Summary
Protamines confer a compact structure to the genome of male gametes. Here, we find that somatic cells can be remodeled by transient expression of protamine 1 (Prm1). Ectopically expressed Prm1 forms scattered foci in the nuclei of fibroblasts, which coalescence into spermatid-like structures, concomitant with a loss of histones and a reprogramming barrier, H3 lysine 9 methylation. Protaminized nuclei injected into enucleated oocytes efficiently underwent protamine to maternal histone TH2B exchange and developed into normal blastocyst stage embryos in vitro. Altogether, our findings present a model to study male-specific chromatin remodeling, which can be exploited for the improvement of somatic cell nuclear transfer.
Graphical Abstract
Highlights
-
•
In vitro protaminization of somatic cell nuclei
-
•
Conversion of interphase somatic nuclei into “spermatid-like” structures
-
•
Protaminization of somatic nuclei that is reversed upon injection into enucleated oocytes
-
•
A simplified model of nuclear remodeling and reprogramming in vitro
Iuso et al. find that exogenous expression of human protamine 1 remodels interphase fibroblast nuclei into spermatid-like structures. The process is reversible upon nuclear transfer into enucleated oocytes. This finding could be exploited as a simplified model for investigating protamine-induced genome compaction and may also boost somatic cell nuclear transfer efficiency.
Introduction
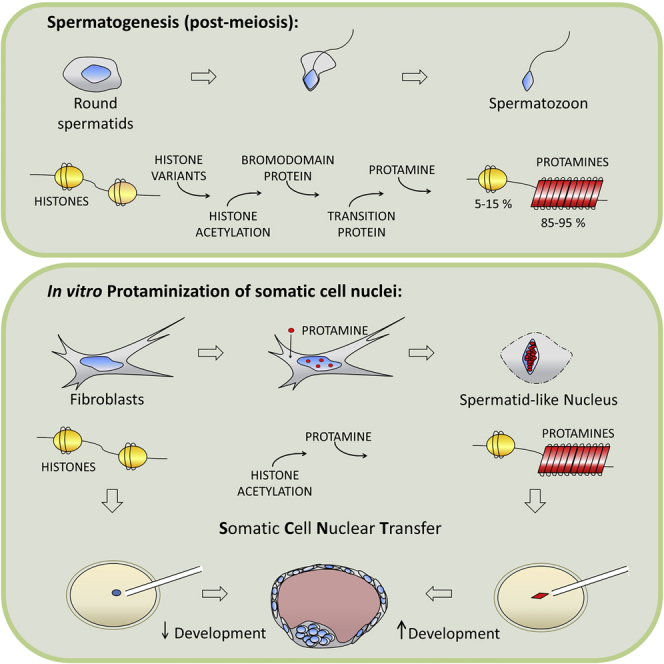
Spermatogenesis is conserved from flies to mammals. Post-meiotic spermatocytes undergo a radical nuclear and cytoplasmic reorganization. Nuclear remodeling relies on a timely translation of stabilized mRNA transcribed at the round spermatid stage (Brock et al., 1980). The first transcripts are testis-specific core and linker histone variants (Gaucher et al., 2010, Kota and Feil, 2010). Testis-specific histones have a lower affinity for DNA than somatic ones (Gaucher et al., 2010), and their subsequent post-translational modification (Goudarzi et al., 2014, Rousseaux and Khochbin, 2015, Morinière et al., 2009) further destabilizes the nucleosome. Subsequently, acetylated histones are recognized by bromodomain testis-specific proteins (Brdt) (Pivot-Pajot et al., 2003, Gaucher et al., 2012), proteins that prepare the ground for the incorporation of transition proteins (TPs). TPs cooperate with topoisomerases to relieve torsional stress (Singh and Rao, 1988) and with DNA repairing enzymes (Akama et al., 1999). The substitution of TPs with protamine (Prm) completes nuclear remodeling, conferring a toroid structure to DNA and the unique shape of male gametes (Miller et al., 2010). Upon fertilization, remodeling is completely reversed. Paternal chromosomes rapidly lose Prm and testis-specific histones (van der Heijden et al., 2005, Wu et al., 2008, Loppin et al., 2005) and regain a nucleosomal organization built upon maternally provided histones.
When, instead, a somatic cell is used to “fertilize” an oocyte, as in somatic cell nuclear transfer (SCNT; Wilmut et al., 1997), the nucleosome organization of the chromatin is only occasionally reversed by the oocyte, often leading to developmental failure (Loi et al., 2013).
The current study set out to explore the possibility of conferring a somatic cell nucleus with a Prm-based toroid organization. Here, we demonstrate that the nuclei of adult somatic cells undergo a dramatic chromatin reorganization following the induced expression of the Prm1 gene, transforming the nuclei into “spermatid-like” structures. The Prm-induced nuclear remodeling is reversible upon nuclear transfer (NT) of protaminized cells into enucleated oocytes, and the resulting embryos develop normally in vitro.
Results
Spermatid-Specific Stepwise Chromatin Remodeling Does Not Function in Somatic Cells
Our first approach to remodel somatic cell nuclei was to induce the co-sequential and sequential expression of four main testis-specific genes in primary cultures of sheep fibroblasts, with the aim to repeat the nuclear remodeling occurring in spermatids. To this end, expression vectors for the bromodomain testis-specific (Brdt-GFP tag), TPs I and II (HA tag), and protamine 1 (Prm1-red variant of GFP tag) were generated and transfected into the fibroblasts. However, our attempts to induce chromatin remodeling of somatic cells following either a co-stepwise or a stepwise transfection of all four expression vectors failed.
