Abstract
RNA-guided gene drives capable of spreading genomic alterations made in laboratory organisms through wild populations in an inheritable way could be used to control populations of organisms that cause environmental and public health problems. However, the possibility of unintended genome editing through the escape of strains from laboratories, coupled with the prospect of unanticipated ecological change, demands caution. We report the efficacy of CRISPR-Cas9 gene drive systems in wild and laboratory strains of the yeast Saccharomyces cerevisiae. Furthermore, we address concerns surrounding accidental genome editing by developing and validating methods of molecular confinement that minimize the risk of unwanted genome editing. We also present a drive system capable of overwriting the changes introduced by an earlier gene drive. These molecular safeguards should enable the development of safe CRISPR gene drives for diverse organisms.
Synthetic gene drive systems have the potential to address diverse ecological problems by altering the traits of wild populations. These genetic elements spread not by improving the reproductive fitness of the organism, but by increasing the odds that they themselves will be inherited. Because this inheritance advantage can overcome the fitness costs associated with the drive itself or adjacent genes carried with the drive, such genetic elements are theoretically capable of ‘driving’ unrelated traits through populations over many generations1.
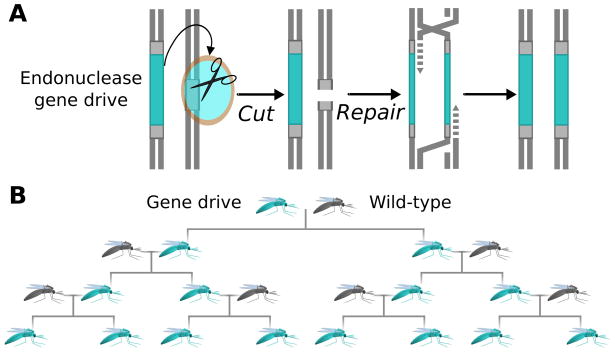
Inheritance-biasing is a common strategy in nature2. One elegant class of inheritance-biasing genes spreads by cutting homologous chromosomes that do not contain them, thereby inducing the cellular repair process to copy them onto the damaged chromosome by homologous recombination (Fig. 1A). This process is known as ‘homing’3. The best-characterised homing endonuclease gene is I-SceI, whose product cuts the gene encoding the large rRNA subunit of S. cerevisiae mitochondria. Most homing endonucleases are extremely efficient, for example, I-SceI is correctly copied 99% of the time4.
Figure 1. Mechanism and population-level effect of endonuclease gene drives.
(A) Homing endonucleases cut competing alleles, inducing the cell to repair the damage by copying the endonuclease gene. (B) By converting heterozygous germline cells into homozygotes containing two copies (teal), gene drives increase the odds that they will be inherited and consequently spread themselves and associated changes through wild populations (grey). Reproduced from 1.
Austin Burt suggested in 2003 that homing endonucleases might form the basis of synthetic gene drives that could alter wild populations of sexually reproducing organisms (Fig. 1B)5. The I-SceI endonuclease gene was subsequently demonstrated to exhibit homing in transgenic laboratory populations of mosquitoes6 and fruit flies7,8 in which an I-SceI recognition site was inserted into the recipient strain at the same locus as the I-SceI gene in the donor strain.. However, the difficulty of retargeting homing endonucleases to cleave specific sequences in wild-type genomes has limited their utility for synthetic gene drive elements 9.
CRISPR-Cas9 which cleaves target sequences specified by single ‘guide RNA’ (sgRNA) molecules, has facilitated attempts to edit the genomes of diverse species10–17. We previously outlined the potential for CRISPR-Cas9 RNA-guided gene drives to alter wild populations. We also described molecular confinement methods robust to human error1 that could be used with such systems and called for public discussions and regulatory reform18. Here we report the validation of our hypotheses by constructing multiple CRISPR-Cas9 RNA-guided gene drive systems for use in S. cerevisiae. The ability of these gene drives to bias inheritance, and the effectiveness of confinement measures, are quantified, and we also present a method to overwrite and restore edited traits.
Results
Before starting our experiments, we established an experimental set-up to minimize the risk of synthetic gene drive elements escaping from the laboratory and altering wild yeast populations. All of our studies used a barrier protocol to reduce contact with wild yeast. The rarity of sexual reproduction in wild S. cerevisiae provides an additional, natural obstacle to synthetic gene drives in the wild (Supplementary Note). Most importantly, all experiments used one of two forms of molecular confinement, allowing us to test the efficacy of this form of safeguard.
Molecularly confined sgRNA-only gene drives
For our initial experiments, we used a form of molecular confinement1 in which the Cas9 based gene drive system was split into two physically separate parts: an episomally encoded Cas9 gene and a gene drive element encoding a guide RNA inserted into the targeted genomic locus. This allowed us to avoid creating a self-sufficient inheritance-biasing cassette while enabling homing in wild-type yeast strains. This simple form of molecular confinement is not vulnerable to human error because even if drive-containing yeast were to escape into the wild, the required and relatively unstable Cas9 episomal plasmid would rapidly be segregated away from the sgRNA-only drive element (Supplementary Fig. 1), thereby preventing the drive from spreading exponentially.
