Abstract
Biofilms are etiologically important in the development of chronic medical and dental infections. The biofilm extracellular polymeric substance (EPS) determines biofilm structure and allows bacteria in biofilms to adapt to changes in mechanical loads such as fluid shear. However, EPS components are difficult to visualize microscopically because of their low density and molecular complexity. Here, we tested potassium permanganate, KMnO4, for use as a non-specific EPS contrast-enhancing stain using confocal laser scanning microscopy in reflectance mode. We demonstrate that KMnO4 reacted with EPS components of various strains of Pseudomonas, Staphylococcus and Streptococcus, yielding brown MnO2 precipitate deposition on the EPS, which was quantifiable using data from the laser reflection detector. Furthermore, the MnO2 signal could be quantified in combination with fluorescent nucleic acid staining. COMSTAT image analysis indicated that KMnO4 staining increased the estimated biovolume over that determined by nucleic acid staining alone for all strains tested, and revealed non-eDNA EPS networks in Pseudomonas aeruginosa biofilm. In vitro and in vivo testing indicated that KMnO4 reacted with poly-N-acetylglucosamine and Pseudomonas Pel polysaccharide, but did not react strongly with DNA or alginate. KMnO4 staining may have application as a research tool and for diagnostic potential for biofilms in clinical samples.
Keywords: biofilm, EPS, confocal microscopy, potassium permanganate staining
The authors describe the use of potassium permanganate as a novel biofilm EPS stain.
Graphical Abstract Figure.
The authors describe the use of potassium permanganate as a novel biofilm EPS stain.
INTRODUCTION
Bacterial biofilms are communities of bacteria that adhere to surfaces and are held together within an extracellular polymeric substance (EPS) matrix. Biofilm formation by pathogens is a recognized virulence factor associated with chronic medical and dental infections (Parsek and Singh 2003; Hall-Stoodley, Costerton and Stoodley 2004; Hu and Ehrlich 2008). The EPS is a hallmark morphological feature of biofilms and serves both structural and protective roles (Wolfaardt et al 1999; Flemming, Neu and Wozniak 2007). While planktonic bacteria of many species may be encapsulated with a glycocalyx, the presence of EPS enveloping multiple bacterial cells is generally regarded as characteristic of biofilm formation. The EPS is a highly hydrated polymeric material with a complex chemistry consisting of extracellular DNA (eDNA), polysaccharides, proteins and lipids (Whitchurch et al. 2002; Staudt et al. 2004; Flemming, Neu and Wozniak 2007; Jurcisek and Bakaletz 2007; Izano et al. 2008). The viscoelastic properties of the EPS allow the biofilm to structurally adapt to changes in fluid flow (Stoodley et al. 1999; Shaw et al. 2004; Rupp, Fux and Stoodley 2005; Ehret and Bol 2012; Bol et al. 2013). The EPS also serves as a physical barrier by which the bacteria are protected from phagocytic cells (Leid et al. 2002, 2005), immunoglobulin (de Beer, Stoodley and Lewandowski 1997), desiccation (Chang et al. 2007), ultraviolet radiation (Elasri and Miller 1999) and exposure to oxidizing agents (Wolfaardt et al 1999). However, the chemical complexity and the low density of EPS make it difficult to image as a gestalt structure in situ (Flemming and Wingender 2010). Fluorophore-conjugated lectins have been used to visualize specific polysaccharide components of biofilm matrices (Johnsen et al. 2000; Neu, Swerhone and Lawrence 2001; Staudt et al. 2004), and lectin cocktails have also been used for more general EPS staining (Hall-Stoodley et al. 2008). More recently, interest has focused on eDNA, an EPS component which has been shown to form structural networks in a number of biofilms formed by human pathogens (Goodman et al. 2011). For instance, intriguing video microscopy has shown how cells in Pseudomonas aeruginosa biofilms form network trails of eDNA on surfaces, which directs bacterial motility within the biofilm (Gloag et al. 2013). Three dimensional (3D) EPS networks have also been reported in biofilms formed by the non-motile Gram-positive pathogen Staphylococcus epidermidis (Schaudinn et al. 2014). These networks were visualized with scanning electron microscopy and stained with crystal violet, but failed to stain with specific fluorescent stains (for DNA, protein, carbohydrates and lipids). Other non-fluorescent stains such as methylene blue, alcian blue and dental plaque disclosing solution have been used to discern biofilm EPS. However, these stains quench fluorescence (Hallden et al. 1991; Usacheva et al. 2008) and therefore are incompatible with fluorescence microscopy, particularly confocal laser scanning microscopy (CLSM), which is routinely used to visualize bacterial biofilm ultrastructure under hydrated conditions. In this report, we investigated the use of potassium permanganate (KMnO4) as a novel stain to enhance 3D visualization of the biofilm structure and EPS under fully hydrated conditions. The KMnO4 staining protocol tested here was adapted from the auramine–rhodamine fluorescence histochemistry method in which KMnO4 is used as counter stain for sputum and tissue in the identification of mycobacteria and other acid-fast bacteria (Koneman et al. 1992). We reasoned that KMnO4 would also counterstain biofilm EPS against bacterial cells labeled with fluorescent nucleic acid probes.
KMnO4 is a strong organic molecule oxidizing agent, and under neutral pH conditions, yields a brown MnO2 crystal precipitate. Therefore, we utilized reflectance confocal microscopy that detected the reflection of laser light off of the MnO2 precipitate deposited on the biofilm EPS of various species and quantified the biovolume using the captured reflected signal. Although reflected imaging is frequently used in material science to identify defects or roughness on relatively smooth, hard surfaces it is rarely used for biological applications. In biology, confocal microscopy is usually conducted in fluorescence mode. Since the reflected light signal is usually much stronger than the fluorescence signal, effort is made to filter out the reflected light which would otherwise overwhelm the fluorescence signal. However, the reflected light that is discarded may contain useful information (Pellacani et al. 2007). Since the filterless detectors of modern confocal microscopes can be set to collect light in multiple detector windows, it is possible to collect reflected light in a separate channel. Reflected confocal microscopy has been previously used to visualize fully hydrated biofilm with no prior staining (Wood et al. 2000; Yawata et al. 2010). Here, we utilized the reflectance capability of the confocal microscope by enhancing the reflectivity of the otherwise very low-density biofilm EPS through staining with KMnO4.
To determine which EPS components reacted with KMnO4, we stained purified DNA and various polysaccharides commonly found in EPS and assessed oxidation by MnO2 and precipitate formation. KMnO4 staining increased the measurement of biofilm biovolume of all clinical and reference strains tested, and revealed previously indiscernible non-eDNA 3D network structures in P. aeruginosa biofilms. Finally, to assess the application of KMnO4 staining for research and clinical specimens, we used a combination of the LIVE/DEAD® BacLight kit and KMnO4 to visualize P. aeruginosa on the mucosal epithelium from chinchilla bullae using multimodal illumination (Ehrlich et al. 2002).
MATERIALS AND METHODS
Bacteria strains and growth conditions
A complete list of strains used in this study can be found in Table 1. Pseudomonas strains were grown on Luria Bertani (LB, Fisher Bioreagents, Fair Lawn, New Jersey) medium, S. aureus strains were grown in brain heart infusion agar (BHI, BD Difco, Sparks, Maryland) and S. mutans was grown in BHI supplemented with 2% sucrose. Primary overnight cultures were grown with aeration at 37°C.
Table 1.
Bacterial strains used in this study.
| Strain Name | Organisms and genotype | Source or reference |
|---|---|---|
| PAO1 | WT P. aeruginosa | Stover et al. (2000) |
| PittD | Pseudomonas aeruginosa clinical isolate from child with otitis externa | (24) |
| WFPA801 | Pseudomonas aeruginosa PBAD-psl | Ma et al. (2006) |
| WFPA831 | Pseudomonas aeruginosa PBAD-pel | Ma et al. (2006) |
| WFPA833 | Pseudomonas aeruginosa PBAD-psl and PBAD-pel | Mann and Wozniak (2012) |
| Seattle 1945 | WT S. aureus | ATCC #25923 |
| UA159 | Streptococcus mutans | ATCC #700610 |
Biofilms
For microscopic imaging, Pseudomonas biofilms were grown in (1/10)th strength LB medium, and Staphylococcus and Streptococcus biofilms were grown in BHI medium. Biofilms were grown in 35 mm glass-bottomed culture dishes (MatTek Corp., Ashland, MA) for between 7 and 10 days at 37°C in a humidified 5% CO2 incubator under static conditions. For KMnO4 staining of EPS components, overnight cultures were subcultured 1:100 in BHI medium and grown statically in Falcon® polystyrene multiwell plates (Corning Incorporated, Tewsbury, MA) for 1–4 days. Mutant overexpressing P. aeruginosa strains were supplemented with 0.2% arabinose to induce PBAD promoter expression, and S. mutans biofilms were supplemented with 2% sucrose to induce glycan production.
