Abstract
Phosphorylation and dephosphorylation events play an important role in the transmission of the ABA signal. Although SnRK2 [sucrose non-fermenting1-related kinase2] protein kinases and group A protein phosphatase type 2C (PP2C)-type phosphatases constitute the core ABA pathway, mitogen-activated protein kinase (MAPK) pathways are also involved in plant response to ABA. However, little is known about the interplay between MAPKs and PP2Cs or SnRK2 in the regulation of ABA pathways. In this study, an effort was made to elucidate the role of MAP kinase kinase kinase18 (MKKK18) in relation to ABA signaling and response. The MKKK18 knockout lines showed more vigorous root growth, decreased abaxial stomatal index and increased stomatal aperture under normal growth conditions, compared with the control wild-type Columbia line. In addition to transcriptional regulation of the MKKK18 promoter by ABA, we demonstrated using in vitro and in vivo kinase assays that the kinase activity of MKKK18 was regulated by ABA. Analysis of the cellular localization of MKKK18 showed that the active kinase was targeted specifically to the nucleus. Notably, we identified abscisic acid insensitive 1 (ABI1) PP2C as a MKKK18-interacting protein, and demonstrated that ABI1 inhibited its activity. Using a cell-free degradation assay, we also established that MKKK18 was unstable and was degraded by the proteasome pathway. The rate of MKKK18 degradation was delayed in the ABI1 knockout line. Overall, we provide evidence that ABI1 regulates the activity and promotes proteasomal degradation of MKKK18.
Keywords: ABA signaling, ABI1 PP2C, Arabidopsis thaliana, MAP kinase cascade, MKKK18, Proteasome
Introduction
The phytohormone ABA recruits many diverse elements for the generation and transmission of endogenous signals (Xiong et al. 2002, Mane et al. 2007, Fujita et al. 2009, Geiger et al. 2009, Ludwików et al. 2009, Nishimura et al. 2009, Santiago et al. 2009, Yoshida et al. 2010). Protein kinases and protein phosphatases are proteins that lead to rearrangements in the network of ABA signal transduction (Ohta et al. 2003, Zhu et al. 2007, Ma et al. 2009, Nakashima et al. 2009, Park et al. 2009, Santiago et al. 2009, Umezawa et al. 2009, Nishimura et al. 2010, Szostkiewicz et al. 2010, Dupeux et al. 2011a, Dupeux et al. 2011b). Both protein kinases and protein phosphatases are obligatory for ABA signal transduction. Sucrose non-fermenting 1 (SNF1)-related protein kinases 2 (SnRK2s), Ca2+-dependent protein kinases (CDPKs) and calcineurin B-like calcium sensor (CBL)-interacting protein kinases (CIPKs) are all known for their involvement in ABA signaling and stress tolerance, and are identified as interacting partners with particular protein phosphatases type 2C (PP2Cs) (Ohta et al. 2003, Zhu et al. 2007, Umezawa et al. 2009). Key players of ABA signaling such as abscisic acid insensitive 1 (ABI1), abscisic acid insensitive 2 (ABI2) and Hypersensitive to Abscisic acid 1 (HAB1) have been shown to interact with RCAR/PYR/PYL (RCAR, Regulatory Component of ABA Receptor/PYR1, Pyrabactin Resistance 1/PYL, PYR1-like), putative ABA receptors that regulate SnRK kinases (Umezawa et al. 2009). ABI1 and ABI2, along with CIPK20, CIPK8, CIPK14 and CIPK15, manage protein–protein interactions and regulate ABA signaling, gene expression and stress response (Guo et al. 2002, Ohta et al. 2003, Ludwików et al. 2013, Babula-Skowrońska et al. 2015). Other PP2C-interacting partners include transcription factors (Himmelbach et al. 2002), antioxidant enzymes (Miao et. al., 2006), constituents of the chromatin remodeling complex (Saez et al. 2008) and components of the ethylene biosynthetic pathway (Ludwików et al. 2014), which have likewise been identified as important to the ABA signaling network.
Despite studies indicating that mitogen-activated protein kinase (MAPK) cascades are important for ABA signaling, little is known about the interplay between MAPKs and PP2Cs in this pathway. A MAPK cascade is a conserved module comprised of at least three elements: MKKKs, MKKs and MAPKs. A group of MKKKKs have also been identified (Jonak et al. 2002, Yoon et al. 2010). MKKKs link upstream receptors with downstream MKKs. MKKKs (as dual specificity kinases) phosphorylate MKKs on Ser/Thr residues in conserved S/T-X3-5-S/T motifs localized in an activation loop (T-loop). Once activated, the MKKs phosphorylate MAPKs at the TxY motif located in their T-loop. Upon activation, the MAPKs phosphorylate downstream substrates, thereby initiating various cellular responses. Among the MAPKs, MPK6 is known to interact with ABI1 PP2C. Both ABI1 and MPK6 are involved in the regulation of biotic and abiotic stress responses (Leung et al. 2006). In addition, MPK3, MPK6, MPK9 and MPK12 have been identified as regulators of ABA signaling in guard cells (Lampard et al. 2009, Brock et al. 2010, Jammes et al. 2011, Salam et al. 2013). MKK1–MPK6 were found to be transiently activated by ABA treatment, to affect downstream generation of H2O2 and to regulate guard cell aperture (Xing et al. 2008). Other kinases, such as MPK9 and MPK12, act upstream of anion channels in guard cell ABA signaling (Jammes et al. 2009). In addition, MKK7 and MKK9 are known to act as regulators during stomatal development (Lampard et al. 2009).
Because MKKKs constitute the largest and most complex group of MAP cascade kinases, only a few MPK modules have been recognized and documented. Reaactive oxygen species (ROS) signaling in plants involves the MEKK1–MKK2–MPK4 phosphorylation pathway (Colcombet and Hirt 2008, Pitzschke et al. 2009). In apple, the cascade MdMKK1–MdMPK1 is activated by ABA (Wang et al. 2010). In Arabidopsis, an unknown MKKK–MKK4/5–MPK3/6 module, and MEKK1–MKK1/2–MPK4, are two MAPK cascades that govern flg22-dependent gene expression (Suarez-Rodriguez et al. 2007, Qiu et al. 2008, Nicaise et al. 2009, Boudsocq et al. 2010, Kong et al. 2012). Another MAPK cascade consists of YODA activating MKK4/5–MKK7/MKK9 and MPK3/6: the YODA module is an important protein complex that regulates stomatal development and affects inflorescence architecture by promoting cell proliferation (Wang et al. 2007, Lampard et al. 2009, Meng et al. 2012). Very recently, a MAPK cascade, MAP3K17/18–MKK3–MPK1/2/7/14, was found to play a role in ABA stress signaling (Danquah et al. 2015) and senescence (Matsuoka et al. 2015).
In this study, we investigated the role of MKKK18 in ABA signaling and response. We find that MKKK18 is regulated by both ABI1 PP2C and the proteasome pathway.
Results
MKKK18 expression is induced by ABA
Previous studies have shown that MKKK18 expression is increased in response to wounding (Taki et al. 2005), pathogen attack (de Torres-Zabala et al. 2007), ozone, mannitol, NaCl and ABA treatment (Hoth et al. 2002, Leonhardt et al. 2004, Ludwików et al. 2009, Danquah et al. 2015). The responsiveness of the MKKK18 promoter to ABA, quinabactin (a sulfonamide ABA agonist) and ASn compounds (ABA analogs) was also demonstrated (Okamoto et al. 2013, Takeuchi et al. 2014). In addition, MKKK18 expression was found to be deregulated in abi1-1 and abi1td mutants (Hoth et al. 2002, Ludwików et al. 2009). The expression of MKKK18 was diminished in abi1td (Ludwików et al. 2009), and abolished in ABA-insensitive abi1-1 (Hoth et al. 2002), quadruple pyr1pyl1pyl2pyr4 and the hab1G246D mutants (Danquah et al. 2015), suggesting that the proper transcription of MKKK18 requires regular ABA signaling.
To gain a more detailed insight into the MKKK18 gene expression profile, we analyzed public transcriptome data compiled by Genevestigator and eFP Browser. The results indicated that MKKK18 exhibits low level expression in a range of plant tissues, with the exception of anthers. A relatively high level of expression was observed in root cells, including the epidermis, endodermis, stele, cortex and lateral root cap. A significant increase in MKKK18 gene expression was observed in guard cells and mesophyll cells in response to ABA treatment.
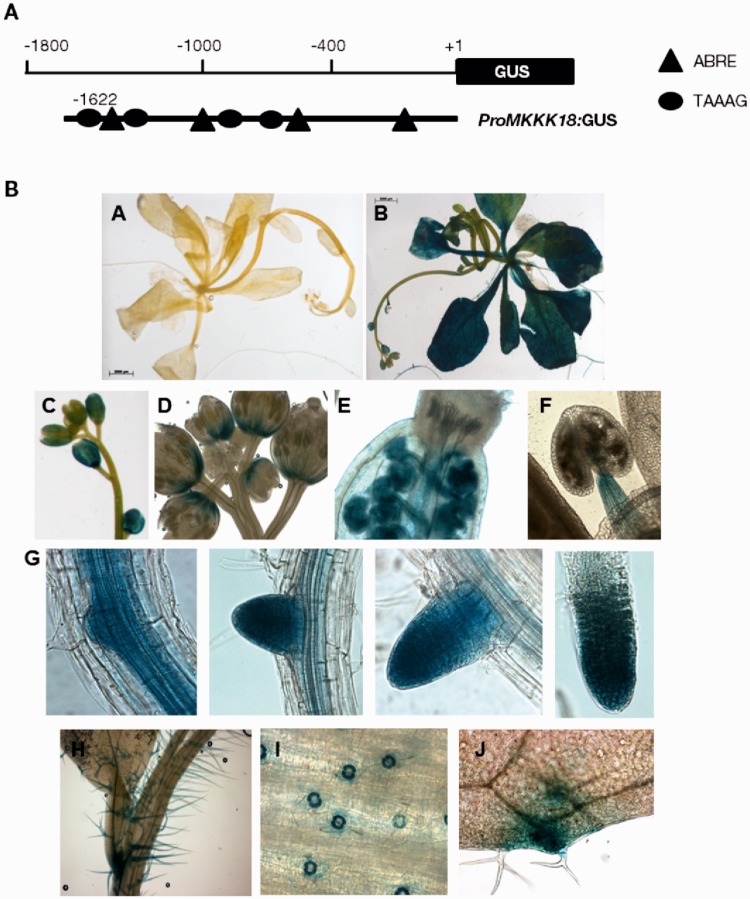
To verify the ABA responsiveness of MKKK18, we studied promoter activity in the 1,622 bp upstream region of the MKKK18 gene in transgenic plants carrying promoters fused to the β-glucuronidase (GUS) reporter genes. A de novo search for cis-acting elements using the PLACE program revealed that the MKKK18 promoter contained a number of cis-regulatory elements. These elements included ABRE (PyACGTGGC) and DRE elements (TACCGACAT) (Yamaguchi-Shinozaki et al. 2005), an ASF1 regulatory sequence (Krawczyk et al. 2002), POLLENLELAT52 (Bate and Twell, 1998), a W-box (Yamasaki et al. 2012) and a TAAAG element required for guard cell-specific gene expression (Plesch et al, 2001). At 24 h after the ABA treatment, a full-length ProMKKK18:GUS construct delivered strong GUS expression in the flowers, leaves and root tissues (Fig. 1). In developing flowers, MKKK18 promoter activity was observed in sepals, anther filaments, ovaries and meristem tissues (Fig. 1C–F). GUS staining was visible in root meristem tissues and in areas of lateral root formation (Fig. 1G). ProMKKK18:GUS staining was observed in guard cells and trichomes (Fig. 1H, I). Our results demonstrate that a 1.6 kb upstream sequence was sufficient for MKKK18 regulation by ABA.
