Abstract
Autophagy is a cellular process to recycle nutrients and has been implicated in cancer treatment. Oral squamous cell carcinoma (OSCC) is the most common oral cancer which ranks 3% of cancers in men and 2% in women. In this study, immunohistochemical staining of OSCC tumor specimens from human subjects and an athymic mouse model demonstrated high levels of autophagy markers LC3-II and ATG5 expression. Further, we identified high levels LC3-II expression in OSCC tumor cell lines (SCC-1, SCC-12 & SCC-14a) compared to normal human epithelial (RWPE-1) cells. OSCC cells express high levels of RANK ligand (RANKL); however a functional role in autophagy is unknown. Interestingly, RANKL stimulation significantly increased autophagosome related gene expressions such as LC3, ATG5, BECN1 and PI3KC3 mRNA expression in OSCC cells. Further, western blot analysis of total cell lysates demonstrated a dose-dependent increase in LC3-II and ATG5 expression in RANKL stimulated cells. In addition, RANKL increased expression of LC3-I and LC3-II, essential for autophagosome formation. Confocal microscopy analysis of LC3-II and localization with lysosome further confirms autophagosome formation in response to RANKL treatment in OSCC cells. Collectively, our results indicate a novel function of RANKL to induce autophagosome formation, and could be a potential therapeutic target to control OSCC tumor progression.
Keywords: RANK ligand, Oral squamous cell carcinoma (OSCC), Autophagosome, LC3, ATG5
Introduction
Head and neck squamous cell carcinoma (HNSCC) is the sixth most common cancer worldwide [Machiels et al., 2014]. Oral squamous cell carcinoma (OSCC) contributes up to 80–90% of all malignant neoplasms of the oral cavity [Johnson et al., 2011]. OSCC ranks 3% of cancers in men and 2% in women. Genetic predisposition and exposure to environmental carcinogens such as tobacco, alcohol, chronic inflammation and viral infection are the major etiologic factors for OSCC tumor development [Choi and Myers, 2008]. A murine model for OSCC tumor on calvarial region of athymic mice showed tumor invasion of bone and osteolysis [Pandruvada et al., 2010]. Also, OSCC cells have been shown to express high levels of receptor activator of nuclear factor-κB ligand (RANKL) [Yuvaraj et al., 2009]. RANKL is a tumor necrosis factor (TNF) superfamily member, produced by the stromal/preosteoblast cells and a variety of tumor cells. RANKL is critical for bone resorbing osteoclast formation and survival [Boyle et al., 2003]. RANKL has also been shown to play a role in prostate tumor cells migration and expression of tumor metastasis genes [Armstrong et al., 2008]. Tumor cells activate bone resorbing osteoclasts and thereby facilitate osteolytic process and bone invasion [Deyama et al., 2008; Tada et al., 2005]. Matrix metalloproteinases such as MMP-7 produced by osteoclasts at the prostate tumor-bone interface in a rodent model has been shown to promote osteolysis through solubilization of RANKL [Lynch et al., 2005]. Inhibition of RANKL in preclinical models has been shown to reduce breast cancer progression and lung metastases [Azim and Azim, 2013; Yoneda et al., 2013]. In addition, RANKL-RANK pathway mediates migration and invasion of breast and prostate cancer cells [Casimiro et al., 2013].
Autophagy is a cellular self-consumption process characterized by sequestration of bulk cytoplasm, long-lived proteins and damaged cellular organelles encapsulated as autophagosomes and delivered for lysosomal degradation to recycle the nutrients [Xie and Klionsky, 2007]. The process is regulated by the conversion of the cytoplasmic microtubule-associated protein 1 light chain 3 (LC3-I) into the membrane form of LC3-II which promotes the autophagosomal degradation. Autophagy plays an important role in tumor progression and maintains the malignant state against anti-cancer therapies [Hippert et al., 2006]. Therefore, autophagy inhibition has emerged as an anticancer therapy, as tumor cells depend on autophagy for survival under anti-cancer drug/radiation induced stress. Signaling pathways such as PI3K, CaMKK, PTEN, TSC1/2, P53, Beclin 1 and DRAM are associated with activation of autophagy. In contrast, PI3K/AKT/mTOR signaling axis inhibits autophagy. However, Ras exhibits dual function as both an inhibitor via PI3K activation and activator via RAF1/MEK1/2/ERK1/2 pathways [Hippert et al., 2006; Janku et al., 2011]. However, the role of RANKL in autophagosome formation in OSCC tumor cells is unknown. In this study, we demonstrated that RANKL stimulates autophagy marker gene expression and autophagosome formation in OSCC tumor cells and could be a potential therapeutic target to control tumor progression.
Materials and Methods
Reagents
Anti-LC3-I&II antibody was purchased from MBL International corporation (Woburn, MA). Anti-LC3-II and anti-ATG5 antibodies were purchased from Cell Signaling Technology (Danvers, MA). Peroxidase-conjugated secondary antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Alexa 488-conjugated anti-rabbit IgG secondary antibody, LysoTracker® Red DND-99 and DRAQ5 were purchased form Life Technologies (Grand Island, NY). BCA protein assay reagent was purchased from Pierce (Rockford, IL). Super signal enhanced chemiluminescence (ECL) reagent was obtained from Amersham Bioscience (Piscataway, NJ) and PVDF membranes were purchased from Millipore (Bedford, MA). HRP-labeled secondary antibody and DAB for immunohistochemical staining were purchased from Vector Laboratories (Burlingame, CA).
Immunohistochemistry (IHC)
OSCC tumor specimens from the human subjects were obtained from the Hollings Cancer Center Tissue Biorepository in accordance with an Institutional Review Board (IRB) approved protocol. Human OSCC tumor derived cells lines (SCC-1, SCC-12 and SCC-14a) were injected over calvaria in athymic mice and tumors developed after 4 weeks period were collected as described earlier [Sambandam et al., 2013]. OSCC tumor specimens were formalin-fixed and processed for paraffin sectioning. Serial 5-µm sections were cut on a modified Leica RM 2155 rotary microtome (Leic Microsystems, Ontario, Canada). Antigen retrieval was performed with Target Retrieval Solution (DAKO, Carpinteria, CA) coupled with steaming. The slides were incubated with primary anti-LC3-II and anti-ATG5 antibodies for 60 min at room temperature. Immunohistochemical staining was performed with HRP labeled secondary antibody and DAB. The slides were briefly counterstained with hematoxylin and dehydrated through graded alcohols to xylene and were cover slipped with a permanent mounting media. LC3-II and ATG5 IHC semi-quantitation was determined using the modified H-score which consists of the sum of the percent of tumor cells staining multiplied by an ordinal value corresponding to the staining intensity level (0=none, 1=weak, 2=moderate, and 3=strong). IHC scores were determined by taking the product of the estimated staining intensity and area of tissue (tumor or normal) stained (<1/3 = 1; 1/3–2/3 = 2; >2/3 = 3), giving a range of possible scores between 0 and 9. IHC scores were averaged to determine a composite score for each case as described [Kraus et al., 2012].
