Abstract
Background:
The human Val66Met polymorphism in brain-derived neurotrophic factor (BDNF), a key factor in neuroplasticity, synaptic function, and cognition, has been implicated in the pathophysiology of neuropsychiatric and neurodegenerative disorders. BDNF is encoded by multiple transcripts with distinct regulation and localization, but the impact of the Val66Met polymorphism on BDNF regulation remains unclear.
Methods:
In BDNF Val66Met knock-in mice, which recapitulate the phenotypic hallmarks of individuals carrying the BDNFMet allele, we measured expression levels, epigenetic changes at promoters, and dendritic trafficking of distinct BDNF transcripts using quantitative PCR, chromatin immunoprecipitation (ChIP), and in situ hybridization.
Results:
BDNF-4 and BDNF-6 transcripts were reduced in BDNFMet/Met mice, compared with BDNFVal/Val mice. ChIP for acetyl-histone H3, a marker of active gene transcription, and trimethyl-histone-H3-Lys27 (H3K27me3), a marker of gene repression, showed higher H3K27me3 binding to exon 5, 6, and 8 promoters in BDNFMet/Met. The H3K27 methyltransferase enhancer of zeste homolog 2 (EZH2) is involved in epigenetic regulation of BDNF expression, because in neuroblastoma cells BDNF expression was increased both by short interference RNA for EZH2 and incubation with 3-deazaneplanocin A, an inhibitor of EZH2. In situ hybridization for BDNF-2, BDNF-4, and BDNF-6 after pilocarpine treatment showed that BDNF-6 transcript was virtually absent from distal dendrites of the CA1 and CA3 regions in BDNFMet/Met mice, while no changes were found for BDNF-2 and BDNF-4.
Conclusions:
Impaired BDNF expression and dendritic targeting in BDNFMet/Met mice may contribute to reduced regulated secretion of BDNF at synapses, and may be a specific correlate of pathology in individuals carrying the Met allele.
Keywords: BDNF, dendritic trafficking, epigenetics, histone, Val66Met polymorphism
Introduction
Brain-derived neurotrophic factor (BDNF) is a key factor in neuroplasticity, synaptic function, and cognition. Due to its pleiotropic effects in the brain, BDNF has been implicated in cognitive functions as well as in the pathogenesis of various neuropsychiatric and neurodegenerative disorders (Duman and Monteggia, 2006; Tardito et al., 2006; Castrén et al., 2007; Martinowich et al., 2007). In particular, the human G196A polymorphism in the BDNF gene, which leads to a substitution of valine with methionine at codon 66 in the prodomain, has been thoroughly investigated for genetic associations with cognitive dysfunction, personality disorders, and neuropsychiatric and neurodegenerative disorders (Egan et al., 2003; Hariri et al., 2003; see Fukumoto et al., 2010; Verhagen et al., 2010 for recent meta-analyses). However, although a number of studies found a significant correlation of the Val66Met polymorphism with select disorders, others were unable to replicate the findings. It has been suggested that these conflicting results may originate from confounding factors such as age, gender, environmental factors, sample size, ethnicity, and phenotype assessment (Hong et al., 2011). In addition, it is likely that the complex genetic background present in human populations can contribute to this problem. The BDNF Val66Met knock-in mouse offers an opportunity to investigate the pathophysiological consequences of the presence of the polymorphism in a constant and reproducible genetic background (Chen et al., 2006).
This knock-in mouse recapitulates the phenotypic hallmarks of the BDNF Val66Met human polymorphism. Indeed, both human and mice BDNFMet allele carriers show reduced hippocampal volume, cognitive deficits, increased anxiety-related behaviour, and impaired extinction of fear conditioning, a type of learning involved in phobias and posttraumatic stress disorder (Chen et al., 2006; Frielingsdorf et al., 2010; Soliman et al., 2010). It has been shown that BDNFMet/Met mice display reduced regulated secretion of BDNF from hippocampal neurons, reduced dendritic arborization in the hippocampal dentate gyrus (DG), and reduced N-methyl-d-aspartate receptor-dependent synaptic plasticity in the hippocampus (Chen et al., 2006; Ninan et al., 2010). Moreover, the binding of the BDNF transcript to the protein translin, required for mRNA export into dendrites, is disrupted by the presence of the Met mutation in the coding region of BDNF mRNA (Chiaruttini et al., 2009). Accordingly, we proposed a hypothesis for the BDNF Val66Met-associated memory deficits as a model of impaired dendritic mRNA trafficking (Baj et al, 2013a).
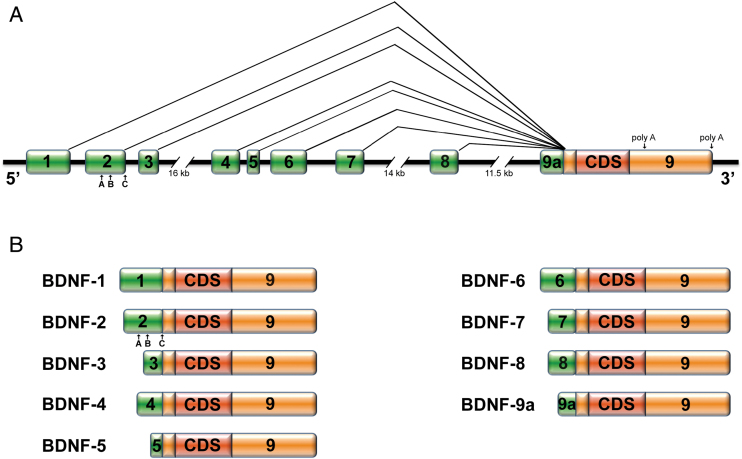
The bdnf rodent gene is composed of at least eight untranslated 5’ exons and one 3’ exon encoding the protein, and different transcript isoforms are expressed by splicing each of the non-coding exons to the coding exon (Aid et al., 2007; Figure 1). The complex structure of the BDNF gene allows the distinct BDNF transcripts to be differentially modulated at subcellular locations, in specific brain regions, and in response to distinct stimuli (Baj et al., 2013b; Musazzi et al., 2014). We have previously shown that BDNF mRNA variants are spatially segregated in response to neuronal activation within three subcellular compartments: the soma (BDNF variants 1, 3, 5, 7, 8, 9a), the proximal (BDNF 4) or the distal dendrites (BDNF 2, 6), thus behaving as a spatial code (Baj et al., 2011, 2013b). Moreover, we have also shown that BDNF-6 trafficking to dendrites is increased by physical exercise and chronic antidepressant treatments (Baj et al., 2012), suggesting a role for a local increase of BDNF-6 mRNA at dendrites in the pro-adaptive effects of these two treatments. Of note, BDNF expression can also be altered by genetic and epigenetic regulations, including histone posttranslational modifications (Martínez-Levy and Cruz-Fuentes, 2014). Accordingly, here we investigated whether the Val66Met mutation differentially affects the expression levels, epigenetic regulation, and dendritic trafficking of the different BDNF transcripts.
Figure 1.
Schematic representation of mouse BDNF (A) gene and (B) transcripts structure. BDNF, brain-derived neurotrophic factor; CDS, coding sequence.
Methods
Animals
Male 3–4 month old BDNFMet/Met and BDNFVal/Val mice were used for all studies (Chen et al., 2006). Mice were kept at 20–22°C on a 12h light/dark cycle (light on at 07:00 hours) with water and food available ad libitum. All animal handling and experimental procedures were performed in accordance with the European Community Council Directive 86/609/EEC and were approved by Italian legislation on animal experimentation (116/1992). All efforts were made to minimize animal distress and to reduce the numbers of animals used in this study.
