Abstract
The exposome is the cumulative measure of environmental influences and associated biological responses throughout the lifespan, including exposures from the environment, diet, behavior, and endogenous processes. A major challenge for exposome research lies in the development of robust and affordable analytic procedures to measure the broad range of exposures and associated biologic impacts occurring over a lifetime. Biomonitoring is an established approach to evaluate internal body burden of environmental exposures, but use of biomonitoring for exposome research is often limited by the high costs associated with quantification of individual chemicals. High-resolution metabolomics (HRM) uses ultra-high resolution mass spectrometry with minimal sample preparation to support high-throughput relative quantification of thousands of environmental, dietary, and microbial chemicals. HRM also measures metabolites in most endogenous metabolic pathways, thereby providing simultaneous measurement of biologic responses to environmental exposures. The present research examined quantification strategies to enhance the usefulness of HRM data for cumulative exposome research. The results provide a simple reference standardization protocol in which individual chemical concentrations in unknown samples are estimated by comparison to a concurrently analyzed, pooled reference sample with known chemical concentrations. The approach was tested using blinded analyses of amino acids in human samples and was found to be comparable to independent laboratory results based on surrogate standardization or internal standardization. Quantification was reproducible over a 13-month period and extrapolated to thousands of chemicals. The results show that reference standardization protocol provides an effective strategy that will enhance data collection for cumulative exposome research. In principle, the approach can be extended to other types of mass spectrometry and other analytical methods.
Keywords: amino acids, metabolomics, analytical chemistry, environment, mass spectrometry
Previous research shows that Fourier-transform mass spectrometry (MS) coupled to liquid chromatography (LC), termed “high-resolution metabolomics (HRM),” provides a practical approach to detail physiological chemistry of an individual (Johnson et al., 2010; Jones et al., 2012; Soltow et al., 2013). With this approach, LC is coupled with ultra-high resolution MS to detect >100 000 ions, characterized by accurate mass, retention time (RT) and ion intensity. Although multiple ions are generated from individual chemicals, single ions are detected for many low-abundance chemicals so that the number of chemicals detected exceeds 20 000. Resolution of low-abundance chemicals with mass accuracy within a few parts-per-million is a key characteristic enabling this unsurpassed coverage of endogenous metabolites and exogenous chemicals in biologic samples.
Advances in LC, MS and data extraction (Chen et al., 2012; Ivanisevic et al., 2013; Uppal et al., 2013) have further expanded the dynamic range for detection of chemicals in biological samples by at least an order of magnitude in the last decade. The use of triplicate analytical replicates, in particular, has transformed reliable measurement of low-abundance ions. Results show that 20 000 ions can be measured in human serum with median coefficient of variation (CV) ≤15% (Uppal and Jones, unpublished), sufficient to measure thousands of chemicals in a single analysis.
Importantly, HRM detects relevant environmental chemicals that are needed to establish the exposome (Jones et al., 2012; Miller and Jones, 2013). For instance, plasticizers, insecticides, fungicides, herbicides, drugs, and bacterial products are detected simultaneously with endogenous metabolites (Cribbs et al., 2014; Frediani et al., 2014; Go et al., 2014a; Jones et al., 2012; Neujahr et al., 2014; Osborn et al., 2013; Park et al., 2012; Roede et al., 2013). Environmental chemicals correlate with plasma phenylalanine (Go et al., 2015) and with health behaviors, eg, cotinine with smoking (Go et al. 2014a; Park et al., 2012). Exposome research is facilitated by use of HRM to study aging (Hoffman et al., 2014; Zhao et al., 2014), Parkinson’s disease (Roede et al., 2013), asthma (Fitzpatrick et al., 2014), human immunodeficiency virus infection (Cribbs et al., 2014), lung transplantation (Neujahr et al., 2014), red blood cells stored for transfusion (Roback et al., 2014), and macular degeneration (Osborn et al., 2013). Human studies are also available on intraindividual variations due to diet (Stamler et al., 2013) and nutritional deficiency (Gregory et al., 2013), and model studies are available for environmental exposures (Go et al. 2014a; Park et al., 2012), molecular mechanisms in immunity (Li et al., 2013; Ravindran et al., 2014; Xu et al., 2014) and complex environmental disease mechanisms (Go et al., 2014a; Roede et al., 2014). This widespread use emphasizes the need for methods to facilitate the integration of data from different studies into common analytic structures to characterize the exposome.
Measurement of 20 000 chemicals in human plasma (Ivanisevic et al., 2013; Uppal et al., 2013) by HRM provides a useful capability for exposome research. However, HRM is limited by reliance upon relative quantification, which allows for the study of chemicals among a set of samples but limits comparisons between platforms or within the same facility at different times of analysis. In a targeted study of aminothiols in human plasma (Johnson et al., 2008), we found that chemicals differing by nearly 4 orders of magnitude were quantifiable by LC-MS with isotopic internal standards, but this approach is impractical for large numbers of analytes. Complex mixtures of external standards provide a possible alternative, but external calibration is usually avoided for LC-MS due to nonlinearity and ion suppression effects (Muller et al., 2002).
The ability to measure chemicals by LC-MS differing in concentration by nearly 4 orders of magnitude suggests that simultaneous quantification of thousands of chemicals with substantial differences in concentration may be possible. This is supported by method-of-addition studies in biological matrices, which show linear increases in intensity with analyte addition (Chelius and Bondarenko, 2002; Wang et al., 2003), suggesting linearity is sufficient to allow single point calibration. Van der Greef et al. (2007) discussed the use of pooled reference standards for nontargeted analysis of large sample batches with significant time between each batch and established feasibility for single point calibration using glucose as an example.
This study builds upon this analytic structure for quantification in exposome research, with the specific goal to develop analytical procedures for HRM over time and between research facilities (Table 1). We analyzed human samples with external calibration, internal standardization, surrogate standardization, and reference standardization. External calibration involved calibration of amino acid (AA) content in 117 human samples against measures obtained by conventional AA analysis. Internal standardization involved measurement of methionine and tyrosine in these samples by stable isotope dilution. Surrogate standardization involved quantification of other AAs against a stable isotopic internal standard using relative response factors (RRFs) (Greizerstein et al., 1997). Reference standardization involved quantification of individual chemicals relative to pooled reference plasma that was calibrated to National Institute of Standards and Technology (NIST) Standard Reference Material for human plasma (SRM 1950) (McGaw et al., 2010). Results of these analyses show that the latter approach, which represents a simple extension of the practice advocated by van der Greef et al. (2007), provides an effective framework to quantitatively compare endogenous metabolites and environmental chemicals for use in cumulative exposome research.
TABLE 1.
Calibration Methods for Exposome Research
| Method | Comments |
|---|---|
| External calibration with method of additions |
|
| Internal standardization with stable isotopic standards |
|
| Surrogate standardization with stable isotopic standards |
|
| Reference standardization using calibrated reference |
|
HRM provides relative quantification of thousands of metabolites. While useful for many metabolomics studies, relative quantification is difficult to combine to create cumulative databases for exposome research. The present research examined 4 methods for quantification of plasma metabolite concentrations for data obtained using HRM.
MATERIALS AND METHODS
Chemicals and HPLC columns
Acetonitrile (HPLC grade), formic acid (HPLC grade), and water (HPLC grade) were obtained from Sigma–Aldrich (St. Louis). A mixture of internal standard stable isotopic chemicals (Soltow et al., 2013) included [15N]-L-tyrosine, [trimethyl-13C3]-caffeine and [15N,13C5]-L-methionine from Cambridge Isotope Laboratories, Inc (Andover, Pennsylvania).
Human plasma samples
Subsets of samples (n = 117) and (n = 157) from 2 studies (ClinicalTrials.gov Identifier: NCT00248638 and NCT00336570, respectively) were used for this methods development research. Details of the complete cohort studies are provided elsewhere (GLND, TR Ziegler, MD, Principal Investigator; PREMED, AA Quyyumi, MD, Principal Investigator). For the present purposes, the demographic and clinical details were not considered relevant to the tests of analytic procedures, and therefore, were not included in the analyses. The use of samples for additional analysis was allowed in the informed consent; both studies were reviewed and approved by the Emory University Investigational Review Board. GLND subjects were participants in a trial (NCT00248638) to determine whether the glutamine (GLN)-containing dipeptide alanyl-GLN protects against morbidity or mortality in surgical intensive care unit patients. The samples analyzed included individuals from treatment and control groups. PREMED subjects were healthy participants between 30 and 90 years who were studied to define a “normal” value or range of values in healthy people and to evaluate tests specifically designed to look for evidence of early multiorgan disease (NCT00336570). Both studies included both sexes and individuals of different races and ethnicities. Plasma was stored at −80°C prior to LC-MS analysis.