Quantitative real-time RT-PCR
Genes encoding autophagosome components such as LC3-II, ATG5 and VPS34 (PI3K class III) and BECN1 mRNA expression levels were measured by real-time RT-PCR as described earlier [Yuvaraj et al., 2009]. Briefly, total RNA was isolated from OSCC cells treated with different concentrations of RANKL (0–100 ng/ml) for 48 h using RNAzol reagent (Biotecx Labs, Houston, TX). To eliminate the residual genomic DNA contamination, total RNA was treated with DNAse I (Sigma) at room temperature for 15 min followed by 10 min at 65 °C with the addition of 25 mM EDTA. The RNA integrity of samples was evaluated based on the intensity of 28S and 18S rRNA bands on agarose gels and A260/A280 ratio between 1.8 and 2.0. The reverse transcription reaction was performed using poly-dT primer and moloney murine leukemia virus reverse transcriptase in a 25 µl reaction volume containing total RNA (2 µg), 1× PCR buffer and 2 mM MgCl2, at 42 °C for 15 min followed by 95 °C for 5 min. The quantitative real-time RT-PCR was performed using IQ™ SYBR Green Supermix in an iCycler (iCycler iQ Single-color real-time-PCR detection system; Bio-Rad, Hercules,CA). The primer sequences used to amplify human glyceraldehyde-3-phosphate dehydrogenase (hGAPDH) mRNA were 5’-CCT ACC CCC AAT GTATCC GTT GTG-3’ (sense) and 5’-GGA GGA ATG GGA GTT GCT GTT GAA-3’ (anti-sense); hATG5 mRNA 5’-TTT GAA TAT GAA GGC ACA CCA-3’ (sense) and 5’-GCA TCC TTA GAT GGA CAG TGC-3’ (anti-sense); hBECN 5’- TTC CTT ACG GAA ACC ATT CA-3’ (sense) and 5’-GGT CAA ACT TGT TGT CCC AGA-3’ (anti-sense); hLC3-A 5’-CAT GTG GAA AAG CAG CTG TG-3’ (sense) and 5’-CCT TGT AGC GCT CGA TGA T-3’ (anti-sense); PI3KC3 (VPS34) mRNA 5’-AAG CAG TGC CTG TAG GAG GA -3 (sense) and 5’-TGT CGA TGA GCT TTG GTG AG-3’ (anti-sense). Thermal cycling parameters were 94 °C for 3 min, followed by 35 cycles of amplifications at 94 °C for 30 s, 60 °C for 1 min, 72 °C for 2 min, and 72 °C for 10 min as the final elongation step. The melt curve analysis was performed from 59–95 °C with 0.5 °C increments. The specificities of PCR amplifications were assessed from the melt curves to confirm the presence of gene specific peaks. Relative levels of mRNA expression were normalized in all the samples analyzed with respect to the levels of GAPDH amplification.
Western blot
OSCC cells were seeded (5×105/well) in 6-well plates and supplemented with DMEM containing 10% FBS. Cells were stimulated with or without RANKL (0–125 ng/ml) and total cell lysates were prepared in a buffer containing 20 mM Tris-HCl at pH 7.4, 1% Triton X-100, 1 mM EDTA, 1.5 mM MgCl2, 10% glycerol, 150 mM NaCl, 0.1 mM Na3VO4 and 1× protease inhibitor cocktail. The protein content of the samples was measured using the BCA protein assay reagent (Pierce, Rockford, IL). Protein (100 µg) samples were then subjected to SDS-PAGE using 4–15% Tris-HCl gels and blot transferred on to a PVDF membrane and immunoblotted with anti-LC3-I&II or LC3-II and ATG5 antibodies. The bands were detected using the enhanced chemiluminescence detection system. The band intensity was quantified by densitometric analysis using the NIH ImageJ Program and full length western blots were provided as supplemental material.
Confocal microscopy
SCC14a cells (1×104 cells/well) were cultured onto 22 mm coverslips in 6-well plates and treated with or without RANKL (100 ng/ml) for 24 h. After treatment, culture medium was replaced with freshly prepared pre-warmed medium containing 500 nM Lysotracker Red, and incubated at 37 °C for 45 min. Following labeling, cells were placed on ice and washed for 10 min in ice-cold phosphate buffered saline (PBS), and fixed with 4% formaldehyde in PBS for 20 min [Fossale et al., 2004]. Cells were then permeabilized with 0.1% Triton X-100 for 10 min at room temperature, which were then blocked for 1 h with PBS containing 2% horse serum. Immunostaining of cells was performed by incubation with primary antibodies against LC3-II in PBS containing 2% horse serum for 3 h. After extensive washing with PBS, cells were incubated with Alexa 488-conjugated anti-rabbit IgG for 1 h at room temperature. Nuclear staining was performed with DRAQ5 and autophagosomes were visualized by IX81 confocal microscope (Olympus, IX81; 40–60 ×).
Statistical analysis
Results are presented as mean ± SD for three independent experiments and were compared by Student’s t-test. Values were considered significantly different for p < 0.05.
