Abstract
Microbial enzyme diversity is a key to understand many ecosystem processes. Whole metagenome sequencing (WMG) obtains information on functional genes, but it is costly and inefficient due to large amount of sequencing that is required. In this study, we have applied a captured metagenomics technique for functional genes in soil microorganisms, as an alternative to WMG. Large-scale targeting of functional genes, coding for enzymes related to organic matter degradation, was applied to two agricultural soil communities through captured metagenomics. Captured metagenomics uses custom-designed, hybridization-based oligonucleotide probes that enrich functional genes of interest in metagenomic libraries where only probe-bound DNA fragments are sequenced. The captured metagenomes were highly enriched with targeted genes while maintaining their target diversity and their taxonomic distribution correlated well with the traditional ribosomal sequencing. The captured metagenomes were highly enriched with genes related to organic matter degradation; at least five times more than similar, publicly available soil WMG projects. This target enrichment technique also preserves the functional representation of the soils, thereby facilitating comparative metagenomics projects. Here, we present the first study that applies the captured metagenomics approach in large scale, and this novel method allows deep investigations of central ecosystem processes by studying functional gene abundances.
Keywords: microbial ecology, functional diversity, sequence capture, 454 pyrosequencing, comparative metagenomics
1. Introduction
The knowledge on the link between functional diversity and species richness is one of the key areas that is lacking for better understanding of ecosystem functioning in soils,1–3 Ecosystems such as soil have vast diverse microorganisms with several thousand complex functional capabilities.4,5 Novel technological advancements are thought to be a way towards the understanding of microbial functional diversity in biogeochemical processes more clearly.6 Several molecular methods like PCR, RFLP, microarrays and sequencing have been utilized in the field of ecology, and in recent years high-throughput sequencing has proven to be efficient in characterizing the diversity of microorganisms in ecological systems.7 Whole metagenomic sequencing is currently limited by its coverage as the interesting regions can form a very low proportion of the whole nucleic acid amounts and will thus not be obtained in the large data set.8,9 Amplicon sequencing through PCR primers is an alternative way to specifically obtain the less abundant genomic regions of interest, which becomes harder when there are several thousand functional targets.10,11 Large-scale functional gene microarrays such as the Geochip12 has been chosen as an alternative method by targeting several specific genes involved in ecosystem functioning. This method was useful but also had problems related to specificity and sensitivity of the fragments of DNA binding to the probes which are difficult to control.13 Although targeted approach could be very informative, it is important to note that these are confined only to known and well-annotated sequence information available in public databases.14
We suggest using a technique, called ‘sequence capture’, to sequence a large number of genes that could be used for analysing the functions of environmental communities at a significantly higher resolution than what has been possible with other approaches. It involves a selection of specific genomic loci through adhesion to probes and sequencing only the DNA fragments that are bound to the probes.15 The main advantage of the method is its ability to efficiently enrich for genes that are in very low abundance in the gene pool.16 It has been predominantly used in the field of medicine to study disease-related point mutations in the human genome.17–20 Sequence capture has also been implemented for use in several non-model organisms such as chipmunks (Tamilas),21 sugarcane (Saccharum)22 and bison (Bos)23 targeting several thousand genomic regions through their closely related species. Sequence capture was also used in a small-scale targeting of two enzymatic regions in a freshwater metagenome from Lake Pavin in France.24
In this study, we aim to demonstrate the application of sequence capture on a large scale for the first time on microbial communities of agricultural soils. The focus was to identify and enhance the capture of functional genes coding for carbohydrate-active enzymes and secretory proteases that are related to organic matter degradation were identified from the public databases were targeted in this method25–27 and subsequently targeted using sequence capture. The probe design was customized to target functional enzymes of soil microorganisms in natural soil communities using MetCap28 and allows for efficient targeting hundreds of thousands of genes. In short, MetCap is a web-based probe-designing pipeline that takes user's sequences of interest and design probes for targeting those sequences optimized for sequence capture. The soil metagenomic DNA was targeted at four different probe hybridization stringencies and sequenced with high-throughput sequencing. Here we determine that: (i) the custom-designed sequence capture is efficient in enriching for functional targets in the metagenomic data sets and the effect of hybridization stringencies was investigated, (ii) the correlation of taxonomic distribution of the targeted metagenomes to their respective traditional ribosomal 16S rDNA libraries is very high and (iii) a comparative metagenomic analyses of the eight captured metagenomes with 22 publicly available soil whole metagenomes from the MG-RAST metagenome database illustrated the efficient enrichment using sequence capture compared with standard shotgun whole metagenome sequencing (WMG).
2. Materials and methods
2.1. Soil samples and DNA extraction
Two soils from Bjornstorp, located in the southern region (Scania) of Sweden that is part of a land-use management study,29 were sampled to test the proposed sequence capture method. The soil samples were from agricultural fields, one from a winter wheat field and the other from grassland nearby the wheat field (Supplementary Table S1). At each field, several sub-samples were collected from different spots (0–15 cm depth) and mixed together. The soils were transported in cold boxes and sieved (2.5 mm) and then stored in a −20°C freezer. Then refrigerated at 4°C before proceeding to DNA extraction. DNA from both samples was extracted using Nucleospin soil DNA isolation kit (Macherey & Nagel, Duren, Germany). The extractions were carried out according to the manufacturer with 0.5 g of soil as the starting material. The extracted DNA was tested for quality (A260/280) and concentration using NanoDrop 2000 spectrophotometer (NanoDrop Technologies, Wilmington, NC, USA). Multiple extractions were carried out for each sample, and the extraction with highest yield and best quality was chosen for amplicon and sequence capture (SeqCap) library preparation steps.
2.2. SeqCap EZ probe generation
Enzymes from Carbohydrate-Active Enzyme database (CAZy)25 and proteases from the MEROPS database27 having a predicted secretion signal based on signalP v430 were chosen. In total, 306,525 sequences were selected for probe generation of which 260,731 were from CAZy and 45,794 were from MEROPS. The nucleotide coding sequences of these genes were used to design probes for sequence capture, and a local sequence database was created with these sequences (subsequently called target database, TDB). The probes were generated based on these sequences using the MetCap pipeline, where the sequences were clustered with 90% sequence similarity and on an average three probes were generated from each cluster and more details of these probes were described in Kushwaha et al.28 In total, 406,277 probes were generated with 351,482 from CAZy and 54,795 from MEROPS. They were generated with melting temperature (50°C) and probe length (50mer) that are suitable to use with protocol based on NimbleGen SeqCap EZ (Roche NimbleGen, Inc.).
