Abstract
This study explores the effect of rearing environment on water bacterial communities (BC) and the association with those present in the gut of Nile tilapia larvae (Oreochromis niloticus, Linnaeus) grown in either recirculating or active suspension systems. 454 pyrosequencing of PCR-amplified 16S rRNA gene fragments was applied to characterize the composition of water, feed and gut bacteria communities. Observed changes in water BC over time and differences in water BCs between systems were highly correlated with corresponding water physico-chemical properties. Differences in gut bacterial communities during larval development were correlated with differences in water communities between systems. The correlation of feed BC with those in the gut was minor compared to that between gut and water, reflected by the fact that 4 to 43 times more OTUs were shared between water and gut than between gut and feed BC. Shared OTUs between water and gut suggest a successful transfer of microorganisms from water into the gut, and give insight about the niche and ecological adaptability of water microorganisms inside the gut. These findings suggest that steering of gut microbial communities could be possible through water microbial management derived by the design and functionality of the rearing system.
Gut microbiota influences a wide range of biological processes of their host, including digestion, innate immunity, proliferation of epithelial cells and structural and functional maturation of the gut in humans1,2, domesticated terrestrial animals3,4 and fish5,6. However, our understanding of the roles and drivers for community diversity and species dominance within the gut is presently limited7.
Fish are exposed to higher microbial loads in the aquatic environment than terrestrial domesticated animals are in air or soil8. Thus, this closer contact with the surrounding water likely affects early gut colonization. Whilst water seems to affect the fish gut microbiota already from mouth opening onwards9, feed microbiota becomes important at a later development stage10,11. However, although water and feed are the two main sources of microorganisms available to fish, the factors underlying the successful colonization of ingested microbes and the community assembly inside the gut are still poorly understood12. For this reason, predictability and repeatability of gut microbiota manipulations is currently limited.
The nutrient rich environment of high density aquaculture is conducive to the proliferation of microbes. To this end, aquaculture management aims to minimize the amount of nutrients in the rearing tank. For example, in recirculating aquaculture systems (RAS) the first step of water purification is solids removal13,14. In contrast, in active suspension (AS) systems, fish excreta and feed left overs are mineralized inside the rearing tank. AS systems are characterized by the formation of bioflocs (aggregates of microorganisms, colloids, organic polymers and dead cells), which contribute to the maintenance of good water quality while providing additional nutrients to the fish15.
In the present study, we hypothesized that water bacterial communities would differ between the two rearing environments (i.e. RAS vs. AS systems) due to differences in systems’ design. Assuming that gut colonization is strongly affected by the composition of the microbiota in the water, we expected that composition of gut communities would differ between systems as well. Compared to RAS, bacteria in the water of AS systems have a higher chance of being successfully transferred in the gut, due to the constant grazing of larvae on bioflocs. Thus, a greater influence of AS water microbiota on the gut is expected. We used 454 pyrosequencing of PCR-amplified bacterial 16S ribosomal RNA (rRNA) gene fragments to characterize for the first time the composition of gut BCs of Nile tilapia Oreochromis niloticus (Linnaeus) reared for 42 days in RAS or AS tanks from the moment of first feeding. The progressive gut colonization in fish reared in the two systems and the similarity of water and feed BCs with those in the gut were evaluated at different time points. Furthermore, the potential role of the most predominant bacteria found in this study is discussed in detail based on a comprehensive literature review.
Materials and Methods
Experimental animals and set up
The experiment was performed in accordance with relevant guidelines and approved by the Ethical Committee of Wageningen University for animal experiments (Registration code: 2009055d). One batch of three days post-fertilization (3 dpf) eggs were washed out from the mouth of a female Nile tilapia and incubated for four days at 27 °C. Eggs were incubated in a tank until swim-up stage and received water from the same source. For rearing the larvae, two different culture systems were used: a recirculating aquaculture system (RAS) and an active suspension (AS) system with two replicates each (Ra&Rb and AS1&AS2), which were initially filled with the same water coming from a well. Replicate systems did not share the same water. Only for RAS systems, each replicate also contained 2 replicate tanks (Ra1, Ra2 & Rb1, Rb2) which were connected to the same water treatment unit (biofilter). In each tank, 100 randomly selected swim-up larvae (7 dpf) were introduced before first feeding and before mouth opening. Mouth opening occurred within the experimental rearing systems. Feeding started 9 dpf (referred to as day 0) and was continued for 42 days. Fish were fed three times a day to apparent satiation with a starter feed (F-0.5 GR Pro Aqua Brut – Trouw Nutrition, France; 57% crude protein, 15% crude fat, 8.5% carbohydrates, 11% crude ash, 0.6% crude fiber, 1.7% phosphorus). A 42 day culture period was considered sufficiently long for all major stages of the gut development16. Water, feed and gut samples were collected on day 7 and 42. Further details about the experimental setup, animals and description of the rearing systems can be found elsewhere17.