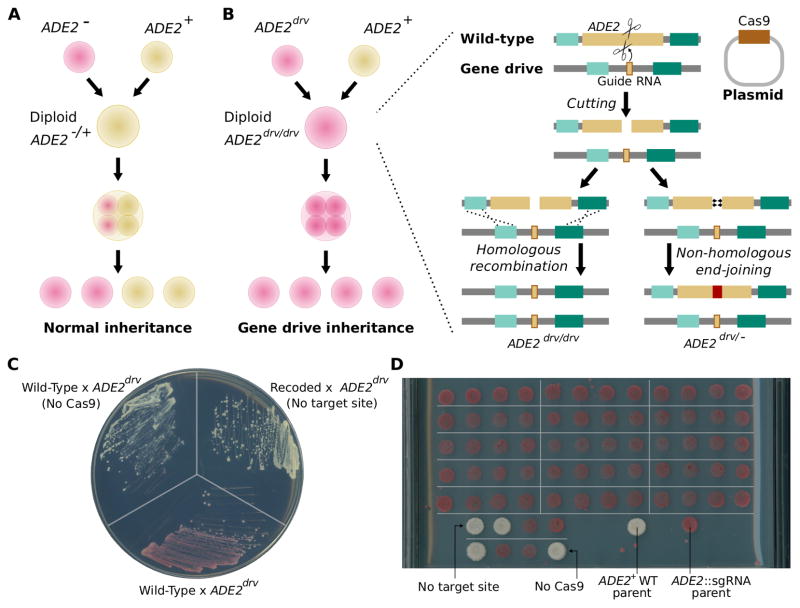
To measure the efficiency of RNA-guided gene drives in yeast, we used the ADE2 gene encoding phosphoribosylaminoimidazole carboxylase as a visual marker19. Cells wild-type for ADE2 are cream colored while ade2Δ mutants are red. If red ade2Δ haploids are mated with cream-colored wild-type haploids, the resulting heterozygous diploids inherit one functional copy of ADE2 and are cream-colored. When these diploids undergo meiosis and reproduce via sporulation, half the resulting haploids inherit the mutated copy and are red; the other half inherit the intact, unmutated copy and are cream-colored (Fig. 2A).
Figure 2.
Biased inheritance of an ADE2 gene drive element is readily visible in S. cerevisiae. (A) Mutations in ADE2 generate a red phenotype on adenine-limiting media due to the buildup of red pigments. Mating a red mutant haploid to a wild-type haploid produces cream-colored diploids, which yield 50% red and 50% cream-colored progeny upon sporulation. (B) When haploids with a gene drive element targeting ADE2 mate with wild-type haploids in the presence of Cas9, cutting and subsequent replacement or disruption of ADE2 produces red diploids that upon meiosis yield exclusively red progeny. (C) Diploids produced by mating wild-type and ade2::sgRNA gene drive haploids yield cream-colored colonies in the absence of Cas9 or when the target site is removed by recoding but uniformly red colonies when both are present, demonstrating Cas9-dependent disruption of the wild-type ADE2 copy. (D) Spores from 15 dissected tetrads produce uniformly red colonies on adenine-limited plates, confirming disruption of the ADE2 gene inherited from the wild-type parent. In the absence of the target site or Cas9, normal 2:2 segregation is observed.
If the red haploids encode a functional gene drive system knocked into the ADE2 locus and designed to target the wild-type ADE2 sequence are mated to wild-type haploids, the drive will cut and replace the intact ADE2 locus that is inherited from the wild-type parent, yielding red diploids. Following meiosis, all haploid progeny will inherit one of the two gene drive alleles and will also be red (Fig. 2B). Thus, the cutting efficiency of a gene drive system that replaces ADE2 can be assessed by mating drive-containing haploids with wild-type haploids, and quantifying the fraction of diploid cells that are red.
We built a split CRISPR-Cas9 gene drive system as described above by inserting a guide RNA targeting the wild-type ADE2 gene into the wild-type ADE2 locus such that ADE2 function was disrupted and the target site removed. We mated these red ade2::sgRNA haploids to wild-type yeast of the opposite mating type in the presence or absence of the episomal Cas9 plasmid and examined the color of the resulting diploid colonies to check for gene drive (Fig 2C). More than 99% of diploid colonies were red when the Cas9 plasmid was present, indicating highly efficient cutting of the ADE2 copy inherited from the wild-type parent. As expected, we did not observe any red diploid colonies in the absence of Cas9 encoding and validating the effectiveness of sgRNA-only drive confinement.
To verify that the ADE2 alleles from drive-containing diploids were disrupted, mated diploids were sporulated and haploid progeny were isolated and examined. Upon dissecting 18 cas9+ diploids, we observed a perfect Mendelian 4:0 ratio of red:cream haploids, confirming that all WT copies of the ADE2 locus were disrupted. In contrast, 18 cream-colored cas9− diploids yielded a 2:2 red:cream ratio, indicating normal Mendelian inheritance of the inactivated drive and the wild-type ADE2 allele (Fig. 2D).
To determine whether ADE2 disruptions in red diploids were the result of successful copying of the drive element, we sequenced all 72 red haploids that we derived from dissected cas9+ diploids. All sequenced haploid colonies contained intact sgRNA-only drive elements without additional mutations. These results indicate that the ade2::sgRNA drive element efficiently cut and replace the native ADE2 locus upon mating with wild-type haploids. Homing took place only in the presence of the unlinked episomal Cas9 plasmid, demonstrating that split gene drive elements cannot spread in the absence of the non-driving element.
