Abstract
With advances in genomics, transcriptomics, metabolomics and proteomics, and more expansive electronic clinical record monitoring, as well as advances in computation, we have entered the Big Data era in biomedical research. Data gathering is growing rapidly while only a small fraction of this data is converted to useful knowledge or reused in future studies. To improve this, an important concept that is often overlooked is data abstraction. To fuse and reuse biomedical datasets from diverse resources, data abstraction is frequently required. Here we summarize some of the major Big Data biomedical research resources for genomics, proteomics and phenotype data, collected from mammalian cells, tissues and organisms. We then suggest simple data abstraction methods for fusing this diverse but related data. Finally, we demonstrate examples of the potential utility of such data integration efforts, while warning about the inherit biases that exist within such data.
Keywords: data integration, bioinformatics, systems biology, systems pharmacology, network biology
1. INTRODUCTION
Big Data does not have to be defined by sheer size, i.e., giga-bytes, tera-bytes, or peta-bytes of data, but by the fact that almost all the variables of a complex system can be measured over time and under different conditions [1]. Computational biology tools and databases rapidly emerge with an attempt to organize and integrate molecular and phenotype data for the ultimate goal of making predictions by performing virtual experiments. Data integration enables imputing missing values given the already existing data, identifying unexpected relationships between variables, mostly through correlation analyses such as unsupervised clustering, learn-to-rank methods such as enrichment analyses, network reconstruction methods, and supervised machine learning algorithms which are used to make predictions for unseen instances. Integrating x-omics data, a.k.a. the integrome, is not as difficult as it may seem because most diverse datasets and resources represent their data in a relatively structured format with common fields such as cells, genes, proteins, drugs, diseases, and assays. Such diverse but structured data can be converted into attribute tables, bi-partite graphs, single-node-type networks, hierarchies and set libraries. Such data structures provide different views of the same data and are useful for different data integration purposes. Combining two or more datasets, if they share common entities such as: genes/proteins, cells, small-molecules/drugs, tissues/tumors/patients, or diseases/phenotypes/side-effects, can lead to new insights. Here we summarize some of the most relevant resources for x-omics data integration for better extracting knowledge from Big Data. We then define the data structures that can be used to combine such resources, and briefly review the primary methods that can be used to operate on the combined data for knowledge discovery, while providing a few examples applied to real data. While we recognize that typically system level data and the methods to integrate and analyze such data were initially developed for model organisms such as yeast, worm, fly and zebra fish, the focus of this review is on data collected from the mammalian system, as well as databases and computation tools applied to the data from mammalian cells, tissues and organisms. Finally, we discuss the concept and implications of the different biases that may exist across the diverse datasets we describe. In this next section we enlist major relevant emergent Big Data resources in computational systems biology.
2. HIGH-CONTENT DATASETS AND RESOURCES
2.1. Organizing and Abstracting Phenotype-Genotype Associations
2.1.1. Mouse Genome Informatics Mammalian Phenotype Ontology (MGI-MPO)
The Mammalian Phenotype Ontology [2] initially developed by the Mouse Genome Informatics group at the Jackson Labs [3] and expanded to an international initiative called KOMP [4] is a useful resource for connecting gene knockouts in mice to phenotypes. The MGI-MPO ontology is a controlled vocabulary of mouse phenotype terms that are related to each other in a hierarchical network, where at each branch-point a term is linked to a set of more specific sub-terms. Each phenotype is annotated with the genotypes of the mice that display the phenotype. Some of the annotated genotypes are from transgenic mice that mimic human diseases. Gene knockout annotations can be pulled from MPO to create an un-weighted attribute table connecting phenotypes to the gene knockouts known to cause the phenotypes. Similarity matrices connecting phenotypes based on shared gene knockouts or connecting gene knockouts based on shared phenotypes can be derived from the attribute table to create single-node-type networks. Similarly, a gene set library can be created by “cutting” the phenotype tree at a specific appropriate and useful level. We previously “cut” the MPO tree at level 3 and 4 to create gene set libraries for Enrichr [5], Lists2Networks [6], Network2Canvas [7], and Expression2Kinases [8].
2.1.2. Online Mendelian Inheritance in Man (OMIM)
The Online Mendelian Inheritance in Man (OMIM) is a database of human diseases with known genetic basis [9, 10]. Each entry in OMIM summarizes the current state of knowledge about gene-phenotype relationships in humans. The content of each entry is obtained by manual curation of peer reviewed biomedical literature. At the time of writing this review there were 14,570 gene entries and 7669 phenotypes. OMIM content is also provided in a condensed form known as the Morbid Map, which lists phenotypes alongside the genes that have mutations known to play a role in manifestation of the phenotype. The Morbid Map is essentially an unweighted gene-set library, and can therefore be converted to a binary attribute table connecting phenotypes to gene mutations known to cause the phenotypes. Similarity networks of phenotypes or genes that cause similar phenotypes can be created.
2.1.3. Genome Wide Association Studies (GWAS)
Genome Wide Association Studies (GWAS) use single nucleotide polymorphism (SNP) microarrays or next generation sequencing to obtain genome wide profiles of DNA sequence variation in human populations with the goal of identifying quantitative trait loci (QTLs), which are locations in the genome where DNA sequence variation is significantly correlated with variation in a quantifiable human trait (phenotype), for example, risk for cardiovascular disease [11]. There are currently many online databases that host the findings of GWAS [12–14]. Although some GWAS results are included in OMIM, GWAS findings generally differ from the associations reported in OMIM in two ways. First, OMIM is focused on associations between phenotypes and genes, whereas GWAS often identify associations between phenotypes and non-coding regions of the genome. Second, OMIM is focused on phenotypes that obey Mendelian rules of inheritance, whereas GWAS often identify QTL for complex phenotypes, such as obesity, which are partially determined by multiple genomic loci and partially determined by environmental factors. GWAS findings can be organized into a weighted attribute table connecting phenotypes to genomic loci where sequence variation is correlated with phenotype variation. Weights can be obtained from the adjusted p-values that quantify the significance of the correlations between phenotype variation and genomic locus sequence variation. Similarity matrices connecting phenotypes based on shared genomic loci or connecting genomic loci based on shared phenotypes can be derived from the attribute tables. Mutated genes can be identified from genomic loci located in or near coding regions of the genome in order to create a weighted attribute table connecting phenotypes to genes, analogous to the OMIM dataset but more expansive.
2.2. Signatures of Differentially Expressed Genes
2.2.1. The Gene Expression Omnibus (GEO)
The Gene Expression Omnibus (GEO) is a database of high-throughput functional genomic data obtained by microarray, next generation sequencing, or other methods that measure gene expression in high throughput [15]. GEO contains primarily gene expression profiles, but also includes genome occupancy profiles, DNA sequence variation profiles, non-coding RNA profiles, and DNA methylation profiles. Datasets are contributed by research laboratories worldwide and the size and popularity of this resource is growing rapidly. Experimental samples in GEO are from primary human tissues and cells, tissues and cells of model organisms, whole organisms, or cell lines. GEO can be mined by focusing on a subset of the data, extracting differentially expressed genes from studies that share the same theme. Perturbations of interest that can be bundled together to create a secondary resource may include: diseases, drugs, environmental toxins, knockouts, knockdown, or over-expression of single genes. A signature of differentially expressed (DE) genes can be obtained from each study by comparing control and perturbed gene expression samples. This requires a statistical method such as the T-test, or the Characteristic Direction [16], as well as data normalization and other data cleaning methods. The signature of DE genes usually takes the form of a list of genes ordered by the direction and significance of the gene expression change [17]. A number that indicates the relative significance of differential expression can be included with each gene. Such numbers can be used as the weights in attribute tables, bi-partite and single entity node networks or for generating fuzzy gene sets.
2.2.2. The Connectivity Map and LINCS
The Connectivity Map (CMAP) is a project undertaken at the Broad Institute to obtain, in high-throughput, signatures of differentially expressed genes following pharmacological or genetic perturbations of human cultured cells, mostly cancer cell lines [17]. The goal of the Connectivity Map is to help researchers find connections between drugs, genes, and diseases by matching patterns across gene expression signatures of DE genes [18]. The original version of CMAP contains over 6000 signatures derived from 1309 small molecule perturbations applied to the breast cancer epithelial cell line MCF7, the prostate cancer epithelial cell line PC3, the leukemia cell line HL60, or the melanoma cell line SKMEL5 where gene expression was measured using Affymetrix microarrays. Most compounds were used at a concentration of 10 µM, and gene expression profiles were measured after 6 hours of treatment. The Library of Integrated Network-Based Cellular Signatures (LINCS) program is a National Institutes of Health Common Fund program that is supporting collection of signatures of cellular states—such as gene expression, protein abundance, post-translational modifications, and phenotypes—for a broad range of conditions—such as drug treatment, ligand treatment, gene knockdown, and gene over-expression—in many different types of human cells—including cell lines and primary cultures [19]. The program is also supporting the development of computational tools and online resources for analysis, integration, visualization, and public dissemination of experimental results. In its initial phase, the LINCS program has funded an expansive version of the CMAP. For the new CMAP, investigators at the Broad Institute developed a new gene expression profiling assay, called the L1000 platform. With the L1000 technology whole genome expression profiles of cultured human cells can be measured in a much higher throughput at a lower cost [20]. The L1000 technology is capable of measuring the relative abundance of ~1000 transcripts while the expression of the remaining 22,000 human genes is inferred using a computational method. So far in the first phase of LINCS, signatures of DE genes were collected for ~15,000 small molecule perturbations and ~6000 genetic perturbations, including knockdowns or over-expressions, in over 45 human cell lines for a total of over a million experiments. Most small molecule perturbations were profiled at multiple time points in a range of concentrations. Multiple signatures of DE genes from the new or old CMAP can be organized into an attribute table where the genes are the rows and the experiments are the columns. Adjacency matrices connecting perturbations based on similarity of DE gene signatures or networks connecting genes based on similarity of gene expression changes across perturbations, i.e., co-expression networks, can be derived from the attribute tables created from the Connectivity Map data.
2.3. Genome Mapping
2.3.1. Encyclopedia of DNA Elements (ENCODE)
The Encyclopedia of DNA Elements (ENCODE) is another large-scale NIH funded project with the goal of annotating the human genome with information about all its functional elements [21]. A major aim of ENCODE is to annotate the genome with information about elements that regulate gene transcription, including: histone modifications, transcription factor binding sites, and DNA methylations [22]. A prioritized list of human cell lines has been selected for profiling. The top priority cell lines that have been profiled in greatest depth and breadth are K562 erythroleukemia cells, EBV B-lymphoblastoid cells, and H1 human embryonic stem cells. So far, the ENCODE project has completed over 1000 assays for mapping transcription factor binding sites [23] using chromatin immuno-precipitation followed by sequencing (ChIP-seq). Transcription factor ChIP-seq data from ENCODE can be processed to create an attribute table where transcription factor/cell-line combinations are the rows, and the genomic loci that are bound by the putative transcription factor binding sites are the columns. Weights can be obtained from the peak height that quantifies the significance of the mapping of sequenced reads to the genomic loci. More abstractedly, the identified genomic loci can be mapped to their nearest genes. Such matrices can be used to identify transcription-factor/transcription-factor-target interactions. Genomic loci-level and/or gene-level representations of transcription-factor binding site data from ChIP-seq studies can be useful for many data integration efforts including enrichment analysis [6]. Such datasets naturally fit with mRNA expression signature data as well as other datasets. For example, the transcription factors that are most likely responsible for the observed differentially expressed genes under a specific condition can be identified by combining genome mapping data with expression data.
2.3.2. The Roadmap Epigenomics Project
The Roadmap Epigenomics project is another NIH Common Fund genome mapping project that seeks to annotate the human genome with information about the distribution of nucleoproteins including histones, DNA binding factors, and accessory proteins, as well as the pattern of reversible covalent modifications on the DNA and the nucleoproteins [24]. The goal of the Roadmap Epigenomics project is to create a set of reference epigenomic maps for stem cells, differentiated cells, and primary tissues. The project is using next generation sequencing to map DNA methylation by several deep sequencing technologies [25], histone modifications by ChIP-seq, and chromatin accessibility by DNase I hypersensitive sites sequencing (DNase-seq). RNA transcripts are also collected by RNA-seq so the relationships between epigenomic features and gene expression can be discerned. The Roadmap Epigenomics datasets can be used to identify epigenetic features involved in gene regulation [26, 27] and to help identify mechanisms by which disease associated SNPs exert their effects [27]. High priority features are DNA methylation, the six most well-studied histone modifications: H3K4me1, H3K4me3, H3K9me3, H3K9ac, H3K27me3 and H3K36me3, chromatin accessibility, and RNA expression. High priority cell types include human embryonic stem cells (hESCs) and their descendants. Other cell types include adult stem cells, differentiated adult cells and induced pluripotent stem cells. The histone modification ChIP-seq data from the Roadmap Epigenomics project can be processed in the same way the transcription factors ChIP-seq data from ENCODE is processed. It should be noted that the ENCODE project also collects ChIP-seq data targeting histone modifications. Thus, it is possible to derive networks that connect histone-modifications/cell-line combinations based on their shared genomic loci. Furthermore, it is possible to convert from genomic loci-level representations to gene-level representations by inferring target genes based on the proximity of genomic loci to transcription start sites of genes [28]. Data integration between ENCODE and Roadmap Epigenomics data is straight forward since many of the datasets can be layered by their genomic coordinates. Recent efforts show that accumulation and integration of such genomic mapping projects can be used to gain new insights, and perform virtual ChIP-seq experiments that can predict binding sites and histone modifications for proteins in cells that were not profiled experimentally for the specific modifications [29, 30].