Sampling of guts, water and feed
On sampling days, three tilapia larvae were randomly collected from Ra1, Ra2, Rb1, Rb2, AS1 and AS2 for gut BC analysis. After euthanization, the larvae were rinsed with 70% ethanol and sterile water, before dissecting the gut. Gut samples were flash-frozen in liquid nitrogen and stored individually at −80 °C until further processing. Furthermore, 250 mL of water was collected from each tank. Water was simultaneously filtered through a 0.45 μm (type HAWP) over a 0.22 μm (type GTTP) membrane filter (Millipore - Isopore™) using a vacuum apparatus. Filters were stored at −80 °C until processing. The 0.45 μm filters were used in order to avoid clogging of the 0.22 μm with large size particles during filtration. However, both filters were used for DNA extraction from water. Finally, also 2 grams of feed were collected and stored at −80 °C until processing. All gut, water and feed samples were stored and analysed individually. Samples were not pooled neither were the corresponding DNA extracts.
On a weekly basis, total ammonia nitrogen (TAN-N), nitrite (NO2−-N), nitrate (NO3−-N), orthophosphate (PO43−), carbon dioxide (CO2), urea, dissolved oxygen (DO), pH, conductivity and temperature (°C) were measured in each tank according to Giatsis et al. (2014).
Isolation of DNA
DNA was extracted from larval gut samples using the DNeasy Blood & Tissue Kit (Qiagen, Venlo, Netherlands), whereas the FastDNA SPIN kit for soil (MP Biomedicals, Ohio, USA) was used for water and feed samples. The use of different physical, mechanical, chemical and enzymatic lysis protocols, different purification and precipitation techniques and different reagents, have shown to affect genomic DNA yield and quality. This was confirmed by Lever et al.18, however, these authors furthermore concluded that the observed effects are almost always sample dependent. This being said, before our study, genomic DNA was extracted from all sample types by using two extraction protocols. Since the DNeasy Blood & Tissue Kit resulted in a very low DNA yield from the water and feed samples and a high DNA yield was obtained with the FastDNA SPIN kit for soil, and hence the latter was used for all water and feed samples in this study. In contrast, the FastDNA SPIN kit for soil showed a low yield for DNA extraction from gut samples, while a high yield was obtained with the DNeasy Blood & Tissue Kit. Therefore, the DNeasy Blood & Tissue Kit was used for all gut samples in this study.
Modifications of the manufacturer’s protocol with respect to enhancement of cell lysis (i.e. bead beating), DNA purification and elution processes, were made as previously described by Giatsis et al. (2014).
PCR and 454 Pyrosequencing
Barcoded amplicons from the V1-V2 region of 16S rRNA genes were generated by PCR using the 27F-DegS primer19 appended with titanium sequencing adaptor A and an 8 nt sample-specific barcode20 at the 5′ end. As reverse primer, an equimolar mix of two primers 338R I and II21,22 was used to carry the titanium adaptor B at the 5′ end. A detailed protocol for the PCR is given by Giatsis et al. (2014). Nucleotide sequences were generated by pyrosequencing using an FLX genome sequencer in combination with titanium chemistry (GATC-Biotech, Konstanz, Germany). These sequence data were submitted to the European Bioinformatics Institute under study accession No PRJEB4462 and sample accession No ERS343984 – ERS344037.
