. However, the rocA mutation cannot be solely responsible given that serotype M18 isolates also have a mutant rocA, yet are not readily associated with cases of invasive infection (Lynskey et al., 2013). While we have not fully characterized the M18 rocA regulon, initial analyses are consistent with the same regulatory pattern as we have discovered for M3 GAS (Figure 4D). Our current model explaining why serotype M3 GAS isolates cause particularly severe invasive infections is shown in figure 10B. Three regulator-encoding genes are disrupted in serotype M3 isolates, each of which results in the alteration of virulence factor expression which modifies the phenotypic characteristics of these isolates. A disrupted FasBCAX regulatory system is only observed in M3 isolates and is a result of an inactivating mutation with the fasC gene (Cao et al., 2014). As a consequence of the fasC mutation the transcription of the sRNA FasX is reduced 200-fold which, given that FasX positively regulates the stability of streptokinase mRNA, results in a reduction in streptokinase expression (Ramirez-Pena et al., 2010). Importantly however, M3 GAS streptokinase expression is actually increased relative to other GAS serotypes, as the increase in streptokinase expression that occurs due to the rocA mutation more than makes up for the decreased expression due to the fasC mutation. The increase in streptokinase expression by M3 GAS isolates is consistent with these isolates having an increased ability to cause severe invasive diseases, given the crucial role of streptokinase in GAS dissemination (Sun et al., 2004). While not yet tested, we hypothesize that M3 isolates also have greater levels of pili on their cell surface as a consequence of the fasC mutation, as FasX reduces pilus expression in other serotypes (Liu et al., 2012b, Danger et al., 2015b). Disruption of RivR activity, which also appears to be unique to M3 isolates, is a result of multiple inactivating mutations within the M3 rivR allele (Cao et al., 2014). The consequence of RivR disruption is an increase in the expression of GRAB which, given that GRAB is a cell-wall anchored protein that binds to the major protease inhibitor found in human plasma (α2-macroglobulin), is expected to result in an inhibition of protease activity at the GAS cell surface (Rasmussen et al., 1999). Finally, a disrupted rocA gene is observed in both serotype M3 and M18 GAS isolates, although the exact mutations differ. The rocA mutation results in the significant up-regulation of immunomodulatory virulence factors that impact multiple facets of the innate and adaptive immune responses (Figures 4B and 9B). Consistent with modulation of the immune response, our rocA complemented M3 GAS isolate was attenuated in its growth and survival in human blood (Figure 6), as well as in our murine bacteremia model of infection (Figure 7). We hypothesize that the unique combination of regulatory mutations present within serotype M3 isolates leads to an M3-specific virulence factor expression profile, and that this profile endows M3 isolates with the ability to cause particularly severe invasive infections. While full characterization of exactly how the M3 GAS virulence factor profile enhances invasive disease severity is ongoing, our finding that isolates of a single GAS serotype can have drastically different expression profiles sheds new light not only on the association of serotype M3 isolates with invasive infections, but also of other GAS-serotype disease-phenotype associations. Furthermore, insights gained from this study are likely to extend to other bacterial pathogens as signaling systems are often identified as “hot-spots” for mutation (Yang et al., 2011, Lieberman et al., 2011, Nasser et al., 2014).
In addition to rocA mutation being a serotype-specific phenomenon a recent report described rocA mutation as contributing to strain-specific variation (Yoshida et al., 2015). Mucoid and non-mucoid serotype M1 GAS isolates were isolated from the cerebrospinal fluid of a patient, and whole-genome sequencing identified that the only genetic difference between the isolates was the introduction of a premature stop codon in rocA (Yoshida et al., 2015). Thus, similar to previous work showing that covR/S mutant strains are positively selected for during invasive GAS infections (Cole et al., 2006, Sumby et al., 2006), the same may also be true for rocA. Given that RocA positively regulates CovR activity, the disruption of rocA generates a phenotype somewhat analogous to a covR mutant strain, and hence is another method by which the “invasive infection phenotype” can be attained in a strain-specific manner.
It is of note that while covR/S mutant strains of multiple GAS serotypes can be selected for in a strain-specific manner these mutations are not maintained in the population (Trevino et al., 2009). This is in contrast to the rocA mutations of M3 and M18 GAS which are present in all contemporary isolates (Lynskey et al., 2013, Lynskey et al., 2015, Miller et al., 2015). As per our model (Figure 10A), we propose that the reduction in Cov activity observed following rocA mutation is not as great as following covS or covR mutation. Thus, while data suggests that covR/S mutants are attenuated in their ability to undergo productive non-invasive (e.g. pharyngeal) infections (Trevino et al., 2009, Alam et al., 2013), the level of attenuation is predicted to be less severe in a rocA mutant strain. Hence, we believe it more likely that rocA mutations may be maintained within a population, since non-invasive infections are the most common type of infection, as is the case for the rocA mutations of M3 and M18 isolates. While serotype M3 isolates are hyper-virulent, serotype M1 isolates are the most successful GAS serotype given that they are the most commonly isolated serotype in many case studies performed in the United States, Canada, and Western Europe since the mid-1980s (Nasser et al., 2014). We propose that part of this success relative to M3 isolates is the greater dynamic range of the Cov system available to M1 isolates, given that they harbor wild-type rocA in addition to wild-type covR/S.