2.3.3. Genotype-Tissue Expression (GTEx) Project
The Genotype-Tissue Expression (GTEx) project is a genome mapping project that seeks to discover expression quantitative trait loci (eQTL) by examining gene expression across human tissues [31]. eQTLs are regions within the genome where DNA sequence variation is correlated with variation in mRNA expression of a gene or a set of genes [32]. Mapping eQTLs is important for identifying phenotype-associated-sequence-variants. This suggests that many of these variants exert their effect through gene expression regulation. The GTEx project collects blood and tissue from post-mortem donors, a sample of blood is used for whole genome single nucleotide polymorphism and copy number variant genotyping, and tissue samples are used for whole genome expression profiling by RNA-seq. Sequence variation and gene expression data from multiple donors are processed and statistical tests are applied to identify eQTLs. The data from the pilot phase of the GTEx project have been released when this review was written, which profiled different tissues from 190 donors. eQTLs from the GTEx project can be combined into a weighted attribute table connecting genomic loci to genes that are putatively regulated by the sequence variation at the genomic loci. Weights can be obtained from adjusted p-values that quantify the significance of the correlations between sequence variation at genomic loci and changes in gene expression. Networks connecting genomic loci based on shared target genes, or networks that connect genes based on their shared regulatory genomic loci, can be derived from this attribute table.
2.4. Drug Induced Cellular Phenotypes
2.4.1. Cancer Target Discovery and Development (CTD2) Network
The Cancer Target Discovery and Development (CTD2) network, organized by the Office of Cancer Genomics (OGC) of the NIH’s National Cancer Institute (NCI), is a cancer phenotyping project that seeks to identify novel cancer drug targets and novel biomarkers for diagnosis of cancer and prediction of drug response [33]. Libraries of small molecules, cDNA over-expression vectors, and shRNA knockdown vectors are used to perturb molecular signaling pathways in cancer cell lines, and cell viability is assayed by measuring the rate of cell proliferation [33–36]. The goal of this line of research, known as cancer functional genomics, is to systematically identify the function of genes/proteins in individual cancers and ultimately discover the genomic alterations that cause cancer cell vulnerability to particular molecular perturbations [37, 38]. Associations between genomic alterations and sensitivity to molecular perturbations are discovered by assaying cell viability across many cancer cell lines that have been genotyped by The Cancer Genome Atlas (TCGA) Research Network [39] and other large scale cancer genomics projects [33]. These associations are potentially informative for drug target and drug identification and prioritization.
2.4.2. Cancer Cell Line Encyclopedia (CCLE) and Genomics of Drug Sensitivity in Cancer (GDSC)
The Cancer Cell Line Encyclopedia (CCLE) is a dataset of gene expression, genotype, and drug sensitivity data for human cancer cell lines [40]. So far, gene expression profiles and sequence variants were measured in 947 cancer cell lines. 479 of the cell lines were treated with a panel of 24 anticancer drugs over a range of concentrations, and drug sensitivity was calculated from the dose-response curves for each cell-line/drug combination. The Genomics of Drug Sensitivity in Cancer (GDSC) project is another large scale effort to profile gene expression, genotype, and drug sensitivity of human cancer cell lines [41]. Gene expression profiles and sequence variants were measured in 639 human cancer cell lines. 130 drugs were selected for screening, and dose-response curves were measured for many cell lines, yielding drug sensitivity data for a total of 48,178 drug/cell-line combinations. Gene expression, genotyping, and drug sensitivity data have also been collected for 77 drugs screened against 49 breast cancer cell lines [42], and 19 drugs were screened against 311 cancer cell lines [43]. As described above for the CTD2 project, these data can be analyzed for patterns of gene expression or sequence variation that may explain differences in drug sensitivity across cell lines. However, some caution must be taken when making use of these data. Meta-analysis of the CCLE and GDSC datasets found that although gene expression measurements were well correlated between the two studies, 471 of the same cell lines were profiled in both studies, drug sensitivity measurements were poorly correlated between the two studies, 15 of the same drugs were profiled in both studies [44]. Cell viability data from CTD2, CCLE, and GDSC can be processed into attribute tables that connect cell lines to perturbations that increase or decrease cell viability. Weights can be obtained from the normalized EC50 values. Networks connecting cell lines based on similarity of sensitivity to perturbations, or networks that connect perturbations based on similarity of their effect on cell viability can be derived from the attribute tables. Gene expression and genotyping data from CCLE and GDSC can also be processed into attribute tables connecting cell lines to the genes that are up- or down-regulated in each cell-line compared to the rest of the cell lines, or to the genes that are mutated or amplified in the cell lines, respectively.
2.5. Properties of Drugs
2.5.1. DrugBank, Pubchem and PharmGKB
DrugBank [45], PubChem [46], and PharmGKB [47] are databases that collect information about drugs, their chemical structure, their effects in cell based assays, their indications, classification, publications, side effects, known targets and more. When writing this review, DrugBank contained 6825 drugs and 4323 proteins that are involved in the drugs’ mechanisms of action, or mechanisms of clearance. 4141 of these proteins are known drug targets, accounting for a total of 14,594 drug-target associations. The database includes the drugs’ molecular structure, pharmacokinetic and pharmacodynamic properties, indications, mechanism of action, and affected molecular pathways. PubChem contains information about the structure, pharmacology, physical properties, and bioactivity of small molecules. The BioAssay branch of PubChem provides access to a collection bioactivity screening studies of small molecules contributed by individual researchers. PharmGKB summarizes what is known about the pharmacokinetic (PK) and pharmacodynamic (PD) pathways of drug distribution, action, and metabolism, with an emphasis on identifying relationships between genetic variants and the PK/PD properties of drugs. Many of the properties of drugs listed in DrugBank, PubChem, and PharmGKB can be processed into attribute tables connecting drugs to their properties. These include: targets, chemical structure, ATC codes, drug-drug interactions from insert labels, and the liver enzymes that are known to process the drugs. Such attributes, together with drug side-effect information as well as drug induced gene expression changes in human cells, can be used to form different types of drug-drug similarity networks that can be used to predict/impute missing data about drug properties for less studied drugs.
2.5.2. Side Effect Resource (SIDER), FDA Adverse Event Reporting System (FAERS) and Offsides
The Side Effect Resource (SIDER) is a database of drug side effects collected from package inserts and other drug documentation [48]. The database currently lists 99,423 drug-side effect interactions covering 4192 side effects and 996 drugs. Side effect terms are standardized using the Medical Dictionary for Regulatory Activities (MedDRA, http://www.meddra.org/), which contains a hierarchy of terms for adverse event reporting. The Food and Drug Administration (FDA) Adverse Event Reporting System (FAERS) is another database of drug/side-effects connections. However, FAERS is made of millions of records created from spontaneous non-mandatory reports by doctors, patients, and drug companies [49–51]. The purpose of FAERS is to help the FDA monitor drugs after they are marketed for adverse events not detected in clinical trials [50, 51]. Each report lists the drugs prescribed to a patient and the adverse event that occurred, which is assigned a term from MedDRA. The FDA collects several hundred thousand reports each year and FAERS has over seven million reports since its inception in 1969 [52]. Because reporting is subjective, and mostly voluntary, many incorrect reports exist within the raw FAERS data. Hence, FAERS data should be considered suggestive [50, 52–54]. Rigorous statistical analysis is required to draw associations with confidence [50, 52–54]. FAERS data suffers from the “missing denominator problem”: It is not known how many patients took the drug and did not experience any side-effects. Offsides is a dataset hosted by PharmGKB [55] to provide drug/side-effect relations mined from FAERS using state-of-the-art statistical methods to correct for such biases [54]. Offsides contains 438,801 drug/side-effect associations covering 1332 drugs and 10097 side effects. These associations, each of which is paired with a p-value to indicate its significance, were derived from 1,851,171 FAERS reports entered from 2004 to 2009. Side effect data from SIDER or FAERS can be processed into attribute tables connecting drugs to side effects that are caused by the drugs. Drug-drug similarity networks can be created based on shared side effects, and side effect similarity networks can be created based on shared drugs.
2.6. Pathway Databases
Pathway databases such WikiPathways [56], BioCarta [57], Kyoto Encyclopedia of Genes and Genomes (KEGG) [58, 59], and Reactome [60, 61] contain databases of cell signaling and metabolic pathways. Each pathway consists of a sub-network of molecules that interact to perform a defined cellular function or process. KEGG contains 456 manually curated pathway maps; WikiPathways currently hosts 1789 pathways covering 29 species that have been manually curated by a community of editors; Reactome has manually curated and peer reviewed protein-, complex-, reaction-, and pathway-level biological knowledge for 21 species, including coverage of 7200 proteins, 6615 complexes, 6849 reactions, and 1491 pathways for human. BioCarta maintains a collection of several hundred manually curated pathways. In addition to these major pathway databases there are many others [62–65]. Sets of genes participating in pathways can be extracted from pathway databases and formatted into a gene set libraries. Networks connecting pathways based on their shared member proteins can also be created. In addition, pathways can be merged to build a composite, mostly directed and signed, PPI regulatory network.
2.7. Physical Interaction Databases
2.7.1. Protein-Protein Interaction Databases
The Biological General Repository for Interaction Datasets (BioGRID) [66], the Human Protein Reference Database (HPRD) [67, 68], the Molecular Interaction Database (MINT) [69, 70], IntAct [71, 72], and STRING [73] are some of the leading databases enlisting protein-protein interactions extracted manually, automatically, or semi-automatically from publications. BioGRID contains over 150,000 genetic interactions and over 200,000 protein interactions from 30 species mostly mined from peer-reviewed publications. HPRD contains 30,047 proteins annotated with 41,327 protein-protein interactions that have been manually curated from the biomedical literature. HPRD also collects information regarding posttranslational modifications, protein abundance, subcellular localization, and protein domains. MINT currently contains 241,458 manually curated protein-protein interactions covering 35,553 proteins. These interactions were used to create HomoMINT, a database of inferred human protein-protein interactions [74]. IntAct contains 81,795 interactors, mostly proteins, but also small molecules, RNA and DNA. At the time of compiling this review IntAct had 287,103 interactions mined from 12,531 publications. Interactions in IntAct are annotated with isoform and posttranslational modification details if available. Roughly 30% of the IntAct data was extracted from assays of human proteins, another 30% from yeast, and the remainder from other model organisms. Unlike the other resources, STRING uses automated algorithms to search the literature for protein-protein interactions, to assign interactions observed in one organism to other organisms, and to calculate a confidence score for each interaction. Extracted interactions in STRING may be physical or functional, known or predicted. The aim of STRING is to be comprehensive and as such STRING has collected over 200,000,000 interactions involving over 5,000,000 proteins for over 1000 organisms. Protein-protein interaction data can be stored in an adjacency matrix. Imputation algorithms can be used to predict undiscovered interactions. One of the caveats of literature-based PPI networks is that they are highly biased toward the most studied proteins. If this bias is not accounted for, reuse and analysis that emerge from such data can be misleading.
2.7.2. The Human Complexome
The view of protein-protein interactions as a binary adjacency matrix is limited because in reality, inside cells, proteins are organized in multi-protein complexes. Typically, such complexes are identified by immuno-precipitations followed by mass spectrometry (IP-MS). This method uses an antibody to “bait” a single protein that is used to pull the rest of the complex. The bait protein is isolated with the complexes that it interacts with and peptides from these complexes are sequenced by a mass spectrometer to identify the complexes’ members. The NIH sponsored long-term project called the Nuclear Receptor Signaling Atlas (NURSA) consists of over 3000 such IP-MS experiments [75]. The NURSA project was designed to characterize complexes made of nuclear gene-regulatory proteins. The NURSA IP-MS dataset is essentially a gene-set library where each term is a pull-down experiment defined by the bait protein and the antibody, and the genes in each set are the co-precipitated proteins identified by mass spectrometry. Each IP-MS experiment provides a snapshot of the entire protein interactome and binary interactions can be inferred from such data [76, 77]. Databases that consolidate knowledge about protein complexes such as CORUM [78] can also be converted to gene-set libraries or used for predicting binary protein interactions.