Results
LC3-II and ATG5 expression in OSCC tumor cells
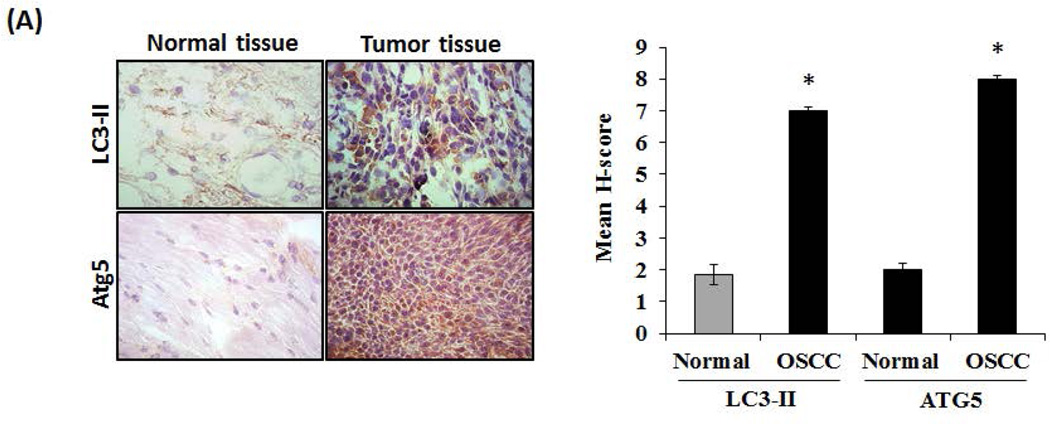
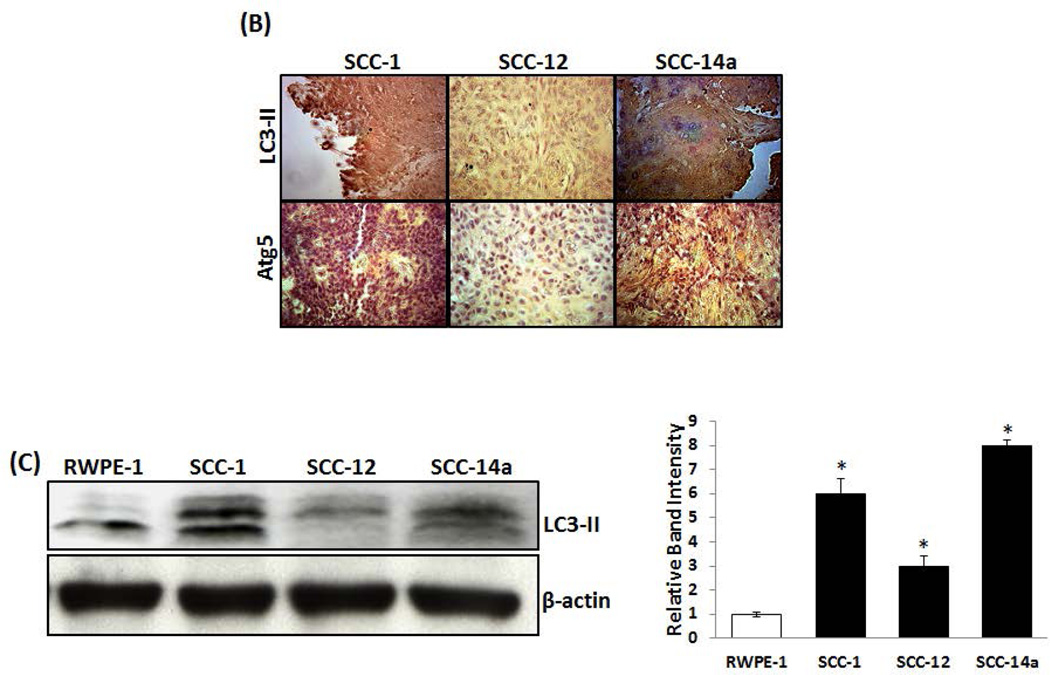
Tumor cells have been shown to express high levels of several autophagy-related genes (Atg), Beclin 1 and LC3 which are associated with longevity [Ahn et al., 2007; Yoshioka et al., 2008]. Previously, OSCC tumor invasion of bone/osteolysis has been demonstrated in an athymic mouse model [Pandruvada et al., 2010]. Since, autophagosome formation was detected in invasive and metastatic melanoma cells [Lazova et al., 2010], we examined the autophagosome marker proteins, LC3-II and ATG5 expression in OSCC tumor specimens. Immunohistochemical analysis of OSCC tumor specimens from human subjects showed high levels of LC3-II and ATG5 expression with an average of H score 7 and 8 respectively. In contrast, immunohistochemistry of normal adjacent tissues demonstrated very low levels of LC3-II and ATG5 (average H score <2) expression (Fig. 1A). Also, OSCC tumor specimens derived from athymic mice injected with the human OSCC cell lines (SCC-1 and SCC-14a) over calvaria stained strong positive for LC3-II and ATG5 expression compared to SCC-12 cells (Fig. 1B). We further examined the expression of LC3-II in OSCC and normal epithelial (RWPE-1) cells by western blot analysis. As shown in Fig. 1C, total cell lysates obtained from SCC-1, SCC-12 and SCC-14a cells demonstrated 6.0, 3.0 and 8.0-fold increase in LC3-II expression respectively, when compared to normal epithelial RWPE-1 cells. These results suggest that autophagosome formation occurs in OSCC tumor cells.
Fig. 1. LC3-II and Atg5 expression in OSCC.
Immunohistochemical staining for LC3-II and Atg5 expression in (A) human primary OSCC tumors specimens compared to normal adjacent tissue, and (B) tumors developed using SCC-1, SCC-12 and SCC-14a cells in an athymic mouse model. The mean H score of OSCC and adjacent normal tissue from 3 specimens respectively is illustrated as a graph. The data are shown as mean ± SD (*p < 0.05). (C) Total cell lysates obtained from SCC-1, SCC-12, SCC-14a and normal epithelial (RWPE-1) cells were analyzed for LC3-II expression by western blot. β-actin expression served as control. Data shown are representative of three replicate studies. The band intensities were quantified by ImageJ program as shown in the right panel. The values are expressed as mean band intensity (LC3-II/β-actin) ± SD of triplicates (*p<0.05).