NIST SRM 1950 and Q-Standard
Our standard operating procedures involves use of 2 pooled human reference plasma samples for each sample set, with quality control and assurance defined by the consistency of data within concurrently analyzed batches of 20 samples (see Go et al., 2014b). The rationale for 2 pooled references was that the SRM1950, although preferable because it has certified concentrations, is costly and of limited supply. Thus, we adopted a procedure to analyze SRM1950 at the beginning and end of each study (or every 25 days) and analyze a second pooled reference sample that was prepared within our laboratory, at the beginning and end of each batch of 20 samples (twice daily). SRM1950 consisted of pooled plasma from an equal number of healthy men and women aged 40–50 years. Our pooled reference Q-Std (Q-Standard, independently prepared pooled reference plasma) consisted of plasma pooled from 2 separate lots, each from an unknown number of males and females without demographic information, from Equitech-Bio, Inc, (Kerrville, Texas); direct comparison of QStd and SRM1950 showed that they were sufficiently similar to allow calibration of the QStd relative to SRM1950 for specific metabolite quantification (see Results).
High-resolution metabolomics
Human plasma samples (see above) were extracted and analyzed as previously described (Soltow et al., 2013; Uppal et al., 2013). Briefly, extractions were performed with acetonitrile containing a mixture of internal standards and maintained in an autosampler at 4°C until injection. LC was performed using a C18 column (Higgins Analytical, Targa, 2.1 × 10 cm) with a short, end-capped C18 precolumn (Higgins Analytical, Targa guard) and an acetonitrile gradient. A flow rate of 0.35 ml/min was used for the first 6 min and then increased to 0.5 ml/min for the remaining 4 min. The first 2-min period consisted of 5% solution A (2% formic acid), 60% water, 35% acetonitrile, followed by a 4-min linear gradient to 5% A, 0% water, 95% acetonitrile. The final 4-min period was maintained at 5% A, 95% acetonitrile. Mass spectrometry was performed using an (Linear Trap Quadropole) LTQ-Velos-Orbitrap mass spectrometer (Thermo Fisher, San Diego, California): HESI probe with S-lens combination for Electrospray Ionization (ESI); MS1 mode scanning m/z (mass to charge, ion) range of 85–2000; resolution, 60 000; maximum number of ions collected, 5.00 × 105. The maximum injection time, 5 ms; capillary temperature, 275°C; source heater; 45°C; voltage, 4.6 kV; sheath gas, 45 (arbitrary units); auxillary gas flow, 5 (arbitrary units); sweep gas flow, 0 (arbitrary units). Each sample was run in triplicate using a 10 μl injection volume. Data were collected continuously over the 10-min chromatographic separation and stored as .raw files.
The MS files were converted using XCalibur file converter software (Thermo Fisher, San Diego, California) to .cdf files for data processing. The .raw files are time and date stamped, and electronically archived as the original data for use in subsequent reanalysis. The data were processed for peak extraction and quantification of ion intensities using xMSanalyzer (MS1 mode) (Uppal et al., 2013) with apLCMS (Yu et al., 2009). The parameter settings used for data extraction determine the number of features retained, where an m/z feature is defined as an accurate mass m/z (defined as ≤10 parts per million mass resolution) with associated RT and intensity. For this study, feature and sample filtering were set to retain features that had a median CV less than 50%, a Pearson correlation greater than 0.7 among 3 replicate injections for each sample, and fewer than 30% missing values. The identities of selected features were validated using MS2 and compared with either authentic reference standards (when available) or online databases (Smith et al., 2005; Wishart et al., 2013). An online supplement (Supplementary Data S1) is provided with details on chemical identification. Statistical analysis included Pearson correlation (r) analyses between analytic platforms. Significance levels were calculated using a standard t test (t = r/sqrt(1 − r2/n − 2). Raw P values are provided, where P < .0025 is significant for 20 comparisons using Bonferroni correction.
Amino acid analysis
AA analysis was performed using either a Beckman System 6300 High Performance Amino Acid Analyzer (Beckman Instruments, Inc. Palo Alto, California) or a Biochrom 30 Amino Acid Analyzer (Biochrom US, Holliston, Massachusetts) at the Emory Genetics Laboratory.
RESULTS
Consistency of Analysis
For 117 human plasma samples, the median CV for intensity among triplicate technical replicates for 22 625 ions (m/z) across all samples was 9.1%. Results for total ion intensity for the entire sample set showed a CV of 39.6%, with only a relatively small decrease to 33.4% obtained by averaging triplicates prior to calculating the standard deviation. Thus, the major variation in total signal intensity was due to differences in characteristics of individual samples resulting from either total chemical content, matrix effects that impact total signal detection, or both. The smaller median CV for individual metabolites suggests that the variation in total ion intensity was due to differences in total metabolite content among samples rather than analytical variations. Differences in total metabolite content could contribute to a matrix effect that impacts quantification, and normalization procedures for—omics data sometime assume that variations in total signal are due to analytic, rather than biologic, differences. Consequently, to determine whether variation in total ion intensity impacted signal detection for individual chemicals, we analyzed intensities for internal standards with and without normalization to total signal intensity for each respective sample. For 15N13C-Met, the mean CV was 6.0% and the median CV was 13.3%; for 15N-Tyr, the mean CV was 8.9% and the median CV was 13.4%. Normalization of signal intensities for 15N13C-Met to the total ion intensity for each respective sample did not improve CV (mean, 14.0; median 17.9%). Similarly, normalization of signal intensities for 15N-Tyr to the respective total ion intensities did not improve coefficients of variation (mean, 28.0%; median 16.2%). Thus, for subsequent analyses, intensities were used without normalization to total ion intensity.
To test whether normalization relative to a surrogate standard decreased variability for replicate analyses, 15N13C-Met was normalized relative to 15N-Tyr. Because these standards were added together during extraction with acetonitrile, they are present in each sample at an exact ratio relative to each other. The results showed that normalization of 15N13C-Met signal relative to 15N-Tyr increased the CV (median, from 8.0 to 16.2%; mean from 13.3% to 28.0%). That is, directly referencing 1 chemical against another chemical amplified the variation of the target chemical, apparently due to the variability in detection of the reference. Consequently, these data indicate that for quantification in HRM, signal intensities for individual ions without normalization to an internal standard, provides smaller CV.
Comparison of HRM Signals for AAs to Results From Conventional AA Analysis
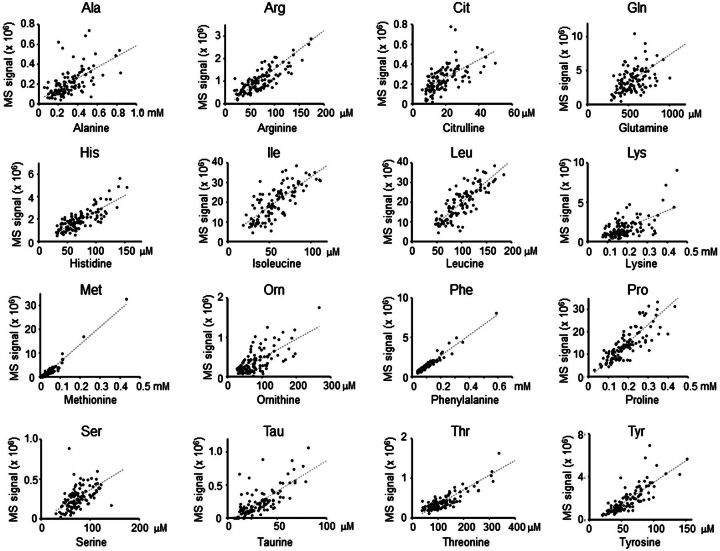
To directly test HRM analyses of individual chemicals, signal intensities for AAs in the 117 samples were compared with measurements obtained by conventional AA analysis performed on matched samples by an independent laboratory. These analyses were performed in random order and operators were blinded to the nature of the samples. Correlations for 17 AA, ranging from relatively poor correlation for cystine (CySS, r = 0.38, P = 2.2 × 10−5; not shown) to high correlations for Met (r = 0.97, P = 1.34 × 10−74) and Phe (r = 0.97, P = 8.28 × 10−73) are shown in Figure 1 and Table 2. To determine whether the correlations for Met were driven by the most extreme high values, analyses were repeated after removal of the top 2 values and top 6 values; respective r and P values were r = 0.90, P = 3.44 × 10−43 and r = 0.90, P = 8.01 × 10−40. For Phe, results for analyses after removal of the top 1 and top 5 values were r = 0.95, P = 1.59 × 10−43 and r = 0.94, P = 4.81 × 10−53.
FIG. 1.
Cross-laboratory comparison of amino acid (AA) quantification. Randomized and blinded analyses of 117 human plasma samples were performed in independent laboratories by HRM and automated AA analysis. Pearson correlation coefficients (r) are provided in Table 2; all correlations were significant at P < .05.
