Abstract
BACKGROUND
Enhanced function of dihydropyridine-sensitive Ca2+ channels (CaV) in hypertensive arterial myocytes (HAM) is well accepted. Increased protein expression of pore forming α1-subunits contributes to this effect, but cannot explain all of the differences in CaV properties in HAM. We hypothesized that differences in expression of CaV subunits and/or their splice variants also contribute.
METHODS
RNA, protein, and myocytes were isolated from small mesenteric arteries (SMA) of 20-week-old male WKY and SHR and analyzed by polymerase chain reaction (PCR), sequencing, immunoblotting, and patch clamp methods.
RESULTS
Cav1.2 α1, β2c, and α2δ1d were the dominant subunits expressed in both WKY and SHR with a smaller amount of β3a. Real-time PCR indicated that the mRNA abundance of β3a and α2δ1 but not total Cav1.2 α1 or β2c were significantly larger in SHR. Analysis of alternative splicing of Cav1.2 α1 showed no differences in abundance of mutually exclusive exons1b, 8, 21 and 32 or alternative exons33 and 45. However, inclusion of exon9* was higher and a 73 nucleotide (nt) deletion in exon15 (exon15Δ73) was lower in SHR. Immunoblot analysis showed higher protein levels of Cav1.2 α1 (1.61±0.05), β3 (1.80±0.32), and α2δ1 (1.80±0.24) but not β2 in SHR.
CONCLUSIONS
The lower abundance of exon15Δ73 transcripts in SHR results in a larger fraction of total Cav1.2 mRNA coding for full-length CaV protein, and the higher abundance of exon9* transcripts and CaVβ3a protein likely contribute to differences in gating and kinetics of CaV currents in SHR. Functional studies of Ca2+ currents in native SMA myocytes and HEK cells transiently transfected with CaV subunits support these conclusions.
Keywords: alternative splicing, blood pressure, Ca2+, channel subunits, gene expression, hypertension, hypertensive rats, protein abundance, small mesenteric arteries, voltage gated Ca2+, channels.
Structural and functional remodeling of arteries is a universally accepted response to chronic hypertension.1 This remodeling sustains and ultimately enhances hypertension leading to the well-described sequelae that contribute to the morbidity and mortality of this disease.2 An important component of this remodeling is increased functional activity of dihydropyridine-sensitive, voltage-gated Ca2+ channels (CaV).3,4 Multiple mechanisms likely contribute to this effect including depolarized membrane potential and larger Ca2+ currents with shifted voltage dependence of activation (about −5 mV) in arterial smooth muscle cells (ASMC).5,6 Notably, differences have also been reported in Ca2+ channel kinetics between normal and hypertensive ASMC.7
CaV are multisubunit complexes composed of pore forming α1 along with accessory β and α2δ subunits.8 Multiple genes code for CaV subunits many of which are subject to alternative splicing that significantly affect channel properties including antagonist sensitivity, gating, kinetics, and regulation.8 The differences in gating and kinetics of CaV cited above suggest that the enhanced activity in hypertension is not simply due to more of the same combination of CaV subunits. Accordingly, this study was performed to test the hypothesis that differences in the expression profile of CaV subunits and/or their splice variants contribute to the enhanced functional activity of CaV in hypertensive ASMC.
METHODS
Animals and tissues
About 20–22 week-old male WKY and SHR (Charles River Labs., Wilmington MA) were used in these studies. Animal protocols were approved by the Lankenau Institute for Medical Research Animal Care and Use Committee. Studies were conducted in accordance with “The Guide for the Care and Use of Laboratory Animals.”
RNA isolation and RT-PCR
ASMCs were dispersed from small mesenteric arteries (SMA) by papain and collagenase treatment from which total RNA was prepared using Tri reagent (RiboPure, Ambion; Austin, TX).9 Total RNA was also prepared from the left ventricle free wall using Tri reagent, and from brain (WKY and SHR) by homogenization in guanidinium isothiocynate.9 First-strand cDNA synthesis and conventional PCR (cPCR) were performed as described.9
Quantitative PCR
Quantitative PCR (qPCR) was performed in triplicate using an Eppendorf ep realplex (Hamburg GR) with qTaq polymerase (Life Technologies; Grand Island, NY) and gene expression assays (Applied Biosystems; Foster City, CA and Operon Labs; Huntsville AL) as described.9 The abundance of potential reference genes was compared in WKY and SHR. As expression of β-actin, Rsp5, and Tpt1 was similar (Supplementary Figure S1) the most abundant (β-actin) was used for normalization.
Sequencing
To analyze exon content of transcripts, target sequences were amplified by PCR using Advantage HF2 polymerase (Clontech; Mountain View, CA), gel purified, A-tailed, cloned into a TOPO TA vector (pCR2.1, Life Technologies), and used to transform competent TOP10 cells (Life Technologies).9,10 Colonies were selected, cultured overnight, used for plasmid DNA isolation (Qiagen Miniprep; Valencia, CA)9,10 and sequenced on both strands at the Kimmel Cancer Center of Thomas Jefferson University.
Colony PCR
Single colonies from plates prepared as above were used as template for PCR with primers that spanned multiple exons. PCR reactions were separated on agarose gels, stained with ethidium bromide, visualized with a MultiFluor S imager and analyzed by Quantity One software (Bio-Rad; Hercules, CA).9,10
Protein analysis
Proteins were isolated from tissue by homogenization in RIPA buffer (Sigma Chemical, St. Louis, MO) with protease inhibitors (Complete mini, Roche Diagnostics; Indianapolis, IN) and analyzed by Western blot as described.9,10
Transfections
Expression constructs for Cav1.2 α1, β2, and α2δ1 in pcDNA3 were kindly provided by Dr Richard Swanson (Merck Research Laboratories; West Point, PA). A construct for Cavβ3 was generated by PCR from SMA RNA using Advantage HF2 polymerase (Clontech Laboratories; Mountain View, CA) and ligated into pcDNA3.1-Directional-V5/His-TOPO (ThermoFisher Scientific). All constructs were sequenced completely on both strands to confirm their accuracy. The Cav1.2 α1 construct used was similar to the smooth muscle variant that includes exons 1b, 8, 15 complete, 21, 32, and 33, but excludes exons 9* and 45.11,12 HEK293 cells (ATCC; Manassas, VA) were transfected with the CaV subunit (1:2:2 molar ratio) and pEGFP plasmids (Clontech) using FuGENE HD (Roche Diagnostics, Basel, Switzerland) and maintained in culture until used 48–72 hours later.