RANKL stimulation of autophagosome associated gene expression
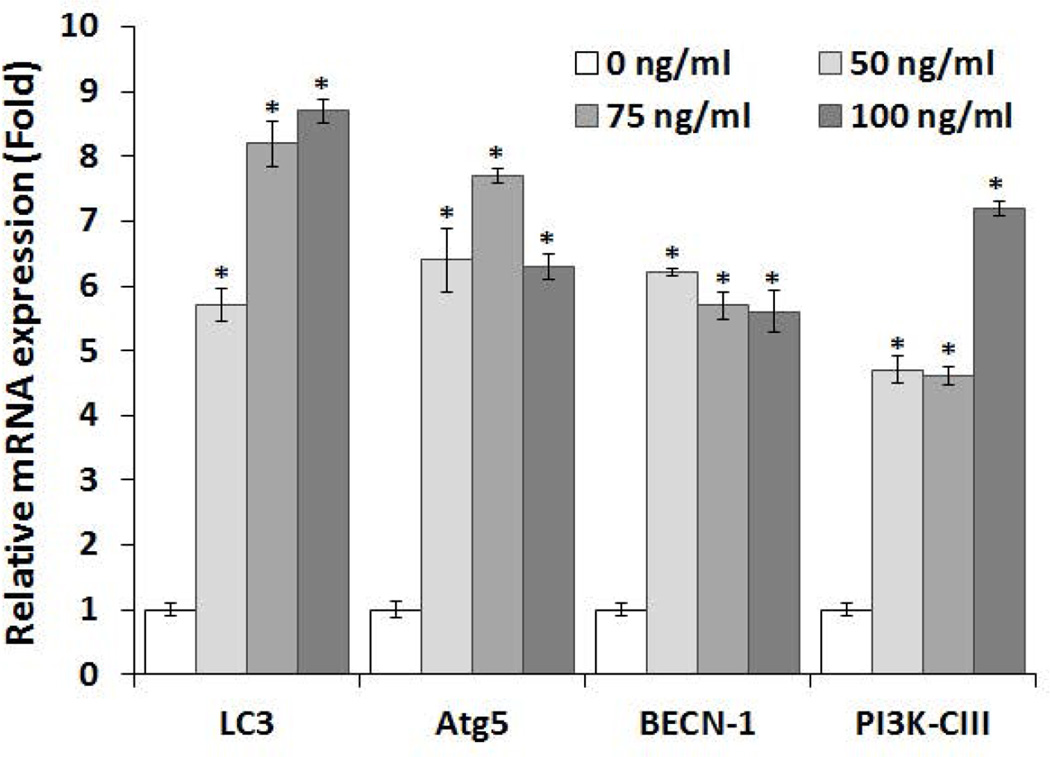
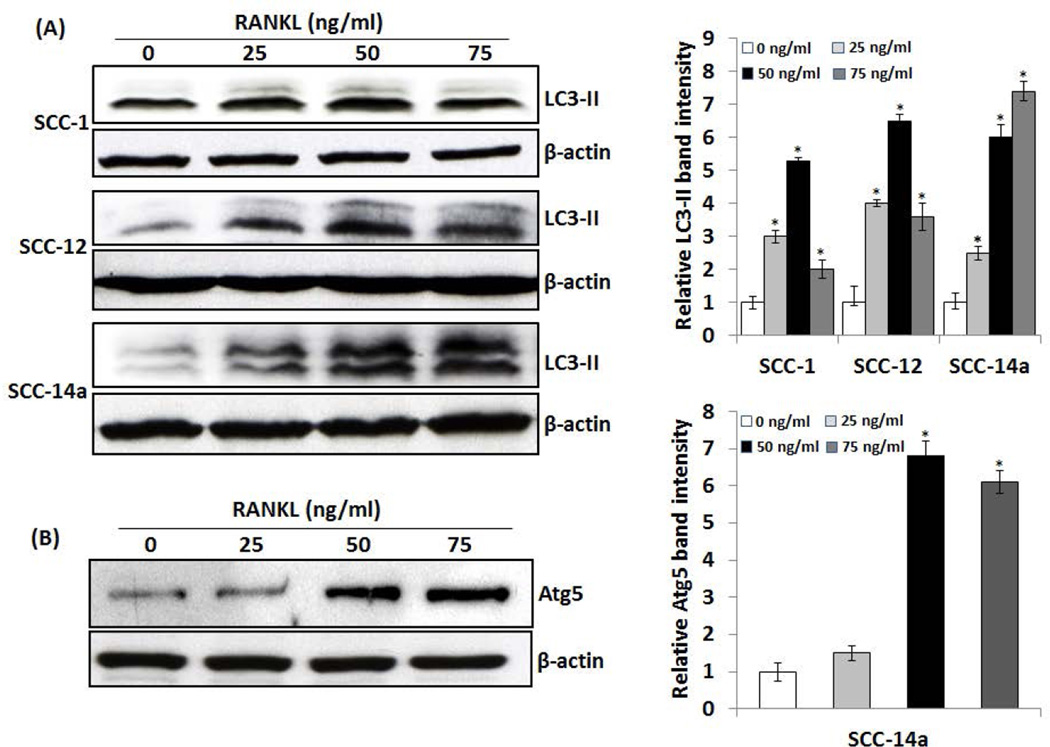
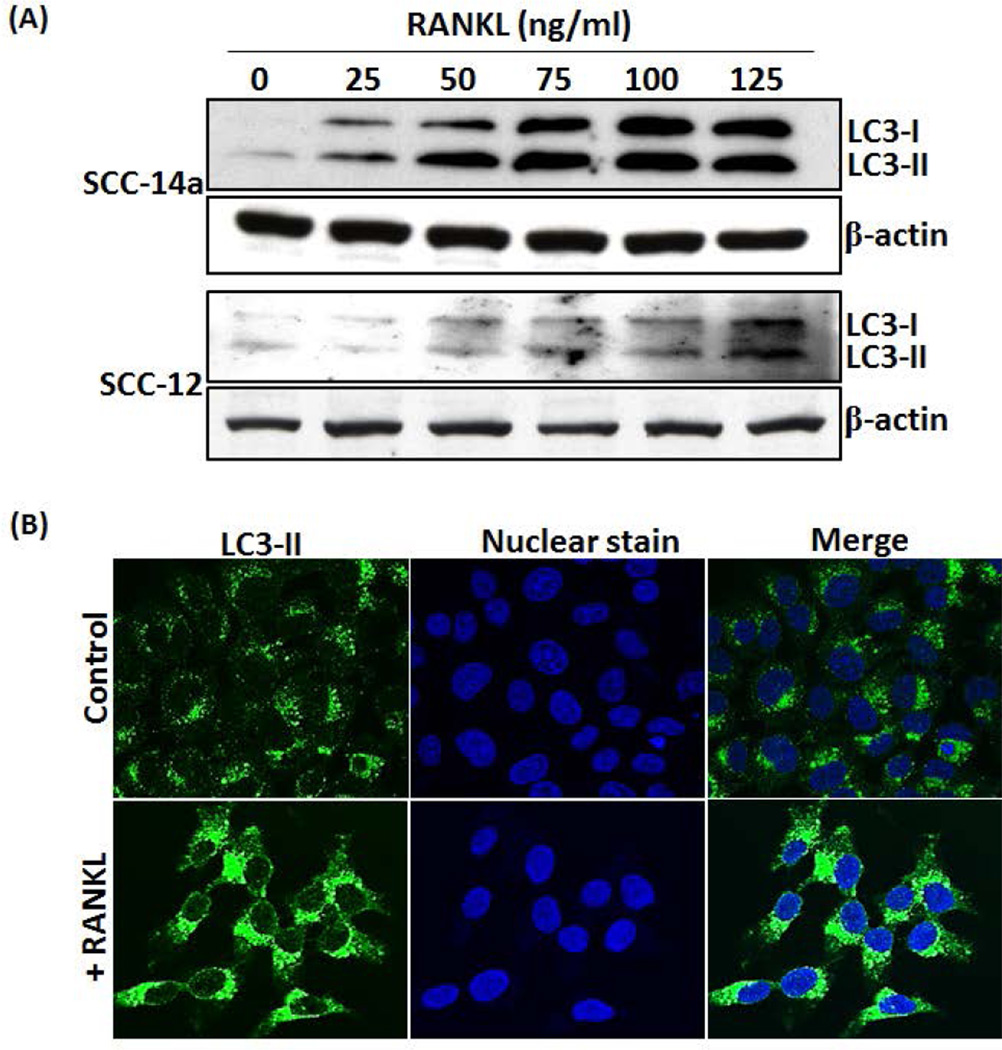
OSCC tumor cells express high levels of RANKL [Yuvaraj et al., 2009]. RANKL has been shown to modulate proliferation and migration of prostate tumor cells [Armstrong et al., 2008; Chen et al., 2006]. RANKL-RANK signaling is involved in mammary cancer development [Yoneda et al., 2013]. Therefore, we further examined the RANKL regulation of autophagosome related gene expression in OSCC cells. Interestingly, real-time RT-PCR analysis of total RNA isolated from SCC14a cells showed a significant increase in ATG5, BECN-1 and VPS34 (PI3KC3) mRNA expression in response to RANKL treatment (Fig. 2). In addition, RANKL dose dependently increased the expression of LC3 mRNA in these OSCC cells. In addition, western blot analysis of total cell lysates obtained from SCC-1, SCC-12 & SCC-14a cells treated with different concentration of RANKL (0–75 ng/ml) showed a significant increase in the autophagosome marker, LC3-II expression in a dose-dependent manner (Fig 3A). It has been shown that recruitment of LC3-II to isolation membranes depends on ATG5 expression [Mizushima et al., 2001]. We therefore examined the status of ATG5 expression in RANKL stimulated OSCC cells. As shown in Fig. 3B, western blot analysis of total cell lysates obtained from SCC-14a cells stimulated with RANKL showed a dose-dependent increase in ATG5 expression. These results implicate a novel function of RANKL in autophagosome formation in OSCC tumor cells.
Fig. 2. Real-time RT-PCR analysis of RANKL stimulated autophagosome associated gene expression in OSCC cells.
The SCC14a cells were stimulated with different concentration of RANKL (0–100 ng/ml) for 48 h. Total RNA isolated from these cells was subjected to real-time RT-PCR analysis for LC3, Atg5, BECN-1 and PI3K-CIII mRNA expression using gene specific primers. The relative levels of mRNA expression were normalized with GAPDH amplification. Data shown are representative of three replicate studies. The values are expressed as mean ± SD (*p<0.05).