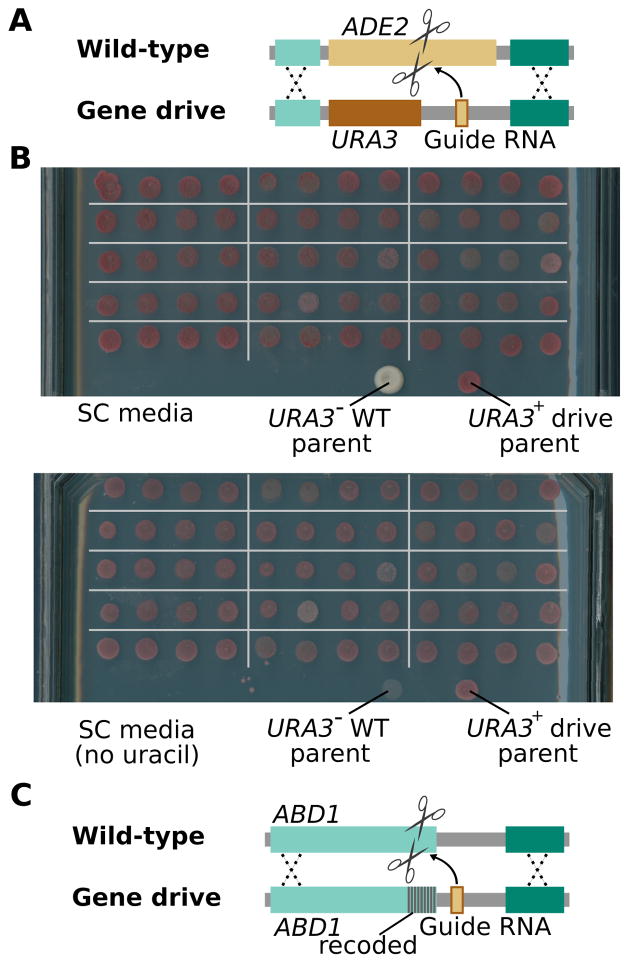
We next tested whether Crispr-Cas9 gene drive systems could be designed to bias the inheritance of not only a minimal drive element, but also any closely associated “cargo” gene(s) whose spread through an existing population might be desirable. As a proof of principle, we inserted the URA3 gene in cis with the ade2::sgRNA drive element. URA3 allows laboratory modified yeast strains to grow in the absence of uracil supplementation (Fig. 3A). We mated URA3-containing drive haploids with wild-type haploids in the presence of an episomal Cas9 plasmid, the resulting diploids (all of which were red) were sporulated and the resulting tetrads were dissected and the phenotype of the resulting haploid colonies was examined. As was the case for the ADE2 gene drive, all of the dissected haploid cells formed red colonies, indicative of biased inheritance. Crucially, all haploids grew without uracil in the growth medium, which demonstrates that URA3 was efficiently copied along with the ADE2 drive (Fig. 3B).
Figure 3.
Gene drives and cargo genes remain intact upon copying and can spread by targeting both non-essential and essential genes. (A) The ADE2-targeting gene drive was modified to carry URA3 as a cargo gene. (B) Diploids produced by mating wild-type URA3− haploid yeast with haploids encoding the gene drive carrying URA3 were sporulated and tetrads dissected to isolate colonies arising from individual spores. Pictures are spores from 15 of these tetrads. All grew when replica-plated onto plates lacking uracil, demonstrating that the drive successfully copied URA3 in all diploids. (C) Depiction of a gene drive designed to cut and recode the 3′ end of the essential ABD1 gene.
We next sought to determine whether CRISPR-cas9 gene drive systems can recode an essential gene but leave its function intact. Especially when targeting multiple sites, such a drive element should be more evolutionarily stable since its target locus cannot readily acquire mutations that prevent homing without disrupting the function of the essential gene and thus compromising viability1. We designed and synthesized a split drive element in which a guide RNA targeting the essential ABD1 gene was encoded adjacent to a recoded version of the same gene (Fig. 3C)20 and mated this haploid strain, with wild-type cells containing the Cas9-expressing episomal plasmid. Isolated diploid cells were sporulated and 18 of the resulting tetrads dissected. The 72 resulting segregants were allowed to form colonies before being sequenced at the ABD1 locus. All 72 haploids contained the drive element and recoded ABD1 locus. The strains with the drive element and recoded ABD1 locus did not exhibit any obvious fitness defects as compared to wild-type strains, supporting the use of essential gene recoding as a potential strategy to build evolutionarily stable CRISPR-Cas9 gene drives.
Gene drive efficacy in diverse genetic backgrounds
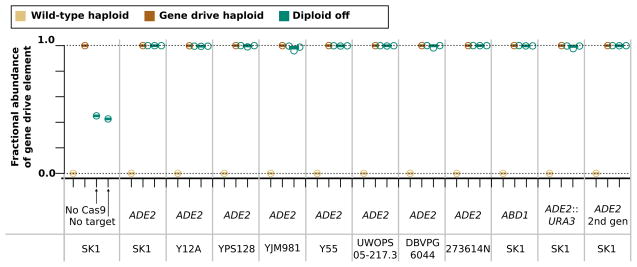
Gene drive efficacy may differ for many reasons, including but not limited to the types of repair machinery available to the cell at the time of the cut, the chromatin status of the locus, and the degree of homology between the sequences flanking the gene drive and the targeted locus. To test gene drive efficacy in different genetic backgrounds, we mated ADE2 drive-containing haploids with 6 phylogenetically and phenotypically diverse wild-type strains of S. cerevisiae. Rather than relying on visual markers of genome editing, we used quantitative PCR on populations of the resulting diploids using one set of primers specific to the drive element and another set designed to amplify either wild-type alleles or those disrupted by non-homologous end-joining.