Electrophysiology
HEK cells were placed in a chamber on an inverted microscope (Diaphot, Nikon Instruments; Melville, NY) and superfused with a solution containing (mmol/l) 140 NaCl, 5 KCl, 1 MgCl2, 2 CaCl2, 10 HEPES, and 10 glucose (pH 7.4 with NaOH). Ca2+ currents (I Ca) were recorded at room temperature (~21 °C) in the whole cell configuration using an Axopatch 200B amplifier controlled by pCLAMP 10.2 software (Molecular Devices, Sunnydale, CA). Micropipettes (WPI; Sarasota, FL) had resistances of 1.5–2.5 MΩ when filled with internal solution containing (in mmol/l) 100 CsCl, 20 TEACl, 5 NaCl, 1 MgCl, 5 Na2ATP, 20 HEPES, 10 BAPTA 0.01 cAMP, and 0.1 NaGTP (pH 7.3 with CsOH). Pipette offset and stray capacitance as well as whole cell capacitance and series resistance were compensated optimally, and series resistance prediction and correction were adjusted to 85% or higher. Voltage and current data were recorded on a computer for offline analysis using Clampfit 10.2 and SigmaPlot 12.0 (Systat, Chicago, IL) software.
Statistics
Statistical analyses were performed with SigmaPlot v12.0 (Systat Software, San Jose, CA). Values of from real-time PCR analysis (Figures 1 and 2) and CaV subunit protein abundance (Figure 4) for SHR/WKY were tested for statistical significance from unity using the one-sample t-test by first subtracting one from the ratio and comparing the result to zero. The Fisher exact test was used to compare the abundance of splice variants (fractions) between WKY and SHR in Figure 3A, whereas the χ2-test was used with the data in Figure 3C. Comparisons of all other data for WKY vs. SHR were made with the unpaired t-test or with the Mann–Whitney Rank Sum Test if data were identified as unsuitable for the t-test by SigmaPlot.
Figure 1.
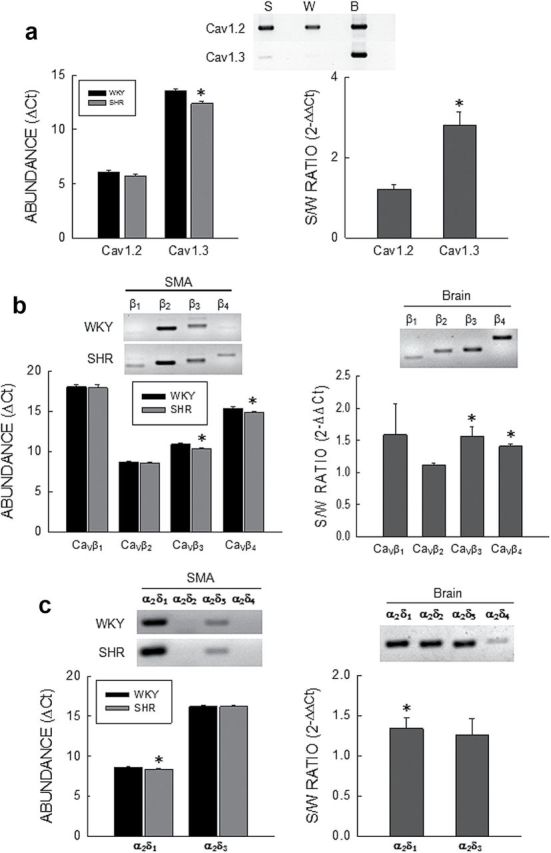
Gene expression of CaV subunits in small mesenteric arteries (SMA). (a) CaV α1 subunits. Top: EtBr stained gel of cPCR products from SHR (S), WKY (W), and brain (B). Left: Quantitative PCR results represented as ΔC t values relative to β-actin. Right: Ratio of abundance of Cav1.2 and Cav1.3 in SHR to WKY represented as . n = 9 for both WKY and SHR. (b) CaV β subunits. Top: EtBr stained gel of PCR products for β1–β4 subunits in SMA from SHR and WKY, and brain. Left: Quantitative PCR results represented as ΔC t values relative to β-actin. Right: Ratio of abundance of β1–β4 in SHR to WKY represented as . n = 4 for both WKY and SHR. (c) CaV α2δ subunits. Top: EtBr stained gel of PCR products for α2δ1–α2δ4 subunits in SMA from SHR and WKY, and brain. Left: Quantitative PCR results represented as ΔC t values relative to β-actin. Right: Ratio of abundance of α2δ1 and α2δ3 in SHR to WKY represented as . n = 4 for both WKY and SHR. Bars and vertical lines represent the mean and 1 SEM, respectively. The star (*) indicates statistically significant differences between WKY and SHR for ΔC t analysis (P < 0.05, unpaired t-test) and statistically significant differences from unity for analysis (P < 0.05, one sample t-test).
Figure 2.
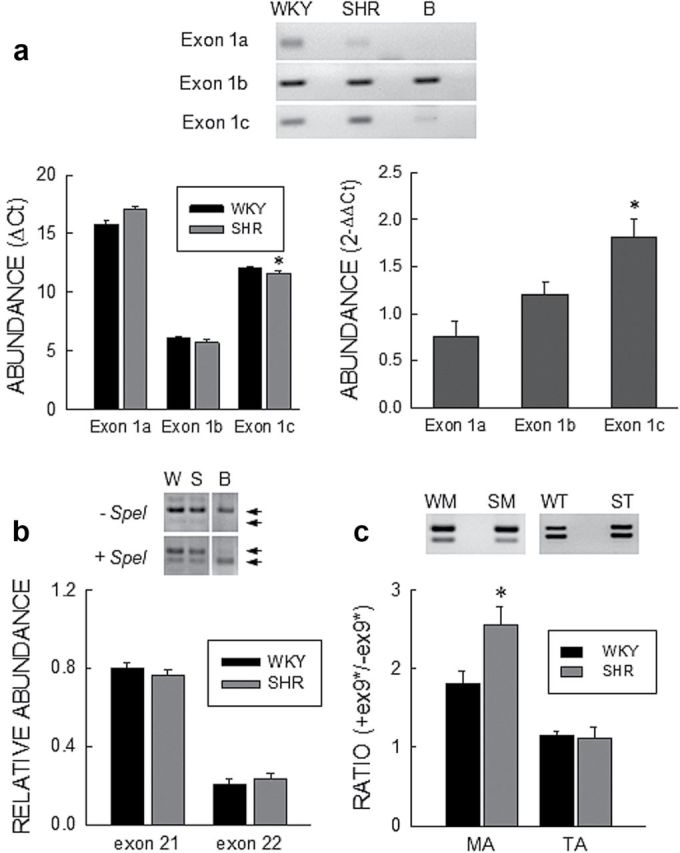
Expression of Cav1.2 splice variants. (a) Exon1 variants. Top: EtBr stained gel of PCR products for Cav1.2 exon1a, 1b, and 1c in SMA from SHR and WKY, and brain. (b) Left: Quantitative PCR results represented as ΔC t values relative to β-actin. Right: Ratio of abundance of Cav1.2 exon1a, 1b, and 1c in SHR to WKY represented as . n = 7 for both WKY and SHR. (b) Exon21/22. Top: EtBr stained gels of PCR products spanning exons21 and 22 with or without treatment with SpeI for WKY (W) and SHR (S), and brain (B). Bottom: Densitometry analysis of gels for exon21 (SpeI-insensitive) and exon22 abundance (SpeI-sensitive minus null fraction); n = 8 for both WKY and SHR. (c) Exon 9*. Top: EtBr stained gel of PCR products for Cav1.2 spanning exon9* in SMA (M) and TA (T) from SHR and WKY. Bottom: Densitometry analysis of the ratio of +exon 9* to –exon9* transcript abundance in MA and TA from WKY (WM and WT) and SHR (SM and ST); n = 8 for WM, SM, WT, and ST. Bars and vertical lines represent the mean and 1 SEM, respectively. The star (*) indicates statistically different between WKY and SHR (P < 0.05). ΔC t data in panel a and data in panel c were compared using the unpaired t-test, while values from the analysis were compared by the one-sample t-Test.