Fig. 3. Western blot analysis of RANKL induction of LC3-II and Atg5 expression in OSCC cells.
(A) SCC-1, SCC-12 and SCC-14a cells were treated with different concentration of RANKL (0–75 ng/ml) for 48 h and total cell lysate obtained from these cells were analyzed by western blot using specific antibodies against LC3-II. (B) Western blot analysis of Atg5 expression in RANKL stimulated SCC14a cells using specific antibodies against Atg5. β-actin expression served as control. The band intensities were quantified by ImageJ program as shown in the right panel. The values are expressed as mean relative band intensity ± SD (*p<0.05).
RANKL increases autophagosome formation in OSCC cells
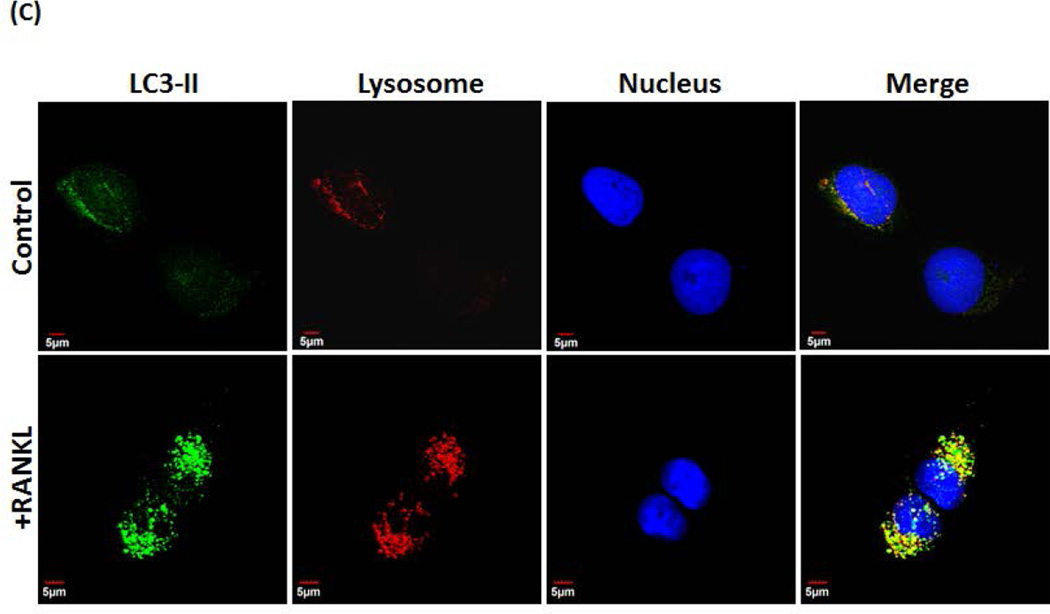
It has been well characterized that conversion of LC3-I to LC3-II serves as an indicator of autophagosome formation [Klionsky et al., 2008]. Therefore, we next examined the effect of RANKL on LC3-I conversion to LC3-II in OSCC tumor cells. Total cell lysate obtained from SCC14a and SCC-12 cells treated with different concentration of RANKL (0–125 ng/ml) for 24 h were subjected to western blot analysis for LC3-I and LC3-II protein expression. As shown in Fig. 4A, RANKL increased the levels of LC3-I and LC3-II expression in OSCC cells. To confirm RANKL stimulation of autophagosome formation in OSCC cells, SCC14a cells were immunostained with LC3-II specific antibody and detected with Alexa 488–conjugated anti-rabbit antibody by confocal microscopy. As shown in Fig. 4B, RANKL markedly increased autophagosome formation in SCC14a cells when compared with unstimulated control cells. To further confirm the autophagosome formation, we have analyzed the co-localization of LC3-II in lysosomes. As shown in Fig. 4C, RANKL treatment elevated co-localization of LC3-II with LysoTracker, a red-fluorescent dye for labeling lysosomes in OSCC tumor cells. These results suggest that RANKL plays an important role in the modulation of autophagosome formation/autophagy in OSCC cells.