Fig. 1.
Analysis of the activity of the MKKK18 promoter in transgenic plants expressing the ProMKKK18:GUS construct. (A) Schematic map of the MKKK18 promoter used for transformation and tissue-specific expression of MKKK18. (B) The ProMKKK18:GUS seedlings were germinated and grown on half-strength MS medium. Five-week-old plants were treated with either 0.1% methanol (mock) or 100 µM ABA. One representative of five independent lines is shown. (A) No changes in GUS activity after mock treatment. The scale bar represents 2,000 µm. (B–J) Plant samples 24 h after ABA treatment. GUS activity is visible in rosette leaves (B), flower buds and sepals (C), meristem tissues (D), pistils (E), anther filaments (F), lateral roots (G), trichomes (H), leaf guard cells (I) and hydathodes (J).
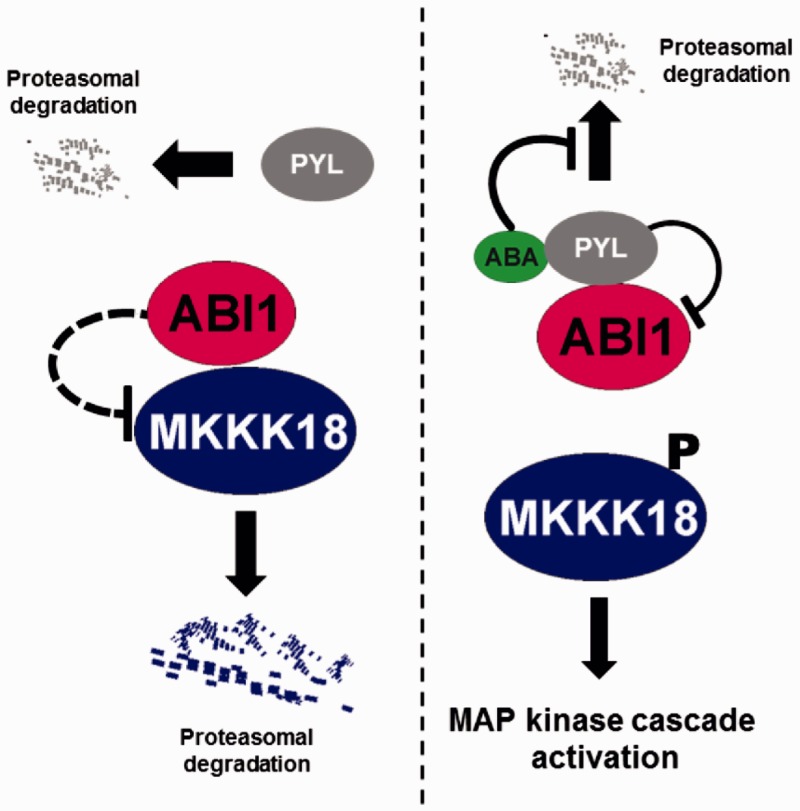
Fig. 10.
Regulation of MKKK18 activity and stability by ABI1 PP2C. Under standard conditions, active ABI1 protein phosphatase inhibits MKKK18 kinase activity and protein stability. ABA receptors and dephosphorylated MKKK18 are directed for degradation via the proteasome pathway. In the presence of ABA, ABI1 PP2C activity is restrained by binding of ABI1 to the ABA–PYL complex (ABA signaling is turned on). Active MKKK18 activates the downstream cascade. ABA inhibits the degradation of ABA receptors by limiting their polyubiquitination.
Active MKKK18 is localized in the nucleus
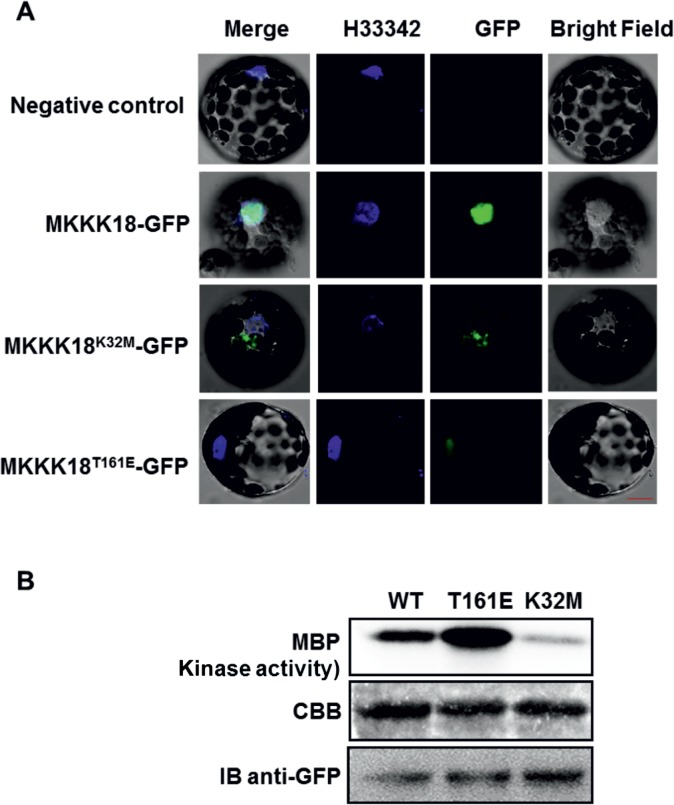
To determine MKKK18 protein localization in vivo, C-terminal green fluorescent protein (GFP) fusions were generated and transiently expressed under the control of the 35S promoter in Arabidopsis thaliana protoplasts. MKKK18–GFP was predominantly localized in the nucleus, whereas, as expected, an empty vector control did not show background fluorescence in any cellular compartment (Fig. 2A). Next, we addressed whether kinase activity affected MKKK18 localization. By multisequence alignment of Arabidopsis MAPKKK, we identified conserved residues important for the activity of MKKK18. Based on this analysis, a kinase-inactive allele (K32M) and a permanently active form of MKKK18 (T161E) were generated. The K32M version of the protein is modified in its ATP-binding loop, while the permanently active, T161E form of MKKK18 is modified in the kinase domain. All versions of MKKK18 were fused to GFP and were again expressed in Arabidopsis protoplasts. Interestingly, the K32M version retained barely detectable kinase activity and was localized outside the nucleus. However, the fluorescent signal of the permanently active T161E version of MKKK18–GFP clearly accumulated in the nucleus of Arabidopsis protoplasts, suggesting a role in mediating the signaling function of MKKK18 in this compartment (Fig. 2A). Immunoblot analysis confirmed the presence of full-length fusion proteins in protoplasts (Fig. 2B).
Fig. 2.
Subcellular localization of the MKKK18 protein in Arabidopsis protoplasts. Active MKKK18 is localized in the nucleus. (A) Microscopy images show nuclear localization of the MKKK18–GFP fusions in Arabidopsis protoplasts. Hoechst H33342 was used as nuclear localization marker. Scale bars are calibrated to 20 µm. (B) MKKK18 protein expression and activity were analyzed by immunoprecipitation using protein-specific anti-MKKK18 antibodies and immunocomplex kinase assay with MBP as a substrate. The immunocomplex assay confirmed residual kinase activity of the MKKK18K32M-GFP allele. Western blot detection using anti-GFP antibodies corroborates the presence of MKKK18–GFP fusion proteins. Coomassie Brilliant Blue (CBB) staining confirmed equal loading.
MKKK18-overexpressing lines show ABA-related phenotypes at the level of germination and root growth
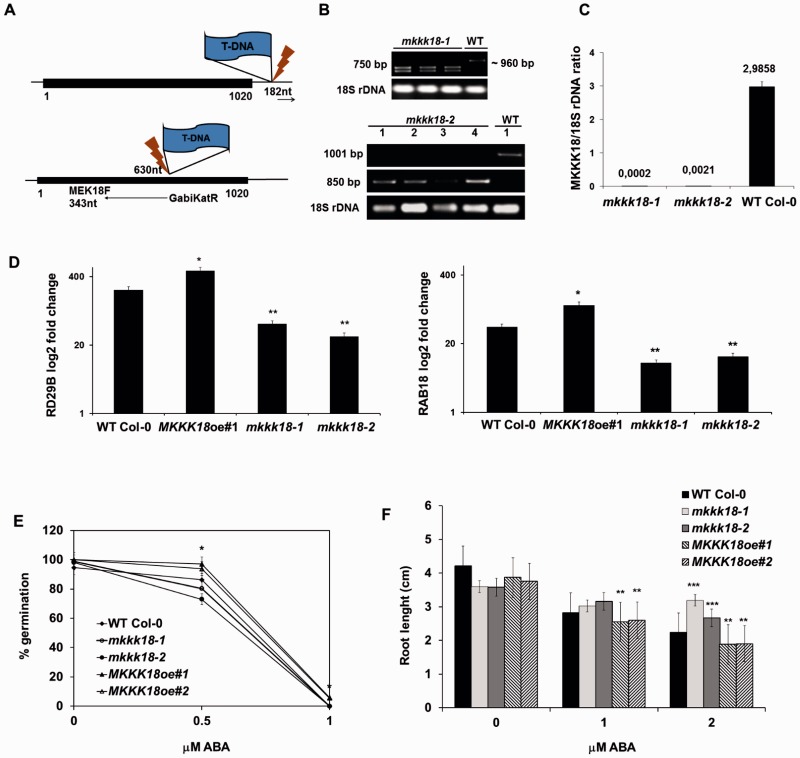
To investigate the functions of MKKK18, six independent MKKK18-overexpressor lines (Col-0/35S:MKKK18-GFP; MKKK18oe1–MKKK18oe6) were generated and two T-DNA insertion lines of MKKK18 in the Columbia (Col-0) background were selected from the SALK and GABI-Kat collections. These knockout lines were designated mkkk18-1 (SALK_087047) and mkkk18-2 (GK-244G02). Homozygous mkkk18-1 and mkkk18-2 plants were identified by PCR-based genotyping, and the insertion site was confirmed by PCR using T-DNA-specific and gene-specific primers (Fig. 3A, B). A quantitative real-time PCR (qPCR) analysis confirmed that expression of MKKK18 was abolished in both mkkk18-1 and mkkk18-2 (Fig. 3C).
Fig. 3.
Germination and root growth assays. (A and B) T-DNA insertion sites derived from the sequencing of genomic DNA isolated from mkkk18-1 and mkkk18-2 mutant lines. Black boxes represent an open reading frame. Homozygous knockout mutants were verified by PCR-based genotyping using the following primers: MKKK18LP plus MKKK18RP plus LBb1 for mkkk18-1 analysis (product size for the WT, ∼960 bp; product for homozygous lines, 750 bp); MEK18F plus GKatTDNA (product for homozygous lines, 850 bp) and GwMK18F plus GwMK18R (product for the WT, 1,001 bp) for mkkk18-2 genotyping. Primer sequences are indicated in Supplementary Table S1. (C) qPCR analysis confirmed a lack of MKKK18 expression in homozygous lines. Each quantification was repeated twice with similar results. The results are given as log2 of the relative MKKK18/18S rDNA expression ratio ± SE (n = 6). (D) RD29B and RAB18 transcript levels in MKKK18 mutants. RD29B and RAB18 expression levels were determined using three biological replicates and were normalized against 18S rDNA. Each quantification was repeated twice on separate plates. The results are displayed as mean log2 fold change ± SE (n = 9) of three independent experiments with consistent results. (E and F) Germination and root growth assays. ABA-mediated inhibition of germination (E) and primary root growth (F) in WT Col-0, MKKK18oe and MKKK18 knockout lines. Both MKKK18oe #1 and #2 lines showed similar results. Values are mean ± SE for three independent experiments (n = 30). *P < 0.01; **P < 0.001; ***P < 0.0001 with respect to the control WT Col-0 line.