Until recently the components of bacterial two-component regulatory systems were largely thought to function in a linear manner, with the kinase activating the response regulator without any involvement from other regulatory proteins. However, studies are now bringing to light the fact that cross-talk between different regulatory systems may be a common mechanism by which multiple signaling inputs can fine-tune regulatory outputs (Guckes et al., 2013). For example, the heme sensing two-component system HssR/S, and cell envelope stress sensing two-component system HitR/S, undergo extensive cross-talk in Bacillus anthracis to integrate a response to these factors (Mike et al., 2014). That the CovR/S system may not function independently of other regulatory proteins was suggested by the finding that covR and covS mutant strains are not phenotypically identical (Trevino et al., 2009), as would be expected if CovS only activated CovR and if CovR was only activated by CovS. This was confirmed by our subsequent identification that Stk phosphorylates CovR on Thr-65 to inhibit CovR activation (Horstmann et al., 2014). Our data presented here suggest another input into the CovR/S regulatory system, RocA, which we hypothesize is through formation of heterodimers with CovS (Figure 10A). Thus, in addition to discoveries made with regard to GAS-serotype disease-phenotype associations, our data also adds to a small but growing number of studies that suggest that regulatory system cross-talk is a common phenomenon, and critical to the ability of pathogens to integrate disparate signals into a unified transcriptional response.
Experimental Procedures
Bacterial strains and culture conditions
The clinical GAS isolates used in this study are listed in table S1. The GAS derivatives constructed and used in this study are listed in table S2. Routine growth of GAS cultures made use of Todd-Hewitt broth with 0.2% yeast extract (THY broth), and cultures were incubated at 37°C (5% CO2). Chloramphenicol (4 μg/ml) and/or spectinomycin (150 μg/ml) were added when required.
Construction of the M3 GAS derivative strains 10870::rocAM1 and 10870::rocAM1-RV-M3
To complement the naturally occurring rocA mutant gene in the representative M3 GAS isolate MGAS10870 we replaced the gene with the wild-type allele present in M1 GAS, creating strain 10870::rocAM1. Allele replacement made use of the suicide vector pBBL740 via standard techniques as previously described (Ramirez-Pena et al., 2010). Briefly, the rocA gene from M1 GAS strain MGAS2221 was PCR amplified and joined, via Gibson Assembly (New England Biolabs), between two 1 kb flanking regions amplified from the M3 GAS strain MGAS10870, and into the PCR-amplified pBBL740 plasmid. The resultant plasmid was PCR amplified and sequenced to ensure that the plasmid was mutation free before transforming it into MGAS10870 competent cells. Selection was on THY agar plates containing chloramphenicol. Transformants were passaged in THY broth without antibiotics to facilitate excision of the plasmid from the chromosome, and this was tested by patching single colonies on THY agar plates with and without chloramphenicol. Phenotypically correct colonies were confirmed as correct via PCR and sequencing. Primers used in strain construction are listed in table S3.
To ensure that any phenotypic differences observed between MGAS10870 and 10870::rocAM1 were a consequence of the rocA alleles, and not spurious mutations, we created a derivative of strain 10870::rocAM1 in which the M3 rocA allele had been re-introduced. This revertant strain, 10870::rocAM1-RV-M3, was created in the same manner as strain 10870::rocAM1 only this time the pBBL740-derivative contained the M3 rocA allele. Phenotypically correct colonies were confirmed as correct via PCR and sequencing. Primers used in strain construction are listed in table S3.
Construction of the M3 GAS derivative strain 10870ΔrocA
A rocA deletion mutant derivative of the parental M3 strain MGAS10870 was created using the primers listed in Table S3. Briefly, primers UNR33/34 and primers UNR36/38 were used to amplify 1kb regions upstream and downstream of the M3 rocA gene. Primers UNR35/37 were used to amplify the 1 kb spectinomycin resistance cassette from plasmid pSL60 (Lukomski et al., 2000a). The three PCR products were joined together, with the spectinomycin resistance cassette in the middle, via over-lap extension PCR using primers UNR33/38. The resultant 3 kb PCR product was TOPO cloned and sequenced to ensure no spurious mutations had been introduced during the PCR process. Subsequently, the 3kb insert was PCR amplified and transformed into MGAS10870. Replacement of the rocA gene with the spectinomycin resistance cassette was confirmed via PCR and sequencing.
Construction of the M1 GAS derivative strains 2221ΔrocA, 2221ΔrocAΔcovR, and 2221rocAComp
The rocA gene of the clinical serotype M1 isolate MGAS2221, and of the MGAS2221-derivative strain 2221ΔcovR, was replaced with a spectinomycin resistance cassette to create strains 2221ΔrocA and 2221ΔrocAΔcovR, respectively. These strains were created as described above for strain 10870ΔrocA, only gDNA from a serotype M1 strain was used as template in the initial PCR reactions. A rocA complemented derivative of 2221ΔrocA, strain 2221rocAComp, was constructed via replacement of the spectinomycin cassette with a functional rocA gene through use of the pBBL740 vector. The functional rocA gene from MGAS2221, plus 1kb flanks either side, was PCR amplified using primers UNR33/38 (Table S3) and cloned into the pBBL740 vector. The resultant plasmid was sequence verified and used to create strain 2221rocAComp as described above for the creation of the M3 GAS derivatives 10870::rocAM1.
Total RNA isolation from GAS
Total RNA was isolated from tested GAS strains as previously described (Sumby et al., 2006). Briefly, the strains of interest were grown to the exponential phase of growth (O.D. 0.5) in THY broth. Two volumes of RNAprotect bacteria reagent (Qiagen Inc) were added to one volume of GAS culture and incubated at room temperature for 5 mins. Following centrifugation (5,000 g for 11 mins at 4°C) the supernatant was discarded, the cell pellets snap frozen in liquid nitrogen, and the frozen pellets placed at −80°C until ready for processing. Cells were processed using a mechanical lysis method with lysing matrix B tubes in conjunction with a FastPrep24 homogenizer (MP Biomedicals). RNA was isolated using the miRNeasy kit (Qiagen) with contaminating DNA being removed with three treatments with TURBO-DNase-free (Life Technologies). The quality and quantity of the purified RNA was determined using a Bioanalyzer system (Agilent Tech).