Figure 4.
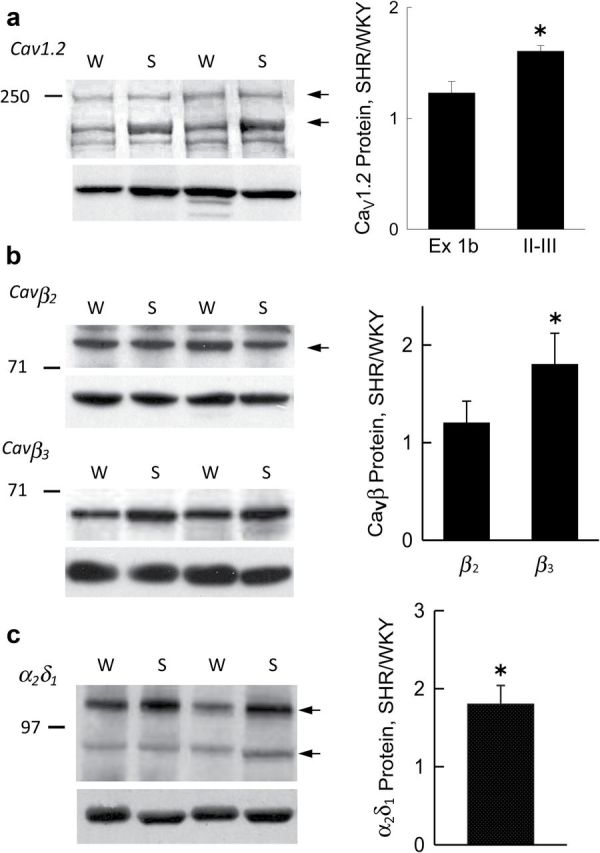
CaV subunit protein expression. Left: Western blot of protein lysates from WKY (W) and SHR (S) probed with antibodies to (a) the II-III intracellular loop of Cav1.2 α1 (top) and β-actin (bottom); (b) CaVβ2 and CaVβ3 (top), and β-actin (bottom); and (c) CaVα2δ1 (top) and β-actin (bottom). Right: Graphs summarizing CaV subunit protein expression determined by densitometry normalized to β-actin in SHR vs. WKY for (A) Cav1.2 α1 with exon1b (n = 5) and II-III loop (n = 10) antibodies; (B) CaVβ2 (n = 8) and CaVβ3 (n = 8), and (c) CaVα2δ1 (n = 8). Bars and vertical lines represent mean and 1 SEM, respectively, and star (*) indicates values statistically different from unity (P < 0.05, one sample t-test).
Figure 3.
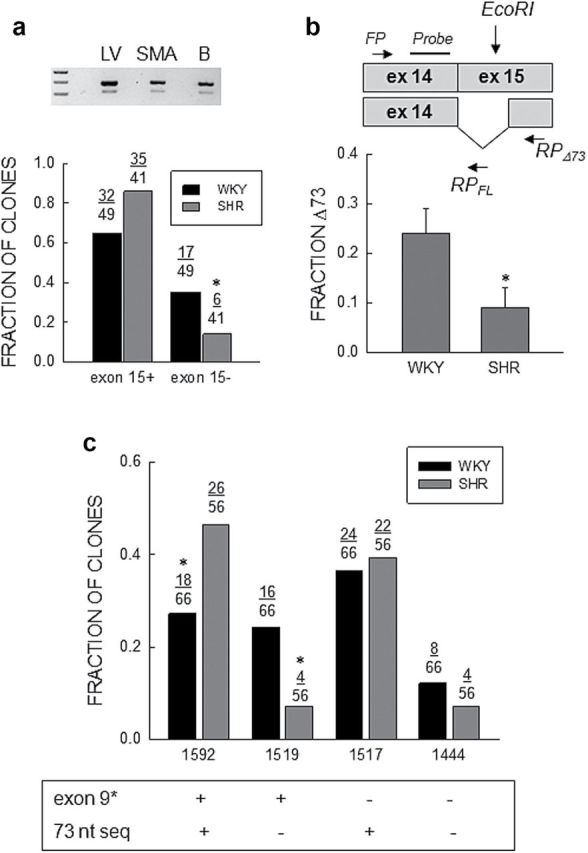
Abundance of Cav1.2Δ73 transcripts. (a) Fraction of clones. Top: EtBr stained gel of PCR products for Cav1.2 spanning exon15 from left ventricle, SMA, and brain. (b) Bottom: Fraction of full length (exon15+) or truncated (exon15−) clones relative to the total number in WKY (n = 49) and SHR (n = 41) SMA analyzed by sequencing. The star (*) indicates a statistically significant difference between WKY and SHR (Fisher exact test; P = 0.05). (b) Quantitative PCR. Top: Schematic representation of the location of the forward (FP) and reverse (RP) qPCR primers, and the probe for the 73 nucleotide (nt) deletion (Δ73) in exon15 plus full length transcripts with the location of the EcoRI site in exon15. Bottom: Analysis of abundance of the 73 nt deletion shown as the ratio of deleted to total Cav1.2 transcripts (Δ73/(Δ73 + FL)). Bars and vertical lines represent the mean and 1 SEM (n = 7), respectively, and star (*) indicates values statistically different from unity (unpaired t-test; P < 0.05). (c) Combinatorial analysis of expression of exons9* and exon15Δ73. The fraction of clones with and/or without exon9* and/or exon15Δ73 are shown for WKY and SHR. The “FL or +” indicate full length clones and the “−” clones with the 73 nt deletion. The star (*) indicates statistically significant difference between WKY and SHR (χ2-test; P = 0.03 for 1592 and P = 0.01 for 1519).