The nuclear reorganization in spermatids relies on a hierarchic translation of synthetized mRNA, and each of the protein prepares the ground for the next one (Barckmann et al., 2013), a scenario we failed to repeat in cultured somatic cells. There are, however, exceptions to the canonical nucleosome disassembly pathway during spermatogenesis. In the male germline of the cephalopod Sepia officinalis, for instance, histones are replaced directly by Prm-like proteins (Martínez-Soler et al., 2007). Hence, we attempted to induce a direct nuclear remodeling in somatic cells through the transient expression of Prm. We selected Prm1 because ram spermatozoa contain Prm1 only and because Prm1 is synthetized as a mature protein with no further processing (de Mateo et al., 2011).
Direct Incorporation of Prm1 into Somatic Chromatin
We transfected sheep adult fibroblasts (SAFs) with pPrm1-RFP, WT Prm1, and RFP tag-only vectors using a lipofectamine transfection kit. Transfected cells approached 40% in all three groups. Expression of mRNA and protein Prm1 was confirmed by RT-PCR, by western blotting, and by tracking the RFP-tag (Figures 1, S1A, and S1B). Prm1-RFP co-localized with nuclei stained by Hoechst (Figure 1), whereas in control, i.e., pRFP transfected fibroblasts, the red signal was diffused in the cytoplasm and nucleus (Figure 1). Fibroblasts transfected with WT Prm1 showed nuclear reorganization similar to that detected in pPrm1-RFP transfected ones (Figures 1 and S2). At 40- to 48-hr post-transfection/trichostatin A (TSA) treatment, 93.3% of Prm1-cells did uptake Prm in nuclei, and 83.3% of Prm1-positive cells completely changed nuclear morphology (Table 1).
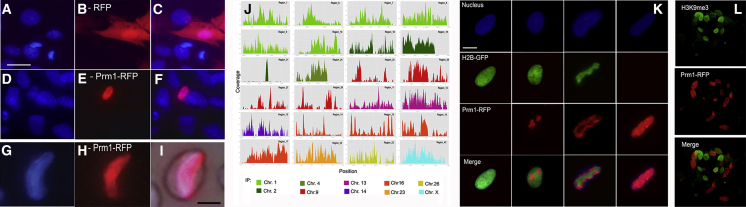
Figure 1.
Ectopic Prm1 Binds to Somatic DNA, Replaces the Histones, and Changes Completely Nuclear Morphology
(A–F) Nuclear incorporation of Prm1 in somatic chromatin. This shows tracking the RFP tag in cells transfected with pTag-RFP (A–C) and pPrm1-RFP (D–F); (A and D) Nuclei. (B) RFP. (E) Prm1-RFP localizations. (C and F) Merge. Scale bar represents 10 μm.
(G–I) Elongated nucleus in pPrm1-RFP-transfected cells. Scale bar represents 10 μm.
(J) ChIP-seq profiles in genomic regions of different chromosomes for pPrm1-RFP in transfected cells (after peak calling the represented genomic regions from pPrm1-RFP sample are enriched than input samples).
(K) Histones/Prm exchange in H2B-GFP fibroblasts: nuclei (blue); H2A.B-GFP (green); Prm1-RFP (red); Merge H2A.B+Prm1. (Left to right) The gradual incorporation (0-, 16-, 24-, 48-hr post-transfection) of Prm1 into somatic nuclei. H3K9m3/Prm exchange (L) was as follows: H3K9me3 (green), Prm1-RFP (red), and Merge H3K9me3+ Prm1.
Table 1.
Incorporation of Prm1 in Somatic Nuclei 16 to 48 hr Post-transfection
| Hours Post-transfection | Group | Prm1 Not in Nucleus | Prm1 Spots in Nucleus | Prm1 in Whole Nucleus |
|---|---|---|---|---|
| 16–20 hr | CTR | 15/147 (10.2%) | 94/147 (63.9%) | 38/147 (25.8 %) |
| TSA | 4/69 (5.8%) | 32/69 (46.3%)a | 33/69 (47.8%)b | |
| G0 | 4/80 (5%) | 52/80 (65%) | 24/80 (30%) | |
| G0 TSA | 2/74 (2.7%) | 40/74 (54%) | 32/74 (43.2%) | |
| 40–48 hr | CTR | 33/191 (17.3%) | 42/191 (21.9%) | 116/191 (60.8%) |
| TSA | 4/60 (6.7%) | 6/60 (10%)c | 50/60 (83.3%)d | |
| G0 | 6/63 (9.5%) | 21/63 (33.3%) | 36/63 (57.14%) | |
| G0 TSA | 6/102 (5.9%) | 21/102 (20.6%) | 75/102 (73.5%)e |
CTR versus TSA, p = 0.0179.
CTR versus TSA, p = 0.0018, Fisher's exact test.
CTR versus TSA p = 0.0399.
CTR versus TSA, p = 0.0010.
G0 versus G0 TSA, p = 0.0401, Fisher's exact test.
Prm1 Binds to DNA
Prm1 contains numerous cysteine residues that might form intra-Prm/inter-Prm disulfide cross-links (Balhorn et al., 1992). To exclude nuclear Prm1 polymerization, chromatin immunoprecipitation (ChIP)-seq assay was carried on of sorted Prm1-positive cells. The ChIP-seq assay confirmed the effective binding of Prm1 to DNA at 42 DNA binding sites on 10 of 27 chromosomes, as early as 16-hr post-pPrm1 transfection (Figure 1J; Table S1).