RNAseq analysis
Duplicate cultures of the M3 GAS strains MGAS10870, 10870ΔrocA, and 10870::rocAM1 were grown to the exponential phase of growth and RNA was isolated as described above. Ribosomal RNAs (rRNAs), which represent ~96% of total RNA, were depleted using the Ribo-Zero Gram-Positive rRNA Removal Kit (Epicentre). The rRNA-depleted RNA was used to generate libraries for sequencing using the ScriptSeq kit (Epicentre), as we have previously described (McClure et al., 2013). Briefly, RNA was fragmented, cDNA was synthesized using random hexamers containing a 5′ tagging sequence, the RNA was hydrolyzed, the cDNA was tagged at the 3′ end, a limited number of PCR cycles (n=14) were used to amplify the libraries via the 5′ and 3′ tags (the libraries will be barcoded by using different primers), and the libraries were size-selected (170 to 300 bp). The six size-selected and barcoded libraries were ran on an Illumina MiSeq flowcell. Data were analyzed using CLC Genomics Workbench and normalized to overall sequencing depth using total mapped reads data. Statistical significance was tested using Kal’s Z-test with a false-discovery rate correction (Kal et al., 1999). The RNAseq data have been deposited at the Gene Expression Omnibus (GEO) database at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/geo) and are accessible through accession number GSE68277.
Quantitative RT-PCR analysis
To determine relative mRNA abundance we used quantitative RT-PCR using Taqman primers and probes. In each experiment, RNA samples from triplicate exponential phase cultures of each GAS strain under investigation were converted into cDNA, using the reverse transcriptase Superscript III (Life Technologies), and used in association with a CFX Connect Real-Time System (Biorad). TaqMan primers and probes for genes of interest and the internal control gene proS are shown in Table S3. Transcript levels were determined using the ΔΔCT method. Each experiment was performed in triplicate with mean values (± standard deviation) shown. Statistical significance was determined via Student’s T-test.
Western blot analysis using secreted GAS protein fractions
Supernatant proteins from exponential phase THY broth GAS cultures were concentrated by ethanol precipitation and resuspended in SDS-PAGE buffer at 1/20th the original volume. Protein samples were separated on 12% Tris-HCl gels before transferring to membrane and using in Western blot analysis with a custom rabbit anti-streptokinase (SKA) polyclonal antibody (made by Pacific Immunology Inc), a commercial rabbit anti-SLO/SPN polyclonal antibody (American Research Products Inc), a custom rabbit anti-GRAB polyclonal antibody (made by Pacific Immunology Inc), and a custom rabbit anti-Spd polyclonal antibody (made by Pacific Immunology Inc), as primary antibodies. After washing, Alexa Fluor 680 donkey anti-rabbit IgG secondary antibody (Molecular Probes) was used (1:10,000 dilution), and the fluorescent signal was detected using a Li-Cor Odyssey Near-Infrared System.
SLO assays
The SLO activity of GAS strains was assayed as described previously (Sumby et al., 2005), with modifications. Briefly, the strains were grown to an O.D.600 of 0.5, cells pelleted by centrifugation at 5000 x g for 10 min, supernatants filter sterilized (pore size 0.22 μm) and divided into aliquots. Before use, the supernatants were diluted (1:10) in PBS. Samples were then incubated with 20 mM dithiothretiol for 10 min at room temperature and separated into 500 μl aliquots into 2 tubes. As a specific SLO inhibitor, 25 μg of water-soluble cholesterol (cholesterol/methyl-cyclodextrin; Sigma-Aldrich) was added to one of the sample aliquots and incubated at 37 °C for 30 min. Following incubation, 250 μl of 2% sheep blood erythrocyte/PBS suspension was added to each sample, mixed by inversion and incubated at 37 °C for 30 min. PBS (500 μl) of was added to each sample and unlysed erythrocytes pelleted by centrifugation at 3000 x g for 5 min. The amount of hemoglobin present in the supernatant was measured by analysis of optical density at 550 nm. As standards, a range of 0% to 2% erythrocyte suspensions were incubated in water, to provide 100% lysis. The standards were treated the same as samples, except that 500 μL of THY was added instead of PBS before final centrifugation.
NADase assays
The NADase activity from the GAS strains was assayed in accordance with a previously described protocol (Sumby et al., 2005). Briefly, the strains were grown to an O.D.600 of 0.5, cells pelleted by centrifugation at 5000 x g for 10 min, supernatants filter sterilized (pore size 0.22 μm) and divided into aliquots. Two-fold serial dilutions of supernatants were performed in microtiter plates with PBS. NAD+ (Sigma-Aldrich) was diluted in PBS and added to wells for a final concentration of 0.67 nM and plates were incubated at 37 °C for 20 min.
Reactions were developed by adding NaOH to final concentration of 2 N, were incubated in the dark for 1 h at room temperature, and read macroscopically by excitation with 360-nm light. Results were recorded as the highest 2-fold dilution capable of hydrolyzing 100 nM of exogenous NAD+, thus resulting in no fluorescence, and converted into relative NADase activity.