Previous reports showed that MKKK18 is an ABA-activated kinase (Danquah et al. 2015, Matsuoka et al. 2015). To monitor the activation of ABA signaling in the leaves of the knockouts and MKKK18-oe lines at the molecular level, the expression of the ABA-induced genes RD29B and RAB18 was examined by qPCR (Fig. 3D). In response to treatment with ABA, RD29B and RAB18 expression was significantly down-regulated in both MKKK18 knockout lines compared with MKKK18oe and wild-type (WT) plants (P < 0.001), suggesting that ABA-mediated induction of these genes is regulated by MKKK18.
In the search for ABA-related phenotypes in the MKKK18 mutants, we first analyzed whether seed germination and root growth were affected by ABA. Seeds of MKKK18oe lines and mkkk18 knockouts were sown on Murashige and Skoog (MS) medium supplemented with ABA, and the germination rate was analyzed and compared with that of WT seeds. As indicated in Fig. 3E, both knockout lines showed similar growth rates to WT Col-0 in the presence of low concentrations of ABA. In contrast, Col-0/35S:MKKK18-GFP germinated better than the WT Col-0 control, which was supported by the t-test results (P < 0.001) between the WT and independent MKKK18oe lines. This observation indicated that the 35S:MKKK18-GFP-overexpressing line displayed insensitivity towards ABA at the level of germination, suggesting that MKKK18 was required for germination. Next, in the search for ABA-related phenotypes, we analyzed ABA-mediated inhibition of root expansion. In the presence of 2 µM ABA, root elongation in seedlings expressing 35S:MKKK18-GFP was reduced by 15% compared with the WT Col-0 control seedlings (n = 46; P < 0.0001). At the same ABA level, the mkkk18-1 and mkkk18-2 knockout lines showed improved root growth (40% and 20%, respectively) compared with the WT Col-0 line (Fig. 3F).
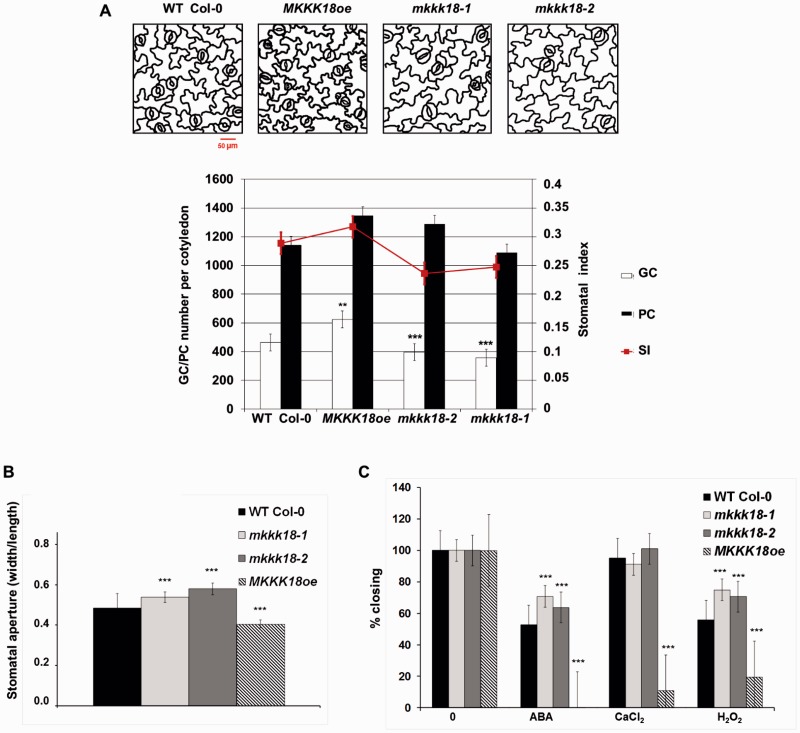
MKKK18 affects stomatal development and function
MAPKs are important elements of signaling cascades that affect stomatal development (Wang et al. 2007, Lampard et al. 2009, Tanaka et al. 2013). Therefore, we compared the number and density of stomata in 6- and 10-day-old seedlings of both MKKK18-oe lines and MKKK18 knockout lines (Fig. 4A). In the cotyledons, the stomatal index (SI) was significantly reduced, by 19% and 26%, in mkkk18-1 (P < 0.001) and mkkk18-2 (P < 0.0001), respectively. Consistent with this trend in SI, the abaxial SI for MKKK18oe lines was 7% higher than that for WT lines. Because GUS expression driven by the MKKK18 promoter was detected in guard cells, we analyzed whether MKKK18 was involved in the regulation of stomatal aperture. Compared with leaves from the WT, leaves from both knockout lines showed increased stomatal aperture under normal growth conditions. However, this effect was more pronounced in the mkkk18-2 mutant (Fig. 4B). Measurements of stomatal aperture in response to ABA revealed that stomata from the knockout lines were again significantly more open than in the WT. Accordingly, MKKK18oe lines were hypersensitive to ABA-induced stomatal closure (Fig. 4C). Next, we analyzed whether MKKK18 acted downstream of responses to CaCl2 or H2O2 in guard cell ABA signaling. Compared with WT Col-0, the knockout lines were significantly, if only slightly, impaired in H2O2-induced stomatal closure but not in response to CaCl2 (Fig. 4C).
Fig. 4.
Stomatal development and movement in MKKK18 knockouts and the MKKK18-overexpressing line. (A) Stomatal development is enhanced in the MKKK18-overexpressing lines. Representative line drawings show the abaxial surface of cotyledons at day 10. Change in stomatal index and the number of guard cells (GCs) and pavement cells (PCs) per cotyledon in WT Col-0, MKKK18oe and MKKK18 knockouts. Data represent the mean ± SD (n = 300) of three independent experiments. Scale bar = 50 µm. (B) Increased stomatal aperture of mkkk18-1 and mkkk18-2 knockouts compared with the WT in standard conditions (three independent experiments, 60–80 stomatal apertures at each data point). (C) The mkkk18-1 and mkkk18-2 knockouts are insensitive to ABA- and H2O2-induced stomatal closure (two independent experiments, 80 stomatal apertures at each data point). MKKK18oe lines were hypersensitive to ABA-, CaCl2- and H2O2-induced stomatal closure (two independent experiments, 100 stomatal apertures at each data point).
MKKK18 is activated by ABA in tobacco
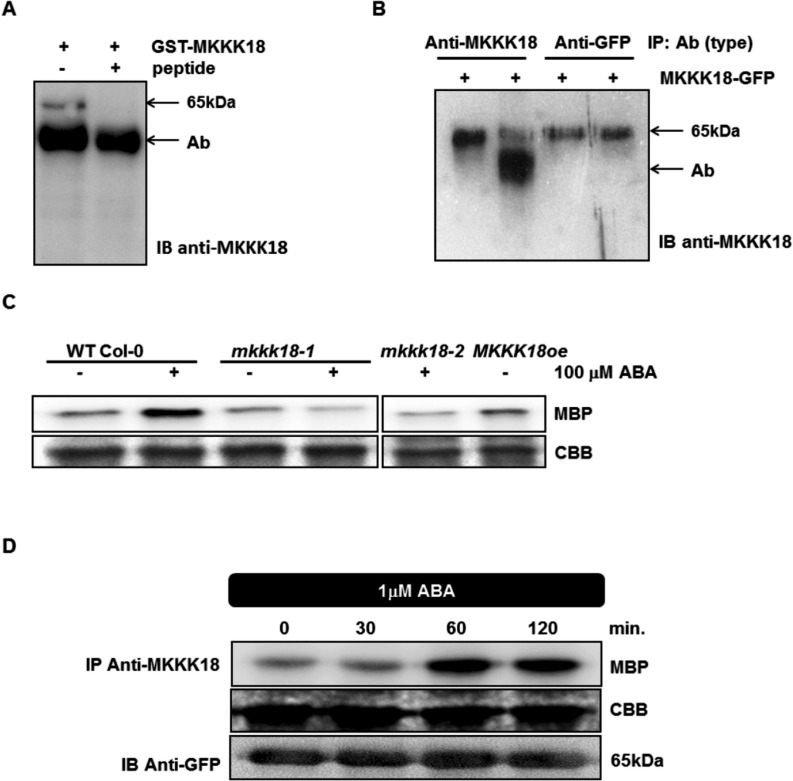
To test the mechanism of MKKK18 activation in response to ABA, we generated a peptide-specific antibody against MKKK18. Using alignment tools, we found a unique N-terminal sequence of MKKK18 with no obvious similarity to any other Arabidopsis protein, including MAPKs of any type. To test the specificity of the generated antibodies, we performed immunoprecipitation of recombinant MKKK18–glutathione S-transferase (GST) with and without an epitope-mimicking peptide. We found that the generated antibody recognized recombinant MKKK18–GST (Fig. 5A, B), while pre-incubation of anti-MKKK18-coupled beads with the peptide blocked immunoprecipitation of MKKK18 (Fig. 5A). We then tested the antibody in Arabidopsis extracts, but were unable to detect the MKKK18 protein directly, either by immunoblot analysis of the total protein extract or in the immunoprecipitates from WT Col-0 and knockout lines. Nevertheless, MKKK18 immunocomplex activity was detectable in ABA-treated WT Col-0, although not in ABA-treated knockout lines (Fig. 5C), demonstrating that both knockout lines are deficient in MKKK18 activity. Overall, these results clearly demonstrate the specificity of the antibody for MKKK18.
Fig. 5.
ABA induces rapid MKKK18 activation in tobacco. (A–C) Generation of MKKK18-specific antibodies. (A) MKKK18 peptide competition assay. Recombinant GST–MKKK18 protein was incubated with anti-MKKK18 with and without 45 µg of blocking peptide. A single band of approximatley 65 kDa specific to GST–MKKK18 is absent in the immunoprecipitates containing the blocking peptide. Immunodetection was performed using anti-MKKK18 antibody. (B) MKKK18–GFP protein immunoprecipitated from tobacco total protein extracts using increasing amounts of anti-MKKK18 and anti-GFP antibodies. The MKKK18–GFP protein was precipitated by 3 µl (lane 1) or 10 µl (lane 2) of MKKK18 antiserum and 0.6 µg (lane 3) or 1.2 µg (lane 4) of anti-GFP antibody, respectively. Immunoblotting with anti-MKKK18 antibodies confirmed the presence of MKKK18–GFP fusion protein. Arrows/Ab indicate anti-MKKK18 antibodies (visible as a band in lane 2) where excess anti-MKKK18 antibody was used in the immunoprecipitation reaction. The 65 kDa band represents MKKK18–GFP protein. (C) Analysis of MKKK18 activity in the mkkk18 knockout lines. MKKK18 protein was immunoprecipitated from 600 µg of total protein extract isolated from ABA-treated WT Col-0, mkkk18-1, mkkk18-2 and MKKK18oe using specific anti-MKKK18 antibodies. Immunocomplex activity was determined using MBP (2 µg) as a substrate. Coomassie Brilliant Blue (CBB) staining of MBP confirmed equal loading. (D) Tobacco plants infiltrated with Agrobacterium strain C58C1 harboring 35S:MKKK18-GFP and 35S:p19 constructs for transient expression. Four to five days after infiltration, the tobacco plants were treated with ABA and tissue samples were collected at the indicated time points. MKKK18 activity was assessed by the immunocomplex assay using anti-MKKK18 antibody and MBP as a substrate. MKKK18–GFP was detected with anti-GFP antibody. CBB staining of MBP confirmed equal loading. The above experiments were repeated several times with similar results.