Protaminization of Somatic Cells’ Nuclei Is Not Cell Cycle Dependent
Cell-cycle analysis by cytofluorimetry of transfected cells revealed that Prm1 was incorporated in all cell-cycle stages (72.6% G1, 4.4% S, 23% G2 stage) (Table S2). Moreover, fibroblasts forced to enter G0 (by serum starvation) were transfected with pPrm1-RFP, and even in this case, Prm1 incorporation and chromatin compaction took place (Table 1). Thus, Prm incorporation on DNA is cell cycle independent. A likely mechanism for Prm1 deposition on chromatin might be the conformational changes that nucleosomes undergo as a result of chromatin remodeling enzymes or thermal fluctuations (Chereji and Morozov, 2015). These nucleosomal dynamics make available free stretches of 11 bp between the dyads, enough for Prm1 docking on DNA (Zhang et al., 1996). The low-off rates of Prm1 (Brewer et al., 2003) might facilitate the prolonged condensation of DNA even after transient Prm1 expression. To indirectly test this hypothesis, we transfected GFP-H2B fibroblasts with Prm1-RFP. A histone-to-Prm exchange was observed in GFP-H2B nuclei 48 hr after Prm1 transfection (Figure 1K). Unequivocally, the red fluorescent signal of Prm replaced the green H2B, confirming the over competition of Prm1 on histones (Figure 1K). Furthermore, we observed that the Prm replaced the histone H3 trimethylated on lysine 9 (H3K9me3), a critical epigenetic barrier of SCNT reprogramming in 30% (36 of 120) of Prm-positive cells (Figure 1L). H3K9me3 was not longer detectable in fully condensed nuclei totally devoid of H3K9me3.
Dynamics of Prm1 Incorporation
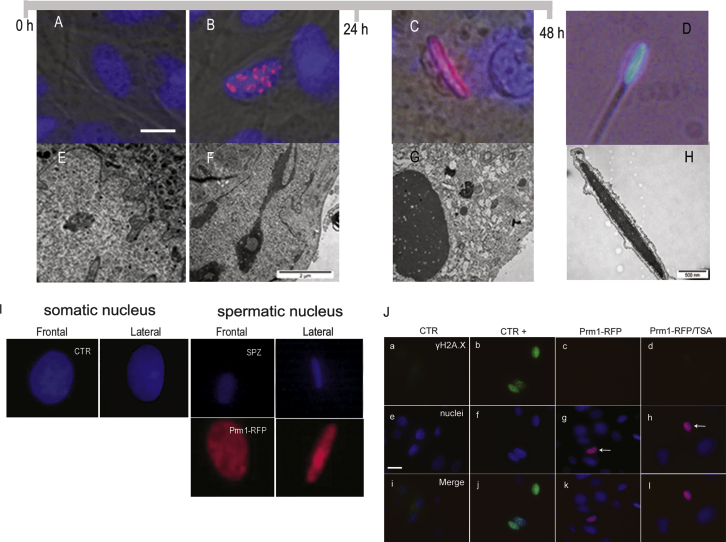
Prm1 assembly on DNA of somatic cells leads to an overlapping of nuclear morphology like the one found in elongating spermatids (Figures 1G–1I). Prm1 appears in foci scattered throughout the nucleus 20-hr post-transfection (Figure 2B) and fully coalescences 48 hr later in an elongated structure (Table 1). The final outcome is an elongated and flattened nucleus, very much similar to the spermatozoa’s, but larger, due to the diploid DNA content (Figure 2I). The degree of nuclear compaction in protaminized somatic cells approaches that of spermatozoa when observed by transmission electron microscopy (TEM) (Figures 2F–2G).
Figure 2.
The Gradual Prm1 Incorporation Leads to Nuclear Compaction Similar to that of Elongated Spermatids without Causing DNA Brakes
(A–C; E–G) Timing of incorporation of Prm in somatic nuclei. This is a representation of post-transfection incorporation of Prm in somatic nuclei. pPrm1-RFP-Prm plasmide is tagged with RFP.
(A) Nuclei (Hoechst, blue) of fibroblasts before the transcription of Prm1.
(B) Incorporation of Prm1 in nucleus 16- to 20-hr post-transfection, visible as red spots.
(C) Complete incorporation of Prm1 in nucleus 40- to 48-hr post-transfection. Prm1 is red, and nuclei are blue.
(E–G) TEM analysis of adult sheep fibroblasts transfected with pPrm1-RFP.
(E) Nucleus of fibroblasts before the transcription of Prm1.
(F) Nucleus of fibroblast after 16- to 20-hr post-transfection. Visible partial compaction of chromatin is seen.
(G) Nucleus of fibroblasts after 40- to 48-hr post-transfection. Visible complete chromatin compaction is seen. Bars represent 8 μm (A) and 2 μm (F).
(D and H) Nucleus of spermatozoa stained with Hoechst (D) and analyzed by TEM (H). Scale bar represents 500 nm.
(I) Representation of nucleus of control (CTR) and Prm-transfected cells (Prm1-RFP) from frontal and lateral view. Nucleus of spermatozoa has been use as a control (SPZ).
(J) DNA double-strand breaks in sheep fibroblasts transfected with pPrm1-RFP visualized by γH2A.X immunostaining. (a, e, and i) Control adult fibroblasts. (b, f, and j) Positive control: adult fibroblasts irradiated with UV light. (c, g, and k) Adult fibroblasts 20-hr post-transfection with pPrm1-RFP (red). (d, h, l) Adult fibroblasts treated with TSA and transfected with pPrm1-RFP. Stained with antibody anti-γH2A.X. Scale bar represents 10 μm.
Histone hyperacetylation opens chromatin structure in spermatids and is a pre-requisite for histone/Prm replacement. Accordingly, transfected cells treated with TSA incorporated more and faster Prm1 than control without TSA (at 16- to 20-hr post-transfection: TSA 47.8% [33 of 69]; without TSA 25.8% [38 of 147], p = 0.0018; at 40–48 hr: TSA 83.3% [50 of 60]; without TSA 60.8% [116 of 191], p = 0.0010; Table 1).
