Abstract
We have developed GeneBase, a full parser of the National Center for Biotechnology Information (NCBI) Gene database, which generates a fully structured local database with an intuitive user-friendly graphic interface for personal computers. Features of all the annotated eukaryotic genes are accessible through three main software tables, including for each entry details such as the gene summary, the gene exon/intron structure and the specific Gene Ontology attributions. The structuring of the data, the creation of additional calculation fields and the integration with nucleotide sequences allow users to make many types of comparisons and calculations that are useful for data retrieval and analysis. We provide an original example analysis of the existing introns across all the available species, through which the classic biological problem of the ‘minimal intron’ may find a solution using available data. Based on all currently available data, we can define the shortest known eukaryotic GT-AG intron length, setting the physical limit at the 30 base pair intron belonging to the human MST1L gene. This ‘model intron’ will shed light on the minimal requirement elements of recognition used for conventional splicing functioning. Remarkably, this size is indeed consistent with the sum of the splicing consensus sequence lengths.
Keywords: NCBI Gene, gene data parsing, computational biology, minimal intron, personal computer software
1. Introduction
The automation of sequencing techniques and the spread of computer use gave rise to a flourishing number of new molecular structures and sequences and to a proliferation of new databases in which to store them.1 The public availability of databases is of inestimable value, because the collective use of data leads to the discovery of new knowledge that goes beyond the results yielded by individual experiments.2
The National Center for Biotechnology Information (NCBI) Gene database (http://www.ncbi.nlm.nih.gov/gene) is a collection of gene records accessible as web page entries. Among the other existing genome browsers, such as the University of California at Santa Cruz (UCSC) Genome Browser (http://genome-euro.ucsc.edu/cgi-bin/hgGateway) and Ensembl (http://www.ensembl.org/index.html), NCBI Gene is the most complete, containing information about more than 2 million genes from more than 350 species (eukaryotic genes with nuclear genome annotations; the total is over 11 million genes from almost 13,000 species). It provides information about gene nomenclature, chromosomal localization, gene transcripts and products, as well as a series of useful links to, among other things, sequences, maps, citations, phenotypes, variation details, interactions and external databases.3 As well as browsing the data, a very efficient way of accessing information is by performing a text search. NCBI Gene is a text searchable database through indexed fields using specific term queries (http://www.ncbi.nlm.nih.gov/books/NBK3841/), although unfortunately not all fields of the database are available for searching.4 Furthermore, NCBI makes some Entrez Programming Utilities (E-Utilities) available that can be combined to form customized data pipelines to extract the desired information from NCBI Gene (http://www.ncbi.nlm.nih.gov/books/NBK25497/). The retrieved matching results can be downloaded in different formats: text, abstract syntax notation one (ASN.1) and extensible markup language (XML), allowing further analyses on a local computer. Parsers able to transform ASN.1- or XML-formatted NCBI Gene data to a relational database exist. They usually give output information that can only be accessed through the structured query language (SQL) without a graphical interface; and some of them also require high-level programming and querying skills.5–8
To address these problems, we have developed GeneBase, a full parser of the NCBI Gene database, which generates a fully structured local and relational database combined with an intuitive user-friendly graphical interface for personal computers. It allows users to do original searches, calculations and analyses of the main information about genes which are fully annotated with the ‘Gene Table’ section in NCBI Gene, i.e. eukaryotic genes. Furthermore, for a subset of gene records, we integrated nucleotide sequences useful for additional elaboration with the corresponding gene-associated meta-information.
GeneBase database contains a wealth of interesting biological information, and we provide an original analysis of the classic biological problem of the ‘minimal intron’ (the minimal DNA sequence element that can function as an intron) across all the available species as an example. A limitation in minimum length below which an intron cannot be spliced must exist.9 In the literature, the minimal intron length is usually given as a range, an average or as a length lesser than a certain number, often without a primary reference, accession number or gene name. Short introns were defined as those not longer than 116 base pairs (bp) in Arabidopsis thaliana (Taxonomy Identifier or ID: 3702).10 In Saccharomyces cerevisiae (Taxonomy ID: 4932), short intron length is ≤191 bp,11 with an average of 92 ± 20 and 49 ± 11 bp, as in Schizosaccharomyces pombe (Taxonomy ID: 4896).12 In Caenorhabditis elegans (Taxonomy ID: 6239), short intron length was on average 51.5 bp,13 and confirmed later with a length of ≤60 bp,11 with a minimum of 48 bp.10 In Drosophila (Taxonomy ID: 7215), the minimum length is 63 bp,10 but the minimum experimentally verified is 74 bp.14 For mouse and human (Taxonomy IDs: 10088 and 9606, respectively), one of the most recent studies defined the minimal intron length range as between 50 and 150 bp, corresponding to the peak value of the intron length distribution,10,15 in contrast with the length <30 bp in Homo sapiens (Taxonomy ID: 9606) hypothesized by Strachan and Read.16
We show that the intron length problem, which still raises researchers' interest,17 may find a solution, regarding all currently available data and canonical introns, through a new tool like GeneBase, which is especially useful for retrieving data with numerical range constraints and with the corresponding gene-associated meta-information. Introns <30 bp were not found in any of the species analysed, shedding light on the minimal sequence requirement elements used by the cell for conventional splicing functioning. Remarkably, the 30 bp size is indeed consistent with the sum of the known 5′/3′ splicing consensus sequence lengths.
2. Materials and methods
2.1. Database construction
GeneBase was developed within the FileMaker Pro Advanced environment (FileMaker, Santa Clara, CA, USA), which has already been proved useful for complex parsing of genomic data.18,19 This is a database management system with an intuitive user-friendly graphical interface for both Macintosh (Mac OS X) and Windows operating systems. Minimum system requirements are: Mac OS X 10.6, Intel-based Mac CPU (Central Processing Unit), 1 GigaByte (GB) of RAM (Random Access Memory), 1024 × 768 or higher resolution video adapter and display; Windows XP Professional, Home Edition (Service Pack 3), 700 MegaHertz (MHz) CPU or faster, 256 MegaBytes (MB) of RAM, 1024 × 768 or higher resolution video adapter and display.