To analyze the time course of MKKK18 activation in response to ABA, we again performed the kinase immunocomplex assay using myelin basic protein (MBP) as a substrate. Nicotiana benthamiana plants were generated that transiently expressed the 35S:MKKK18-GFP construct, and the MKKK18–GFP protein was immunoprecipitated using the MKKK18-specific antibody (Fig. 5D). As expected, treatment with 1 µM ABA caused an increase in the activity of MKKK18, which was observed within 60 min of treatment. Importantly, before ABA treatment we did not observe MKKK18–GFP kinase activity on MBP. Overall, our results demonstrate that MKKK18 activity is indeed regulated by ABA.
ABI1 PP2C interacts with MKKK18 and affects its activation by ABA
Our previous results showed that MKKK18 expression was significantly affected in the abi1td mutant (Ludwików et al. 2009). Because MKKK18 is also ABA responsive, and ABI1 is known to be a negative regulator of ABA signaling, we then asked whether ABI1 would affect MKKK18 function. We transformed abi1td protoplasts with WT and mutated forms of MKKK18 fused to GFP, and examined their subcellular localization. We observed that the distribution of the various forms of MKKK18–GFP in abi1td mesophyll protoplasts was similar to that in WT protoplasts, suggesting that ABI1 did not affect MKKK18 localization (Supplementary Fig. S1).
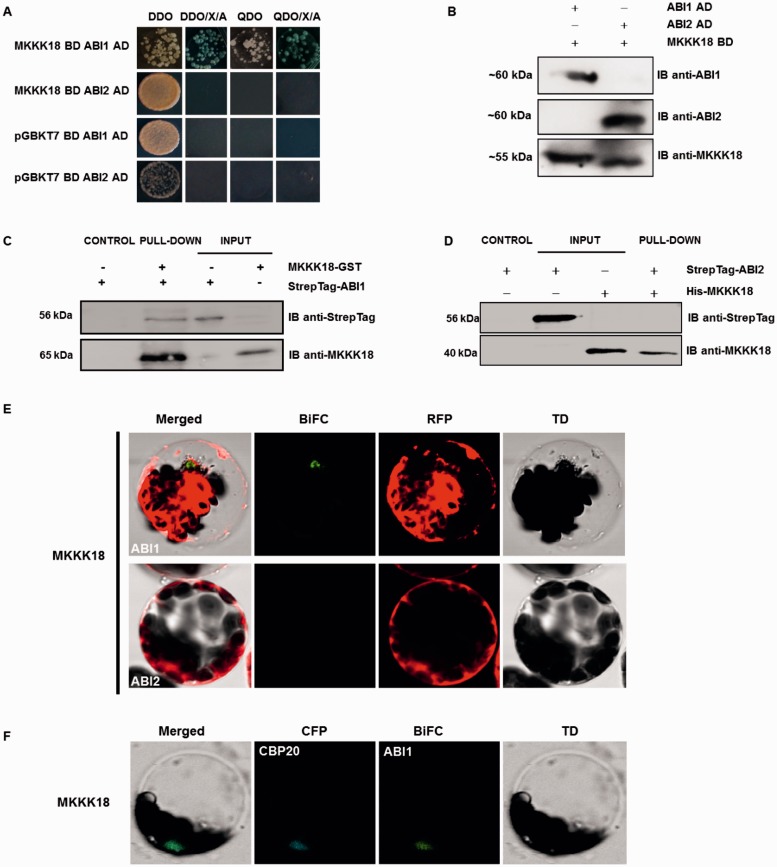
We next tested whether ABI1 would interact with MKKK18 using yeast two-hybrid, GST pull-down and bimolecular fluorescence complementation (BiFC) assays. Full-length ABI1 (and homologous ABI2 PP2C as a control) and MKKK18 cDNA were fused to either the binding domain (BD) or the activation domain (AD) for yeast two-hybrid analysis and double-transformed yeast lines were generated. Positive results, indicating interaction, were seen only for the ABI1–MKKK18 pair (Fig. 6A, B; Supplementary Fig. S2). The binding of ABI1 to MKKK18 was then analyzed using pull-down assays. Streptag-ABI1, and Streptag-ABI2 as a control, were purified from plant extracts and incubated with recombinant MKKK18–GST and His-MKKK18, respectively. Again, only ABI1 was capable of binding MKKK18, indicating substrate specificity of ABI1 PP2Cs for interaction with MKKK18 (Fig. 6C, D). This result was further confirmed by BiFC experiments using ABI1 (or ABI2 as a control) with MKKK18, which also revealed that ABI1 and MKKK18 were capable of interacting (Fig. 6E, F; Supplementary Fig. S3).
Fig. 6.
MKKK18 interacts with the ABI1 protein phosphatase. (A) Yeast two-hybrid analysis of the interaction between MKKK18 and ABI1/2 PP2Cs. Diploid yeast colonies were grown on double (DDO-SD medium without Leu and Trp) or quadruple selective medium (QDO-SD medium without Leu, Trp, His or Ade) with or without supplemented X-α-Gal and aureobasidin. The bait (MKKK18) did not autoactivate the reporter genes in yeast. (B) Detection of ABI1/2 and MKKK18 expression in diploid yeast strains. Expression of BD and AD fusion proteins in yeast was determined by immunoblotting using specific anti-ABI1, anti-ABI2 and anti-MKKK18 antibodies. (C and D) Pull-down assays to verify the interaction of ABI1/2 with MKKK18. Input lines represent 100% of the ABI1/2. Recombinant GST–MKKK18 or His-MKKK18 was pre-coupled to glutathione–Sepharose, and incubated with StrepTag-ABI1 or StrepTag-ABI2, respectively. Pulled-down StrepTagged ABI1 protein was detected (IB) with the epitope tag antibody. The presence of recombinant protein was confirmed using anti-MKKK18 antibody. (E and F) ABI1–MKKK18 interaction occurs within the nucleus. BiFC analysis in Arabidopsis protoplasts expressing full-length ABI1/2 and MKKK18 fused to cECFP or nVenus, respectively. RFP (E) was used as a transformation control. CFP–CBP20 (cyan fluorescent protein–Cap Binding Protein 20) (F) was used as a marker of nuclear localization.
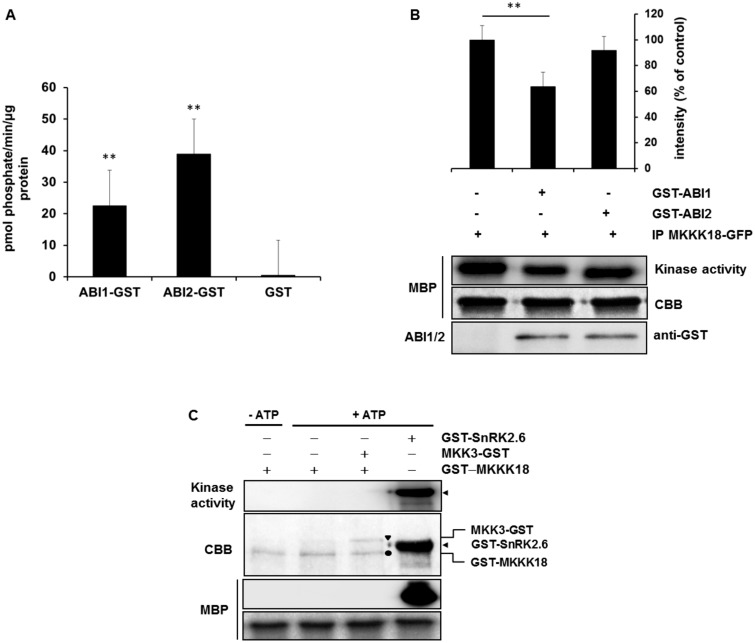
Consequently, we analyzed the effect of ABI1 PP2C activity on MKKK18 activation. Arabidopsis mesophyll protoplasts were transfected with 35S:MKKK18–GFP constructs, and immunocomplex MKKK18 activity was determined by in vitro kinase assays in the presence of recombinant ABI1 or ABI2 proteins (Fig. 7A, B). ABI1 but not ABI2 inhibited MKKK18 kinase activity. As a control treatment, the activities of both PP2Cs were assessed using a non-radioactive phosphatase assay (Fig. 7A). Previous studies revealed that ABI1 can inhibit the autoactivation of SnRK2 kinases (Ng et al. 2011). To test the mechanism of MKKK18 activation and the role of ABI1 in this process, we purified recombinant GST–MKKK18 and analyzed the kinase activity in vitro in the presence of the downstream in vivo substrate, MKK3 (Danquah et al. 2015, Matsuoka et al. 2015), and an artificial substrate, MBP. As a positive control for the kinase activity assay, SnRK2.6 was used (Fig. 7C). Interestingly, we observed no autophosphorylation activity of MKKK18 and as a consequence no phosphorylation of the substrates. These results indicate that MKKK18 activity is precisely controlled and that ABI1 does not regulate autoactivation of MKKK18.
Fig. 7.
ABI1 inhibits MKKK18 activity. (A) Phosphatase activity of recombinant ABI1 and ABI2 proteins. The enzyme reactions were performed in a 50 µl final volume containing 3–5 µg pf GST–ABI1 or GST–ABI2. The results presented are the means from three independent biological replicates. (B) ABI1 inactivates MKKK18. The MKKK18–GFP immunocomplex was incubated with 3 µg of GST–ABI1 and GST–ABI2 (as a negative control), after which the kinase activity was determined with MBP as a substrate. Equal loading was confirmed by Coomassie Brilliant Blue (CBB) staining of MBP. The 32P-labeled MBP bands were quantified and then normalized against the intensity of the corresponding control band using ImageJ software. Data are means ± SD of the relative band intensities from three independent experiments. An asterisk (*) indicates statistically significant changes determined using Student’s t-test. (C) Recombinant MKKK18 has no autophosphorylation activity in vitro. GST–MKKK18 was incubated with MBP or/and MKK3, without or in the presence of [32P]ATP. Bands of GST–MKKK18, MKK3–GST and GST–SnRK2.6 are indicated by a filled circle, a triangle and an arrow, respectively. Protein loading was confirmed by CBB staining. The blots shown are representative of three independent trials.
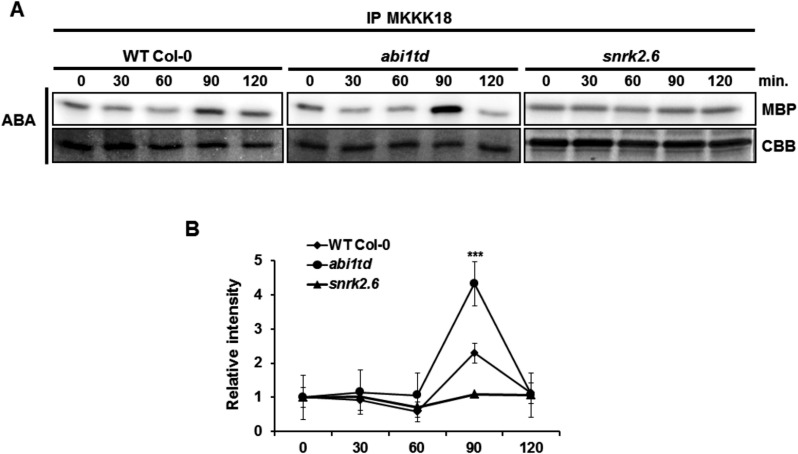
Finally, to confirm the role of ABI1 in the regulation of MKKK18 activity, we examined MKKK18 activation in WT Col-0 and abi1td after ABA treatment. Previously, it was reported that MKKK18 expression is absent in the ABA receptor quadruple pyr1pyl1pyl2pyl4 mutant (Danquah et al. 2015). Therefore, SnRK2.6 kinase, another crucial element of the ABA pathway, was also included in this analysis. Under the conditions indicated, we observed a stronger induction of MKKK18 activity in the abi1td mutant than in the WT Col-0 plants after 90 min of ABA treatment (Fig. 8A, B). Under the same conditions, ABA-induced MKKK18 activation was abolished in the snrk2.6 mutant. These experiments demonstrated that both ABI1 PP2C and SnRK2.6 affect functional activation of MKKK18 in response to ABA treatment.
