Abstract
In an analytical study of microbial broths, the actinomycete strain Kitasatospora sp. P07101 was found to produce three new congeners, which were designated hazimycins B (1), C (2), and D (3), together with the previously reported hazimycin (renamed hazimycin A (4)). The structures of these hazimycins were examined using various spectroscopic methods including nuclear magnetic resonance (NMR), and the results revealed that 1–3 were analogues of hazimycin with the replacement of one of the two isonitrile groups in 4 by an NH-formyl group in 1, the two isonitrile groups and an amide group by two NH-formyl groups and a nitrile group in 2, and the two isonitrile groups and two amide groups by two NH-formyl groups and two nitrile groups in 3. Only hazimycin A exhibited moderate antimicrobial activities against Gram-positive bacteria and Candida albicans. These results indicated that the presence of two isonitrile groups in the hazimycin structure is essential for antimicrobial activity.
KEY WORDS: Hazimycin, Isonitrile, Nitrile, Microbial metabolites, Kitasatospora
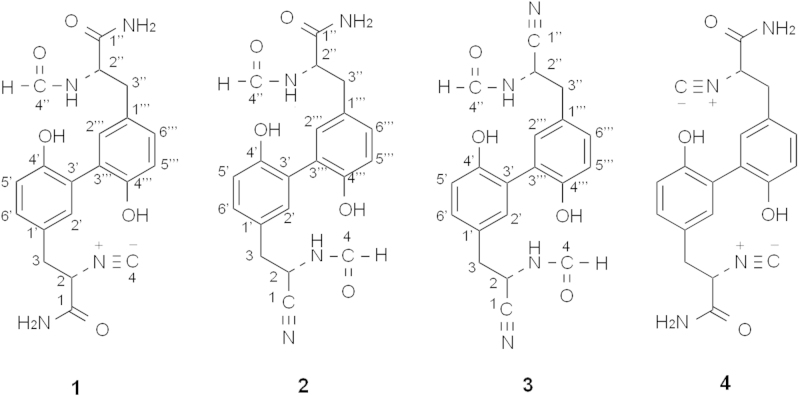
Graphical abstract
In an analytical study of microbial broths, the actinomycete strain Kitasatospora sp. P07101 was found to produce three new congeners, which were designated hazimycins B (1), C (2), and D (3), together with the previously reported hazimycin (renamed hazimycin A). Only hazimycin A exhibited moderate antimicrobial activities against Gram-positive bacteria and yeast. These results indicated that the presence of two isonitrile groups in the hazimycin structure is essential for antimicrobial activity.
1. Introduction
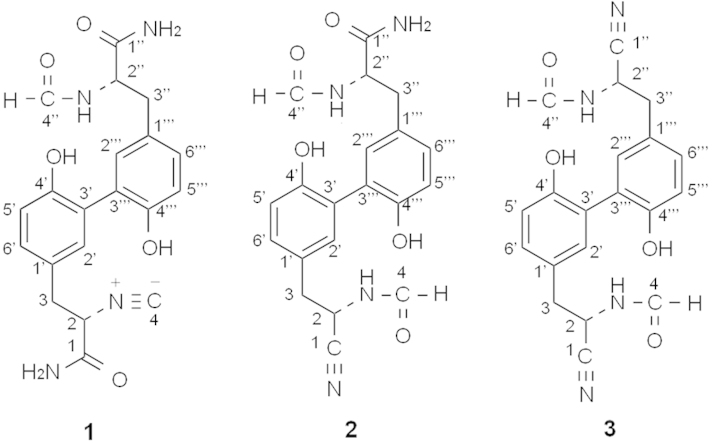
Our research group has focused on discovering novel compounds from microbial metabolites1, 2, 3, 4. Compounds were screened from our original culture collection using LC–UV and LC–MS/MS instruments. During this chemical screening program, the actinomycete strain Kitasatospora sp. P07101 was found to produce unidentified compounds. Novel hazimycins, hazimycins B (1), C (2), and D (3), were recently isolated from the fermentation broth along with the known antibiotic hazimycin5 (renamed hazimycin A (4), Fig. 1). These new congeners possessed a diaryl skeleton that contained isonitrile and nitrile groups, which are rare among microbial metabolites. The isolation, structure elucidation, and biological activities of 1–3 have been described in the present study.