RESULTS
Basic animal characteristics
Systolic blood pressure measured by indirect tail cuff methods was significantly higher is SHR compared with WKY (Table 1). There were no significant differences in body weight while total heart weight and heart weight to body weight ratio were significantly higher in SHR (Table 1).
Table 1.
General animal characteristics
| Group | N | BW (g) | HW (g) | HW/BW (mg/g) | SBP (mm Hg) |
|---|---|---|---|---|---|
| WKY | 21 | 418±11 | 1.33±0.04 | 3.17±0.03 | 127±4 |
| SHR | 21 | 422±8 | 1.65±0.04* | 3.91±0.10* | 181±4* |
Abbreviations: n = number of animals; BW = body weight in g; HW = heart weight in g; HW/BW = ratio of heart weight to body weight in mg/g; SBP = indirect systolic blood pressure in mm Hg;
*P < 0.05 (unpaired t-test).
CaV subunit expression
Ca2+ currents recorded from SMA myocytes were previously shown to be completely inhibited by 10 µmol/l nifedipine suggesting the presence of only Cav1 α1-subunits.6 cPCR showed high levels of Cav1.2 and trace levels of Cav1.3 expression in mRNA from WKY and SHR myocytes (Figure 1A, top). No expression of Cav1.1 or Cav1.4 was found. qPCR showed no significant differences in gene expression for Cav1.2 between WKY and SHR by ΔC t (unpaired t-test) and analysis (SHR/WKY = 1.18±0.09; n = 9, one sample t-test), whereas gene expression of Cav1.3 was significantly higher in SHR by both ΔC t and analysis (SHR/WKY = 2.81±0.33; n = 9, one sample t-test) (Figure 1A).
Using primers common to all splice variants of individual CaVβs, cPCR showed gene expression of all four β subunits in WKY and SHR mRNA with high levels of β2 and β3, low levels of β4, and trace levels of β1 (Figure 1B). This profile was significantly different from brain mRNA expression where cPCR showed expression of CaVβs in the following abundance order: β4>β3>β2>β1. qPCR by both ΔC t (unpaired t-test) and analysis showed that the mRNA abundance of β3 (1.56±0.15; n = 4, one sample t-test) and β4 (1.41±0.04; n = 4, one sample t-test) were significantly higher in SHR but not β1 or β2 (Figure 1B).
cPCR showed gene expression of α2δ1 and α2δ3 at what appeared to be high and low levels, respectively, with no apparent expression of either α2δ2 or α2δ4 in either WKY or SHR mRNA (Figure 1C), which was significantly different from the brain expression profile. qPCR (ΔC t and analysis) confirmed this and showed that α2δ1 represented about 98% of the total in both WKY and SHR (Figure 1C). mRNA abundance of α2δ1 (1.34±0.13; n = 4, one sample t-test) but not α2δ3 was significantly higher in SHR (Figure 1C).
CaV α1 subunit splice variants
To determine splice variant gene expression profile in SMA, the entire open reading frame of Cav1.2 α1 was amplified by cPCR using multiple RNA preparations of both WKY and SHR. Assembled sequences from multiple clones (Sequencer v4.8, Gene Codes Corp., Ann Arbor, MI) showed the presence of four mutually exclusive and four alternative exons.
For the mutually exclusive exons, we found gene expression of all three exon1 variants in SMA mRNA (Figure 2A). However, qPCR (ΔC t) showed exon1b to be the most abundant with expression of exon1c at low and exon1a at trace levels (Figure 2A). The ratio of abundance of exon1b to exon1c was about 60:1, whereas the ratio of exon1b to exon1a was more than 1000:1 for both WKY and SHR. qPCR ( analysis) showed that mRNA abundance of exon1c (1.81±0.19; n = 7, one sample t-test) but not exon1b (1.21±0.12; n = 7 in one sample t-test) or exon1a (0.76±0.16; n = 7 in one sample t-test) was significantly different between SHR and WKY (Figure 2A).
For exons8/8a, analysis of multiple clones showed that exon8 represented the majority of the total clones with no difference between WKY (94% ± 2%, n = 8) and SHR (93% ± 2%, n = 8). For exons21/22,13 cPCR analysis showed a small fraction of transcripts to be null for both which was not different between WKY (4% ± 1%, n = 8) and SHR (3% ± 1%, n = 8). To distinguish between exon21 and 22, we took advantage of the presence of a SpeI restriction site in exon22. SpeI insensitive clones (exon21) analyzed by densitometry represented the majority with a similar abundance in WKY (80% ± 3%, n = 8) and SHR SMA (76% ± 3%, n = 8) (Figure 2B). For comparison, the majority of clones in brain contained exon22 (Figure 2B). Analysis of multiple clones for exons31/32, showed that exon31 was present in 100% of clones for both WKY and SHR.
Of the variable exons in Cav1.2 α1, the abundance of transcripts containing exon9* was significantly higher than those lacking it in both WKY (n = 8) and SHR (n = 8) SMA (Figure 2C) with a significantly larger ratio in SHR. A similar analysis using RNA from tail artery myocytes showed a lower abundance of exon9* containing transcripts compared to SMA with no significant difference between WKY (n = 8) and SHR (n = 8).
Sequencing cDNAs that included Domain II revealed clones with a 73 nt deletion at the proximal end of exon15 that were less frequent in SHR (Figure 3A). To quantitate the abundance of transcripts with and without the 73 nt deletion (exon15Δ73), we took advantage of an EcoRI restriction site within the 73 nt sequence (Figure 3B). We first amplified a fragment (205 nt) of Cav1.2 α1 that completely spanned exon15 with 10 cycles of cPCR. We then incubated 5 µl aliquots of this reaction with active or heat inactivated (15 minutes at 65 °C empirically determined) EcoRI at 37 °C for 2 hour in a reaction volume of 10 µl. We then took two 5 µl aliquot of each “digest” for two qPCR reactions with a common forward primer and probe, and two different reverse primers. One was located in exon15 upstream from the EcoRI site for full length transcripts (RPFL) and the other in exon15 downstream from the alternative splice acceptor site used to splice out the 73 nt fragment (RPΔ73) for Δ73 transcripts (Supplementary Figure S2). Results of this analysis confirmed a lower mRNA level of Cav1.2Δ73 in SMA from SHR (n = 7) compared with WKY (n = 7) (Figure 3B).