Physiologically, the massive DNA compaction proper of spermatozoa is achieved through the induction of double strand DNA breaks and repair (Rathke et al., 2014). Therefore, we tested whether the same would apply to our forced conversion of somatic chromatin to a protaminized structure. We analyzed via immunofluorescence the nuclei for the presence of the histone variant γH2A.X (a marker of double-strand DNA breaks) around the Prm1 condensed foci, but no positive γH2A.X signal was detected (H2A.X positive: CTR 10.2% [6 of 59]; CTR + [UV irradiated] 63.2% [48 of 76]; Prm1-RFP 10% [6 of 60]; Prm1-RFP/TSA 12.5% [8 of 64]; Figure 2J). A comet assay also excluded the presence of DNA fragmentation in fully protaminized fibroblasts (Figure S3).
Protaminized cells showed signs of degeneration on the third day after transfection, likely a consequence of the global transcription shut down resulting from chromatin compaction (Figure 2C).
Prm-to-Histone Transition after NT
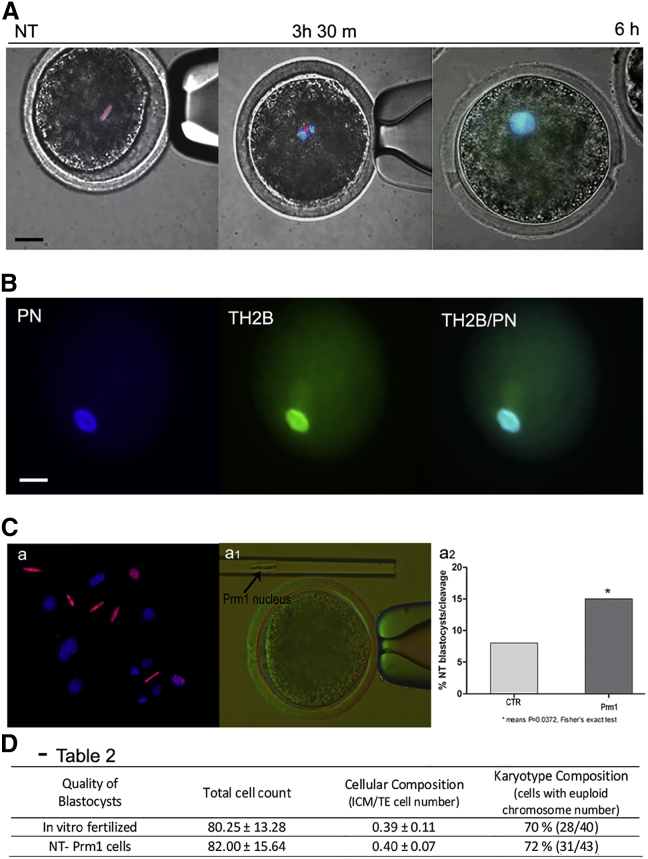
Finally, we wanted to verify the reversibility of genome protaminization using NT as a biological assay (Figure 3C, a and a1). A large pronucleus was observed in the injected oocytes starting 6 hr after activation and swelling in size (16.2 ± 2.3 μm) by 8 hr. Meanwhile, Prm progressively disappeared in the pronuclei of 77% of injected oocytes (28 of 36; Figure 3A) and replaced by TH2B (Figure 3B), an oocyte-specific histone variant that plays a key role in nuclear reprogramming (Shinagawa et al., 2014, Montellier et al., 2013). Hence, protaminized somatic nuclei decondense and reacquire a nucleosomal organization following a physiological path after artificial activation of the injected oocyte.
Figure 3.
Protaminized Somatic Nuclei Re-acquire a Nucleosome Organization after Nuclear Transfer
(A) Displacement Prm1 during pronucleus formation after nuclear transfer (NT). Scale bar represents 20 μm. Prm1 is red, and nucleus is blue.
(B) Incorporation TH2B in pronucleus (PN) after NT of protaminized nuclei: pronucleus (PN, blue), TH2B (green), Merge (TH2B/PN). Scale bar represents 20 μm.
(C) SCNT of fibroblast transfected with pPrm1-RFP. (a) Picture represents Prm1 (red)-positive fibroblasts used as donors for SCNT. (a1) Protaminized nucleus in injected capillary before nuclear transfer into enucleated MII sheep oocyte. (a2) In vitro development of nuclear transfer embryos.
(D) Table 2. Quality of blastocysts is as follows: IVF, in vitro fertilized; NT Prm1 cells. Total cell number is mean ± SD. Cellular composition is the number of inner cell mass/trophectoderm cell number. Karyotype composition is the cell with euploid chromosome number divided by total cells (%).
The enucleated oocytes reconstructed with protaminized somatic nuclei were further cultured in vitro in seven separated replicates, indicating their full competence to direct early embryonic cleavages until the blastocyst stage (Figure 3C, a2). The quality of the embryos (total cell count and karyotype) was comparable to that of normal in vitro fertilized embryos (Figure 3D).
Discussion
Here, we have found that ectopic expression of Prm remodels the interphase chromatin of somatic cells, leading to a nuclear compaction strikingly similar to that of elongated spermatids. The absence of γH2AX immune localization was surprising, for it is hardly conceivable a genome-wide protaminization without torsional stress alleviation through DNA double-strand breaks (Laberge and Boissonneault, 2005). Probably, other DNA repairing enzymes not tested here might be involved; accordingly, γH2AX has not been detected during chromatin remodeling in human spermatids (De Vries et al., 2012). Another possibility is that the histone variant γH2AX might have been evicted by Prm binding; however, a comet assay of fully protaminized cells excluded major DNA damage (Figure S3).