SKA assays
SKA activity from GAS supernatants was measured indirectly via the SKA-induced cleavage of human plasminogen to plasmin and subsequently the breakdown of the chromogenic substrate S2-2251 (Diapharma) by plasmin. GAS strains were grown to an O.D.600 of 0.5, cells were pelleted by centrifugation at 5000 x g for 10 min, and supernatants were filter sterilized (pore size 0.22 μm) and divided into aliquots. From these stocks, 170 μL of supernatant was added to the wells of a microtiter plate (3 wells per sample). Then to each sample, 2 μg of human plasminogen and S-2251 to a final concentration of 200 nM were added. For the first 5 hr, the plate was incubated at 37 °C, then transferred to a temperature controlled spectrophotometer (VersaMax) and incubated at 37 °C. Between the 5 to 9 hr incubation, the absorbance at 405 nm was read every 5 min. For the standards, known amounts of purified streptokinase from group C Streptococcus (Sigma-Aldrich) were used and treated exactly as the experimental samples.
Isolation of cytoplasmic GAS protein fractions
GAS cell pellets were gained from exponential phase THY broth cultures were resuspended in PBS containing protease inhibitors (Complete Mini Protease Inhibitor Cocktail, Roche). Cells were then lysed via mechanical disruption (FastPrep Machine, MP Biomedicals) using speed 6 for 15 seconds and repeated three times. After centrifugation to pellet the cell debris the supernatants, containing cytoplasmic proteins, were removed to a clean tube containing 2X SDS-PAGE buffer. Protein concentrations were determined by Qubit (LifeTechnologies) according to manufacturer’s recommendations.
Phos-tag electrophoresis
Protein samples were separated on a standard 10% SDS-PAGE gel with the addition of 100 μM Phos-tag™ Acrylamide AAL-107 (Wako Pure Chemicals) and 100 μM MgCl2 (Sigma-Aldrich). Equal amounts of cytoplasmic GAS protein samples (70 μg) were loaded onto the SDS-PAGE gel and electrophoresis set at 100 volts for >2 hr at 4 °C. Samples were then transferred to PVDF membrane via wet transfer at 400 constant mA. Analysis of proteins was accomplished with use of a custom rabbit anti-CovR antibody as previously described (Horstmann et al., 2015). After washing, Alexa Fluor 680 donkey anti-rabbit IgG secondary antibody (Molecular Probes) was used (1:10,000 dilution), and the fluorescent signal was detected using a Li-Cor Odyssey Near-Infrared System.
Bactericidal assays
GAS strains were grown to the early exponential phase of growth (O.D.600 ~ 0.2) and diluted 1:10,000 in PBS. Subsequently, 50μl of the diluted bacterial suspension was added to 450μl of heparinized human whole blood and incubated at 37°C for 3 hours with end-to-end rotation. In addition, 100μl of the diluted bacterial suspension was plated onto three separate blood agar plates to enable enumeration of the initial inoculum. Following the 3 hour incubation the blood/GAS mixtures were plated onto duplicate blood agar plates, with both undiluted and 1:10 diluted aliquots being plated. All plates were incubated at 37°C overnight before calculating the initial inoculum and the surviving inoculum CFU plates. Data are presented as percent survival ([surviving CFUs / initial CFUs] x 100). The experiment was performed in triplicate with mean values (± standard deviation) shown. Statistical significance was determined via T-test (P < 0.05).
Murine soft tissue infection model
The clinical isolates MGAS2221 (M1), MGAS10870 (M3), and MGAS6180 (M28) were chosen for use in this study as they are all representative of their respective serotypes (Sumby et al., 2005, Beres et al., 2004, Green et al., 2005). However, while MGAS2221 and MGAS10870 harbor wild-type alleles of the regulator-encoding genes covR/S and ropB, MGAS6180 does not (Stetzner et al., 2015). Thus, it should be stated that it is possible that the regulator mutations in MGAS6180 could confound the data from our soft tissue infection model assay (the mutations in MGAS6180 were not discovered until after the experiment was performed). Each GAS strain was used to infect ten SKH1 immunocompetent hairless mice (Charles River). Mice were infected with 1 × 106 GAS colony forming units (CFUs) in 100μl PBS via subcutaneous injection between the shoulder blades. The lesion volume was determined daily for up to 3 weeks by using the formula for a spherical ellipsoid (Lukomski et al., 2000b). The percent of mice with skin lesions that ulcerated, and changes in weight, were also calculated daily. Statistical significance was determined via repeated measures ANOVA (lesion volume), Fishers exact test (percent ulceration), or Student’s Test (24h weight loss). This experiment was performed in strict accordance to the guidelines set forth in our protocol approved by The University of Nevada’s Institutional Animal Care & Use Committee (IACUC).
Murine bacteremia model of infection
Parental, rocA complemented, and revertant M3 GAS isolates were each used to infect twenty CD-1 mice (Charles River). GAS cells for infection were grown to the exponential phase of growth, washed twice with PBS, resuspended in PBS, snap frozen in liquid nitrogen in six 2 ml aliquots, and titers calculated from three of the six frozen aliquots. Mice were infected with either 2.5 × 107 GAS colony forming units (CFUs; experiment 1) or 3.5 × 107 CFUs (experiment 2) in 250 μl PBS via intraperitoneal injection. Mice were monitored every six hours for the first 24 hours, every 2 hours between 24 and 72 hours, and then three times daily until the end of the experiment. The experiment was performed twice and the combined data were analyzed for statistical significance via the Log-rank test. This experiment was performed in strict accordance to the guidelines set forth in our protocol approved by The University of Nevada’s Institutional Animal Care & Use Committee (IACUC).
Supplementary Material
Acknowledgments
This research was funded in part by grant R01AI087747 from the National Institute of Allergy and Infectious Diseases (to P.S.). Additional support was provided by the School of Medicine, University of Nevada.