PCR analysis showed that transcripts including exon33 predominated (about 90%) in both WKY (90.1±1.5, n = 8) and SHR (89.4±2.5, n = 8) with no significant difference between groups. Finally, PCR and sequencing indicated that exon45 was absent in 100% of clones from WKY (n = 8) and SHR (n = 8). Sequences from all reactions were aligned (Sequencher v4.8, Gene Codes Corp., Ann Arbor, MI) to produce a consensus sequence which was similar to one reported for cerebral artery (DQ538522) with the exception of exon1.
Alternative splicing of exons in Cav1.2 has been suggested to be coupled producing preferential exon combinations.11 From the results described above, the Cav1.2 exons differentially expressed in WKY and SHR are between segments IS5 and IIS6.11 This region was amplified by cPCR using RNA from multiple preparations and clones sequenced. Four contigs were found after alignment that were the result of the inclusion or exclusion of exon9* and the proximal 73 nt of exon15 (Figure 3C). Compared to WKY (n = 66 clones), there was a larger fraction of cDNA clones containing both exon9* and full-length exon15, and a smaller number of clones with exon15Δ73 with or without exon9* in SHR (n = 56 clones). No difference in the fraction of clones containing the 73 nt cassette but lacking exon9* was found between the two groups.
CaV accessory subunit splice variants
In addition to α1 subunits, β, and α2δ1 subunits are also alternatively spliced.14,15 Using cPCR and sequencing we found gene expression of the a, b, c and e variants of β2 in the brain, but only β2c (AF394941) was present in mRNA of both WKY and SHR (Supplementary Figure S3).
α2δ1 subunits have been shown to contain regions of alternative splicing in exon19 (15 nt) and exon24 (21 nt).15 cPCR showed exon24 to be absent in both WKY and SHR while gene expression of transcripts with (α2δ1d) and without (α2δ1e) exon19 were present in mRNA from both WKY and SHR. The former appeared to be more abundant as it was present after fewer PCR cycles in both WKY and SHR (Supplementary Figure S3). qPCR primers to quantitate gene expression of these splice variants that met our quality control criteria could not be found due to the short length of exon 19.
Protein expression
Two forms of functional Cav1.2 α1-subunit protein previously identified16 were both expressed at higher levels in SHR compared with WKY in blots probed with a II-III loop antibody but not with an exon1b antibody. The smaller molecular weight form was dominant in both (Figure 4A). The difference in antibodies likely reflects the differences in transcripts containing exon15Δ73 between WKY and SHR, which do not code for full length (loop II-III containing) protein. Expression of CaVβ3 but not CaVβ2 protein was significantly higher in SHR (Figure 4B). For α2δ1 subunits two distinct components were found one of which was glycosylated (Supplementary Figure S4) and represented 80% of total α2δ1 protein in both WKY and SHR. The fully glycosylated α2δ1 protein which likely reflects plasma membrane located subunits15 was significantly higher in SHR (Figure 4C).
Functional effects
To assess the functional significance of differences in CaV mRNA and protein expression between WKY and SHR reported herein, we compared the properties of Ca2+ currents from native SMA myocytes (Figure 5A, C, and E) with those of cells heterologously expressing CaV subunits (Figure 5B, D, E and F). For this purpose, we used data previously published (Figure 5A–D) as well as newly analyzed (Figure 5E) and newly performed experiments (Figure 5F). In a previous study,6 we found that peak Ca2+ current density (I Ca) was larger in SMA myocytes from SHR compared to WKY (Figure 5A). This difference is associated with higher protein levels of FL-Cav1.2 α1, β3, and α2δ1 in SHR, and a lower abundance of transcripts coding for the truncated Cav1.2Δ73 vs. the full-length forms of Cav1.2 α1 reported herein. By comparison we previously reported that increasing expression of Cav1.2Δ73 in ASMC decreases I Ca (Figure 5B) demonstrating an inverse relation between Cav1.2Δ73 expression and peak I Ca.17
Figure 5.
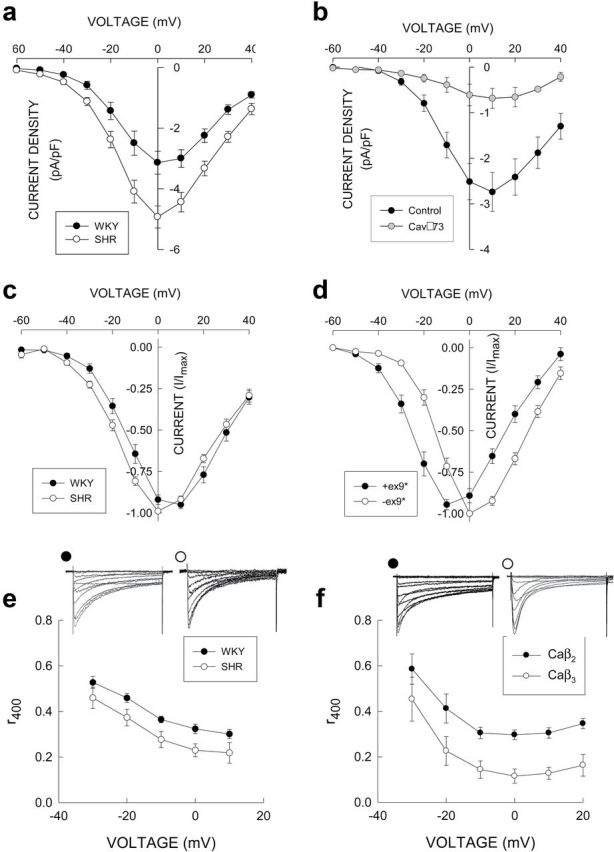
Functional effects of CaV subunit differences. (a) I Ca (pA/pF) was larger in isolated SHR myocytes that have lower expression levels of Cav1.2Δ73 compared to WKY (data were replotted from Figure 3 in ref.6 with permission). (b) Increasing expression of Cav1.2Δ73 decreased I Ca (pA/pF) in ASMC (data reproduced from Figure 5 in ref.17). (c) Voltage dependence of I Ca normalized to the maximum value is shifted in the negative voltage direction in SHR myocytes which have a higher level of expression of exon9* containing transcripts (data were replotted from ref.6 with permission). (d) Voltage dependence of I Ca in HEK cells transfected with Cav1.2 constructs with (77–9*) or without (77-WT) exon9* (data were replotted from Figure 4 in ref.18). (e) I Ca inactivation represented as r400 (current measured 400 milliseconds after the voltage clamp step divided by the peak value) was faster in SHR myocytes that have a higher level of CaVβ3 expression. (f) In transiently transfected HEK cells inactivation was faster (smaller r400) in cells expressing CaVβ3 compared to CaVβ2c along with the same Cav1.2 α1 and α2δ1 constructs. For (e) and (f), families of Ca2+ currents recorded at voltages from −60 to +40 mV in 10 mV, 500-ms-long voltage steps from a holding potential of −80 mV are shown for each condition. For all panels, symbols and vertical lines represent the mean and ±1 SEM, respectively.