The ChIP-seq dataset demonstrated the effective binding of Prm1 to DNA on 42 DNA binding sites on 10 out of 27 chromosomes (2n = 54 in sheep) (Figure 2J; Table S1). Binding occurred in 28 gene-rich domains—14 (50%) genic and 14 (50%) intergenic—while the other 14 were classified as scaffold (Table S1).
Early nuclear remodeling in round human spermatids is marked by the appearance of a single doughnut-like structure. The doughnut is the morphological expression of nucleosome disassembly, for nucleosome-destabilizing histones (H4K8ac and H4K16ac, H3K9me2) co-localize with it (De Vries et al., 2012). These findings suggest that genome remodeling starts at a defined site in the genome and spreads in a spatially regulated manner, ending up with the nuclear compaction found universally in male gametes. Live imaging of the histone-to-Prm transition could be a powerful tool to test this hypothesis, but it is only available in Drosophila (Awe and Renkawitz-Pohl, 2010), not in mice or man. Also, currently unavailable are in vitro models to monitor the whole process of spermatogenesis.
The protaminization of somatic cells might provide unique insights on the early steps of chromatin remodeling, allowing the identification of the very first genome domain(s) binding with Prm. The simplified model for nucleosome-to-Prm exchange reported here might also add to the ongoing debate on nucleosomal retention in male gametes (Samans et al., 2014, Weiner et al., 2015). It would be interesting to map on a genome-wide scale, the Prm-histones’ footprints on Prm1-expressing somatic cells, to see whether the pattern described in spermatozoa is conserved.
Our findings also impact SCNT (Wilmut et al., 1997). SCNT is a promising technology whose full implementation is on hold because of its low efficiency. Recent successes on human embryonic stem cells isolated from somatic cell-derived cloned embryos have boosted interest in SCNT (Tachibana et al., 2013, Chung et al., 2014). This renewed interest in SCNT goes along with targeted nuclear reprogramming strategies, like RNAi-mediated downregulation of Xist (Ogura et al., 2013, Matoba et al., 2011) or the depletion of H3K9 methyltransferases in somatic cells before NT (Matoba et al., 2014). In vitro protaminization of somatic cells simplifies the nuclear structure of a somatic cell enormously, formatting it in a way that is easily readable by the oocytes and thus holds potential to improve SCNT efficiency.
Experimental Procedures
Cell Culture
SAFs were derived from ear biopsy of three female Sarda breed sheep (2 years old). SAFs (between second and eighth passage) were cultured in DMEM (GIBCO) containing 10% fetal bovine serum (FBS), 2 mM glutamine, 3.7 g/L NaHCO3, and 0.5% gentamicin.
Plasmids Construction
Sperm Prm1 cDNA (GenBank: NM_002761.2) was amplified from a human testis cDNA library with appropriate primes and cloned into a pTagRFP vector (Evrogen). The identity of the cloned cDNA and its in-frame cloning C-terminal to RFP was verified by sequencing. Mouse TP1 and TP2 cDNAs were amplified by PCR from testis total RNA and cloned in a Ha-tagged pcDNA vector (derived from a Life Technologies pcDNA His-tagged vector). The GFP-Brdt construct is described in Pivot-Pajot et al. (2003) and Gaucher et al. (2012).
Transfection
SAFs at 80% confluence were transfected with 3 μg of pBrdt-GFP, pTP1, pTP2, pPrm1-RFP, pPrm1, or pRFP by Lipofectamine 2000 (Invitrogen), according to the manufacturer’s instructions. At 4-hr post-transfection, medium was changed for DMEM containing 5 nM TSA or control (no TSA), and SAFs were cultured for additional 16, 20, 40, 48 hr. For cell-cycle analysis, SAFs before transfection were starved with 0.5% FBS for 5 days.
Histone Exchange Visualization
To visualize histone-to-Prm exchange, SAFs were first transfected with construct expressed GFP fused to histone 2B using BacMam 2.0 Technology (C10594, Celllight Histone 2B-GFP, BacMam 2.0, Life Technologies) according to the manufacturer’s instructions and then with pPrm1-RFP, as described above. Histone 2B-GFP and Prm1-RFP behavior in live cells using fluorescent imaging were observed.
TEM
Cells were fixed in glutaraldehyde (2.5% in 0.1 M cacodylate buffer [pH 7.4]) for 24 hr. After washing in ddH2O, cells were post-fixed in 2% OsO4 in ddH2O for 2 hr. Next, cells were dehydrated through a graded series of ethanol solutions (30%, 10 min; 50%, 15 min; 70%, 24 hr; 80%, 10 min; 96%, 10 min; 100%, 10 min; acetone, twice for 15 min) and were infiltrated with graded concentrations of Epoxy 812 Resin (EPON) resin in 100% acetone (1:3, 20 min; 1:1, 24 hr; 3:1, 2 hr), infused twice for 1 hr in pure EPON resin, and polymerized at 65°C for 24 hr. Next, 60-nm sections were prepared and examined using a LEO 912AB electron microscope. Images were captured using a Slow Scan CCD camera (Proscane) and EsiVision Pro 3.2 software (Soft Imaging Systems GmbH).