2.8. Molecular Patient Data
2.8.1. The Cancer Genome Atlas (TCGA)
The Cancer Genome Atlas (TCGA) is a cancer profiling project that seeks to collect genomic and clinical data from many different cancer patients [79, 80]. One of the aims of the project is to enable researchers to mine TCGA datasets for associations between genomic characteristics of cancer tissues and clinical outcomes of patients [79], while another aim is to search for genomic similarities between cancers that may suggest similar pathologic mechanisms and similar treatment strategies [80]. Exomes, copy number variations, DNA methylations, mRNA expression and miRNA expression are profiled for most tumors. Whole genome sequencing and protein abundance are measured for a subset of tumors. Clinical data, such as survival, recurrence, and therapeutic regimens are collected to identify correlations between epigenetic, DNA, RNA, and protein data with patient outcomes. So far, nearly 9000 tumor samples covering 29 tumor types have been profiled under the TCGA program. The TCGA Research Network has published many analyses of these datasets [79–88]. Genotype and gene expression data from TCGA can be processed in much the same way as the data from CCLE and GDSC. Genotype data from TCGA can be processed into an attribute table connecting tumors to the genes that are mutated or amplified in the tumors. Networks connecting patients based on shared mutated genes, gene expression, methylation patterns, or any other molecular data, or by combining datasets from across regulatory layers can be used to create patient similarity networks. Similarly, networks that connect genes based on mutations or co-expression can be created from the same data. Outcome data can be brought in to interpret analyses of the tumor profiling datasets. For example, unsupervised or supervised clustering of patients may be used to identify groups of patients with similar outcome [89]. Kaplan-Meier survival curves [90] could then be computed for the distinct clusters to test if patient outcomes differ by tumor cluster. Such clusters, besides being biomarkers for patient classification, can also explain the molecular mechanisms that drive tumor development in those patients.
2.9. Resources Overview
Table 1 summarizes the attribute tables that can be obtained from the resources described in this section. Each attribute table defines connections between a pair of the following classes of entities: genes, genomic loci, drugs, phenotypes, cell lines, and tissues. In principle, any two attribute tables that share a class in common are amenable for data fusion. In the next section we discuss data structures that can be used to abstract the data from the resources so they can be easily integrated.
Table 1.
Attribute tables that can be derived from high-content datasets from systems biology and systems pharmacology. Nearly all of the data can be represented as connections between pairs of the following entities: phenotypes, genes, genomic loci, drugs, cell lines, and tissues. DE- differentially expressed; DS- differentially sensitive; DM- differentially methylated. These are only examples of such sources but there are many more databases and projects.
| Row Labels | Column Labels | Resource |
|---|---|---|
| Phenotypes | Genes (knockouts) | MGI-MPO |
| Phenotypes | Genes (mutations) | OMIM, GWASdb |
| Phenotypes | Genomic loci (QTL) | GWASdb |
| Phenotypes (diseases) | Genes (DE) | GEO |
| Drugs | Genes (DE) | GEO, CMAP |
| Genes (knockdowns) | Genes (DE) | GEO, CMAP |
| Genes (over-expressions) | Genes (DE) | GEO, CMAP |
| Transcription factors | Genomic loci (binding sites) | ENCODE, ChEA |
| Transcription factors | Genes (targets) | ENCODE, ChEA |
| Histone modifications | Genomic loci (binding sites) | Roadmap Epigenomics, ENCODE |
| Histone modifications | Genes (targets) | Roadmap Epigenomics, ENCODE |
| Genomic loci (eQTL) | Genes (targets) | GTEx |
| Drugs | Cell lines (DS) | CCLE, GDSC, CTD2 |
| Genes (knockdowns) | Cell lines (DS) | CTD2 |
| Genes (over-expressions) | Cell lines (DS) | CTD2 |
| Cancer Cell lines | Genes (DE) | CCLE, GDSC |
| Cancer Cell lines | Genes (mutations) | CCLE, GDSC |
| Drugs | Phenotypes (side effects) | SIDER, FAERS, Offsides |
| Drugs | Phenotypes (indications) | DrugBank, PubChem, PharmGKB |
| Drugs | Genes (target proteins) | DrugBank, PubChem, PharmGKB |
| Pathways | Genes (member proteins) | WikiPathways, BioCarta, KEGG, Reactome |
| Genes (proteins) | Genes (interacting proteins) | BioGRID, HPRD, MINT, IntAct, STRING |
| Kinases | Genes (substrates) | Phosphosite Plus, NetworKIN, Phospho.ELM, KEA |
| Genes (IP target proteins) | Genes (co-precipitated proteins) | NURSA IP-MS |
3. DATA STRUCTURES
3.1. Attribute tables and bi-partite graphs
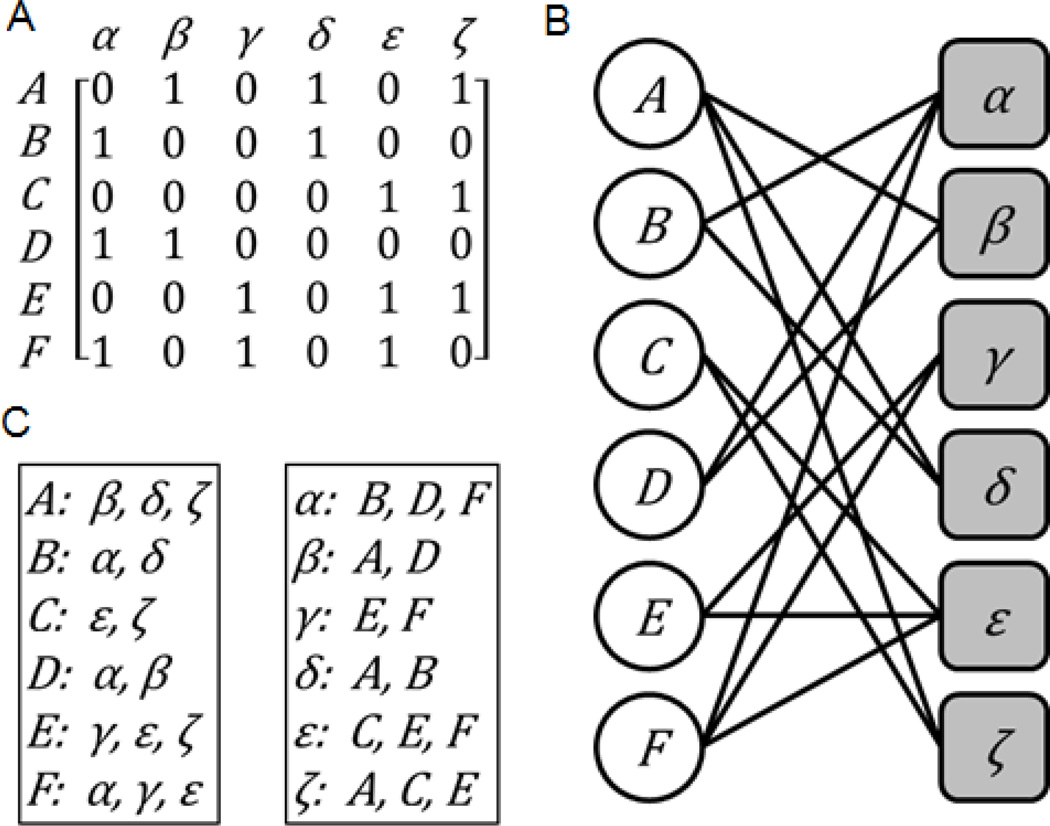
An attribute table is the most common and raw form for organizing high-content experimental data. Computationally, attribute tables can be represented as a matrix that defines the relationships between entities of two different classes (Fig. 1A) [91]. The row labels correspond to entities of one class and the column labels correspond to entities of the other class. Typically, the rows are the variables of a system, and measuring their level captures some aspect of the entities they represent. The columns on the other hand are typically the experimental conditions, or any other aspect that can be considered attributes of the system’s variables. The columns can be entities too, for example, the rows can represent genes and the columns represent different cell lines. Data entries in the matrix, which define the relationships between the variables and their attributes, can be discrete, binary, contain continuous positive values only, or both positive and negative values.
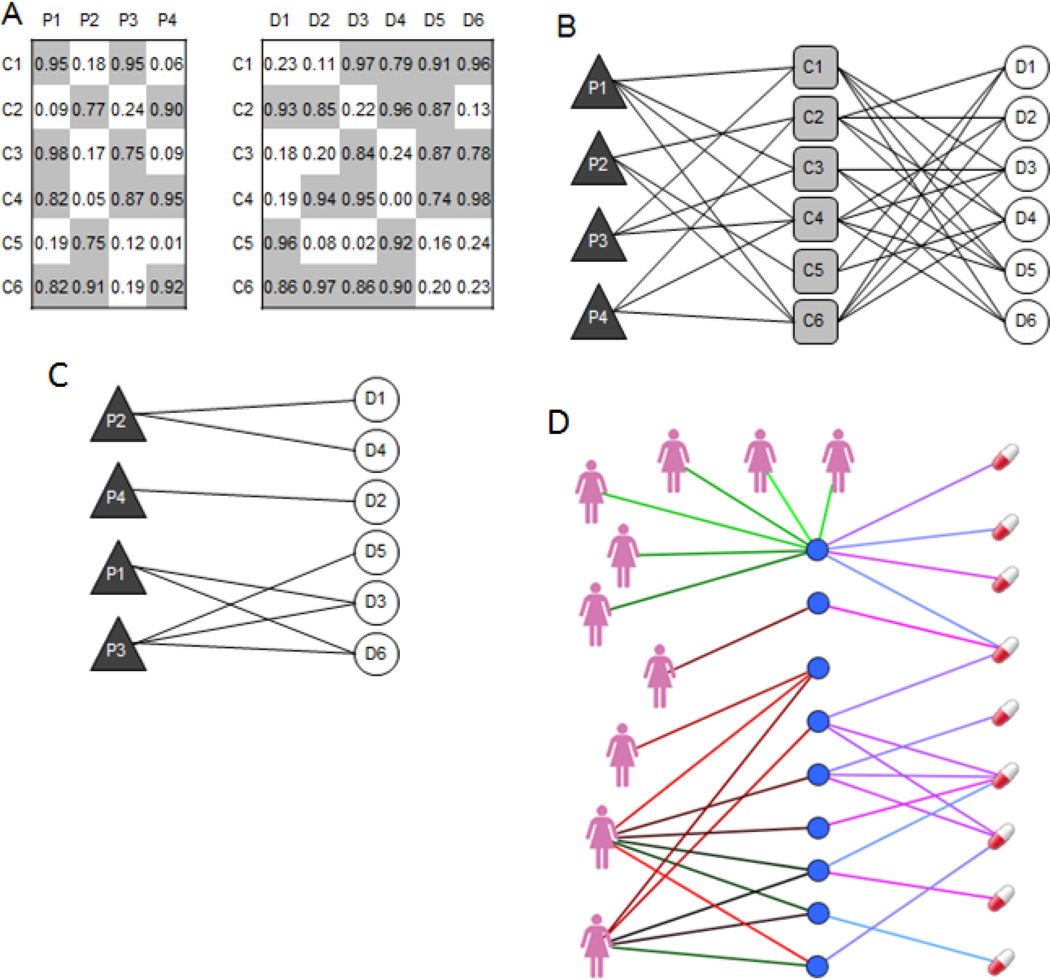
Fig. 1.
Illustration of (A) an attribute table, (B) its corresponding bi-partite graph, and (C) the corresponding set libraries.
Another related data structure is a bi-partite graph. Bi-partite graphs have two types of nodes representing the entities of two different classes. Connections between nodes are only allowed for linking nodes from the different two classes. The links in bi-partite graphs can be directly derived from an attribute table by setting a threshold so the bi-partite graph remains relatively sparse (Fig. 1B) [91]. Links between nodes in bi-partite graphs can also be binary, discrete, signed, or have continuous values which can be considered the links’ weights.
To make these concepts more concrete, next are some examples. Side effects of approved drugs can be represented as an attribute table, or a bi-partite graph, where the side effects belong to one class and the drugs belong to the other class. In this bi-partite graph representation, links connect drugs to the side effects that are known or suspected to be caused by the drugs [48]. In the attribute table representation, drugs are the variables and side-effects are the attributes and the matrix values are binary, with 1 indicating side-effect/drug association and zero indicating no association. Signatures of differentially expressed genes can also be represented in the form of an attribute table or a bi-partite graph [92]. A signature of differentially expressed genes is a vector where each entry contains a signed value that indicates the significance and direction (up-regulation or down-regulation) of the change in expression for a gene’s mRNA level between two states [17]. For example, a signature of differentially expressed genes can be obtained by comparing gene expression profiles measured after exposing cultured cancer cell lines to a drug, and measuring gene expression before and after exposure to the drug. Many related signatures of differentially expressed genes, such as signatures for different drug treatments applied to different cell lines in different concentrations, can be grouped into an attribute table. In this case, genes belong to one class of nodes, and drugs belong to the other class. The links in this bipartite graph represent the associations between drugs and the genes they influence. These links can be positive or negative and directed, indicating whether the drug treatment caused up- or down-regulation of the gene. The relationship between a drug and the mRNAs of the gene the drug affect is context dependent. It depends on the cell type and concentration of the drug, and the time point when gene expression was measured after drug exposure. Understanding this complex relationship is one of the current challenges in the field.