The mean fraction of diploid chromosomes containing the ADE2 gene drive element was more than 99% regardless of wild-type parent strain used (Fig. 4), attesting to the robustness of the drive in diverse backgrounds. The copying efficiency was identical for the URA3-carrying gene drive element and the drive element that targeting and recoding the essential ABD1 gene, suggesting that CRISPR-Cas9 gene drives are robust to variance in genetic background, cargo size, and target site selection. In addition, the drive-containing haploid progeny of an ADE2 gene drive exhibited equivalent copying efficiency in a subsequent cross to the original parental wild-type yeast strain, demonstrating that the drive construct is stable during homing (Fig. 4).
Figure 4.
Quantitative PCR demonstrates highly efficient inheritance biasing by split drives across diverse yeast strains in the presence of Cas9. Results depict the relative abundance of wild-type and drive-containing alleles in diploids arising from matings between SK1 haploids bearing gene drives and diverse wild-type haploid strains. “No Cas9” and “No Target” refer to haploid cells containing the ADE2 drive mated to wild-type haploids in the absence of Cas9 or to an otherwise wild-type strain with Cas9 that has a mutation in the targeted sequence that blocks cutting. “2nd gen” refers to the haploid progeny of an earlier mating. Data points are from independent cultures or mating events and represent the mean of 3 technical replicates.
Molecularly confined autonomous gene drives
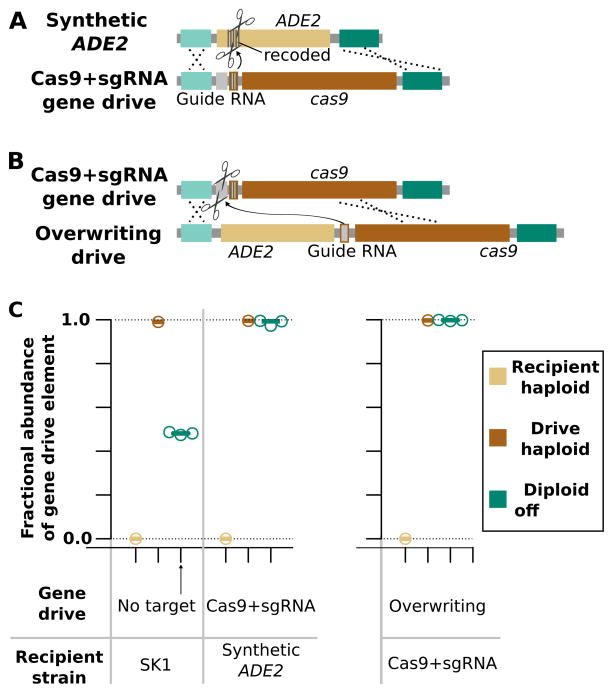
Our first experiments used a split-drive molecular confinement strategy in which Cas9 was encoded on an unlinked episomal plasmid and the gene drive element contained only the sgRNA. Because the required Cas9 gene is unlinked from the drive and wild yeast populations do not encode Cas9, the sgRNA-only drive is unable to spread in wild organisms lacking Cas9. In contrast, an autonomous drive element capable of propagating in wild populations would encode the Cas9 gene and sgRNA in cis. This ~5kb genetic construct is much larger than the ~500 bp sgRNA-only system that we had observed driving at a rate >99%. We hypothesized that the larger autonomous drive cassette might be copied less efficiently than the smaller sgRNA-only split-drive cassette. To safely test an autonomous Cas9+sgRNA drive under molecular confinement, we created a recipient yeast strain with a recoded ADE2 gene containing a synthetic DNA sequence that is not present in wild populations. We subsequently built a Cas9+sgRNA gene drive construct targeting the synthetic sequence such that homing will disrupt the ADE2 gene. Diploids produced by crossing autonomous drive-carrying haploids and wild-type haploids were universally red. Quantitative PCR analysis of populations of diploid colonies confirmed that the Cas9+sgRNA drive system was copied at an average efficiency of more than 99%, demonstrating that inclusion of the large cas9 gene in cis does not impede gene drive function in yeast (Fig. 5B). Crucially, crossing drive-carrying haploids to wild-type cells lacking the synthetic target sequence did not yield detectable homing, demonstrating the efficacy of synthetic site targeting as a molecular confinement strategy.
Figure 5.
Gene drives can be safely tested by targeting synthetic sites and drive-spread phenotypic changes reversed with a subsequent gene drive. (A) An autonomous Cas9+sgRNA gene drive that cuts and replaces the recoded ADE2 gene. (B) Quantitative PCR results depicting the relative abundance of wild-type and drive-containing alleles in diploids arising from matings between SK1 haploids bearing the above gene drive and wild-type SK1 yeast. Data points are from independent cultures (n=3 technical replicates). (C) A drive that cuts the autonomous drive and restores ADE2. (D) Quantitative PCR results for diploids arising from matings between SK1 haploids bearing the ADE2-disrupting and ADE2-restoring gene drives.
Reversibility of driven genome edits but not transgenesis
We next sought to determine whether the loss of ADE2 function induced by this Cas9+sgRNA drive might be corrected using another gene drive to overwrite the earlier change1. In principle, such an overwriting drive can restore the original phenotype - in this case the loss of ADE2 - but the resulting organisms will still be transgenic due to the presence of a residual Cas9 gene and guide RNA. We designed a Cas9+sgRNA gene drive element (~7 kb) to cleave a synthetic sequence contained within the first ADE2-disrupting drive and restore an intact copy of the ADE2 gene upon homing (Fig. 5C). Crosses between ADE2-disrupting drive haploids and ADE2-restoring drive haploids yielded all white diploid colonies. Quantitative PCR analysis of diploids (Fig. 5C) showed that the ADE2-restoring drive overwrote the earlier drive at >99% efficiency. Finally, when diploid colonies were sporulated and the phenotype of their haploid progeny examined all cells showed restoration of ADE2 function
Discussion
We have shown that CRISPR-Cas9 gene drives can bias inheritance in diverse wild yeast strains over successive generations at efficiencies over 99%. A previous report describing a method to produce biallelic loss-of-function mutations using a CRISPR-Cas9 gene drive construct in the fruit fly Drosophila melanogaster exhibited 97% copying efficiency, with noteworthy implications for potential applications in mosquitoes21. Although it is impossible to generalize from two studies, that neither study employed any optimization suggests that RNA-guided gene drives may function in many of the sexually reproducing organisms that have to date been engineered with Cas9.