RNA Isolation and Reverse Transcription
Total RNA was extracted from hippocampi of BDNFMet/Met and BDNFVal/Val mice, using the RNeasy Lipid Tissue Mini Kit (Qiagen) according to manufacturer’s instructions, and then quantified by absorption at A260nm, measured by UV spectrophotometry (NanoVue, GE Healthcare Europe GmbH). cDNA was synthesized from 1 μg of DNase-treated total RNA using a QuantiTect Reverse Transcription kit (Qiagen), following manufacturer’s directions.
Quantitative Real-Time PCR
Quantitative real time polymerase chain reaction (qPCR) analysis of mRNA expression levels was performed on a 7900HT Fast PCR System (Applied Biosystems by Life Technologies Italia) by using specific primers (Table 1) and QuantiFast SYBR Green master mix (Qiagen). All primers were as shown previously (Ieraci et al., 2015). To measure the distinct BDNF transcripts, we designed one specific forward primer for each 5’ exon and one common reverse primer in the 5’ BDNF coding sequence. To measure BDNF-2, BDNF-7, and total BDNF mRNA, both forward and reverse primers were within the respective exon region. PCR cycling conditions were: 10min at 95°C, 40 cycles of 15 s at 95°C, and 1min at 60°C. Data from qPCR were normalized on the mean of two reference genes (β-actin and glyceraldehyde-3-phosphate dehydrogenase). An analysis of melting curve verified the specificity of the PCR products. Relative amounts were determined using the comparative Cq method (Schmittgen and Livak, 2008).
Table 1.
List and Sequence of Primers Used in This Study
| Gene | Forward | Reverse |
|---|---|---|
| Real time PCR | ||
| BDNF-1 a | CCTGCATCTGTTGGGGAGAC | CGCCTTCATGCAACCGAAGTAT |
| BDNF-2 a | ACCTTTTCCTCCTCCTGCG | TGGATGAAGTACTACCACCTCGG |
| BDNF-3 a | TGAGACTGCGCTCCACTCCC | CGCCTTCATGCAACCGAAGTAT |
| BDNF-4 a | CAGAGCAGCTGCCTTGATGTTT | CGCCTTCATGCAACCGAAGTAT |
| BDNF-6 a | ACAATGTGACTCCACTGCCGG | CGCCTTCATGCAACCGAAGTAT |
| BDNF-7 a | ACTTACAGGTCCAAGGTCAACG | GGACAGAGGGTCGGATACAG |
| BDNF-8 a | ATGACTGTGCATCCCAGGAGAAA | CGCCTTCATGCAACCGAAGTAT |
| Total BDNF a | TCGTTCCTTTCGAGTTAGCC | TTGGTAAACGGCACAAAAC |
| EZH2 a | CAACCCTGTGACCATCCACG | CGACATCCAGGAAAGCGGTT |
| ß-actin a | GCCAGAGCAGTAATCTCCTTCT | AGTGTGACGTTGACATCCGTA |
| Gapdh a | CGTGCCGCCTGGAGAAACC | TGGAAGAGTGGGAGTTGCTGTTG |
| ChIP | ||
| pBDNF-1 b | TGATCATCACTCACGACCACG | CAGCCTCTCTGAGCCAGTTACG |
| pBDNF-2 b | CCGTCTTGTATTCCATCCTTTG | CCCAACTCCACCACTATCCTC |
| pBDNF-3 b | GTGAGAACCTGGGGCAAATC | ACGGAAAAGAGGGAGGGAAA |
| pBDNF-4 b | CTTCTGTGTGCGTGAATTTGCT | AGTCCACGAGAGGGCTCCA |
| pBDNF-5 | GCTGAAGGCGTGCGAGTATT | GGCCGATATGTACTCCTGTTCTG |
| pBDNF-6 b | ACTCACACTCGCTTCCTCCT | GCACTGGCTTCTCTCCATTT |
| pBDNF-7 | GGAAGGGTGGTGAGAGAGATATAGAG | GTGCCTAAGCCGGTGGTAAG |
| pBDNF-8 | TGAAAGGAAAAAGGAGAGGCTTT | AGAAAATGCCACTCCCACATAAG |
aPrimer sequences as in Ieraci et al. (2015).
bPrimer sequences as in Tsankova et al. (2006).
BDNF, brain-derived neurotrophic factor; ChIP, chromatin immunoprecipitation; EZH2, enhancer of zeste homolog 2; Gapdh, glyceraldehyde-3-phosphate dehydrogenase; PCR, polymerase chain reaction.
Chromatin Immunoprecipitation Assay
Total hippocampi of 15 BDNFMet/Met and 15 BDNFVal/Val mice were used. Hippocampal tissue was cut into 1mm3 cubes and fixed in 1% formaldehyde at room temperature for 15min to cross-link protein to DNA. Reaction was stopped by adding glycine (125mM final concentration) for 5min. Tissue was homogenized in sodium dodecyl sulfate (SDS) nuclei lysis buffer (50mM Tris, pH 8.1, 10mM EDTA, 1% SDS; Weinmann et al., 2001). Shearing of chromatin was performed with 13 cycles (20sec on, 40sec off) in a Microson sonicator (Misonix Inc.). Chromatin sonication sheared cross-linked DNA to the optimal size of 200–800 base pairs in length. Then, 150 µl chromatin was diluted 1:10 in chromatin immunoprecipitation (ChIP) dilution buffer (16.7mM Tris, pH 8.1, 1.2mM EDTA, 167mM NaCl, 1.1% Triton X-100, 0.01% SDS). ChIPs were performed by using the MagnaChIP kit (Merck Millipore), following manufacturer’s instructions. Briefly, chromatin was immunoprecipitated at 4°C with 10 µg each of the following antibodies: anti-H3K27me3 (Merck Millipore, catalog # 07-449), anti-acetyl histone H3 (acH3, Merck Millipore, catalog # 06-599), or normal rabbit immunoglobulin G as control antibody (MagnaChIP kit, Merck Millipore). One hundredth of the pre-immunoprecipitated diluted chromatin was saved as input DNA for ChIP normalization. Immune complexes were sequentially washed with low salt immune buffer, high salt buffer, LiCl immune complex buffer, and Tris-EDTA buffer. Protein-DNA cross-links were reversed by heating at 62°C for 2 hours. After proteinase K digestion, DNA was purified by spin filter columns (MagnaChIP kit, Merck Millipore). To quantify histone-associated gene promoters, immunoprecipitated DNA samples were subjected to qPCR on a 7900HT Fast PCR System (Applied Biosystems) using gene-specific primers and QuantiFast SYBR Green (Qiagen). The primer sequences used were either de novo designed using Primer Express software (Applied Biosystems) or previously published elsewhere (Tsankova et al., 2006). PCR cycling conditions were: 10min at 95°C, 40 cycles of 15 s at 95°C, and 1min at 60°C. DNA quantities were estimated from the quantification cycle (Cq) and then expressed as a percentage of corresponding input to normalize for differences in ChIP sample aliquots before immunoprecipitation. Briefly, percentage of input was calculated by 100 x 2(Cq-adjusted Input — Cq-enriched). Input DNA Cq was adjusted from 1% to 100% equivalent by subtracting 6.644 Cqs or Log2 100. An analysis of melting curve verified the specifity of the PCR products.
Cell Cultures, 3-Deazaneplanocin A Treatment, and Short Interference RNA Transfection
Mouse neuroblastoma N2A cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Euroclone), containing 10% fetal bovine serum (Euroclone) and 1% penicillin/streptomycin (Euroclone) at 37oC and 5% CO2. 3-deazaneplanocin A (DZNep) was purchased from Sequoia Research. DZNep was dissolved in distilled water and stored at -20°C.