Figure 1.
Structures of 1–4.
2. Results and discussion
2.1. Structure elucidation of 1–3
The physicochemical properties of compounds 1–3 are summarized in Table 1. Compounds 1–3 showed UV absorption between approximately 212 nm and 289 nm, which was identical to that of 4. The IR absorption at 2150–2300 cm–1 suggested the presence of isonitrile and/or nitrile groups in their structures. These results indicated that the basic skeleton of 1–3 was similar to that of 4.
Table 1.
Physicochemical properties of 1–3.
| Parameter | 1 | 2 | 3 |
|---|---|---|---|
| Appearance | Pale yellow powder | Colorless oil | Colorless oil |
| Molecular formula | C20H20N4O5 | C20H20N4O5 | C20H18N4O4 |
| Molecular weight | 396 | 396 | 378 |
| HR-ESI-MS (m/z) | |||
| Found | 419.1339 [M+Na]+ | 419.1338 [M+Na]+ | 401.1227 [M+Na]+ |
| Calcd. | 419.1331 | 419.1331 | 401.1226 |
| UV (MeOH) λmax (nm)/ε | 212/30,254 | 212/38,945 | 214/36,931 |
| 289/4514 | 289/4672 | 289/5065 | |
| [α]D28 (c=0.1, MeOH) | −1.4° | −1.6° | −10.6° |
| IR (KBr) vmax (cm−1) | 3400, 3205 | 3315, 3027 | 3379, 3021 |
| 3086, 2150 | 2252, 1674 | 2248, 1673 | |
| 1671, 1621 | 1614, 1504 | 1612, 1500 |
The structure of 1 was elucidated from various spectral data including NMR experiments. The molecular formula of 1 was determined to be C20H20N4O5 based on HR-ESI-MS measurements, which indicated that the molecular formula of 1 has one oxygen atom and two hydrogen atoms more than that of 4. The 13C-NMR spectrum showed 20 resolved signals, which were classified into two sp3 methylene carbons, two sp3 methine carbons, six sp2 methine carbons, four sp2 quaternary carbons, two sp2 quaternary oxycarbons, one sp carbon, two sp2 carbonyl carbons, and one sp2 formyl carbon by 1H–13C heteronuclear single-quantum correlation (HSQC) analysis. The 1H NMR spectrum (in DMSO-d6) displayed 18 proton signals. The connectivity of the proton and carbon atoms was established from the 1H–13C HSQC spectrum (Table 2). A comparison of the NMR spectra of 1 and 4 indicated that they both possessed a dihydroxydiaryl skeleton. However, most double signals were observed in the 1H and 13C NMR spectra of 1, suggesting that 1 was a heterodimer, while 4 was a homodimer. A formyl proton signal (δ 7.92) and amide proton signal (δ 8.17) were observed in 1, but were absent in 4, which indicated that one of two isonitrile groups was converted to an NH-formyl group in 1. Cross peaks were observed from H-2″ (δ 4.43) to C-4″ (δ 160.9) as well as from NH-2″ (δ 8.17) to C-4″ in the 13C–1H heteronuclear multiple-bond correlation (HMBC) experiments (Fig. 2A). The structure satisfied the unsaturation number, UV spectra, and molecular formula. These results indicated that compound 1 was a 2″-NH-formyl hazimycin, as shown in Fig. 1.
Table 2.