Our potential ability to construct synthetic gene drive elements in multiple species demands caution. Scientists share a fundamental obligation to ensure that the products of laboratory experiments are confined to the laboratory. Because a single escaped organism carrying a synthetic CRISPR-Cas9 gene drive system could alter a substantial fraction of the wild population with unpredictable ecological consequences the decision to deploy a gene drive must be made collectively by society. Any accidental release could severely damage public trust in scientists, transgenics, and the field of gene drive research. It is therefore imperative for scientists performing experiments with gene drive constructs to use stringent confinement measures in order to minimize the risk of accidentally altering wild populations.
Barrier protocols are the traditional confinement measure used in laboratories working with dangerous organisms. However, human pathogen research has conclusively demonstrated that barrier protocols are vulnerable to human error and will eventually fail22. The CRISPR-Cas9 gene drive construct tested in D. melanogaster relied exclusively on a barrier protocol, causing concerns that led to recommendations that future experiments employ additional confinement strategies 23.
We report two easy-to-use methods of molecular confinement that are robust to human error. For example, studies requiring cutting or replacement of wild-type genes could use a sgRNA-only split drive element in a Cas9-expressing strain. Although the episomal Cas9 plasmid used in this study cannot be used in all eukaryotic species, there is no known reason why a chromosomally-encoded copy of the Cas9 gene could not be used for split-drive molecular confinement. Strains in which chromosomally-integrated Cas9 is expressed under one of several of different housekeeping and germline promoters are available in multiple species24–26. Similarly, experiments intended to study intact Cas9+sgRNA gene drives in model systems could exclusively target synthetic sequences that are only present in laboratory populations.
We recommend that future gene drive experiments use at least one form of molecular confinement unless they aim to create a candidate for eventual release. A combination of molecular and barrier confinement may suffice in yeast, the nematode worm Caenorhabditis elegans, and other organisms that rarely reproduce sexually because gene drives in such organisms can impose only a tiny fitness cost if they are to spread (Supplementary Note). That said, natural endonuclease gene drives such as I-SceI do exist in yeast. Whether CRISPR-cas9 gene drives will constitute this level of burden is as yet unknown. We openly acknowledge that the barrier protocol used for the experiments in this study should have been stronger; we now perform such experiments using a more stringent protocol involving biological containment hoods.
Gene drive experiments in species that always reproduce sexually pose a greater risk of spread if any organisms should escape from the laboratory. We therefore suggest that gene drive experiments in sexually-reproducing organisms use multiple confinement measures that are not vulnerable to human error. For example, laboratories might use both forms of molecular confinement (an sgRNA-only drive element that targets a synthetic sequence), molecular as well as ecological confinement (in which experiments take place in geographic areas where the organism in question cannot survive or find mates)1, or molecular as well as reproductive confinement (the use of genetic backgrounds that are entirely unable to reproduce with wild-type organisms, such as Drosophila strains with compound autosomes27). These precautions are most important in flying insects and other species that most easily escape physical confinement.
Even scientists not intending to work with gene drives should consider taking precautions, since any unintended insertion of the cas9 gene and guide RNAs near a targeted site could produce a gene drive element. Fortunately, a simple and free precaution is available and already used for different reasons by many laboratories, namely, researchers avoid delivering the Cas9 gene on a DNA cassette that also encodes a guide RNA. As we have shown, guide RNAs alone cannot bias inheritance in the absence of Cas9 and therefore cannot spread through wild populations (Fig. 4).
Our results lay the groundwork for future efforts to design highly efficient and stable gene drives intended for real-world applications. These experiments could initially create and test sgRNA-only gene drive systems, but must eventually create a gene drive capable of spreading in the wild, at which point they are likely limited to ecological and barrier confinement methods. Our demonstration that a subsequently released gene drive can correct changes made by an earlier gene drive constitutes an additional safeguard: in the event of a premature release or unanticipated ecological effect, the override drive could mitigate the damage. We recommend that ongoing gene drive efforts consider constructing an appropriate corrective drive element in parallel with the candidate gene drive construct. In addition, the success of our ABD1 gene drive shows that it is feasible to target and recode genes important for fitness, a strategy that may improve the evolutionary stability of gene drive systems1.
More generally, our findings suggest that yeast could be a useful platform for rapid testing of CRISPR gene drives before moving them into multicellular organisms. The power of yeast genetics and the ease of genome manipulation could facilitate combinatorial investigations into gene drive optimization. For example, studies might explore how biasing repair pathway choice28 affects the efficiency of copying for gene drives of various sizes. Because the factors involved in these pathways are broadly conserved, these experiments could guide gene drive optimization in other organisms29.
Lastly, the confinement strategies we report will enable researchers to exploit the power of gene drives for facile genome engineering. For example, mating libraries of organisms in which sgRNA-only cassettes have been used to replace non-essential target genes could quickly generate all viable combinations of knockouts to better understand interaction networks and potentially identify therapeutic relevant pathways pathogenic organisms. Furthermore, the successive combinatorial knockouts created by our outlined gene drive approach could eventually enable the identification of candidate minimal genomes.