The pre-loaded version of GeneBase was obtained by first downloading all the available Animalia (Metazoa, Taxonomy ID: 33208), Fungi (Taxonomy ID: 4751) and Plant (Viridiplantae, Taxonomy ID: 33090) kingdom gene entries from NCBI Gene. Specific text queries were used to fragment the download according to the three kingdoms and to retrieve all current (alive/live) records with a genomic gene source, excluding gene models (generated by annotation pipelines), as described in detail in the GeneBase guide. The initial download was performed on 22 April 2015 choosing the ASN.1 format, as it is the data reference representation format used by NCBI, providing smaller file sizes, fewer errors and complete data, while avoiding problems encountered by the FileMaker Pro XML parsing engine with large data files. We have developed a Python (http://www.python.org/, version 2.7) executable script to quickly parse ASN.1-formatted downloaded gene entries and thus obtain three tab-delimited files suitable for import into the three main related tables of GeneBase (corresponding to NCBI sections): ‘Gene_Summary’, ‘Gene_Table’ and ‘Gene_Ontology’. ‘Gene_Summary’ table contains one record for each gene and collects details such as the official gene symbol, the official gene full name, the organism's name and a brief summary description of the gene and its cellular localization and function (when available). ‘Gene_Table’ consists of one record for each exon including the corresponding intron if an intron follows that exon, representing the exon/intron structure of each transcript isoform as annotated on the indicated genomic Reference Sequence (RefSeq).20 Each record contains details such as RefSeq GenBank accession numbers of chromosome, messenger RNA (mRNA) and protein (when available), and the genomic coordinates of exons and introns. Additional calculation fields were created to extract further data not present in the original NCBI gene entries from the available information and are highlighted in red text. Furthermore, additional boxes were created to show useful related fields of other related software tables, giving the opportunity to perform crossed searches. Different buttons were developed to facilitate navigation through the software tables, the retrieval of features in popular databases and the launch of online BLAST (Basic Local Alignment Search Tool)21 comparisons. Finally, text fields were specifically designed to contain exon and intron sequences. ‘Gene_Ontology’ table contains the specific Gene Ontology (GO) attributions for each gene when available.22 A specific table named ‘Transcripts’ is available, showing the RefSeq status provided for each gene as well as for each of its individual transcripts. An additional table named ‘Reports’ is generated to provide statistics such as the mean length of exons and introns. In agreement with general criteria in database design, we decided to fragment information into distinct fields as much as possible, to facilitate an independent management of data. All database fields are indexed to ensure efficient data retrieval through the query options.
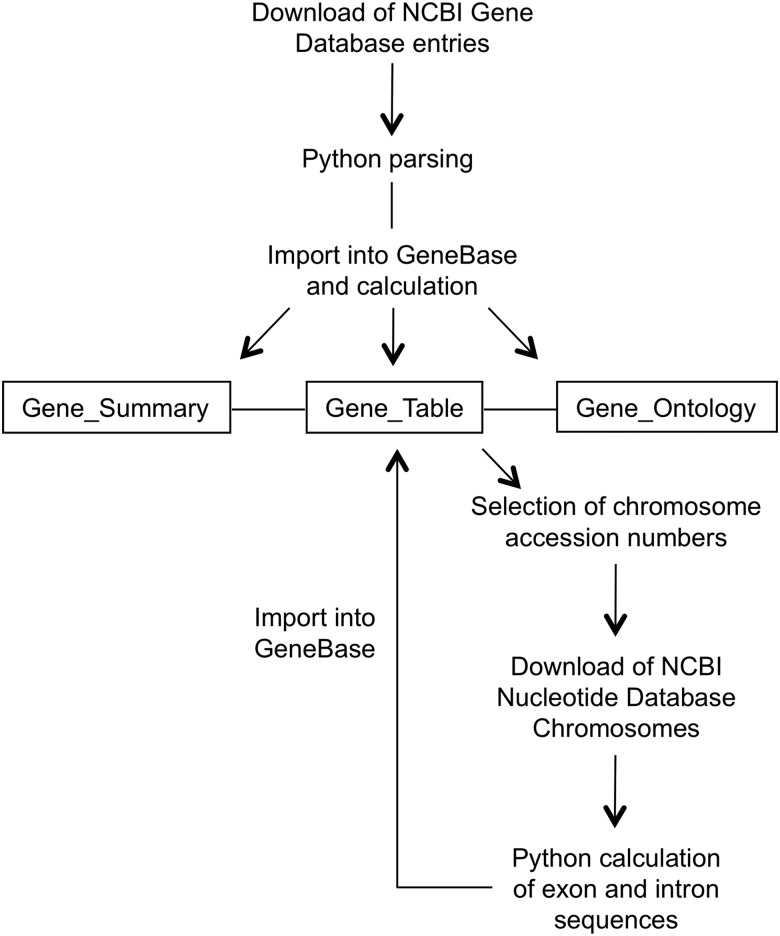
A specific feature of GeneBase is the integration with nucleotide sequences. To achieve this, a Python executable script was developed to extract exon (both coding and not coding) and intron sequences of each entry from any chromosome sequences in FASTA format and, thus, obtain a tab-delimited file suitable for import into GeneBase, if desired (Fig. 1).
Figure 1.
Flow diagram illustrating the data parsing involved in the GeneBase development. ‘Gene_Summary’, ‘Gene_Table’ and ‘Gene_Ontology’ are the three main related software tables.
For the example application that we have presented here, we have selected ‘Gene_Table’ exon/intron records of GeneBase database belonging to genes with a ‘REVIEWED’ or ‘VALIDATED’ RefSeq status, whose corresponding RNA has an ‘NM_’ or ‘NR_’ RefSeq accession number, to exclude ‘XM_’ or ‘XR_’ model Refseq records generated by automated pipelines.20 We downloaded the corresponding chromosome sequences from the NCBI Nucleotide database using Batch Entrez (http://www.ncbi.nlm.nih.gov/sites/batchentrez) in FASTA format on 5 May 2015. Then the tab-delimited file containing the corresponding exon and intron sequences obtained from script parsing was imported into the specific ‘Gene_Table’ fields.
We provide here a version of GeneBase pre-loaded with eukaryotic gene data updated on 22 April 2015, along with an empty template that may be used at any time to load ab initio the latest version or any desired subset of NCBI Gene data by any user, following parsing by our scripts. The import of parsed NCBI Gene entries can be performed also using a pre-loaded version of GeneBase without deleting previously imported records, if desired. In any case, we plan to release an updated version of our eukaryote GeneBase each year, for the convenience of users.
We made stand-alone software (of both GeneBase pre-loaded and empty versions), including the FileMaker runtime with a user guide included and the relative Python scripts for the initial data pre-processing and sequence calculations, freely available to basic users at http://apollo11.isto.unibo.it/software/. The freely distributed licensed runtime application allows full data import, records export in diverse file formats, as well as full record management and analysis and script execution. The user can also export sequence data contained in GeneBase in an automatically generated FASTA formatted text file, allowing further processing of these data, e.g. their use as a target database by a locally installed version of BLAST. The downloading, parsing and import of gene entries, the downloading of chromosome sequences and the calculation of exon and intron sequences are described in detail in the software documentation.
Only for the creation of new fields, further calculation or additional relationship definition an original copy of FileMaker Pro version 12 (or higher) is required.
2.2. Example application
The fully structured local database obtained (Supplementary Fig. S1) allows users to make many types of comparisons and do a variety of calculations that are useful for data retrieval and analysis.