Data handling
Pyrosequencing data were analyzed using the QIIME 1.5.0 pipeline23 and quality filtering was performed as follows: low quality sequences were removed using default parameters. Removed reads included (i) reads with less than 200 or more than 1000 nucleotides; (ii) reads with more than 6 ambiguous nucleotides, homopolymer runs exceeding 6 bases, reads with missing quality scores and reads with a mean quality score lower than 25; and (iii) reads with mismatches in the primer sequence. Operational taxonomic units (OTUs) were identified at the 97% identity level. Representative sequences from the OTUs were aligned using PyNAST24. The taxonomic affiliation of each OTU was determined using the RDP classifier at a confidence threshold of 80% against the 12_10 Greengenes core set25. Possible chimeric OTUs were identified using QIIME’s ChimeraSlayer and removed from the initially generated OTU list, producing a final set of non-chimeric OTUs.
To visualize possible commonalities and anti-correlations between gut, water and feed BCs, physical samples and OTUs were plotted as nodes in a bipartite network. In the network analysis, the average relative abundance of OTUs in replicate samples was represented. To cluster the OTUs and hosts in this network, the Edge-weighted Spring Embedded algorithm as implemented in Cytoscape 3.0.2 was used26.
Statistical analysis
A repeated measure ANOVA was applied for water quality measurements on day 7 and 42 with “system” (RAS or AS) as main factor and “day” (7 or 42) as repeated measure. Based on Draftsman plots (variable pair-wise scatter plots) of water quality data, the TAN values were log-transformed (Figure S1). Subsequently, all environmental variables were normalized. The Euclidean distance of normalized environmental variables was used as dissimilarity measure. Gut, water and feed BC data were expressed as relative abundance of all OTUs in each sample, and Bray Curtis similarity was calculated based on square root transformed data.
A distance-based linear model (distLM) was used for analysing and modelling the relationship between the bacterial community composition, as described by a resemblance matrix, and the predictor variables (i.e. water parameters). When not enough possible permutations were available, P-values for testing the null hypothesis of no relationship between the two datasets were obtained by Monte Carlo permutations (a sample was drawn from the theoretical asymptotic permutation distribution)27. Forward selection of the environmental variables was applied and distance-based redundancy analysis (dbRDA) was used for visualization28,29.
Based on Bray Curtis similarity of relative abundance data, RELATE analysis evaluated the relatedness between water and gut BC by calculating Spearman’s ρ correlation coefficient between all elements. Similarity percentages (SIMPER) analysis was used to determine the OTUs which contributed most to the discrimination or relatedness of samples in each group. To measure alpha-diversity of the bacterial communities: the total number of observed species (S), Pielou’s evenness (J′ = H′/log(S)) and Shannon (H′ = −SUM(Pi*ln(Pi))) were calculated (Pi is the proportion of the total count (abundance) arising from the ith species). As species richness and evenness can only be compared between samples when sample sizes are equal30, resulting reads were randomly selected so as to standardize to the sample with the least number of obtained reads (n = 1719). To visualize community evenness, dominance plots were created based on relative abundance data. OTU relative abundance (y-axis) was averaged per sample type (gut and water), for each system (AS and RAS) per day (7 and 42). Cumulative relative abundance was plotted against the increasing species rank (x-axis).
Statistical analyses were performed using Primer 6 (version 6.1.11) (Primer-E Ltd., Plymouth, United Kingdom) and its PERMANOVA add-on package31. Venn diagrams were produced in Venny online freeware32.
Phylogenetic analysis
The first 150 OTUs contributing most to differences between groups were considered for a more thorough phylogenetic analysis. Representative sequences of these OTUs were aligned using the online SINA alignment service of the ARB-Silva database33. Aligned sequences were imported into ARB in the Silva release 115 SSU NR 99 tree34 using the ARB parsimony method without changing the tree topology. Three near neighbors of all 150 OTUs were selected according to the following criteria: (i) they were at least 800 nucleotides long and included the entire sequenced amplicon, and (ii) neighbors per OTU were picked from different published studies. An alignment containing only the neighbors was exported before constructing a Bayesian tree. Ambiguous regions of the alignment were systematically removed using the program Gblocks v.0.91b35. Default parameters were used, except allowing a minimum block length of 5 and gaps in 50% of positions. Phylogenetic trees were calculated by Bayesian analysis, using a locally installed version of MrBayes 3.236. All parameters were treated as unknown variables with uniform prior-probability densities at the beginning of each run, and their values were estimated from the data during the analysis. All Bayesian analyses were initiated with random starting trees and were run for 107 generations. The number of chains was set to four and Markov chains were sampled every 1000 iterations. Points prior to convergence were determined graphically and discarded. Calculated trees were imported into ARB, and short sequences obtained in this study were subsequently added by use of the ARB parsimony method without changing the tree topology.