2.3. SeqCap design
The design of the SeqCap protocol has been modified and tested for the hybridization stringency on target sequence binding specificity, to account for the combination of variability in metagenomic samples and the number of target regions. To test for this stringency, ‘hybridization time’ has been chosen as a factor, since it is effective, and it is also least dependent on other parameters of the protocol making it easy to handle according to the recommendations of Roche NimbleGen, Inc. The metagenomic SeqCap setup was designed to test both soil samples at four different hybridization times. Along with the recommended 72 h (Roche NimbleGen, Inc.) hybridization time for probes and DNA fragments at 47°C, three (24, 16 and 8 h) less-stringent hybridization times have been tested, where 72 h was expected to be most stringent and 8 h to be the least stringent in binding specificity against the target genes.
2.4. SeqCap library preparation
Four libraries were constructed from each soil DNA sample (4 hybridization times × 2 soil samples), following manufacturer's instructions from GS FLX rapid library preparation method (Roche). A total of 500 ng of each DNA sample was fragmented using a Nebulizer (Roche) along with nebulization buffers at 2.1 bar pressure to get the average fragment length in the range of 700 bp. This length is the recommended fragment length for the combination of sequence captures using SeqCap EZ probes coupled with 454 GS FLX sequencing (NimbleGen, Roche). The cleaned fragments were then subjected to the end repair treatment (Roche) followed by 454-adapter ligation. To make multiplexing possible in both cases of sequence capture and sequencing, each library was prepared with a specific MID (Multiplexing Identifier) on its 454 adapters (Lib-L). Each of the eight libraries contained a unique MID, and the qualities of the libraries were checked with High Sensitivity DNA chip (Bioanalyzer, Agilent) at the end of the library preparation step. Before subjecting the fragments to hybridization with probes, a ligation-mediated (LM) PCR (pre-capture) with the 454 adapters as primers was done with FastStart High Fidelity PCR System (Roche Applied Science) with only 12 cycles and purified with QIAquick PCR purification kit (Qiagen, Limburg, The Netherlands), so as to increase the amount of DNA fragments for hybridization step that passed adapter ligation step.
2.5. Sequence capture
The actual sequence capture process has been carried out with the capture protocol (Roche NimbleGen, Inc.) with adjustments in hybridization time as described in the design. The following steps are according to the standard SeqCap EZ protocol (Roche NimbleGen, Inc.). The hybridization step was multiplexed and carried out for each hybridization time as the libraries contain MIDs. For each hybridization time, 1 µg (500 ng from each soil DNA) of amplified libraries together with hybridization-enhancing oligos was dried in a DNA vacuum concentrator. The corresponding DNA libraries were dried, so that there were four dried samples representing each hybridization time. Hybridization buffers were added to the libraries and heated at 95°C for 10 min to denature the DNA fragments. Then the probes (6.5 µl/reaction) were added to the denatured libraries immediately and incubated at 47°C in a thermal cycler with a heated cover at 57°C for respective hybridization times. After the hybridization, the libraries were washed with buffers specified in the SeqCap EZ protocol (Roche NimbleGen, Inc.) along with streptavidin dynabeads at 47°C and magnetic device to retain just the hybridized fragments with probes and remove unbound fragments. These captured DNA fragments bound to probes that were attached to the dynabeads were used as template in a LM-PCR (post-capture) reaction by which the captured DNA fragments were amplified and also separated from the beads/probes. These samples were checked again with a Bioanalyzer to ensure that the DNA fragments were captured and to check for primer dimers. The captured DNA fragments were purified two times using AmpPure Bead (Beckman Coulter Inc., Brea, USA) purification method for the removal of any primer dimers from the post-capture LM PCR that could hinder in the sequencing step. The quantities of double-stranded DNA in these four captured DNA libraries were measured using Quant-it Pico Green kit (Invitrogen, Carlsbad, USA).
2.6. 16s rDNA amplicon library preparation
As part of the sample analysis, bacterial species composition was studied through amplicon sequencing of ribosomal DNA from these soil samples. Amplicon libraries were prepared by running a PCR with fusion primers optimized for 454 sequencing. Fusion primers for bacterial 16S rDNA V3-V4 region were designed using the forward primer B341F [‘CCTACGGGNGGCWGCAG’] and 454 (Lib-A) adapter-A and the reverse primer B805R [‘GACTACHVGGGTATCTAATCC’] preceded by 454 (Lib-A) adapter-B with a MID.31 All amplicon libraries were prepared using reagents based on Phire Hot Start DNA Polymerase (Thermo Fisher Scientific Inc., Waltham, MA, USA). The pre-PCR mix was prepared in the same proportion for each sample totaling to 25 µl (5 µl-5× Buffer; 0.5 µl-10 mM dNTPs; 1 µl-10 µM forward primer; 2 µl-5 µM reverse primer; 0.5 µl-DNA polymerase; 0.5 µl-bovine serum albumin; 2.5 µl-Template DNA; 13 µl-MilliQ water). The PCR conditions for 16S rDNA amplification were as follows: initial denaturation step at 98°C for 30 s; 27 cycles of denaturation at 98°C for 5 s, annealing at 56°C for 5 s, extension at 72°C for 10 s; final extension at 72°C for 60 s. Three 25 µl PCR reactions were run separately for each sample and pooled together. These PCR amplicons were purified with QIAquick PCR purification kit (Qiagen), and the quantity was measured using Quant-it Pico Green kit (Invitrogen).
2.7. 454 sequencing
The SeqCap and amplicon libraries were sequenced separately with Lib-L and Lib-A chemistry, respectively (Roche 454, Shirley, NY, USA). The multiplexing (MID) option used in both SeqCap and amplicon libraries facilitate sequencing them as respective pools to yield more sequencing depth and then to separate the libraries computationally. For SeqCap pool, equal amounts of captured DNA from all four different hybridizations were pooled together. The pooled captured libraries were sequenced in the sequencing facility at Lund University on a whole plate of GS FLX Titanium series with Lib-L chemistry in two regions. For amplicon libraries, both 16S rDNA amplicon libraries were sequenced as part of an amplicon pool made from equal amounts of 24 other amplicon libraries. This amplicon pool was sequenced in a 1/4 plate using a GS FLX Titanium series with Lib-A chemistry also sequenced at the same facility. The sequence data related to the captured metagenomes can be found on the MG-RAST server with their specific Metagenome IDs (WS-72: 4527652.3, WS-24: 4529373.3, WS-16: 4529786.3, WS-8: 4528934.3, GL-72: 4527653.3, GL-24: 4529374.3, GL-16: 4529787.3 and GL-8: 4528937.3). The 16S rDNA amplicon sequencing data can be found in the study accession number PRJEB9530 at EMBL (WS-16S: ERS743453 and GL-16S: ERS743454).