In conclusion, we hope that reversible and molecularly confined gene drives in yeast will inform, safeguard, and accelerate efforts to build CRISPR-Cas9 gene drives in other organisms. Given the considerable potential for this technology to address global problems in health, agriculture and conservation, these results underscore the urgent need for formal guidelines specifying requisite safeguards, inclusive public discussions, and regulatory reform18 in order to build a reliable foundation for future humanitarian applications.
Online Methods
Physical Confinement of Gene Drives
All experiments were carried out using a barrier protocol to supplement the molecular confinement strategies. Plates and tubes containing gene drive strains were kept in separate areas of bench surfaces, cold room surfaces and freezer space. All plates were secures with Parafilm and bench surfaces were cleaned daily using a 70% ethanol solution. Exposed glassware was first soaked for 24 hours in Wescodyne solution before cleaning. All waste was bagged in biohazard specific bags, sealed and incinerated. Future experiments will use a stricter barrier protocol in which all work is carried out in a biological hood as a further precaution.
Plasmids and genomic cassettes
Gene drive cassettes were synthesized from gBlocks (Integrated DNA Technologies, Coralville, IA) and inserted into SK1 cells via Cas9-mediated genome modification as follows. Guide RNAs for each drive were cloned into p416-Cas9 containing plasmids with expression driven by the SNR52 promoter16. 60 base pair homology arms to the target locus were added on both ends of the gene drive cassette via PCR and 5 ug of PCR product was co-transformed with the p416-Cas9-gRNA plasmids. Correctly integrated gene drives were verified by sequencing and p416-Cas9-gRNA plasmids were cured using 5-Fluoroorotic Acid (5-FOA) selection.
To create the URA3-containing ADE2 gene drive, the ADE2 gene drive was cloned next to the Candida albicans URA3 gene in the pAG60 plasmid. The entire URA3 cassette and gene drive were PCR amplified and inserted using Cas9-mediated genome modification into the ADE2 locus of haploid SK1 cells.
The recoded C-terminus of the ABD1 gene and corresponding gene drive were synthesized as a gBlock to remove homology and generate mutations in the seed sequence via synonymous changes. The TEF1 terminator was inserted at the 3′ end of the recoded ABD1 gene between the gene and the gRNA as ABD1 shares a terminator with the VHC1 gene. The entire cassette was integrated into the haploid SK1 genome using Cas9-mediated genome modification.
The ADE2 gene was recoded by cotransforming a double stranded oligonucleotide and a p416 plasmid containing Cas9 and a gRNA targeting the ADE2 region to recode. The oligonucleotide silently recoded the ADE2 gene and included an orthogonal target and PAM sequence. The complete gene drive (Cas9 and gRNA, targeting the recoded ADE2 gene) was generated by cloning a gRNA into the p416-Cas9 plasmid. An orthogonal genomic target was also included in the complete gene drive to later be targeted by the reversal drive. The Cas9 and gRNA linear construct was amplified by PCR using the same homology arms as the sole gRNA gene drive construct. The construct was co-transformed into By4723 cells with the plasmid it was amplified from and the cells were selected for uracil prototrophy. Correct integrations were screened via colony PCR. This plasmid was later removed using 5-FOA.
The overwriting drive (Cas9 and gRNA integrated upstream of the ADE2 gene) was generated by cloning an alternately encoded gRNA into a p414 plasmid containing Cas9. This alternatively endoded gRNA contains less homology to previously used gRNAs and would reduce the chance of unwanted recombination when used to replace the complete gene drive. This gRNA targets a 20bp region inserted with the complete gene drive. The TRP1 gene with the Cas9 and gRNA were PCR amplified with homology arms to the 5′ region of the ADE2 and the product was transformed into SK1 A cells. Cells were selected for tryptophan prototrophy and screened via PCR for correct integrations.
The p416-Cas9-gRNA plasmid (conferring uracil prototrophy) is a variant of the previously described p414-Cas9-gRNA plasmid (conferring tryptophan prototrophy)16 (Addgene #43802). One or the other was used in each mating experiment. The pRS413 vector was transformed into select cell types to confer histidine prototrophy as a marker to select for diploid cells.
Strain Genotypes:
| Strain | Genotype |
|---|---|
| SK1A | MATa ho::LYS2 lys2 ura3 leu2::hisG his3::hisG trp1::hisG |
| SK1 α | MATα ho::LYS2 lys2 ura3 leu2::hisG his3::hisG trp1::hisG |
| Y12A | MATa ho::HygMX ura3::KanMX |
| YPS128 | MATa ho::HygMX ura3::KanMX |
| YJM981 | MATa ho::HygMX ura3::KanMX |
| Y55 | MATa ho::HygMX ura3::KanMX |
| UWOPS05-217-3 | MATa ho::HygMX ura3::KanMX |
| DBVPG6044 | MATa ho::HygMX ura3::KanMX |
| 273614N | MATa ho::HygMX ura3::KanMX |
Yeast mating experiments
Haploid drive-containing SK1 yeast and haploid wild-type strains of the opposite mating type were mixed in equal amounts in YPAD liquid media and incubated overnight. The resulting diploids were washed in sterile water and plated on selective media for both parental genotypes. Specific crosses and selection conditions are detailed in Supplementary Table 1.