Fig. 8.
MKKK18 activity increases in the abi1td mutant in response to ABA treatment. (A) Seedlings of WT Col-0 and the abi1td and snrk2.6 mutants were treated with 100 µM ABA for the indicated times. A 600 µg aliquot of total protein was used for the immune complex MKKK18 activity assay using anti-MKKK18 antibody. Coomassie Brilliant Blue (CBB) staining of MBP confirmed equal loading. Due to low abundance, the endogenous MKKK18 protein was not detectable by immunoblot analysis. The results shown are representative of three independent experiments (n = 9) with consistent results. (B) 32P-labeled MBP bands were quantified using ImageJ software and normalized by taking the radioactivity of the band in the absence of ABA as 1. Data are means ± SD of the relative band intensities from three independent experiments (n = 9).
MKKK18 accumulation is regulated by ABI1 and the ubiquitin–proteasome pathway
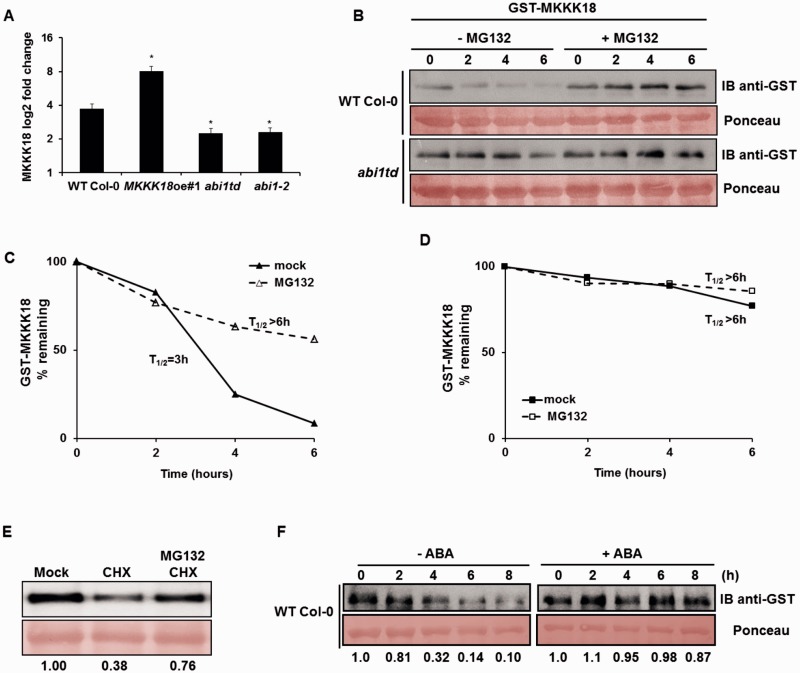
The MKKK18 protein was found to be in low abundance and, indeed, undetectable in total protein extracts from WT Col-0 plants. This feature of MKKK18 resembles other short-lived proteins, such as aminocyclopropane-1-carboxylic acid synthases (ACC synthases) 2 and 6, whose degradation is mediated by the ubiquitin–proteasome pathway (Liu and Zhang 2004, Joo et al. 2008). Recently, we demonstrated that ABI1 controls turnover of ACC synthase 6 (Ludwików et al. 2014, Ludwików, 2015); therefore, we hypothesize that ABI1 might also be involved in the regulation of MKKK18 stability. To test this hypothesis, we first examined MKKK18 transcript levels in WT plants and two independent ABI1 knockout lines, abi1td (Ludwikow et al. 2009) and abi1-2 (Saez et al. 2006). Importantly, in response to 90 min of 50 µM ABA treatment, MKKK18 expression was significantly lower in both ABI1 knockout lines compared with WT plants (P < 0.0001) (Fig. 9A). Because the decrease in MKKK18 transcript accumulation does not explain the increased MKKK18 kinase activity in the ABI1 knockout, we examined the turnover of MKKK18. Cell-free degradation assays showed that recombinant GST–MKKK18 was degraded more slowly in abi1td than in WT cell extracts (Fig. 9B). The degradation was blocked by MG132 (a proteasome inhibitor) in both abi1td and WT plant extracts. The half-life of GST–MKKK18 in the assay was substantially longer in abi1td protein extracts compared with WT protein extracts (Fig. 9C, D). Overall, these results demonstrate that ABI1 PP2C regulates MKKK18 turnover.
Fig. 9.
Proteasome-dependent degradation of MKKK18 is ABA-dependent and regulated by ABI1. (A) qPCR analysis of MKKK18 transcript accumulation in response to treatment with 50 µM ABA for 90 min in WT Col-0, MKKK18oe, abi1td and abi1-2 strains. MKKK18 expression levels were determined using three biological replicates and were normalized against 18S rDNA. The results are displayed as mean log2 fold change ± SE (n = 9) of three independent experiments with consistent results. (B) MKKK18 stability in the cell-free degradation assay. GST–MKKK18 was incubated with 100 µg of protein extract from either WT Col-0 or abi1td protoplasts incubated with or without 100 µM MG132 for 6 h in the dark. GST–MKKK18 protein levels at the indicated time points were determined by immunoblotting using anti-GST antibodies. Ponceau S staining confirmed equal loading. (C and D) Half-life plot for cell-free degradation of MKKK18 in WT Col-0 (C) and abi1td (D) extracts. Immunoblot images from each experiment were recorded simultaneously using a G:BOX Chemi XR5 fluorescence and chemiluminescence imaging system (Syngene), and the results were quantified using ImageJ software. (E) CHX treatment suppresses accumulation of MKKK18. Arabidopsis protoplasts expressing 35S:MKKK18-GFP were treated with 3 mM CHX, 3 mM CHX and 100 µM MG132, or given a mock treatment. The blot is representative of four experiments. MKKK18–GFP protein levels were determined by immunoblotting using anti-GFP antibodies. Ponceau S staining confirmed equal loading. MKKK18 protein bands were quantified using ImageJ software and normalized to the control (mock) band (set as 1). (F) MKKK18 protein levels are modified by ABA. GST–MKKK18 was incubated with 100 µg of total protein extract isolated from WT Col-0 incubated with or without 50 µM ABA for 3 h in the dark. GST–MKKK18 protein levels at the indicated time points were determined by immunoblotting using GST antibodies. Ponceau S staining confirmed equal loading. MKKK18 protein bands were quantified using ImageJ software and normalized to the control band (set as 1).
Next, we investigated MKKK18–GFP abundance in Arabidopsis protoplasts transiently expressing 35S:MKKK18-GFP. Under such conditions, the degradation rate of MKKK18 could be affected by its increased synthesis; therefore, to distinguish between translational or post-translational effects, we used a protein synthesis inhibitor, cycloheximide (CHX) (Fig. 9E). As expected, CHX treatment significantly reduced the accumulation of MKKK18 protein. Notably, simultaneous CHX and MG132 treatment prevents a reduction in MKKK18 protein levels, demonstrating that MKKK18 is degraded by the proteasome pathway.
Because both MKKK18 transcript level and kinase activity are regulated by ABA, we asked whether ABA affects the MKKK18 protein level. Again, the cell-free degradation assay was performed to test whether MKKK18 stability is affected by ABA. GST–MKKK18 recombinant protein was incubated with protein extracts prepared from WT protoplasts treated with 50 µM ABA or mock treated. GST–MKKK18 was rapidly degraded when incubated with WT protein extracts and this degradation was blocked by ABA (Fig. 9F). These results indicate that ABA regulates MKKK18 protein stability.
Discussion
Previously it was shown that ABA activates several MAPKs to perform various functions in the cell (Danquah et al. 2014, Danquah et al. 2015). In this study, we demonstrate that MKKK18 is also an element of the ABA response pathway and that it is specifically expressed in guard cells (Fig. 1I) and root meristem tissues (Fig. 1G). Interestingly, active MKKK18 is targeted specifically to the nucleus. We also show that MKKK18 kinase activity is increased by ABA treatment. Consistent with this, we show that MKKK18 is involved in ABA-related responses including stomatal development (Fig. 4), stomatal movement (Fig. 4), ABA inhibition of germination and root growth (Fig. 3). Notably, we identify ABI1 PP2C as an MKKK18-interacting protein (Fig. 6) and we demonstrate that ABI1 inhibits MKKK18 activity (Fig. 7B). We also establish that MKKK18 is unstable and is degraded by the 26S proteasome, providing evidence that ABI1 promotes proteasomal degradation of MKKK18 (Fig. 9).
ABA plays a major role in various aspects of plant growth and development, and is pivotal in stomatal function (Cai et al. 2014, Danquah et al. 2014, Smekalova et al. 2014). In this study, we show that loss of MKKK18 function leads to impaired stomatal development and a constitutive phenotype with more open stomata. As previously documented, ABA inhibits initiation of stomatal development (Tanaka et al. 2013), reducing the number of stomata per leaf and decreasing the SI (Franks and Farquhar 2001, Tanaka et al. 2013). Our results show that the SI of the MKKK18oe lines is significantly higher than in the WT Col-0 line (Fig. 4). Accordingly, the SIs of both MKKK18 knockouts are significantly lower (Fig. 4), suggesting that MKKK18 acts as a positive regulator of stomatal development. Interestingly, another MKKK, YODA, which fine-tunes this process, acts as a negative regulator in the early stages, but acts as a positive regulator in the final step, of stomatal development (Wang et al. 2007, Lampard et al. 2009). According to Danquah et al. (2015) and Matsuoka et al. (2015), the MKKK17/18 kinase cascade recruits MKK3–MPK1/2/7/14. YODA activates the MKK4/5–MKK7/MKK9 and MPK3/6 pathways during stomatal development. Thus we may hypothesize that the MKKK18 pathway is largely independent of YODA, although it may co-operate in the final stages of stomatal development. The involvement of different MAPK cascades in the same process was reviewed recently (Danquah et al. 2014), while participation of the same cascades in different processes has also been reported (Wang et al. 2007, Meng et al. 2012).
As is clear from the phenotypes described in this study, MKKK18oe lines show different responses to ABA stimulus depending on developmental stage and cell type. For example, germinating MKKK18oe seedlings exhibit a weak ABA-insensitive phenotype (Fig. 3E), while overexpression of MKKK18 also causes ABA-hypersensitive stomatal closing and root growth (Figs. 3C, 4F). This suggests that similar MKKK18-based signaling pathways may operate in these processes, but the opposite phenotypic responses to ABA stimulus may also be observed, indicating that MKKK18 signaling integrates various pathways.
Protein kinase autoactivation, via phosphorylation of the activation loop, is a common regulatory mechanism that switches the status of the signaling cascades to standby. Very recently, it was shown that recombinant GST–MKKK18 is able to autophosphorylate in vitro (Matsuoka et al. 2015). Our data appear to contradict this result. Compared with a positive control, SnRK2.6 kinase (Ng et al. 2011), we conclude that MKKK18 does not undergo autoactivation (Fig. 7). In our hands, GST–MKKK18 autophosphorylates neither during production in bacterial cells nor in in vitro kinase assays (Fig. 7C). Our results suggest, therefore, that MKKK18 must be activated by an upstream kinase (possibly a receptor kinase or another type of protein kinase) in response to ABA. Future studies are necessary to determine specific sites of regulatory significance in MKKK18.
To determine the exact mechanism underlying MKKK18 regulation, we tested the interaction between MKKK18 and the homologous phosphatases ABI1 and ABI2 (Fig. 6). ABI1/2 are known to be negative regulators of ABA signaling that interact with the ABA receptors RCAR/PYR/PYL and SnRK2s (Ma et al. 2009, Park et al. 2009, Umezawa et al. 2009, Vlad et al. 2009). Importantly, ABA-induced MKKK18 kinase activity is significantly increased in the ABI1 knockout (Fig. 8), while MKKK18 transcript levels were significantly reduced (Fig. 9A) compared with the WT. Our results suggest that the ABI1 protein phosphatase is needed for down-regulation of the pathway mediated by MKKK18. Interaction between MKKK18 and ABI1 supports the notion that the MKKK18 cascade operates downstream of the core ABA signaling pathway.