Results
The effect of rearing system and time on water quality
Analysis of environmental data showed significant differences in water quality between RAS and AS. Conductivity, NO3-N and PO4-P were higher, whereas CO2, DO and pH were lower in RAS (P < 0.05) than in AS (Table 1). Conductivity, NO3-N and PO4-P increased, and CO2, DO and pH dropped between day 7 and 42 (P < 0.05). In contrast, temperature and concentrations of NO2-N, urea and TAN were neither significantly different (P > 0.05) between RAS and AS, nor between day 7 and 42.
Table 1. ANOVA results with system as main factor and day as repeated measure.
| Parameter | System |
Day |
Syst*Day | ||||||
|---|---|---|---|---|---|---|---|---|---|
| RAS |
AS |
7 |
42 |
||||||
| 7 | 42 | 7 | 42 | RAS | AS | RAS | AS | ||
| TAN (mg L−1) | 0.03 | 0.05 | 0.06 | 0.04 | 0.03 | 0.06 | 0.05 | 0.04 | 0.047 |
| NO2 (mg L−1) | 0.03 | 0.11 | 0.04 | 0.10 | 0.303 | ||||
| NO3 (mg L−1) | 12.4 | 54.3 | 4.5 | 28.2 | 12.4 | 4.5 | 54.3 | 28.2 | 0.000 |
| PO4 (mg L−1) | 1.46 | 1.04 | 0.37 | 2.13 | 0.098 | ||||
| CO2 (mg L−1) | 59.1 | 9.4 | 72.9 | 39.0 | 59.1 | 72.9 | 9.4 | 39.0 | 0.003 |
| Urea (mg L−1) | 0.02 | 0.07 | 0.03 | 0.06 | 0.465 | ||||
| T (oC) | 27.3 | 27.5 | 27.2 | 27.6 | 0.884 | ||||
| pH | 8.06 | 7.28 | 8.15 | 8.10 | 8.06 | 8.15 | 7.28 | 8.10 | 0.002 |
| DO (mg L−1) | 7.72 | 8.00 | 8.18 | 7.55 | 0.150 | ||||
| Cond (μS cm−1) | 402 | 820 | 260 | 456 | 402 | 260 | 820 | 456 | 0.002 |
Average water quality parameters calculated for recirculating aquaculture system (RAS) or active suspension (AS) and for day 7 and 42 (n = 4) are given. The right column gives the “System * Day” interaction P values (bold for P < 0.05). Within system or day, bold values indicate significant difference (P < 0.05). TAN: Total ammonia nitrogen; NO2-N: Nitrite nitrogen; NO3-N: Nitrate nitrogen; PO43-P: Phosphate phosphorous; CO2: Carbon dioxide; T: Temperature; DO: Dissolved oxygen; Cond.: Conductivity.