For DZNep treatments, 2 x 105 cells were seeded in six multiwell plates 24 hours before the treatments. DZNep was added to cell cultures with a range of concentration (0.5; 1 and 2 μM) and cells were harvested 24 hours later. RNA was extracted and processed for qPCR analysis.
For short interference RNA (siRNA) knock-down studies, N2A cells were transfected with EZH2 (GeneSolution siRNA 1027416, Qiagen) or scramble siRNA (Negative Control siRNA 1022076, Qiagen) according to the optimized protocol for N2A Cells (Amaxa Cell Line Nucleofector Kit V, Lonza). Briefly, 1 x 105 cells were resuspended in 100 μl of Mouse NSC Nucleofector Solution containing 100nM of siRNA and nucleofected using the Program T-024 on an Amaxa Nucleofector II (Lonza). Cells were harvested 24 and 48 hours after transfection, and RNA was extracted and processed for qPCR analysis.
Pilocarpine Treatment
In order to measure dendritic targeting of BDNF by in situ hybridization and BDNF-2, BDNF-4, BDNF-6 and total BDNF mRNA expression, BDNFMet/Met and BDNFVal/Val mice were treated with pilocarpine (150mg/kg, i.p., Sigma-Aldrich) dissolved in 0.9% NaCl. To reduce peripheral cholinergic effects, mice were pre-treated with methylscopolamine (1mg/kg, i.p., Sigma-Aldrich) dissolved in PBS 20min before receiving pilocarpine. For the in situ hybridization experiment, six hours after pilocarpine administration, mice were anaesthetized with ketamine (100mg/kg, i.p., Sigma-Aldrich) and transcardially perfused with freshly made 4% paraformaldehyde (PFA, in PBS pH 7.3). The brains were then removed and kept in 4% PFA/20% sucrose (in PBS pH 7.3) at 4°C for at least 3 days before sectioning in 40 µm slices using a CO2-freezing microtome (Leica Microsystems). For BDNF transcripts expression, the vehicle groups were treated in sequence with PBS (as vehicle for methylscopolamine) and after 20min with saline (as vehicle for pilocarpine). Six hours after either vehicle or pilocarpine administration mice were sacrificed, their hippocampi were collected and RNA was extracted and processed for qPCR analysis.
In Situ Hybridization
Five BDNFMet/Met and five BDNFVal/Val mice were used. In situ hybridization on free-floating 40 µm coronal brain sections cut at the level of the dorsal hippocampus was performed as described previously (Tongiorgi et al., 1998, 2004). Specific probes against BDNF transcript containing BDNF-2, BDNF-4, or BDNF-6 were prepared from exon 2 nt 318–586 (Accession #NM_001270631), exon 4 nt 703–901 (Accession #X67107), or exon 6 nt 279–573 (Accession #S71211) cloned in pBSKS vector as previously described (Chiaruttini et al. 2008) using the DIG-RNA labeling kit according to manufacturer’s instructions (Roche Diagnostics, Monza, Italy). Probe specificity for BDNF-2, BDNF-4, and BDNF-6 was previously demonstrated (Chiaruttini et al., 2008). Hybridization with antisense probe was performed at 57°C followed by high stringency washes with 0.01X sodium saline citrate buffer containing 0.1% Tween-20 (SSCT) at 62°C. In situ hybridizations on brain sections from BDNFMet/Met and BDNFVal/Val mice were conducted in parallel. To obtain reproducible and comparable results and avoid saturation of the reaction, alkaline phosphatase development was always performed for 5h at room temperature. The use of non-radioactive in situ hybridization with digoxigenin-labeled probes for semi-quantitative evaluation of the in situ staining has been previously validated to study localization of dendritic mRNAs in vivo (Steward et al., 1998; Tongiorgi et al., 2004). In the present study, no saline-injected mice were included because in our previous work the BDNFMet/Met and the BDNFVal/Val mice differed in dendritic targeting of BDNF only following pilocarpine-induced neuronal activity, since in the basal condition BDNF mRNA is proximally confined (Chiaruttini et al., 2009). In addition, we previously found no difference between saline-injected or naive animals regarding the distribution of total BDNF mRNA (Tongiorgi et al., 2004) or exon 1, 2C, 4, and 6 mRNAs (Chiaruttini et al, 2008).
Statistical Analysis for ChIP and qPCR
For ChIP data, the percent input values from each data set were used in two-tailed unpaired t-tests to determine statistical significance. For mRNA data, fold changes relative to the respective control group (∆∆Cq method) were used in two-tailed unpaired t-tests (when two experimental groups were analyzed), in a Kruskal-Wallis test followed by Dunn’ multiple comparison test (when more than two experimental groups were analyzed), or in a two-way ANOVA followed by Bonferroni’s multiple comparison test (when the effect of the independent factors treatment and genotype were considered) to determine statistical significance. For all analyses, a value of p < 0.05 was considered statistically significant. Statistical analysis of the data was carried out using GraphPad Prism4 (GraphPad Software).
Quantitative Image Analysis and Statistics
Non-radioactive in situ hybridization was analyzed with a Nikon E800 microscope (X20 and X60 magnification, Nikon) and a CCD camera (DXM-1200, Nikon). Images were captured with a fixed illumination using a procedure described previously (Tongiorgi et al., 2004) and analyzed with ImageJ (Schneider et al., 2012). Student’s t-test was used for two-sample comparisons using the SigmaStat software (Systat Software).
Results
Expression of Splice Variants Containing BDNF-4 and BDNF-6 is Lower in BDNFMet/Met Mice
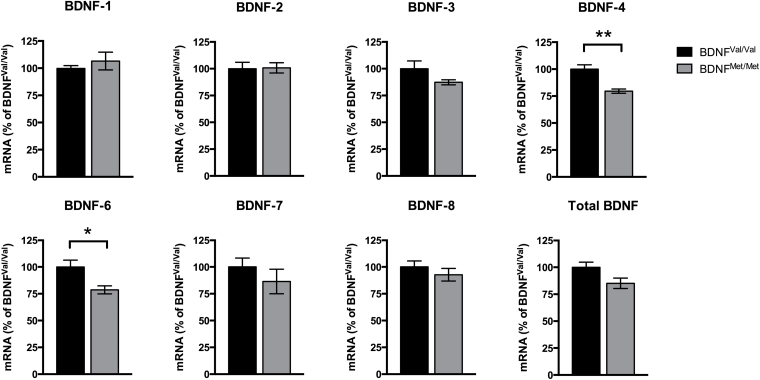
We assessed the levels of BDNF mRNAs containing distinct transcripts of non-coding exons (BDNF-1 to BDNF-8) and total BDNF mRNA (all transcripts containing the coding exon 9) in the hippocampi of BDNFVal/Val and BDNFMet/Met mice. qPCR analysis showed lower levels of BDNF-4 (-20.45% vs. BDNFVal/Val; p < 0.01) and BDNF-6 (-21.33% vs. BDNFVal/Val; p < 0.05; Figure 2) in BDNFMet/Met mice, while no significant differences were observed for the other BDNF variants (Figure 2). Interestingly, levels of total BDNF showed a trend toward reduction, although this was not statistically significant (-14.88%; p = 0.054; Figure 2). BDNF-5 transcript levels were very low in both BDNFVal/Val and BDNFMet/Met mice, which did not allow a reliable measurement of these mRNAs (not shown).
Figure 2.