Fig. 4. RANKL increases LC3-I, LC3-II and autophagosome formation in OSCC cells.
(A) SCC-14a and SCC-12 cells were stimulated with different concentrations of RANKL (0–125 ng/ml) for 24 h and total cell lysate were subjected to western blot analysis for LC3 proteins using the anti-LC3 antibody which recognize both LC3-I and LC3-II molecules. β-actin expression served as control. (B) Confocal microscopy analysis of autophagosome formation in OSCC cells. SCC14a cells were stimulated with RANKL (100 ng/ml) for 24 h and immunostained for LC3-II. Nuclear staining was performed by DRAQ5. Merged image demonstrates cytosolic localization of autophagosomes. (C) Co-localization of LC3-II in lysosome. SCC14a cells were stimulated with RANKL and immunostained for LC3-II. Also, label lysosome using LysoTracker® Red DND-99, a red-fluorescent dye. Nuclear staining was performed by DRAQ5. Representative images of cells demonstrate co-localization of lysosome with LC3-II (magnification 60×).
Discussion
Autophagy plays an important role in tumor progression and maintains the malignant state against anti-cancer therapies [Hippert et al., 2006]. Autophagy is shown to be up regulated in colorectal cancer and associated with poor prognosis, drug resistance [Lai et al., 2014]. This study demonstrated that OSCC tumors from human subjects and an athymic mouse model express high levels of LC3-II and ATG5 expression which indicates the autophagy in OSCC tumor cells. RANKL has been shown to directly affect proliferation, migration in prostate tumor cells [Armstrong et al., 2008; Chen et al., 2006]. Also, RANKL has been implicated in mammary cancer development [Schramek et al., 2011; Yoneda et al., 2013]. RANKL-RANK signaling has been shown to play an important role in OSCC tumor invasion of bone [Shin et al., 2011]. Herein, we demonstrated that RANKL treatment of OSCC tumors cells enhances the levels of several autophagy-related genes (ATG), BECN 1 and LC3 which are associated with longevity of the cancer cells [Ahn et al., 2007; Yoshioka et al., 2008]. LC3 conversion from LC3-I to LC3-II correlated with the autophagosome formation [Klionsky et al., 2008]. Our results that RANKL treatment significantly increased LC3-II in OSCC cells suggest that RANKL modulates autophagy in OSCC cells. RANKL has been reported to be involved in post-translational modifications of signaling molecules [Kim et al., 2011]. Therefore, RANKL could be involved in the conversion of LC3-I to LC3-II. Confocal microscopy analysis used in this study further confirmed RANKL induction of autophagosome formation essential for autophagy in OSCC tumor cells. This is consistent with the previous report that constitutive formation of autophagosomes has been detected in invasive and metastatic melanoma cells [Lazova et al., 2010]. Thus, the present study identified that RANKL expression in OSCC cells plays a novel role in modulation of autophagy, and may have implications in malignant progression of tumors. Recently, it has also been shown that OSCC tumor cells production of chemokines such as CXCL13 induce RANKL expression in bone marrow stromal/preosteoblast cells [Sambandam et al., 2013]. Therefore, tumor derived cytokines/chemokines may contributes to elevated levels of RANKL expression in the OSCC and autophagosome formation in the tumor microenvironment. RANKL has been shown to be expressed in metastatic bone cancer cells and play a key role in cell migration and metastatic behavior [Lee et al., 2011]. Recently, it has been shown that impaired autophagy suppressed pulmonary metastasis of hepatocellular carcinoma [Peng et al., 2013], development of hepatoblastoma [Chang et al., 2011] and augment apoptosis in human OSCC under nutrient depletion [Jiang et al., 2014]. Also, increased RANKL expression is implicated in renal carcinoma tumor migration and metastasis [Mikami et al., 2009]. Therefore, RANKL induced autophagy may play a role in tumor cell migration/metastasis. The p62 (sequestosome 1) is a multifunctional ubiquitin-binding protein which binds to LC3-II on the autophagosome membrane for degradation [Pankiv et al., 2007; Wooten et al., 2008]. Autophagic modulation of p62 has been shown to play a role in tumorogenesis [Mathew et al., 2009]. A correlation between LC3 or p62/SQSTM1 and the infiltration of T-cells suggests a role for autophagy in malignancy through immune cells mobilization to the cancer microenvironment [Sakakura et al., 2014]. In this scenario, it is possible that RANKL may regulate autophagic process through p62 in OSCC tumor cells.
Autophagy inhibition has emerged as an anti-cancer treatment, as tumor cells depend on autophagy for survival under anti-cancer drug/radiation induced stress. Ionizing radiation has been shown to induce autophagy in human OSCC [Wu et al., 2014]. Therefore, impaired autophagy could promote the anti-cancer effects of radiotherapy in patients with OSCC tumor. Impaired autophagy has been shown to suppress the development of hepatoblastoma [Chang et al., 2011]. Malignant OSCC tumors are known to have a potent activity of local bone invasion. OSCC tumor progression involves invasion of bone/osteolysis [Pandruvada et al., 2010]. Thus, it is necessary to develop therapeutic measures to control OSCC tumor progression. Tamoxifen has been shown to inhibit OSCC cell growth in vitro [Nelson et al., 2007]. Also, Cisplatin chemotherapy displayed antitumor activity against OSCC tumors [Hiraishi et al., 2008]. However, recurrence of OSCC tumor after chemotherapy is observed. Therefore, inhibition of RANKL expression in OSCC cells in conjunction with anti-cancer drugs could be a potential therapy to control OSCC tumor progression.