1H and 13C NMR chemical shifts of 1–3.
| Position |
1 |
2 |
Position |
3 |
|||
|---|---|---|---|---|---|---|---|
| δc | δH | δc | δH | δc | δH | ||
| 1 | 167.1s | – | 119.0s | – | 1, 1″ | 119.0s | – |
| 1-NH2 | – | 7.48 (1H, s), 7.71 (1H, s) | – | – | 1-NH2, 1″-NH2 | – | – |
| 2 | 58.9d | 4.49 (1H, dd, J=4.8, 4.4) | 40.1d | 4.98 (1H, dd, J=8.0, 7.6) | 2, 2″ | 40.4d | 4.90 (1H, dd, J=8.0, 7.6) |
| 2-NH | – | – | – | 8.86 (1H, d, J=7.6) | 2-NH, 2″-NH | – | 8.86 (1H, d, J=7.6) |
| 3 | 37.8t | 2.86 (1H, m), 3.03 (1H, dd, J=8.8, 4.8) | 36.5t | 2.98 (1H, m) | 3, 3″ | 36.5t | 2.98 (1H, m) |
| 4 | 158.0s | – | 161.1d | 8.06 (1H, s) | 4, 4″ | 161.1s | 8.06 (1H, s) |
| 1′ | 126.0s | – | 125.3s | – | 1′. 1‴ | 125.2s | – |
| 2′ | 132.2d | 7.07 (1H, s) | 132.5d | 7.07 (1H, s) | 2′, 2‴ | 132.5d | 7.08 (1H, s) |
| 3′ | 125.4s | – | 126.0s | – | 3′, 3‴ | 125.7s | – |
| 4′ | 153.7s | – | 153.7s | – | 4′, 4‴ | 153.9s | – |
| 5′ | 115.7d | 6.80 (1H, d, J=8.0) | 115.8d | 6.81 (1H, d, J=8.8) | 5′, 5‴ | 115.8d | 6.80 (1H, d, J=8.4) |
| 6′ | 128.9d | 7.03 (1H, d, J=8.0) | 128.9d | 7.05 (1H, d, J=8.8) | 6′, 6‴ | 129.0d | 7.05 (1H, d, J=8.4) |
| 1″ | 172.8s | – | 172.8s | – | |||
| 1″-NH2 | – | 7.02 (1H, s), 7.48 (1H, s) | – | 7.04 (1H, s), 7.48 (1H, s) | |||
| 2″ | 52.7d | 4.43 (1H, ddd, J=8.4, 4.8, 4.0) | 52.7d | 4.44 (1H, ddd, J=8.4, 4.8, 4.0) | |||
| 2″-NH | – | 8.17 (1H, d, J=8.4) | – | 8.16 (1H, d, J=8.4) | |||
| 3″ | 37.0t | 2.65 (1H, m), 2.90 (1H, m) | 36.7t | 2.60 (1H, m), 2.91 (1H, m) | |||
| 4″ | 160.9d | 7.92 (1H, s) | 160.8d | 7.92 (1H, s) | |||
| 1‴ | 127.8s | – | 127.8s | – | |||
| 2‴ | 132.2d | 7.02 (1H, s) | 132.2d | 7.02 (1H, s) | |||
| 3‴ | 126.0s | – | 125.3s | – | |||
| 4‴ | 153.1s | – | 153.0s | – | |||
| 5‴ | 115.5d | 6.74 (1H, d, J=8.0) | 115.4d | 6.75 (1H, d, J=8.0) | |||
| 6‴ | 128.8d | 6.98 (1H, d, J=8.0) | 128.8d | 6.98 (1H, d, J=8.0) | |||
Figure 2.
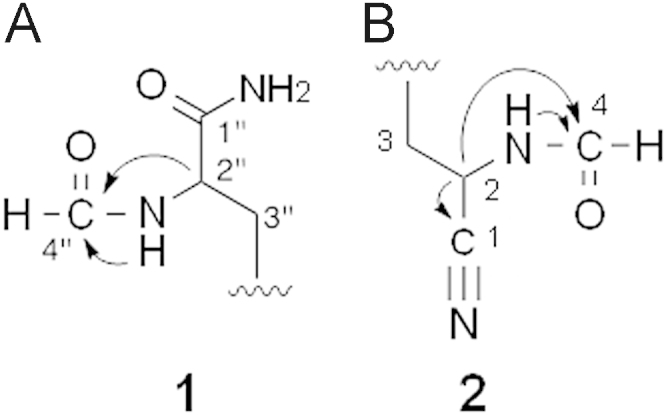
Key HMBCs of 1 and 2.