Correlation of environmental parameters with water bacterial communities
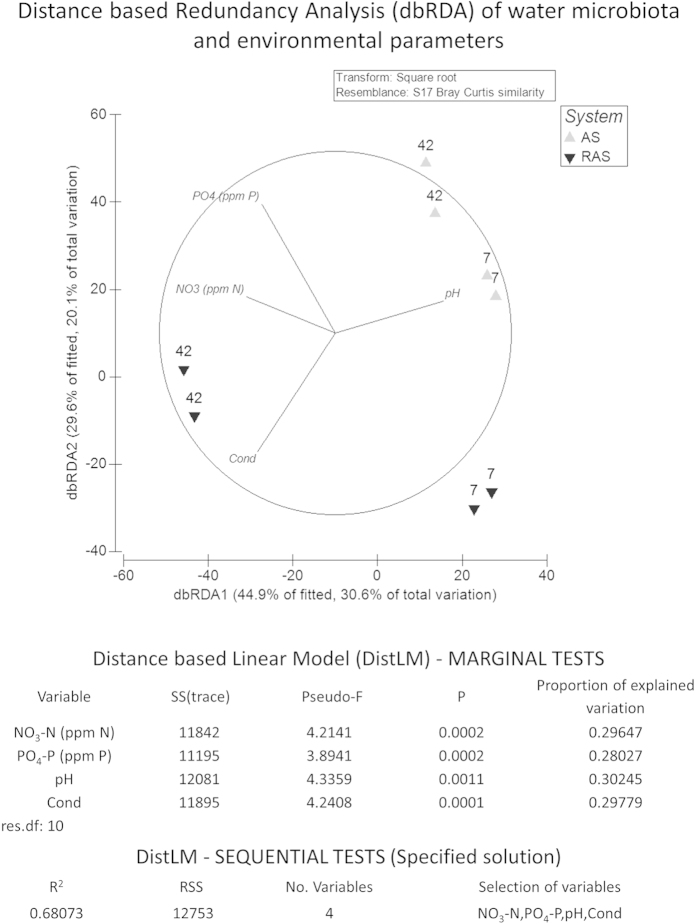
Distance-based linear model (DistLM) analysis revealed that pH, conductivity, NO3-N and PO43-P together explained 68% (R2 sequential) of the observed total variation in the composition of the bacterial community in the water. Conductivity and pH strongly correlated with differences in BCs between RAS and AS, while PO4-P and NO3-N correlated with differences in the communities between day 7 and 42 within system (Fig. 1). The same parameters showed a highly significant system * day interaction with repeated measures ANOVA of water quality data (Table 1).
Figure 1. Distance based Redundancy Analysis (dbRDA) of water microbiota.
Relative position of water samples in the biplot is based on Bray Curtis similarity of square root transformed relative abundance at the OTU level. Vectors indicate the weight and direction of those water quality parameters that were best predictors of water bacterial composition as suggested by the results of the distance-based linear model (distLM). The dbRDA axes describe the percentage of the fitted or total variation explained by each axis while being constrained to account for group differences. Sample IDs indicate the sampling day (7 and 42) and the rearing system (AS & RAS: Active suspension and recirculating aquaculture system with two replicates each). NO3−-N: Nitrate nitrogen; PO43−-P: Phosphate phosphorous; Cond.: Conductivity.
Similarity of water bacterial communities between replicate systems over time
Overall, 11,658 OTUs were identified in all samples. At OTU level, the average similarity of water BC within AS on day 7 was 64% (Table 2). The 5 OTUs (SIMPER analysis), in order of relative abundance, explaining most of the average similarity were OTU 9828 (order Sphingomonadales), 4552 (genus Limnohabitans), 8066 (genus Sediminibacterium), 2756 (genus Rhodobacter) and 11945 (genus Nitrospira). Combined, these 5 OTUs accounted for 49% of the bacterial community in AS water on day 7 (Table S1).
Table 2. Percentage average Bray Curtis similarity between gut, water and feed bacterial communities.
| % Average Bray Curtis similarity | ||||||
|---|---|---|---|---|---|---|
| Gut |
Water |
|||||
| Between replicate AS (AS1, AS2) | Between replicate RAS (Ra, Rb) | Between AS & RAS | Between replicate AS (AS1, AS2) | Between replicate RAS (Ra, Rb) | Between AS & RAS | |
| Day 7 | 68.9 (±7.6) | 55.1 (±15.9) | 21.6 | 63.8 | 59.0 | 23.9 |
| Day 42 | 59.9 (±18.1) | 56.4 (±11.0) | 10.8 | 25.8 | 60.8 | 1.8 |
| Between day 7 & 42 of gut | Between day 7 & 42 of water | |||||
| AS | 21.6 | 17.3 | ||||
| RAS | 27.2 | 3.5 | ||||
| Day 7 | Day 42 | |||||
| Between gut & water | Between gut & feed | Between water & feed | Between gut & water | Between gut & feed | Between water & feed | |
| AS | 4.1 | 0.1 | 0.1 | 8.5 | 0.1 | 0.1 |
| RAS | 3.9 | 0.1 | 0.1 | 8.1 | 0.1 | 0.1 |
Values are based on SIMPER analysis of square root transformed relative abundance data at the OTU level. Values within brackets indicate the standard deviation of the average gut Bray Curtis similarity within group, as a measure of multivariate within group dispersion. AS & RAS: Active suspension and recirculating aquaculture systems respectively; AS1 & AS2: Replicate active suspension system 1 & 2; Ra & Rb: Replicate recirculating aquaculture system a & b; 7 and 42: Sampling day 7 and 42.