Immunostaining
γH2A.X and Trimethyl-HistoneH3
Cells were fixed in 4% paraformaldehyde (PFA) for 15 min, permeabilized with 0.1% (v/v) Triton X-100 in PBS for 30 or 20 min, and blocked (0.1% BSA/0.05% Tween 20 [v/v] in PBS) for 1 hr at room temperature (RT). Then cells were incubated with the anti-phospho-Histone H2A.X (Ser139) (γH2A.X) mouse antibody (1:100) (05-636, Millipore) at RT for 2 hr or anti-Trimethyl-HistoneH3 (Lys9) rabbit antibody (1:200) (07-523, Millipore) at 4°C overnight. Subsequently, the secondary anti-mouse IgG-FITC (1:100) or anti-rabbit IgG-FITC (1:200) was added for 2 hr or 50 min at RT. Nuclei were counterstained with 5 μg/ml of 4′,6-diamidin-2-fenilindolo (DAPI).
ChIP-seq Assay
Chromatin Immunoprecipitation
ChIP assay was performed with the EZ-Magna ChIP kit (Millipore) according to the manufacturer’s instructions with minor modification. Protein and DNA complexes from pPrm1-RFP-transfected cells were cross-linked with 0.5% formaldehyde for 10 min at RT. Sonications were done in nuclear buffer (six 30-s pulses, power setting 10 and six 30-s pulses, power setting 15 in ice with 50-s rest between pulses; Bandelin Sonopuls). Soluble chromatin was immunoprecipitated with anti-RFP antibody (Evrogen) directly conjugated with Magnetic Protein A beads. DNA and protein immune complexes were eluted and reverse cross-linked, and then DNA was extracted using a spin filter column. DNA obtained from pRFP, pPR1-RFP, and Input were subjected to ChIP sequencing.
ChIP Sequencing
Next-generation sequencing experiments, comprising samples quality control, were performed. Indexed libraries were prepared from 10 ng/each ChIP DNA with TruSeq ChIP Sample Prep Kit (Illumina) according to the manufacturer’s instructions (for details see Supplemental Experimental Procedures).
NT
In vitro embryos production was adapted from those previously described (Ptak et al., 2002). Oocytes manipulation was carried out with a piezo-driven enucleation and injection pipette (PiezoXpert), as previously described. Enucleate oocytes were injected with a nucleus, either from CTR or a Prm1-RFP fibroblasts suspended in PBS with 6% polyvinylpyrrolidone (Sigma-Aldrich). Activation and culture of reconstructed oocytes were processed as described (Ptak et al., 2002, Iuso et al., 2013a). Qualities of obtained blastocysts were performed as described by Iuso et al. (2013b).
TH2B Immunostaining on Pronuclear Stage Embryos
The zona pellucida of embryos at pronuclear stage (10- to 12-hr post-activation) was removed by incubation in 0.5% (w/v) pronase and acid Tyrode’s solution for 30 s. Then embryos were fixed in 4% PFA for 15 min and subjected to immunofluorescence analysis as described in Torres-Padilla et al. (2006) with the rabbit anti-TH2B antibody (1:700, ab23913 Abcam). Finally, zygotes were mounted with VectaShield mounting medium with 5 μg/ml of DAPI.
Statistical Analyses
The Fisher’s exact were used to compare quantitative data on nuclear incorporation of Prm1 and in vitro development. Statistical analyses were performed using GraphPad Prism 5.0 software.
Author Contributions
P.L., D.I., M.C., and S.K. conceived the experiments. D.I. and M.C. performed all NT and characterization work of Prm cells, and M.C. performed the vector expressions and F.Z. did the embryological work. G.E.P. supervised the embryological work and discussed the results. P.T. and A.F. performed the molecular work. S.H. cloned Prm. T.B., S.C., and H.S. performed TH2B immunofluorescence and microscopy of the injected eggs. R.F. designed and interpreted the data. P.L., D.I., M.C., S.K., and R.F. wrote the manuscript.
Acknowledgments
Work in SK’s laboratory is supported by ANR EpiSperm2, INCa. H.S. is a recipient of a Marie Curie Initial Training Network grant funded by the European Commission (FP7-PEOPLE-2011-ITN and PITN-GA-289880). This work was supported by the European Research Council (FP7/2007-2013) programme IDEAS GA 210103 to G.E.P; the European Research Council, program FP7-KBBE-2012.1.3-04 GA 312097 (FECUND) to GP; and MIUR/CNR, program FIRB GA B81J12002520001 (GenHome) to P.L. The authors are participating in the COST action FA 1201 ‘‘Epiconcept’’ Epigenetic and Peri-conception Environment. A warm acknowledgment goes to Prof. Heiner Niemann and Dr. Bjoern Petersen (Institute of Farm Animal Genetics, FLI Mariensee) for plasmid amplification at the very beginning of this study and to Dr. Francesco Mosca (University of Teramo) for his assistance in flow cytometric analysis.
Published: November 25, 2015
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Supplemental Information includes Supplemental Experimental Procedures, three figures, and two tables and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2015.10.066.
Supplemental Information
Binding occurred in 28 gene-rich domains—14 (50%) genic and 14 (50%) intergenic—whereas the other 14 were classified as scaffold (2n = 54 in sheep).