Reduced expression of brain-derived neurotrophic factor (BDNF) transcripts in BDNFMet/Met mice. BDNF transcript expression levels were analyzed by quantitative real-time PCR (qPCR). BDNFMet/Met mice showed lower levels of BDNF-4 and BDNF-6 compared to BDNFVal/Val mice. Expression levels were normalized to β-actin and Gapdh and expressed as a percentage relative to BDNFVal/Val mice. Data are presented as mean ± standard error of the mean (n= 6 independent biological replicates). Student’s t-test, *p < 0.05, **p < 0.01.
Lys27 Trimethylation of H3 Histone at Select BDNF Promoters is Higher in Hippocampus of BDNFMet/Met Mice
To understand whether epigenetic changes are involved in BDNF expression in mice carrying the BDNF Val66Met polymorphism, we investigated the regulation of chromatin architecture at different BDNF promoters. To this aim, we used ChIP assay combined with qPCR to assess levels of acH3 and H3K27me3 at promoters of all non-coding BDNF transcripts (BDNF-1 to -8). AcH3 and H3K27me3 are known markers of transcriptional activation and repression, respectively (Tsankova et al., 2006). AcH3 was unchanged for all BDNF promoters (Table 2). Instead, we found significantly higher level of H3K27me3 at BDNF-5 (p < 0.005), BDNF-6 (p < 0.005), and BDNF-8 (p < 0.05) promoters in BDNFMet/Met compared to BDNFVal/Val mice (Table 2). These results show that reduced expression of BDNF-6 in BDNFMet/Met mice may be accounted for by this epigenetic change at the corresponding promoter. No changes were found at the BDNF-4 promoter, although expression of the corresponding transcript was reduced in BDNFMet/Met.
Table 2.
Levels of Occupancy of acH3 and H3K27me3 at BDNF Gene Promoters
| Gene promoter | BDNF Val/Val | BDNF Met/Met | p -value |
|---|---|---|---|
| acH3 | |||
| pBDNF-1 | 0.430±0.0996 | 0.572±0.1044 | 0.3819 |
| pBDNF-2 | 0.219±0.0252 | 0.339±0.0447 | 0.0799 |
| pBDNF-3 | 0.442±0.0834 | 0.697±0.1766 | 0.2616 |
| pBDNF-4 | 0.491±0.0478 | 0.493±0.0938 | 0.9858 |
| pBDNF-5 | 0.517±0.0668 | 0.673±0.1110 | 0.2948 |
| pBDNF-6 | 0.812±0.0888 | 1.059±0.0698 | 0.0946 |
| pBDNF-7 | 0.874±0.1258 | 1.118±0.1974 | 0.3561 |
| pBDNF-8 | 0.099±0.0249 | 0.1323±0.0313 | 0.4561 |
| H3K27me3 | |||
| pBDNF-1 | 3.250±0.3227 | 3.717±0.3250 | 0.3658 |
| pBDNF-2 | 3.658±0.1721 | 4.487±0.3230 | 0.0861 |
| pBDNF-3 | 3.555±0.4545 | 4.374±0.5488 | 0.3143 |
| pBDNF-4 | 3.196±0.2194 | 3.300±0.4021 | 0.8321 |
| pBDNF-5 | 2.421±0.0428 | 3.156±0.079 | 0.0012* |
| pBDNF-6 | 3.954±0.0832 | 4.543±0.0557 | 0.0042* |
| pBDNF-7 | 2.992±0.1374 | 3.657±0.574 | 0.0851 |
| pBDNF-8 | 1.645±0.1478 | 2.331±0.079 | 0.0150* |
Data are expressed as percentage of input and presented as mean ± standard error of the mean. Student’s t-test. *significant difference.
BDNF, brain-derived neurotrophic factor.
EZH2 Modulates BDNF Gene Expression
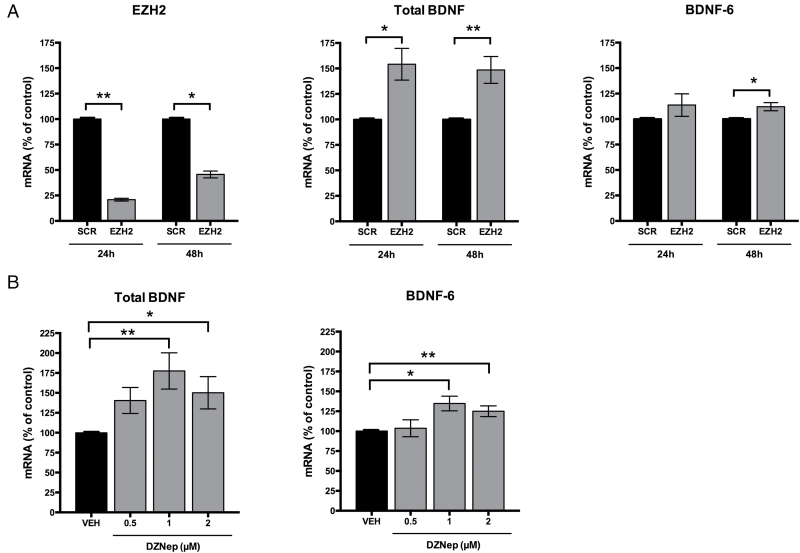
H3K27 di- and trimethylation, epigenetic modifications leading to chromatin condensation and transcriptional repression, are mediated by the multiprotein complex Polycomb Repressive Complex 2 (Pasini et al., 2004; Margueron and Reinberg, 2011). The minimum core components necessary for the catalytic activity in vitro are EZH2, embryonic ectoderm development, and suppressor of zeste 12, with EZH2 being the actual histone methyltransferase in K27 trimethylation (Laugesen and Helin, 2014). EZH2 has been involved in epigenetic reprogramming related to carcinogenesis (Chang and Hung, 2012; Deb et al., 2014). To understand whether EZH2 is able to modulate the transcription of either total BDNF mRNA or BDNF-6 transcript, we selectively down-regulated EZH2 expression in vitro using siRNA. Nucleofection of EZH2 siRNA in N2A neuroblastoma cells resulted in about 79.10±1.35% (p < 0.01) and 54.36±3.37% (p < 0.05) EZH2 mRNA reduction after 24 and 48 hours, respectively (Figure 3A). This reduction of EZH2 was accompanied by up-regulation of total BDNF of 54.10±15.53% (p < 0.05) and 48.50±13.18% (p < 0.01) after 24 and 48 hours, respectively (Figure 3A). At the same time points, the increase of BDNF-6 revealed a significant difference only after 48 hours (12.10±3.95%; p < 0.05; Figure 3A).
Figure 3.
Modulation of total brain-derived neurotrophic factor (BDNF) and BDNF-6 expression by EZH2. (A) Knockdown of EZH2 in N2A cells reduced expression of EZH2 (left panel). Knockdown of EZH2 induced up-regulation of total BDNF mRNA 24 and 48 hours after the nucleofection, and up-regulation of BDNF-6 after 48 hours (central and right panels). Expression levels were normalized to β-actin and Gapdh and results were expressed as mean ± standard error of the mean (SEM; n = 5 independent biological replicates). *p < 0.05; **p < 0.01 compared to scramble siRNA (SCR) by Student’s t-test. (B) mRNA levels of total BDNF and BDNF-6 in N2A cells were increased after 24 hours of treatment with 3-deazaneplanocin A (DZNep) at 0.5, 1, and 2 μM. Expression levels were normalized to β-actin and Gapdh and results are expressed as mean ± SEM (n = 5–6 independent biological replicates). *p < 0.05; **p < 0.01 compared to vehicle (VEH) by Kruskal-Wallis test followed by Dunn’s multiple comparison test.