On day 42, the water BC average similarity within AS decreased to 26% (Table 2). Almost 60% of this similarity was attributed to OTU 8066 (genus Sediminibacterium), 14254 (family Comamonadaceae), and 4686 (order Sphingobacteriales) (Table S1). Other predominant OTUs of AS water BC on day 42 included: OTU 5688 (genus Fimbriimonas) and 7261 (genus Armatimonas).
Overall, water bacterial communities in AS systems on day 7 was 17% similar with BC present on day 42 since most of the predominant OTUs on one day were absent or rare on the other (Table 2). Despite the low similarity, 85 OTUs were shared between the two dates, with a cumulative relative abundance of 63% on day 7 and 52% on day 42 (Table 3). OTU 8066 contributed most to the similarity, whereas OTU 9828 contributed most to the dissimilarity between the two days (Table S1).
Table 3. Number of unique and shared OTUs and relative abundance of shared OTUs between days.
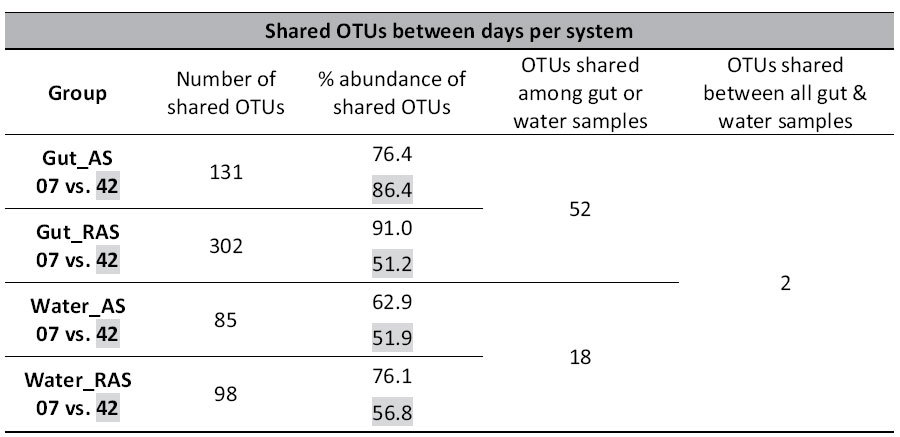
Shaded abundance values correspond to shaded sampling group in the first column. Sampling groups are compared between sampling days for each system. OTU: Operational taxonomic unit; AS & RAS: Active suspension and recirculating aquaculture systems; 07 & 42: Sampling days 7 and 42 respectively.
In RAS, the average similarity of water BC on day 7 was 59% (Table 2). More than half of this similarity was attributed to only two predominant OTUs: OTU 7333 (genus Polynucleobacter) and 9828 (family Sphingomonadales), which together had a cumulative relative abundance of 46.8% on that day (Table S1).
On day 42, similarity of water BC within RAS remained at levels similar to day 7. This similarity was mostly attributed to OTU 7090 (family Rhodocyclaceae), 14286 (genus Limnohabitans), and 4377 (genus Rhodobacter). Other predominant OTUs of RAS water BC on day 42 included: OTU 5688 (genus Fimbriimonas) and 7261 (genus Armatimonas). The aforementioned five OTUs accounted for 55.2% of the total bacterial community on that day (Table S1).
Bacterial communities in the RAS water differed considerably between day 7 and 42 (3.5% similarity) (Table 2). Predominant OTUs in water on day 7 were almost absent on day 42 and vice versa. For example, OTU 7090 was absent on day 7 whilst accounted for 35% of the community on day 42, thus contributing about 20% to the difference between the two days (Table S1). Despite the low similarity, water BC in RAS shared 98 OTUs between day 7 and 42, with a cumulative relative abundance of 76% and 57%, respectively (Table 3).