Sporulation and tetrad dissection
After mating in liquid YPAD and selection for diploids on selection plates, the selection plates were scraped into 10 mL selective media and grown overnight at 30°C. A fresh 5 mL YPAD culture was then inoculated to and OD=0.1 and grown 4–5 hours at 30°C. The entire culture was then washed twice in 10 mL water, inoculated into 2 mL of sporulation media(1% potassium acetate), and incubated at room-temperature for 3 days or until spores were visible. Sporulated cells were suspended in 50 μL of a stock solution of zymolyase (50 μg/mL in 1M sorbitol) and incubated at 30C for 5 minutes, transferred to ice, diluted with 150 μL cold H2O, microdissected using a Zeiss tetrad dissection microscope, and isolated spores grown on YPAD plates.
Selection for URA3 function
Dissected spores were grown in synthetic complete (SC) media and then spotted onto SC medium as well as SC medium without uracil. To enhance red color, all SC solid media used for plate images contained 0.5 X adenine hemisulfate (final concentration of 0.08 mM).
Verification of chromosomal segregation
Three genes on chromosome 15 flanking the ADE2 gene were sequenced in two dissected tetrads of the complete gene drive cross. VAM3, TRS33, and DPP1 were amplified using PCR from colonies and sequenced using Sanger sequencing to verify that homologous recombination copied only the gene drive cassette rather than the entire chromosome.
Measuring mitotic plasmid loss
BY4722 cells containing p416-Cas9 plasmids were grown overnight in SC-uracil media. 50 μl of saturated cultured was then used to inoculate 10 mL cultures of YPAD. The YPAD cultures were grown overnight and then plated. Overnight dilutions were plated on YPAD and 5-FOA plates and the difference in colony counts between the two medias were used to determine plasmid loss rate. To determine the total number of population doublings after overnight growth cell counts before overnight growth and after overnight growth were measured.
Measuring meiotic plasmid loss
A URA3 gene placed under a haploid specific promoter (STE5) was used to select for diploids and haploids. SK1α ho::pSTE5-CaURA3 ade2::gRNA containing p413-Cas9 plasmid was mated with SK1 A ho::pSTE5-CaURA3. After mating in YPAD, cells were passaged an additional day in YPAD media. Diploids were then selected for in SC-histidine+5-FOA. After diploid selection, cells were grown to log phase in YPAD and then placed in sporulation media (2% Potassium Acetate) and allowed to sporulated for 3 days. After visual confirmation of asci formation, asci were disrupted using 0.5 mg/mL zymolyase (20-T) for 20 minutes and then vortexing for 5 minutes. Disrupted sporulated cultures were then plated to either SC-uracil (haploid selection) or SC-uracil-histidine (haploid+Cas9 plasmid selection) and the difference between colony numbers was calculated.
Quantitative PCR
Candidate primer pairs were designed to amplify short regions specific to each drive or the wild-type sequence replaced by the drive, as well as the ACT1 gene as a control. All sequences are included in the supplementary information. Genomic DNA was extracted using Method A as described in Looke et al.29
KAPA SYBR FAST qPCR Master Mix (2X) was used to perform the qPCR reaction along with 25 ng of genomic DNA. The amplification efficiency and relative specificity of each primer pair were measured by amplifying dilutions of genomic DNA from wild-type and drive haploids, respectively, and the best-performing and well-matched pairs selected for use (see below for all primers used). Quantitative PCR reactions were performed on genomic DNA isolated from each parental haploid as well as from diploids arising from three independent mating events. Three reactions (technical replicates) were performed per sample on a LightCycler 96 machine by Roche.
Calculations
Results from three technical replicates were averaged for calculations. In order to directly calculate the ratio of alleles before PCR amplification, we first determined the efficiencies of the different primer pairs. Efficiencies were calculated from qPCR runs of serial dilutions (6 orders of magnitude) as:
R2 values were higher than 0.99 in all cases except for one pair (ade2::URA3+sgRNA).
The allelic ratios were calculated as:
with
xa and xb being the initial concentration of drive and wt DNA,
Ea and Eb the efficiency of the respective primer pairs and
Ct,a and Ct,b the Ct values for each sample.
Figures 4 and 5 were generated using BoxPlotR30.
Raw data and calculations are available in the Supplementary Data.
Sequencing traces
Trace files for sequences corresponding to the spores of the 18 tetrads dissected for the ADE2 KO gene drive and the ABD1 recoding gene drive were uploaded to the NCBI Trace Archive with TI numbers 2342918196-2342918339. Templates corresponding to expected sequences used for alignments are available as GenBank Accession KT876200 (ADE2 KO) and KT876201 (ABD1 recoding).
Oligonucleotides
Sequences of oligonucleotides used in the study (Integrated DNA Technologies, Coraville IA) are listed in Supplementary Table 2.
Supplementary Material
Acknowledgments
We are very grateful to Steve Doris, Dan Spatt and Fred Winston for their incredible patience, generosity and expertise in tetrad dissection. We also thank Fred Winston for providing us with SK1 strains and members of the Church laboratory for insightful discussions.
This work was supported by grants from the DOE (DE-FG02-02ER63445 to G.M.C.), NSF (SynBERC SA5283-11210 and MCB-1330914 to G.M.C.), NCI (5T32CA009216-34 to A.C.), NIDDK (1K99DK102669-01 to K.M.E.), and the Wyss Institute for Biologically Inspired Engineering (Technology Development Fellowship to K.M.E.).
Footnotes
Author contributions:
S.L.D. initiated the study; J.E.D., A.C., S.L.D., and K.M.E. designed the experiments; J.E.D. performed the experiments with assistance from A.C.; J.E.D., A.C., S.L.D., and K.M.E. analyzed the data; and K.M.E. wrote the paper with A.C. and contributing input from J.E.D., S.L.D., and G.M.C.