In addition, functional activation of the MKKK18 pathway is also constrained at the transcriptional level. MKKK18 expression is nearly abolished in ABA-insensitive abi1-1 (Hoth et al. 2002), hab1G246D and quadruple pyr1pyl1pyl2pyr4 ABA receptor mutants (Danaquah et al. 2015). Importantly, the hab1G246D mutant confers an equivalent hypermorphic mutation to abi1-1 (abi1G180D). Consistent with this, we observed that MKKK18 activity is abolished in the snrk2.6 mutant and another ABA-insensitive mutant (Fig. 8). Because SnRK2.6 activity is abolished or nearly abolished in snrk2.6, abi1-1 and pyr1pyl1pyl2pyr4 mutants, it is most likely that MKKK18 transcription is controlled by SnRK2.6. In the abi1td mutant, decreased MKKK18 transcription compensates for its increased stability. Overall, we may conclude that components of the core ABA pathway regulate MKKK18 function at multiple levels.
Another element of kinase activity regulation, especially in signal transduction pathways, is protein degradation via the proteasome pathway (Coulombe et al. 2003, Wang and Dohlman 2006, Peng et al. 2008, Fragoso et al. 2009, Lyzenga et al. 2013). Ubiquitinated proteins are present at low abundance and are unstable: in mammalian cells, MEKK1 and several isoforms of extracellular signal-regulated kinases (ERKs) are degraded via the proteasome pathway, leading to inactivation of ERK1/2 and c-Jun N-terminal kinase (JNK) pathways mediated by MEKK1 (Wang and Dohlman 2006). In Arabidopsis, well-documented examples of protein kinase degradation include SnRK1.1, SnRK1.2, GSK3-like kinase and CIPK26 (Peng et al. 2008, Fragoso et al. 2009, Lyzenga et al. 2013). Here we demonstrate that the interaction between ABI1 and MKKK18 is necessary for the destruction of the MKKK18 kinase (Fig. 9B–D). As shown recently, ABI1 promotes the degradation of ACS6 protein (Ludwików et al. 2014). This new role of ABI1 phosphatase is not restricted to a single protein, but also extends to the regulation of MKKK18 activity. We show that ABI1 inhibits MKKK18 activity (Fig. 7B) and, using a cell-free assay, we reveal that ABI1 promotes its degradation (Fig. 9B–D). Consistent with this, we also show that ABA treatment increases MKKK18 stability in cell-free assays (Fig. 9F), presumably due to inhibition of the ABI1 PP2C by ABA. However, it is possible that other mechanisms also regulate MKKK18 turnover. For example, the accumulation of the ABA-responsive transcription factor abscisic acid insensitive5 (ABI5) is achieved by ABA-dependent regulation of KEG E3 ligase turnover (Liu and Stone 2010). In conclusion, our results suggest that ABI1 PP2C can be considered a key player in the ABA-dependent feedback mechanism for resetting various signaling pathways to pre-stimulatory status.
In summary, an important aspect of the current research is the identification of key regulators of the ABA response and their signaling networks. Our current findings demonstrate that phosphorylation and dephosphorylation events play an important role in the transmission of the ABA signal. Although the SnRK2 protein kinases and group A PP2C-type phosphatases constitute a core ABA signaling pathway, the involvement of other groups of proteins, including MAP cascade kinases, has been recognized and documented in earlier studies (for the most recent review, see Danquah et al. 2014). The critical outstanding questions facing researchers are: which MAPKs are involved in ABA signal transduction, and what are the molecular details of the response to this signal? The analyses presented here demonstrate that MKKK18 is an ABA-activated kinase regulated by ABI1 and the proteasome. Based on the results presented in this study and on previously published data (Irigoyen et al. 2014), we propose a schematic model (Fig. 10) to describe the effect of ABI1 regulation on MKKK18. Further analysis will help to elucidate novel regulatory mechanisms involving MKKK18.
Materials and Methods
Plant growth and treatment
Arabidopsis thaliana Col-0 seeds were surface sterilized by treatment with 70% ethanol for 10 min, followed by four washes with sterile distilled water. The seeds were germinated on half-strength MS medium including 1% sucrose. After a 3 d stratification at 4°C, the seeds were transferred to a growth chamber and grown as described in Ludwików et al. (2009). Seeds of mkkk18-1 and mkkk18-2 were obtained from the Nottingham Arabidopsis Stock Centre. The T-DNA insertion line for abi1td was described previously (Ludwików et al. 2009).
Plasmid construction
For protein interaction analysis in yeasts, full-length ABI1 BD constructs and the PP2C catalytic domain (ΔN-ABI1) were used as described in Ludwików et al. (2009) and Ludwików et al. (2014), respectively. MKKK18 AD constructs were PCR-amplified using specific primer pairs (MKKK18fEcoRI and MKKK18rSacI), cloned into the pGEM-T Easy vector (Promega), excised as EcoRI–SacI restriction fragments, and ligated into the pACT2-AD vector. To generate the MKKK18 BD construct, a pUNI clone for MKKK18 (U18652) was recombined with the pACT2 vector using Cre recombinase (Liu et al. 1998). To generate the pENTR™/SD/D-TOPO® vector for facilitating the generation of C- or N-terminal in-frame MKKK18 fusions, cDNA for MKKK18 was amplified using Pfu polymerase, cloned into pENTR and then sequenced. To generate the recombinant GST- or His-tagged vectors, the pENTR-MKKK18 constructs were recombined with the Gateway® pDEST™15, Gateway® pDEST™17 or Gateway® pDEST™24 vectors using Gateway® LR Clonase® II Enzyme Mix (Invitrogen). For the BiFC assay, cDNAs for ABI1, ABI2 and MKKK18 were amplified using Pfu polymerase and then cloned into the pENTR/SD/D-TOPO vector (Invitrogen) (see Supplementary Methods S1 for details on the construction of the BiFC vector). The clones generated were digested with MluI, and the cDNA-containing restriction fragments were recombined with the pSAT3-cCFP-DEST and the pSAT5-DEST-nVenus vectors using Gateway® LR Clonase® II Enzyme Mix (Invitrogen), resulting in ncECFP–ABI1/2 and cnVenus–MKKK18 constructs, respectively. The DNA construct for MKKK18 localization and plant transformation (35S:MKKK18-GFP) was prepared in pEarleyGate 103 (Earley et al. 2006) using Gateway technology. Two out of six independent Col-0/35S:MKKK18-GFP transgenic lines were tested: #1 and #2. Site-directed mutagenesis of MKKK18 was performed using a QuikChange II XL Site-Directed Mutagenesis Kit (Agilent) according to the manufacturer’s protocol, and using the following oligonucleotides: K32M For 5′-CACTCGCCGTAATGTCCGCCGAGT-3′ and Rev 5′-ACTCGGCGGACATTACGGCGAGTG-3′; and T161E For 5′-GGTTGAACCGGAAATAGAGGAACCGGTTAGAGGAAC-3′ and Rev 5′-GTTCCTCTAACCGGTTCCTCTATTTCCGGTTCAACC-3′.
Construction of the MKKK18 promoter–GUS vector
The 1,622 bp MKKK18 promoter region was generated from genomic Arabidopsis DNA by PCR amplification using the primers indicated in Supplementary Table S1. The promoter sequence was verified by sequencing. BLAST analysis confirmed that isolated fragments had 99% identity with the known MKKK18 promoter region deposited in the TAIR database. To produce the 1,622 bp MKKK18 promoter region (the D::GUS construct), the PCR products were cloned into the pGEM T-Easy vector system (Promega), sequenced and cloned into the KpnI–PstI sites of the pBI101 vector. The promoter–reporter fusion construct was transformed into Agrobacterium tumefaciens strain LBA4404 and used for A. thaliana (Col-0) plant transformations using the floral dip method (Clough and Bent 1998). More than five independent transgenic lines were produced and tested for responsiveness by GUS histochemical assays. Three representative lines were selected for detailed characterization and showed the same expression pattern under all conditions tested.
Histochemical localization of GUS expression
For histochemical analysis of GUS expression, T2 seeds of Col-0/ProMKKK18:GUS were grown as described and exposed to 1 µM ABA for 24 h. Tissue samples were immersed in staining buffer [100 mM sodium phosphate, pH 7.0, 10 mM EDTA, 0.5 mM K4Fe(CN)6, 0.5 mM K3Fe(CN)6, 0.1% Triton X-100 and 1 mM X-gluc], and incubated overnight at 37°C. After the GUS reaction, plant samples were incubated in 80% ethanol to remove chlorophyll from the green tissues. GUS staining patterns were recorded using a Zeiss Stereo Lumar V12 microscope.
Analyses of the mkkk18-1 and mkkk18-2 T-DNA insertion lines
The mkkk18-1 and the mkkk18-2 T-DNA insertion lines were isolated from SALK insertion lines (SALK_087047) and from the GABI-Kat collection (GK-244G02), respectively. The qPCR analyses were performed as described in Ludwików et al. (2009).
Stomatal index and stomatal aperture
The SI was calculated based on the method described in Tanaka et al. (2013). Counts of stomata and pavement cells were performed on five areas of each cotyledon from 10 separate seedlings, giving 100 measurements per experiment, using a Nikon Eclipse Ti microscope with a Nikon Digital Sight DS-Fi1c camera. The SI was calculated individually for each cotyledon using the formula: SI = [number of stomata/(number of other epidermal cells + number of stomata)]. For statistical analysis, Student’s t-test was performed. For guard cell aperture measurements, rosette leaves of 5-week-old plants were floated in opening buffer (10 mM KCl, 7.5 mM EGTA, 10 mM MES-KOH, at pH 6.15) and incubated at 20°C under light conditions for 2 h to open the stomata. Subsequently, the same opening buffer was supplemented with either 15 µM ABA, 1 mM H2O2 or 1 mM CaCl2, and the leaves were incubated for another 2 h. The stomatal apertures (width divided by length) along three different epidermal transects were measured using a Nikon Eclipse Ti (with Nikon Digital Sight DS-Fi1c digital camera) and Nikon NIS Elements AR software. No less than 60 mature stomata were analyzed per transect.
Protoplast isolation, transformation and transient expression assays
Transient expression assays were performed using protoplasts from Arabidopsis mesophyll cells. Protoplast isolation, transformation and transient expression assays were performed based on methods described in Ludwików et al. (2014). Protoplasts were transiently transformed with 5 µg of plasmid, collected by centrifugation and resuspended in lysis buffer for further analysis. Transient protein expression in tobacco leaves was performed as described in Voinnet et al. (2003). Agrobacterium tumefaciens C58C1 cells with different MKKK18 constructs were mixed with an equal volume of Agrobacterium C58C1 (pCH32 35S:p19) expressing the silencing suppressor p19. The bacteria were injected into fully expanded leaves of tobacco plants, and were examined after 5 days.
Kinase activity assays
For the in vitro kinase assays, the MKKK18 immunocomplex assay was performed based on methods described in Ludwików et al. (2014). Briefly, tissue extracts containing 500 µg of total protein were immunoprecipitated for 1 h at 4°C with 5 µg of anti-GFP or anti-MKKK18 antibody pre-coupled to Dynabeads Protein-A (Invitrogen). This mixture was washed three times with wash buffer I (20 mM Tris–HCl, 5 mM EDTA, 100 mM NaCl, 1% Triton X-100), once with the same buffer but containing 1 M NaCl, and once with kinase buffer [20 mM HEPES, pH 7.5, 10 mM MgCl2, 1 mM dithiothreitol (DTT)]. A half-aliquot of MKKK18 was used to phosphorylate 5 µg of substrate (MBP; Sigma) in kinase buffer containing 25 µM ATP and [γ-32P]ATP (2 µCi per reaction) at 30°C. Another half-aliquot of activated MKKK18 was assayed under the same conditions without radioactive ATP. The reactions were stopped by the addition of SDS-loading buffer after 60 min. SDS–PAGE reaction products were analyzed using autoradiography or Western blotting.