As an example of how to use GeneBase, we have provided an original analysis of the existing introns across all the available species. To define the minimal intron (the minimal known DNA sequence element that can function as an intron), we queried GeneBase for introns of between 1 and 40 bp, considering only genes currently annotated on the most recent genome annotation (excluding records with ‘Genome_Annontation_Status’: ‘not in current annotation release’). Among the retrieved records, we only considered for bioinformatic validation introns belonging to gene entries with a ‘REVIEWED’ or ‘VALIDATED’ RefSeq status, with an ‘NM_’ or ‘NR_’ type of corresponding RefSeq RNA accession number and with the canonical splicing sites (GT and AG for donor and acceptor sites, respectively), thus focusing on the conventional splicing mechanism.23 We excluded from the analysis sequences predicted only by annotation of genomic sequence and thus lacking experimental evidence. We used BLASTN, the standard Nucleotide BLAST (https://blast.ncbi.nlm.nih.gov/?Blast.cgiCMD=Web&PAGE_TYPE=BlastDocs&DOC_TYPE=References),21 to find at least two independent expressed sequence tag (EST) sequences, or one manually curated RNA sequence, that encompass the intron, as a proof of existence of intron sequences. We queried the chromosome accession of each considered intron, using the previous exon start and the following exon end coordinate range as ‘Query subrange’, limiting the searches to the ‘Nucleotide collection nr/nt’ and then to the ‘Expressed sequence tags (est)’ databases, and to the organism to which the intron sequence belongs. The analysis was carried out in May 2015.
The validated intron sequences were studied for the presence of the well-known functionally important sites, e.g. the branch point and the poly-pyrimidine tract, using the following eukaryotic consensus reference sequence: MAGGTRAGT…YNYYRAYY…YYYYYYYYYYYNYCAGG, where M = A/C, R = A/G, Y = C/T, N = any base.16,24
Furthermore, the 20 shortest human canonical intron sequences available in GeneBase, with a ‘REVIEWED’ or ‘VALIDATED’ RefSeq status and an ‘NM_’ or ‘NR_’ type of corresponding RefSeq RNA accession number, were analysed for the guanine (G) content.25
Where indicated, the number of organisms was calculated by sorting records by organism and then exporting them to generate a summary report of the organisms present in the database.
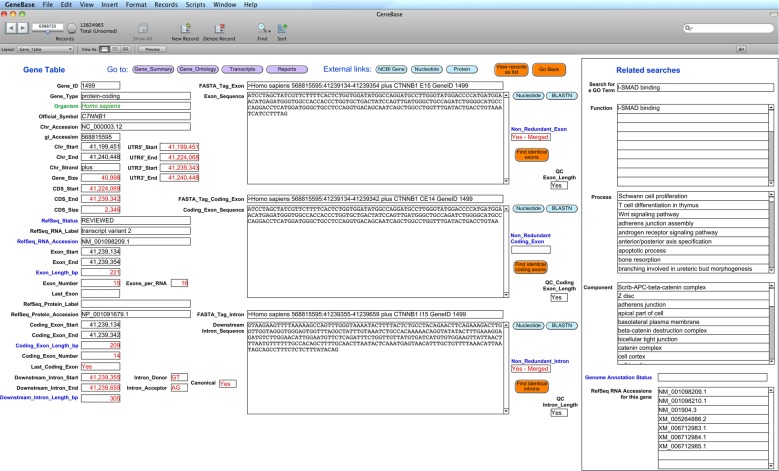
The total number of currently annotated exon, coding exon or intron records, with a ‘REVIEWED’ or ‘VALIDATED’ RefSeq status and with an ‘NM_’ or ‘NR_’ type of corresponding RefSeq RNA accession number, were retrieved typing: ‘REVIEWED’ or ‘VALIDATED’ in the ‘RefSeq_Status’ field of ‘Gene_Table’ table, ‘N*’ in the ‘RefSeq_RNA_Accession’ field, ‘>0’ in the corresponding length fields and omitting entries marked as ‘not in current annotation release’ in the ‘Genome_Annotation_Status’ field. A set of non-redundant intron records can be retrieved typing ‘Yes’ in the ‘Non_Redundant_Intron’ field (Fig. 2).
Figure 2.
Screen shot of GeneBase ‘Gene_Table’ interface. Fields are described in detail in the software documentation. Exon and coding exon sequences are in distinct fields to allow an independent management of data (see Materials and methods). This figure is available in black and white in print and in colour at DNA Research online.
Intron length distribution graphs and correlation analyses between intron and genome lengths among selected organisms were done using the R package (http://www.r-project.org/).
3. Results
3.1. Database construction
We downloaded from NCBI Gene all current (alive/live) eukaryotic records with a genomic gene source (excluding gene models) available up to 22 April 2015. We obtained 679,451 entries for Animalia (Metazoa, Taxonomy ID: 33208), 1,203,082 for Fungi (Taxonomy ID: 4751) and 534,875 for Plants (Viridiplantae, Taxonomy ID: 33090), for a total of 359 organisms. Among the 2,417,408 total gene entries, 76,182 are ‘REVIEWED’, 41,862 ‘VALIDATED’, 2,245,205 ‘PROVISIONAL’, 31,464 ‘INFERRED’, 22,691 ‘PREDICTED’, 1 ‘MODEL’ and 1 ‘WITHDRAWN’ (despite the gene model exclusion performed using the web search described in the GeneBase guide; the remaining two gene entries are not specified).
After the initial parsing and importing steps, the three main tables in GeneBase database are constituted as follows: ‘Gene_Summary’ contains 2,417,408 records (one for each NCBI Gene entry), ‘Gene_Table’ (Fig. 2) contains 13,824,965 records (one record for each gene exon, included the corresponding intron if an intron follows that exon) and ‘Gene_Ontology’ contains 149,064 records in all (one for each gene with GO information available). Due to the lack of annotated transcribed products, a gene structure was not available for 86,824 Gene IDs (explaining the presence of ‘Gene_Summary’ gene entries without or with incomplete corresponding ‘Gene_Table’ records, for example Gene ID: 105667210 and Gene ID: 100121140, respectively).
Among the total gene entries, 2,368,726 are protein-coding, 25,796 pseudogenes (pseudo), 21,247 non-coding RNA (ncRNA), 527 coding for small nucleolar RNA (snoRNA), 137 for small nuclear RNA (snRNA), 86 for ribosomal RNA (rRNA) and 6 for cytoplasmic RNA genes (scRNA) (the remaining are not specified).
Then, to integrate nucleotide sequences, from the ‘Gene_Table’ table of our database, we selected 861,550 records with the ‘REVIEWED’ RefSeq status and 534,578 with the ‘VALIDATED’ RefSeq status (in both cases having an ‘NM_’ or ‘NR_’ type of RefSeq RNA accession number, to exclude ‘XM_’ or ‘XR_’ model Refseq records generated by automated pipelines) for a total of 1,396,128 exon entries. Using Batch Entrez, we were able to retrieve and download 1,336 records out of the 1,338 corresponding chromosome sequences. This selection gave rise to a total of 1,385,944 ‘Gene_Table’ records, which represent 10% of all available entries, updated with exon, coding exon (for protein-coding genes) and the corresponding downstream intron sequences up to 5 May 2015. The whole database including sequences has a size of 25.1 GB following decompression.