2.8. Data analysis
The sequencing output from both regions in the captured libraries was separated based on their MID tags into their respective eight different data sets. The sequence reads from each of these captured data sets were processed through MG-RAST,32 an online tool that is mainly used for functional annotations of metagenomic sequences. The default parameters for quality filtering in MG-RAST were used to remove reads with bad quality and to remove artificial sequence duplicates.33 The filtered sequence reads were used in further analysis. The optimal hybridization time with respect to this particular set of probe design method was deduced based on the enrichment efficiency (fraction of on-targets) for each hybridization time. On-targets were identified through sequence similarity (blastx, E-value of 10−5) with the target database (TDB) consisting of carbohydrate-active enzymes and secreted proteases.25–27
The functional assignment of sequences from captured metagenomes was also carried out in MG-RAST based on sequence similarity matches with different databases along with SEED34 database where reads were annotated with different subsystems (E-value of 10−5). The CAZy domains were predicted from the captured metagenomes using CAZy Analysis Toolkit (CAT) with E-value cut-off of 10−5.35 The taxonomic analysis of captured metagenomes has been analysed mainly based on counts for each taxa obtained from MG-RAST.32 The amplicon metagenomes were processed with QIIME36 in combination with Greengenes37 for 16S rDNA amplicons as a resource for taxonomy assignments. The taxonomic distributions from the amplicon metagenomes were compared with that of the captured metagenomes. A comparative metagenomic analyses between the eight captured metagenomes along with 22 publicly available whole metagenomes (WMG) (Supplementary Table S6) also from soil sequenced with 454 were achieved through the R package ‘matR’ in MG-RAST.38 These metagenomes were also normalized with log transformation and centred for each sample using ‘matR’. STAMP39 was used for the visualization and the statistics of the comparative metagenomic data sets with Bonferroni correction for P-values. The CAZy domains were predicted for two of the intensively sequenced, publicly available whole metagenomes and compared with the captured metagenomes from our study.
3. Results
In total, 914,996 sequence reads were obtained from the SeqCap pool. The number of sequences with good quality (after QC and dereplication) for all eight samples was uniform and ranged between 89,281 and 129,198 per sample (Supplementary Table S2). The targeted metagenomes that were sequenced with 454 pyrosequencing generated >850,000 reads with an average read length of 348 after quality control (QC). The fraction of reads removed through QC was consistent (∼7%) among all eight captured metagenomes. In total from the amplicon pool, 4,990 sequence reads were obtained from 16S rDNA amplicons after QC from both the wheat soil (WS) and grassland soil (GL), respectively.
3.1. Capture efficiency
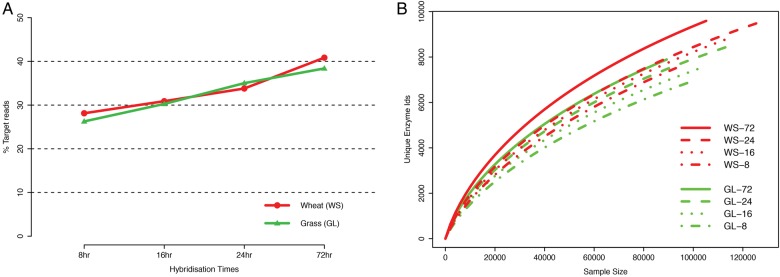
The efficiency of different hybridization times to enrich for the sequences of interest was measured as the proportion of reads with significant similarity to proteins sequences in the TDB (Fig. 1A). The fraction of reads that matched (blastx) to TDB varied from 27 ± 1% for the shortest hybridization time (8 h) to 40 ± 2% for the longest hybridization time (72 h). The total number of reads that belonged to the carbohydrate subsystem from SEED34 and the fraction of reads that have a predicted CAZy domain from CAT35 also increased with increase in hybridization times for both samples (Supplementary Fig. S1). The capture efficiencies of the different hybridization times obtained through three independent methods (Supplementary Fig. S1) showed that 72 h hybridization time was most efficient in enriching targets from metagenomes.
Figure 1.
(A) The capture efficiency in relation to different hybridization times from the wheat (WS) and grassland (GL) soils calculated as fraction (%) of total filtered reads that had a significant sequence match (blastx) to the protein sequences from TDB. (B) Rarefaction curves of unique enzyme IDs (UniProt) from random sampling of filtered sequence reads from the captured metagenomes. This figure is available in black and white in print and in colour at DNA Research online.
The abundance of each enzyme (Uniprot enzyme ID) based on the number of reads that matched the enzymes in TDB sequences through blastx was obtained for each metagenome. The rarefaction curve (Fig. 1B) for unique enzymes in each metagenome also depicts the variation between hybridization times for both soil samples. The metagenomes in terms of enzyme abundance counts were significantly explained by hybridization time when they were homogenized for soil types (ANOSIM: P = 0.04). The 72 h hybridization time had more unique enzymes and more on-target matches than the other times in any given amount of random sampling of sequences.
3.2. Taxonomic distribution of the metagenomes
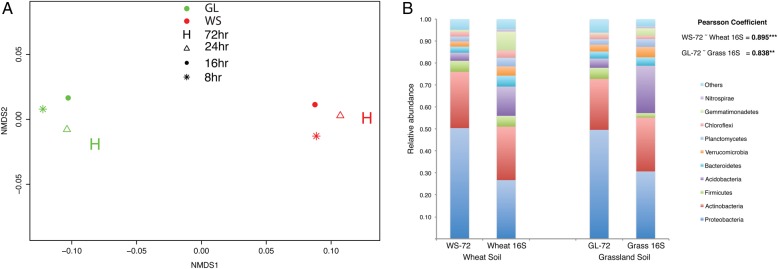
The captured metagenomes were dominated by sequences of the bacterial phyla Actinobacteria and Proteobacteria in both soil samples. Comparative taxonomic analysis of metagenomes was based on the relative abundances of different genera (both bacteria and fungi). The taxonomic groups of the captured metagenomes from the same soil type were significantly similar (PERMANOVA: P < 0.001, R2 = 0.74; ANOSIM: P = 0.03), while the hybridization time did not have any effect on their taxonomic distribution (PERMANOVA: P = 0.45; ANOSIM: P = 0.22) (Fig. 2A). The high-stringency (72 h) metagenomes were used for the taxonomic comparison to the taxonomic distribution obtained from the 16S rDNA amplicon sequencing of respective soil samples (WS-16 and GL-16). The relative abundances of bacterial taxa in the captured metagenomes (72 h) from MG-RAST were well correlated with their respective 16S rDNA amplicons (Fig. 2B). The taxonomic representations of the captured metagenomes were similar to that of the representations obtained from the ribosomal amplicon metagenomes. The taxonomy distributions between amplicon libraries and the captured metagenomes were significantly correlated at different levels of taxa for both soil types (Supplementary Table S3).