BacLight LIVE/DEAD staining and the potassium permanganate EPS contrast enhancer
Prior to staining, biofilms were rinsed twice with HBSS with CaCl2 and MgCl2 and without phenol red (Cat. # 14025, Invitrogen, Carlsbad, CA). The rationale for using the formulation with Ca++ and Mg++ was to help preserve biofilm structure through the cross-linking of negative residues associated with the cell wall and EPS components (Kang et al. 2006). The solution was then stained with Molecular Probes Syto 9 nucleic acid dye or with the BacLight LIVE/DEAD kit according to the manufacturer's instructions. The biofilm was rinsed again and stained with a freshly prepared 0.5% KMnO4 solution in HBSS for 2 min. Excess stain was removed by rinsing twice with HBSS.
Confocal laser scanning microscopy
CLSM is used to obtain non-fluorescent information from biological specimens by employing the reflectance mode (RM), which can be used to image unstained specimens (Wood et al. 2000; Yawata et al. 2010). We used the RM with potassium permanganate to enhance the reflected signal from non-fluorescently labeled components of the biofilm. Biofilms were observed using a Leica DM RXE microscope attached to a TCS SP2 Acousto Optical Beam Splitter (AOBS) confocal system (Leica Microsystems, Exton, PA) as previously described (Nistico et al. 2009). Briefly, specimens were examined with a Leica TCS SP2 AOBS confocal microscope attached to an upright Leica microscope using a ×63 long working distance 0.90 NA water immersion objective operating in sequential scanning mode. Live (Syto 9) and dead (propidium iodide -PI) stained cells were excited with the 488 and 594 nm lasers and imaged in separate green and red channels, respectively. The EPS was imaged by collecting the reflected signal from the 488 laser using the AOBS optimization so that the EPS appeared in a third channel (designated blue for optimal contrast with red and green).
The reflected imaging mode was used to detect the brown MnO2 reflective precipitate in the specimen with the 488 nm laser and the reflectance detector set to collect reflected 488 nm light. The reflected image was then optimized using the AOBS function. Optimal contrast was achieved by using high gain and high background settings. The effect of KMnO4 on enhancing the reflected signal was compared by measuring the relative proportion of EPS volume to cell volume by subtracting the difference of non-colocalized reflected signal in biofilms with and without KMnO4 staining. A single area in the center region of the plate was chosen at random for the collection of a stack of images from between four and six independently grown replicate biofilms.
Image analysis
Each channel of the 3D confocal stacks was quantified for biovolume using COMSTAT (Heydorn et al. 2000) to determine the relative proportions of live and dead cells and EPS to the biofilm volume. The degree of colocalization (Imaris Coloc) between the blue and the green signal from the cells was used to determine the relative volume of the EPS to cells. For this measurement, the threshold was set to exclude the low-intensity signal, which was presumed to come from the staining of eDNA, rather than background ‘noise’ which was filtered out by frame averaging. Reflected light that colocalized with the nucleic acid stain was assumed to have come from bacterial cells, and was not included in the EPS volume measurement. The total volume of EPS was found by subtracting reflected light signal from the bacterial cells from the total reflected light volume. COMSTAT was also used to construct a depth profile of signals from the Syto 9, PI and KMnO4 to show the vertical distribution of live and dead cells and EPS in the PAO1 biofilm. The roughness and maximum thickness of the biofilms were found by COMSTAT analysis of the reflected channel after KMnO4 staining, since it included signal from both cells and EPS.
Assessment of KMnO4 reaction (and MnO2 formation) with individual EPS components
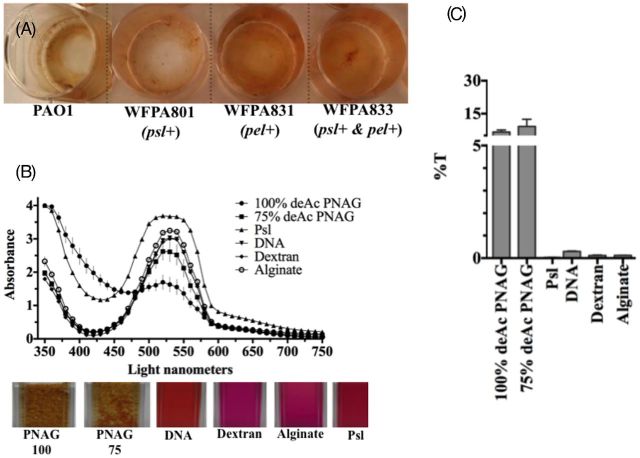
We applied the fact that manganese ions exhibit various colorimetric readings depending on their oxidation state. For this method, we measured absorbance spectra from 350 to 750 nm (10 nm increments) for KMnO4 reacting with 1 mg mL−1 of the various EPS components (Table 2) dissolved in water using a SpectraMax M2 multiwell plate reader (Molecular Devices, LLC, Sunnyvale, CA). DNA, dextran and alginate are all soluble in water; however, PNAG and P. aeruginosa Psl are not. Thus, for this experiment, we acquired 75% and 100% deacetylated PNAG (PNAG deacetylation ≥ 75% enables water solubility) for testing. Pseudomonas aeruginosa Psl is also insoluble in water, although when homogenized efficiently, Psl remains suspended in solution. Most of the EPS components tested were commercially available (Table 3), but deacetylated PNAG was a gift from Dr John Vournakis and the Pseudomonas PAO1 Psl extract was made in-house. All EPS components were filter sterilized with a 0.22 μm filter and performed in triplicate in 96-well polystyrene plates at room temperature (RT). In addition, we repeated the reactions in larger individual spectroscopy cuvettes to document the formation of the brown precipitate via digital photography. To assess the spectrometric formation of MnO2 formation, we analyzed absorbance at 525 nm (Jaganyi, Altaf and Wekesa 2013). Because we lacked a Pseudomonas Pel extract, or the protocol to extract Pel, we tested KMnO4 reactivity with Pel using P. aeruginosa strains overexpressing Pel (or Psl) polysaccharide. To do this, we grew biofilms of P. aeruginosa wild type (WT) or strains overexpressing pel, psl or both from an arabinose-inducible plasmid in multiwell polystyrene plates with 0.2% arabinose to induce polysaccharide overproduction. After 4 days, the culture medium from biofilms was removed from each well and discarded, and an equal volume of RT sterile PBS was gently added (to avoid dislodging the biofilm) and removed once to rinse the biofilms of broth medium (broth reacts positively with KMnO4). An equal volume of 0.05% solution of KMnO4 in PBS was then added to each well. The excess stain was removed from each well after 60-min exposure. Digital images of the remaining brown crystal precipitates were captured after the plates were dried.
Table 2.
EPS components used for KMnO4 staining and their sources.
| Rep. EPS component | Compound used | Source |
|---|---|---|
| PNAG | 75% and 100% deacetylated PNAG (water soluble) | John Vournakis, Marine Poly Technologies, Inc., Danvers, MA |
| DNA | Salmon sperm DNA | Ambion, Inc. (Austin, TX) |
| Glycan | Dextran from Leuconostoc spp. | Sigma-Aldrich |
| Alginate | Alginic acid from Brown algae | Sigma-Aldrich |
| Psudomonas aeruginosa Psl | Psl crude extract | This study |
Table 3.