Supplementary Material
Acknowledgement
This work was supported by the National Center for Research Resources and the Office of the Director of the National Institutes of Health through Grant Number C06 RR015455 and grants from the National Institutes of Health (R01 CA129560) to A. Haque. The research presented in this article was also supported in part by the Tissue Biorepository Shared Resource as part of the Hollings Cancer Center at the Medical University of South Carolina which is funded by a Cancer Center Support Grant P30 CA138313. We also thank Dr. S.V. Reddy for critical reading of the manuscript and helpful discussions.
Footnotes
Disclosures
The authors have declared that no conflict of interest exists.
References
- Ahn CH, Jeong EG, Lee JW, Kim MS, Kim SH, Kim SS, Yoo NJ, Lee SH. Expression of beclin-1, an autophagy-related protein, in gastric and colorectal cancers. APMIS. 2007;115:1344–1349. doi: 10.1111/j.1600-0463.2007.00858.x. [DOI] [PubMed] [Google Scholar]
- Armstrong AP, Miller RE, Jones JC, Zhang J, Keller ET, Dougall WC. RANKL acts directly on RANK-expressing prostate tumor cells and mediates migration and expression of tumor metastasis genes. Prostate. 2008;68:92–104. doi: 10.1002/pros.20678. [DOI] [PubMed] [Google Scholar]
- Azim H, Azim HA., Jr Targeting RANKL in breast cancer: bone metastasis and beyond. Expert Rev Anticancer Ther. 2013;13:195–201. doi: 10.1586/era.12.177. [DOI] [PubMed] [Google Scholar]
- Boyle WJ, Simonet WS, Lacey DL. Osteoclast differentiation and activation. Nature. 2003;423:337–342. doi: 10.1038/nature01658. [DOI] [PubMed] [Google Scholar]
- Casimiro S, Mohammad KS, Pires R, Tato-Costa J, Alho I, Teixeira R, Carvalho A, Ribeiro S, Lipton A, Guise TA, Costa L. RANKL/RANK/MMP-1 molecular triad contributes to the metastatic phenotype of breast and prostate cancer cells in vitro. PLoS One. 2013;8:e63153. doi: 10.1371/journal.pone.0063153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y, Chen L, Liu Y, Hu L, Li L, Tu Q, Wang R, Wu M, Yang J, Wang H. Inhibition of autophagy may suppress the development of hepatoblastoma. FEBS J. 2011;278:4811–4823. doi: 10.1111/j.1742-4658.2011.08381.x. [DOI] [PubMed] [Google Scholar]
- Chen G, Sircar K, Aprikian A, Potti A, Goltzman D, Rabbani SA. Expression of RANKL/RANK/OPG in primary and metastatic human prostate cancer as markers of disease stage and functional regulation. Cancer. 2006;107:289–298. doi: 10.1002/cncr.21978. [DOI] [PubMed] [Google Scholar]
- Choi S, Myers JN. Molecular pathogenesis of oral squamous cell carcinoma: implications for therapy. J Dent Res. 2008;87:14–32. doi: 10.1177/154405910808700104. [DOI] [PubMed] [Google Scholar]
- Deyama Y, Tei K, Yoshimura Y, Izumiyama Y, Takeyama S, Hatta M, Totsuka Y, Suzuki K. Oral squamous cell carcinomas stimulate osteoclast differentiation. Oncol Rep. 2008;20:663–668. [PubMed] [Google Scholar]
- Fossale E, Wolf P, Espinola JA, Lubicz-Nawrocka T, Teed AM, Gao H, Rigamonti D, Cattaneo E, MacDonald ME, Cotman SL. Membrane trafficking and mitochondrial abnormalities precede subunit c deposition in a cerebellar cell model of juvenile neuronal ceroid lipofuscinosis. BMC Neurosci. 2004;5:57. doi: 10.1186/1471-2202-5-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hippert MM, O'Toole PS, Thorburn A. Autophagy in cancer: good, bad, or both? Cancer Res. 2006;66:9349–9351. doi: 10.1158/0008-5472.CAN-06-1597. [DOI] [PubMed] [Google Scholar]
- Hiraishi Y, Wada T, Nakatani K, Tojyo I, Matsumoto T, Kiga N, Negoro K, Fujita S. EGFR inhibitor enhances cisplatin sensitivity of oral squamous cell carcinoma cell lines. Pathol Oncol Res. 2008;14:39–43. doi: 10.1007/s12253-008-9020-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janku F, McConkey DJ, Hong DS, Kurzrock R. Autophagy as a target for anticancer therapy. Nat Rev Clin Oncol. 2011;8:528–539. doi: 10.1038/nrclinonc.2011.71. [DOI] [PubMed] [Google Scholar]
- Jiang LC, Xin ZY, Deborah B, Zhang JS, Yuan DY, Xu K, Liu XB, Jiang HQ, Fan QC, Zhang B, Li KY. Inhibition of autophagy augments apoptosis in human oral squamous cell carcinoma under nutrient depletion. J Oral Pathol Med. 2014 doi: 10.1111/jop.12250. [DOI] [PubMed] [Google Scholar]
- Johnson NW, Jayasekara P, Amarasinghe AA. Squamous cell carcinoma and precursor lesions of the oral cavity: epidemiology and aetiology. Periodontol 2000. 2011;57:19–37. doi: 10.1111/j.1600-0757.2011.00401.x. [DOI] [PubMed] [Google Scholar]
- Kim JH, Kim K, Youn BU, Jin HM, Kim JY, Moon JB, Ko A, Seo SB, Lee KY, Kim N. RANKL induces NFATc1 acetylation and stability via histone acetyltransferases during osteoclast differentiation. Biochem J. 2011;436:253–262. doi: 10.1042/BJ20110062. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus JA, Dabbs DJ, Beriwal S, Bhargava R. Semi-quantitative immunohistochemical assay versus oncotype DX((R)) qRT-PCR assay for estrogen and progesterone receptors: an independent quality assurance study. Mod Pathol. 2012;25:869–876. doi: 10.1038/modpathol.2011.219. [DOI] [PubMed] [Google Scholar]
- Lai K, Killingsworth MC, Lee CS. The significance of autophagy in colorectal cancer pathogenesis and implications for therapy. J Clin Pathol. 2014 doi: 10.1136/jclinpath-2014-202529. [DOI] [PubMed] [Google Scholar]