Figure 2.
(A) In an NMDS plot based on Bray–Curtis distances, the taxonomic distributions from the eight captured metagenomes show that the metagenomes from the same soil wheat (WS) and grassland (GL) cluster together (NMDS: stress = 0.04; PERMANOVA: R2 = 0.74, P < 0.001). (B) The relative abundances of different taxonomic groups (phyla) from the captured metagenomes (WS-72 and GL-72) and 16S rDNA amplicon metagenomes (WS-16S and GL-16S). This figure is available in black and white in print and in colour at DNA Research online.
3.3. Functional distribution of the metagenomes
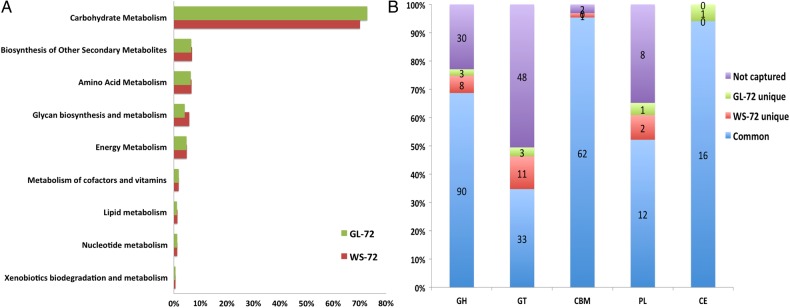
The functional annotations based on the different databases available in MG-RAST showed that the fraction of reads annotated with functional proteins increased with longer hybridization time for both soil samples (Supplementary Fig. S2). However, the fraction of reads that had neither protein similarity nor functional annotation was constant in all captured metagenomes (12.5 ± 1%). The subgroup of reads with KEGG orthology annotations (KO) (identity ≥60% and length ≥15 aa) showed that >85% of these proteins were involved in metabolism (Supplementary Table S4). Also, >70% of those metabolic reads were particularly involved in carbohydrate metabolism in both wheat and grassland soil with high-stringent hybridization time (WS-72: 18,628 reads; GL-72: 15,900 reads) (Fig. 3A) and are part of >200 different KO functional groups. This is followed by synthesis of secondary metabolites, amino acid metabolism and glycan biosynthesis and metabolism. These four metabolic categories constitute >90% of all the metabolic annotations.
Figure 3.
(A) The distribution of different metabolic functions (%) obtained and based on KEGG orthology (KO) annotations. (B) The number of different CAZy enzyme families captured under the different enzyme classes based on three different annotation sources (CAT, MG-RAST and blastx against TDB) from each or both the captured metagenomes WS-72 and GL-72. This figure is available in black and white in print and in colour at DNA Research online.
Among the different annotation sources in the MG-RAST server, the SEED-based subsystem had the most abundant annotations with 943 different subsystems (Level 3) annotated at least two times among the eight captured metagenomes. Comparing the abundances of broader subsystems (Level 1), the carbohydrates subsystem was the most abundant (WS-72: 35% and GL-72: 36%) followed by the clustering-based subsystem (functionally coupled genes) (WS-72: 33% and GL-72: 35%). The three most abundant finer (Level 3) subsystems that were found in both wheat (WS-72) and grassland soil (GL-72) were trehalose biosynthesis, glycogen metabolism and glycogen metabolism cluster which form ∼50% of the entire subsystems. The blastx matches obtained against the TDB sequences showed that ∼75% of the matches were annotated to CAZy enzymes and the rest were proteases for both soils (WS-72, GL-72). Among the different enzyme families, the most abundant bacterial family was the glycosyl hydrolase 13 (GH13) in the CAZy database (WS-72: 41%, GL-72: 45%). This was followed by metalloproteases (WS-72: 13%, GL-72: 11%) and serine proteases (WS-72: 13%, GL-72: 10%) in both soils.
3.4. CAZy analysis
Out of 331 different CAZy enzyme families, 243 were found in both soil samples (WS-72 and GL-72) at least through one of the three methods of annotation (CAT, MG-RAST and blastx) (Supplementary Table S5). The number of different enzyme families captured in each CAZy enzyme class varied between the soil samples and the enzyme class (Fig. 3B). Larger number of enzyme families from glycosyl transferase (GT) class was not found in both metagenomes (WS-72 and GL-72) compared with other enzyme classes. Glycosyl hydrolases were the most abundant enzyme class from all three methods in both metagenomes (WS-72 and GL-72). The most abundant glycosyl hydrolase enzyme family GH13 was predicted along with carbon binding motif CBM48 from the domain predictor (CAT). The fractions of reads with GH13 domain along with CBM48 (WS-72: 39%, GL-72: 46%) were the most abundant next to GH13 domains (WS-72: 17%, GL-72: 17%) that were predicted independently.
3.5. Comparative metagenomics
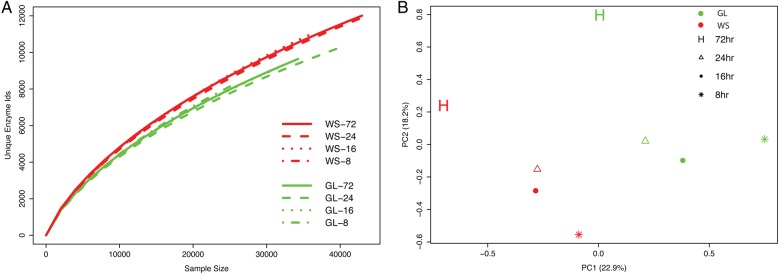
As all the captured metagenomes in this study represented two soils, the metagenomes from the different hybridization times were treated as replicates and the different enzyme IDs among the portion of on-targets (removing the off-targets) that matched (blastx) to TDB was very similar within each soil type (Fig. 4A). Similarly, the abundances of different SEED-based subsystems (Level 3) from MG-RAST after normalization were analysed for the eight captured metagenomes (Fig. 4B). The subsystem abundances in these eight captured metagenomes were significantly affected by both soil type (PERMANOVA: P < 0.001) and hybridization times (PERMANOVA: P < 0.01). Enzymes such as amino-peptidisases, GH10 and GH36 for example were highly enriched in grassland (GL) while enzymes related to peptidoglycan biosynthesis and GH46 were highly enriched in the wheat field (WS) (Supplementary Figs S3 and S4).