TABLE 2.
Comparison of AA Detection by HRM to Conventional AA Analysis
| AA | AA Analysis Mean (µM) | LC-FTMS C18 Mean Intensity | R | P | m/z | Adduct | RT (s) | Median CV (%) |
|---|---|---|---|---|---|---|---|---|
| Alanine | 321 | 216 532 | 0.70 | 2.97E-18 | 90.0543 | H+ | 61 | 15.3 |
| Arginine | 73 | 1 095 024 | 0.84 | 4.76E-32 | 219.0818 | 2Na+-H+ | 47 | 5.4 |
| 405 164 | 0.79 | 6.34E-26 | 197.1000 | Na+ | 46 | 10.4 | ||
| 1 825 927 | 0.79 | 1.10E-25 | 175.1180 | H+ | 50 | 6.0 | ||
| Aspartic acid | 7 | No highly associated identifiable m/z | ||||||
| Citrulline | 19 | 252 184 | 0.61 | 3.16E-13 | 176.1024 | H+ | 71 | 17.1 |
| Cystine | 46 | 277 006 | 0.38 | 2.23E-05 | 284.9939 | 2Na+-H+ | 52 | 8.6 |
| Glutamate acid | 58 | No highly associated identifiable m/z | ||||||
| Glutamine | 554 | 6 305 193 | 0.68 | 4.54E-17 | 191.0393 | 2Na+-H+ | 49 | 2.5 |
| 295 356 | 0.66 | 1.35E-15 | 192.0427 | 2Na+-H+ (13C) | 51 | 8.7 | ||
| 7 382 949 | 0.59 | 4.67E-12 | 147.0756 | H+ | 60 | 5.4 | ||
| Glycine | 242 | Not within m/z range measured | ||||||
| Histidine | 71 | 1 914 683 | 0.81 | 5.97E-28 | 200.0398 | 2Na+-H+ | 52 | 5.4 |
| 546 666 | 0.76 | 2.72E-23 | 178.0578 | Na+ | 48 | 5.3 | ||
| Isoleucine | 59 | 20 296 291 | 0.79 | 5.17E-26 | 132.1012 | H+ | 57 | 6.6 |
| 1 158 099 | 0.61 | 5.45E-13 | 133.1046 | H+ (13C) | 56 | 8.6 | ||
| Leucine | 104 | 20 296 291 | 0.84 | 1.36E-32 | 132.1012 | H+ | 57 | 6.6 |
| 1 158 099 | 0.68 | 1.02E-16 | 133.1046 | H+ (13C) | 56 | 8.6 | ||
| 301 942 | 0.57 | 3.37E-11 | 176.0649 | 2Na+-H+ | 53 | 10.1 | ||
| Lysine | 187 | 1 505 166 | 0.73 | 1.58E-20 | 191.0759 | 2Na+-H+ | 49 | 7.7 |
| 1 715 298 | 0.61 | 6.81E-13 | 147.1120 | H+ | 59 | 13.0 | ||
| Methionine | 41 | 2 047 964 | 0.97 | 1.34E-74 | 150.0575 | H+ | 55 | 6.2 |
| 247 136 | 0.82 | 1.12E-29 | 194.0213 | 2Na+-H+ | 52 | 16.7 | ||
| Ornithine | 82 | 385 853 | 0.69 | 1.29E-17 | 177.0599 | 2Na+-H+ | 47 | 14.3 |
| 646 111 | 0.62 | 1.88E-13 | 133.0967 | H+ | 63 | 9.3 | ||
| Phenylalanine | 108 | 1 455 188 | 0.97 | 8.28E-73 | 210.0490 | 2Na+-H+ | 53 | 3.4 |
| 17 603 867 | 0.86 | 1.91E-35 | 166.0854 | H+ | 61 | 5.5 | ||
| 158 595 | 0.77 | 2.91E-24 | 211.0524 | H+ (13C) | 54 | 17.6 | ||
| Proline | 192 | 14 676 899 | 0.84 | 8.79E-32 | 116.0700 | H+ | 57 | 4.9 |
| 401 834 | 0.65 | 2.32E-15 | 160.0337 | 2Na+-H+ | 49 | 8.6 | ||
| Serine | 74 | 273 417 | 0.58 | 1.26E-11 | 106.0494 | H+ | 61 | 17.8 |
| 235 221 | 0.58 | 1.29E-11 | 150.0132 | 2Na+-H+ | 42 | 17.3 | ||
| Taurine | 34 | 277 462 | 0.83 | 2.91E-31 | 148.0033 | Na+ | 57 | 10.5 |
| 175 084 | 0.79 | 2.14E-26 | 126.0213 | H+ | 70 | 9.1 | ||
| Threonine | 124 | 427 113 | 0.88 | 9.01E-39 | 164.0286 | 2Na+-H+ | 49 | 6.0 |
| 908 010 | 0.81 | 3.03E-28 | 120.0648 | H+ | 55 | 7.6 | ||
| Tyrosine | 59 | 653 421 | 0.92 | 3.45E-49 | 226.0441 | 2Na+-H+ | 52 | 3.3 |
| 1 817 830 | 0.81 | 4.03E-28 | 182.0802 | H+ | 56 | 11.1 | ||
| Tryptophan | Not detected by AA analysis | |||||||
| Valine | 226 | No highly associated identifiable m/z | ||||||
Data are summarized for 117 GLND serum samples. Pearson correlation (r) with respective P is given for accurate mass ions (m/z) detected at indicated RT. The median CV is given for the triplicate analyses by HRM.
Poor correlation for CySS was found to be due to partial oxidation to sulfoxide and sulfone forms, and efforts to improve accuracy by summation of the respective forms was not useful, apparently due to the additive error of the individual measurements. Leucine (Leu) and Isoleucine (Ile) were not resolved by LC-MS, but AA analyses for each were correlated with the combined LC-MS signal. Based upon “r” values, the reliability of the LC-MS quantification from best to worst relative to conventional AA analysis followed the order: Met=Phe>Tyr>Thr>Pro=Arg=Leu/Ile>Tau>His>Lys>Ala>Orn>Gln>Citrulline>Ser>CySS (Table 2). Importantly, the plots show little evidence for nonlinearity of response between analytical platforms and reasonable extrapolation to the origin (Fig. 1).
Poor correlation (r < 0.7) is likely due to combined analytic variation of the 2 methods. The conventional AA analyses were performed only once so reproducibility of analyses for these individual samples is unknown. The LC-MS analyses, which were performed in triplicate, allowed determination of CV for AA detection in individual samples. With the exception of alanine and serine, all had at least 1 detected m/z ion, ie, H+ adduct, Na+ adduct, with CV < 10% (Table 2). Signals with median CV > 10% had lower r, indicating that lower analytic variability of detection by LC-MS was associated with better correlation. For glutamine, the median CV for LC-MS data was small, but correlation was relatively poor, perhaps reflecting a poor reproducibility of conventional AA analysis of glutamine. Correlations of lysine (Lys) and arginine (Arg) within and between platforms further indicate contributions of both platforms to analytic variation. Lys and Arg are transported into and out of plasma by common transport systems, and as expected, are correlated within the AA analysis (r = 0.84) and within the HRM analysis (r = 0.70). The similar correlations of Lys (r = 0.73) and Arg (r = 0.84) between methods (Table 2) indicate that similar extents of analytical variation exist for both platforms. Based upon the median CV values, the LC-MS reliability from best to worst follows the order: Gln>Phe>Tyr>Pro>His>Arg>Thr>Met>Leu/Ile>Lys>CySS>Tau>Orn>Ala>Citrulline>Ser. Importantly, the results show that external standardization against data obtained from an independent, authenticated method can be used for absolute HRM quantification.
Absolute Quantification Based Upon Stable Isotope Internal Standardization
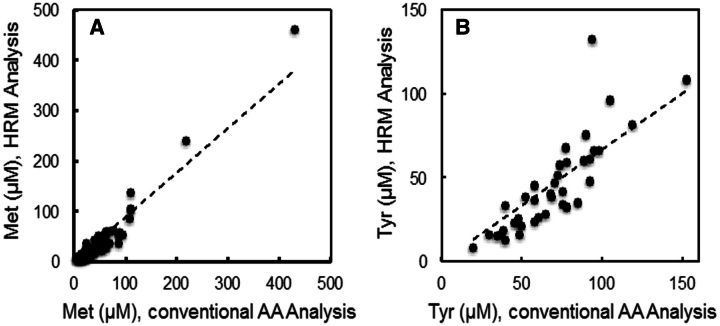
15N,13C-Met and 15N-Tyr were included in each sample, so that these AAs could be directly determined in the 117 samples relative to their respective internal standard. Results showed correlations between the measurements using internal standardization and the independent measurements by conventional AA analysis (Figs. 2A and 2B), although the absolute value was lower for Met. As for Figure 1, the correlations remained significant after removal of the most extreme values. Specifically, the mean value for Met was 29.0 µM (Table 3), compared with 41 µM by conventional AA analysis (Table 2). The respective Tyr values were 62 µM (Table 3) and 59 µM (Table 2). For comparison, values for Met and Tyr in NIST1950 were 22.5 and 57.3 µM, respectively (Table 3) (McGaw et al., 2010).