References
- Alam FM, Turner CE, Smith K, Wiles S, Sriskandan S. Inactivation of the CovR/S virulence regulator impairs infection in an improved murine model of Streptococcus pyogenes naso-pharyngeal infection. PLoS One. 2013;8:e61655. doi: 10.1371/journal.pone.0061655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aracil B, Slack M, Perez-Vazquez M, Roman F, Ramsay M, Campos J. Molecular epidemiology of Haemophilus influenzae type b causing vaccine failures in the United Kingdom. J Clin Microbiol. 2006;44:1645–1649. doi: 10.1128/JCM.44.5.1645-1649.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastiat-Sempe B, Love JF, Lomayesva N, Wessels MR. Streptolysin O and NAD-glycohydrolase prevent phagolysosome acidification and promote group a streptococcus survival in macrophages. mBio. 2014;5:e01690–01614. doi: 10.1128/mBio.01690-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beres SB, Carroll RK, Shea PR, Sitkiewicz I, Martinez-Gutierrez JC, Low DE, McGeer A, Willey BM, Green K, Tyrrell GJ, Goldman TD, Feldgarden M, Birren BW, Fofanov Y, Boos J, Wheaton WD, Honisch C, Musser JM. Molecular complexity of successive bacterial epidemics deconvoluted by comparative pathogenomics. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:4371–4376. doi: 10.1073/pnas.0911295107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beres SB, Musser JM. Contribution of exogenous genetic elements to the group A Streptococcus metagenome. PloS one. 2007;2:e800. doi: 10.1371/journal.pone.0000800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beres SB, Richter EW, Nagiec MJ, Sumby P, Porcella SF, DeLeo FR, Musser JM. Molecular genetic anatomy of inter- and intraserotype variation in the human bacterial pathogen group A Streptococcus. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:7059–7064. doi: 10.1073/pnas.0510279103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beres SB, Sylva GL, Sturdevant DE, Granville CN, Liu M, Ricklefs SM, Whitney AR, Parkins LD, Hoe NP, Adams GJ, Low DE, DeLeo FR, McGeer A, Musser JM. Genome-wide molecular dissection of serotype M3 group A Streptococcus strains causing two epidemics of invasive infections. Proc Natl Acad Sci U S A. 2004;101:11833–11838. doi: 10.1073/pnas.0404163101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas I, Scott JR. Identification of rocA, a positive regulator of covR expression in the group A streptococcus. Journal of bacteriology. 2003;185:3081–3090. doi: 10.1128/JB.185.10.3081-3090.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao TN, Liu Z, Cao TH, Pflughoeft KJ, Trevino J, Danger JL, Beres SB, Musser JM, Sumby P. Natural disruption of two regulatory networks in serotype M3 group A Streptococcus isolates contributes to the virulence factor profile of this hypervirulent serotype. Infection and immunity. 2014;82:1744–1754. doi: 10.1128/IAI.01639-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll RK, Beres SB, Sitkiewicz I, Peterson L, Matsunami RK, Engler DA, Flores AR, Sumby P, Musser JM. Evolution of diversity in epidemics revealed by analysis of the human bacterial pathogen group A Streptococcus. Epidemics. 2011;3:159–170. doi: 10.1016/j.epidem.2011.07.001. [DOI] [PubMed] [Google Scholar]
- Cheung GY, Wang R, Khan BA, Sturdevant DE, Otto M. Role of the accessory gene regulator agr in community-associated methicillin-resistant Staphylococcus aureus pathogenesis. Infection and immunity. 2011;79:1927–1935. doi: 10.1128/IAI.00046-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole JN, McArthur JD, McKay FC, Sanderson-Smith ML, Cork AJ, Ranson M, Rohde M, Itzek A, Sun H, Ginsburg D, Kotb M, Nizet V, Chhatwal GS, Walker MJ. Trigger for group A streptococcal M1T1 invasive disease. FASEB J. 2006;20:1745–1747. doi: 10.1096/fj.06-5804fje. [DOI] [PubMed] [Google Scholar]
- Croucher NJ, Harris SR, Fraser C, Quail MA, Burton J, van der Linden M, McGee L, von Gottberg A, Song JH, Ko KS, Pichon B, Baker S, Parry CM, Lambertsen LM, Shahinas D, Pillai DR, Mitchell TJ, Dougan G, Tomasz A, Klugman KP, Parkhill J, Hanage WP, Bentley SD. Rapid pneumococcal evolution in response to clinical interventions. Science. 2011;331:430–434. doi: 10.1126/science.1198545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham MW. Pathogenesis of group A streptococcal infections. Clin Microbiol Rev. 2000;13:470–511. doi: 10.1128/cmr.13.3.470-511.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danger JL, Cao TN, Cao TH, Sarkar P, Trevino J, Pflughoeft KJ, Sumby P. The small regulatory RNA FasX enhances group A Streptococcus virulence and inhibits pilus expression via serotype-specific targets. Mol Microbiol. 2015a doi: 10.1111/mmi.12935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danger JL, Cao TN, Cao TH, Sarkar P, Trevino J, Pflughoeft KJ, Sumby P. The small regulatory RNA FasX enhances group A Streptococcus virulence and inhibits pilus expression via serotype-specific targets. Mol Microbiol. 2015b;96:249–262. doi: 10.1111/mmi.12935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougherty BA, Ivan de Rijn Molecular characterization of hasA from an operon required for hyaluronic acid synthesis in group A streptococci. J Biol Chem. 1994;269:169–175. [PubMed] [Google Scholar]