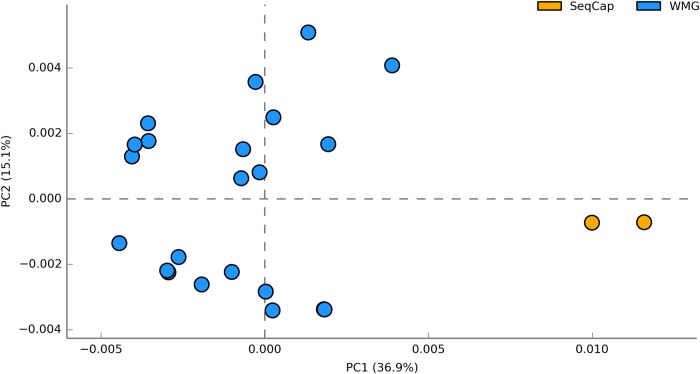
Figure 4.
(A) Rarefaction of unique enzyme IDs (UniProt) obtained from random sampling (sequences) of on-targets from the blastx matches against TDB shows that the enzyme diversity of samples converges based on the soil type. (B) A PCA based on normalized subsystem (Level 3) abundances between the captured metagenomes showing that the metagenomes from wheat (WS) and grassland (GL) cluster separately (PERMANOVA: P < 0.001). This figure is available in black and white in print and in colour at DNA Research online.
To elucidate the ability of captured metagenomics, a number of whole metagenomic data sets from soil (WMG) that were deeply sequenced by pyrosequencing and were publicly available in MG-RAST were chosen for comparison. Some of these public metagenomes were sequenced to a depth >10 times that of the captured metagenomes in our study. Two of WMG have been analysed for CAT, since it only allows 50,000 sequences to be analysed per run. The fraction of reads that contained CAZy domains in the captured metagenomes was more than five times higher than the possible untargeted metagenomes despite the coverage of sequencing (Table 1). The most abundant enzyme family GT2 in the two public metagenomes formed only ∼0.8 and 0.2% of the total reads, whereas in our targeted approach the two most common enzyme families were GH13 together with CBM48 and represented 12% of the WS-72 and 13% of the GL-72 metagenomes.
Table 1.
The fraction of reads predicted with CAZy domains from the captured metagenomes is at least five times more than intensively sequenced public WMG
| Metagenomes | Filtered reads | CAZy domains (reads) | %CAZy | Expected CAZy domains (per million reads) |
|---|---|---|---|---|
| WS-72 | 105,190 | 30,999 | 29.50 | 294,695 |
| GL-72 | 89,281 | 24,637 | 27.60 | 275,949 |
| Public-A | 937,368 | 52,817 | 5.60 | 56,346 |
| Public-2M | 354,345 | 4,692 | 1.30 | 13,241 |
CAZy, Carbohydrate-Active Enzymes; WMG, whole metagenome sequencing.
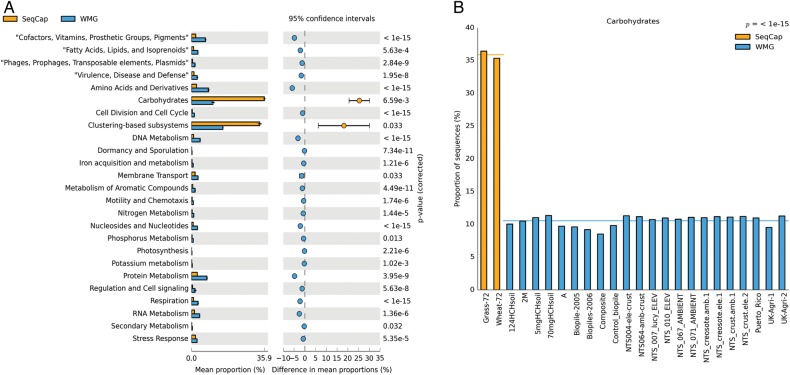
In total, 22 publicly available whole metagenomes were used for comparison towards the two captured metagenomes (WS-72 and GL-72). After normalization of these 24 metagenomes, there were 1,124 subsystems (Level 3) whose abundances were compared between the metagenomes. Among these subsystems, there were 390 subsystems that had a significant difference in the relative frequencies (P < 0.05, q < 0.05) between the whole metagenomes and captured metagenomes (Fig. 5) (PERMANOVA: P < 0.001, R2 = 0.59). The most common subsystems that were significantly enriched in the captured metagenomes (Supplementary Table S7) belonged to: carbohydrates (43), clustering-based subsystem (45) and protein metabolism (18). The captured metagenomes are highly efficient in enriching for sequences related to carbohydrate subsystem as expected through design compared with the whole metagenomes (Fig. 6).
Figure 5.
A PCA based on normalized subsystem (Level 3) abundances between the captured metagenomes (SeqCap) and whole metagenomes from the MG-RAST database (WMG) showing that the captured metagenomes are clustered completely different from the whole metagenomes (P < 0.001). This figure is available in black and white in print and in colour at DNA Research online.
Figure 6.
(A) The mean proportions of the significantly different subsystems (Level 1) in the captured metagenomes (SeqCap) from the 22 whole metagenomes from the MG-RAST database (WMG) with their respective 95% CI and corrected P-values. (B) The proportion of carbohydrate subsystem in the captured metagenomes (SeqCap) compared with 22 whole metagenomes (WMG). This figure is available in black and white in print and in colour at DNA Research online.
4. Discussion
This study has for the first time applied a targeted metagenomic approach to study genes of more than hundreds of different enzyme families in environmental samples. The approach was successfully demonstrated for soil from two different land-use types, grass and wheat, but is applicable to any metagenome. The soil functional diversity can be efficiently investigated by using the sequence capture technique that has been developed with an online web tool for probe design28 for genes encoding enzymes regulating specific functional mechanisms in ecosystems. This method circumvents the inefficiency due to coverage or high labour demand for gene targeting, of the techniques that are commonly applied for the study of community functions.8,9,13,40 We have applied this technique successfully for two different soil metagenomes to target enzymes involved in organic matter degradation.