3.2. Statistics
In GeneBase, an additional table named ‘Reports’ is generated to easily provide statistics such as the mean length of exons (both coding and not coding) and introns (Table 1). These values were obtained considering all the available records in the ‘Gene_Table’ table of GeneBase database belonging to gene entries currently annotated on the most recent genome annotation, with a ‘REVIEWED’ or ‘VALIDATED’ RefSeq status and with an ‘NM_’ or ‘NR_’ type of the corresponding RefSeq RNA entry accession number. The mean exon length for all organisms is 308 base pairs (bp) with a standard deviation (SD) of 613 (range 1–91,671). In total, 1,252,462 GeneBase records are related to coding exons. The mean coding exon length is 206 bp with a SD of 325 (range 1–27,708). The mean intron length for all organisms is 3,820 bp with a SD of 15,693 (range 1–1,160,411), and the overall intron length distribution is shown in Supplementary Fig. S2. Other intron length statistics for some representative organisms are provided in Table 2 and Supplementary Fig. S3, considering a non-redundant set. The correlation coefficient between intron and genome lengths among these organisms is 0.957 with a P-value of 5.302e−05 (Supplementary Fig. S4) while, considering Vertebrata (Taxonomy ID: 7742), the correlation coefficient is 0.968 with a P-value of 0.007 (Supplementary Fig. S5).
Table 1.
Statistical analysis of exon and intron lengths
| Number of recordsa | Mean length (bp) | Standard deviation (bp) | Minimum length (bp)b | Maximum length (bp) | |
|---|---|---|---|---|---|
| Exons | 1,396,026 | 308 | 613 | 2 | 91,671 |
| Coding exons | 1,252,462 | 206 | 325 | 2 | 27,708 |
| Introns | 1,219,806 | 3,820 | 15,693 | 30 | 1,160,411 |
The analysis was carried out considering only ‘Gene_Table’ records belonging to gene entries with a ‘REVIEWED’ or ‘VALIDATED’ RefSeq status and with an ‘NM_’ or ‘NR_’ type of corresponding RefSeq RNA accession number, omitting entries marked as ‘not in current annotation release’ in the ‘Genome_Annotation_Status’ field (see Materials and methods). Mean and standard deviation values were obtained from the ‘Reports’ database table calculation fields. Lengths are given in base pairs (bp).
aCommon exons and introns belonging to multiple transcript variants are counted multiple times. The existence of intronless genes and the fact that terminal exons are not followed by an intron account for a reduced number of introns in comparison with exons.
bMinimum exon and intron length determination is subject to the annotation artifacts described in the text, so only the manually verified data are shown here.
Table 2.
Statistical analysis of intron length of some representative organisms
| Organism (Taxonomy ID) | Number of introns | Mean length (bp) | Standard deviation (bp) | Minimum length (bp)a | Maximum length (bp) |
|---|---|---|---|---|---|
| Arabidopsis thaliana (3702) | 124,533 | 169 | 194 | — | 11,602 |
| Caenorhabditis elegans (6239) | 107,605 | 324 | 803 | 39 | 100,913 |
| Tribolium castaneum (7070) | 434 | 1,883 | 5,592 | 35 | 53,400 |
| Drosophila melanogaster (7227) | 58,480 | 1,657 | 5,841 | 40 | 189,627 |
| Xenopus tropicalis (8364) | 4,174 | 2,849 | 7,646 | — | 160,644 |
| Danio rerio (7955) | 10,306 | 2,281 | 5,578 | — | 170,685 |
| Gallus gallus (9031) | 14,706 | 2,856 | 9,719 | — | 351,090 |
| Mus musculus (10090) | 85,507 | 5,622 | 20,369 | — | 1,041,985 |
| Homo sapiens (9606) | 155,222 | 7,386 | 24,002 | 30 | 1,160,411 |
The analysis was carried out considering a set of non-redundant ‘Gene_Table’ records belonging to gene entries with a ‘REVIEWED’ or ‘VALIDATED’ RefSeq status and with an ‘NM_’ or ‘NR_’ type of corresponding RefSeq RNA accession number, omitting entries marked as ‘not in current annotation release’ in the ‘Genome_Annotation_Status’ field (see Materials and methods). Mean and standard deviation values were obtained from the ‘Reports’ database table calculation fields. Lengths are given in base pairs (bp).
aMinimum length determination is subject to the annotation artifacts described in the text, so only the manually verified data for C. elegans, T. castaneum, D. melanogaster and H. sapiens are shown here.
3.3. Search for the minimal intron
We found 118,942 ‘Gene_Table’ records related to an intron sequence with a length range between 1 and 40 bp (Table 3). Among the entries with ‘REVIEWED’ or ‘VALIDATED’ RefSeq status, 587 intron records belong to an RNA with an ‘NM_’ or ‘NR_’ RefSeq type accession number (Supplementary Table S1). Considering the presence of the canonical GT-AG splicing sites as a prerequisite, a subset of 170 introns was used for the bioinformatic validation of the minimal intron (Supplementary Tables S1 and S2). Through BLASTN software, we were able to find at least two independent EST sequences, or one manually curated RNA sequence, encompassing the intron as validation of the existence of intron sequences, for 15 introns only (Table 4 and Supplementary Tables S2 and S3).
Table 3.
Number of retrieved GeneBase records and corresponding intron lengths
| Intron length queried (bp) | Number of retrieved records |
|||||
|---|---|---|---|---|---|---|
| REVIEWED | VALIDATED | PREDICTED | PROVISIONAL | INFERRED | EMPTY | |
| 1–10 | 21 | 5 | 2 | 2,822 | 1 | 954 |
| 11–20 | 22 | 1 | 4 | 8,250 | 1 | 7,291 |
| 21–30 | 63 | 13 | 37 | 32,487 | 2 | 4,269 |
| 31–40 | 442 | 20 | 38 | 58,242 | 8 | 3,947 |
| Total | 548 | 39 | 81 | 101,801 | 12 | 16,461 |
Lengths are given in base pairs (bp). The total number of ‘Gene_Table’ records with a currently annotated intron length between 1 and 40 bp is 118,942. Common introns belonging to multiple transcript variants are counted multiple times.
Table 4.
Minimal introns validated through bioinformatic analysis (one representative intron for each available organism; see Supplementary Tables S1, S2 and S3 for more details)
| Gene symbol (Gene ID) | Organism (Taxonomy ID) | RefSeq RNA accession number | Intron number | Positiona | Intron length (bp) | Previous exon 3′ | Intron sequence 5′-3′b | Following exon 5′ | GenBank accession numbersc |
|---|---|---|---|---|---|---|---|---|---|
| MST1L (11223) | Homo sapiens (9606) | NM_001271733.1 | 9 | CDS | 30 | gca | gtgagtccctggtgctcccggccccgccag ****** ***** | g * |
AY192149.1 |
| nAChRb1 (657999) | Tribolium castaneum (7070) | NM_001162528.1 | 8 | CDS | 35 | aag *** |
gtaaaaatctaatcacatacccacccccattcaag **** ****:** ** | c | EU937812.1 |
| CELE_B0348.6 (178536) | Caenorhabditis elegans (6239) | NM_070722.3 | 2 | CDS | 39 | caa ** |
gtacgttttgagaaatatattttattcaatgaatcatag *** ** *** :** * ** | a | CK586343.1 CK586324.1 CK586322.1 CK579517.1 |
| bcd (40830) | Drosophila melanogaster (7227) | NM_176410.3 | 2 | CDS | 40 | cag *** |
gtgagctcaaagccaacaaagtcagccatcgtcttatcag ***** ** * :** ***** | a |
X14459.1 BT021332.1 CO301329.1 BI163743.1 AA941658.2 |
aCDS: coding sequence.
bBold: donor (5′) and acceptor (3′) splice sites. Underline: possible pyrimidine-rich region. Colon: possible branch point. Asterisks: eukaryotic conserved bases (eukaryotic consensus sequence taken as reference: MAGGTRAGT…YNYYRAYY…YYYYYYYYYYYNYCAGG, where M = A/C, R = A/G, Y = C/T, N = any base).16,24
cSome example independent RNA sequences encompassing the intron and thus validating its existence.