- Federle MJ, McIver KS, Scott JR. A response regulator that represses transcription of several virulence operons in the group A streptococcus. Journal of bacteriology. 1999;181:3649–3657. doi: 10.1128/jb.181.12.3649-3657.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer J, Guiney DG. Diverse virulence traits underlying different clinical outcomes of Salmonella infection. J Clin Invest. 2001;107:775–780. doi: 10.1172/JCI12561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green NM, Zhang S, Porcella SF, Nagiec MJ, Barbian KD, Beres SB, LeFebvre RB, Musser JM. Genome sequence of a serotype M28 strain of group a streptococcus: potential new insights into puerperal sepsis and bacterial disease specificity. J Infect Dis. 2005;192:760–770. doi: 10.1086/430618. [DOI] [PubMed] [Google Scholar]
- Guckes KR, Kostakioti M, Breland EJ, Gu AP, Shaffer CL, Martinez CR, 3rd, Hultgren SJ, Hadjifrangiskou M. Strong cross-system interactions drive the activation of the QseB response regulator in the absence of its cognate sensor. Proc Natl Acad Sci U S A. 2013;110:16592–16597. doi: 10.1073/pnas.1315320110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusa AA, Gao J, Stringer V, Churchward G, Scott JR. Phosphorylation of the group A Streptococcal CovR response regulator causes dimerization and promoter-specific recruitment by RNA polymerase. Journal of bacteriology. 2006;188:4620–4626. doi: 10.1128/JB.00198-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heithoff DM, Shimp WR, House JK, Xie Y, Weimer BC, Sinsheimer RL, Mahan MJ. Intraspecies variation in the emergence of hyperinfectious bacterial strains in nature. PLoS pathogens. 2012;8:e1002647. doi: 10.1371/journal.ppat.1002647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollands A, Pence MA, Timmer AM, Osvath SR, Turnbull L, Whitchurch CB, Walker MJ, Nizet V. Genetic switch to hypervirulence reduces colonization phenotypes of the globally disseminated group A streptococcus M1T1 clone. J Infect Dis. 2010;202:11–19. doi: 10.1086/653124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horstmann N, Sahasrabhojane P, Saldana M, Ajami NJ, Flores AR, Sumby P, Liu CG, Yao H, Su X, Thompson E, Shelburne SA. Characterization of the effect of the histidine kinase CovS on response regulator phosphorylation in group A Streptococcus. Infection and immunity. 2015;83:1068–1077. doi: 10.1128/IAI.02659-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horstmann N, Saldana M, Sahasrabhojane P, Yao H, Su X, Thompson E, Koller A, Shelburne SA., 3rd Dual-site phosphorylation of the control of virulence regulator impacts group a streptococcal global gene expression and pathogenesis. PLoS pathogens. 2014;10:e1004088. doi: 10.1371/journal.ppat.1004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikebe T, Ato M, Matsumura T, Hasegawa H, Sata T, Kobayashi K, Watanabe H. Highly frequent mutations in negative regulators of multiple virulence genes in group A streptococcal toxic shock syndrome isolates. PLoS pathogens. 2010;6:e1000832. doi: 10.1371/journal.ppat.1000832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y, McLandsborough L, Kondagunta A, Cleary PP. C5a peptidase alters clearance and trafficking of group A streptococci by infected mice. Infection and immunity. 1996;64:503–510. doi: 10.1128/iai.64.2.503-510.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kal AJ, van Zonneveld AJ, Benes V, van den Berg M, Koerkamp MG, Albermann K, Strack N, Ruijter JM, Richter A, Dujon B, Ansorge W, Tabak HF. Dynamics of gene expression revealed by comparison of serial analysis of gene expression transcript profiles from yeast grown on two different carbon sources. Molecular biology of the cell. 1999;10:1859–1872. doi: 10.1091/mbc.10.6.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappeler KV, Anbalagan S, Dmitriev AV, McDowell EJ, Neely MN, Chaussee MS. A naturally occurring Rgg variant in serotype M3 Streptococcus pyogenes does not activate speB expression due to altered specificity of DNA binding. Infect Immun. 2009;77:5411–5417. doi: 10.1128/IAI.00373-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreikemeyer B, McIver KS, Podbielski A. Virulence factor regulation and regulatory networks in. Streptococcus pyogenes and their impact on pathogen-host interactions. Trends Microbiol. 2003;11:224–232. doi: 10.1016/s0966-842x(03)00098-2. [DOI] [PubMed] [Google Scholar]
- Lei B, DeLeo FR, Hoe NP, Graham MR, Mackie SM, Cole RL, Liu M, Hill HR, Low DE, Federle MJ, Scott JR, Musser JM. Evasion of human innate and acquired immunity by a bacterial homolog of CD11b that inhibits opsonophagocytosis. Nature medicine. 2001;7:1298–1305. doi: 10.1038/nm1201-1298. [DOI] [PubMed] [Google Scholar]
- Levin JC, Wessels MR. Identification of csrR/csrS, a genetic locus that regulates hyaluronic acid capsule synthesis in group A Streptococcus. Mol Microbiol. 1998;30:209–219. doi: 10.1046/j.1365-2958.1998.01057.x. [DOI] [PubMed] [Google Scholar]