FIG. 2.
Comparison of absolute concentration measurements for methionine (Met) and tyrosine (Tyr) by HRM and amino acid (AA) analysis. Met (A) and Tyr (B) concentrations were obtained by HRM using stable isotope dilution and directly compared to corresponding measurements by conventional AA analysis.
TABLE 3.
AA Quantification in NIST1950 by HRM Using Surrogate Internal Standards (15N13C-Met and 15N-Tyr)
| AA | Adduct | McGaw et al. (µM) | RRF | vs 15N13C-Met (µM) | RRF | vs 15N-Tyr (µM) | Detected N | Median CV |
|---|---|---|---|---|---|---|---|---|
| Alanine | H+ | 300 | 74 | — | 16 | — | 0 | — |
| Arginine | 2Na+-H+ | 81.4 | 3.3 | — | 0.74 | — | 0 | — |
| Na+ | 9.0 | 90 ± 24 | 2.0 | 62 ± 16 | 6 | 24 | ||
| H+ | 2.0 | 71 ± 12 | 0.44 | 48 ± 8 | 9 | 13 | ||
| Citrulline | H+ | 3.8 | 33 ± 7 | 0.83 | 22 ± 5 | 5 | 23 | |
| Cystine | 2Na+-H+ | 7.78 | 8.3 | 52 ± 8 | 1.8 | 36 ± 5 | 9 | 33 |
| Glutamine | 2Na+-H+ | 4.4 | 503 ± 44 | 0.97 | 344 ± 30 | 12 | 4 | |
| 2Na+-H+ (13C) | 94 | 294 ± 132 | 21 | 201 ± 90 | 4 | 22 | ||
| H+ | 3.8 | 422 ± 36 | 0.83 | 289 ± 24 | 12 | 10 | ||
| Histidine | 2Na+-H+ | 72.6 | 1.8 | 68 ± 11 | 0.41 | 47 ± 8 | 12 | 8 |
| Na+ | 6.5 | 58 ± 9 | 1.4 | 40 ± 6 | 10 | 21 | ||
| Isoleucine | H+ | 55.5 | 0.15 | 65 ± 6 | 0.03 | 45 ± 4 | 8 | 6 |
| H+ (13C) | 2.5 | 100 ± 10 | 0.56 | 69 ± 7 | 12 | 24 | ||
| Leucine | H+ | 100 | 0.26 | 115 ± 11 | 0.06 | 79 ± 7 | 8 | 6 |
| H+ (13C) | 4.5 | 177 ± 17 | 0.99 | 121 ± 12 | 12 | 24 | ||
| 2Na+-H+ | 17 | 252 ± 26 | 3.8 | 172 ± 18 | 11 | 11 | ||
| Lysine | 2Na+-H+ | 140 | 6.2 | 135 ± 18 | 1.38 | 93 ± 12 | 11 | 15 |
| H+ | 5.4 | 139 ± 20 | 1.21 | 95 ± 14 | 12 | 26 | ||
| Methionine | H+ | 22.5 | 1.0 | 29 ± 2 | 0.22 | 20 ± 2 | 12 | 10 |
| 2Na+-H+ | 8.3 | — | 1.84 | — | 0 | — | ||
| Ornithine | 2Na+-H+ | 52.1 | 10.6 | 53 ± 6 | 2.4 | 37 ± 4 | 7 | 19 |
| H+ | 6.3 | 82 ± 23 | 1.4 | 56 ± 16 | 4 | 35 | ||
| Phenylalanine | 2Na+-H+ | 50.8 | 3.7 | 93 ± 4 | 0.82 | 64 ± 3 | 11 | 9 |
| H+ | 0.31 | 101 ± 8 | 0.07 | 69 ± 5 | 12 | 13 | ||
| H+ (13C) | 34 | 62 ± 18 | 7.5 | 43 ± 12 | 4 | 47 | ||
| Proline | H+ | 177 | 0.65 | 221 ± 18 | 0.14 | 151 ± 13 | 12 | 11 |
| 2Na+-H+ | 24 | 190 ± 23 | 5.3 | 130 ± 15 | 12 | 20 | ||
| Serine | H+ | 95.9 | 14 | — | 3.0 | — | 0 | — |
| 2Na + -H+ | 16 | 55 ± 19 | 3.5 | 38 ± 13 | 8 | 49 | ||
| Taurine | Na+ | 6.1 | 37 ± 10 | 1.4 | 25 ± 7 | 12 | 24 | |
| H+ | 9.7 | 24 ± 3 | 2.2 | 17 ± 2 | 9 | 28 | ||
| Threonine | 2Na+-H+ | 119 | 14 | 129 ± 10 | 3.2 | 89 ± 7 | 12 | 15 |
| H+ | 6.8 | 131 ± 13 | 1.5 | 90 ± 9 | 12 | 11 | ||
| Tyrosine | 2Na+-H+ | 57.3 | 4.5 | 91 ± 8 | 1.0 | 62 ± 6 | 12 | 6 |
| H+ | 1.6 | — | 0.36 | — | 0 | — |
Intensity data for individual adducts were used with RRFs calculated from data in Table 2. Summary data for quantification of AAs by 5 different platforms from McGaw et al (McGaw et al., 2010) are provided for comparison. Four aliquots of NIST were analyzed in triplicate; the number of times that adduct was detected is expressed as “Detected N.” The median CV is the variation for triplicate analyses.
Absolute Quantification Based Upon Stable Isotope Surrogate Internal Standardization
Surrogate standardization was based upon the methods of Greizerstein et al. (1997). RRFs for ionization and detection of specific AA ions in Table 2 were calculated relative to Met [+H]+ and Tyr [+2Na+-H+]+ (Table 3). Using these values, concentrations of AA in NIST1950 were determined on 4 different weeks with 15N13C-Met and 15N-Tyr as surrogate standards. Results show quantification for all detected AA except cystine and serine, which had poor CV (Table 3). The m/z for Gly [+H]+ is less the operational range for m/z (85–850) used for the measurements, and the Na+ and 2Na+-H+ adducts were not detected. H+ adduct was not detected in the NIST samples for either Ala or Ser. Thus, for the detected AA, these results support previous findings (Greizerstein et al., 1997) indicating that surrogate standardization can be used for absolute quantification.
Use of NIST SRM1950 (NIST1950) as an External Reference for Absolute Quantification
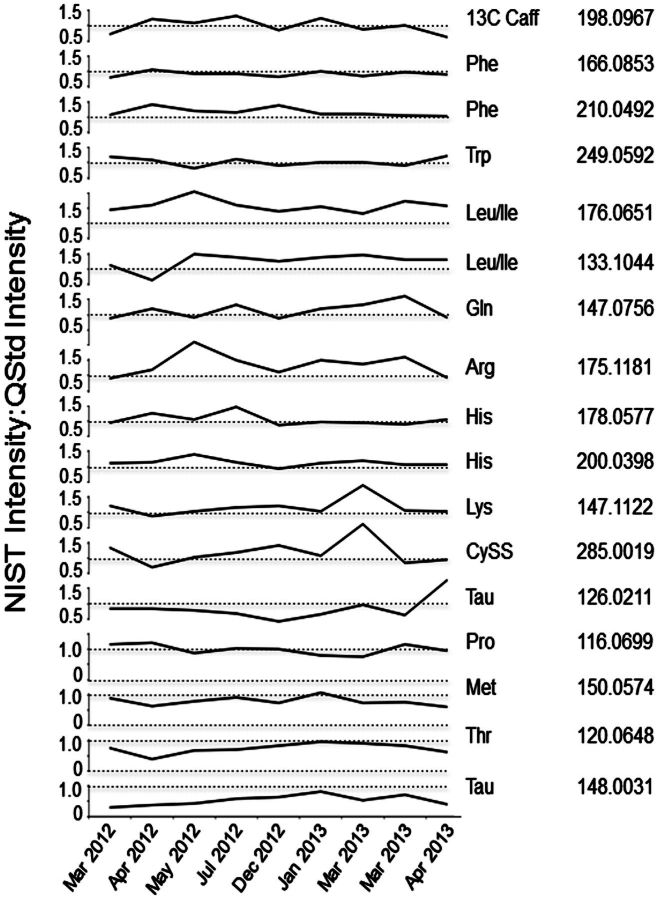
The data in Table 3 provide independent confirmation of the AA measurements of McGaw (McGaw et al., 2010), and additionally suggest that NIST1950 can be used as a reference standard for quantification of AA. The stability of NIST1950 for quantification was verified by comparing the signals of AAs in NIST1950 and 13C-caffeine added on the day of analysis to signals of AAs in pooled reference plasma (QStd) prepared independently and analyzed together over a period of 13 months (Fig. 3). Results showed that signals for AAs in NIST1950 and Qstd over this time period were stable relative to each other and also relative to the added 13C-caffeine internal standard. These results establish that pooled reference plasma can be calibrated relative to NIST1950 (Table 4), and that such preparations are stable at −80°C for at least 13 months. To test use of Qstd for absolute quantification, AA concentrations in the 117 human serum samples were calculated based upon corresponding AA signals in Qstd, analyzed concurrently. The results were comparable to the direct measurements by AA analysis (Table 4). Consequently, the results demonstrate that “Reference Standardization” can be used as an alternative to external standardization, stable isotope dilution, or surrogate standardization for quantification of HRM data.