Degree of colocalization (by volume) of bacterial cells (stained with nucleic acid stains) and EPS (measured by reflected confocal imaging) for the various biofilms (mean and 1 SE). With no KMnO4 staining the reflected signal came from the bacteria themselves since there was colocalization between the Live/Dead stain signal and the reflected signal. The increase in reflected signal after staining with KMnO4 which was not co-localized was the contribution from the EPS.
| Degree of colocalization | PAO1 | PittD | Streptococcus aureus | Streptococcus mutans |
|---|---|---|---|---|
| Without KMnO4 (cells alone) | 69 ± 3% | 33 ± 8% | 80 ± 7% | 71 ± 7% |
| Without KMnO4 (EPS alone) | 31 ± 3% | 67 ± 8% | 20 ± 7% | 29 ± 7% |
| With KMnO4 (cells alone) | 20 ± 5% | 20 ± 7% | 41 ± 10% | 67 ± 10% |
| With KMnO4 (EPS) | 80 ± 5% | 80 ± 7% | 59 ± 10% | 33 ± 10% |
| P (comparing % of reflected signal from EPS with and without KMnO4 staining. | <0.01 | 0.24 | 0.02 | 0.69 |
Preparation of surface-associated Psl
Pseudomonas aeruginosa strains WFPA801 and WFPA800 were grown in 500 mL of chemically defined Jensen's medium (Jensen, Fecycz and Campbell 1980) with the addition of 0.2% L-arabinose. Biomass was collected by centrifugation at 27 000 × g and the resultant bacterial pellets were suspended in 0.9% NaCl (Baxter, Deerfield, IL). Cell-surface-associated polymers were detached by mild sonication using a Sonic Dismembrator 100 probe sonicator (Fisher Scientific, Pittsburgh, PA) (three cycles of 1 min sonication and 1 min rest at 50% power). The released material was collected by centrifugation at 27 000 × g for 1 h and the supernatant was transferred to a sterile container. The bacterial pellets were suspended and extracted twice more, and the supernatants combined and lyophilized. To separate the cell-associated polysaccharides from the rest of the crude matrix extract, the dried pellets were suspended in cold 75% ethanol and precipitated at 4°C overnight. The precipitate was collected by centrifugation at 27 000 × g for 1 h and the supernatant was discarded. The precipitate was again suspended in cold 95% ethanol and dislodged from the side of the centrifuge tube using a metal spatula. The suspension was centrifuged as previously described and the supernatant was discarded. Pelleted material was washed once more with cold 95% ethanol, followed by one wash with cold absolute ethanol, and the resulting pellet was allowed to air dry at RT overnight. To completely remove any residual nucleic acid or protein contaminates, the crude polysaccharides were suspended in a minimal volume of sterile, endotoxin-free water (Baxter, Deerfield, IL). DNAse I stock solution (Sigma-Aldrich, St Louis, MO) and RNAse A stock solution (USB Corporation, Cleveland, OH) were each added to a final concentration of [0.1 mg mL−1]. The samples were incubated overnight at 37°C with agitation. Following nucleic acid digestion, proteinase K stock solution (Qiagen, Valencia, CA) was added to a final concentration of [0.1 mg mL−1] and incubated overnight at 37°C with agitation. After proteolysis, the remaining crude exopolysaccharides were transferred to a 3500 MWCO Slide-A-Lyzer dialysis cassette (Thermo Pierce, Rockford, IL) and dialyzed for three complete exchanges using sterile water at 4°C. When fully exchanged into water, the EPS was transferred from the dialysis cassette into a sterile centrifuge tube, lyophilized and the Psl content was determined by ELISA.
Chinchilla bullae
To assess the applicability of KMnO4 to stain the EPS of biofilms on clinical specimens, we used mucosal epithelium covering the bullae from Chinchilla laniger (Ehrlich et al. 2002). These experiments were conducted in accordance with institutional approval from Allegheny–Singer Research Institute according to IACUC protocol #798. An overnight culture of P. aeruginosa PAO1 was diluted to approximately 10 CFU in 100 μL PBS and inoculated directly into each bulla of four young adult acclimatized chinchillas as previously described (Ehrlich et al. 2002). Two control animals received 100 μL of a PBS mock infection. After either 2 or 3 days, the animals were sacrificed and the bullae were excised. A small piece of each bulla (∼5 × 5 mm) was cut from the bottom of the bulla ensuring that the mucosa was still attached to the bone. The outside of the bone was blotted dry and then mounted in the bottom of a 35-mm petri plate by either gently pressing onto Lubreseal grease, avoiding the central area intended for viewing, or allowing to set in 1% cooled molten agar to prevent movement during imaging. The specimen was stained with BacLight LIVE/DEAD kit as before for 10–15 min and rinsed with HBSS with CaCl2 and MgCl2 without phenol red. In some cases, the host nuclei stained so brightly with PI it was difficult to observe the surrounding bacteria. To minimize this problem, we used the LIVE/DEAD at (1/10)th strength for a final concentration to reduce the brightness of the host nuclei allowing the bacteria to be readily visible. Specimens from infected and mock-infected animals were stained with 0.5% KMnO4 for 2 min, rinsed twice to remove excess KMnO4, submerged in HBSS, and examined using a ×63 water immersion objective and confocal microscopy. Images of KMnO4 stained infected and mock-infected specimens were qualitatively compared with specimens stained with LIVE/DEAD kit only.
Statistical evaluation
Data were reported as mean ±1 SD or 1 SE. Statistical comparisons between mean values were made using the one way analysis of variance (Anova) function in Microsoft. Differences were considered significant when P > 0.05.
RESULTS
First, as proof of principle, we wanted to qualitatively observe KMnO4 reactivity with P. aeruginosa biofilms (Fig. 1). A brown MnO2 precipitate formed on all strains although the intensity of the reaction varied; however, it was not known whether this was due to the amount of EPS or to the reactivity of the EPS with KMnO4. Pseudomonas aeruginosa WFPA831 overexpressing Pel polysaccharide resulted in the greatest oxidation and MnO2 formation compared with wild-type (PAO1) (Fig. 1A). Since KMnO4 appeared to react with all biofilms, but varied in spectrophotometric intensity across strains, we reasoned that KMnO4 reacted with some, but not all components of the EPS. To determine which EPS components reacted with KMnO4, reactivity was quantified using spectrophotometry with purified individual EPS components dissolved in water in vitro. The KMnO4 absorbance peak was maximal at 525 nm and the formation of MnO2 resulted in a marked decrease in absorbance at 525 nm (Jaganyi, Altaf and Wekesa 2013). A PA01 Psl crude extract, DNA, dextran and alginate all exhibited peak absorbance at 525 nm (Fig. 1B). After 60 min, purified PNAG showed decreased absorbance at 525 nm (Fig. 1C) corresponding with MnO2 formation. Brown MnO2 precipitate formation was evident in cuvettes containing PNAG after 60 min (Fig. 1A bottom images) and a graph of the percent transmission as a function of the absorbance data confirmed the reactivity of PNAG with KMnO4 (Fig. 1B). Notably, after 48 h, MnO2 also formed when mixed with dextran (data not shown), but this reaction did not occur with the 60-min spectrometric analysis. Interestingly, P. aeruginosa WFPA831 overexpressing pel appeared to react more strongly with KMnO4 compared with the WT or P. aerugionsa overpressing psl only (Fig. 1A). These data suggest that KMnO4 reacted with Pel, PNAG, and to a lesser extent dextran, DNA, alginate or Pseudomonas Psl.
Figure 1.
KMnO4 reacts strongly with PNAG and protein but weakly with Psl, DNA, dextran or alginate. (A) KMnO4 reacting with 4-day-old P. aeruginosa WT and mutant strains overexpressing psl, pel or both from arabinose-inducible PBAD promoter. (B) Absorbance spectra (top) for each individual EPS component-KMnO4 reaction with digital images (below) of individual EPS components solubilized in water and stained with KMnO4 in cuvettes (60 min). (C) Percent transmission of light of individual EPS component-KMnO4 as a function of absorbance at 525 nm. Data points and columns in B and C (respectively) represent the means and error bars represent the standard error.