Immunoblotting
Proteins were separated by 10% SDS–PAGE (BioRad) and were transferred onto Immobilon-P membranes (Millipore). The membranes were blocked for 1 h in PBS-T containing a 3% blocking solution, washed three times and incubated for 1 h with rabbit anti-GFP (1 : 200; sc-8334, Santa Cruz Biotechnology), StrepMAB-Classic (1 : 3,000; IBA BioTAGnology), anti-GST-Tag (1 : 5,000; Sigma), anti-MKKK18 antibody (AS13 2673, Agrisera), anti-ABI1 (AS12 1861, Agrisera) and anti-ABI2 (AS12 1871, Agrisera). After washing three times, the membranes were incubated for 1 h with the appropriate secondary antibody. Detection was performed with ECL (Thermo Scientific) according to the manufacturer’s instructions.
GST-tagged protein overexpression, purification, pull-down and cell-free degradation assays
Escherichia coli BL21(DE3) competent cells were transformed with recombinant MKKK18–GST, GST–ABI1 and GST–ABI2 expression constructs. Bacterial growth, isolation of recombinant GST fusion proteins, pull-down assays and cell-free degradation assays were conducted according to Ludwików et al. (2014). The abundance of MKKK18–GST was determined using anti-GST antibodies.
Bimolecular fluorescence complementation (BIFC) in Arabidopsis protoplasts
For the BiFC analysis, various combinations of plasmids encoding cECFP, nVenus fusion proteins and red fluorescent proteins (RFPs) as an internal control were mixed at a 1 : 1 : 1 (w/w) ratio, and the mixture of plasmid DNA was used for polyethylene glycol-mediated transformation as described by Ludwików et al. (2014).
Supplementary data
Supplementary data are available at PCP online.
Funding
This work was supported by the Dean of the Faculty of Biology [‘Grants for Interdisciplinary Grants’ No. GIWB-02/2011]; the National Science Centre [grants No. DEC-2012/05/B/NZ3/00352 to A.L. and DEC-2011/03/N/NZ3/01796 to M.M.]; KNOW Poznan RNA Centre [01/KNOW2/2014].
Supplementary Material
Acknowledgments
We thank David Baulcombe for providing the p19 vector construct and the Agrobacterium C58C1 strain, Olga Sztatelman for providing seeds of the snrk2.6 mutant, and Hanna Korcz-Szatkowska for excellent technical assistance.
Glossary
Abbreviations
- ABI1
abscisic acid insensitive 1
- ABI2
abscisic acid insensitive 2
- ABRE
ABA-responsive element
- ACS/ACC synthase
aminocyclopropane-1-carboxylic acid synthase
- AD
activation domain
- BD
binding domain
- BiFC
bimolecular fluorescence complementation
- CHX
cycloheximide
- CIPK
calcium-induced protein kinase
- GFP
green fluorescent protein
- GST
glutathione S-transferase
- GUS
β-glucuronidase
- HAB1
Hypersensitive to Abscisic acid 1
- MAPK/MPK
mitogen-activated protein kinase
- MAPKKK18
MAPK kinase kinase 18
- MKK
MAPK kinase
- MKKK/MEKK
MAPK kinase kinase
- MKKK18oe
transgenic overexpresor line
- MBP
myelin basic protein
- MS
Murashige and Skoog
- PP2C
protein phosphatase type 2C
- PYR/PYL/RCAR
Pyrabactin resistance1 (PYR1)/PYR1-like (PYL)/Regulatory Components of Abscisic acid Receptors (RCAR)
- qPCR
quantitative real-time PCR
- ROS
reactive oxygen species
- SI
stomatal index
- SnRK
sucrose non-fermenting (SNF)-related kinase
- WT
wild type
Disclosures
The authors have no conflicts of interest to declare.
References
- Babula-Skowrońska D., Ludwików A., Cieśla A., Olejnik A., Cegielska-Taras T., Bartkowiak-Broda I., et al. (2015) Involvement of genes encoding ABI1 protein phosphatases in the response of Brassica napus L. to drought stress. Plant Mol. Biol. 88: 445–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bate N., Twell D. (1998) Functional architecture of a late pollen promoter: pollen-specific transcription is developmentally regulated by multiple stage-specific and co-dependent activator elements. Plant Mol. Biol. 37: 859–869. [DOI] [PubMed] [Google Scholar]
- Boudsocq M., Willmann M.R., McCormack M., Lee H., Shan L., He P., et al. (2010) Differential innate immune signaling via Ca(2+) sensor protein kinases. Nature 464: 418–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock A.K., Willmann R., Kolb D., Grefen L., Lajunen H.M., Bethke G., et al. (2010) The Arabidopsis mitogen-activated protein kinase phosphatase PP2C5 affects seed germination, stomatal aperture, and abscisic acid-inducible gene expression. Plant Physiol. 153: 1098–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai G., Wang G., Wang L., Pan J., Liu Y., Li D. (2014) ZmMKK1, a novel group A mitogen-activated protein kinase kinase gene in maize, conferred chilling stress tolerance and was involved in pathogen defense in transgenic tobacco. Plant Sci. 214: 57–73. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Colcombet J., Hirt H. (2008) Arabidopsis MAPKs: a complex signaling network involved in multiple biological processes. Biochem. J. 413: 217–226. [DOI] [PubMed] [Google Scholar]
- Coulombe P., Rodier G., Pelletier S., Pellerin J., Meloche S. (2003) Rapid turnover of extracellular signal-regulated kinase 3 by the ubiquitin–proteasome pathway defines a novel paradigm of mitogen-activated protein kinase regulation during cellular differentiation. Mol. Cell. Biol. 23: 4542–4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danquah A., de Z.A., Colcombet J., Hirt H. (2014) The role of ABA and MAPK signaling pathways in plant abiotic stress responses. Biotechnol. Adv. 32: 40–52. [DOI] [PubMed] [Google Scholar]
- Danquah A., de Zélicourt A., Boudsocq M., Neubauer J., Frei dit Frey N., Leonhardt N., et al. (2015), Identification and characterization of an ABA-activated MAP kinase cascade in Arabidopsis thaliana. Plant J. 82: 232–244. [DOI] [PubMed] [Google Scholar]
- de Torres-Zabala M., Truman W., Bennett M.H., Lafforgue G., Mansfield J.W., Rodriguez E.P., et al. (2007) Pseudomonas syringae pv. tomato hijacks the Arabidopsis abscisic acid signaling pathway to cause disease. EMBO J. 26: 1434–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupeux F., Antoni R., Betz K., Santiago J., Gonzalez-Guzman M., Rodriguez L., et al. (2011a) Modulation of abscisic acid signaling in vivo by an engineered receptor-insensitive protein phosphatase type 2C allele. Plant Physiol. 156: 106–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupeux F., Santiago J., Betz K., Twycross J., Park S.Y., Rodriguez L., et al. (2011b) A thermodynamic switch modulates abscisic acid receptor sensitivity. EMBO J. 30: 4171–4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley K.W., Haag J.R., Pontes O., Opper K., Juehne T., Song K., et al. (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45: 616–629. [DOI] [PubMed] [Google Scholar]
- Fragoso S., Espindola L., Paez-Valencia J., Gamboa A., Camacho Y., Martinez-Barajas E., et al. (2009) SnRK1 isoforms AKIN10 and AKIN11 are differentially regulated in Arabidopsis plants under phosphate starvation. Plant Physiol. 149: 1906–1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks P.J., Farquhar G.D. (2001) The effect of exogenous abscisic acid on stomatal development, stomatal mechanics, and leaf gas exchange in Tradescantia virginiana. Plant Physiol. 125: 935–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y., Nakashima K., Yoshida T., Katagiri T., Kidokoro S., Kanamori N., et al. (2009) Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in Arabidopsis. Plant Cell Physiol. 50: 2123–2132. [DOI] [PubMed] [Google Scholar]
- Geiger D., Scherzer S., Mumm P., Stange A., Marten I., Bauer H., et al. (2009) Activity of guard cell anion channel SLAC1 is controlled by drought-stress signaling kinase–phosphatase pair. Proc. Natl. Acad. Sci. USA 106: 21425–21430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Xiong L., Song C.P., Gong D., Halfter U., Zhu J.K. (2002) A calcium sensor and its interacting protein kinase are global regulators of abscisic acid signaling in Arabidopsis. Dev. Cell 3: 233–244. [DOI] [PubMed] [Google Scholar]
- Himmelbach A., Hoffmann T., Leube M., Hohener B., Grill E. (2002) Homeodomain protein ATHB6 is a target of the protein phosphatase ABI1 and regulates hormone responses in Arabidopsis. EMBO J. 21: 3029–3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoth S., Morgante M., Sanchez J.P., Hanafey M.K., Tingey S.V., Chua N.H. (2002) Genome-wide gene expression profiling in Arabidopsis thaliana reveals new targets of abscisic acid and largely impaired gene regulation in the abi1-1 mutant. J. Cell Sci. 115: 4891–4900. [DOI] [PubMed] [Google Scholar]
- Irigoyen M.L., Iniesto E., Rodriguez L., Puga M.I., Yanagawa Y., Pick E., et al. (2014) Targeted degradation of abscisic acid receptors is mediated by the ubiquitin ligase substrate adaptor DDA1 in Arabidopsis. Plant Cell 26: 712–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jammes F., Song C., Shin D., Munemasa S., Takeda K., Gu D., et al. (2009) MAP kinases MPK9 and MPK12 are preferentially expressed in guard cells and positively regulate ROS-mediated ABA signaling. Proc. Natl. Acad. Sci. USA 106: 20520–20525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jammes F., Yang X., Xiao S., Kwak J.M. (2011) Two Arabidopsis guard cell-preferential MAPK genes, MPK9 and MPK12, function in biotic stress response. Plant Signal. Behav. 6: 1875–1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonak C., Okrész L., Bögre L., Hirt H. (2002) Complexity, cross talk and integration of plant MAP kinase signalling. Curr. Opin. Plant Biol. 5: 415–424. [DOI] [PubMed] [Google Scholar]
- Joo S., Liu Y., Lueth A., Zhang S. (2008) MAPK phosphorylation-induced stabilization of ACS6 protein is mediated by the non-catalytic C-terminal domain, which also contains the cis-determinant for rapid degradation by the 26S proteasome pathway. Plant J. 54: 129–140. [DOI] [PubMed] [Google Scholar]
- Kong Q., Qu N., Gao M., Zhang Z., Ding X., Yang F., et al. (2012) The MEKK1–MKK1/MKK2–MPK4 kinase cascade negatively regulates immunity mediated by a mitogen-activated protein kinase kinase kinase in Arabidopsis. Plant Cell 24: 2225–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawczyk S., Thurow C., Niggeweg R., Gatz C. (2002) Analysis of the spacing between the two palindromes of activation sequence-1 with respect to binding to different TGA factors and transcriptional activation potential. Nucleic Acids Res. 30: 775–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampard G.R., Lukowitz W., Ellis B.E., Bergmann D.C. (2009) Novel and expanded roles for MAPK signaling in Arabidopsis stomatal cell fate revealed by cell type-specific manipulations. Plant Cell 21: 3506–3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonhardt N., Kwak J.M., Robert N., Waner D., Leonhardt G., Schroeder J.I. (2004) Microarray expression analyses of Arabidopsis guard cells and isolation of a recessive abscisic acid hypersensitive protein phosphatase 2C mutant. Plant Cell 16: 596–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung J., Orfanidi S., Chefdor F., Meszaros T., Bolte S., Mizoguchi T., et al. (2006) Antagonistic interaction between MAP kinase and protein phosphatase 2C in stress recovery. Plant Sci. 171: 596–606. [Google Scholar]