FIG. 3.
Comparison of signal intensities for amino acids (AAs) in NIST SRM1950 and Qstd with repeated analyses over 13 months. To evaluate reproducibility of reference standardization over time, AA signals were compared for analyses performed during different time periods. The results confirm that long-term referencing is robust. The results also showed that variability for different AAs occurred with triplicate analyses, providing a basis to change standard operating procedures to perform 6 technical replicates of the reference standards after each batch of 20 samples.
TABLE 4.
Use of NIST-Calibrated QStd for Calculation of AA Concentrations in 117 Human Samples
| AA | Adduct | Reference Concentration QStd | Reference Standardization 117 samples | Conventional AA Analysis 117 Samples |
|---|---|---|---|---|
| Alanine | H+ | 295 | 322 ± 228 | 321 ± 196 |
| Arginine | H+ | 60 ± 8 | 59 ± 41 | 73 ± 37 |
| Aspartic Acid | — | — | — | 7 ± 5 |
| Citrulline | H+ | 33 | 19 ± 9 | 19 ± 9 |
| Cystine | H+ | 15 | 52 ± 63 | 46 ± 27 |
| Glutamic Acid | — | — | — | 58 ± 34 |
| Glutamine | 2Na+-H+ | 367 | 559 ± 225 | 554 ± 163 |
| Glycine | — | — | — | 242 ± 90 |
| Histidine | Na+ | 61 ± 9 | 85 ± 39 | 71 ± 28 |
| Leucine/Isoleucine | H + | 127 ± 17 | 148 ± 82 | 175 ± 60 |
| Lysine | H+ | 133 ± 38 | 123 ± 94 | 187 ± 75 |
| Methionine | H+ | 30 ± 5 | 45 ± 78 | 41 ± 47 |
| Ornithine | 2Na+-H+ | 28 ± 4 | 36 ± 28 | 82 ± 42 |
| Phenylalanine | H+ | 51 ± 15 | 98 ± 58 | 108 ± 72 |
| Proline | H+ | 191 ± 28 | 231 ± 134 | 192 ± 88 |
| Serine | H+ | 108 | 75 ± 42 | 74 ± 24 |
| Taurine | H+ | 37 | 24 ± 18 | 34 ± 25 |
| Threonine | 2Na+-H+ | 112 ± 37 | 141 ± 47 | 124 ± 67 |
| Tyrosine | 2Na+-H+ | 52 ± 6 | 49 ± 20 | 59 ± 24 |
| Valine | — | — | — | 226 ± 73 |
General Applicability of Reference Standardization
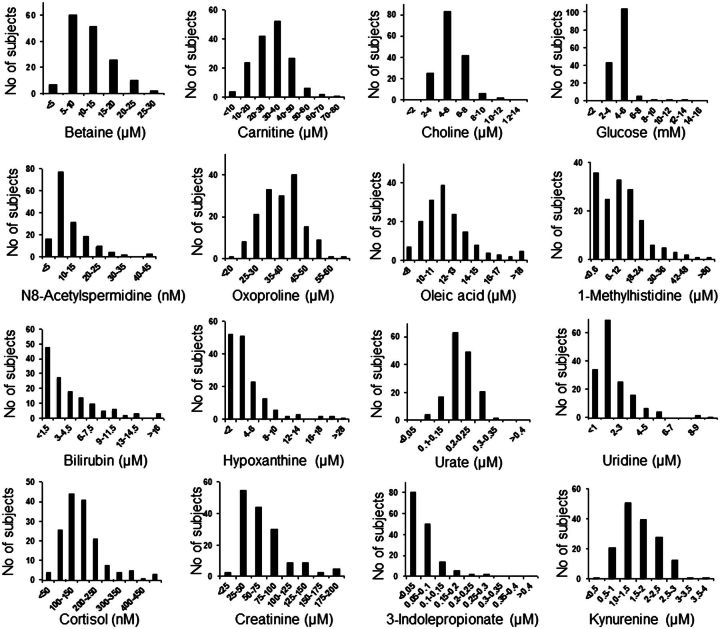
To test the general applicability of the method, we selected metabolites with confirmed identifications for which concentrations in NIST1950 are available (http://srm1950.nist.gov/srm_search.php?cert=on) and/or for which we had performed independent external method-of-addition calibration of values in the Qstd. Analysis of metabolite concentrations in plasma from 157 healthy adults showed distributions comparable to data in Human Metabolomics Database (HMDB), Version 3.0 (Fig. 4 and Table 5). Thus, the evaluation shows that reference standardization can be used to convert HRM data to absolute concentrations for metabolites that have been calibrated in NIST1950 or in a comparable pooled reference material.
FIG. 4.
Histograms of selected metabolite concentrations in 157 healthy adults. Samples were analyzed in triplicate by HRM and quantification was performed using reference standardization.
TABLE 5.
Comparison of Metabolite Concentrations in 157 Healthy Adults Obtained Using Reference Standardization to Metabolite Data Summarized in HMDB
| Reference Standardization |
HMDB |
||||
|---|---|---|---|---|---|
| Metabolite | Median | Mean | SD | Mean | SD |
| N8-Acetylspermidine (nM) | 9.1 | 11.0 | 6.7 | 50 | 14 |
| Betaine (μM) | 11 | 11.7 | 5.1 | 33.6 | — |
| Bilirubin (μM) | 2.5 | 4.1 | 4.4 | 8.0 | 0.9 |
| Carnitine (μM) | 32 | 31.6 | 12.1 | 30 | 7.6 |
| Choline (μM) | 2.5 | 5.4 | 1.6 | 6.0 | 0.3 |
| Cortisol (nM) | 151 | 169 | 89 | 320 | 190 |
| Cotinine (nM) | 3.9 | 5.5 | 4.2 | 1400 | 900 |
| Creatinine (μM) | 59 | 71 | 36 | 70 | 10 |
| Glucose (mM) | 4.4 | 4.6 | 1.3 | 5.3 | 1.2 |
| Hypoxanthine (μM) | 2.9 | 4.4 | 5.4 | 4.87 | 0.36 |
| 3-Indolepropionate (μM) | 0.48 | 0.067 | 0.070 | 0.48 | — |
| Kynurenine (μM) | 1.55 | 1.63 | 0.6 | 1.6 | 0.1 |
| Oleic acid (μM) | 47.4 | 49.7 | 12.9 | 49 | 19 |
| Oxoproline (μM) | 38 | 38.4 | 13.7 | 19.5 | 3.7 |
| 1-Methylhistidine (μM) | 12.2 | 14.9 | 12.2 | 12.7 | 2.9 |
| Urate (μM) | 190 | 200 | 51 | 272 | 43 |
| Uridine (μM) | 1.64 | 2.00 | 1.37 | 3.12 | 1.31 |
The subject characteristics were not well matched between the present study and those for the HMDB data, likely accounting for many of the differences observed. MS/MS data and notes concerning identification of metabolites listed in this table are provided in Supplementary Data S1.