References
- Akama K., Kondo M., Sato H., Nakano M. Transition protein 4 from boar late spermatid nuclei is a topological factor that stimulates DNA-relaxing activity of topoisomerase I. FEBS Lett. 1999;442:189–192. doi: 10.1016/s0014-5793(98)01649-4. [DOI] [PubMed] [Google Scholar]
- Awe S., Renkawitz-Pohl R. Histone H4 acetylation is essential to proceed from a histone- to a protamine-based chromatin structure in spermatid nuclei of Drosophila melanogaster. Syst Biol Reprod Med. 2010;56:44–61. doi: 10.3109/19396360903490790. [DOI] [PubMed] [Google Scholar]
- Balhorn R., Corzett M., Mazrimas J.A. Formation of intraprotamine disulfides in vitro. Arch. Biochem. Biophys. 1992;296:384–393. doi: 10.1016/0003-9861(92)90588-n. [DOI] [PubMed] [Google Scholar]
- Barckmann B., Chen X., Kaiser S., Jayaramaiah-Raja S., Rathke C., Dottermusch-Heidel C., Fuller M.T., Renkawitz-Pohl R. Three levels of regulation lead to protamine and Mst77F expression in Drosophila. Dev. Biol. 2013;377:33–45. doi: 10.1016/j.ydbio.2013.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer L., Corzett M., Lau E.Y., Balhorn R. Dynamics of protamine 1 binding to single DNA molecules. J. Biol. Chem. 2003;278:42403–42408. doi: 10.1074/jbc.M303610200. [DOI] [PubMed] [Google Scholar]
- Brock W.A., Trostle P.K., Meistrich M.L. Meiotic synthesis of testis histones in the rat. Proc. Natl. Acad. Sci. USA. 1980;77:371–375. doi: 10.1073/pnas.77.1.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chereji R.V., Morozov A.V. Functional roles of nucleosome stability and dynamics. Brief. Funct. Genomics. 2015;14:50–60. doi: 10.1093/bfgp/elu038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung Y.G., Eum J.H., Lee J.E., Shim S.H., Sepilian V., Hong S.W., Lee Y., Treff N.R., Choi Y.H., Kimbrel E.A. Human somatic cell nuclear transfer using adult cells. Cell Stem Cell. 2014;14:777–780. doi: 10.1016/j.stem.2014.03.015. [DOI] [PubMed] [Google Scholar]
- de Mateo S., Ramos L., de Boer P., Meistrich M., Oliva R. Protamine 2 precursors and processing. Protein Pept. Lett. 2011;18:778–785. doi: 10.2174/092986611795713998. [DOI] [PubMed] [Google Scholar]
- De Vries M., Ramos L., Housein Z., De Boer P. Chromatin remodelling initiation during human spermiogenesis. Biol. Open. 2012;1:446–457. doi: 10.1242/bio.2012844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaucher J., Reynoird N., Montellier E., Boussouar F., Rousseaux S., Khochbin S. From meiosis to postmeiotic events: the secrets of histone disappearance. FEBS J. 2010;277:599–604. doi: 10.1111/j.1742-4658.2009.07504.x. [DOI] [PubMed] [Google Scholar]
- Gaucher J., Boussouar F., Montellier E., Curtet S., Buchou T., Bertrand S., Hery P., Jounier S., Depaux A., Vitte A.L. Bromodomain-dependent stage-specific male genome programming by Brdt. EMBO J. 2012;31:3809–3820. doi: 10.1038/emboj.2012.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudarzi A., Shiota H., Rousseaux S., Khochbin S. Genome-scale acetylation-dependent histone eviction during spermatogenesis. J. Mol. Biol. 2014;426:3342–3349. doi: 10.1016/j.jmb.2014.02.023. [DOI] [PubMed] [Google Scholar]
- Iuso D., Czernik M., Zacchini F., Ptak G., Loi P. A simplified approach for oocyte enucleation in mammalian cloning. Cell. Reprogram. 2013;15:490–494. doi: 10.1089/cell.2013.0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuso D., Czernik M., Di Egidio F., Sampino S., Zacchini F., Bochenek M., Smorag Z., Modlinski J.A., Ptak G., Loi P. Genomic stability of lyophilized sheep somatic cells before and after nuclear transfer. PLoS ONE. 2013;8:e51317. doi: 10.1371/journal.pone.0051317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kota S.K., Feil R. Epigenetic transitions in germ cell development and meiosis. Dev. Cell. 2010;19:675–686. doi: 10.1016/j.devcel.2010.10.009. [DOI] [PubMed] [Google Scholar]
- Laberge R.M., Boissonneault G. On the nature and origin of DNA strand breaks in elongating spermatids. Biol. Reprod. 2005;73:289–296. doi: 10.1095/biolreprod.104.036939. [DOI] [PubMed] [Google Scholar]
- Loi P., Czernik M., Zacchini F., Iuso D., Scapolo P.A., Ptak G. Sheep: the first large animal model in nuclear transfer research. Cell. Reprogram. 2013;15:367–373. doi: 10.1089/cell.2013.0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loppin B., Bonnefoy E., Anselme C., Laurençon A., Karr T.L., Couble P. The histone H3.3 chaperone HIRA is essential for chromatin assembly in the male pronucleus. Nature. 2005;437:1386–1390. doi: 10.1038/nature04059. [DOI] [PubMed] [Google Scholar]
- Martínez-Soler F., Kurtz K., Ausió J., Chiva M. Transition of nuclear proteins and chromatin structure in spermiogenesis of Sepia officinalis. Mol. Reprod. Dev. 2007;74:360–370. doi: 10.1002/mrd.20515. [DOI] [PubMed] [Google Scholar]