The shortest intron sequence is 30 bp long and belongs to the MST1L gene of H. sapiens (Gene ID: 11223, Taxonomy ID: 9606), encoding for the putative macrophage stimulating 1-like protein. The following validated intron length is 35 bp and belongs to the nAChRb1 gene of Tribolium castaneum (Gene ID: 657999, Taxonomy ID: 7070), encoding for the nicotinic acetylcholine receptor beta-1 subunit. Regarding other organisms, also validated were the 39 bp intron of CELE_B0348.6 gene of C. elegans (Gene ID: 178536, Taxonomy ID: 6239) and the 40 bp intron of bcd gene of Drosophila melanogaster (Gene ID: 40830, Taxonomy ID: 7227), encoding for the eukaryotic translation initiation factor 4E-3 and for bicoid, respectively.
The analysis of the validated intron sequences revealed the presence of many conserved bases compared with the well-known consensus sequence (Table 4 and Supplementary Table S3).
The G content of the 20 shortest ‘REVIEWED’ or ‘VALIDATED’ human canonical intron sequences is shown in Supplementary Table S4.
4. Discussion
We have described a full parsing of all eukaryotic gene entries annotated in NCBI Gene and generated a relational and fully indexed local database named GeneBase. It consists of three main related tables containing information—such as gene nomenclature, structure and transcripts—in indexed fields about more than 2 million genes from more than 350 species, ranging from Plants and Fungi to Animals. This compares very favourably to, for example, the 91 species available through the Gene Table download utility provided by UCSC (http://genome-euro.ucsc.edu/cgi-bin/hgTables) or the 69 available in Ensembl through the download tool BioMart (http://www.ensembl.org/biomart/). The choice we made this time of considering NCBI Gene data, in contrast with our other sequence parsing tools based on UCSC data,26,27 also gave us maximum flexibility in the choice of search parameters. However, the parsing of such a high number of gene entries inevitably needs a considerable amount of disk space and time to perform calculations. We chose not to provide a GeneBase web tool, because FileMaker Pro is particularly useful due to its structure and interface but has limitations for web publication of full features of the local file. In addition, a web version would not be able to give users the freedom to customize the database and apply it to local files. Therefore, along with the eukaryote pre-loaded version of the database (for users interested in all the available entries), we have made an empty version available. It can be loaded by any user by importing subsequent versions or desired subsets of NCBI Gene data downloaded using specific text queries and parsed by our provided scripts, according to the GeneBase guide.
Among all the genes that are present in the pre-loaded version of GeneBase (based on NCBI Gene records available up to 22 April 2015), only 3.15 and 1.73% are ‘REVIEWED’ and ‘VALIDATED’ records, respectively, while almost all of the entries are ‘PROVISIONAL’ (92.88%), according to the entry status as provided by RefSeq. Among all the ‘Gene_Table’ exon/intron records, 16.9% are derived from an ‘NM_’ or ‘NR_’ RefSeq RNA type sequence. This implies that not only ‘REVIEWED’ and ‘VALIDATED’ but also ‘PROVISIONAL’, ‘INFERRED’ or ‘PREDICTED’ genes can present a gene product derived from these types of sequences. In addition, there are some discrepancies regarding the ‘live’ status provided by NCBI even for genes not annotated on the most recent genome annotation, addressed in GeneBase with the ‘Genome_Annotation_Status’ field, which is useful to filter out these entries. We also observed that in some cases, a ‘REVIEWED’ gene (for example Gene ID: 405306) with an ‘NM_’ RNA type product has an intron of 0 bp (or even −1 bp) length which is biologically not sound, but combined with the ‘low-quality sequence region’ markup in the corresponding RefSeq genome sequence, it indicates a compensation for genome assembly defects. The structuration of data of our database (e.g. numbers are seen as numeric fields) along with an intuitive user-friendly graphic interface allows users to perform many types of searches, even to exclude these kind of artifacts, and to make comparisons and calculations that are useful for data retrieval and analysis. The use of numeric fields has the specific advantage of making it possible to search for the exact number (or range), thus avoiding, querying ‘1’, for example, other unwanted results, such as 10, 11, 12 and so forth.28,29 In addition, calculation fields were created to generate further data that are not present in the original NCBI gene entries or in other databases, making the analysis capability of GeneBase unique. For example, the 3′-untranslated or 3′-translated and untranslated terminal exons30 are well known for their particular characteristics (e.g. the presence of the termination codon, the polyadenylation site and an average length) and sometimes for their implications in pathological nonsense mutations escaping nonsense-mediated decay.31 They cannot usually be retrieved with other databases, while GeneBase shows specific indexed calculation fields, labelling these exons as ‘Last_Exon’ and ‘Last_Coding_Exon’ (Fig. 2), to highlight their importance and to make them easily retrievable. In addition, specific fields showing related information from relational software tables allow users to perform original crossed searches of many entry features in a single organism of interest as well as across a full range of species. Finally, the implementation with exon (both coding and not coding) and intron sequences (Fig. 2) gives rise to original and previously unexplored gene information and sequence correlations.
GeneBase database contains a wealth of interesting biological information, and we provide an original analysis of the existing introns across all the available species as an example application to confirm its usefulness.
Although several exon and/or intron databases32–38 and NCBI Gene parsers5–8 exist, to our knowledge none is able to dynamically generate a fully structured relational database that could be updated (regenerated) by any user, with an intuitive user-friendly graphic interface that allows any type of further calculations and analysis. Use of NCBI Gene data has the advantage of allowing users to avoid GenBank redundancy purging procedures used to develop an Exon-Intron Database (EID),32 the Intron Exon-Knowledge base (IE-Kb),33 another Exon-Intron Database (ExInt)35 and the database of eukaryotic protein-encoding genes (Xpro).37 In the particular case of multiple transcript variants presenting the same exon or intron, the ‘Non_Redundant’ fields are useful for searching for unique exons and introns. On the other hand, the decision to keep the exon/intron structure of each transcript isoform for each gene, allows the option of, for example, alternative splicing studies.32,37,38 In addition, unlike more specialised databases such as, among others, the database on intron less/single exonic genes (SEGE)36 and the Intron and Intron Evolution Database (IDB and IEDB),34 GeneBase contains exon/intron structure of 5′UTR (untranslated region), coding region and 3′UTR of coding and non-coding nuclear genes,33,34,37 as well as intronless genes.36 In the NCBI Gene subset parsed and loaded into GeneBase, the majority of gene entries does not belong to C. elegans or to A. thaliana (Taxonomy IDs: 6239 and 3702, respectively), unlike for example in ExInt35 and in IDB and IEDB,34 but to H. sapiens (Taxonomy ID: 9606), in agreement with the current choice to exclude gene entries with a ‘MODEL’ RefSeq status.32 In any case, the user can select and download any NCBI Gene entry set for parsing.