In a previous publication, we reported that the voltage dependence of activation of I Ca in SHR myocytes (that have a higher level of expression of exon9*) was shifted approximately −5 mV (Figure 5C) compared to WKY.6 Based upon heterologous expression of CaV subunits in HEK cells, Liao et al. 18 reported that the voltage dependence of activation of Ca2+ channels containing Cav1.2 α1 subunits that included exon9* was shifted approximately −10 mV compared to α1 subunits lacking exon9* (Figure 5D).
This difference in voltage dependence of I Ca current–voltage curves between SHR and WKY produces a similar difference in the voltage dependence of conductance (Supplementary Figure S5A in supplementary results section). Combined with no difference in the voltage dependence of inactivation this difference shifts the voltage dependence of window currents to more negative voltages and increase its peak value (Supplementary Figure S5B in supplementary results section). When the differences in peak Ca2+ current density between WKY and SHR are included an even larger difference exists in window currents especially in the physiological range of voltages (−60 to −30 mV).
In order to assess all inactivation processes19 and to eliminate the subjectiveness of analyzing time constants of I Ca inactivation, the latter was represented as the ratio of Ca2+ current 400ms after the initiation of the voltage clamp step (to 0 mV from a holding potential of −80 mV) to the peak value (i.e., r400). We applied this analysis to unpublished data from experiments previously performed in this laboratory and found that I Ca inactivation was faster (smaller r400) in SHR myocytes (with higher levels of expression of CaVβ3) compared to WKY (Figure 5E). We transiently transfected HEK cells with CaVβ2c (n = 17) or CaVβ3 (n = 11) plus Cav1.2 and α2δ1 and found that CaVβ3 conferred a faster inactivation (lower r400) compared to CaVβ2c (Figure 5F). Values of inactivation rate for ICa at 0 mV in both WKY and SHR myocytes were intermediate between those of HEK cells expressing CaVβ2c and CaVβ3 reflecting the expression of both subunits in native cells. Interestingly, we also found that conductance-voltage curves for Ca2+ currents in HEK cells expressing CaVβ3 were shifted to the right compared to cells expressing CaVβ2c (Supplementary Figure S6A). Combining these results with the differences in CaVβ protein expression between WKY and SHR predicts a small shift in the conductance-voltage curve to the right with a more prominent effect at negative voltages (Supplementary Figure S6B). Since this shift is opposite to that found in native myocytes, it must be concluded that the increase in exon9* containing α1 subunits in SHR is the primary mechanism responsible for the differences in voltage dependence of activation.
DISCUSSION
Increased functional activity of CaV in arteries3,4 and larger Ca2+ current density in isolated ASMC of hypertensive subjects is well documented.6,20,21 We found greater Cav1.2 α1-protein without increased total mRNA in hypertensive arterial myocytes (HAM) in agreement with others,21–23 and a lower abundance of Cav1.2 α1-transcripts containing exon15Δ73 (produced by alternative splicing24) that code for a truncated, nonfunctional subunit.21 Thus, a larger fraction of total α1 transcripts code for full length protein in SHR (~90%) compared with WKY (~70%) which could contribute to larger Cav1.2 α1-subunit protein expression. Indeed, full length Cav1.2 α1 protein (II-III loop containing subunits) was found to be significantly larger in SHR. It is important to note that transcript abundance (total mRNA) and transcript diversity (splice variants) involve distinct processes that can be independently regulated,25 and other factors such as mRNA and/or protein stability could also be involved in the enhanced Cav1.2 α1 protein expression in HAM.
Notably, both CaVβ3a and α2δ1 were equivalently higher at both the transcript and protein level in SHR. Both CaVβ3 14 and α2δ1 15 subunits increase assembly of CaV complexes enhancing trafficking to the plasma membrane, and β subunits decrease proteosomal degradation of α1 subunits26 effectively increasing α1 protein half life. Also, both accessory subunits have been shown to be involved in the increased functional activity of CaV in hypertensive ASMC.20,21 The finding that protein expression of all three CaV subunits is higher in hypertensive SMA is consistent with the suggestion that cell surface expression of α1/β/α2δ channel complexes may be regulated by cotranslation and ER assembly mechanisms.27
Heterologous expression studies have shown that expression of CaVβ3 with α1 and α2δ1 subunits produces a faster rate of inactivation of Ca2+ currents compared to expression with CaVβ2, whereas coexpression of a mixture of CaVβ2 and CaVβ3 produces an intermediate rate of inactivation.28 These findings were confirmed in this study. In addition, the rate of inactivation of CaV in mice following CaVβ3 deletion is slower than wild type mice.28 This suggests that the higher level of CaVβ3 protein in SHR is likely responsible for the faster rate of CaV current inactivation in SHR.7 It should be noted that differences do exist in the voltage dependence of r400 between native myocytes (Figure 5E) and transfected HEK cells (Figure 5F). This is likely due to differences in other inactivating currents such as Ca2+ sensitive Cl− currents between the cell types. Notably, at 0 mV where Cl− currents are expected to negligible due to the symmetrical Cl− conditions of the patch clamp experiments there is a close correspondence with the expected results. It is also interesting that protein expression of CaVβ2 is not different between WKY and SHR and is also not altered with CaVβ3 deletion in the mouse.28 This suggests that regulation of CaVβ3 protein expression is independent of CaVβ2 and linked to blood pressure in some manner, and that CaVβ2 and CaVβ3 protein expression are not reciprocally regulated.28
Along with a smaller abundance of Cav1.2 α1 subunit transcripts with exon15Δ73, we found a larger abundance of exon9* in SHR myocytes. Furthermore, combinatorial analysis (Figure 3C) showed that the fraction of exon9* transcripts that also contained exon15Δ73 (−/+) was larger in WKY (24%) than in SHR (7%). Since exon15Δ73 containing transcripts are translated into a protein truncated in the IIS6 region and shown to be nonfunctional,17 about 47% of exon9* containing transcripts in WKY are predicted not to code for functional channels compared to only 13% in SHR. Inclusion of exon9* in 100% of the channels in heterologously transfected cells produces a −10 mV shift in the voltage dependence of activation in exon1b containing Cav1.2 channels.18 The difference in the V 0.5 of ICa conductance between WKY and SHR6 is expected to be smaller than this value as less than 100% of the channels contain exon9*.