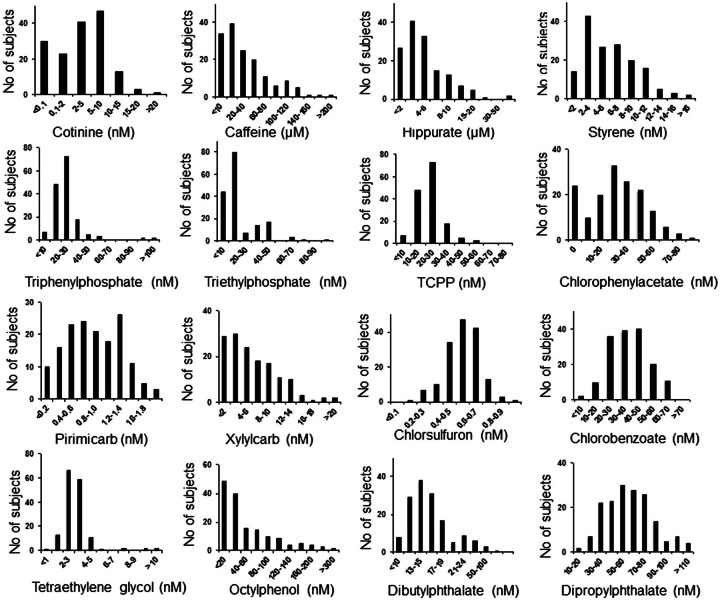
Application of Reference Standardization to Measurement of Environmental Chemicals
In contrast to most of the endogenous metabolites measured above, environmental chemicals were detected in the nanomolar and subnanomolar concentration ranges (Fig. 5 and Table 6). Of those quantified, octylphenol (57 ± 61 nM) and dipropylphthalate (60 ± 24 nM) were present at highest concentrations. Dibutylphalate was present in all samples, but calibration of standards was inconsistent, and 6 samples had levels calculated to be in the micromolar range and were excluded because of possible contamination. Styrene, flame retardants (triethylphosphate, triphenylphosphate), and some insecticides and related metabolites were also detected in the nanomolar range (Fig. 5 and Table 6). Pirimicarb was present in most samples at subnanomolar concentration. The 157 healthy individuals were all nonsmokers and extensively screened to assure absence of disease; accordingly, measured values of cotinine (4.6 ± 4.2 nM; median 4.0 nM; Fig. 5) were lower than pooled reference NIST (10 nM) and Qstd (15 nM), but similar to HMDB values for nonsmokers. Caffeine, which is directly derived from diet, and hippurate, which is formed by glycine conjugation of dietary benzoic acid, were present with micromolar concentrations consistent with HMDB data.
FIG. 5.
Histograms of selected environmental and dietary chemicals in 157 healthy adults. Samples were analyzed in triplicate by HRM and quantification was performed using reference standardization.
TABLE 6.
Environmental Chemicals Concentrations in Plasma of 157 Healthy Adults Obtained Using Reference Standardization
| Reference Standardization |
Literature Values |
||||
|---|---|---|---|---|---|
| Metabolite | Median | Mean | SD | Mean or Range | References |
| Caffeine (μM) | 22 | 36 | 46 | 26–129 | HMDB |
| Chlorobenzoic acid (nM) | 38 | 38 | 13 | — | — |
| Chlorophenylacetic acid (nM) | 28 | 34 | 18 | — | — |
| Chlorsulfuron (nM) | 0.56 | 0.55 | 0.13 | — | — |
| Cotinine (nM) | 3.9 | 5.5 | 4.2 | 6.7–13.5 | HMDB |
| Dibutylphthalate (nM) | 20.5 | 25.4 | 16.4 | 15–989 | ToxNet |
| Dipropylphthalate (nM) | 57 | 60 | 24 | — | — |
| Hippuric acid (μM) | 4.7 | 7.4 | 11.7 | 17 ± 11 | HMDB |
| Octylphenol (nM) | 34 | 57 | 61 | 0.14–2.2a | (Qin et al., 2013) |
| Pirimicarb (nM) | 0.82 | 0.79 | 0.36 | — | — |
| Styrene (nM) | 5.3 | 6.0 | 3.6 | 0.4–5.3 | HMDB |
| Tetraethylene glycol (nM) | 2.9 | 3.2 | 1.6 | — | — |
| Triethylphosphate (nM) | 6.9 | 10.0 | 8.1 | — | — |
| Triphenylphosphate (nM) | 22 | 25 | 12 | 0.6 nM | (Shah et al., 2006) |
| Tris(2-chloropropyl)phosphate (nM) | 45 | 51 | 23 | — | — |
| Xylylcarb (nM) | 4.6 | 6.4 | 8.2 | — | — |
Previous literature values are provided for serum or plasma where available. For dibutylphalate, 6 samples were in the micromolar range and were excluded because of possible contamination. Additionally, standardization was inconsistent for dibutylphalate. ToxNet, http://toxnet.nlm.nih.gov (Shah et al., 2006). MS/MS data and notes concerning identification of metabolites listed in this table are provided in Supplementary Data S1.
a Value for urinary 4-n-octylphenol, expressed as nmole/l urine
Application of Reference Standardization to Unidentified Metabolites
HRM detects large numbers of ions that have not been characterized. To determine the suitability of reference standardization to support quantification of currently unidentified metabolites in plasma, including those derived from microbiome, diet, or environmental exposures, we compared high-resolution m/z features detected in NIST1950 with those detected in QStd. Results showed that >8000 ions were detected in at least 50% of replicates for both NIST and QStd across 13 months of analyses. A similar examination of data for the 117 plasma samples showed that >14 000 of the 22 629 metabolites detected were also detected at least 50% of the time in QStd; respective values for the 157 healthy subjects were >10 000 out of 16 665 metabolites. These data suggest that as chemicals are identified and quantified in NIST SRM1590, reference standardization will support quantification of thousands of chemicals using HRM methods.
DISCUSSION
Exposome research to study links between disease and exposure history across a lifespan requires new analytic methods to address both the range of exposures and the time frame between exposure and outcome. Human biomonitoring with HRM provides means to capture chemical exposure information in terms of measured body burden and simultaneously measure metabolic responses to current or prior exposures (Park et al., 2012; Soltow et al., 2013). This study shows that multiple standardization methods can be applied to HRM data to create cumulative exposome research databases (Table 1). The most salient finding is that reference standardization, a simple modification of analytic workflows commonly used for metabolomics, provides a framework for creation of cumulative exposome reference databases. The results demonstrate that by calibrating the concentrations of chemicals in a pooled reference sample, data can be converted to absolute concentrations for direct comparisons to data collected with different platforms and/or at different times. In practical use, the method is critically dependent upon detection of low abundance chemicals in the reference standard. We have adopted standard operating procedures to analyze the reference standard 12 times daily, 6 preceding and 6 following batches of 20 samples, to facilitate quantification of metabolites that are inconsistently detected upon repeat analysis. This strategy overcomes limitations of external calibration and internal standardization to measure thousands of chemicals in a routine and affordable manner. At the same time, environmental chemicals that are present in only a small fraction of people are diluted in pooled reference samples. Thus, for optimization of the approach, pooled reference samples with known, relevant concentrations of environmental chemicals will be needed.
External calibration with a method of additions is a labor-intensive approach that is useful for determination of environmental chemical concentrations in samples but requires a suitable quality standard and care in determination. In the current evaluation, we used additions of relevant small amounts of standards into a constant small volume of diluent added to the Qstd. At least 3 different amounts were used to increase target signal from 1.3- to 20-fold. This approach allows for extrapolation back to signal obtained with only the diluent to obtain an estimate of the concentration in Qstd. For exposome research, this methodology is useful for calibrating the Reference Standard.
Internal standardization is the “gold standard” for targeted analyses by LC-MS and provides quantitatively accurate data. However, this approach is impractical for use to measure thousands of chemicals in a high-throughput format because of the cost of large numbers of stable isotopic standards (Table 1). Nonetheless, inclusion of a small number of stable isotopic standards in every sample provides means for quality control and for surrogate standardization of chemicals that are unstable in the reference standard. Additionally, internal standardization provides a reliable means to calibrate the Reference Standard.
Surrogate standardization provides a useful alternative to reference standardization for absolute quantification. Surrogate standardization requires concentrations of known metabolites to obtain RRFs, but this can be done retrospectively in the same manner as suggested above for reference standardization. Both reference standardization and surrogate standardization are expected to lose reliability for absolute quantification for values that deviate substantially from the mean. Additionally, surrogate standardization is limited when the RRF is calculated from a regression curve with a nonzero intercept (data not shown).
Whether reference standardization is better than surrogate standardization is not clearly evident for the metabolites analyzed.
The present analysis of quantification strategies shows that reference standardization, with only a modest change in analytic workflow for nontargeted metabolomics, can enable use of HRM for cumulative exposome research. Nontargeted metabolomic work flows typically include a pooled reference sample to provide a means to ensure analytical quality control and batch correction (Bijlsma et al., 2006; Dunn et al., 2011; Masson et al., 2010; O'Kane et al., 2013; Want et al., 2010). The resulting data demonstrate that by calibrating this pooled reference sample to a calibration reference, such as NIST SRM1950, one creates an analytic structure that is simple and quantitatively reliable. Although validated for only a small number of chemicals, these data suggest suitability for use with thousands of chemicals. In principle, reference standardization also provides a means for batch correction by calibrating each chemical within each sample to the same chemical analyzed concurrently in the reference material. At present, the number of quantifiable analytes based upon NIST is approximately 100 chemicals, ie, too small to make this highly useful for exposome research. However, quantification using traditional methods of addition with authentic standards (Bueschl et al., 2013; Hewavitharana, 2011; Niessen et al., 2006) can readily expand this capability. Additionally, availability of certified reference materials with relevant low environmental concentrations of chemicals of interest would facilitate interlaboratory comparisons of exposome research for various populations, analytical platforms and phenotypes.