Exploiting our ‘Gene_Table’ database table, which collects exon/intron structure for each gene product, our aim was to find the minimal intron, namely the minimal known DNA sequence element that can function as an intron. The overall distribution of intron (Supplementary Fig. S2) and the high standard deviation (Tables 1 and 2) reflects the extreme variation in intron lengths.33 Regarding the chosen representative organisms, A. thaliana, C. elegans, T. castaneum and D. melanogaster (Taxonomy IDs: 3702, 6239, 7070 and 7227, respectively) have only one major narrow peak around 100 bp, while Danio rerio (Taxonomy ID: 7955) has two similar peaks (Supplementary Fig. S3). Other chosen representative vertebrates (Xenopus tropicalis, Gallus gallus, Mus musculus and H. sapiens, Taxonomy IDs: 8364, 9031, 10090 and 9606, respectively) have one initial minor narrow peak and a subsequent higher and wider one (Supplementary Fig. S3). It is interesting to note that M. musculus and H. sapiens (Taxonomy ID: 9606) trends are remarkably overlapping (Supplementary Fig. S3), in accordance with an earlier report.10 Mean intron lengths of these organisms significantly correlate with their genome length and a noteworthy correlation can be found considering only Vertebrata (Taxonomy ID: 7742, Supplementary Figs S3 and S4).
The intron length problem, which currently raises researchers’ interest,17 has recently also been addressed by Sasaki-Haraguchi et al.,39 limiting their study to the human genome. They demonstrate an alternative splicing leading to low efficiency to the removal of an ultra-short 43 bp intron of the ESRP2 gene (Gene ID: 80004), following transfection of a construct containing the intron and the two flanking exons in human cultured neoplastic cells. This 43 bp intron is not currently annotated in the NCBI Gene database.
Uncertainty in defining the minimal intron length can be due to sequencing errors in genomes, to artifacts in intron size determination by annotation pipelines and to the lack of a tool like GeneBase, which is especially useful for retrieving data with numerical range constraints. As other authors previously supposed40 and verified,38 annotation errors and mismatches between the sequenced transcript and the reference genome can even generate one-nucleotide long introns, which we still came across too (Table 3 and Supplementary Table S1). Despite the additional exclusion procedure of gene ‘MODEL’ and the decision to consider only ‘REVIEWED’ or ‘VALIDATED’ gene entries with an ‘NM_’ or ‘NR_’ type of corresponding RefSeq RNA accession number, we were compelled to validate our results through a further bioinformatic analysis,37,38 which yielded 8.8% of actual intron sequences in this subset (Supplementary Table S2). The validation process was also hindered by genomic sequence repetitions, but with manual analysis, it was possible to identify intron sequences that had been erroneously annotated (Supplementary Table S2). In addition, 141 sequences derived from an annotated genomic sequences belonging to A. thaliana (Taxonomy ID: 3702). These lack experimental evidence as indicated in RefSeq Nucleotide entries and by the gene confidence ranking provided by TAIR (The Arabidopsis Information Resource at https://www.arabidopsis.org/index.jsp)41 and were not considered further.
Following our analyses based on all currently available data, the shortest eukaryotic GT-AG intron length belongs to the human MST1L gene (MST1-like, Gene ID: 11223), encoding for the putative macrophage stimulating 1-like protein, setting the physical limit of the intron size at 30 bp (Table 4). Remarkably, the 30 bp size is indeed consistent with the sum of the 5′/3′ splicing consensus site lengths.16,24,42 Unlike MST1 (Gene ID: 4485), for which 153 orthologs are reported in NCBI Gene, MST1L (Gene ID: 11223, which is the MST1 human paralog) lacks experimentally confirmed orthologs in other species. This is probably due to the fact that MST1L (Gene ID: 11223) appears to be an interesting case of ‘resurrected’ pseudogene in humans, so that it is now classified as a protein-coding gene with mass spectrometry evidence for the polypeptide product.43 Another human minimal intron validated through our application is the AQP12A 37 bp intron (Gene ID: 375318, Supplementary Table S3). Its sequence existence was also confirmed by a work published while this study was being submitted for publication.25 Finally, the inclusion in our analysis of organisms not present in other genome browsers also gave us the opportunity to validate four introns shorter than 40 bp of T. castaneum (Taxonomy ID: 7070).
Thanks to their small size, our ‘model introns’ will help identify the basic and crucial minimal requirement elements of recognition used by the cell for conventional splicing functioning. We were able to locate possible pyrimidine-rich regions and possible consensus branch points, although only one intron (LOC656453, Gene ID: 656453, Supplementary Table S4) presents the whole classic eukaryotic consensus sequence YNYYRAYY.16 The shortest validated human introns (MST1L 30 bp intron, Gene ID: 11223 and AQP12A 37 bp intron, Gene ID: 375318) present a G-rich sequence, as shown for other ultra-short human introns (Supplementary Table S4).25 The typical alternative splicing sites GYNGYN44 or NAGNAG for ‘subtle splicing’45–47 were not identified. Recent advances in the study of alternative splicing show that part of the maturation process of the primary transcript produces errors and creates stochastic noise.48,49 This would increase the difficulties faced by intron prediction tools taking into account all the well-known factors: 5′/3′ splice site, branch point, splicing regulatory elements such as exonic/intronic splicing enhancers/silencers, the spliceosome and other trans-acting elements.50 In the light of the observed data, the formation of secondary structures of the primary transcript might intervene in intron identification by the cell.51 More in-depth analysis of minimal introns to test this possibility will be necessary in the future.
Our analysis presented here confirms the difficulties still encountered in working with genomic sequences and is a starting point for further studies. Furthermore, it depends on the chosen gene entry subset and on the RefSeq classification system and is subject to the accuracy of the input dataset. On the other hand, our example application shows how a simple biological question such as how long the minimal GT-AG intron is (a numeric datum combined with a sequence feature) in all eukaryotic validated genes (which means a selection of a common gene characteristic in different organisms) is easily achievable with a single search, thanks to the GeneBase architecture. To our knowledge, GeneBase is a unique example of a database which relationally correlates and allows the complete elaboration of gene-associated meta-information data and the corresponding sequences across different organisms. Our tool's strength is also to allow large-scale analysis of genes, considerably increasing the possibilities for the study of the 2.4 million genes available in the NCBI Gene databank. Furthermore, GeneBase is useful for studying other characteristic intron lengths and sequences such as, for example, the unusual length of 5′-end first introns52 in terms of evolution and gene expression levels.53–55 The implementations of different databases using the same platform (FileMaker) recently led to intriguing results.56 In conjunction with quantitative transcriptome mapping in normal57 and pathological58 human cell types, GeneBase may represent the nucleus for a novel relational multi-purpose and user-friendly modular platform for the analysis of biological data, from meta-information of genes to their sequences and expression values. It will especially be used in the context of our current reanalysis of human chromosome 21 gene content to identify new targets for trisomy 21.59
Availability
GeneBase (both pre-loaded and empty versions), the ‘Database Design Report’, the user guide and the relative Python scripts for the initial data pre-processing and sequence calculations are publicly available at http://apollo11.isto.unibo.it/software/.