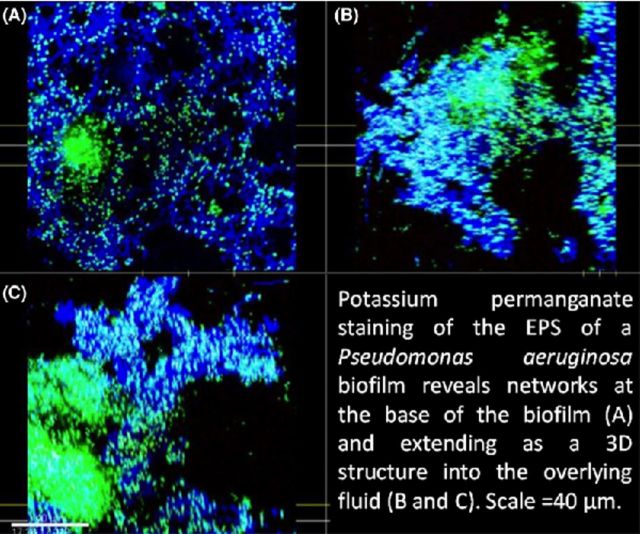
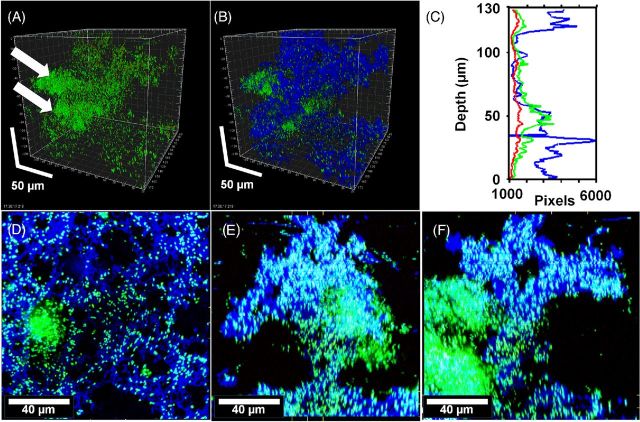
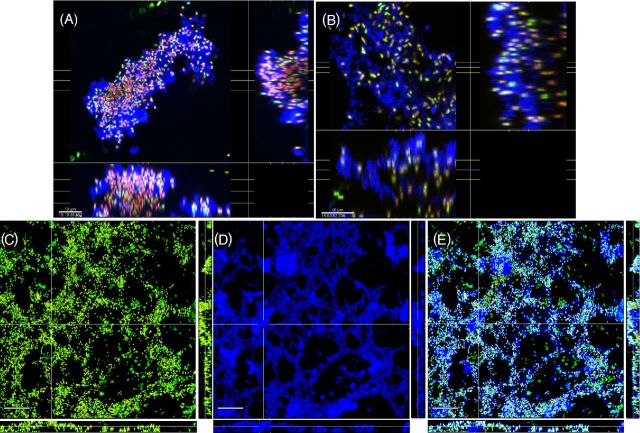
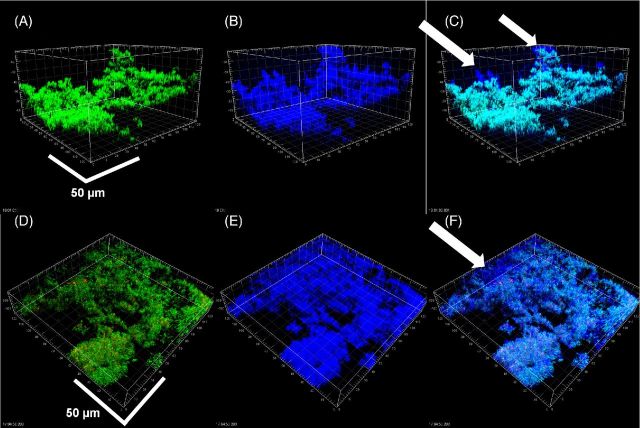
To determine if KMnO4 could be used as an EPS stain for CLSM, biofilms of a clinical strain of P. aeruginosa PittD stained with BacLight LIVE/DEAD were examined with CLSM using the RM. CSLM clearly identified bacterial cells in P. aeruginosa PittD biofilms, which exhibited 3D cell clusters and concentrated aggregates of cells separated by interstitial voids and channels in the fluorescent channel (Fig. 2). Strikingly, upon staining with KMnO4, voluminous amounts of EPS, which were otherwise invisible, were visible using the reflected signal from the KMnO4-treated samples (Fig. 2B). In some areas, the biofilm exhibited two distinct layers; a network of EPS connecting individual cells at the base of the biofilm (Fig. 2D-F), and amorphous cumulus-like structures at the top of the biofilm the EPS (Fig. 2B). Small cell clusters and single cells were connected to larger cell clusters by strands of EPS, often devoid of cells within the strands. The vertical profile of green, red and blue (reflectance) signal from the biofilm suggested that the greatest concentration of live cells was at a depth of 50 μm in the 130 μm biofilm (Fig. 2C), while the EPS was most abundant in the base layer (0–40 μm) and the top layer (110–130 μm), indicating a base biofilm with protruding mushroom-shaped cell clusters typical of P. aeruginosa biofilms. A few dead cells were evenly spread through the depth of the biofilm. In some areas, we noted that there were regions in the interior of the biofilm that were not stained with KMnO4, but gave a high Syto9 signal (Fig. 3). Some of the signal was from the cells themselves, but between the cells there was a diffuse signal that we interpreted as being areas of concentrated eDNA. The exclusion of staining eDNA is consistent with the spectrophotometer data, which showed that only minimal brown colloid or precipitate formed with DNA (Fig. 1B and C). The structure of the PAO1 biofilms was similar to that of the PittD biofilms, but more compact. However, network-like structures of EPS components and bacterial cells were seen at the base of the biofilm (Fig. 4). KMnO4 contrast staining also enhanced the 3D visualization of S. aureus and S. mutans biofilm EPS (Fig. 5). However, unlike the P. aeruginosa biofilms, the EPS was tightly colocalized with bacteria and no network-like structures were observed.
Figure 2.
Pseudomonas aeruginosa PittD biofilm showing cells stained with Molecular Probes Live (green)/Dead (red) BacLight LIVE/DEAD Kit and reflected signal from the EPS stained with KMnO4 contrast enhancer (blue). (A) Signal from green and red channels showing a conventional LIVE/DEAD view of the biofilm, eDNA staining is indicated by the white arrows. (B) Same image as ‘A’ with blue reflected signal from KMnO4 revealed large amounts of EPS between individual cells. Scale = 50 μm. (C) The biomass distribution depth profile showing the vertical distribution of live (green), dead (red) and EPS (blue) signal. (D) XY plan view of the base of the biofilm revealed a network of EPS connecting individual bacteria. (E) and (F) XZ and XY side views, respectively, with the base of the biofilm at the bottom showing the EPS forming convoluted structures growing into the overlying liquid medium. There was a dense cluster of cells (shown by arrows in panel (A) and to the left in panel (E) that did not stain with KMnO4 but appeared to contain eDNA (diffuse green). Panels (E), (F) and (G): scale = 40 μm, sections are projections of 10 μm thick.
Figure 3.
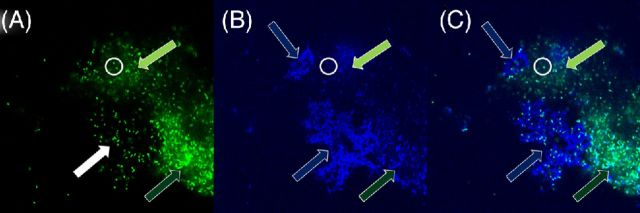
Potassium permanganate does not strongly stain biofilm eDNA. Pseudomonas aeruginosa PittD biofilm stained with BacLIght LIVE/DEAD and KMnO4. (A) The green signal from the Syto 9 shows brightly stained individual bacterial rods (a representative group indicated by the white filled arrow and two cells inside the white circle) as well as diffuse, less intense, staining between the cells (light and dark green arrows indicate less and more intense areas of diffuse staining, respectively). The diffuse staining surrounding cells is consistent with that of eDNA. (B) The blue reflected signal from the KMnO4 shows a weak reflected signal in these same areas, but shows strong signal in areas where there are no significant amounts of eDNA (blue arrows). There is a weak reflected signal from the individual cells (white circle). (C) Overlay of the two images show the eDNA and KMnO4 staining are not colocalized. Images A and B are raw tiff files.
Figure 4.
Pseudomonas aeruginosa PAO1 biofilms showing cells stained with Molecular Probes Live (green) /Dead (red) BacLight Kit and reflected signal from the EPS stained with KMnO4 contrast enhancer (blue). (A) and (B) plan (XY) and side (XZ and YZ) views of two different biofilms showing EPS connecting cells and extending into the liquid growth medium. Scale = 10 μm. Web-like structures at the base of the biofilms were also observed in the PAO1 biofilms (D–F). (D) ‘Conventional’ Live / dead image without KMnO4, (E) reflected signal from KMnO4 staining, (F) combined image showed that cells were connected along webs of EPS. Panels show square XY plan view, and XZ and YZ side views below and to the right of the plan view. Scale = 30 μm.
Figure 5.
Biofilms of S. mutans (A–C) and S. aureus (D–F) shown by conventional LIVE/DEAD kit staining (A, D), reflection from KMnO4 staining (B, E) and the combined images (C, F). Colocalization of green/red and blue channels (bacteria which were stained with LIVE/DEAD kit and also giving a strong reflected signal appeared light blue. In contrast with the P. aeruginosa biofilms, the EPS was more localized to the bacteria, however, in both biofilms there were some areas that had EPS, but were relatively devoid of bacteria (arrows). Scale bars = 50 μm.