The abundances of these target enzymes in each captured metagenome were significantly affected by the hybridization time. However, the diversity of these targeted enzymes was consistent within each soil regardless of their hybridization time. This shows that stringent hybridization, in terms of hybridization time, increases the enrichment of target genes while maintaining the diversity. It could also be inferred that this technique is reproducible as there is an increase in target enrichment while maintaining diversity at higher hybridization stringencies for both metagenomes. Lower hybridization stringencies also increase the fraction of proteins without any annotations in the public databases in the metagenome. This is mainly due to the random hybridizations of DNA fragments to probes (i.e. increased number of off-targets), which would be less likely at higher stringencies. Since this technique involves sequencing of the hybridized fragments, it provides the advantage of separating out the randomly bound fragments from the ones that contain functional annotation compared with microarray-based techniques.13 It is also important to note that these fragments were sequenced with 454 for longer sequence reads. As 454 frequently does not sequence the entire DNA fragment, adjacent non-coding region instead of the region bound to the probe might have been sequenced, which could also hinder their functional annotations. In addition, 454 also has higher per-base sequencing errors like homopolymer errors41 that could cause problems while matching sequence reads for homology in databases. These issues could be solved with other less error-prone sequencing technologies such as Illumina.
Most of the metabolic gene predictions in the captured metagenomic datasets for both soils were annotated to carbohydrate metabolism. Other major metabolic components (e.g. amino acid metabolism) of the captured metagenomes are also related to organic matter degradation. This clearly illustrates the efficiency of using our customized probes to enrich for regions of interest in a complex metagenome and is a highly suitable approach to increase the understanding of a particular environmental process. Similarly in the subsystem analysis, the most abundant subsystem (Level 1) was carbohydrates, also showing the efficiency of this capture technique. The second most abundant subsystem, clustering-based subsystems, are the genetic regions that are collocated to functional genes in the genomes of different taxa, but their functions are not well known.42 We suggest that these genes are abundant, because they are sequenced along with the targeted genetic DNA fragments. The functional subsystems distribution between the captured metagenomes was mainly influenced by their soil type rather than their hybridization stringency, although both properties significantly affected their distribution. This shows that this targeted metagenomics approach does not bias the functional representation of the soils even at very high stringency. The results clearly show the efficiency of applying this method to provide insights into important ecological questions, such as understanding key processes in complex environments.
The number of different CAZy enzyme families found from these metagenomes also shows the ability of this target enrichment strategy to obtain reliable data for >200 enzyme families at different taxonomic levels. It is to be noted that an efficient enrichment of proteases was also obtained along with CAZy enzymes, although there are common enzymes between the two databases CAZy and MEROPS. Due to our approach, the numbers of CAZy enzyme families found in our metagenomes are much higher to what has been reported in earlier findings.43,44 Despite this, some enzyme families from certain enzyme class like GT were not acquired completely. This could be due to the inefficiency of probes targeting of these families or the very low abundance of these enzyme classes in these soils. Also, these enzyme classes could co-occur with other enzyme families and the annotations like blast would predict one of those with higher scores. For example, the glycosyl hydrolase 13, one of the major CAZy enzyme families, frequently co-occur with CBM4845 and is the most abundant enzyme family in captured metagenomes. GH13 enzyme catalysis is known to be an important step in trehalose biosynthesis,46 and this biosynthesis is also the most abundant subsystem in the captured metagenomes.
The taxonomic analysis of the captured metagenomes from MG-RAST showed that the target enrichment strategy applied here does not appear to bias the targeting towards any particular taxonomic group. The taxonomic distribution between all captured metagenomes was significantly explained by their soil type rather than the hybridization times. It is evident that even at the high stringency, captured metagenomes have a similar taxonomic distribution as the other metagenomes from the same soil. Hence, the developed sequence capture method does not have any taxonomic bias. This was further supported by the significant correlations between captured metagenomes (WS-72 and GL-72) and the amplicon metagenomes based on the relative abundances of taxa at different levels. The changes in certain taxa between captured metagenomes and amplicon sequencing could be explained through the availability of functional information of particular taxa. The phylum acidobacteria, for example, is represented highly in 16S rDNA amplicon data but not in case of the captured metagenomes. This can be explained partially that it is well known for its abundance in soil communities but has not been extensively studied for their functions.47,48 The fraction of genes coding for enzymes from Acidobacteria used for our probe were measured to be only 0.6% in TDB, also only 0.9% among the other bacterial enzymes. This limitation is not due to the capture technique, but rather due to biases in the public databases. Several ongoing projects are aiming at increasing the resolution of these under-studied organisms through genome sequencing,49,50 which will be helpful in understanding the ecosystems better in the future.
The comparison of CAZy domains from captured metagenomes to publicly available whole metagenomes showed that enrichment through probes was more efficient than intensive sequencing to identify lowly abundant functional genes. Even the most abundant CAZy domains were <1% in WMG, showing the inability of whole metagenomic approach to obtain these important genes.51 However, our targeted approach efficiently enriched for genes that were targeted, at considerably lower sequencing depths. The abundance of enzymes related to carbon cycling from captured metagenomes was much higher than the whole metagenomes (WMG) despite that the WMG was very deeply sequenced. These 22 WMG were obtained from different soils at different conditions, including metagenomes from places like rain forest in Puerto Rico, permafrost and high arctic soils which are known to have communities with higher organic matter degradation capabilities.51–53
The differences between two soil communities (WS and GL) were deducible with the availability of abundance and richness of the enzymes in captured metagenomes. It was clear that enzyme families such as GH10 coding for xylanases10 were abundant in grassland as expected since it is related to degradation of plant cell-wall material. Similarly amino peptidases are also found abundant in grassland, as the soil does not receive any free nitrogen as in the case of wheat soil through fertilization. This also shows the ability of captured metagenomics to explain soil functionality. Although, it is important to note that the probes used in this method depend on known functions/enzymes in databases and the information related to unknown functions/enzymes in communities may only be obtained if they were genetically similar. WMG could still be a way of obtaining such information in these particular cases but not to forget that annotating them could still be a subject of biases.54
This study has for the first time implemented the sequence capture technique to study the functional diversity of enzymes degrading organic matter in natural soil communities. This approach has the ability to solve the coverage issues with the WMG to get enough amounts of representative sequences.55 We also argue that it is superior to the large-scale microarrays56 since it is able to detect unspecific binding of DNA fragments to the probes and facilitates the discovery of novel genes. It has also been clearly shown, based on multiple hybridization stringencies, that this technique is highly stable and reproducible. As this method represents the taxonomic diversity very well, it could be used to understand the relation between the taxonomic and functional diversity present in the environmental communities.1 As mentioned in Fierer et al.,40 this approach would be an excellent tool for measuring ‘community aggregated traits’ (CAT) and hence proved an integrated understanding of the functional capabilities of complex microbial ecosystems. The relative functional gene abundances from different samples, as traits irrespective of its taxonomic origin, are a better way to determine the functional capabilities of the community. Overall, this method demonstrates its ability to improve our understanding of those community functions in different ecological processes of interest and is applicable even to the most complex metagenomes.