Similarity of water bacterial communities between systems
Already on day 7, the average similarity between AS and RAS was only 24% (Table 2). The two systems shared 113 OTUs with a cumulative relative abundance of 62.5% and 73% in AS and RAS systems, respectively (Table 4). On day 42, the similarity of water BC between the two systems decreased to 2%, since almost all predominant OTUs in AS water, were absent in RAS and vice versa. Overall, water from the two systems shared 40 OTUs with a cumulative relative abundance of 30% in AS and only 4% in RAS (Table 4).
Table 4. Number of unique and shared OTUs within and between systems.
| Shared OTUs between systems per day | |||
|---|---|---|---|
| Group | Total number of OTUs | Number of shared OTUs | Within group % abundance of shared OTUs |
| Gut_07_AS | 732 | 247 | 70.7 |
| Gut_07_RAS | 1003 | 71.4 | |
| Gut_42_AS | 279 | 90 | 43.9 |
| Gut_42_RAS | 879 | 50.7 | |
| Water_07_AS | 683 | 113 | 62.5 |
| Water_07_RAS | 815 | 73.0 | |
| Water_42_AS | 394 | 40 | 30.3 |
| Water_42_RAS | 552 | 3.6 | |
Percentages indicate the relative abundance of shared OTUs within each system. Operational taxonomic unit; AS & RAS: Active suspension and recirculating aquaculture systems; 07 & 42: Sampling days 7 and 42 respectively.
In terms of evenness, an overall system (AS & RAS) * day (7 & 42) interaction was observed. On day 7, 17 OTUs had a cumulative relative abundance of 60% in AS water, whereas on day 42, the same percentage was attributed to six OTUs. In RAS water, 60% of the relative abundance was attributed to 6 OTUs on day 7 and three OTUs on day 42 (Table S1). Additionally, over time species richness decreased from 683 to 394 OTUs in AS water and from 815 to 552 in RAS. This observation is also in agreement with the results of the dominance plots and the biodiversity indices which showed that the richness and evenness of water bacterial communities in AS decreased between day 7 and 42, whereas this difference was less pronounced in RAS (Figure S2 & Table S2).
Similarity of gut bacterial communities between replicate systems over time
On day 7, average similarity of gut bacterial communities within AS was 69% (Table 2). Half of this similarity was attributed to OTUs 4790 (species Mycobacterium llatzerense), 1651 (genus Rhodobacter), 10562 (genus Gordonia), 10599 (family Bradyrhizobiaceae) and 8299 (genus Sporocytophaga). Those five OTUs accounted for a cumulative relative abundance of 42.2% in AS (Table S1). On day 42, the similarity of gut BC between samples dropped to 60%. More than 80% of this similarity was attributed to OTUs 4563 (family Isosphaeraceae), OTU 12537 (genus Arthrobacter) and OTU 4790 (species Mycobacterium llatzerense). Those predominant OTUs were encountered on both days, albeit at different relative abundances (Table 5). M. llatzerense (OTU 4790) was the only OTU with a high relative abundance on both sampling days. Despite the 131 shared OTUs between the two days, gut BC was 21.6% similar between days (Table 2, Table 3).
Table 5. Heatmap of the most predominant gut, water and feed OTUs on day 7 and 42.
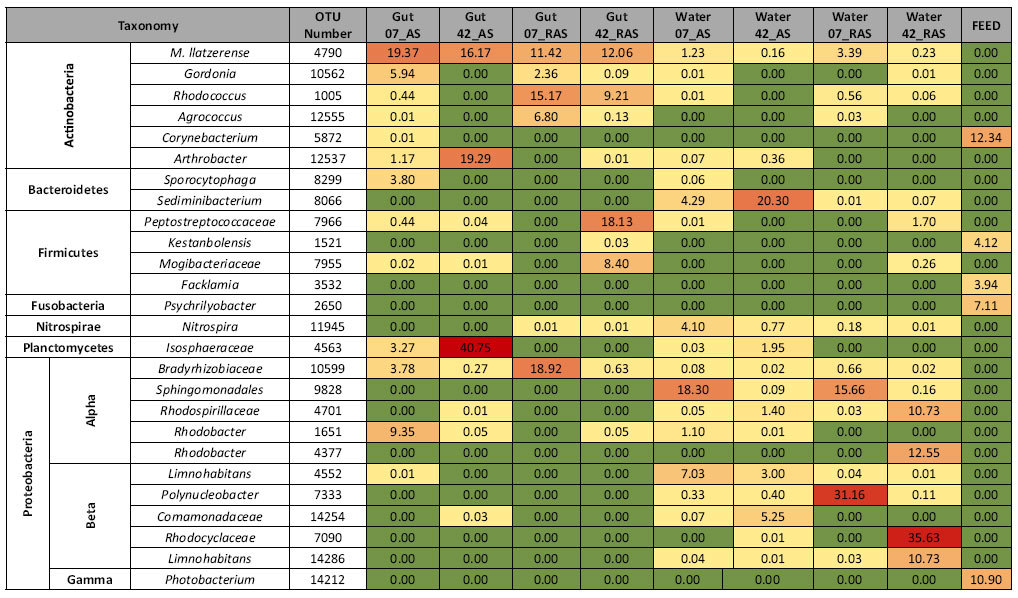
Values represent the % average relative abundance of each OTU within each sample group and colour codes are proportional to increasing OTU abundance (from green: lower, to red: higher relative abundance). 07 and D42: sampling day 7 & 42; AS & RAS: Active suspension and recirculating aquaculture systems respectively.