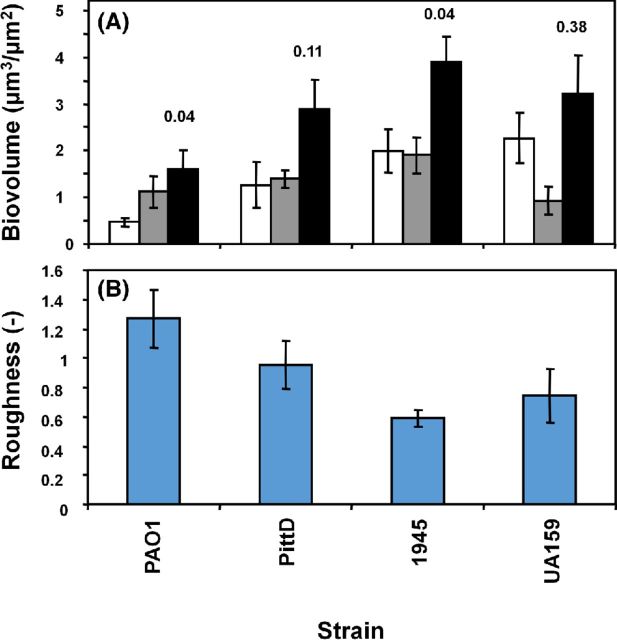
COMSTAT quantification
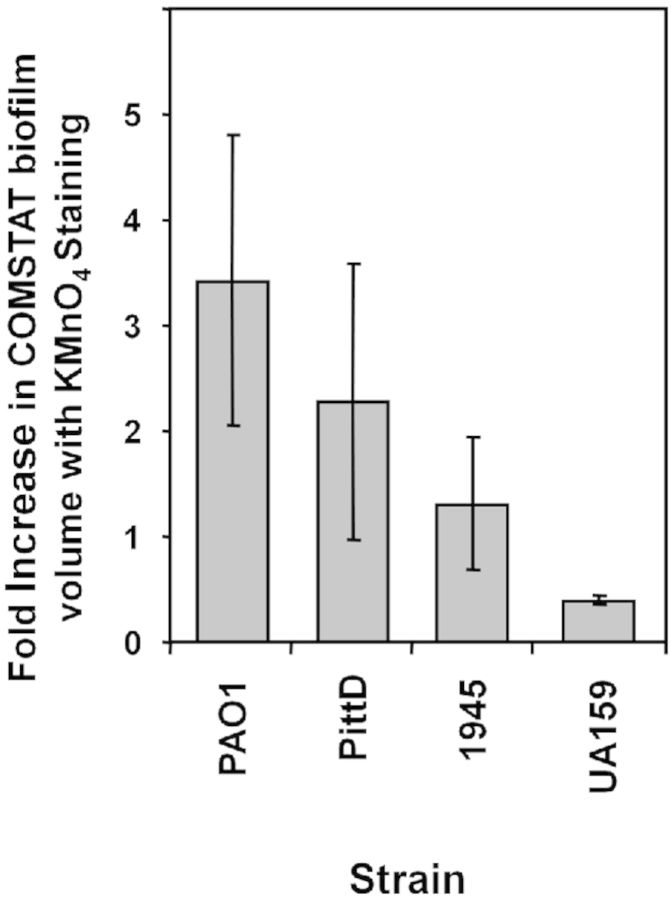
COMSTAT was used to quantify the biofilm volume made up of bacterial cells and EPS stained with KMnO4 (Fig. 6). The PittD, S. mutans, and S. aureus strains formed similar amounts of biofilm as measured by total volume per unit area of colonized surface (Fig. 6A). While the PAO1 biofilm formed the least biofilm, it had a significantly greater volume of EPS compared with other strains. The mean maximum thickness of the biofilms ranged from 18 μm for PAO1 to 31 μm for S. mutans (data not shown). The PAO1 and PittD strains exhibited a higher roughness coefficient than the S. aureus and S. mutans biofilms (Fig. 6B) suggesting that they were more structurally heterogeneous and contained a greater proportion of towers and interstitial channels. The increase in biofilm volume measured by COMSTAT when the reflected signal from the KMnO4 EPS was included in the measurement ranged from 342% for PAO1 biofilms to 32% for S. mutans biofilm (Fig. 7). The degree of colocalization between reflected and green signal (cells alone) with and without KMnO4 staining is shown in Table 3. There was no significant difference between the volume of live cells (all P > 0.36) between the KMnO4-stained and unstained samples suggesting that biofilms in both groups were comparable, and that KMnO4 staining did not adversely affect the viability as assessed by the LIVE/DEAD kit. We did note that in some biofilms, over time a minority of bacterial cells appeared swollen, possibly due to damage to the cell wall by KMnO4 oxidation.
Figure 6.
COMSTAT analysis of the various biofilms. (A) Volume of cells (white bars), EPS (grey bars) and cells + EPS (black bars), mean ±1 SE. Pixels colocalizing with the bacteria and the reflected signal were subtracted out. P values for the volume estimated from cells alone compared to cells and EPS are given above the black bars. (B) Biofilm roughness. Data represents mean and 1 S.E. from 4, 5 or 6 independent replicates.
Figure 7.
KMnO4 staining increased the estimate of biofilm volume when the reflected signal from the EPS was included in the COMSTAT analysis compared to the volume of biofilm estimated from the fluorescent nucleic acid staining of the cells alone. Data represent mean and 1 S.E. from 4, 5 or 6 independent replicates.
Staining of Chinchilla middle ear biofilms
These results suggested the KMnO4 staining protocol might be a good EPS research tool and useful as a diagnostic technique for biofilm-associated infections ex vivo. Therefore, we tested its applicability using a P. aeruginosa infection of mucosal epithelium. Staining of Chinchilla mucosal epithelium showed that both bacteria and host nuclei took up Syto 9, and appeared green after 20 min (Fig. 8). However, over longer time periods, the host nuclei incorporated PI turning the nuclei first yellow (green + red) and then predominantly red. Therefore, the BacLight LIVE/DEAD kit could be used to assess the viability of the bacteria at the time of staining, but not as a viability indicator for eukaryotic cells. In mock-infected bullae, there was no effusion and the mucosal epithelial cells revealed a thin layer of healthy cells (Fig. 8A). In contrast, P. aeruginosa-infected mucosal epithelium was inflamed and exhibited areas with high concentrations of bacterial rods as single cells and larger cell clusters over 20 μm diameter (Fig. 8B). Biofilms were patchy and up to 20 μm thick in certain areas. Increased thickness (z) of the mucosa due to inflammation made it difficult to image to the underlying bone, but bacterial rods in biofilms were visible interspersed beneath the mucosal cell epithelial layer and bone. The nuclei of host cells were also seen in the upper layers of the biofilm and their morphology and size was consistent with that of inflammatory cells. There was no evidence of EPS between the individual rods stained with the LIVE/DEAD kit.
Figure 8.
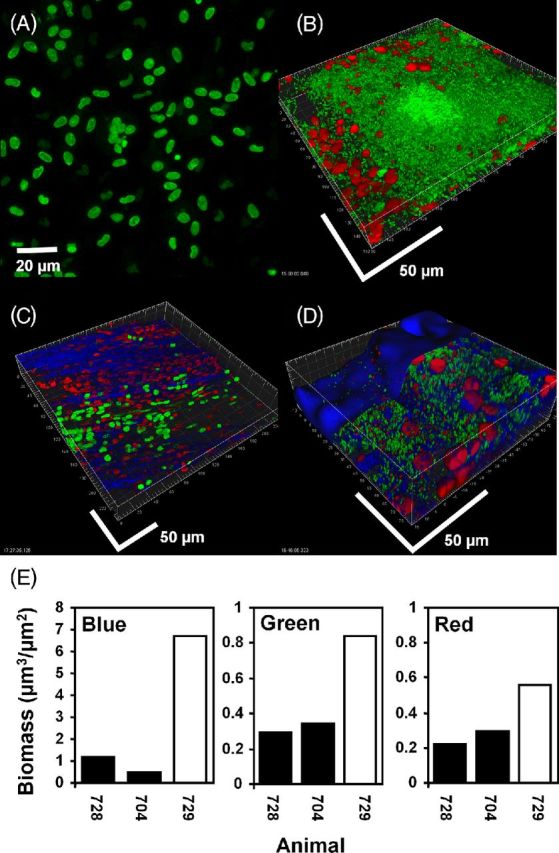
Mucosal P. aeruginosa PAO1 biofilm in the bulla in the chinchilla stained for cells with live (green) / dead (red) kit and KMnO4 (blue). (A) and (C): uninoculated controls. The nuclei of host epithelial cells stained initially primarily with Syto 9 (green) but over the imaging time became red as they absorbed PI. Cells with approximate equal proportions of the two stains appeared yellow. KMnO4 revealed matrix material between the cells. (B) and (D): bullae from two animals inoculated with PAO1. Bacterial rods stained green indicating that they were viable. Unlike the host cells, bacterial cells did not integrate PI, making the majority of bacteria readily distinguishable from the nuclei of host cells on the basis of size, morphology and differential staining. (D) After staining with KMnO4 material between and colocalized with the biofilm cell clusters was visible (blue). Biofilm bacteria and the nuclei of host cells did not appear morphologically or structurally different from the specimen stained only with the BacLight kit. Scale bars = 50 μm. (E) COMSTAT analysis of confocal images from the bullae of two uninoculated animals (black bars) and an animal infected with PAO1 (open bar) stained with BacLight LIVE/DEAD kit and KMnO4 and RM. Reflected signal from the KMnO4 (blue), live bacteria (green) and host nuclei (red).