Conflict of interest statement
None declared.
Supplementary data
Supplementary data are available at www.dnaresearch.oxfordjournals.org.
Funding
This work was supported by The Swedish Research Council for Environment, Agricultural Sciences and Spatial Planning (215-2010-1679). Funding to pay the Open Access publication charges for this article was provided by Lund University library and The Swedish Research Council for Environment, Agricultural Sciences and Spatial Planning (215-2010-1679).
Supplementary Material
Acknowledgements
We thank Helene Bracht Jørgensen (Lund University, Sweden) for helping us with the sampling of soils and Dimitrios Floudas (Lund University, Sweden) for the valuable discussions. We also thank Xavier Miro (Roche) for his suggestions and help with the sequence capture experimental procedures.
References
- 1.Gilbert J.A., Field D., Swift P. et al. 2010, The taxonomic and functional diversity of microbes at a temperate coastal site: a ‘multi-omic’ study of seasonal and diel temporal variation, PLoS ONE, 5, e15545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Diaz S., Cabido M.. 2001, Vive la difference: plant functional diversity matters to ecosystem processes, Trends Ecol. Evol., 16, 646–55. [Google Scholar]
- 3.Gravel D., Bell T., Barbera C., Combe M., Pommier T., Mouquet N.. 2012, Phylogenetic constraints on ecosystem functioning, Nat. Commun., 3, 1117. [DOI] [PubMed] [Google Scholar]
- 4.Fitter A.H., Gilligan C.A., Hollingworth K. et al. 2005, Biodiversity and ecosystem function in soil, Funct. Ecol., 19, 369–77. [Google Scholar]
- 5.Coleman D.C., Whitman W.B.. 2005, Linking species richness, biodiversity and ecosystem function in soil systems, Pedobiologia, 49, 479–97. [Google Scholar]
- 6.Burns R.G., DeForest J.L., Marxsen J. et al. 2013, Soil enzymes in a changing environment: current knowledge and future directions, Soil Biol. Biochem., 58, 216–34. [Google Scholar]
- 7.Gilbert J.A., Dupont C.L.. 2011, Microbial metagenomics: beyond the genome, Annu. Rev. Mar. Sci., 3, 347–71. [DOI] [PubMed] [Google Scholar]
- 8.Kunin V., Copeland A., Lapidus A., Mavromatis K., Hugenholtz P.. 2008, A bioinformatician's guide to metagenomics, Microbiol. Mol. Biol. Rev., 72, 557–78, Table of Contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wooley J.C., Godzik A., Friedberg I.. 2010, A primer on metagenomics, PLoS Comput. Biol., 6, e1000667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang G.Z., Wang Y.R., Yang P.L. et al. 2010, Molecular detection and diversity of xylanase genes in alpine tundra soil, Appl. Microbiol. Biot., 87, 1383–93. [DOI] [PubMed] [Google Scholar]
- 11.Rotthauwe J.H., Witzel K.P., Liesack W.. 1997, The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations, Appl. Environ. Microbiol., 63, 4704–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He Z.L., Van Nostrand J.D., Wu L.Y., Zhou J.Z.. 2008, Development and application of functional gene arrays for microbial community analysis, T. Nonferr. Metal. Soc., 18, 1319–27. [Google Scholar]
- 13.Barton L.L., Mandl M., Loy A., Nostrand J., He Z., Zhou J.. 2010, Analysis of microbial communities by functional gene arrays. In: Geomicrobiology: molecular and environmental perspective. Springer: The Netherlands, pp.109–26. [Google Scholar]
- 14.Suenaga H. 2012, Targeted metagenomics: a high-resolution metagenomics approach for specific gene clusters in complex microbial communities, Environ. Microbiol., 14, 13–22. [DOI] [PubMed] [Google Scholar]
- 15.Albert T.J., Molla M.N., Muzny D.M. et al. 2007, Direct selection of human genomic loci by microarray hybridization, Nat. Methods, 4, 903–5. [DOI] [PubMed] [Google Scholar]
- 16.Burbano H.A., Hodges E., Green R.E. et al. 2010, Targeted investigation of the Neandertal genome by array-based sequence capture, Science, 328, 723–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ng S.B., Turner E.H., Robertson P.D. et al. 2009, Targeted capture and massively parallel sequencing of 12 human exomes, Nature, 461, 272–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rehman A.U., Morell R.J., Belyantseva I.A. et al. 2010, Targeted capture and next-generation sequencing identifies C9orf75, encoding taperin, as the mutated gene in nonsyndromic deafness DFNB79, Am. J. Hum. Genet., 86, 378–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ross J.S., Wang K., Gay L.M. et al. 2014, A high frequency of activating extracellular domain ERBB2 (HER2) mutation in micropapillary urothelial carcinoma, Clin. Cancer Res., 20, 68–75. [DOI] [PubMed] [Google Scholar]
- 20.Duncavage E.J., Abel H.J., Szankasi P., Kelley T.W., Pfeifer J.D.. 2012, Targeted next generation sequencing of clinically significant gene mutations and translocations in leukemia, Modern Pathol., 25, 795–804. [DOI] [PubMed] [Google Scholar]
- 21.Bi K., Vanderpool D., Singhal S., Linderoth T., Moritz C., Good J.M.. 2012, Transcriptome-based exon capture enables highly cost-effective comparative genomic data collection at moderate evolutionary scales, BMC Genomics, 13, 403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bundock P.C., Casu R.E., Henry R.J.. 2012, Enrichment of genomic DNA for polymorphism detection in a non-model highly polyploid crop plant, Plant Biotechnol. J., 10, 657–67. [DOI] [PubMed] [Google Scholar]
- 23.Cosart T., Beja-Pereira A., Chen S., Ng S.B., Shendure J., Luikart G.. 2011, Exome-wide DNA capture and next generation sequencing in domestic and wild species, BMC Genomics, 12, 347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Denonfoux J., Parisot N., Dugat-Bony E. et al. 2013, Gene capture coupled to high-throughput sequencing as a strategy for targeted metagenome exploration, DNA Res., 20, 185–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cantarel B.L., Coutinho P.M., Rancurel C., Bernard T., Lombard V., Henrissat B.. 2009, The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics, Nucleic Acids Res., 37, D233–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levasseur A., Piumi F., Coutinho P.M. et al. 2008, FOLy: an integrated database for the classification and functional annotation of fungal oxidoreductases potentially involved in the degradation of lignin and related aromatic compounds, Fungal Genet. Biol., 45, 638–45. [DOI] [PubMed] [Google Scholar]