The molecular formula of 2 was identical to that of 1. However, two proton signals of an NH-formyl group (δ 8.06 and 8.86) were newly observed, and one of the amide proton signals of the two carboxamide groups (δ 7.48 and 7.71) disappeared in the 1H NMR spectrum of 2. Furthermore, a new sp carbon signal (δ 119.0) was observed in place of one of the two carboxamide carbon signals (δ 167.1) in the 13C NMR spectrum of 2. These results indicated the formylation of another isonitrile group of 1 and the conversion of one of the two carboxamide groups of 1 to a nitrile group in 2. The position of the nitrile group was confirmed by 13C–1H HMBC experiments (Fig. 2B): cross peaks were observed from H-2 (δ 4.98) to C-1 (δ 119.0) and C-4 (δ 161.1). Thus, compound 2 was elucidated to be 2,2″-NH-formyl and 2-nitrle hazimycin (Fig. 1).
As listed in Table 1, the molecular formula of 3 has one oxygen atom and two hydrogen atoms fewer than that of 2. Its 1H-NMR spectrum revealed homodimer-type proton signals, and was almost identical to that of 2 except for the disappearance of the amide proton signals of the carboxamide groups (δ 7.04 and 7.48) in 3. Furthermore, the presence of a nitrile carbon signal (δ 119.0) was confirmed as well as 2 in the 13C-NMR spectrum, which indicated that another carboxamide group of 2 was converted to a nitrile group in 3. Finally, cross peaks were observed from H-2″ (δ 4.90) to C1″ (δ 119.0) and C4″ (δ 161.1) as well as from NH-2″ (δ 8.86) to C4″ in the 13C–1H HMBC experiments. Thus, compound 3 was elucidated to be a 2,2″-NH-formyl and 2,2″-nitrile hazimycin (Fig. 1)
Regarding the absolute stereochemistry of the novel hazimycin analogs, dityrosine was prepared by hydrolyzing 4 under acidic conditions because its optical rotation has already been accurately described in previous studies6. The results obtained in this study were consistent with those of l,l-dityrosine. Thus, the absolute stereochemistry of 2 and 2″ of 4 was defined as S. Similarly, the hazimycin congeners 1–3 are considered to be derived from l,l-dityrosine because they are generated via common biosynthetic pathway. Thus, compounds 1–3 should have the same absolute stereochemistry as 4.
Waitz et al.7 previously reported that hazimycin A was biosynthesized from tyrosine and methionine based on 14C-labeling experiments. They postulated that an isonitrile group was formed through the NH-formyl residue generated due to the methylation, reduction, and oxidation of an amino group of tyrosine. In the present study, we demonstrated the presence and structure of not only the NH-formyl product, but also the new family of nitrile-type hazimycins in the culture broth of the actinomycete strain.
2.2. Antimicrobial activities of 1–3
We examined antimicrobial activity against 7 test microorganisms using the paper disk method8. As shown in Table 3, compounds 1–3 and dityrosine did not inhibit the growth of these microorganisms; only compound 4 exhibited antimicrobial activity against Mycobacterium smegmatis, Bacillus subtilis, Staphylococcus aureus, Micrococcus luteus, and Candida albicans with inhibition zones of 19, 14, 23, 26 and 20 mm, respectively. These results indicated that the attachment of both isonitrile groups in the side chain of the dihydroxydiaryl skeleton was crucial for antimicrobial activity.
Table 3.