Competing financial interests statement:
K.M.E. and G.M.C. are authors of a patent filed on CRISPR gene drive (PCT/US2015/010550).
K.M.E. is author of a provisional patent filed on CRISPR gene drive (serial no. 62/236,545).
References
- 1.Esvelt KM, Smidler AL, Catteruccia F, Church GM. Concerning RNA-guided gene drives for the alteration of wild populations. eLife. 2014:e03401. doi: 10.7554/eLife.03401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burt A, Trivers R. Genes in Conflict: The Biology of Selfish Genetic Elements. Harvard University Press; 2009. [Google Scholar]
- 3.Burt A, Koufopanou V. Homing endonuclease genes: the rise and fall and rise again of a selfish element. Curr Opin Genet Dev. 2004;14:609–615. doi: 10.1016/j.gde.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 4.Macreadie IG, Scott RM, Zinn AR, Butow RA. Transposition of an intron in yeast mitochondria requires a protein encoded by that intron. Cell. 1985;41:395–402. doi: 10.1016/s0092-8674(85)80012-x. [DOI] [PubMed] [Google Scholar]
- 5.Burt A. Site-specific selfish genes as tools for the control and genetic engineering of natural populations. Proc Biol Sci. 2003;270:921–928. doi: 10.1098/rspb.2002.2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Windbichler N, et al. A synthetic homing endonuclease-based gene drive system in the human malaria mosquito. Nature. 2011;473:212–215. doi: 10.1038/nature09937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan YS, Naujoks DA, Huen DS, Russell S. Insect population control by homing endonuclease-based gene drive: an evaluation in Drosophila melanogaster. Genetics. 2011;188:33–44. doi: 10.1534/genetics.111.127506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan YS, Huen DS, Glauert R, Whiteway E, Russell S. Optimising homing endonuclease gene drive performance in a semi-refractory species: the Drosophila melanogaster experience. PloS One. 2013;8:e54130. doi: 10.1371/journal.pone.0054130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takeuchi R, Choi M, Stoddard BL. Redesign of extensive protein–DNA interfaces of meganucleases using iterative cycles of in vitro compartmentalization. Proc Natl Acad Sci. 2014;111:4061–4066. doi: 10.1073/pnas.1321030111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jinek M, et al. RNA-programmed genome editing in human cells. eLife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang H, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friedland AE, et al. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat Methods. 2013;10:741–743. doi: 10.1038/nmeth.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gratz SJ, et al. Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics. 2013;194:1029–1035. doi: 10.1534/genetics.113.152710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DiCarlo JE, et al. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 2013;41:4336–4343. doi: 10.1093/nar/gkt135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang W, et al. Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 2013;41:e188. doi: 10.1093/nar/gkt780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oye KA, et al. Regulating gene drives. Science. 2014;345:626–628. doi: 10.1126/science.1254287. [DOI] [PubMed] [Google Scholar]
- 19.Chamberlain N, Cutts NS, Rainbow C. The formation of pigment and arylamine by yeasts. J Gen Microbiol. 1952;7:54–60. doi: 10.1099/00221287-7-1-2-54. [DOI] [PubMed] [Google Scholar]
- 20.Mao X, Schwer B, Shuman S. Mutational analysis of the Saccharomyces cerevisiae ABD1 gene: cap methyltransferase activity is essential for cell growth. Mol Cell Biol. 1996;16:475–480. doi: 10.1128/mcb.16.2.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gantz VM, Bier E. The mutagenic chain reaction: A method for converting heterozygous to homozygous mutations. Science. 2015:aaa5945. doi: 10.1126/science.aaa5945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Akbari OS, et al. Safeguarding gene drive experiments in the laboratory. Science. 2015;349:927–929. doi: 10.1126/science.aac7932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henkel RD, Miller T, Weyant RS. Monitoring Select Agent Theft, Loss and Release Reports in the United States—2004–2010. Appl Biosaf. 2012;17:171–180. [Google Scholar]
- 24.Kondo S, Ueda R. Highly improved gene targeting by germline-specific Cas9 expression in Drosophila. Genetics. 2013;195:715–721. doi: 10.1534/genetics.113.156737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Port F, Chen HM, Lee T, Bullock SL. Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc Natl Acad Sci U S A. 2014;111:E2967–2976. doi: 10.1073/pnas.1405500111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ablain J, Durand EM, Yang S, Zhou Y, Zon LI. A CRISPR/Cas9 Vector System for Tissue-Specific Gene Disruption in Zebrafish. Dev Cell. 2015;32:756–764. doi: 10.1016/j.devcel.2015.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Novitski E, Grace D, Strommen C. The entire compound autosomes of Drosophila melanogaster. Genetics. 1981;98:257–273. doi: 10.1093/genetics/98.2.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jain S, et al. A recombination execution checkpoint regulates the choice of homologous recombination pathway during DNA double-strand break repair. Genes Dev. 2009;23:291–303. doi: 10.1101/gad.1751209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Costantino L, et al. Break-Induced Replication Repair of Damaged Forks Induces Genomic Duplications in Human Cells. Science. 2014;343:88–91. doi: 10.1126/science.1243211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Looke M, Kristjuhan K, Kristjuhan A. Extraction of genomic DNA from yeasts for PCR-based applications. BioTechniques. 2011;50:325–328. doi: 10.2144/000113672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spitzer M, Wildenhain J, Rappsilber J, Tyers M. BoxPlotR: a web tool for generation of box plots. Nat Methods. 2014;11:121–122. doi: 10.1038/nmeth.2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.