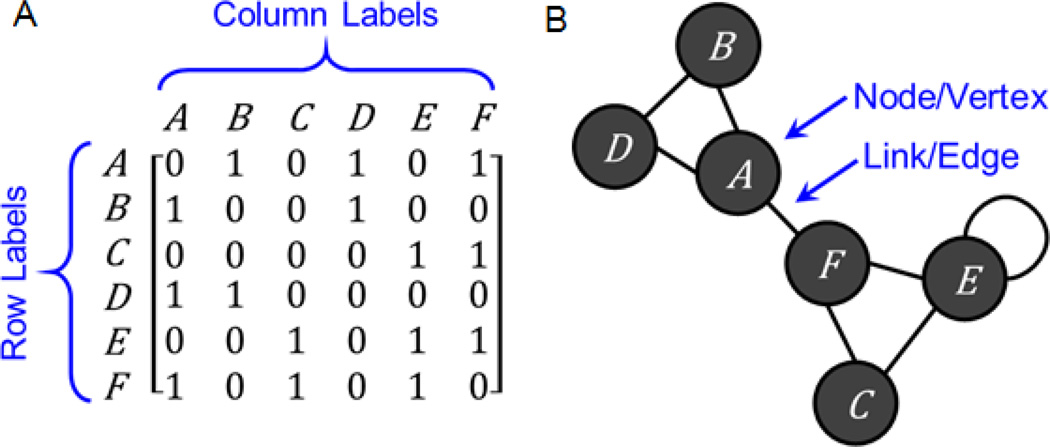
3.2. Adjacency matrices to represent single entity networks
A related data structure to the attribute table and the bi-partite graph, which is commonly applied for performing data integration tasks, is the single entity network, also named functional association networks (FANs) [93]. Such networks can be represented computationally as adjacency matrices. An adjacency matrix defines the connections between entities of a single class (Fig. 2A) [91]. The row labels are identical to the column labels and each label corresponds to a distinct entity. Thus, entry aij defines the relationship between entity i and entity j. If the connections between entities are undirected, then the adjacency matrix is symmetric. An adjacency matrix can be binary, contain discrete or continuous values that can be signed or unsigned. The meaning of values in the matrix depends on the network that the adjacency matrix represents. Single entity-type networks can be visualized as ball-and-stick diagrams (Fig. 1B) [91]. Each entry in the adjacency matrix corresponds to a link that connects nodes in the network. Here too thresholds are typically applied to keep the network sufficiently sparse. There is generally no best way for visualizing networks and many algorithms exist for computing layouts that attempt to minimize clutter [94, 95].
Fig. 2.
Illustration of (A) an adjacency matrix, and (B) its corresponding network.
One popular example for single node entity network is the representation of protein-protein interactions (PPI). PPI networks are the most typical and obvious FANs [96]. In such networks the nodes represent proteins and the edges represent physical interactions between proteins. Any protein-protein functional similarity network can also be represented in the same form. Here again, the nodes represent proteins, whereas the links represent functional similarity between the proteins, and the link weights represent the degree of similarity between the proteins, or the level of confidence such interactions carry. The degree of functional similarity between proteins can be calculated using one or several attributes of the proteins. For example, similarity scores can be computed from pair-wise amino acid sequence alignments, shared structural domains, shared Gene Ontology terms, co-expression, co-regulation by the same transcription factors, co-regulation by the same microRNAs, and more [77]. This concept is further elaborated later.
3.3. Set libraries
The third and slightly overlooked data structure that can be used for data integration and for extracting knowledge from big data is a set library. A set library consists of a collection of sets, where each set contains entities of the same class. Members of a single set are related in some way beyond the general class membership, and entities can belong to multiple sets in the library. Each set is assigned a label that defines why the entities in the set are related (Fig. 1C) [28, 97]. A set library, similarly to an attribute table, a single-entity network, or a bi-partite graph, is an alternative data structure that can be used to define relationships between entities of two different classes. The set labels correspond to entities of one class, and the sets contain entities of the other class. Set libraries can also be weighted or un-weighted [98]. For a weighted set library, each entity in a set is paired with a value, usually ranging from 0 to 1 to indicate the level of membership each entity has within the set. For the typical un-weighted set library, no values are paired with the entities, and a value of 1, representing full membership, is implied. Set libraries can store the same information as attribute tables or bi-partite graphs and are not just useful for organizing gene sets. For example, side effects for drugs can be represented in the form of a drug-set library, where side effects are assigned as set labels, and drugs, known or suspected to cause the side effects, are assigned to each set [7]. Other examples of gene-set libraries are transcription factors and their putative targets from ChIP-seq studies [6], kinases and their known substrates extracted from literature [99], pathways and the proteins that compose each pathway [100], genes that are putative targets of microRNAs [101] and more. Single node-type networks can also be converted to a set library by making the hubs, the highly connected nodes in the network, the set labels, while the elements of each set are the direct neighbors of each hub. We created such gene set library from a literature-based PPI network for the enrichment analysis software tools Enrichr [5], Lists2Networks [6], Network2Canvas [7], and Expression2Kinases [8]. In the next section we begin to describe how the processed and abstracted datasets can be analyzed for knowledge extraction.
4. DATA ANALYSIS AND DATA INTEGRATION
4.1. Supervised and unsupervised learning
4.1.1. Unsupervised Clustering
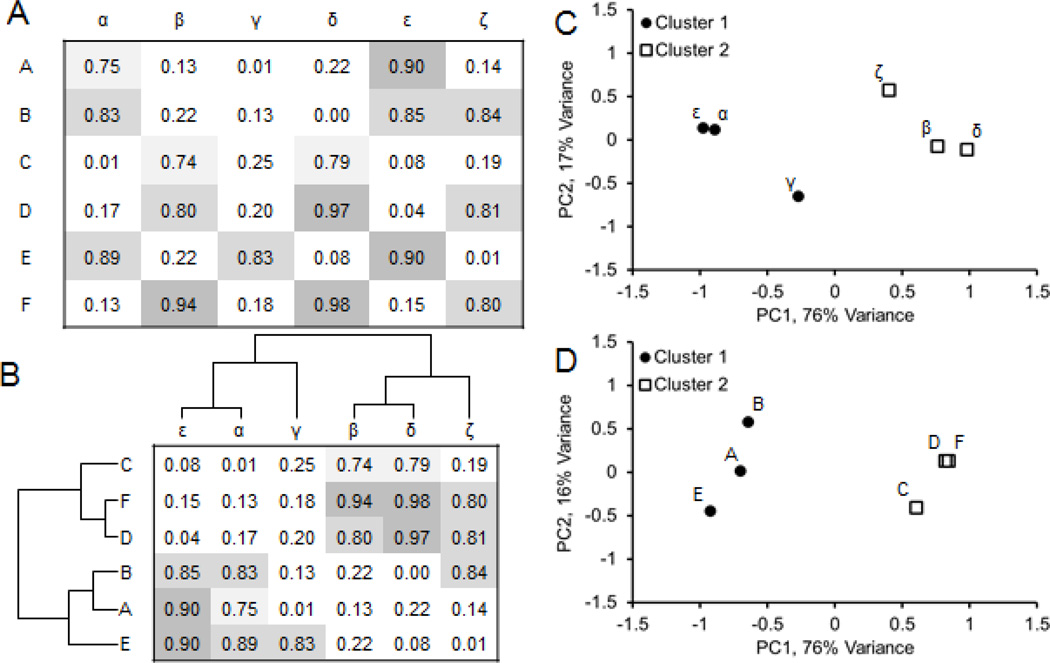
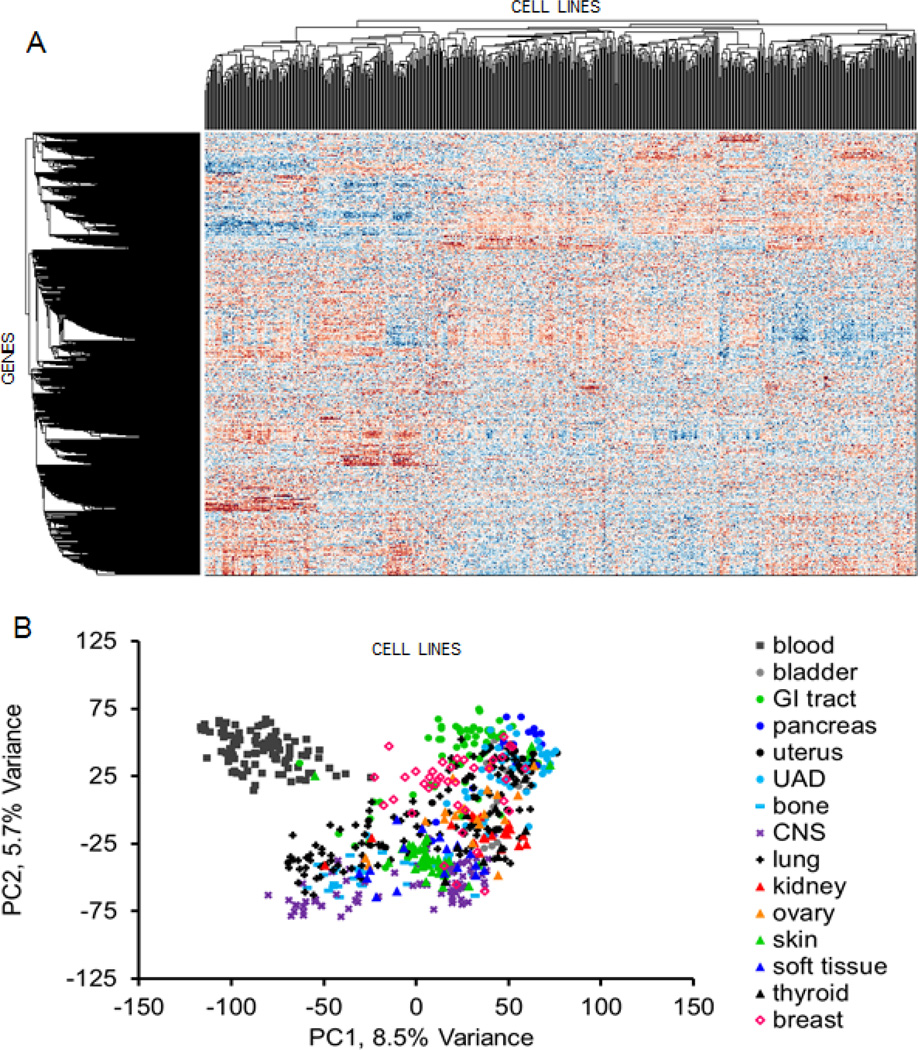
In the previous section we discussed how the information content of many open online resources can be converted into the simple data structures of attribute tables, bi-partite graphs, networks and set libraries. By organizing all this data into these formats, the task of data integration becomes straightforward. Entities in attribute tables, bi-partite graphs, networks and set libraries can be clustered based on entity similarity. Clustering is an unsupervised machine learning task for which many algorithms exist [102–104]. Hierarchical clustering is one of the most popular methods (Fig. 3a and 3b). It takes an attribute table or an adjacency matrix as an input and outputs a structure of branching connections, a dendrogram, that defines a hierarchy of groupings of entities based on how similar they are to each other, and also defines an ordering of entities such that similar entities are near each other [104, 105]. Bi-clustering refers to the application of hierarchical clustering to both the rows and columns of a matrix [92, 106, 107]. The bi-clustered matrix can then be visualized as a heatmap, known as a clustergram, which can show interesting patterns of connectivity that may lead to new hypotheses about how entities function or interact [108–111]. For example, if we create a bi-clustered attribute table connecting cancer cell-lines to genes based on mRNA expression, we can see groups of cells and genes with similar expression patterns (Fig. 4). Another, unsupervised clustering method is principle component analysis (PCA). PCA can be applied to the rows or columns of a dataset. The method rotates the axis of the data to reduce its dimensionality with minimal information loss. The results are typically visualized in two or three-dimensions to show an estimated distance between entities based on their data vector similarity in a reduced dimension and where each entity is represented as a point in the new and reduced PCA space (Fig. 3c and 3d). A tutorial that explains PCA step-by-step is provide here [112]. Another, unsupervised clustering method is to arrange the entities on a canvas [7, 113]. With this method the entities are randomly arranged on a grid and then shuffled, using simulated annealing [114] with the aim of placing similar entities near each other, while reducing the overall geometrical distance between entities on the grid. The entity representation on the grid /canvas can be made of any geometrical shape that tessellates, though hexagons provide the optimal tessellation. The canvas has no axis and the edges are made to fold on themselves to form a three dimensional shape such as a torus. The canvas visualization method clusters entities of the same type, so in essence the canvas is a way to visualize a clustered single-node-type network. We used this approach to visualize drug-drug and gene-gene similarity networks [7], display similarity between gene expression signatures after different drugs were applied to the same human cell lines for the LINCS project [115], and for clustering cancer cell lines based on their response to drugs, basal expression, or molecular structure similarity [116].