KMnO4 staining of specimens from mock-infected animals on the other hand, showed that the tissue was stained brown (data not shown) and CSLM demonstrated a reflected signal primarily from the bone (Fig. 8C). LIVE/DEAD staining of infected bullae showed biofilms consisting of bacterial rods embedded with host cells, and the biofilms appeared similar in both non-KMnO4 and KMnO4-stained specimens. Some nuclei produced a stronger reflected signal colocalized with the red from the PI staining and therefore appeared pink. However, the reflected signal that surrounded the bacteria in the KMnO4 stained specimens was much stronger than that of the host nuclei and suggested the presence of biofilm EPS (Fig. 8D). COMSTAT analysis of LIVE/DEAD and KMnO4-stained fields demonstrated greater biovolume for the green, red and blue (reflectance) channels in infected specimens compared with those from mock-infected bullae. CSLM images showed that the red fluorescence derived principally from PI staining of host nuclei and green fluorescence was derived principally from viable bacteria stained with Syto 9 (Fig. 8E).
DISCUSSION
We report the development of a novel biofilm staining method utilizing KMnO4 and RM CSLM. A strong reflected signal was obtained from the bacteria and the EPS between bacterial cells in the biofilm after KMnO4 staining, while preserving fluorescence from the BacLight LIVE/DEAD staining for all of the bacterial strains tested. We were concerned about the potential destruction of biofilm structure from the oxidizing action of KMnO4, which is used as a disinfectant (Bryant, Fulton and Budd 1992) and antiseptic agent (Stalder et al. 1992). However, viability staining suggested that this was not problematic when specimens were rinsed within a few minutes following addition of KMnO4. In some cases with longer KMnO4 contact times no obvious signs of disruption to the biofilm itself was observed, but swelling of individual bacterial cells suggested permeabilization of the cell wall which indicated that sequential staining with the LIVE/DEAD kit and rinsing excess stain before adding KMnO4 was necessary to avoid previously live cells taking up PI if viability assessment is required. Nevertheless, there are a number of potential limitations to this technique, which require further characterization. The first is that redox reactions with potassium permanganate are complex and pH dependent since MnO2 is formed under neutral pH. In our work, we did not take special precautions regarding pH other than using physiological buffers routinely used for tissue culture. For future work, and work with environmental samples, pH should be taken into consideration. A second limitation is that staining with KMnO4 requires additional rinse steps that likely removed biomass, particularly from low-density biofilms. Thirdly, it is not clear whether the particle size and distribution of MnO2 particles deposited on and within the EPS may overestimate or underestimate the volume of EPS. The degree of the artifact will be a function of the particle size, the size of the object being studied and the resolving power of the confocal optics. It would be interesting to further characterize the interaction of the KMnO4 with biofilm components using high-resolution microscopy such as SEM or TEM. In terms of distribution artifacts if the stain coated the outer part of the biofilm it might ‘shield’ or attenuate signal from deeper in the biofilm and thus underestimate the volume of EPS. Such a phenomenon is seen with fluorescent imaging where often the biofilm clusters can appear to have an outer ‘shell-like’ structure due to attenuation of fluorescence signal from deeper in the biofilm. However, our images suggest that this is not the case since we did not see well-defined reflective planes or ‘shells’ but rather a uniform signal throughout the biofilm clusters with no obvious attenuation with depth. In fact, the reflected signal was generally much stronger than the fluorescent signal which allowed imaging deeper into the specimen. Lastly, while we used 448 nm for our reflected imaging since it was the most powerful laser on our microscope, in ongoing studies, we have used the longer 633 nm red light, which has provided even greater laser penetration (depth).
KMnO4 staining of EPS was most striking in the clinical PittD strain and PAO1 P. aeruginosa biofilms. Although the base biofilm was relatively flat in the towers, the reflected signal revealed voluminous EPS enveloping individual bacterium with strands of EPS containing few cells that covered distances tens to hundreds of microns connecting satellite clusters of cells to the biofilm. Pseudomonas aeruginosa biofilms had two distinct layers; a densely packed base layer consisting of a branched network of EPS connecting individual bacterial cells, and a voluminous amorphous upper layer with a cumulus appearance. Data from KMnO4 staining of purified components, extracts and mutant strains suggest that the main component of this voluminous EPS was not eDNA. Figures 3 and 4 clearly show that areas of the biofilm with diffuse Syto9 staining between cells (indicating denser areas of eDNA) did not colocalize with MnO2 deposition. The oxidative reaction demonstrated by the colorimetric change with individual components and the psl and pel overexpressing strains (WFPA801 and WFPA831, respectively) suggest that Pel may be a major component of the EPS network. While the structure of Pel is not known, our data suggest it has a similar oxidation profile to PNAG based on the strength of its reactivity with KMnO4. While good staining of the EPS was obtained with our non-mucoid P. aeruginosa isolates, the fact that the reaction was weak with alginate suggests the technique might be limited for visualization of the EPS of mucoid P. aeruginosa strains, which commonly develop in CF lung infections (Hoiby, Ciofu and Bjarnsholt 2010). Calcofluor white has also been successfully used to stain alginate in mucoid P. aeruginosa biofilms (Peterson et al. 2014) and calcofluor white may be useful as an additional stain along with KMnO4 in mucoid strains.
The discovery of 3D networks formed by the EPS suggests that biofilms develop complex higher order structures, and similar complex structures have been reported in S. epidermidis biofilms (Schaudinn et al. 2007, 2014) and in marine biofilms (Thar and Kuhl 2002). Schaudinn and colleagues speculated that specialized biofilm structures provide a framework scaffold for the biofilm bacteria, facilitating their response to differential shear and directional fluid changes in flowing medium (Schaudinn et al. 2007). Indeed, eDNA structures have since been demonstrated to play a role in critical biofilm processes such as adhesion, aggregation and architecture (Whitchurch et al. 2002; Qin et al. 2007; Das et al. 2010, 2011), but also in the mechanical stability of the biofilm in response to physical and chemical stress (Mulcahy, Charron-Mazenod and Lewenza 2008; Flemming and Wingender 2010; Huseby et al. 2010; Das et al. 2011). Previously, Gloag et al. (2013) used video microscopy to show that P. aeruginosa cells could deposit trails of eDNA as they moved over surfaces, thus creating a network of trails that reflected motility. Similar networks on the glass surface were observed but, since KMnO4 did not react strongly with DNA, these networks appear to result from something other than eDNA. Psl, a polysaccharide produced by P. aeruginosa PAO1 has been shown to demonstrate a similar network of trails on the surface (Zhao et al. 2013) to those formed by eDNA (Gloag et al. 2013). While these studies demonstrated motility networks on a surface, our observations suggest that networks may extend into the overlying liquid to create 3D structure.
These results raise the question of how such networks might be secreted without support of a substratum. Recently Ehret and Bol (2012) have shown that a polymer network model can explain many of the mechanical properties of biofilms. Although the model is based on individual polymer strands, the model might be able to be scaled to account for larger ‘quaternary’ network structures in the biofilm. The function of such 3D structures is not clear, but ultrastructure may promote 3D networks, which facilitate bacterial movement through the biofilm EPS matrix that is unrestricted to the surface. We have previously postulated that 3D networks of viscoelastic polymers would allow biofilms to behave like an open cell memory foam structure where the biofilm can readily deform under transient mechanical loading but return to its previous configuration when the load is removed (Schaudinn et al. 2014). The KMnO4 contrast staining method may therefore be useful as a visualization research tool in further elucidating the role of biofilm EPS in biofilm structure and function.