Supplementary data
Supplementary data are available at www.dnaresearch.oxfordjournals.org.
Funding
This work was funded by ‘RFO’ grants from Alma Mater Studiorum—University of Bologna to P.S. and L.V. M.C.'s and A.P.'s fellowships were supported by Fondazione Umano Progresso, Milano, Italy. The software was run on the Apple Mac Pro ‘Multiprocessor Server’ available at the DIMES Department and funded by ‘Fondazione CARISBO’, Bologna, Italy. Funding to pay the Open Access publication charges for this article was provided by donations to our Laboratory of Genomics for the study of trisomy 21.
Supplementary Material
Acknowledgements
We specially thank the Fondazione Umano Progresso, Milano, Italy, for supporting the research on trisomy 21 conducted at the DIMES Department. We are grateful to Danielle Mitzman for her expert revision of the manuscript.
References
- 1.Smith T.F. 1990, The history of the genetic sequence databases, Genomics, 6, 701–7. [DOI] [PubMed] [Google Scholar]
- 2.Kennard O. 1997, From private data to public knowledge. In: Butterworth I., (ed.), The Impact of Electronic Publishing on the Academic Community, an International Workshop Organised by the Academia Europaea and the Wenner-Gren Foundation, Wenner-Gren Center, Stockholm, 16–20 April, 1997 Portland Press Ltd, London, UK. [Google Scholar]
- 3.Brown G.R., Hem V., Katz K.S. et al. 2014, Gene: a gene-centered information resource at NCBI, Nucleic Acids Res., 43, D36–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lewitter F. 1998, Text-based database searching, TRENDS Biotechnol., 16, 3–5. [Google Scholar]
- 5.Hart K.W., Searls D.B., Overton G.C.. 1994, SORTEZ: a relational translator for NCBI's ASN.1 database, Comput. Appl. Biosci., 10, 369–78. [DOI] [PubMed] [Google Scholar]
- 6.Liu M., Grigoriev A.. 2005, Fast parsers for Entrez Gene, Bioinformatics, 21, 3189–90. [DOI] [PubMed] [Google Scholar]
- 7.Shah S.P., Huang Y., Xu T., Yuen M.M.S., Ling J., Ouellette B.F.F.. 2005, Atlas—a data warehouse for integrative bioinformatics, BMC Bioinformatics, 6, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hagen M.S., Lee E.K.. 2010, BIOSPIDA: a relational database translator for NCBI, AMIA Annu. Symp. Proc., 20,10, 422–6. [PMC free article] [PubMed] [Google Scholar]
- 9.Deutsch M., Long M.. 1999, Intron-exon structures of eukaryotic model organisms, Nucleic Acids Res., 27, 3219–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu J., Xiao J., Wang L. et al. 2013, Systematic analysis of intron size and abundance parameters in diverse lineages, Sci. China. Life Sci., 56, 968–74. [DOI] [PubMed] [Google Scholar]
- 11.Lim L.P., Burge C.B.. 2001, A computational analysis of sequence features involved in recognition of short introns, Proc. Natl Acad. Sci. USA, 98, 11193–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu J., Yang Z., Kibukawa M., Paddock M., Passey D.A., Wong G.K.-S.. 2002, Minimal introns are not ‘junk’, Genome Res., 12, 1185–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fields C. 1990, Information content of Caenorhabditis elegans splice site sequences varies with intron length, Nucleic Acids Res., 18, 1509–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mount S.M., Burks C., Hertz G., Stormo G.D., White O., Fields C.. 1992, Splicing signals in Drosophila: intron size, information content, and consensus sequences, Nucleic Acids Res., 20, 4255–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu J., He F., Wang D. et al. 2010, A novel role for minimal introns: routing mRNAs to the cytosol, PLoS One, 5, e10144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Strachan T., Read A.. 2010, Human molecular genetics, 4th edition Garland Science: New York. [Google Scholar]
- 17.Hubé F., Francastel C.. 2015, Mammalian introns: when the junk generates molecular diversity, Int. J. Mol. Sci., 16, 4429–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lenzi L., Frabetti F., Facchin F. et al. 2006, UniGene Tabulator: a full parser for the UniGene format, Bioinformatics, 22, 2570–1. [DOI] [PubMed] [Google Scholar]
- 19.Lenzi L., Facchin F., Piva F. et al. 2011, TRAM (Transcriptome Mapper): database-driven creation and analysis of transcriptome maps from multiple sources, BMC Genomics, 12, 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Agarwala R., Barrett T., Beck J. et al. 2015, Database resources of the National Center for Biotechnology Information, Nucleic Acids Res., 43, D6–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Altschul S.F., Madden T.L., Schäffer A.A. et al. 1997, Gapped BLAST and PSI-BLAST: a new generation of protein database search programs, Nucleic Acids Res., 25, 3389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gene Ontology Consortium Blake J., Dolan M. et al. 2013, Gene Ontology annotations and resources, Nucleic Acids Res., 41, D530–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burset M., Seledtsov I.A., Solovyev V.V.. 2000, Analysis of canonical and non-canonical splice sites in mammalian genomes, Nucleic Acids Res., 28, 4364–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alberts B., Johnson A., Lewis J. et al. 2014, Molecular biology of the cell, 6th edition Garland Science: New York. [Google Scholar]
- 25.Shimada M.K., Sasaki-Haraguchi N., Mayeda A.. 2015, Identification and validation of evolutionarily conserved unusually short pre-mRNA introns in the human genome, Int. J. Mol. Sci., 16, 10376–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Casadei R., Piovesan A., Vitale L. et al. 2012, Genome-scale analysis of human mRNA 5′ coding sequences based on expressed sequence tag (EST) database, Genomics, 100, 125–30. [DOI] [PubMed] [Google Scholar]
- 27.Piovesan A., Caracausi M., Pelleri M.C. et al. 2014, Improving mRNA 5′ coding sequence determination in the mouse genome, Mamm. Genome, 25, 149–59. [DOI] [PubMed] [Google Scholar]
- 28.Etzold T., Ulyanov A., Argos P.. 1996, SRS: information retrieval system for molecular biology data banks, Methods Enzymol., 266, 114–28. [DOI] [PubMed] [Google Scholar]
- 29.D'Addabbo P., Lenzi L., Facchin F. et al. 2004, GeneRecords: a relational database for GenBank flat file parsing and data manipulation in personal computers, Bioinformatics, 20, 2883–5. [DOI] [PubMed] [Google Scholar]
- 30.Zhang M.Q. 1998, Statistical features of human exons and their flanking regions, Hum. Mol. Genet., 7, 919–32. [DOI] [PubMed] [Google Scholar]