These analytic procedures do not address sample quality, which is largely determined by patient characteristics, sample collection procedures, and sample storage prior to delivery to an analytical facility. However, the information-rich nature of HRM data allows retrospective evaluation of sample quality in databases. For instance, signals for EDTA, citrate, and lithium adducts directly show whether a plasma sample was collected as described in the metadata. Loss of cystine and presence of oxidation products (cystine sulfoxide and cystine sulfone) may indicate adverse or prolonged storage of samples.
The results presented here establish feasibility for use of reference standardization to build cumulative databases. This approach needs to be validated by application to multiple datasets and datasets from multiple analytic facilities. At least 3 conditions are needed to assure reliability: authenticated chemical identification and/or accurate m/z and calibrated LC-RT; quantitative accuracy of reference material calibration; and similarity of instrument response characteristics over a similar dynamic range. In this regard, multicenter cooperation and blinded cross-validation will be needed to assure integrity of data.
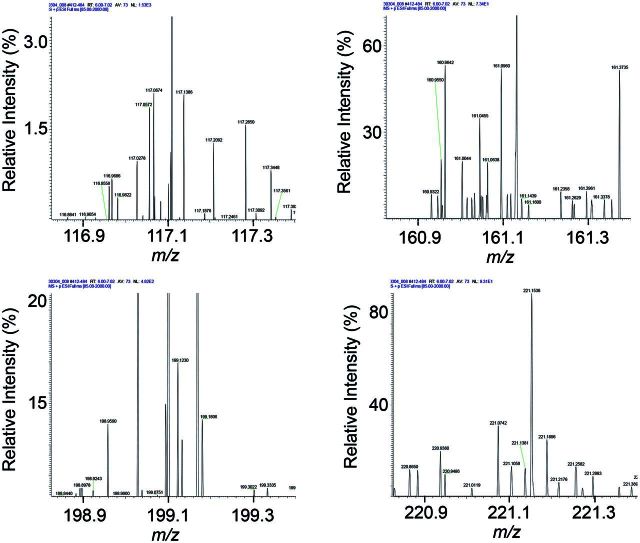
This study was performed using ultra-high resolution MS and comparable studies will be needed to test suitability of the approach for other mass spectrometers and other analytic approaches. Analyses were performed using an analytic structure optimized for 10-min analyses of plasma (Johnson et al., 2010; Soltow et al., 2013); the LTQ-Velos-Orbitrap was operated at 60 000 resolution, which is not the maximal resolution of this or other Fourier transform instruments (Marshall and Hendrickson, 2008). Commonly used Q-TOF MS operate at 35 000 resolution, and similar quantitative reliability using a reference standardization protocol can be expected, at least for relatively high abundance chemicals. For lower abundance chemicals, which include many environmental chemicals, quantification may be more problematic due to the presence of multiple low-intensity ions within the ion selection window for ion dissociation (Fig. 6).
FIG. 6.
Representative ultra-high resolution MS1 scans of human plasma show that many low abundance ions are present within a 0.5 atomic mass unit (AMU) window. Four examples of mass spectra are given and are representative of nominal mass <1000.
Cross-laboratory comparisons of AA measurements in NIST1950 show that differences occur among values obtained from competent analytic laboratories (McGaw et al., 2010). This implies that regardless of interlaboratory comparisons, computational methods will be required to normalize metabolomics data entered into cumulative databases. Such integration was extensively addressed for gene expression microarray data, and this experience suggests that despite analytic hurdles, computational methods can be developed to manage such differences in metabolomics research. Consequently, one can anticipate that reference standardization, perhaps with a concurrent surrogate standardization structure, will facilitate development of cumulative exposome databases. The resulting databases will serve to foster exposome-related metabolomics research for detailed examination of cumulative environmental as well as dietary, microbiome, and therapeutic influences on health and disease.
FUNDING
This study was supported in part by grants from the National Institutes of Health (R01 ES019776, R01 AG038746, R01 HL113451, R21 ES025632, R01 ES023485 and S10 OD018006, T32 GM095442, K24 DK096574, and UL1 TR000454).
SUPPLEMENTARY DATA
Supplementary data are available online at http://toxsci.oxfordjournals.org/.
REFERENCES
- Bijlsma S., Bobeldijk I., Verheij E. R., Ramaker R., Kochhar S., Macdonald I. A., van Ommen B., Smilde A. K. (2006). Large-scale human metabolomics studies: A strategy for data (pre-) processing and validation. Anal. Chem. 78, 567–574. [DOI] [PubMed] [Google Scholar]
- Bueschl C., Krska R., Kluger B., Schuhmacher R. (2013). Isotopic labeling-assisted metabolomics using LC-MS. Anal. Bioanal. Chem. 405, 27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelius D., Bondarenko P. V. (2002). Quantitative profiling of proteins in complex mixtures using liquid chromatography and mass spectrometry. J. Proteome Res. 1, 317–323. [DOI] [PubMed] [Google Scholar]
- Chen Q., Park H. C., Goligorsky M. S., Chander P., Fischer S. M., Gross S. S. (2012). Untargeted plasma metabolite profiling reveals the broad systemic consequences of xanthine oxidoreductase inactivation in mice. PLoS One 7, e37149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cribbs S. K., Park Y., Guidot D. M., Martin G. S., Brown L. A., Lennox J., Jones D. P. (2014). Metabolomics of bronchoalveolar lavage differentiate healthy HIV-1-infected subjects from controls. AIDS Res. Hum. Retroviruses 30, 579–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn W. B., Broadhurst D., Begley P., Zelena E., Francis-McIntyre S., Anderson N., Brown M., Knowles J. D., Halsall A., Haselden J. N., et al. (2011). Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat. Protoc. 6, 1060–1083. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick A. M., Park Y., Brown L. A., Jones D. P. (2014). Children with severe asthma have unique oxidative stress-associated metabolomic profiles. J. Allergy Clin. Immunol. 133, 258–261 e251-e258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frediani J. K., Jones D. P., Tukvadze N., Uppal K., Sanikidze E., Kipiani M., Tran V. T., Hebbar G., Walker D. I., Kempker R. R., et al. (2014). Plasma metabolomics in human pulmonary tuberculosis disease: A pilot study. PLoS One 9, e108854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Go Y. M., Roede J. R., Orr M., Liang Y., Jones D. P. (2014a). Integrated redox proteomics and metabolomics of mitochondria to identify mechanisms of cd toxicity. Toxicol. Sci. 139, 59–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Go Y. M., Uppal K., Walker D.I., Tran V., Dury L., Strobel F.H., Baubichon-Cortay H., Pennell K.D., Roede J.R., Jones D.P. (2014b). Mitochondrial Metabolomics Using High-Resolution Fourier-Transform Mass Spectrometry. Methods. Mol. Biol. 1198, 43–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Go Y. M., Walker D. I., Soltow Q. A., Uppal K., Wachtman L. M., Strobel F. H., Pennell K., Promislow D. E., Jones D. P. (2015). Metabolome-wide association study of phenylalanine in plasma of common marmosets. Amino Acids 47, 589–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory J. F., III, Park Y., Lamers Y., Bandyopadhyay N., Chi Y. Y., Lee K., Kim S., da Silva V., Hove N., Ranka S., et al. (2013). Metabolomic analysis reveals extended metabolic consequences of marginal vitamin B-6 deficiency in healthy human subjects. PLoS One 8, e63544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greizerstein H. B., Gigliotti P., Vena J., Freudenheim J., Kostyniak P. J. (1997). Standardization of a method for the routine analysis of polychlorinated biphenyl congeners and selected pesticides in human serum and milk. J. Anal. Toxicol. 21, 558–566. [DOI] [PubMed] [Google Scholar]
- Hewavitharana A. K. (2011). Matrix matching in liquid chromatography-mass spectrometry with stable isotope labelled internal standards—is it necessary? J. Chromatogr. A 1218, 359–361. [DOI] [PubMed] [Google Scholar]