Staphylococcus aureus and S. mutans biofilms were more compact and the EPS was more closely localized with the bacteria. By subtracting the reflected data from the cells using the Imaris colocalization software, we were able to quantify the volume ratio of biofilm bacteria to EPS separately using COMSTAT analysis. For each biofilm species, the volume of biofilm estimated from the fluorescent signal from bacteria cells alone underestimated the total biofilm volume; ranging from an increase of ∼32% for S. mutans biofilms to over 340% for PAO1 biofilms. In all cases, the reflected imaging with KMnO4 increased the estimate of biofilm biovolume, ranging from a factor of 1.2 for S. mutans to 12.0 for PAO1 (although these differences were not statistically significant). We believe that this variability may reflect inherent differences in the density of EPS in the replicate biofilms. It is known that S. mutans grown on sucrose forms a dense EPS due to the production of insoluble glucans (Pratten et al. 2000). For S. mutans, KMnO4 contrast enhancement only slightly increased the estimate of EPS compared to the use of reflected imaging alone, and is in agreement with in vitro staining results indicating that dextran was not oxidized by KMnO4 in the time frame of the biofilm experiments.
The present study also suggests that KMnO4 has the potential to be used as a biofilm stain for clinical specimens. KMnO4 in combination with nucleic acid stains were successfully used to visualize patchy mucosal PAO1 biofilms on inflamed chinchilla bullar epithelia. The mucosal biofilms varied in structure ranging from patches of mucosa containing many single cells to well-defined cell clusters which formed mounds of P. aeruginosa covering thousands of μm2 with a thickness of up to 30 μm. Interestingly, while in vitro grown P. aeruginosa biofilms were also heterogeneous, we did not see the distinctive structures (i.e. mushrooms) reported in in vitro studies (Barken et al. 2008). However these results were similar to in vitro experiments in the presence of leukocytes (Leid et al. 2002) with little evidence of phagocytic engulfment of live bacteria in the biofilm in the immediate vicinity of the host cells.
KMnO4 chiefly stained the area surrounding the biofilm bacteria and its colocalization with bacterial cells provides empirical evidence of EPS by applying the criteria used to identify biofilm EPS by ruthenium red in TEM images (Gristina et al. 1985; Marrie and Costerton 1985). These results suggest that KMnO4 may be an inexpensive and effective contrast enhancing method to demonstrate biofilm EPS in fully hydrated samples using the CSLM reflective signal to allow 3D imaging compatible with conventional fluorescent staining.
Acknowledgments
The authors thank Mary O'Toole for help in the preparation of this manuscript. The authors would also like to thank Dr John Vournakis (Marine Polymer Technologies, Inc., Danvers, MA) for so graciously donating 100% and 75% deacetylated poly-N-acetylglucosamine preparations for this study.
FUNDING
This work was supported by The Ohio State University College of Medicine, Allegheny General Hospital, Allegheny–Singer Research Institute, and grants from the National Institute on Deafness and Other Communication Disorders DC 05659 (J.C. Post), DC 02148 and DC 04173 (G.D. Ehrlich). Public Health Service awards AI097511 and NR013898 (DJW) and NIH/NIAID award # 1-T32-AI-112542, a NRSA training grant administered by the Center for Microbial Interface Biology (CMIB), at The Ohio State University supported this work.
Conflict of interest. None declared.
REFERENCES
- Barken KB, Pamp SJ, Yang L, et al. Roles of type IV pili, flagellum-mediated motility and extracellular DNA in the formation of mature multicellular structures in Pseudomonas aeruginosa biofilms. Environ Microbiol. 2008;10:2331–43. doi: 10.1111/j.1462-2920.2008.01658.x. [DOI] [PubMed] [Google Scholar]
- Bol M, Ehret AE, Bolea Albero A, et al. Recent advances in mechanical characterisation of biofilm and their significance for material modelling. Crit Rev Biotechnol. 2013;33:145–71. doi: 10.3109/07388551.2012.679250. [DOI] [PubMed] [Google Scholar]
- Bryant EA, Fulton GP, Budd GC. Disinfection Alternatives for Safe Drinking Water. New York: Van Nostrand Reinhold; 1992. [Google Scholar]
- Chang WS, van de Mortel M, Nielsen L, et al. Alginate production by Pseudomonas putida creates a hydrated microenvironment and contributes to biofilm architecture and stress tolerance under water-limiting conditions. J Bacteriol. 2007;189:8290–9. doi: 10.1128/JB.00727-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das T, Krom BP, van der Mei HC, et al. DNA-mediated bacterial aggregation is dictated by acid-base interactions. Soft Matter. 2011:2927–35. [Google Scholar]
- Das T, Sharma PK, Busscher HJ, et al. Role of extracellular DNA in initial bacterial adhesion and surface aggregation. Appl Environ Microb. 2010;76:3405–8. doi: 10.1128/AEM.03119-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Beer D, Stoodley P, Lewandowski Z. Measurement of local diffusion coefficients in biofilms by microinjection and confocal microscopy. Biotechnol Bioeng. 1997;53:151–8. doi: 10.1002/(SICI)1097-0290(19970120)53:2<151::AID-BIT4>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Ehret AE, Bol M. Modelling mechanical characteristics of microbial biofilms by network theory. J Roy Soc Interface. 2012;10:20120676. doi: 10.1098/rsif.2012.0676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich GD, Veeh R, Wang X, et al. Mucosal biofilm formation on middle-ear mucosa in the chinchilla model of otitis media. J Am Med Assoc. 2002;287:1710–5. doi: 10.1001/jama.287.13.1710. [DOI] [PubMed] [Google Scholar]
- Elasri MO, Miller RV. Study of the response of a biofilm bacterial community to UV radiation. Appl Environ Microb. 1999;65:2025–31. doi: 10.1128/aem.65.5.2025-2031.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemming HC, Neu TR, Wozniak DJ. The EPS matrix: the “house of biofilm cells”. J Bacteriol. 2007;189:7945–7. doi: 10.1128/JB.00858-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemming HC, Wingender J. The biofilm matrix. Nat Rev Microbiol. 2010;8:623–33. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- Gloag ES, Turnbull L, Huang A, et al. Self-organization of bacterial biofilms is facilitated by extracellular DNA. P Natl Acad Sci USA. 2013;110:11541–6. doi: 10.1073/pnas.1218898110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman SD, Obergfell KP, Jurcisek JA, et al. Biofilms can be dispersed by focusing the immune system on a common family of bacterial nucleoid-associated proteins. Mucosal Immunol. 2011;4:625–37. doi: 10.1038/mi.2011.27. [DOI] [PubMed] [Google Scholar]
- Gristina AG, Price JL, Hobgood CD, et al. Bacterial colonization of percutaneous sutures. Surgery. 1985;98:12–9. [PubMed] [Google Scholar]
- Hallden G, Skold CM, Eklund A, et al. Quenching of intracellular autofluorescence in alveolar macrophages permits analysis of fluorochrome labelled surface antigens by flow cytofluorometry. J Immunol Methods. 1991;142:207–14. doi: 10.1016/0022-1759(91)90108-r. [DOI] [PubMed] [Google Scholar]
- Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol. 2004;2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- Hall-Stoodley L, Nistico L, Sambanthamoorthy K, et al. Characterization of biofilm matrix, degradation by DNase treatment and evidence of capsule downregulation in Streptococcus pneumoniae clinical isolates. BMC Microbiol. 2008;8:173. doi: 10.1186/1471-2180-8-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heydorn A, Nielsen AT, Hentzer M, et al. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology. 2000;146:2395–407. doi: 10.1099/00221287-146-10-2395. [DOI] [PubMed] [Google Scholar]
- Hoiby N, Ciofu O, Bjarnsholt T. Pseudomonas aeruginosa biofilms in cystic fibrosis. Future Microbiol. 2010;5:1663–74. doi: 10.2217/fmb.10.125. [DOI] [PubMed] [Google Scholar]
- Hu FZ, Ehrlich GD. Population-level virulence factors amongst pathogenic bacteria: relation to infection outcome. Future Microbiol. 2008;3:31–42. doi: 10.2217/17460913.3.1.31. [DOI] [PubMed] [Google Scholar]