- Lazova R, Klump V, Pawelek J. Autophagy in cutaneous malignant melanoma. J Cutan Pathol. 2010;37:256–268. doi: 10.1111/j.1600-0560.2009.01359.x. [DOI] [PubMed] [Google Scholar]
- Lee JA, Jung JS, Kim DH, Lim JS, Kim MS, Kong CB, Song WS, Cho WH, Jeon DG, Lee SY, Koh JS. RANKL expression is related to treatment outcome of patients with localized, high-grade osteosarcoma. Pediatr Blood Cancer. 2011;56:738–743. doi: 10.1002/pbc.22720. [DOI] [PubMed] [Google Scholar]
- Lynch CC, Hikosaka A, Acuff HB, Martin MD, Kawai N, Singh RK, Vargo-Gogola TC, Begtrup JL, Peterson TE, Fingleton B, Shirai T, Matrisian LM, Futakuchi M. MMP-7 promotes prostate cancer-induced osteolysis via the solubilization of RANKL. Cancer Cell. 2005;7:485–496. doi: 10.1016/j.ccr.2005.04.013. [DOI] [PubMed] [Google Scholar]
- Machiels JP, Lambrecht M, Hanin FX, Duprez T, Gregoire V, Schmitz S, Hamoir M. Advances in the management of squamous cell carcinoma of the head and neck. F1000Prime Rep. 2014;6:44. doi: 10.12703/P6-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen HY, Bray K, Reddy A, Bhanot G, Gelinas C, Dipaola RS, Karantza-Wadsworth V, White E. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137:1062–1075. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikami S, Katsube K, Oya M, Ishida M, Kosaka T, Mizuno R, Mochizuki S, Ikeda T, Mukai M, Okada Y. Increased RANKL expression is related to tumour migration and metastasis of renal cell carcinomas. J Pathol. 2009;218:530–539. doi: 10.1002/path.2567. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yamamoto A, Hatano M, Kobayashi Y, Kabeya Y, Suzuki K, Tokuhisa T, Ohsumi Y, Yoshimori T. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol. 2001;152:657–668. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson K, Helmstaedter V, Lage H. The influence of tamoxifen on growth behavior and cell-cell adhesion in OSCC in vitro. Oral Oncol. 2007;43:720–727. doi: 10.1016/j.oraloncology.2006.09.005. [DOI] [PubMed] [Google Scholar]
- Pandruvada SN, Yuvaraj S, Liu X, Sundaram K, Shanmugarajan S, Ries WL, Norris JS, London SD, Reddy SV. Role of CXC chemokine ligand 13 in oral squamous cell carcinoma associated osteolysis in athymic mice. Int J Cancer. 2010;126:2319–2329. doi: 10.1002/ijc.24920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G, Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- Peng YF, Shi YH, Ding ZB, Ke AW, Gu CY, Hui B, Zhou J, Qiu SJ, Dai Z, Fan J. Autophagy inhibition suppresses pulmonary metastasis of HCC in mice via impairing anoikis resistance and colonization of HCC cells. Autophagy. 2013;9:2056–2068. doi: 10.4161/auto.26398. [DOI] [PubMed] [Google Scholar]
- Sakakura K, Takahashi H, Kaira K, Toyoda M, Oyama T, Chikamatsu K. Immunological significance of the accumulation of autophagy components in oral squamous cell carcinoma. Cancer Sci. 2014 doi: 10.1111/cas.12559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambandam Y, Sundaram K, Liu A, Kirkwood KL, Ries WL, Reddy SV. CXCL13 activation of c-Myc induces RANK ligand expression in stromal/preosteoblast cells in the oral squamous cell carcinoma tumor-bone microenvironment. Oncogene. 2013;32:97–105. doi: 10.1038/onc.2012.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramek D, Sigl V, Penninger JM. RANKL and RANK in sex hormone-induced breast cancer and breast cancer metastasis. Trends Endocrinol Metab. 2011;22:188–194. doi: 10.1016/j.tem.2011.02.007. [DOI] [PubMed] [Google Scholar]
- Shin M, Matsuo K, Tada T, Fukushima H, Furuta H, Ozeki S, Kadowaki T, Yamamoto K, Okamoto M, Jimi E. The inhibition of RANKL/RANK signaling by osteoprotegerin suppresses bone invasion by oral squamous cell carcinoma cells. Carcinogenesis. 2011;32:1634–1640. doi: 10.1093/carcin/bgr198. [DOI] [PubMed] [Google Scholar]
- Tada T, Jimi E, Okamoto M, Ozeki S, Okabe K. Oral squamous cell carcinoma cells induce osteoclast differentiation by suppression of osteoprotegerin expression in osteoblasts. Int J Cancer. 2005;116:253–262. doi: 10.1002/ijc.21008. [DOI] [PubMed] [Google Scholar]
- Wooten MW, Geetha T, Babu JR, Seibenhener ML, Peng J, Cox N, Diaz-Meco MT, Moscat J. Essential role of sequestosome 1/p62 in regulating accumulation of Lys63-ubiquitinated proteins. J Biol Chem. 2008;283:6783–6789. doi: 10.1074/jbc.M709496200. [DOI] [PubMed] [Google Scholar]
- Wu SY, Liu YW, Wang YK, Lin TH, Li YZ, Chen SH, Lee YR. Ionizing radiation induces autophagy in human oral squamous cell carcinoma. J BUON. 2014;19:137–144. [PubMed] [Google Scholar]
- Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- Yoneda T, Tanaka S, Hata K. Role of RANKL/RANK in primary and secondary breast cancer. World J Orthop. 2013;4:178–185. doi: 10.5312/wjo.v4.i4.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka A, Miyata H, Doki Y, Yamasaki M, Sohma I, Gotoh K, Takiguchi S, Fujiwara Y, Uchiyama Y, Monden M. LC3, an autophagosome marker, is highly expressed in gastrointestinal cancers. Int J Oncol. 2008;33:461–468. [PubMed] [Google Scholar]
- Yuvaraj S, Griffin AC, Sundaram K, Kirkwood KL, Norris JS, Reddy SV. A novel function of CXCL13 to stimulate RANK ligand expression in oral squamous cell carcinoma cells. Mol Cancer Res. 2009;7:1399–1407. doi: 10.1158/1541-7786.MCR-08-0589. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.