Although the general pattern of exon use found in SMA of both WKY and SHR was similar to most previous studies,12 one interesting difference that did exist was in exon1.12 Of the three transcript variants of exon1, exon1a is highly expressed in cardiac myocytes.11 Of the other two, transcripts containing the newly identified exon1c predominate in cerebral arteries12 while those with exon1b predominate in SMA. These findings support the concept of regional regulation of CaV mRNA alternative splicing, and expression.
The lower levels and lack of a difference in exon9* abundance between WKY and SHR in RNA derived from TA myocytes compared to SMA is of interest. The TA is primarily involved in temperature regulation in the rat and in a normal laboratory environment the tail vasculature is relatively vasoconstricted which would be expected to lower transmural pressure in that area. This finding suggests that local factors not circulating blood borne agents are involved in this difference. It could be speculated that the lower abundance of exon9* in TA myocytes is related to the lower arterial pressure in the tail vasculature. Alternatively, the TA is highly innervated with neurotransmitters possibly playing a role in regulating the abundance of exon9*. This suggests that blood pressure or sympathetic nerve activity or their surrogates could be part of a signaling pathway that regulates exon9* inclusion.29
Two lines of evidence suggest that the differences in CaV gene and protein expression reported herein are related to blood pressure rather than being differences between two genetically distinct animal groups. First, we showed that systolic blood pressure and peak CaV current density (pA/pF) in SMA from both WKY and SHR determined in animals of different ages (6, 12, and 20 months) fall on the same linear regression curve.6 Furthermore, chronic treatment of both WKY and SHR with ramipril decreases both systolic blood pressure and CaV current density to points on the same curve.30 This suggests that a deterministic relation exists between blood pressure and CaV current in WKY and SHR SMA. Second, experimental hypertension produced by abdominal aorta constriction between the origins of the renal arteries produced an increase in Cav1.2 α1-subunit protein and Ca2+ current but not mRNA (gene) expression in the “hypertensive” compared with the “normotensive” renal artery.31 Also, experimental hypertension produced by angiotensin II infusion produced an increase in CaVβ3 protein expression and peak Ca2+ current.21 Thus, in models of experimental hypertension similar changes in CaV subunit gene and protein expression and peak CaV currents occur as in the SHR. Together, these results suggest that the differences in CaV subunit gene and protein expression including splice variants reported herein represent an effect related to blood pressure or some surrogate as previously suggested21,31 but cannot exclude a contribution from genetic differences.
SUMMARY
The increased functional activity of CaV in HAM is associated with increased Ca2+ currents with altered gating and kinetics.6,7 The former is associated with an increased number of CaV channels, and the latter suggests differences in the molecular composition of CaV complexes. This study reports for the first time a detailed study of gene and protein expression of CaV subunits and their splice variants in SMA myocytes of mature SHR and WKY. The lower abundance of exon15Δ73 containing Cav1.2 transcripts plus the higher abundance of Cavβ3 and α2δ1 protein likely contribute to the greater functional activity of Cav1.2 channels in SHR. In SHR, the higher abundance of exon9* in full length Cav1.2 transcripts likely contributes to the difference in voltage dependence of CaV activation, and the higher expression of CaVβ3 protein to the faster current inactivation. Finally, since Cav1.2Δ73 is produced by alternative splicing24 and alternative splicing is a regulated process,25 it is possible that the mechanisms that regulate the production of Cav1.2Δ73 are themselves regulated.32 It is speculated that such processes could act to fine tune expression of Cav1.2 α1 protein in arterial myocytes.33 Further studies are required to test this hypothesis, to determine the mechanisms involved in this process, and to determine their significance.
SUPPLEMENTARY MATERIALS
Supplementary materials are available at American Journal of Hypertension (http://ajh.oxfordjournals.org).
FUNDING
This work was supported by National Institutes of Health grant HL28476 (to R.H.C), American Heart Association grant 0350084N (to R.H.C.), and W. W. Smith Charitable Trust grant H0604 (to R.H.C.).
DISCLOSURE
The authors declared no conflict of interest.
Supplementary Material
REFERENCES
- 1. Sonoyama K, Greenstein A, Price A, Khavandi K, Heagerty T. Vascular remodeling: implications for small artery function and target organ damage. Ther Adv Cardiovasc Dis 2007; 1:129–137. [DOI] [PubMed] [Google Scholar]
- 2. Drazner MH. The progression of hypertensive heart disease. Circulation 2011; 123:327–334. [DOI] [PubMed] [Google Scholar]
- 3. Cox RH, Rusch NJ. New expression profiles of voltage-gated ion channels in arteries exposed to high blood pressure. Microcirculation 2002; 9:243–257. [DOI] [PubMed] [Google Scholar]
- 4. Matsuda K, Lozinskaya I, Cox RH. Augmented contributions of voltage-gated Ca2+ channels to contractile responses in spontaneously hypertensive rat mesenteric arteries. Am J Hypertens 1997; 10:1231–1239. [DOI] [PubMed] [Google Scholar]
- 5. Stekiel WJ. Electrophysiological mechanisms of force development by vascular smooth muscle in hypertension. In Lee RMKW. (ed), Blood Vessel Changes in Hypertension: Structure and Function, Vol 2 CRC Press, Inc: Boca Raton, FL, 1989, pp. 127–170. [Google Scholar]
- 6. Lozinskaya IM, Cox RH. Effects of age on Ca2+ currents in small mesenteric artery myocytes from Wistar-Kyoto and spontaneously hypertensive rats. Hypertension 1997; 29:1329–1336. [DOI] [PubMed] [Google Scholar]
- 7. Cox RH, Lozinskaya IM. Ca2+ channel inactivation in small mesenteric arteries of WKY and SHR. Am J Hypertens 2008; 21:406–412. [DOI] [PubMed] [Google Scholar]