Fig. 3.
Hierarchical clustering and PCA as examples of unsupervised clustering.
(A) Hypothetical attribute-table connecting two classes of entities. (B) Hierarchical clustering can find groups of similar entities based on their attributes. In this example, we find two clusters. (C) Principal component analysis treating the column labels as entities and the row labels the attributes. The first two principal components capture 93% of the variance of the attributes across entities. (D) Principal component analysis treating the row labels the entities and the column labels the attributes.
Fig. 4.
The Cancer Genome Project [41] has profiled gene expression of hundreds of cancer cell lines. The data from 650 cell lines was quantile-normalized and log transformed, and then converted to z-scores for each row. (A) Hierarchical clustering applied to this data revealed distinct groups of cell lines and genes. (B) PCA treating the cell lines as entities and the genes the attributes coloring the cell lines by cancer type shows non-random ordering.
Unsupervised clustering becomes even more powerful when bringing together multiple datasets. For example, if we have a bi-partite graph connecting entities of class A to entities of class B, and a bi-partite graph connecting entities of class B to entities of class C, then we can find relationships between clusters of entities of class A and clusters of entities of class C simply by merging the two graphs (Fig. 5A–B). A tri-partite graph, however, may be difficult to interpret, so we may want to drop the intermediate entities (class B) from the picture, substituting edges that connect from A to B to C with edges that connect A to C (Fig. 5C). The integrated dataset is now in the form of a new bi-partite graph. At that stage, we can perform another round of data integration, for example, to find relationships between entities of class A and entities of class D given a bi-partite graph connecting entities of class C to entities of class D. However, as we join datasets this way, we expect to lose accuracy and introduce errors with longer chains. One example of merging two bi-partite graphs for discovering non-trivial relationships is to merge cell viability data after drug treatment from CCLE and GDP with patient data from profiling of tumors from TCGA. Such tri-partite graphs can suggest drugs that would be most effective to treat subsets of cancer patients (Fig. 5D) [117].
Fig. 5.
(A) Hypothetical attribute tables connecting patients (P1, P2, etc.) to cell lines (C1, C2, etc.) on the left, and connecting drugs (D1, D2, etc.) to cell lines on the right. Because these attribute tables share entities (cell lines) they can be integrated to form indirect connections between the disjoint entities (patients and drugs). (B) The datasets are integrated by merging their bi-partite graphs into a tri-partite graph. (C) Application of this concept on real data was previously published by Duan et al. [117] to integrate drug sensitivity data from CCLE with gene expression data from TCGA to connect patients to drugs. (D) The tri-partite graph in (B) can be made into a bi-partite graph by removing the intermediate nodes.
Many integrative analyses of this kind are made possible by the datasets described in Section 2. Side effect associations for drugs (e.g. from SIDER or Offsides) and drug-target interactions (e.g. from DrugBank [45], STITCH [118] or PubChem) have been integrated to identify proteins likely to cause side effects when perturbed [119]. GWAS data connecting diseases to genomic loci have been integrated with eQTL mapping data connecting genomic loci to regulated genes to find disease genes for complex disorders such as type 2 diabetes, crohn’s disease, and chronic obstructive pulmonary disease [120, 121]. GWAS data connecting diseases to genomic loci have also been integrated with ChIP-seq data mapping transcription factor binding sites to find annotated SNPs associated with various diseases as potential transcription factor binding sites [122]. These data can be integrated to identify sequence variants that regulate transcription factor binding and histone modifications [123]. Multi-omics profiling of cell lines, which is central to several large-scale NIH funded projects including LINCS can be useful for connecting transcription factors, histone modifications, sequence variants, and gene expression with proteomics, protein-protein interactions, and upstream cell signaling pathways.
4.1.2. Supervised Classification and Learn-to-Rank Algorithms
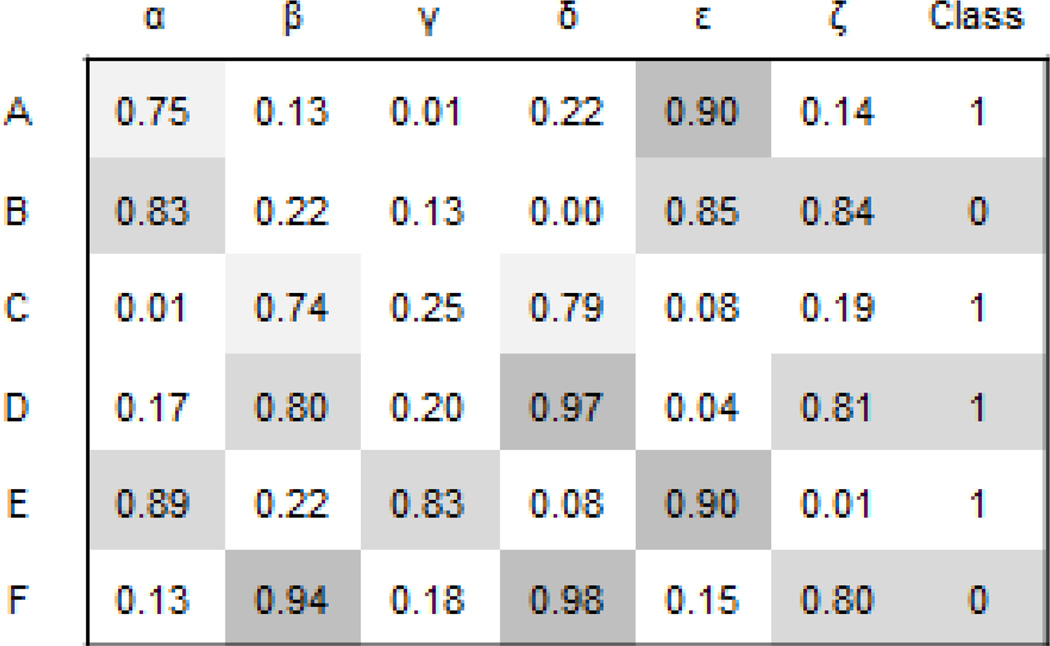
Conceptually, most supervised machine learning tasks are based on the “guilty by association” concept and as such this method is useful for filling knowledge gaps, imputing attribute values, ranking predicted interactions, and performing virtual experiments. Attribute tables with a known class are ideal for supervised learning tasks (Fig. 6). The goal is to learn the class for entities without known class but with known attributes. For this, classifiers are constructed. Classifiers infer the class for entities given their known attributes based on known attributes of similar entities with a known class [104, 105]. There are many types of classifiers with increased computational and mathematical sophistication and each method has its pros and cons. Some of the most commonly applied classifiers are naïve Bayes [124], logistic regression [125], support vector machine [126], neural networks [127], and random forests [128]. To train the classifiers and to evaluate their performance cross-validation methods are applied [129–131]. Cross-validation means that we train the classifier on some of the data instances with known class and hide some for testing. A full description of how to set up a classification problem, choosing classifier functions, training the classifier, selecting attributes, and cross-validating and testing classifiers is beyond the scope of this review. Many resources are available for learning the details of how to execute supervised machine learning tasks [104]. However, here we briefly present few applications of supervised machine learning for data integration tasks that can be used to extract more knowledge from public Big Data resources in biomedical research. These applications and methods can be used to benchmark computational and experimental methods, and to predict the most likely novel interactions in networks created from the fusion of several datasets.
Fig. 6.
Hypothetical attribute-table with known class.
4.1.3. Predicting PPI links
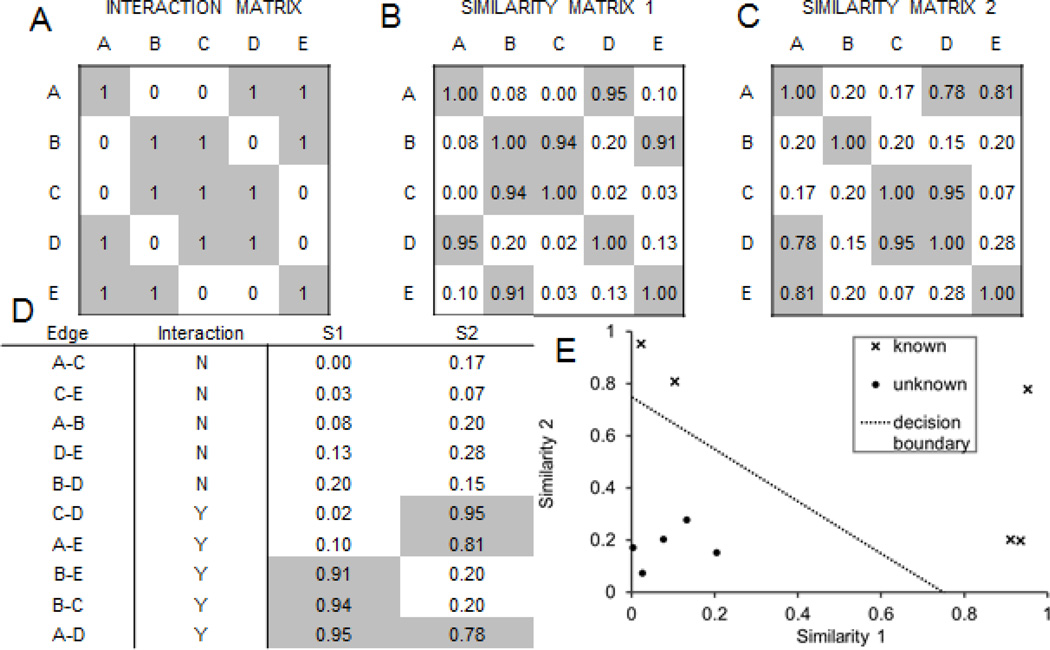
In section 3, we discussed how a single-node-type network can be derived from an attribute table or a bi-partite graph. We can combine several networks that connect the same entities through different types of links to predict new links and validate existing links. In other words, and for example, if we have a network with weighted ranked interactions where the weights represent likelihood of true interactions, and another network that we consider a gold standard true interaction network, and we believe that the two networks are related but represent different aspects of the entities they connect, we can use this setup for supervised learning: predicting and validating links (Fig. 7). This data abstraction for data integration application is powerful for benchmarking various computational methods that are used to collect data, process it, and eventually construct networks from the data. The quality of those final networks can be assessed by evaluating the networks using other networks, created from yet another independent source for connecting the same entities.
Fig. 7.
Similarity matrices can be integrated to classify interactions between entities. (A) Matrix of known interactions between entities, e.g. physical interactions between proteins. (B) Similarity matrix derived from an attribute table for the entities, e.g. protein similarity based on signatures of differentially expressed genes after knockdown. (C) Similarity matrix derived from another attribute table for the entities, e.g. protein similarity based on shared phenotypes in human disease or knockout mice. (D) List of edges, their class labels from the interaction matrix, and similarity scores for each edge. (E) Location of known (X) and unknown (filled circles) interactions in two-dimensional space defined by the two similarity measures. The two classes of interactions cannot be separated by either the similarity measures alone, but can be separated using a combination of the similarity measures.