- Liu Q., Li M.Z., Leibham D., Cortez D., Elledge S.J. (1998) The univector plasmid-fusion system, a method for rapid construction of recombinant DNA without restriction enzymes. Curr. Biol. 8: 1300–1309. [DOI] [PubMed] [Google Scholar]
- Liu H., Stone S.L. (2010) Abscisic acid increases Arabidopsis ABI5 transcription factor levels by promoting KEG E3 ligase self-ubiquitination and proteasomal degradation. Plant Cell 22: 2630–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Zhang S. (2004) Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. Plant Cell 16: 3386–3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwikow A., Kierzek D., Gallois P., Zeef L., Sadowski J. (2009) Gene expression profiling of ozone-treated Arabidopsis abi1td insertional mutant: protein phosphatase 2C ABI1 modulates biosynthesis ratio of ABA and ethylene. Planta 230: 1003–1017. [DOI] [PubMed] [Google Scholar]
- Ludwików A., Babula-Skowrońska D., Szczepaniak M., Belter N., Dominiak E., Sadowski J. (2013) Expression profiles and genomic organisation of group A protein phosphatase 2C genes in Brassica oleracea. Ann. Appl. Biol. 163: 124–134. [Google Scholar]
- Ludwikow A., Ciesla A., Kasprowicz-Maluski A., Mitula F., Tajdel M., Galganski L., et al. (2014) Arabidopsis protein phosphatase 2C ABI1 interacts with type I ACC synthases and is involved in the regulation of ozone-induced ethylene biosynthesis. Mol. Plant. 7: 960–976. [DOI] [PubMed] [Google Scholar]
- Ludwików A. (2015) Targeting proteins for proteasomal degradation—a new function of Arabidopsis ABI1 protein phosphatase 2C. Front. Plant Sci. 6: 310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyzenga W.J., Liu H., Schofield A., Muise-Hennessey A., Stone S.L. (2013) Arabidopsis CIPK26 interacts with KEG, components of the ABA signaling network and is degraded by the ubiquitin–proteasome system. J. Exp. Bot. 64: 2779–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Szostkiewicz I., Korte A., Moes D., Yang Y., Christmann A., et al. (2009) Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324: 1064–1068. [DOI] [PubMed] [Google Scholar]
- Mane S.P., Vasquez-Robinet C., Sioson A.A., Heath L.S., Grene R. (2007) Early PLDalpha-mediated events in response to progressive drought stress in Arabidopsis: a transcriptome analysis. J. Exp. Bot. 58: 241–252. [DOI] [PubMed] [Google Scholar]
- Matsuoka D., Yasufuku T., Furuya T., Nanmori T. (2015) An abscisic acid inducible Arabidopsis MAPKKK, MAPKKK18 regulates leaf senescence via its kinase activity. Plant Mol. Biol. 87: 565–575. [DOI] [PubMed] [Google Scholar]
- Meng X., Wang H., He Y., Liu Y., Walker J.C., Torii K.U., et al. (2012) A MAPK cascade downstream of ERECTA receptor-like protein kinase regulates Arabidopsis inflorescence architecture by promoting localized cell proliferation. Plant Cell 24: 4948–4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y., Lv D., Wang P., Wang X.C., Chen J., Miao C., et al. (2006) An Arabidopsis glutathione peroxidase functions as both a redox transducer and a scavenger in abscisic acid and drought stress responses. Plant Cell 18: 2749–2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K., Fujita Y., Kanamori N., Katagiri T., Umezawa T., Kidokoro S., et al. (2009) Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol. 50: 1345–1363. [DOI] [PubMed] [Google Scholar]
- Ng L.M., Soon F.F., Zhou X.E., West G.M., Kovach A., Suino-Powell K.M., et al. (2011) Structural basis for basal activity and autoactivation of abscisic acid (ABA) signaling SnRK2 kinases. Proc. Natl Acad. Sci. USA 108: 21259–21264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicaise V., Roux M., Zipfel C. (2009) Recent advances in PAMP-triggered immunity against bacteria: pattern recognition receptors watch over and raise the alarm. Plant Physiol. 150: 1638–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura N., Hitomi K., Arvai A.S., Rambo R.P., Hitomi C., Cutler S.R., et al. (2009) Structural mechanism of abscisic acid binding and signaling by dimeric PYR1. Science 326: 1373–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura N., Sarkeshik A., Nito K., Park S.Y., Wang A., Carvalho P.C., et al. (2010) PYR/PYL/RCAR family members are major in-vivo ABI1 protein phosphatase 2C-interacting proteins in Arabidopsis. Plant J. 61: 290–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta M., Guo Y., Halfter U., Zhu J.K. (2003) A novel domain in the protein kinase SOS2 mediates interaction with the protein phosphatase 2C ABI2. Proc. Natl. Acad. Sci. USA 100: 11771–11776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto M., Peterson F.C., Defries A., Park S.Y., Endo A., Nambara E., et al. (2013) Activation of dimeric ABA receptors elicits guard cell closure, ABA-regulated gene expression, and drought tolerance. Proc. Natl. Acad. Sci. USA 110: 12132–12137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.Y., Fung P., Nishimura N., Jensen D.R., Fujii H., Zhao Y., et al. (2009) Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324: 1068–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng P., Yan Z., Zhu Y., Li J. (2008) Regulation of the Arabidopsis GSK3-like kinase BRASSINOSTEROID-INSENSITIVE 2 through proteasome-mediated protein degradation. Mol. Plant 1: 338–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitzschke A., Djamei A., Bitton F., Hirt H. (2009) A major role of the MEKK1–MKK1/2–MPK4 pathway in ROS signaling. Mol. Plant 2: 120–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plesch G., Ehrhardt T., Mueller-Roeber B. (2001) Involvement of TAAAG elements suggests a role for Dof transcription factors in guard cell-specific gene expression. Plant J. 28: 455–464. [DOI] [PubMed] [Google Scholar]
- Qiu J.L., Zhou L., Yun B.W., Nielsen H.B., Fiil B.K., Petersen K., et al. (2008) Arabidopsis mitogen-activated protein kinase kinases MKK1 and MKK2 have overlapping functions in defense signaling mediated by MEKK1, MPK4, and MKS1. Plant Physiol. 148: 212–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez A., Rodrigues A., Santiago J., Rubio S., Rodriguez P.L. (2008) HAB1–SWI3B interaction reveals a link between abscisic acid signaling and putative SWI/SNF chromatin-remodeling complexes in Arabidopsis. Plant Cell 20: 2972–2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salam M.A., Jammes F., Hossain M.A., Ye W., Nakamura Y., Mori I.C., et al. (2013) Two guard cell-preferential MAPKs, MPK9 and MPK12, regulate YEL signaling in Arabidopsis guard cells. Plant Biol. (Stuttg.) 15: 436–442. [DOI] [PubMed] [Google Scholar]
- Santiago J., Dupeux F., Round A., Antoni R., Park S.Y., Jamin M., et al. (2009) The abscisic acid receptor PYR1 in complex with abscisic acid. Nature 462: 665–668. [DOI] [PubMed] [Google Scholar]
- Smekalova V., Luptovciak I., Komis G., Samajova O., Ovecka M., Doskocilova A., et al. (2014) Involvement of YODA and mitogen activated protein kinase 6 in Arabidopsis post-embryogenic root development through auxin up-regulation and cell division plane orientation. New Phytol. 203: 1175–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez-Rodriguez M.C., Adams-Phillips L., Liu Y., Wang H., Su S.H., Jester P.J., et al. (2007) MEKK1 is required for flg22-induced MPK4 activation in Arabidopsis plants. Plant Physiol. 143: 661–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostkiewicz I., Richter K., Kepka M., Demmel S., Ma Y., Korte A., et al. (2010) Closely related receptor complexes differ in their ABA selectivity and sensitivity. Plant J. 61: 25–35. [DOI] [PubMed] [Google Scholar]
- Takeuchi J., Okamoto M., Akiyama T., Muto T., Yajima S., Sue M., et al. (2014) Designed abscisic acid analogs as antagonists of PYL–PP2C receptor interactions. Nat. Chem. Biol. 10: 477–482. [DOI] [PubMed] [Google Scholar]
- Taki N., Sasaki-Sekimoto Y., Obayashi T., Kikuta A., Kobayashi K., Ainai T., et al. (2005) 12-Oxo-phytodienoic acid triggers expression of a distinct set of genes and plays a role in wound-induced gene expression in Arabidopsis. Plant Physiol. 139: 1268–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y., Nose T., Jikumaru Y., Kamiya Y. (2013) ABA inhibits entry into stomatal-lineage development in Arabidopsis leaves. Plant J. 74: 448–457. [DOI] [PubMed] [Google Scholar]
- Umezawa T., Sugiyama N., Mizoguchi M., Hayashi S., Myouga F., Yamaguchi-Shinozaki K., et al. (2009) Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc. Natl. Acad. Sci. USA 106: 17588–17593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlad F., Rubio S., Rodrigues A., Sirichandra C., Belin C., Robert N., et al. (2009) Protein phosphatases 2C regulate the activation of the Snf1-related kinase OST1 by abscisic acid in Arabidopsis. Plant Cell 21: 3170–3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O., Rivas S., Mestre P., Baulcombe D. (2003) An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33: 949–956. [DOI] [PubMed] [Google Scholar]
- Wang H., Ngwenyama N., Liu Y., Walker J.C., Zhang S. (2007) Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell 19: 63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.J., Zhu S.Y., Lu Y.F., Zhao R., Xin Q., Wang X.F., et al. (2010) Two coupled components of the mitogen-activated protein kinase cascade MdMPK1 and MdMKK1 from apple function in ABA signal transduction. Plant Cell Physiol. 51: 754–766. [DOI] [PubMed] [Google Scholar]
- Wang Y., Dohlman H.G. (2006) Regulation of G protein and mitogen-activated protein kinase signaling by ubiquitination: insights from model organisms. Circ. Res. 99: 1305–1314. [DOI] [PubMed] [Google Scholar]
- Xing Y., Jia W., Zhang J. (2008) AtMKK1 mediates ABA-induced CAT1 expression and H2O2 production via AtMPK6-coupled signaling in Arabidopsis. Plant J. 54: 440–451. [DOI] [PubMed] [Google Scholar]
- Xiong L., Lee H., Ishitani M., Zhu J.K. (2002) Regulation of osmotic stress-responsive gene expression by the LOS6/ABA1 locus in Arabidopsis. J. Biol. Chem. 277: 8588–8596. [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K., Shinozaki K. (2005) Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci. 10: 88–94. [DOI] [PubMed] [Google Scholar]
- Yamasaki K., Kigawa T., Watanabe S., Inoue M., Yamasaki T., Seki M., et al. (2012) Structural basis for sequence-specific DNA recognition by an Arabidopsis WRKY transcription factor. J. Biol. Chem. 287: 7683–7691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon J.H., Choi E.J., Parker R. (2010) Dcp2 phosphorylation by Ste20 modulates stress granule assembly and mRNA decay in Saccharomyces cerevisiae. J. Cell Biol. 189: 813–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T., Fujita Y., Sayama H., Kidokoro S., Maruyama K., Mizoi J., et al. (2010) AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J. 61: 672–685. [DOI] [PubMed] [Google Scholar]
- Zhu S.Y., Yu X.C., Wang X.J., Zhao R., Li Y., Fan R.C., et al. (2007) Two calcium-dependent protein kinases, CPK4 and CPK11, regulate abscisic acid signal transduction in Arabidopsis. Plant Cell 19: 3019–3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.