- 31.Isidor B., Lindenbaum P., Pichon O. et al. 2011, Truncating mutations in the last exon of NOTCH2 cause a rare skeletal disorder with osteoporosis, Nat. Genet., 43, 306–8. [DOI] [PubMed] [Google Scholar]
- 32.Saxonov S., Daizadeh I., Fedorov A., Gilbert W.. 2000, EID: the Exon-Intron Database-an exhaustive database of protein-coding intron-containing genes, Nucleic Acids Res., 28, 185–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sakharkar M.K., Kangueane P., Woon T.W. et al. 2000, IE-Kb: intron exon knowledge base, Bioinformatics, 16, 1151–2. [DOI] [PubMed] [Google Scholar]
- 34.Schisler N.J., Palmer J.D.. 2000, The IDB and IEDB: intron sequence and evolution databases, Nucleic Acids Res., 28, 181–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sakharkar M., Passetti F., de Souza J.E., Long M., de Souza S.J.. 2002, ExInt: an exon intron database, Nucleic Acids Res., 30, 191–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sakharkar M.K., Kangueane P., Petrov D.A., Kolaskar A.S., Subbiah S.. 2002, SEGE: a database on ‘intron less/single exonic’ genes from eukaryotes, Bioinformatics, 18, 1266–7. [DOI] [PubMed] [Google Scholar]
- 37.Gopalan V., Tan T.W., Lee B.T.K., Ranganathan S.. 2004, Xpro: database of eukaryotic protein-encoding genes, Nucleic Acids Res., 32, D59–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shepelev V., Fedorov A.. 2006, Advances in the exon-intron database (EID), Brief Bioinform., 7, 178–85. [DOI] [PubMed] [Google Scholar]
- 39.Sasaki-Haraguchi N., Shimada M.K., Taniguchi I., Ohno M., Mayeda A.. 2012, Mechanistic insights into human pre-mRNA splicing of human ultra-short introns: potential unusual mechanism identifies G-rich introns, Biochem. Biophys. Res. Commun., 423, 289–94. [DOI] [PubMed] [Google Scholar]
- 40.Spingola M., Grate L., Haussler D., Ares M.. 1999, Genome-wide bioinformatic and molecular analysis of introns in Saccharomyces cerevisiae, RNA, 5, 221–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huala E., Dickerman A.W., Garcia-Hernandez M. et al. 2001, The Arabidopsis Information Resource (TAIR): a comprehensive database and web-based information retrieval, analysis, and visualization system for a model plant, Nucleic Acids Res., 29, 102–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burge C.B., Tuschl T., Sharp T.A.. 1999, Splicing of precursors to mRNAs by the spliceosomes. In: Gesteland R., Cech T., Atkins J. (eds), The RNA World, 2nd edition Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, pp.525–60. [Google Scholar]
- 43.Pei B., Sisu C., Frankish A. et al. 2012, The GENCODE pseudogene resource, Genome Biol., 13, R51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sinha R., Lenser T., Jahn N. et al. 2010, TassDB2—a comprehensive database of subtle alternative splicing events, BMC Bioinformatics, 11, 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hiller M., Huse K., Szafranski K. et al. 2004, Widespread occurrence of alternative splicing at NAGNAG acceptors contributes to proteome plasticity, Nat. Genet., 36, 1255–7. [DOI] [PubMed] [Google Scholar]
- 46.Vitale L., Lenzi L., Huntsman S.A. et al. 2006, Differential expression of alternatively spliced mRNA forms of the insulin-like growth factor 1 receptor in human neuroendocrine tumors, Oncol. Rep., 15, 1249–56. [PubMed] [Google Scholar]
- 47.Vitale L., Frabetti F., Huntsman S.A. et al. 2007, Sequence, ‘subtle’ alternative splicing and expression of the CYYR1 (cysteine/tyrosine-rich 1) mRNA in human neuroendocrine tumors, BMC Cancer, 7, 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pickrell J.K., Pai A.A., Gilad Y., Pritchard J.K.. 2010, Noisy splicing drives mRNA isoform diversity in human cells, PLoS Genet., 6, e1001236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang M., Zhang P., Shu Y. et al. 2014, Alternative splicing at GYNNGY 5′ splice sites: more noise, less regulation, Nucleic Acids Res., 42, 13969–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jian X., Boerwinkle E., Liu X.. 2014, In silico prediction of splice-altering single nucleotide variants in the human genome, Nucleic Acids Res., 42, 13534–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Parada G.E., Munita R., Cerda C.A., Gysling K.. 2014, A comprehensive survey of non-canonical splice sites in the human transcriptome, Nucleic Acids Res., 42, 10564–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chorev M., Carmel L.. 2013, Computational identification of functional introns: high positional conservation of introns that harbor RNA genes, Nucleic Acids Res., 41, 5604–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Castillo-Davis C.I., Mekhedov S.L., Hartl D.L., Koonin E.V., Kondrashov F.A.. 2002, Selection for short introns in highly expressed genes, Nat. Genet., 31, 415–8. [DOI] [PubMed] [Google Scholar]
- 54.Pozzoli U., Menozzi G., Comi G.P., Cagliani R., Bresolin N., Sironi M.. 2007, Intron size in mammals: complexity comes to terms with economy, Trends Genet., 23, 20–4. [DOI] [PubMed] [Google Scholar]
- 55.Stival Sena J., Giguère I., Boyle B. et al. 2014, Evolution of gene structure in the conifer Picea glauca: a comparative analysis of the impact of intron size, BMC Plant Biol., 14, 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Piovesan A., Vitale L., Pelleri M.C., Strippoli P.. 2013, Universal tight correlation of codon bias and pool of RNA codons (codonome): the genome is optimized to allow any distribution of gene expression values in the transcriptome from bacteria to humans, Genomics, 101, 282–9. [DOI] [PubMed] [Google Scholar]
- 57.Caracausi M., Vitale L., Pelleri M.C., Piovesan A., Bruno S., Strippoli P.. 2014, A quantitative transcriptome reference map of the normal human brain, Neurogenetics, 15, 267–87. [DOI] [PubMed] [Google Scholar]
- 58.Pelleri M.C., Piovesan A., Caracausi M., Berardi A.C., Vitale L., Strippoli P.. 2014, Integrated differential transcriptome maps of Acute Megakaryoblastic Leukemia (AMKL) in children with or without Down Syndrome (DS), BMC Med. Genomics, 7, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Strippoli P., Pelleri M.C., Caracausi M. et al. 2013, An integrated route to identifying new pathogenesis-based therapeutic approaches for trisomy 21 (Down Syndrome) following the thought of Jérôme Lejeune, Sci. Postprint, 1, e00010. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.