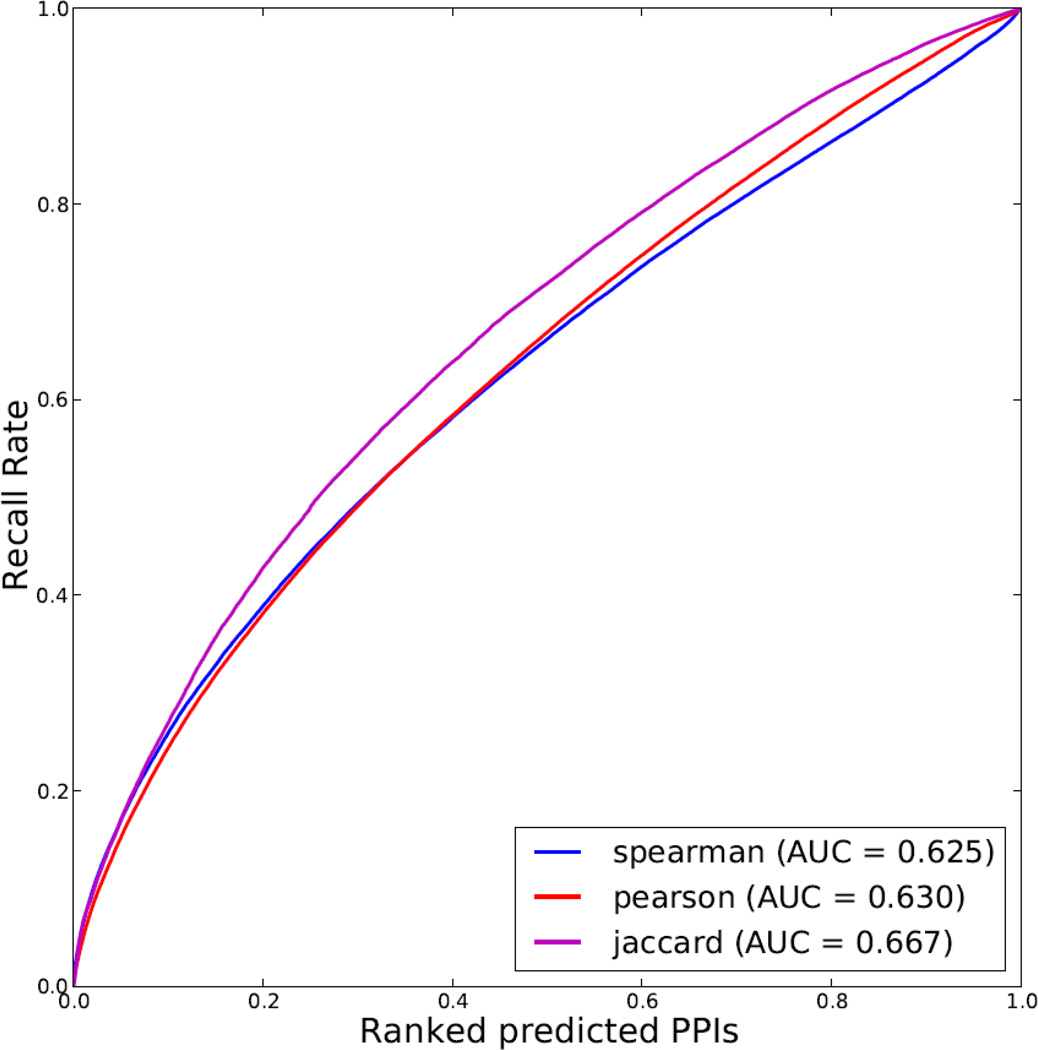
To better understand this concept, real examples are useful. The most obvious and typical example is to attempt to predict physical protein-protein interactions. Two recent studies used mass-spectrometry proteomics to measure protein expression in different human cells and tissues [132, 133]. One of those studies [132] showed that protein co-expression is more predictive of known protein-protein interactions as compared with protein-protein interactions predicted based on gene mRNA co-expression. To show this, the authors used a receiver operating characteristic (ROC) curve plot. Here we processed the data from the second study [133], and evaluated the ability of using the data from this study to predict protein-protein interactions by three computational methods that define similarity between proteins by quantifying co-expression (Fig. 8). The results show that the Jaccard Index is a better method than the other two measures of similarity: Spearman or Pearson correlation. The gold standard that was used to evaluate the predictions of protein-protein interactions is based on a literature-derived protein interaction network that we created for our tool Genes2Networks by merging 18 of the most established protein interaction databases [134]. This literature based protein-protein interaction network covers 15,630 human proteins connected through 185,068 interactions from 37,015 publications. Although this protein-protein interaction network is used as gold standard, it suffers from false positives and research focus biases. Hence, the top predicted interactions that are not confirmed by the gold standard network represent good candidates for experimental validation for real and novel protein-protein interactions.
Fig. 8.
Protein co-expression data to predict binary protein-protein interactions (PPI). Receiver Operating Characteristic (ROC) curves are plotted for evaluating predictions of binary protein-protein interactions based on protein co-expression across multiple tissues and cell lines measured by Pearson correlation coefficient (blue), Jaccard index (red) and Spearman's rank correlation coefficient (magenta). The area-under-the-curve (AUC) values are indicated in the legend. Protein expression data for 18,097 proteins across 207 different human tissues and cell lines were downloaded from proteomicsdb.org. Protein expression quantifications were log transformed. Proteins with expression values in at least seven tissues or cell lines were retained. Predicted protein interactions were sorted by the similarity measures in descending order, and validated against a literature-based protein interaction network created by integrating interactions from 18 databases and datasets.
4.1.4. Benchmarking computational methods
The example provided above, to predict protein-protein interactions, is just one of many possibilities. Recently, we used a similar approach to benchmark computational methods that can be used to identify differentially expressed genes from microarrays and RNA-seq data [135]. We combined transcription factor knockdowns followed by genome-wide expression with ChIP-seq data, profiling the binding sites of the same transcription factors on the human and mouse genomes. We used the ChIP-seq data as the silver standard and evaluated different methods that call differentially expressed genes for overlap with the matched transcription factor ChIP-seq inferred target genes. We consider the ChIP-seq data to be a silver standard and not a gold standard because we know that it contains many false positives. However, such data is still useful for benchmarking computational methods that identify differential expression because we expect more overlap between differentially expressed genes after transcription factor perturbation with putative targets for a transcription factor as determined by ChIP-seq, if the methods to identify such genes are better. This might not be true for a single study, particularly if the experiments were done in different cell types, but is likely to be true for a large collection of studies that profiled the same factors.
The rapid emergence of Big Data in biomedical research opens the opportunity for performing method validation and predictions at an unprecedented level. The ability to benchmark tools and datasets this way is related to gene set enrichment analysis [136] which belongs to a family of machine learning methods under the umbrella of learn-to-rank [137]. The Google search engine is known to combine many attributes to learn-to-rank best search results for its users. Related to this are multi-label-classifiers [138]. Multi-label-classifiers try to learn many class attributes at once. All these methods and applications are still in essence supervised machine learning methods that only differ in the problem setup. Supervised and unsupervised machine learning algorithms are central for continued progress with many exciting applications that involve data integration and method benchmarking. Much creativity can be applied to the processing of such data with the opportunity of discovering many hidden patterns, new biology and novel therapeutics. Crowdsourcing challenges such as DREAM are typically setup this way where participants compete for developing the best predictive models [139].
4.1.5. Predicting transcription-factor/target–gene and drug/target interactions
Another example of integrating transcription factor related data, using a semi-supervised approach, is the study by Ciofani et al. who integrated four datasets to infer Th17 cell specific gene regulatory networks [140]. The four datasets were: transcription factor binding site profiles measured by ChIP-seq, signatures of gene expression changes following transcription factor knockdown measured by RNA-seq, gene expression profiles for helper T cells measured by RNA-seq, and gene expression profiles for diverse immune cell types measured by microarray. On each of the latter two datasets, the Inferelator was applied to learn a regulatory network connecting transcription factors to their target genes. The Inferelator is a supervised method that uses Lasso regression [141] to find, for each gene or cluster of strongly-co-regulated genes, a parsimonious collection of transcription factors whose expression pattern across a number of conditions best explains the expression pattern of the gene cluster. After learning these networks, the four datasets were organized into weighted bipartite graphs connecting transcription factors to putative target genes, edge weights were converted to edge ranks for each graph, and then the edge ranks were averaged across graphs. The quality of the integrated (averaged) regulatory network was assessed by computing a ROC curve for a literature-curated Th17 specific transcription factor-target gene interaction network.
In addition to predicting protein-protein interactions, many attempts have been made to predict drug-protein interactions using supervised learning algorithms. Campillos et al. used similarity of drug side effects to predict drug targets [142]. Atias et al. used drug side effects and drug chemical structure to predict drug targets [143]. Takarabe et al. used drug side effects, drug chemical structure, and protein amino acid sequence to predict drug targets [144]; whereas Perlman et al. used drug side effects, drug chemical structure, signatures of DE genes after exposure of cell lines to drugs, drug ATC class, protein amino acid sequence, protein closeness in PPI, and Gene Ontology terms to predict drug targets [145]. Kernel methods were used for several of these approaches. That is, a kernel function was used to compute a drug-drug or protein-protein similarity matrix for each data type considered, and these matrices were integrated by, for example, summation, multiplication, or concatenation, and then supplied as input to a classifier such as a support vector machine.
4.1.5. Graphical Models
Another approach for integrating multiple types of data is to build a graphical model that explicitly accounts for relationships between data types. The tool Pathway Representation and Analysis by Direct reference on Graphical Models (PARADIGM) integrates copy number, gene expression, and protein expression in a Bayesian graphical model [146]. PARADIGM attempts to infer activation of pathways given some or all of the three data types just mentioned. First, a pathway is modeled as a directed acyclic graph. Edges are defined as activating or deactivating their downstream nodes and a function is defined for each node that combines all input signals to determine the activation of the node. Then each node that corresponds to a protein is assigned a parent node that corresponds to an mRNA transcript, which is assigned a parent node that corresponds to a gene sequence. Copy number variation data are prescribed at gene sequence nodes, gene expression data are prescribed at mRNA transcript nodes, and protein expression data are prescribed at protein nodes. The expectation-maximization algorithm is used to solve for the model parameters and all node activations given the prescribed data.
4.2 Biases within Big Biomedical Datasets
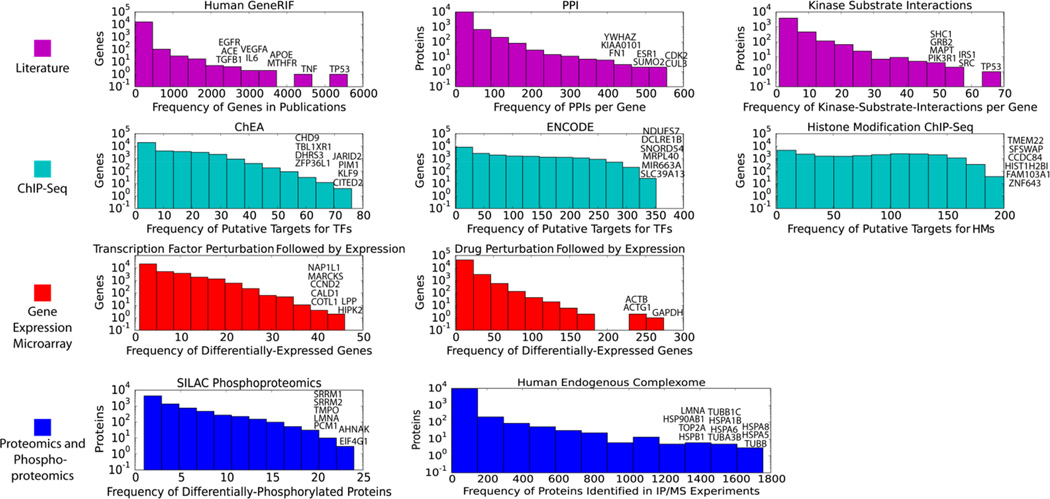
While abstracting and integrating datasets with the purpose of better knowledge extraction, attention should be given to gene-occurrence distribution biases that exist within each type of data. Such biases exist in data collected from both high content experiments and aggregation of results from low content studies in the literature. These biases origins are in the limits of experimental methods and human research focus biases. These biases are visible by the skewed distribution of the frequency of genes/proteins in gene set libraries, for example. To illustrate the skewed distribution of gene occurrence in various gene set libraries, we plotted the frequency of genes/proteins for gene set libraries created from different origins, ranging from literature curation abstracting results from low content experiment to high content experiment including ChIP-Seq, cDNA microarray and proteomics (Fig. 9). It should be noted that all of the histograms are in logarithm scale for the y-axis, demonstrating that the distribution of the frequencies of genes/proteins in these gene set libraries are highly skewed. The fact that some genes/proteins are overrepresented in gene set libraries does not necessarily means that biases exist, but we highly suspect them. If not considered and potentially corrected, this can lead to misinterpretations that potentially mask the true underlying associations between genes, cells, drugs, diseases and other phenotypes.
Fig. 9.
Frequency of gene/protein appearance in various biological networks and datasets. The most frequent genes/proteins from each datasets are highlighted. HM- histone modifications.