- Li J, Liu G, Feng W, Zhou Y, Liu M, Wiley JA, Lei B. Neutrophils select hypervirulent CovRS mutants of M1T1 group A Streptococcus during subcutaneous infection of mice. Infection and immunity. 2014;82:1579–1590. doi: 10.1128/IAI.01458-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Diep BA, Villaruz AE, Braughton KR, Jiang X, DeLeo FR, Chambers HF, Lu Y, Otto M. Evolution of virulence in epidemic community-associated methicillin-resistant Staphylococcus aureus. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:5883–5888. doi: 10.1073/pnas.0900743106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Z, Zhang Y, Agrahari G, Chandrahas V, Glinton K, Donahue DL, Balsara RD, Ploplis VA, Castellino FJ. A natural inactivating mutation in the CovS component of the CovRS regulatory operon in a pattern D Streptococcal pyogenes strain influences virulence-associated genes. J Biol Chem. 2013;288:6561–6573. doi: 10.1074/jbc.M112.442657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman TD, Michel JB, Aingaran M, Potter-Bynoe G, Roux D, Davis MR, Jr, Skurnik D, Leiby N, LiPuma JJ, Goldberg JB, McAdam AJ, Priebe GP, Kishony R. Parallel bacterial evolution within multiple patients identifies candidate pathogenicity genes. Nature genetics. 2011;43:1275–1280. doi: 10.1038/ng.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Zhu H, Li J, Garcia CC, Feng W, Kirpotina LN, Hilmer J, Tavares LP, Layton AW, Quinn MT, Bothner B, Teixeira MM, Lei B. Group A Streptococcus secreted esterase hydrolyzes platelet-activating factor to impede neutrophil recruitment and facilitate innate immune evasion. PLoS pathogens. 2012a;8:e1002624. doi: 10.1371/journal.ppat.1002624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Trevino J, Ramirez-Pena E, Sumby P. The small regulatory RNA FasX controls pilus expression and adherence in the human bacterial pathogen group A Streptococcus. Mol Microbiol. 2012b;86:140–154. doi: 10.1111/j.1365-2958.2012.08178.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukomski S, Hoe NP, Abdi I, Rurangirwa J, Kordari P, Liu M, Dou SJ, Adams GG, Musser JM. Nonpolar inactivation of the hypervariable streptococcal inhibitor of complement gene (sic) in serotype M1 Streptococcus pyogenes significantly decreases mouse mucosal colonization. Infection and immunity. 2000a;68:535–542. doi: 10.1128/iai.68.2.535-542.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukomski S, Nakashima K, Abdi I, Cipriano VJ, Ireland RM, Reid SD, Adams GG, Musser JM. Identification and characterization of the scl gene encoding a group A Streptococcus extracellular protein virulence factor with similarity to human collagen. Infection and immunity. 2000b;68:6542–6553. doi: 10.1128/iai.68.12.6542-6553.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynskey NN, Goulding D, Gierula M, Turner CE, Dougan G, Edwards RJ, Sriskandan S. RocA truncation underpins hyper-encapsulation, carriage longevity and transmissibility of serotype M18 group A streptococci. PLoS pathogens. 2013;9:e1003842. doi: 10.1371/journal.ppat.1003842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynskey NN, Turner CE, Heng LS, Sriskandan S. A Truncation in the Regulator RocA Underlies Heightened Capsule Expression in Serotype M3 Group A Streptococci. Infection and immunity. 2015;83:1732–1733. doi: 10.1128/IAI.02892-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maripuu L, Eriksson A, Norgren M. Superantigen gene profile diversity among clinical group A streptococcal isolates. FEMS Immunol Med Microbiol. 2008;54:236–244. doi: 10.1111/j.1574-695X.2008.00469.x. [DOI] [PubMed] [Google Scholar]
- McArthur JD, Cook SM, Venturini C, Walker MJ. The role of streptokinase as a virulence determinant of Streptococcus pyogenes--potential for therapeutic targeting. Current drug targets. 2012;13:297–307. doi: 10.2174/138945012799424589. [DOI] [PubMed] [Google Scholar]
- McClure R, Balasubramanian D, Sun Y, Bobrovskyy M, Sumby P, Genco CA, Vanderpool CK, Tjaden B. Computational analysis of bacterial RNA-Seq data. Nucleic Acids Res. 2013;41:e140. doi: 10.1093/nar/gkt444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIver KS. Stand-alone response regulators controlling global virulence networks in streptococcus pyogenes. Contrib Microbiol. 2009;16:103–119. doi: 10.1159/000219375. [DOI] [PubMed] [Google Scholar]
- Mike LA, Choby JE, Brinkman PR, Olive LQ, Dutter BF, Ivan SJ, Gibbs CM, Sulikowski GA, Stauff DL, Skaar EP. Two-component system cross-regulation integrates Bacillus anthracis response to heme and cell envelope stress. PLoS pathogens. 2014;10:e1004044. doi: 10.1371/journal.ppat.1004044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller EW, Pflughoeft KJ, Sumby P. Reply to “A Truncation in the Regulator RocA Underlies Heightened Capsule Expression in Serotype M3 Group A Streptococci”. Infection and immunity. 2015;83:1734. doi: 10.1128/IAI.03162-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell ES. Frequency of serotypes of Streptococcus pyogenes in different diseases. J Clin Pathol. 1962;15:231–234. doi: 10.1136/jcp.15.3.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasser W, Beres SB, Olsen RJ, Dean MA, Rice KA, Long SW, Kristinsson KG, Gottfredsson M, Vuopio J, Raisanen K, Caugant DA, Steinbakk M, Low DE, McGeer A, Darenberg J, Henriques-Normark B, Van Beneden CA, Hoffmann S, Musser JM. Evolutionary pathway to increased virulence and epidemic group A Streptococcus disease derived from 3,615 genome sequences. Proc Natl Acad Sci U S A. 2014;111:E1768–1776. doi: 10.1073/pnas.1403138111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicol MP, Wilkinson RJ. The clinical consequences of strain diversity in Mycobacterium tuberculosis. Transactions of the Royal Society of Tropical Medicine and Hygiene. 2008;102:955–965. doi: 10.1016/j.trstmh.2008.03.025. [DOI] [PubMed] [Google Scholar]