- Huseby MJ, Kruse AC, Digre J, et al. Beta toxin catalyzes formation of nucleoprotein matrix in staphylococcal biofilms. P Natl Acad Sci USA. 2010;107:14407–12. doi: 10.1073/pnas.0911032107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izano EA, Amarante MA, Kher WB, et al. Differential roles of poly-N-acetylglucosamine surface polysaccharide and extracellular DNA in Staphylococcus aureus and Staphylococcus epidermidis biofilms. Appl Environ Microb. 2008;74:470–6. doi: 10.1128/AEM.02073-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaganyi D, Altaf M, Wekesa I. Synthesis and characterization of whisker-shaped MnO2 nanostructure at room temperature. Appl Nanosci. 2013;3:329–33. [Google Scholar]
- Jensen SE, Fecycz IT, Campbell JN. Nutritional factors controlling exocellular protease production by Pseudomonas aeruginosa. J Bacteriol. 1980;144:844–7. doi: 10.1128/jb.144.2.844-847.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsen AR, Hausner M, Schnell A, et al. Evaluation of fluorescently labeled lectins for noninvasive localization of extracellular polymeric substances in Sphingomonas biofilms. Appl Environ Microb. 2000;66:3487–91. doi: 10.1128/aem.66.8.3487-3491.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurcisek JA, Bakaletz LO. Biofilms formed by nontypeable Haemophilus influenzae in vivo contain both double-stranded DNA and type IV pilin protein. J Bacteriol. 2007;189:3868–75. doi: 10.1128/JB.01935-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang SY, Bremer PJ, Kim KW, et al. Monitoring metal ion binding in single-layer Pseudomonas aeruginosa biofilms using ATR-IR spectroscopy. Langmuir. 2006;22:286–91. doi: 10.1021/la051660q. [DOI] [PubMed] [Google Scholar]
- Koneman EW, Allen SD, Janda WM, et al. Color Atlas and Textbook of Diagnostic Microbiology. Philadelphia: J.B. Lippincott Company; 1992. [Google Scholar]
- Leid JG, Shirtliff ME, Costerton JW, et al. Human leukocytes adhere to, penetrate, and respond to Staphylococcus aureus biofilms. Infect Immun. 2002;70:6339–45. doi: 10.1128/IAI.70.11.6339-6345.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leid JG, Willson CJ, Shirtliff ME, et al. The exopolysaccharide alginate protects Pseudomonas aeruginosa biofilm bacteria from IFN-gamma-mediated macrophage killing. J Immunol. 2005;175:7512–8. doi: 10.4049/jimmunol.175.11.7512. [DOI] [PubMed] [Google Scholar]
- Ma L, Jackson KD, Landry RM, et al. Analysis of Pseudomonas aeruginosa conditional psl variants reveals roles for the psl polysaccharide in adhesion and maintaining biofilm structure post attachment. J Bacteriol. 2006;188:8213–21. doi: 10.1128/JB.01202-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann EE, Wozniak DJ. Pseudomonas biofilm matrix composition and niche biology. FEMS Microbiol Rev. 2012;36:893–916. doi: 10.1111/j.1574-6976.2011.00322.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrie TJ, Costerton JW. Mode of growth of bacterial pathogens in chronic polymicrobial human osteomyelitis. J Clin Microbiol. 1985;22:924–33. doi: 10.1128/jcm.22.6.924-933.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulcahy H, Charron-Mazenod L, Lewenza S. Extracellular DNA chelates cations and induces antibiotic resistance in Pseudomonas aeruginosa biofilms. PLoS Pathog. 2008;4:e1000213. doi: 10.1371/journal.ppat.1000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neu T, Swerhone GD, Lawrence JR. Assessment of lectin-binding analysis for in situ detection of glycoconjugates in biofilm systems. Microbiology. 2001;147:299–313. doi: 10.1099/00221287-147-2-299. [DOI] [PubMed] [Google Scholar]
- Nistico L, Gieseke A, Stoodley P, et al. Fluorescence “in situ” hybridization for the detection of biofilm in the middle ear and upper respiratory tract mucosa. Methods Mol Biol. 2009;493:191–213. doi: 10.1007/978-1-59745-523-7_12. [DOI] [PubMed] [Google Scholar]
- Parsek MR, Singh PK. Bacterial biofilms: an emerging link to disease pathogenesis. Annu Rev Microbiol. 2003;57:677–701. doi: 10.1146/annurev.micro.57.030502.090720. [DOI] [PubMed] [Google Scholar]
- Pellacani G, Guitera P, Longo C, et al. The impact of in vivo reflectance confocal microscopy for the diagnostic accuracy of melanoma and equivocal melanocytic lesions. J Invest Dermatol. 2007;127:2759–65. doi: 10.1038/sj.jid.5700993. [DOI] [PubMed] [Google Scholar]
- Peterson BW, Busscher HJ, Sharma PK, et al. Visualization of microbiological processes underlying stress relaxation in Pseudomonas aeruginosa biofilms. Microsc Microanal. 2014;20:912–5. doi: 10.1017/S1431927614000361. [DOI] [PubMed] [Google Scholar]
- Pratten J, Andrews CS, Craig DQ, et al. Structural studies of microcosm dental plaques grown under different nutritional conditions. FEMS Microbiol Lett. 2000;189:215–8. doi: 10.1111/j.1574-6968.2000.tb09233.x. [DOI] [PubMed] [Google Scholar]
- Qin Z, Ou Y, Yang L, et al. Role of autolysin-mediated DNA release in biofilm formation of Staphylococcus epidermidis. Microbiology. 2007;153:2083–92. doi: 10.1099/mic.0.2007/006031-0. [DOI] [PubMed] [Google Scholar]
- Rupp CJ, Fux CA, Stoodley P. Viscoelasticity of Staphylococcus aureus biofilms in response to fluid shear allows resistance to detachment and facilitates rolling migration. Appl Environ Microb. 2005;71:2175–8. doi: 10.1128/AEM.71.4.2175-2178.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaudinn C, Stoodley P, Hall-Stoodley L, et al. Death and transfiguration in static Staphylococcus epidermidis cultures. PLoS One. 2014;9:e100002. doi: 10.1371/journal.pone.0100002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaudinn C, Stoodley P, Kainovic A, et al. Bacterial biofilms, other structures seen as mainstream concepts. Microbe. 2007;2:231–7. [Google Scholar]
- Shaw T, Winston M, Rupp CJ, et al. Commonality of elastic relaxation times in biofilms. Phys Rev Lett. 2004;93:098102. doi: 10.1103/PhysRevLett.93.098102. [DOI] [PubMed] [Google Scholar]
- Stalder JF, Fleury M, Sourisse M, et al. Comparative effects of two topical antiseptics (chlorhexidine vs KMnO4) on bacterial skin flora in atopic dermatitis. Acta Derm Venereol Suppl (Stockh) 1992;176:132–4. [PubMed] [Google Scholar]
- Staudt C, Horn H, Hempel DC, et al. Volumetric measurements of bacterial cells and extracellular polymeric substance glycoconjugates in biofilms. Biotechnol Bioeng. 2004;88:585–92. doi: 10.1002/bit.20241. [DOI] [PubMed] [Google Scholar]
- Stoodley P, Lewandowski Z, Boyle JD, et al. Structural deformation of bacterial biofilms caused by short-term fluctuations in fluid shear: an in situ investigation of biofilm rheology. Biotechnol Bioeng. 1999;65:83–92. [PubMed] [Google Scholar]
- Stover CK, Pham XQ, Erwin AL, et al. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature. 2000;406:959–64. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- Thar R, Kuhl M. Conspicuous veils formed by vibrioid bacteria on sulfidic marine sediment. Appl Environ Microb. 2002;68:6310–20. doi: 10.1128/AEM.68.12.6310-6320.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usacheva MN, Teichert MC, Usachev YM, et al. Interaction of the photobactericides methylene blue and toluidine blue with a fluorophore in Pseudomonas aeruginosa cells. Laser Surg Med. 2008;40:55–61. doi: 10.1002/lsm.20593. [DOI] [PubMed] [Google Scholar]
- Whitchurch CB, Tolker-Nielsen T, Ragas PC, et al. Extracellular DNA required for bacterial biofilm formation. Science. 2002;295:1487. doi: 10.1126/science.295.5559.1487. [DOI] [PubMed] [Google Scholar]
- Wolfaardt GM, Lawrence JR, Korber DR. Function of EPS. In: Wingender J, Neu TR, Flemming H-C., editors. Microbial Extracellular Polymeric Substances: Characterization, Structure and Function. New York: Springer-Verlag; 1999. pp. 171–200. [Google Scholar]
- Wood SR, Kirkham J, Marsh PD, et al. Architecture of intact natural human plaque biofilms studied by confocal laser scanning microscopy. J Dent Res. 2000;79:21–7. doi: 10.1177/00220345000790010201. [DOI] [PubMed] [Google Scholar]
- Yawata Y, Toda K, Setoyama E, et al. Monitoring biofilm development in a microfluidic device using modified confocal reflection microscopy. J Biosci Bioeng. 2010;110:377–80. doi: 10.1016/j.jbiosc.2010.04.002. [DOI] [PubMed] [Google Scholar]
- Zhao K, Tseng BS, Beckerman B, et al. Psl trails guide exploration and microcolony formation in Pseudomonas aeruginosa biofilms. Nature. 2013;497:388–91. doi: 10.1038/nature12155. [DOI] [PMC free article] [PubMed] [Google Scholar]