Antimicrobial activities of 1–4a.
| Test organism | Inhibition zone (diameter, mm) |
||||
|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | Dityrosine | |
| Gram-positive bacteria | |||||
| Mycobacterium smegmatis | –b | – | – | 19 | – |
| Bacillus subtilis | – | – | – | 14 | – |
| Staphylococcus aureus | – | – | – | 23 | – |
| Micrococcus luteus | – | – | – | 26 | – |
| Gram-negative bacteria | |||||
| Escherichia coli | – | – | – | – | – |
| Pseudomonas aeruginosa | – | – | – | – | – |
| Yeast | |||||
| Candida albicans | – | – | – | 20 | – |
10 μg/6 mm disk.
Can not detect antimicrobial activity.
3. Material and methods
3.1. General procedures
The actinomycete strain Kitasatospora sp. P07101 was originally isolated from a soil sample collected at Minato-ku, Tokyo, Japan. The genus was determined based on taxonomic studies and genetic analysis of 16S rRNA by the identification services of TechnoSuruga Laboratory Co., Ltd., Shizuoka, Japan. This strain was used to produce 1–3. UV spectra were recorded on a spectrophotometer (8453 model, Agilent). IR spectra were recorded on a Fourier transform infrared spectrometer (FT-710, Horiba). Optical rotations were measured with a digital polarimeter (DIP-370, JASCO). FAB-MS spectra and HR-FAB-MS spectra were recorded on a mass spectrometer (JMS-AX505HA, JEOL). The various NMR spectra were collected with a spectrometer (XL-400, Varian).
3.2. Isolation of 1–3
The three-day-old fermentation broth (6.5 L) of Kitasatospora sp. P07101 was centrifuged to separate the mycelia and supernatant. A part of the supernatant (4.0 L) was then added to a Diaion HP-20 column (volume: 0.2 L, Mitsubishi Chemical Co.) After washing with distilled water (1.0 L), the desired substances were eluted with MeOH (1.0 L) and concentrated in vacuo to dryness to produce red brown materials (2.3 g), which were then dissolved in MeOH and purified using high performance liquid chromatography (HPLC, column: DevelosilC30, 250 mm × 20 mm, Nomura Scientific Co.; solvent: a gradient system of 40 min from 5% CH3CN containing 0.05% trifluoroacetic acid (TFA) to 55% CH3CN containing 0.05% TFA; detection: UV at 210 nm; flow: 8.0 mL/min). Under these conditions, the peaks eluted at retention time of 18.8 and 27.8 min were repeatedly collected and concentrated to dryness to give 1 (1.0 mg) and 4 (110 mg) as pale yellow powders, respectively.
A residual supernatant (2.5 L) was extracted twice with an equal volume of ethylacetate, and concentrated in vacuo to dryness to produce red brown materials (1.2 g). This sample was dissolved in MeOH, and then purified using HPLC (column: PEGASIL ODS, 250 mm×20 mm, Senshu Scientific Co.; solvent: a gradient system of 60 min from 10% CH3CN containing 0.05% TFA to 45% CH3CN containing 0.05% TFA; detection: UV at 210 nm; flow: 6.0 mL/min). Under these conditions, the peaks eluted at retention time of 27 and 40 min were repeatedly collected and concentrated to dryness to give 2 (44.4 mg) and 3 (55.2 mg) as colorless oils, respectively.
3.3. Preparation of dityrosine
Dityrosine was prepared according to a previous method6 by hydrolyzing hazimycin A under acidic conditions. Hazimycin A (20.0 mg) was dissolved in 12 mol/L HCl (1.0 mL), and then hydrolyzed at 60°C for 12 h. The reaction solution was neutralized with 10 mol/L NaOH, and then centrifuged to precipitate the resulting salts. Finally, the collected supernatant was purified using HPLC (column: Develosil C30, 250 mm×20 mm; solvent: 3.0% CH3CN; detection: UV at 210 nm; flow: 6.0 mL/min). The peak eluted at 16.7 min was repeatedly collected and concentrated to dryness to give dityrosine (10.2 mg) as a white powder. The spectroscopic data listed below were consistent with the findings of a previous study6.