To further confirm the role of EZH2 in the regulation of BDNF gene expression, N2A cells were treated with different concentrations of DZNep, which has been shown to deplete and inhibit EZH2 (Kikuchi et al., 2012). DZNep increased the expression of both total BDNF and BDNF-6, with the higher response observed at 1 μM (total BDNF: 77.50±22.79% of Veh; BDNF-6: 34.70±9.22% of Veh; p < 0.01; Figure 3B). Therefore, the present results show that EZH2 may modulate BDNF transcription through H3K27 methylation.
BDNF-6 Dendritic Trafficking is Impaired in Hippocampi of BDNFMet/Met Mice
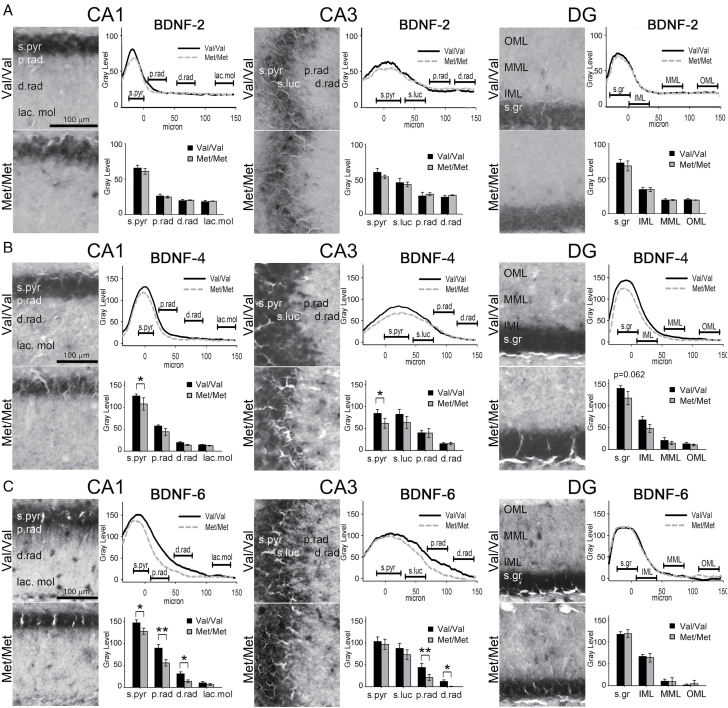
The splice variants containing BDNF-2, BDNF-6, and, to a lesser extent, BDNF-4, are the main ones targeted to dendrites in hippocampal neurons upon activation (Baj et al., 2011, 2013b). We have previously shown that trafficking of total BDNF in dendrites upon pilocarpine treatment is abolished in the CA1 and CA3 hippocampal areas of BDNFMet/Met mice (Chiaruttini et al., 2009). Therefore, in the present study we sought to assess whether pilocarpine-induced dendritic targeting of these mRNA variants was also reduced in BDNFMet/Met mice. We employed in situ hybridization to measure intracellular BDNF-2, BDNF-4, and BDNF-6 mRNA after pilocarpine stimulation in the CA1, CA3, and DG areas of the hippocampi of both mouse genotypes, as done previously for total BDNF mRNA (Chiaruttini et al., 2009; Figure 4A–C). In situ hybridization experiments were carried out at 6h post-pilocarpine because BDNF mRNA targeting in CA3 is at a maximum at this time in rodents (Tongiorgi et al., 2004) and CA3 is the region where the deficit in BDNF mRNA targeting is at a maximum in BDNFMet/Met mice (Chiaruttini et al., 2009).
Figure 4.
Reduced BDNF-6 dendritic targeting in BDNFMet/Met mice after pilocarpine treatment. High-magnification pictures of the in situ hybridization and densitometric analysis of dendrites at progressive distances from soma for: (A) BDNF2-mRNA, (B) BDNF-4, and (C) BDNF-6 mRNAs in mouse hippocampal CA1, CA3, and dental gyrate (DG) regions of BDNFVal/Val and BDNFMet/Met mice. Data are expressed by the gray level of the staining; the distance is expressed in µm from the point of emergence of the apical dendrites from the soma (=0 µm), and the bar represents the mean ± standard error of the mean (n= 5 independent biological replicates). *p < 0.05; **p < 0.01. d.rad, distal radiatum; IML, inner molecular layer; lac.mol, lacunosum molecular layer; MML, medial molecular layer; OML, outer molecular layer; p.rad, proximal radiatum; s.gr, stratum granularis; s.luc, stratum lucidum; s.pyr, stratum pyramidale.
Densitometric analysis of in situ labeling, to measure the labeling intensity at different distances from the cell body layer, showed no differences in BDNF-2 subcellular localization in all three areas of the hippocampi in BDNFMet/Met mice with respect to BDNFVal/Val (Figure 4A). However, for BDNF-4, a significant reduction was found in the stratum pyramidale of CA1 and CA3 (Figure 4B), while no changes were found in the other laminas. BDNF-6 mRNA labeling was significantly reduced in BDNFMet/Met mice in the proximal and distal stratum radiatum of both CA1 and CA3 (Figure 4C). No difference in pilocarpine-induced BDNF-6 trafficking was found in DG between the two genotypes. These results showed that the reduced trafficking of BDNF mRNA in hippocampal areas, previously found in BDNFMet/Met mice (Chiaruttini et al., 2009), is accounted for mostly, if not exclusively, by BDNF-6 containing splice variant.
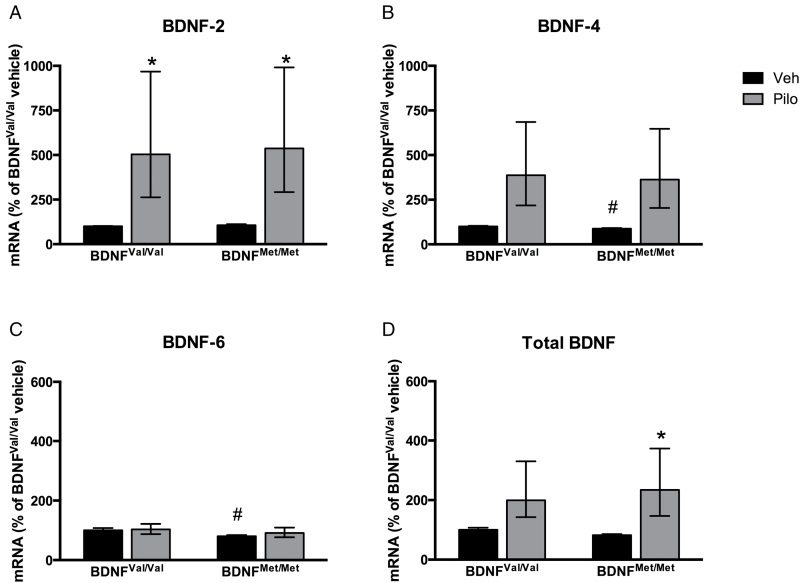
Pilocarpine has been shown to induce BDNF mRNA expression (Poulsen et al., 2004; Baj et al., 2013b), and since defective dendritic trafficking of BDNF transcripts could be due in principle to changes in either the trafficking mechanism or expression or both, we also sought to investigate whether expression of BDNF-2, BDNF-4, BDNF-6, and total BDNF were altered by pilocarpine treatment, in both genotypes. We found that pilocarpine increased the expression of BDNF-2, BDNF-4, and total BDNF similarly in both BDNFVal/Val and BDNFMet/Met mice (Figure 5A, B, and D), but did not affect significantly the expression of BDNF-6 (Figure 5C). Therefore, our present and previous results combined show that impaired trafficking of BDNF-6 in BDNFMet/Met mice is not due to reduced expression but to a selective defect in translocation.