5. CONCLUSIONS
One of the most important aspects of integrating data for converting Big Data to knowledge is dealing with identifiers. IDs are used by different resources to represent entities with the same but sometimes also partially overlapping meaning. One example is gene IDs and protein IDs. ID mapping is a critical aspect of the data integration process which we ignored in our discussions. Another important aspect for data integration is data normalization. Because data is collected by different experimental methods, and high-content experiments are typically done in batches, data normalization strategies may have significant effect on the quality of the data fusion results. Here we avoided the discussion of specific data normalization strategies. However, we note that various normalization methods can be benchmarked as described in section 4.1.2. Another important consideration is that in order to integrate datasets of different types, for example, gene expression data with ChIP-seq data, some dataset-specific information must be sacrificed. For example, when creating a gene set library for transcription factors and their putative targets, we may lose the exact binding sites, peak height, number of peaks and distance to the transcription start site. It is expected that tools that consider such information for performing enrichment analyses, for example, will be more accurate that tools that use gene sets alone [147]. An important aspect of the data integration strategy that we propose here is that computational methods that operate on standard data structures can be reused for analyzing and extracting knowledge from the various datasets listed in section 2. The same functions and methods that operate on molecular networks can be applied to networks of drugs, patients, cells and phenotypes. For successful data integration applications across resources, deep understanding is required. We must ask the right questions, and aim to add value to the existing body of knowledge which typically is concerned with direct evidence and the most striking correlations within a single data type. Here we laid out a simple but effective foundation for integrating many of the leading resources in the field. Following such strategy is expected to shed new light on many undiscovered biological and pharmacological processes, and eventually lead us to improved personalized therapeutic strategies.
Highlights.
A small fraction of biomedical Big Data is converted to useful knowledge or reused.
Overview of a collection of structured mostly molecular mammalian biomedical Big Data resources.
Biases within data from these resources are suspected.
Data abstraction to attribute tables, networks and gene-sets enables reuse of biomedical datasets for integrative analyses.
Once data is abstracted it can be integrated and analyzed using supervised, unsupervised and integrative methods.
ACKNOWLEDGEMENTS
Funding: This work was supported in part by grants from the NIH: U54HL127624, U54CA189201, R01GM098316 and T32HL007824.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Mayer-Schönberger V, Cukier K. Big data: A revolution that will transform how we live, work, and think. Houghton Mifflin Harcourt; 2013. [Google Scholar]
- 2.Smith CL, Goldsmith CW, Eppig JT. The Mammalian Phenotype Ontology as a tool for annotatinganalyzing and comparing phenotypic information. Genome Biol. 2004;6:R7. doi: 10.1186/gb-2004-6-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blake JA, et al. The Mouse Genome Database: integration of and access to knowledge about the laboratory mouse. Nucleic Acids Res. 2014;42(1):D810–D817. doi: 10.1093/nar/gkt1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Austin CP, et al. The knockout mouse project. Nature genetics. 2004;36(9):921–924. doi: 10.1038/ng0904-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen E, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14(1):128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lachmann A, et al. ChEA: transcription factor regulation inferred from integrating genome-wide ChIP-X experiments. Bioinformatics. 2010;26(19):2438–2444. doi: 10.1093/bioinformatics/btq466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tan CM, et al. Network2Canvas: network visualization on a canvas with enrichment analysis. Bioinformatics. 2013;29(15):1872–1878. doi: 10.1093/bioinformatics/btt319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen EY, et al. Expression2Kinases: mRNA profiling linked to multiple upstream regulatory layers. Bioinformatics. 2012;28(1):105–111. doi: 10.1093/bioinformatics/btr625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amberger J, Bocchini C, Hamosh A. A new face and new challenges for Online Mendelian Inheritance in Man (OMIM(R)) Hum Mutat. 2011;32(5):564–567. doi: 10.1002/humu.21466. [DOI] [PubMed] [Google Scholar]
- 10.Amberger J, et al. McKusick's Online Mendelian Inheritance in Man (OMIM) Nucleic Acids Res. 2009;37(Database issue):D793–D796. doi: 10.1093/nar/gkn665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lara-Pezzi E, Dopazo A, Manzanares M. Understanding cardiovascular disease: a journey through the genome (and what we found there) Dis Model Mech. 2012;5(4):434–443. doi: 10.1242/dmm.009787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Becker KG, et al. The genetic association database. Nature Genetics. 2004;36:431–432. doi: 10.1038/ng0504-431. [DOI] [PubMed] [Google Scholar]
- 13.Li MJ, et al. GWASdb: a database for human genetic variants identified by genome-wide association studies. Nucleic Acids Res. 2012;40(Database issue):D1047–D1054. doi: 10.1093/nar/gkr1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thorisson GA, Muilu J, Brookes AJ. Genotype-phenotype databases: challenges and solutions for the post-genomic era. Nat Rev Genet. 2009;10(1):9–18. doi: 10.1038/nrg2483. [DOI] [PubMed] [Google Scholar]
- 15.Barrett T, et al. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 2013;41(Database issue):D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clark NR, et al. The characteristic direction: a geometrical approach to identify differentially expressed genes. BMC Bioinformatics. 2014;15:79. doi: 10.1186/1471-2105-15-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lamb J, et al. The Connectivity Map: using gene-expression signatures to connect small molecules genes and disease. Science. 2006;313(5795):1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 18.Lamb J. The connectivity map: a new tool for biomedical research. Nature Reviews Cancer. 2007;7 doi: 10.1038/nrc2044. [DOI] [PubMed] [Google Scholar]
- 19.Vempati UD, et al. Metadata Standard Data Exchange Specifications to Describe Model and Integrate Complex and Diverse High-Throughput Screening Data from the Library of Integrated Network-based Cellular Signatures (LINCS) J Biomol Screen. 2014;19(5):803–816. doi: 10.1177/1087057114522514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duan Q, et al. LINCS Canvas Browser: interactive web app to query, browse and interrogate LINCS L1000 gene expression signatures. Nucleic Acids Res. 2014 doi: 10.1093/nar/gku476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Consortium EP. The ENCODE (ENCyclopedia of DNA elements) project. Science. 2004;306(5696):636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- 22.Consortium TEP. A User’s Guide to the Encyclopedia of DNA Elements (ENCODE) PLoS Biology. 2011;9:e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rosenbloom KR, et al. ENCODE data in the UCSC Genome Browser: year 5 update. Nucleic Acids Res. 2013;41(Database issue):D56–D63. doi: 10.1093/nar/gks1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bernstein BE, et al. The NIH Roadmap Epigenomics Mapping Consortium. Nat Biotechnol. 2010;28(10):1045–1048. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chadwick LH. The NIH roadmap epigenomics program data resource. Epigenomics. 2012;4:317–324. doi: 10.2217/epi.12.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karnik R, Meissner A. Browsing (Epi)genomes: a guide to data resources and epigenome browsers for stem cell researchers. Cell Stem Cell. 2013;13(1):14–21. doi: 10.1016/j.stem.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rivera CM, Ren B. Mapping human epigenomes. Cell. 2013;155(1):39–55. doi: 10.1016/j.cell.2013.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen EY, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14(128) doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ernst J, Kellis M. Large-scale imputation of epigenomic datasets for systematic annotation of diverse human tissues. Nat Biotech. 2015;33(4):364–376. doi: 10.1038/nbt.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoffman MM, et al. Integrative annotation of chromatin elements from ENCODE data. Nucleic Acids Research. 2012 doi: 10.1093/nar/gks1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Consortium GT. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45(6):580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gilad Y, Rifkin SA, Pritchard JK. Revealing the architecture of gene regulation: the promise of eQTL studies. Trends Genet. 2008;24(8):408–415. doi: 10.1016/j.tig.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Network T.C.T.D.a.D. Towards patient-based cancer therapeutics. Nat Biotechnol. 2010;28:904–906. doi: 10.1038/nbt0910-904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Basu A, et al. An interactive resource to identify cancer genetic and lineage dependencies targeted by small molecules. Cell. 2013;154(5):1151–1161. doi: 10.1016/j.cell.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheung HW, et al. Systematic investigation of genetic vulnerabilities across cancer cell lines reveals lineage-specific dependencies in ovarian cancer. Proc Natl Acad Sci U S A. 2011;108(30):12372–12377. doi: 10.1073/pnas.1109363108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim HS, et al. Systematic identification of molecular subtype-selective vulnerabilities in non-small-cell lung cancer. Cell. 2013;155(3):552–66. doi: 10.1016/j.cell.2013.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boehm JS, Hahn WC. Towards systematic functional characterization of cancer genomes. Nat Rev Genet. 2011;12(7):487–498. doi: 10.1038/nrg3013. [DOI] [PubMed] [Google Scholar]
- 38.McDermott U, et al. Identification of genotype-correlated sensitivity to selective kinase inhibitors by using high-throughput tumor cell line profiling. Proc Natl Acad Sci U S A. 2007;104(50):19936–19941. doi: 10.1073/pnas.0707498104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weinstein JN, et al. The cancer genome atlas pan-cancer analysis project. Nature genetics. 2013;45(10):1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barretina J, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483(7391):603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garnett MJ, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483(7391):570–575. doi: 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heiser LM, et al. Subtype and pathway specific responses to anticancer compounds in breast cancer. Proc Natl Acad Sci U S A. 2012;109(8):2724–2729. doi: 10.1073/pnas.1018854108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Greshock J, et al. Molecular target class is predictive of in vitro response profile. Cancer Res. 2010;70(9):3677–3686. doi: 10.1158/0008-5472.CAN-09-3788. [DOI] [PubMed] [Google Scholar]
- 44.Haibe-Kains B, et al. Inconsistency in large pharmacogenomic studies. Nature. 2013;504(7480):389–393. doi: 10.1038/nature12831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wishart DS, et al. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic acids research. 2006;34(suppl 1):D668–D672. doi: 10.1093/nar/gkj067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Y, et al. PubChem: a public information system for analyzing bioactivities of small molecules. Nucleic acids research. 2009;37(suppl 2):W623–W633. doi: 10.1093/nar/gkp456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hewett M, et al. PharmGKB: the pharmacogenetics knowledge base. Nucleic acids research. 2002;30(1):163–165. doi: 10.1093/nar/30.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kuhn M, et al. A side effect resource to capture phenotypic effects of drugs. Mol Syst Biol. 2010;6:343. doi: 10.1038/msb.2009.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moore TJ, Cohen MR, Furberg CD. Serious Adverse Drug Events Reported to the Food, Drug Administration 1998–2005. Arch Intern Med. 2007;167:1752–1759. doi: 10.1001/archinte.167.16.1752. [DOI] [PubMed] [Google Scholar]
- 50.Sakaeda T, et al. Data mining of the public version of the FDA Adverse Event Reporting System. Int J Med Sci. 2013;10(7):796–803. doi: 10.7150/ijms.6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weiss-Smith S, et al. The FDA Drug Safety Surveillance Program: Adverse Event Reporting Trends. Arch Intern Med. 2011;171(591–592) doi: 10.1001/archinternmed.2011.89. [DOI] [PubMed] [Google Scholar]
- 52.Harpaz R, et al. Performance of pharmacovigilance signal-detection algorithms for the FDA adverse event reporting system. Clin Pharmacol Ther. 2013;93(6):539–546. doi: 10.1038/clpt.2013.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bate A, Evans SJ. Quantitative signal detection using spontaneous ADR reporting. Pharmacoepidemiol Drug Saf. 2009;18(6):427–36. doi: 10.1002/pds.1742. [DOI] [PubMed] [Google Scholar]
- 54.Tatonetti NP, et al. Data-driven prediction of drug effects and interactions. Sci Transl Med. 2012;4(125):125ra31. doi: 10.1126/scitranslmed.3003377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hewett M, et al. PharmGKB: the pharmacogenetics knowledge base. Nucleic Acids Res. 2002;30:163–165. doi: 10.1093/nar/30.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kanehisa M, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36(Database issue):D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nishimura D. BioCarta. Biotech Software & Internet Report: The Computer Software Journal for Scient. 2001;2(3):117–120. [Google Scholar]
- 58.Kanehisa M, et al. Data binformation knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 2014;42(1):D199–D205. doi: 10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ogata H, et al. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Croft D, et al. The Reactome pathway knowledgebase. Nucleic Acids Res. 2014;42(1):D472–D477. doi: 10.1093/nar/gkt1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vastrik I, et al. Reactome: a knowledge base of biologic pathways and processes. Genome Biol. 2007;8(3):R39. doi: 10.1186/gb-2007-8-3-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gough NR. Science's Signal Transduction Knowledge Environment. Annals of the New York Academy of Sciences. 2002;971(1):585–587. doi: 10.1111/j.1749-6632.2002.tb04532.x. [DOI] [PubMed] [Google Scholar]