- 8. Varadi G, Mori Y, Mikala G, Schwartz A. Molecular determinants of Ca2+ channel function and drug action. Trends Pharmacol Sci 1995; 16:43–49. [DOI] [PubMed] [Google Scholar]
- 9. Cox RH, Fromme SJ, Folander KL, Swanson RJ. Voltage gated K+ channel expression in arteries of Wistar-Kyoto and spontaneously hypertensive rats. Am J Hypertens 2008; 21:213–218. [DOI] [PubMed] [Google Scholar]
- 10. Cox RH, Folander K, Swanson R. Differential expression of voltage-gated K(+) channel genes in arteries from spontaneously hypertensive and Wistar-Kyoto rats. Hypertension 2001; 37:1315–1322. [DOI] [PubMed] [Google Scholar]
- 11. Tang ZZ, Hong X, Wang J, Soong TW. Signature combinatorial splicing profiles of rat cardiac- and smooth-muscle Cav1.2 channels with distinct biophysical properties. Cell Calcium 2007; 41:417–428. [DOI] [PubMed] [Google Scholar]
- 12. Cheng X, Liu J, Asuncion-Chin M, Blaskova E, Bannister JP, Dopico AM, Jaggar JH. A novel Ca(V)1.2 N terminus expressed in smooth muscle cells of resistance size arteries modifies channel regulation by auxiliary subunits. J Biol Chem 2007; 282:29211–29221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Soldatov NM, Bouron A, Reuter H. Different voltage-dependent inhibition by dihydropyridines of human Ca2+ channel splice variants. J Biol Chem 1995; 270:10540–10543. [DOI] [PubMed] [Google Scholar]
- 14. Birnbaumer L, Qin N, Olcese R, Tareilus E, Platano D, Costantin J, Stefani E. Structures and functions of calcium channel beta subunits. J Bioenerg Biomembr 1998; 30:357–375. [DOI] [PubMed] [Google Scholar]
- 15. Klugbauer N, Marais E, Hofmann F. Calcium channel alpha2delta subunits: differential expression, function, and drug binding. J Bioenerg Biomembr 2003; 35:639–647. [DOI] [PubMed] [Google Scholar]
- 16. Gerhardstein BL, Gao T, Bünemann M, Puri TS, Adair A, Ma H, Hosey MM. Proteolytic processing of the C terminus of the alpha(1C) subunit of L-type calcium channels and the role of a proline-rich domain in membrane tethering of proteolytic fragments. J Biol Chem 2000; 275:8556–8563. [DOI] [PubMed] [Google Scholar]
- 17. Cox RH, Fromme SJ. A naturally occurring truncated Cav1.2 α1-subunit inhibits Ca2+ current in A7r5 cells. Am J Physiol Cell Physiol 2013; 305:C896–C905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liao P, Yu D, Lu S, Tang Z, Liang MC, Zeng S, Lin W, Soong TW. Smooth muscle-selective alternatively spliced exon generates functional variation in Cav1.2 calcium channels. J Biol Chem 2004; 279:50329–50335. [DOI] [PubMed] [Google Scholar]
- 19. Cens T, Restituito S, Galas S, Charnet P. Voltage and calcium use the same molecular determinants to inactivate calcium channels. J Biol Chem 1999; 274:5483–5490. [DOI] [PubMed] [Google Scholar]
- 20. Bannister JP, Bulley S, Narayanan D, Thomas-Gatewood C, Luzny P, Pachuau J, Jaggar JH. Transcriptional upregulation of α2δ-1 elevates arterial smooth muscle cell voltage-dependent Ca2+ channel surface expression and cerebrovascular constriction in genetic hypertension. Hypertension 2012; 60:1006–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kharade SV, Sonkusare SK, Srivastava AK, Thakali KM, Fletcher TW, Rhee SW, Rusch NJ. The β3 subunit contributes to vascular calcium channel upregulation and hypertension in angiotensin II-infused C57BL/6 mice. Hypertension 2013; 61:137–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pratt PF, Bonnet S, Ludwig LM, Bonnet P, Rusch NJ. Upregulation of L-type Ca2+ channels in mesenteric and skeletal arteries of SHR. Hypertension 2002; 40:214–219. [DOI] [PubMed] [Google Scholar]
- 23. Wang WZ, Saada N, Dai B, Pang L, Palade P. Vascular-specific increase in exon 1B-encoded CAV1.2 channels in spontaneously hypertensive rats. Am J Hypertens 2006; 19:823–831. [DOI] [PubMed] [Google Scholar]
- 24. Soldatov NM. Genomic structure of human L-type Ca2+ channel. Genomics 1994; 22:77–87. [DOI] [PubMed] [Google Scholar]
- 25. Moore MJ, Silver PA. Global analysis of mRNA splicing. RNA 2008; 14:197–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li Q, Sarna SK. Chronic stress targets posttranscriptional mechanisms to rapidly upregulate α1C-subunit of Cav1.2b calcium channels in colonic smooth muscle cells. Am J Physiol Gastrointest Liver Physiol 2011; 300:G154–G163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Brust PF, Simerson S, McCue AF, Deal CR, Schoonmaker S, Williams ME, Veliçelebi G, Johnson EC, Harpold MM, Ellis SB. Human neuronal voltage-dependent calcium channels: studies on subunit structure and role in channel assembly. Neuropharmacology 1993; 32:1089–1102. [DOI] [PubMed] [Google Scholar]
- 28. Murakami M, Yamamura H, Suzuki T, Kang MG, Ohya S, Murakami A, Miyoshi I, Sasano H, Muraki K, Hano T, Kasai N, Nakayama S, Campbell KP, Flockerzi V, Imaizumi Y, Yanagisawa T, Iijima T. Modified cardiovascular L-type channels in mice lacking the voltage-dependent Ca2+ channel beta3 subunit. J Biol Chem 2003; 278:43261–43267. [DOI] [PubMed] [Google Scholar]
- 29. Tang ZZ, Zheng S, Nikolic J, Black DL. Developmental control of CaV1.2 L-type calcium channel splicing by Fox proteins. Mol Cell Biol 2009; 29:4757–4765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cox RH, Lozinskaya I, Matsuda K, Dietz NJ. Ramipril treatment alters Ca(2+) and K(+) channels in small mesenteric arteries from Wistar-Kyoto and spontaneously hypertensive rats. Am J Hypertens 2002; 15:879–890. [PubMed] [Google Scholar]
- 31. Pesic A, Madden JA, Pesic M, Rusch NJ. High blood pressure upregulates arterial L-type Ca2+ channels: is membrane depolarization the signal? Circ Res 2004; 94:e97–e104. [DOI] [PubMed] [Google Scholar]
- 32. Chen M, Manley JL. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat Rev Mol Cell Biol 2009; 10:741–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chiu YH, Alvarez-Baron C, Kim EY, Dryer SE. Dominant-negative regulation of cell surface expression by a pentapeptide motif at the extreme COOH terminus of an Slo1 calcium-activated potassium channel splice variant. Mol Pharmacol 2010; 77:497–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