Figure 5.
Expression of BDNF-6 is not altered by pilocarpine. (A) Pilocarpine treatment increased BDNF-2 expression levels in the hippocampi of BDNFVal/Val and BDNFMet/Met mice [two-way ANOVA, treatment effect: F(1,19) = 13,83, p = 0.0015; genotype effect: F(1,19) = 0,02023, p = 0,8884; interaction: F(1,19) = 1,289e-005, p = 0,9972]. (B) Pilocarpine treatment increased BDNF-4 expression levels in the hippocampi of BDNFVal/Val and BDNFMet/Met mice [two-way ANOVA, treatment effect: F(1,18) = 11.31, p = 0,0035; genotype effect: F(1,18) = 0,0538, p = 0,8192; interaction: F(1,18) = 0,00645, p = 0,9369]. An additional post hoc analysis with Student’s t-test confirmed lower BDNF-4 levels in BDNFMet/Met vs. BDNFVal/Val mice (p = 0.0325). (C) BDNF-6 expression levels were analyzed by quantitative real-time PCR (qPCR) in the hippocampi of BDNFVal/Val and BDNFMet/Met mice. Treatment with pilocarpine did not modify BDNF-6 levels [two-way ANOVA, treatment effect: F(1,19) = 0,4259, p = 0,5218; genotype effect: F(1,19) = 1.870, p = 0,1874; interaction: F(1,19) = 0,1794, p = 0,6767]. An additional post hoc analysis with Student’s t-test confirmed lower BDNF-6 levels in BDNFMet/Met vs. BDNFVal/Val mice (p = 0.0309). (D) Pilocarpine treatment increased total brain-derived neurotrophic factor (BDNF) expression levels in the hippocampi of BDNFVal/Val and BDNFMet/Met mice [two-way ANOVA, treatment effect: F(1,18) = 11.93, p = 0,0028; genotype effect: F(1,18) = 0,1005, p = 0,7548; interaction: F(1,18) = 0,1471, p = 0,7059]. Expression levels were normalized to β-actin and Gapdh and expressed as a percentage relative to BDNFVal/Val control mice. Data are presented as mean ± standard error of the mean (n= 5–7 independent biological replicates). Two-way ANOVA of vehicle (Veh) groups vs. pilocarpine treated (Pilo) groups followed by Bonferroni’s multiple comparison test (*p < 0.05 compared to respective vehicle group) or Student’s t-test (# p < 0.05 compared to BDNFVal/Val vehicle).
Discussion
BDNF has been implicated in the pathophysiology of brain disorders as well as in the action of therapeutic drugs, particularly antidepressants. We have previously reported that different BDNF transcripts code for different neuronal localization of BDNF (Chiaruttini et al., 2008), and that BDNF-6 is the major BDNF transcript targeted to dendrites upon neuronal activation (Baj et al., 2011, 2013b). More recently, we have shown that physical exercise and chronic antidepressant treatments not only increase BDNF expression in rodent hippocampi, but also enhance targeting of BDNF mRNA in hippocampal dendrites, and that this increased trafficking is accounted for by the BDNF-6–containing splice variant (Baj et al., 2012; Ieraci et al., 2015).
The results of the present work can be summarized as follows: (1) homozygous mice carrying the human BDNF Val66Met polymorphism of BDNF (BDNFMet/Met) display repressive epigenetic changes (increased H3K27me3) at BDNF promoters 5, 6, and 8; (2) the repressive epigenetic change at the BDNF-6 promoter is accompanied by reduced transcription of BDNF-6; (3) neuronal activation–induced trafficking of BDNF-6 is also impaired in BDNFMet/Met mice. This impairment is not due to reduced BDNF-6 expression, but to a defect in translocation.
In a recent genome-wide (ChIP-Seq) analysis we found differential epigenetic changes in a large number of gene promoters of BDNFMet/Met versus BDNFVal/Val mice (Popoli, 2012; ms. in preparation). Here we show that the presence of the human polymorphism also induces epigenetic changes in the BDNF gene itself. In particular, we have found an increase of the H3K27me3 level at the BDNF-6 promoter, consistent with the decrease of BDNF-6 transcript expression. Moreover, in cell cultures we have shown that RNA silencing or pharmacological inhibition of EZH2, the methyltransferase that induces histone H3 trimethylation of Lys 27, increased the expression of total BDNF and BDNF-6 transcripts. These results are in line with recent data showing that EZH2 increases the amount of H3K27me3 at BDNF promoters and decreases BDNF expression in endothelial cells and primary cortical neurons, indicating a possible common mechanism of BDNF regulation in different cell types (Qi et al., 2014; Mitić et al., 2015). The present results strongly suggest that increased H3K27me3 induces transcriptional repression of BDNF-6, although additional epigenetic changes may be involved. We have measured the expression of total BDNF in the hippocampi of BDNFMet/Met mice and found no significant changes (although a trend for reduction was found), in agreement with previous studies comparing BDNFVal/Met and BDNFMet/Met mice (Li et al., 2010; Yu et al., 2012). This lack of effect of the Val/Met polymorphism on total BDNF may suggest that the reduction in expression of BDNF-4 and BDNF-6 splice variants is not enough to significantly reduce the total expression of BDNF (including all splice variants). However, it is noteworthy that, although total BDNF mRNA was found to be unchanged in BDNFMet allele carrier mice, the BDNF protein has been previously found to be significantly reduced in both BDNFVal/Met and BDNFMet/Met mice (Li et al., 2010; Bath et al., 2012; Yu et al., 2012).
In the present study (except for BDNF-6) we often found a lack of correlation between expression levels of transcripts and epigenetic changes at corresponding promoters. For instance, we found a lower expression of BDNF-4 in BDNFMet/Met mice, although no changes were found in H3 histone modifications. Also, differently from BDNF-4 and BDNF-6, we found no changes in the expression of BDNF-8 in BDNFMet/Met, although this promoter showed increased K27 trimethylation of associated H3 histone. However, the level of expression of a gene is the combinatorial result of several epigenetic changes. It is not surprising to find a lack of correlation between a single histone mark at the gene promoter and expression levels of the corresponding gene (see Tsankova et al., 2004). It is possible that additional epigenetic changes regulate transcription of BDNF-8, explaining why in this case there is no direct correspondence between increased H3K27 trimethylation and reduced transcription.
We have previously shown that dendritic targeting of total BDNF is defective in BDNFMet/Met mice (Chiaruttini et al., 2009). Interestingly, as shown here, BDNFMet/Met mice display a selective impairment of BDNF-6 targeting to dendrites in the CA1 and CA3 hippocampal areas. Combined with our previous results, showing that increased BDNF dendritic trafficking induced by physical exercise and chronic antidepressants is accounted for by BDNF-6 (Baj et al., 2012), the present results suggest a crucial role for BDNF-6 in mediating some effects of the neurotrophin at synaptic locations. It can be envisaged that defective dendritic targeting of BDNF mRNA in BDNFMet/Met mice is mainly due to changes in BDNF-6. Of note, we found that at 6h post-pilocarpine treatment, BDNF-2 mRNA was not detectable in hippocampal laminas containing dendrites either in BDNFVal/Val or in BDNFMet/Met mice. This finding suggests that BDNF-2 mRNA targeting may have a different time-course with respect to BDNF-6.