- Perez N, Trevino J, Liu Z, Ho SC, Babitzke P, Sumby P. A genome-wide analysis of small regulatory RNAs in the human pathogen group A Streptococcus. PLoS One. 2009;4:e7668. doi: 10.1371/journal.pone.0007668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Pena E, Trevino J, Liu Z, Perez N, Sumby P. The group A Streptococcus small regulatory RNA FasX enhances streptokinase activity by increasing the stability of the ska mRNA transcript. Mol Microbiol. 2010;78:1332–1347. doi: 10.1111/j.1365-2958.2010.07427.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen M, Muller HP, Bjorck L. Protein GRAB of streptococcus pyogenes regulates proteolysis at the bacterial surface by binding alpha2-macroglobulin. J Biol Chem. 1999;274:15336–15344. doi: 10.1074/jbc.274.22.15336. [DOI] [PubMed] [Google Scholar]
- Smoot JC, Korgenski EK, Daly JA, Veasy LG, Musser JM. Molecular analysis of group A Streptococcus type emm18 isolates temporally associated with acute rheumatic fever outbreaks in Salt Lake City, Utah. Journal of clinical microbiology. 2002;40:1805–1810. doi: 10.1128/JCM.40.5.1805-1810.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steer AC, Law I, Matatolu L, Beall BW, Carapetis JR. Global emm type distribution of group A streptococci: systematic review and implications for vaccine development. The Lancet Infectious diseases. 2009;9:611–616. doi: 10.1016/S1473-3099(09)70178-1. [DOI] [PubMed] [Google Scholar]
- Stetzner ZW, Li D, Feng W, Liu M, Liu G, Wiley J, Lei B. Serotype M3 and M28 Group A Streptococci Have Distinct Capacities to Evade Neutrophil and TNF-alpha Responses and to Invade Soft Tissues. PLoS One. 2015;10:e0129417. doi: 10.1371/journal.pone.0129417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suerbaum S, Josenhans C. Helicobacter pylori evolution and phenotypic diversification in a changing host. Nature reviews Microbiology. 2007;5:441–452. doi: 10.1038/nrmicro1658. [DOI] [PubMed] [Google Scholar]
- Sumby P, Porcella SF, Madrigal AG, Barbian KD, Virtaneva K, Ricklefs SM, Sturdevant DE, Graham MR, Vuopio-Varkila J, Hoe NP, Musser JM. Evolutionary origin and emergence of a highly successful clone of serotype M1 group a Streptococcus involved multiple horizontal gene transfer events. J Infect Dis. 2005;192:771–782. doi: 10.1086/432514. [DOI] [PubMed] [Google Scholar]
- Sumby P, Whitney AR, Graviss EA, DeLeo FR, Musser JM. Genome-wide analysis of group a streptococci reveals a mutation that modulates global phenotype and disease specificity. PLoS pathogens. 2006;2:e5. doi: 10.1371/journal.ppat.0020005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Ringdahl U, Homeister JW, Fay WP, Engleberg NC, Yang AY, Rozek LS, Wang X, Sjobring U, Ginsburg D. Plasminogen is a critical host pathogenicity factor for group A streptococcal infection. Science. 2004;305:1283–1286. doi: 10.1126/science.1101245. [DOI] [PubMed] [Google Scholar]
- Syrovets T, Lunov O, Simmet T. Plasmin as a proinflammatory cell activator. Journal of leukocyte biology. 2012;92:509–519. doi: 10.1189/jlb.0212056. [DOI] [PubMed] [Google Scholar]
- Trevino J, Liu Z, Cao TN, Ramirez-Pena E, Sumby P. RivR is a negative regulator of virulence factor expression in group A Streptococcus. Infection and immunity. 2013;81:364–372. doi: 10.1128/IAI.00703-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevino J, Perez N, Ramirez-Pena E, Liu Z, Shelburne SA, 3rd, Musser JM, Sumby P. CovS simultaneously activates and inhibits the CovR-mediated repression of distinct subsets of group A Streptococcus virulence factor-encoding genes. Infection and immunity. 2009;77:3141–3149. doi: 10.1128/IAI.01560-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlemann AC, Otto M, Lowy FD, DeLeo FR. Evolution of community- and healthcare-associated methicillin-resistant Staphylococcus aureus. Infection, genetics and evolution : journal of molecular epidemiology and evolutionary genetics in infectious diseases. 2014;21:563–574. doi: 10.1016/j.meegid.2013.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems RJ, Hanage WP, Bessen DE, Feil EJ. Population biology of Gram-positive pathogens: high-risk clones for dissemination of antibiotic resistance. FEMS Microbiol Rev. 2011;35:872–900. doi: 10.1111/j.1574-6976.2011.00284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmers MJ, Cunliffe AC, Williams RE. Type-12 streptococci associated with acute haemorrhagic nephritis. Lancet. 1954;267:17–18. doi: 10.1016/s0140-6736(54)90004-5. [DOI] [PubMed] [Google Scholar]
- Yang L, Jelsbak L, Marvig RL, Damkiaer S, Workman CT, Rau MH, Hansen SK, Folkesson A, Johansen HK, Ciofu O, Hoiby N, Sommer MO, Molin S. Evolutionary dynamics of bacteria in a human host environment. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:7481–7486. doi: 10.1073/pnas.1018249108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, Ishigaki Y, Takizawa A, Moro K, Kishi Y, Takahashi T, Matsui H. Comparative Genomics of the Mucoid and Nonmucoid Strains of Streptococcus pyogenes, Isolated from the Same Patient with Streptococcal Meningitis. Genome Announc. 2015;3 doi: 10.1128/genomeA.00221-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinkernagel AS, Timmer AM, Pence MA, Locke JB, Buchanan JT, Turner CE, Mishalian I, Sriskandan S, Hanski E, Nizet V. The IL-8 protease SpyCEP/ScpC of group A Streptococcus promotes resistance to neutrophil killing. Cell host & microbe. 2008;4:170–178. doi: 10.1016/j.chom.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.