- 63.Mueller LA, Zhang P, Rhee SY. AraCyc: a biochemical pathway database for Arabidopsis. Plant Physiology. 2003;132(2):453–460. doi: 10.1104/pp.102.017236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bader GD, Cary MP, Sander C. Pathguide: a pathway resource list. Nucleic acids research. 2006;34(suppl 1):D504–D506. doi: 10.1093/nar/gkj126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Karp PD, et al. The metacyc database. Nucleic acids research. 2002;30(1):59–61. doi: 10.1093/nar/30.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chatr-Aryamontri A, et al. The BioGRID interaction database: 2013 update. Nucleic Acids Res. 2013;41(Database issue):D816–D823. doi: 10.1093/nar/gks1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Peri S, et al. Development of human protein reference database as an initial platform for approaching systems biology in humans. Genome Res. 2003;13(10):2363–2371. doi: 10.1101/gr.1680803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Keshava Prasad TS, et al. Human Protein Reference Database--2009 update. Nucleic Acids Res. 2009;37(Database issue):D767–D772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Licata L, et al. MINT, the molecular interaction database: 2012 update. Nucleic Acids Res. 2012;40(Database issue):D857–D861. doi: 10.1093/nar/gkr930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zanzoni A, et al. MINT: a molecular interaction database. FEBS Lett. 2002;513:135–140. doi: 10.1016/s0014-5793(01)03293-8. [DOI] [PubMed] [Google Scholar]
- 71.Hermjakob H, et al. IntAct: an open source molecular interaction database. Nucleic Acids Res. 2004;32:D452–D455. doi: 10.1093/nar/gkh052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kerrien S, et al. The IntAct molecular interaction database in 2012. Nucleic Acids Res. 2012;40(Database issue):D841–D846. doi: 10.1093/nar/gkr1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Franceschini A, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41(Database issue):D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Persico M, et al. HomoMINT: an inferred human network based on orthology mapping of protein interactions discovered in model organisms. BMC bioinformatics. 2005;6(Suppl 4):S21. doi: 10.1186/1471-2105-6-S4-S21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Malovannaya A, et al. Analysis of the human endogenous coregulator complexome. Cell. 2011;145(5):787–799. doi: 10.1016/j.cell.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mazloom AR, et al. Recovering protein-protein and domain-domain interactions from aggregation of IP-MS proteomics of coregulator complexes. PLoS computational biology. 2011;7(12):e1002319. doi: 10.1371/journal.pcbi.1002319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Clark NR, et al. Sets2Networks: network inference from repeated observations of sets. BMC systems biology. 2012;6(1):89. doi: 10.1186/1752-0509-6-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ruepp A, et al. CORUM: the comprehensive resource of mammalian protein complexes. Nucleic acids research. 2008;36(suppl 1):D646–D650. doi: 10.1093/nar/gkm936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cancer Atlas Research, N. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cancer Genome Atlas Research, N et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 2013;45(10):1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cancer Genome Atlas Research, N. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474(7353):609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cancer Genome Atlas, N. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cancer Genome Atlas, N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cancer Genome Atlas Research, N. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489(7417):519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cancer Genome Atlas Research, N. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499(7456):43–49. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cancer Genome Atlas Research, N. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368(22):2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cancer Genome Atlas Research, N et al. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497(7447):67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.The Cancer Genome Atlas Research, N. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014 doi: 10.1038/nature12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Duan Q, et al. Metasignatures identify two major subtypes of breast cancer. CPT: pharmacometrics & systems pharmacology. 2013;2(3):1–10. doi: 10.1038/psp.2013.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kaplan EL, Meier P. Nonparametric estimation from incomplete observations. J. Amer. Statist.Assn. 1958;53(282):457–481. [Google Scholar]
- 91.Balakrishnan R, Ranganathan K. In: A textbook of graph theory. Universitext. Axler S, et al., editors. New York: Springer; 2012. [Google Scholar]
- 92.Madeira SC, Oliveira AL. Biclustering algorithms for biological data analysis: a survey. IEEE Transactions on Computational Biology and Bioinformatics. 2004;1:24–45. doi: 10.1109/TCBB.2004.2. [DOI] [PubMed] [Google Scholar]
- 93.Dannenfelser R, Clark NR, Ma'ayan A. Genes2FANs: connecting genes through functional association networks. BMC bioinformatics. 2012;13(1):156. doi: 10.1186/1471-2105-13-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Suderman M, Hallett M. Tools for visually exploring biological networks. Bioinformatics. 2007;23(20):2651–2659. doi: 10.1093/bioinformatics/btm401. [DOI] [PubMed] [Google Scholar]
- 95.Gehlenborg N, et al. Visualization of omics data for systems biology. Nat Methods. 2010;7(3 Suppl):S56–S68. doi: 10.1038/nmeth.1436. [DOI] [PubMed] [Google Scholar]
- 96.Fung DC, et al. Visualization of the interactome: what are we looking at? Proteomics. 2012;12(10):1669–1686. doi: 10.1002/pmic.201100454. [DOI] [PubMed] [Google Scholar]
- 97.Liberzon A, et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27(12):1739–1740. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Qureshi R, Sacan A. Weighted set enrichment of gene expression data. BMC Syst Biol. 2013;7(Suppl 4):S10. doi: 10.1186/1752-0509-7-S4-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lachmann A, Ma'ayan A. KEA: kinase enrichment analysis. Bioinformatics. 2009;25(5):684–686. doi: 10.1093/bioinformatics/btp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Liberzon A, et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27(12):1739–1740. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Steinfeld I, et al. miRNA target enrichment analysis reveals directly active miRNAs in health and disease. Nucleic acids research. 2013;41(3):e45–e45. doi: 10.1093/nar/gks1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bandyopadhyay S, Saha S. Unsupervised Classification. Heidelberg: Springer-Verlag; 2013. [Google Scholar]
- 103.Jain AK. Data clustering: 50 years beyond K-means. Pattern Recognition Letters. 2010;31(8):651–666. [Google Scholar]
- 104.Hastie T, Tibshirani R, Friedman J. The Elements of Statistical Learning. 2 ed. Springer; 2008. [Google Scholar]
- 105.Larranaga P. Machine learning in bioinformatics. Briefings in Bioinformatics. 2006;7(1):86–112. doi: 10.1093/bib/bbk007. [DOI] [PubMed] [Google Scholar]
- 106.Oghabian A, et al. Biclustering methods: biological relevance and application in gene expression analysis. PLoS One. 2014;9(3):e90801. doi: 10.1371/journal.pone.0090801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Eren K, et al. A comparative analysis of biclustering algorithms for gene expression data. Brief Bioinform. 2013;14(3):279–292. doi: 10.1093/bib/bbs032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Harpaz R, et al. Biclustering of adverse drug events in the FDA's spontaneous reporting system. Clin Pharmacol Ther. 2011;89(2):243–50. doi: 10.1038/clpt.2010.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Choi H, et al. Analysis of protein complexes through model-based biclustering of label-free quantitative AP-MS data. Mol Syst Biol. 2010;6:385. doi: 10.1038/msb.2010.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ghasemi O, et al. A biclustering approach to analyze drug effects on extracellular matrix remodeling post-myocardial infarction. IEEE International Conference on Bioinformatics and Biomedicine Workshops. 2012:143–150. [Google Scholar]
- 111.Wu C, et al. A Biclustering Algorithm to Discover Functional Modules from ENCODE ChIP-Seq Data. 2013. pp. 96–103. [Google Scholar]
- 112.Clark NR, Ma’ayan A. Introduction to statistical methods to analyze large data sets: Principal components analysis. Science signaling. 2011;4(190):tr3. doi: 10.1126/scisignal.2001967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.MacArthur BD, et al. GATE: software for the analysis and visualization of high-dimensional time series expression data. Bioinformatics. 2010;26(1):143–144. doi: 10.1093/bioinformatics/btp628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Aarts E, Korst J. Simulated annealing and Boltzmann machines. 1988. [Google Scholar]
- 115.Duan Q, et al. LINCS Canvas Browser: interactive web app to query, browse and interrogate LINCS L1000 gene expression signatures. Nucleic acids research. 2014:gku476. doi: 10.1093/nar/gku476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Duan Q, et al. Drug/Cell-line Browser: interactive canvas visualization of cancer drug/cell-line viability assay datasets. Bioinformatics. 2014 doi: 10.1093/bioinformatics/btu526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Duan Q, et al. Metasignatures identify two major subtypes of breast cancer. CPT: Pharmacometrics & Systems Pharmacology. 2013;2(3):e35. doi: 10.1038/psp.2013.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kuhn M, et al. STITCH 3: zooming in on protein–chemical interactions. Nucleic acids research. 2012;40(D1):D876–D880. doi: 10.1093/nar/gkr1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kuhn M, et al. Systematic identification of proteins that elicit drug side effects. Molecular systems biology. 2013;9(1) doi: 10.1038/msb.2013.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.He X, et al. Sherlock: detecting gene-disease associations by matching patterns of expression QTL and GWAS. The American Journal of Human Genetics. 2013;92(5):667–680. doi: 10.1016/j.ajhg.2013.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lamontagne M, et al. Refining susceptibility loci of chronic obstructive pulmonary disease with lung eqtls. PloS one. 2013;8(7):e70220. doi: 10.1371/journal.pone.0070220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bryzgalov LO, et al. Detection of regulatory SNPs in human genome using ChIP-seq ENCODE data. PloS one. 2013;8(10):e78833. doi: 10.1371/journal.pone.0078833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kilpinen H, et al. Coordinated effects of sequence variation on DNA binding, chromatin structureand transcription. Science. 2013;342(6159):744–747. doi: 10.1126/science.1242463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lewis DD. Machine learning: ECML-98. Springer; 1998. Naive (Bayes) at forty: The independence assumption in information retrieval; pp. 4–15. [Google Scholar]
- 125.Bishop CM. Pattern recognition and machine learning. Vol. 4. New York: springer; 2006. [Google Scholar]
- 126.Cortes C, Vapnik V. Support-vector networks. Machine learning. 1995;20(3):273–297. [Google Scholar]
- 127.Russell S, Norvig P, Intelligence A. Artificial Intelligence. Vol. 25 Egnlewood Cliffs: Prentice-Hall; 1995. A modern approach. [Google Scholar]
- 128.Breiman L. Random forests. Machine learning. 2001;45(1):5–32. [Google Scholar]
- 129.Shao J. Linear Model Selection by Cross-validation. Journal of the American Statistical Association. 1993;88(422):486–494. [Google Scholar]
- 130.Schaffer C. Selecting a classification method by cross-validation. Machine Learning. 1993;13:135–143. [Google Scholar]
- 131.Zhang P. Model selection via multifold cross validation. Annals of Statistics. 1993;21:299–313. [Google Scholar]
- 132.Kim M-S, et al. A draft map of the human proteome. Nature. 2014;509(7502):575–581. doi: 10.1038/nature13302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wilhelm M, et al. Mass-spectrometry-based draft of the human proteome. Nature. 2014;509(7502):582–587. doi: 10.1038/nature13319. [DOI] [PubMed] [Google Scholar]
- 134.Berger S, Posner J, Ma'ayan A. Genes2Networks: connecting lists of gene symbols using mammalian protein interactions databases. BMC Bioinformatics. 2007;8(1):372. doi: 10.1186/1471-2105-8-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Clark NR, et al. The characteristic direction: a geometrical approach to identify differentially expressed genes. BMC bioinformatics. 2014;15(1):79. doi: 10.1186/1471-2105-15-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Liu T-Y. Learning to rank for information retrieval. Foundations and Trends in Information Retrieval. 2009;3(3):225–331. [Google Scholar]
- 138.Tsoumakas G, Katakis I. Multi-label classification: An overview. International Journal of Data Warehousing and Mining (IJDWM) 2007;3(3):1–13. [Google Scholar]
- 139.Marbach D, et al. Wisdom of crowds for robust gene network inference. Nature methods. 2012;9(8):796–804. doi: 10.1038/nmeth.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ciofani M, et al. A validated regulatory network for Th17 cell specification. Cell. 2012;151(2):289–303. doi: 10.1016/j.cell.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Tibshirani R. Regression shrinkage and selection via the lasso. Journal of the Royal Statistical Society. Series B (Methodological) 1996:267–288. [Google Scholar]
- 142.Campillos M, et al. Drug target identification using side-effect similarity. Science. 2008;321(5886):263–266. doi: 10.1126/science.1158140. [DOI] [PubMed] [Google Scholar]
- 143.Atias N, Sharan R. An algorithmic framework for predicting side effects of drugs. Journal of Computational Biology. 2011;18(3):207–218. doi: 10.1089/cmb.2010.0255. [DOI] [PubMed] [Google Scholar]
- 144.Takarabe M, et al. Drug target prediction using adverse event report systems: a pharmacogenomic approach. Bioinformatics. 2012;28(18):i611–i618. doi: 10.1093/bioinformatics/bts413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Perlman L, et al. Combining drug and gene similarity measures for drug-target elucidation. Journal of computational biology. 2011;18(2):133–145. doi: 10.1089/cmb.2010.0213. [DOI] [PubMed] [Google Scholar]
- 146.Vaske CJ, et al. Inference of patient-specific pathway activities from multi-dimensional cancer genomics data using PARADIGM. Bioinformatics. 2010;26(12):i237–i245. doi: 10.1093/bioinformatics/btq182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Welch RP, et al. ChIP-Enrich: gene set enrichment testing for ChIP-seq data. Nucleic Acids Research. 2014 doi: 10.1093/nar/gku463. [DOI] [PMC free article] [PubMed] [Google Scholar]