The present findings add to the significance of the BDNF Val66Met knock-in mice as an interesting model that recapitulates several aspects of neuropsychiatric pathology (Frielingsdorf et al., 2010). Reduced expression and dendritic targeting of BDNF-6 could be a causal factor for reduced regulated release of BDNF at synapses, found in this knock-in mouse (Chen et al., 2006).
It is not known how the presence of the BDNF polymorphism may affect epigenetic regulation of BDNF transcription. At present, a likely explanation is that BDNF and other neurotrophins are capable of regulating their own transcription (Mallei et al., 2004; Wibrand et al. 2006; Yasuda et al., 2007). This regulation is selective for different BDNF transcripts, as we have shown previously (Musazzi et al., 2014). Reduced availability of BDNF at synapses may in turn contribute to reduced dendritic development and hippocampal volume, which are recognized features of BDNF Val66Met knock-in mice. This could be a distinct feature of pathology not only in mice, but also in humans carrying the BDNF Val66Met polymorphism that display reduced hippocampal volume (Frodl et al., 2007). In addition, because dendritic targeting of BDNF-6 is involved in the antidepressant mechanism (Baj et al., 2012), the impairment of this mechanism in BDNFMet/Met mice could be a reason for the lack of response to selective serotonin reuptake inhibitors in these mice (Chen et al., 2006; Bath et al., 2012).
It has been proposed that dysfunction in the glutamate system, including altered glutamate release/transmission, clearance, and metabolism, has a central role in neuropsychiatric pathophysiology (Sanacora et al., 2012). BDNF has a crucial role in the regulation of transmission and plasticity at glutamate synapses, and BDNFMet/Met mice have been shown to have impaired synaptic plasticity in the hippocampus (Ninan et al., 2010). Further studies are warranted to investigate the effects of BDNF-6 overexpression or silencing on synaptic function and behavior, as well as on the response to stress. It will also be interesting to assess the behavioral and cellular/molecular effects of chronic antidepressants or physical exercise in Val66Met knock-in mice, considering the specific influence exerted by these environmental factors on BDNF-6 expression and targeting (Baj et al., 2012) and its ability to modify the number of dendritic branches in the distal portion of the dendritic arbor (Baj et al., 2011).
Statement of Interest
None.
Acknowledgments
This work was supported by a grant from MIUR (PRIN 2010N8PBAA) to Drs Popoli and Tongiorgi and is part of the collaborative consortium “Italian Network on BDNF” (InBDNF). Dr Baj’s fellowship is supported by Fondazione Kathleen Foreman Casali-Trieste, FRA-UniTS 2012, and Beneficentia Stiftung-Vaduz (Lichtenstein).
References
- Aid T, Kazantseva A, Piirsoo M, Palm K, Timmusk T. (2007) Mouse and rat BDNF gene structure and expression revisited. J Neurosci Res 85:525–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baj G, Leone E, Chao MV, Tongiorgi E. (2011) Spatial segregation of BDNF transcripts enables BDNF to differentially shape distinct dendritic compartments. Proc Natl Acad Sci USA 108:16813–16818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baj G, D’Alessandro V, Musazzi L, Mallei A, Sartori CR, Sciancalepore M, Tardito D, Langone F, Popoli M, Tongiorgi E. (2012) Physical exercise and antidepressants enhance BDNF targeting in hippocampal CA3 dendrites: further evidence of a spatial code for BDNF splice variants. Neuropsychopharmacology 37:1600–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baj G, Carlino D, Gardossi L, Tongiorgi E. (2013a) Toward a unified biological hypothesis for the BDNF Val66Met-associated memory deficits in humans: a model of impaired dendritic mRNA trafficking. Front Neurosci 7:188. 10.3389/fnins.2013.00188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baj G, Del Turco D, Schlaudraff J, Torelli L, Deller T, Tongiorgi E. (2013b) Regulation of the spatial code for BDNF mRNA isoforms in the rat hippocampus following pilocarpine-treatment: a systematic analysis using laser microdissection and quantitative real-time PCR. Hippocampus 23:413–423. [DOI] [PubMed] [Google Scholar]
- Bath KG, Jing DQ, Dincheva I, Neeb CC, Pattwell SS, Chao MV, Lee FS, Ninan I (2012) BDNF Val66Met impairs fluoxetine-induced enhancement of adult hippocampus plasticity. Neuropsychopharmacology 37:1297–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castrén E, Võikar V, Rantamäki T. (2007) Role of neurotrophic factors in depression. Curr Opin Pharmacol 7:18–21. [DOI] [PubMed] [Google Scholar]
- Chang CJ, Hung MC. (2012) The role of EZH2 in tumour progression. Br J Cancer 106:243–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZY, Jing D, Bath KG, Ieraci A, Khan T, Siao CJ, Herrera DG, Toth M, Yang C, McEwen BS, Hempstead BL, Lee FS. (2006) Genetic variant BDNF (Val66Met) polymorphism alters anxiety-related behavior. Science 314:140–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiaruttini C, Sonego M, Baj G, Simonato M, Tongiorgi E. (2008) BDNF mRNA splice variants display activity-dependent targeting to distinct hippocampal laminae. Mol Cell Neurosci 37:11–19. [DOI] [PubMed] [Google Scholar]
- Chiaruttini C, Vicario A, Li Z, Baj G, Braiuca P, Wu Y, Lee FS, Gardossi L, Baraban JM, Tongiorgi E. (2009) Dendritic trafficking of BDNF mRNA is mediated by translin and blocked by the G196A (Val66Met) mutation. Proc Natl Acad Sci USA 106:16481–16486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deb G, Singh AK, Gupta S. (2014) EZH2: not EZHY (easy) to deal. Mol Cancer Res 12:639–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duman RS, Monteggia LM. (2006) A neurotrophic model for stress-related mood disorders. Biol Psychiatry 59:1116–1127. [DOI] [PubMed] [Google Scholar]
- Egan MF, Kojima M, Callicott JH, Goldberg TE, Kolachana BS, Bertolino A, Zaitsev E, Gold B, Goldman D, Dean M, Lu B, Weinberger DR. (2003) The BDNF val66met polymorphism affects activity-dependent secretion of BDNF and human memory and hippocampal function. Cell 112:257–269. [DOI] [PubMed] [Google Scholar]
- Frielingsdorf H, Bath KG, Soliman F, Difede J, Casey BJ, Lee FS. (2010) Variant brain-derived neurotrophic factor Val66Met endophenotypes: implications for posttraumatic stress disorder. Ann NY Acad Sci 1208:150–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frodl T, Schüle C, Schmitt G, Born C, Baghai T, Zill P, Bottlender R, Rupprecht R, Bondy B, Reiser M, Möller HJ, Meisenzahl EM. (2007) Association of the brain-derived neurotrophic factor Val66Met polymorphism with reduced hippocampal volumes in major depression. Arch Gen Psychiatry 64:410–416. [DOI] [PubMed] [Google Scholar]
- Fukumoto N, et al. (2010) Sexually dimorphic effect of the Val66Met polymorphism of BDNF on susceptibility to Alzheimer’s disease: new data and meta-analysis. Am J Med Genet B Neuropsychiatr Genet 153B:235–242. [DOI] [PubMed] [Google Scholar]