- Hoffman J. M., Soltow Q. A., Li S., Sidik A., Jones D. P., Promislow D. E. (2014). Effects of age, sex, and genotype on high-sensitivity metabolomic profiles in the fruit fly, Drosophila melanogaster. Aging Cell 13, 596–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanisevic J., Zhu Z. J., Plate L., Tautenhahn R., Chen S., O'Brien P. J., Johnson C. H., Marletta M. A., Patti G. J., Siuzdak G. (2013). Toward ‘omic scale metabolite profiling: A dual separation-mass spectrometry approach for coverage of lipid and central carbon metabolism. Anal. Chem. 85, 6876–6884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J. M., Strobel F. H., Reed M., Pohl J., Jones D. P. (2008). A rapid LC-FTMS method for the analysis of cysteine, cystine and cysteine/cystine steady-state redox potential in human plasma. Clin. Chim. Acta. 396, 43–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J. M., Yu T., Strobel F. H., Jones D. P. (2010). A practical approach to detect unique metabolic patterns for personalized medicine. Analyst 135, 2864–2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D. P., Park Y., Ziegler T. R. (2012). Nutritional metabolomics: Progress in addressing complexity in diet and health. Annu. Rev. Nutr. 32, 183–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Park Y., Duraisingham S., Strobel F. H., Khan N., Soltow Q. A., Jones D. P., Pulendran B. (2013). Predicting network activity from high throughput metabolomics. PLoS Comput. Biol. 9, e1003123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall A. G., Hendrickson C. L. (2008). High-resolution mass spectrometers. Annu. Rev. Analyt. Chem. 1, 579–599. [DOI] [PubMed] [Google Scholar]
- Masson P., Alves A. C., Ebbels T. M., Nicholson J. K., Want E. J. (2010). Optimization and evaluation of metabolite extraction protocols for untargeted metabolic profiling of liver samples by UPLC-MS. Anal. Chem. 82, 7779–7786. [DOI] [PubMed] [Google Scholar]
- McGaw E. A., Phinney K. W., Lowenthal M. S. (2010). Comparison of orthogonal liquid and gas chromatography-mass spectrometry platforms for the determination of amino acid concentrations in human plasma. J. Chromatogr. A 1217, 5822–5831. [DOI] [PubMed] [Google Scholar]
- Miller G. W., Jones D. P. (2013). The nature of nurture: refining the definition of the exposome. Toxicol. Sci. 137, 1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller C., Schafer P., Stortzel M., Vogt S., Weinmann W. (2002). Ion suppression effects in liquid chromatography-electrospray-ionisation transport-region collision induced dissociation mass spectrometry with different serum extraction methods for systematic toxicological analysis with mass spectra libraries. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 773, 47–52. [DOI] [PubMed] [Google Scholar]
- Neujahr D. C., Uppal K., Force S. D., Fernandez F., Lawrence C., Pickens A., Bag R., Lockard C., Kirk A. D., Tran V., et al. (2014). Bile acid aspiration associated with lung chemical profile linked to other biomarkers of injury after lung transplantation. Am. J. Transplant. 14, 841–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niessen W. M., Manini P., Andreoli R. (2006). Matrix effects in quantitative pesticide analysis using liquid chromatography-mass spectrometry. Mass Spectrom Rev. 25, 881–899. [DOI] [PubMed] [Google Scholar]
- O'Kane A. A., Chevallier O. P., Graham S. F., Elliott C. T., Mooney M. H. (2013). Metabolomic profiling of in vivo plasma responses to dioxin-associated dietary contaminant exposure in rats: Implications for identification of sources of animal and human exposure. Environ. Sci. Technol. 47, 5409–5418. [DOI] [PubMed] [Google Scholar]
- Osborn M. P., Park Y., Parks M. B., Burgess L. G., Uppal K., Lee K., Jones D. P., Brantley M. A., Jr (2013). Metabolome-wide association study of neovascular age-related macular degeneration. PLoS One 8, e72737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y. H., Lee K., Soltow Q. A., Strobel F. H., Brigham K. L., Parker R. E., Wilson M. E., Sutliff R. L., Mansfield K. G., Wachtman L. M., et al. (2012). High-performance metabolic profiling of plasma from seven mammalian species for simultaneous environmental chemical surveillance and bioeffect monitoring. Toxicology 295, 47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y., Chen M., Wu W., Xu B., Tang R., Chen X., Du G., Lu C., Meeker J. D., Zhou Z., et al. (2013). Interactions between urinary 4-tert-octylphenol levels and metabolism enzyme gene variants on idiopathic male infertility. PloS One 8, e59398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravindran R., Khan N., Nakaya H. I., Li S., Loebbermann J., Maddur M. S., Park Y., Jones D. P., Chappert P., Davoust J., et al. (2014). Vaccine activation of the nutrient sensor GCN2 in dendritic cells enhances antigen presentation. Science 343, 313–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roback J. D., Josephson C. D., Waller E. K., Newman J. L., Karatela S., Uppal K., Jones D. P., Zimring J. C., Dumont L. J. (2014). Metabolomics of ADSOL (AS-1) red blood cell storage. Transfus Med. Rev. 28, 41–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roede J. R., Uppal K., Park Y., Lee K., Tran V., Walker D., Strobel F. H., Rhodes S. L., Ritz B., Jones D. P. (2013). Serum metabolomics of slow vs. rapid motor progression Parkinson's disease: A pilot study. PLoS One 8, e77629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roede J. R., Uppal K., Park Y., Tran V., Jones D. P. (2014). Transcriptome–metabolome wide association study (TMWAS) of maneb and paraquat neurotoxicity reveals network level interactions in toxicologic mechanism. Toxicol. Rep. 1, 435–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah M., Meija J., Cabovska B., Caruso J. A. (2006). Determination of phosphoric acid triesters in human plasma using solid-phase microextraction and gas chromatography coupled to inductively coupled plasma mass spectrometry. J. Chromatogr.A. 1103, 329–336. [DOI] [PubMed] [Google Scholar]
- Smith C. A., O'Maille G., Want E. J., Qin C., Trauger S. A., Brandon T. R., Custodio D. E., Abagyan R., Siuzdak G. (2005). METLIN: A metabolite mass spectral database. Ther. Drug Monit. 27, 747–751. [DOI] [PubMed] [Google Scholar]
- Soltow Q. A., Strobel F. H., Mansfield K. G., Wachtman L., Park Y., Jones D. P. (2013). High-performance metabolic profiling with dual chromatography-Fourier-transform mass spectrometry (DC-FTMS) for study of the exposome. Metabolomics 9, 132–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamler J., Brown I. J., Yap I. K., Chan Q., Wijeyesekera A., Garcia-Perez I., Chadeau-Hyam M., Ebbels T. M., De Iorio M., Posma J., et al. (2013). Dietary and urinary metabonomic factors possibly accounting for higher blood pressure of black compared with white Americans: Results of International Collaborative Study on macro-/micronutrients and blood pressure. Hypertension 62, 1074–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uppal K., Soltow Q. A., Strobel F. H., Pittard W. S., Gernert K. M., Yu T., Jones D. P. (2013). xMSanalyzer: Automated pipeline for improved feature detection and downstream analysis of large-scale, non-targeted metabolomics data. BMC bioinformatics 14, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Greef J., Martin S., Juhasz P., Adourian A., Plasterer T., Verheij E. R., McBurney R. N. (2007). The art and practice of systems biology in medicine: Mapping patterns of relationships. J. Proteome Res. 6, 1540–1559. [DOI] [PubMed] [Google Scholar]
- Wang W., Zhou H., Lin H., Roy S., Shaler T. A., Hill L. R., Norton S., Kumar P., Anderle M., Becker C. H. (2003). Quantification of proteins and metabolites by mass spectrometry without isotopic labeling or spiked standards. Anal. Chem. 75, 4818–4826. [DOI] [PubMed] [Google Scholar]
- Want E. J., Wilson I. D., Gika H., Theodoridis G., Plumb R. S., Shockcor J., Holmes E., Nicholson J. K. (2010). Global metabolic profiling procedures for urine using UPLC-MS. Nat. Protoc. 5, 1005–1018. [DOI] [PubMed] [Google Scholar]
- Wishart D. S., Jewison T., Guo A. C., Wilson M., Knox C., Liu Y., Djoumbou Y., Mandal R., Aziat F., Dong E., et al. (2013). HMDB 3.0—The Human Metabolome Database in 2013. Nucleic Acids Res. 41, D801–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Araki K., Li S., Han J. H., Ye L., Tan W. G., Konieczny B. T., Bruinsma M. W., Martinez J., Pearce E. L., et al. (2014). Autophagy is essential for effector CD8(+) T cell survival and memory formation. Nat. Immunol. 15, 1152–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu T., Park Y., Johnson J. M., Jones D. P. (2009). apLCMS—adaptive processing of high-resolution LC/MS data. Bioinformatics 25, 1930–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Zhu Y., Uppal K., Tran V. T., Yu T., Lin J., Matsuguchi T., Blackburn E., Jones D., Lee E. T., et al. (2014). Metabolic profiles of biological aging in American Indians: The Strong Heart Family Study. Aging. 6, 176–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.