- 27.Rawlings N.D., Barrett A.J., Bateman A.. 2012, MEROPS: the database of proteolytic enzymes, their substrates and inhibitors, Nucleic Acids Res., 40, D343–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kushwaha S.K., Manoharan L., Meerupati T., Hedlund K., Ahrén D.. 2015, MetCap: a bioinformatics probe design pipeline for large-scale targeted metagenomics, BMC Bioinformatics, 16, 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsiafouli M.A., Thebault E., Sgardelis S.P. et al. 2014, Intensive agriculture reduces soil biodiversity across Europe, Global Chang. Biol., 21, 973–85. [DOI] [PubMed] [Google Scholar]
- 30.Emanuelsson O., Brunak S., von Heijne G., Nielsen H.. 2007, Locating proteins in the cell using TargetP, SignalP and related tools, Nat. Protoc., 2, 953–71. [DOI] [PubMed] [Google Scholar]
- 31.Herlemann D.P.R., Labrenz M., Jurgens K., Bertilsson S., Waniek J.J., Andersson A.F.. 2011, Transitions in bacterial communities along the 2000km salinity gradient of the Baltic Sea, Isme J., 5, 1571–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meyer F., Paarmann D., D'Souza M. et al. 2008, The metagenomics RAST server - a public resource for the automatic phylogenetic and functional analysis of metagenomes, BMC Bioinformatics, 9, 386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gomez-Alvarez V., Teal T.K., Schmidt T.M.. 2009, Systematic artifacts in metagenomes from complex microbial communities, Isme J., 3, 1314–7. [DOI] [PubMed] [Google Scholar]
- 34.Overbeek R., Begley T., Butler R.M. et al. 2005, The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes, Nucleic Acids Res., 33, 5691–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park B.H., Karpinets T.V., Syed M.H., Leuze M.R., Uberbacher E.C.. 2010, CAZymes Analysis Toolkit (CAT): Web service for searching and analyzing carbohydrate-active enzymes in a newly sequenced organism using CAZy database, Glycobiology, 20, 1574–84. [DOI] [PubMed] [Google Scholar]
- 36.Caporaso J.G., Kuczynski J., Stombaugh J. et al. 2010, QIIME allows analysis of high-throughput community sequencing data, Nat. Methods, 7, 335–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McDonald D., Price M.N., Goodrich J. et al. 2012, An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea, Isme J., 6, 610–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Braithwaite D., Keegan K.. 2014, matR: Metagenomics Analysis Tools for R, http://CRAN.R-project.org/package=matR (1 August 2014, date last accessed).
- 39.Parks D.H., Tyson G.W., Hugenholtz P., Beiko R.G.. 2014, STAMP: statistical analysis of taxonomic and functional profiles, Bioinformatics, 30, 3123–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fierer N., Barberan A., Laughlin D.C.. 2014, Seeing the forest for the genes: using metagenomics to infer the aggregated traits of microbial communities, Front. Microbiol., 5, 614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Balzer S., Malde K., Jonassen I.. 2011, Systematic exploration of error sources in pyrosequencing flowgram data, Bioinformatics, 27, i304–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gerdes S., El Yacoubi B., Bailly M. et al. 2011, Synergistic use of plant-prokaryote comparative genomics for functional annotations, BMC Genomics, 12 (Suppl 1), S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tasse L., Bercovici J., Pizzut-Serin S. et al. 2010, Functional metagenomics to mine the human gut microbiome for dietary fiber catabolic enzymes, Genome Res., 20, 1605–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barbi F., Bragalini C., Vallon L. et al. 2014, PCR primers to study the diversity of expressed fungal genes encoding lignocellulolytic enzymes in soils using high-throughput sequencing, PloS ONE, 9, e116264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Janecek S., Svensson B., MacGregor E.A.. 2011, Structural and evolutionary aspects of two families of non-catalytic domains present in starch and glycogen binding proteins from microbes, plants and animals, Enzyme Microb. Tech., 49, 429–40. [DOI] [PubMed] [Google Scholar]
- 46.Jiang L., Lin M., Zhang Y. et al. 2013, Identification and characterization of a novel trehalose synthase gene derived from saline-alkali soil metagenomes, PLoS ONE, 8, e77437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ward N.L., Challacombe J.F., Janssen P.H. et al. 2009, Three genomes from the phylum Acidobacteria provide insight into the lifestyles of these microorganisms in soils, Appl. Environ. Microb., 75, 2046–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Quaiser A., Ochsenreiter T., Lanz C. et al. 2003, Acidobacteria form a coherent but highly diverse group within the bacterial domain: evidence from environmental genomics, Mol. Microbiol., 50, 563–75. [DOI] [PubMed] [Google Scholar]
- 49.Rinke C., Schwientek P., Sczyrba A. et al. 2013, Insights into the phylogeny and coding potential of microbial dark matter, Nature, 499, 431–7. [DOI] [PubMed] [Google Scholar]
- 50.Wu D.Y., Hugenholtz P., Mavromatis K. et al. 2009, A phylogeny-driven genomic encyclopaedia of Bacteria and Archaea, Nature, 462, 1056–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yergeau E., Hogues H., Whyte L.G., Greer C.W.. 2010, The functional potential of high Arctic permafrost revealed by metagenomic sequencing, qPCR and microarray analyses, Isme J., 4, 1206–14. [DOI] [PubMed] [Google Scholar]
- 52.DeAngelis K.M., Gladden J.M., Allgaier M. et al. 2010, Strategies for enhancing the effectiveness of metagenomic-based enzyme discovery in lignocellulolytic microbial communities, Bioenerg. Res., 3, 146–58. [Google Scholar]
- 53.Yergeau E., Sanschagrin S., Beaumier D., Greer C.W.. 2012, Metagenomic analysis of the bioremediation of diesel-contaminated Canadian high arctic soils, PLoS ONE, 7, e30058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schnoes A.M., Ream D.C., Thorman A.W., Babbitt P.C., Friedberg I.. 2013, Biases in the experimental annotations of protein function and their effect on our understanding of protein function space, PLoS Comput. Biol., 9, e1003063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Howe A.C., Jansson J.K., Malfatti S.A., Tringe S.G., Tiedje J.M., Brown C.T.. 2014, Tackling soil diversity with the assembly of large, complex metagenomes (vol 111, pg 4904, 2014), Proc. Natl Acad. Sci. USA, 111, 6115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.He Z.L., Gentry T.J., Schadt C.W. et al. 2007, GeoChip: a comprehensive microarray for investigating biogeochemical, ecological and environmental processes, Isme J., 1, 67–77. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.