- Hariri AR, Goldberg TE, Mattay VS, Kolachana BS, Callicott JH, Egan MF, Weinberger DR. (2003) Brain-derived neurotrophic factor val66met polymorphism affects human memory-related hippocampal activity and predicts memory performance. J Neurosci 23:6690–6694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong CJ, Liou YJ, Tsai SJ. (2011) Effects of BDNF polymorphisms on brain function and behavior in health and disease. Brain Res Bull 86:287–297. [DOI] [PubMed] [Google Scholar]
- Ieraci A, Mallei A, Musazzi L, Popoli M. (2015) Physical exercise and acute restraint stress differentially modulate hippocampal brain-derived neurotrophic factor transcripts and epigenetic mechanisms in the mouse. Hippocampus. Advance online publication. Retrieved 14 May 2015. 10.11002/hipo.22458 [DOI] [PubMed] [Google Scholar]
- Kikuchi J, Takashina T, Kinoshita I, Kikuchi E, Shimizu Y, Sakakibara-Konishi J, Oizumi S, Marquez VE, Nishimura M, Dosaka-Akita H. (2012) Epigenetic therapy with 3-deazaneplanocin A, an inhibitor of the histone methyltransferase EZH2, inhibits growth of non-small cell lung cancer cells. Lung Cancer 78:138–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laugesen A, Helin K. (2014) Chromatin repressive complexes in stem cells, development, and cancer. Cell Stem Cell 14:735–751. [DOI] [PubMed] [Google Scholar]
- Li WJ, Yu H, Yang JM, Gao J, Jiang H, Feng M, Zhao YX, Chen ZY. (2010) Anxiolytic effect of music exposure on BDNFMet/Met transgenic mice. Brain Res 1347:71–79. [DOI] [PubMed] [Google Scholar]
- Mallei A, Rabin SJ, Mocchetti I. (2004) Autocrine regulation of nerve growth factor expression by Trk receptors. J Neurochem 90:1085–1093. [DOI] [PubMed] [Google Scholar]
- Margueron R, Reinberg D. (2011) The Polycomb complex PRC2 and its mark in life. Nature 469:343–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Levy GA, Cruz-Fuentes CS. (2014) Genetic and epigenetic regulation of the brain-derived neurotrophic factor in the central nervous system. Yale J Biol Med 87:173–186. [PMC free article] [PubMed] [Google Scholar]
- Martinowich K, Manji H, Lu B. (2007) New insights into BDNF function in depression and anxiety. Nat Neurosci 10:1089–1093. [DOI] [PubMed] [Google Scholar]
- Mitić T, Caporali A, Floris I, Meloni M, Marchetti M, Urrutia R, Angelini GD, Emanueli C. (2015) EZH2 modulates angiogenesis in vitro and in a mouse model of limb ischemia. Mol Ther 23:32–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musazzi L, Rimland JM, Ieraci A, Racagni G, Domenici E, Popoli M. (2014) Pharmacological characterization of BDNF promoters I, II and IV reveals that serotonin and norepinephrine input is sufficient for transcription activation. Int J Neuropsychop 17:779–91. [DOI] [PubMed] [Google Scholar]
- Ninan I, Bath KG, Dagar K, Perez-Castro R, Plummer MR, Lee FS, Chao MV. (2010) The BDNF Val66Met polymorphism impairs NMDA receptor-dependent synaptic plasticity in the hippocampus. J Neurosci 30:8866–8870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Helin K. (2004) Polycomb group proteins in cell cycle progression and cancer. Cell Cycle 3:396–400. [PubMed] [Google Scholar]
- Popoli M. (2012) Multi-method approaches to pharmacogenomics in animal models. Int J Neuropsychop 15(Supp 1):p. 25. [Google Scholar]
- Poulsen FR, Lauterborn J, Zimmer J, Gall CM. (2004) Differential expression of brain-derived neurotrophic factor transcripts after pilocarpine-induced seizure-like activity is related to mode of Ca2+ entry. Neuroscience 126:665–676. [DOI] [PubMed] [Google Scholar]
- Qi C, Liu S, Qin R, Zhang Y, Wang G, Shang Y, Wang Y, Liang J. (2014) Coordinated regulation of dendrite arborization by epigenetic factors CDYL and EZH2. J Neurosci 34:4494–4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanacora G, Treccani G, Popoli M. (2012) Towards a glutamate hypothesis of depression: an emerging frontier of neuropsychopharmacology for mood disorders. Neuropharmacology 62:63–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3:1101–1108. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soliman F, Glatt CE, Bath KG, Levita L, Jones RM, Pattwell SS, Jing D, Tottenham N, Amso D, Somerville LH, Voss HU, Glover G, Ballon DJ, Liston C, Teslovich T, Van Kempen T, Lee FS, Casey BJ. (2010) A genetic variant BDNF polymorphism alters extinction learning in both mouse and human. Science 327:863–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steward O, Wallace CS, Lyford GL, Worley PF. (1998) Synaptic activation causes the mRNA for the IEG Arc to localize selectively near activated postsynaptic sites on dendrites. Neuron 21:741–751. [DOI] [PubMed] [Google Scholar]
- Tardito D, Perez J, Tiraboschi E, Musazzi L, Racagni G, Popoli M. (2006) Signaling pathways regulating gene expression, neuroplasticity, and neurotrophic mechanisms in the action of antidepressants: a critical overview. Pharmacol Rev 58:115–134. [DOI] [PubMed] [Google Scholar]
- Tongiorgi E, Righi M, Cattaneo A. (1998) A non-radioactive in situ hybridization method that does not require RNAse-free conditions. J Neurosci Methods 85:129–139. [DOI] [PubMed] [Google Scholar]
- Tongiorgi E, Armellin M, Giulianini PG, Bregola G, Zucchini S, Paradiso B, Steward O, Cattaneo A, Simonato M. (2004) Brain-derived neurotrophic factor mRNA and protein are targeted to discrete dendritic laminas by events that trigger epileptogenesis. J Neurosci 24:6842–6852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsankova NM, Kumar A, Nestler EJ. (2004) Histone modifications at gene promoter regions in rat hippocampus after acute and chronic electroconvulsive seizures. J Neurosci 24:5603–5610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsankova NM, Berton O, Renthal W, Kumar A, Neve RL, Nestler EJ. (2006) Sustained hippocampal chromatin regulation in a mouse model of depression and antidepressant action. Nat Neurosci 9:519–525. [DOI] [PubMed] [Google Scholar]
- Verhagen M, van der Meij A, van Deurzen PA, Janzing JG, Arias-Vásquez A, Buitelaar JK, Franke B. (2010) Meta-analysis of the BDNF Val66Met polymorphism in major depressive disorder: effects of gender and ethnicity. Mol Psychiatry 15:260–271. [DOI] [PubMed] [Google Scholar]
- Weinmann AS, Bartley SM, Zhang T, Zhang MQ, Farnham PJ. (2001) Use of chromatin immunoprecipitation to clone novel E2F target promoters. Mol Cell Biol 21:6820–6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wibrand K, Messaoudi E, Håvik B, Steenslid V, Løvlie R, Steen VM, Bramham CR. (2006) Identification of genes co-upregulated with Arc during BDNF-induced long-term potentiation in adult rat dentate gyrus in vivo. Eur J Neurosci 23:1501–1511. [DOI] [PubMed] [Google Scholar]
- Yasuda M, Fukuchi M, Tabuchi A, Kawahara M, Tsuneki H, Azuma Y, Chiba Y, Tsuda M. (2007) Robust stimulation of TrkB induces delayed increases in BDNF and Arc mRNA expressions in cultured rat cortical neurons via distinct mechanisms. J Neurochem 103:626–636. [DOI] [PubMed] [Google Scholar]
- Yu H, Wang DD, Wang Y, Liu T, Lee FS, Chen ZY. (2012) Variant brain-derived neurotrophic factor Val66Met polymorphism alters vulnerability to stress and response to antidepressants. J Neurosci 32:4092–4101. [DOI] [PMC free article] [PubMed] [Google Scholar]