Dityrosine: FAB-MS-positive; [M+H]+=361, 1H NMR in DMSO-d6 (600 MHz) δH; 2.90 (2H), 3.03 (2H), 3.93 (2H), 6.80 (2H), 7.01 (2H), 7.16 (1H), 7.19 (1H), 13C NMR in DMSO-d6 (150 MHz) δC; 34.8, 35.4, 54.0, 54.5, 115.8, 116.0, 125.6, 126.0, 126.3, 129.5, 130.1, 133.2, 133.4, 153.2, 153.3, 170.6, 170.8, [α]D28; –3.54 (c=0.1, 1 mol/L HCl).
3.4. Assay for antimicrobial activity
Antimicrobial activity against 7 test microorganisms as listed in Table 3 was measured using paper disks (6 mm, ADVANTEC) containing a sample, according to our established method8.
Acknowledgments
This study was supported in part by a Kakenhi Grant 23790020 (to Nobuhiro Koyama), and a Kitasato University Research Grant for Young Researchers (to Nobuhiro Koyama). We express our thanks to Ms. Noriko Sato and Dr. Kenichiro Nagai, the School of Pharmacy, Kitasato University for their help measuring NMR and MS spectra, respectively.
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
References
- 1.Inokoshi J., Shigeta N., Fukuda T., Uchida R., Nonaka K., Masuma R. Epi-trichosetin, a new undecaprenyl pyrophosphate synthase inhibitor, produced by Fusarium oxysporum FKI-4553. J Antibiot. 2013;66:549–554. doi: 10.1038/ja.2013.44. [DOI] [PubMed] [Google Scholar]
- 2.Fukuda T., Tomoda H., Tylopilusin C. a new diphenolic compound from the fruiting bodies of Tylopilus eximinus. J Anitibiot. 2013;66:355–357. doi: 10.1038/ja.2013.23. [DOI] [PubMed] [Google Scholar]
- 3.Kawaguchi M., Uchida R., Ohte S., Miyachi N., Kobayashi K., Sato N. New dinapinone derivatives, potent inhibitors of triacylglycerol synthesis in mammalian cells, produced by Talaromyces pinophilus FKI-3864. J Antibot. 2013;66:179–189. doi: 10.1038/ja.2012.127. [DOI] [PubMed] [Google Scholar]
- 4.Koyama N., Tokura Y., Takahashi Y., Tomoda H. Discovery of nosokophic acid, a predicted intermediate of moenomycins, from nosokomycin-producing Streptomyces sp. K04-0144. Bioorg Med Chem Lett. 2013;23:860–863. doi: 10.1016/j.bmcl.2012.11.044. [DOI] [PubMed] [Google Scholar]
- 5.Marquez J.A., Horan A.C., Kalyanpur M., Lee B.K., Loebenberg D., Miller G.H. The hazimicins, a new class of antibiotics. Taxonomy, fermentation, isolation, characterization and biological properties. J Anitibot. 1983;36:1101–1108. doi: 10.7164/antibiotics.36.1101. [DOI] [PubMed] [Google Scholar]
- 6.Skaff O., Jolliffe K.A., Hutton C.A. Synthesis of the side chain cross-linked tyrosine oligomers dityrosine, trityrosine, and pulcherosine. J Org Chem. 2005;70:7353–7363. doi: 10.1021/jo051076m. [DOI] [PubMed] [Google Scholar]
- 7.Puar M.S., Munayyer H., Hedge V., Lee B.K., Waitz J.A. The biosynthesis of hazimicins: possible origin of isonitrile carbon. J Antibiot. 1985;38:530–532. doi: 10.7164/antibiotics.38.530. [DOI] [PubMed] [Google Scholar]
- 8.Koyama N., Kojima S., Nonaka K., Masuma R., Matsumoto M., Omura S. Calpinactam, a new anti-mycobacterial agent, produced by Mortierella alpina FKI-4905. J Antibiot. 2010;63:183–186. doi: 10.1038/ja.2010.14. [DOI] [PubMed] [Google Scholar]