- Matoba S., Inoue K., Kohda T., Sugimoto M., Mizutani E., Ogonuki N., Nakamura T., Abe K., Nakano T., Ishino F., Ogura A. RNAi-mediated knockdown of Xist can rescue the impaired postimplantation development of cloned mouse embryos. Proc. Natl. Acad. Sci. USA. 2011;108:20621–20626. doi: 10.1073/pnas.1112664108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matoba S., Liu Y., Lu F., Iwabuchi K.A., Shen L., Inoue A., Zhang Y. Embryonic development following somatic cell nuclear transfer impeded by persisting histone methylation. Cell. 2014;159:884–895. doi: 10.1016/j.cell.2014.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller D., Brinkworth M., Iles D. Paternal DNA packaging in spermatozoa: more than the sum of its parts? DNA, histones, protamines and epigenetics. Reproduction. 2010;139:287–301. doi: 10.1530/REP-09-0281. [DOI] [PubMed] [Google Scholar]
- Montellier E., Boussouar F., Rousseaux S., Zhang K., Buchou T., Fenaille F., Shiota H., Debernardi A., Héry P., Curtet S. Chromatin-to-nucleoprotamine transition is controlled by the histone H2B variant TH2B. Genes Dev. 2013;27:1680–1692. doi: 10.1101/gad.220095.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morinière J., Rousseaux S., Steuerwald U., Soler-López M., Curtet S., Vitte A.L., Govin J., Gaucher J., Sadoul K., Hart D.J. Cooperative binding of two acetylation marks on a histone tail by a single bromodomain. Nature. 2009;461:664–668. doi: 10.1038/nature08397. [DOI] [PubMed] [Google Scholar]
- Ogura A., Inoue K., Wakayama T. Recent advancements in cloning by somatic cell nuclear transfer. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2013;368:20110329. doi: 10.1098/rstb.2011.0329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pivot-Pajot C., Caron C., Govin J., Vion A., Rousseaux S., Khochbin S. Acetylation-dependent chromatin reorganization by BRDT, a testis-specific bromodomain-containing protein. Mol. Cell. Biol. 2003;23:5354–5365. doi: 10.1128/MCB.23.15.5354-5365.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptak G., Clinton M., Tischner M., Barboni B., Mattioli M., Loi P. Improving delivery and offspring viability of in vitro-produced and cloned sheep embryos. Biol. Reprod. 2002;67:1719–1725. doi: 10.1095/biolreprod.102.006171. [DOI] [PubMed] [Google Scholar]
- Rathke C., Baarends W.M., Awe S., Renkawitz-Pohl R. Chromatin dynamics during spermiogenesis. Biochim. Biophys. Acta. 2014;1839:155–168. doi: 10.1016/j.bbagrm.2013.08.004. [DOI] [PubMed] [Google Scholar]
- Rousseaux S., Khochbin S. Histone Acylation beyond Acetylation: Terra Incognita in Chromatin Biology. Cell J. 2015;17:1–6. doi: 10.22074/cellj.2015.506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samans B., Yang Y., Krebs S., Sarode G.V., Blum H., Reichenbach M., Wolf E., Steger K., Dansranjavin T., Schagdarsurengin U. Uniformity of nucleosome preservation pattern in Mammalian sperm and its connection to repetitive DNA elements. Dev. Cell. 2014;30:23–35. doi: 10.1016/j.devcel.2014.05.023. [DOI] [PubMed] [Google Scholar]
- Shinagawa T., Takagi T., Tsukamoto D., Tomaru C., Huynh L.M., Sivaraman P., Kumarevel T., Inoue K., Nakato R., Katou Y. Histone variants enriched in oocytes enhance reprogramming to induced pluripotent stem cells. Cell Stem Cell. 2014;14:217–227. doi: 10.1016/j.stem.2013.12.015. [DOI] [PubMed] [Google Scholar]
- Singh J., Rao M.R. Interaction of rat testis protein, TP, with nucleosome core particle. Biochem. Int. 1988;17:701–710. [PubMed] [Google Scholar]
- Tachibana M., Amato P., Sparman M., Gutierrez N.M., Tippner-Hedges R., Ma H., Kang E., Fulati A., Lee H.S., Sritanaudomchai H. Human embryonic stem cells derived by somatic cell nuclear transfer. Cell. 2013;153:1228–1238. doi: 10.1016/j.cell.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Padilla M.E., Bannister A.J., Hurd P.J., Kouzarides T., Zernicka-Goetz M. Dynamic distribution of the replacement histone variant H3.3 in the mouse oocyte and preimplantation embryos. Int. J. Dev. Biol. 2006;50:455–461. doi: 10.1387/ijdb.052073mt. [DOI] [PubMed] [Google Scholar]
- van der Heijden G.W., Dieker J.W., Derijck A.A., Muller S., Berden J.H., Braat D.D., van der Vlag J., de Boer P. Asymmetry in histone H3 variants and lysine methylation between paternal and maternal chromatin of the early mouse zygote. Mech. Dev. 2005;122:1008–1022. doi: 10.1016/j.mod.2005.04.009. [DOI] [PubMed] [Google Scholar]
- Weiner A., Hsieh T.H., Appleboim A., Chen H.V., Rahat A., Amit I., Rando O.J., Friedman N. High-resolution chromatin dynamics during a yeast stress response. Mol. Cell. 2015;58:371–386. doi: 10.1016/j.molcel.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmut I., Schnieke A.E., McWhir J., Kind A.J., Campbell K.H. Viable offspring derived from fetal and adult mammalian cells. Nature. 1997;385:810–813. doi: 10.1038/385810a0. [DOI] [PubMed] [Google Scholar]
- Wu F., Caron C., De Robertis C., Khochbin S., Rousseaux S. Testis-specific histone variants H2AL1/2 rapidly disappear from paternal heterochromatin after fertilization. J. Reprod. Dev. 2008;54:413–417. doi: 10.1262/jrd.20067. [DOI] [PubMed] [Google Scholar]
- Zhang X., Balhorn R., Mazrimas J., Kirz J. Mapping and measuring DNA to protein ratios in mammalian sperm head by XANES imaging. J. Struct. Biol. 1996;116:335–344. doi: 10.1006/jsbi.1996.0051. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Binding occurred in 28 gene-rich domains—14 (50%) genic and 14 (50%) intergenic—whereas the other 14 were classified as scaffold (2n = 54 in sheep).