Within RAS, average similarity of gut BC between samples was 55% on day 7 (Table 2). More than 60% of this similarity was due to OTUs 10599 (family Bradyrhizobiaceae), 1005 (genus Rhodococcus), 4790 (Mycobacterium llatzerense) and 12555 (genus Agrococcus) (Table S1). On day 42, average similarity between gut samples remained similar. This similarity was mostly attributed to OTUs 7966 (family Peptostreptococcaceae), 4790 (Mycobacterium llatzerense), 7955 (family Mogibacteriaceae), 1005 (genus Rhodococcus) and 2787 (Cetobacterium somerae). These five OTUs accounted for a cumulative relative abundance of 59.5% in RAS. All predominant gut OTUs of day 7 were also present on day 42, albeit at lower relative abundance, whereas the OTUs most predominant on day 42 were absent on day 7 (Table S1). Similar as for AS, M. llatzerense was the shared OTU with a high relative abundance on both sampling days. In RAS, 302 gut OTUs with a cumulative relative abundance of 91% and 51% respectively, were shared between days (Table 3).
Similarity of gut bacterial communities between systems
On day 7, similarity of gut BC between systems was 22% (Table 2). Most of the predominant OTUs were present in both systems, although at different relative abundance (Table 5). OTUs 1651 and 8299 were predominant in AS but absent in RAS and OTU 3934 was predominant in RAS though absent in AS. Despite the low similarity in gut BC, the two systems shared 247 OTUs with a cumulative relative abundance of about 71% in both systems (Table 4).
On day 42, similarity between systems decreased to 11% (Table 2). This was partly due to the difference in the abundance of OTU 4563 (family Isosphaeraceae) between the two systems (40.8% in AS and absent in RAS) (Table 5). Among the shared OTUs on that day, M. llatzerense was the OTU with high relative abundance in both systems. Overall, the number of shared OTUs between systems decreased over time from 247 to 90 (Table 4).
In terms of biodiversity of the BCs, a system (AS & RAS) * day (7 & 42) interaction was observed. On day 7, 60% of cumulative relative abundance in AS was attributed to 13 OTUs whereas on day 42 to only two; this is clearly depicted also in the large decrease of the Shannon index over time (Table S2). On the contrary, in RAS, the 60% of the abundance was attributed to five and six OTUs on day 7 and 42, respectively (Table S1) while the biodiversity indices remained on similar levels. Over time, in AS, the total number of observed gut species decreased from 732 to 279, whereas this decrease was not so evident in RAS. This observation is in agreement with the results of the dominance plots for day 7 (Figure S2).
Predominant OTUs of feed bacterial communities
Since only one feed sample was analyzed per sampling day, no statistical comparison between sampling days was performed. OTUs 5872 (genus Corynebacterium), 14212 (genus Photobacterium), 3747 (genus Sporosarcina), 2650 (genus Psychrilyobacter), 3532 (genus Facklamia) and 1521 (species Anoxybacillus kestanbolensis), were among the most abundant OTUs on both days (Table S1, Table 5).
Relationship between gut, water and feed bacterial communities
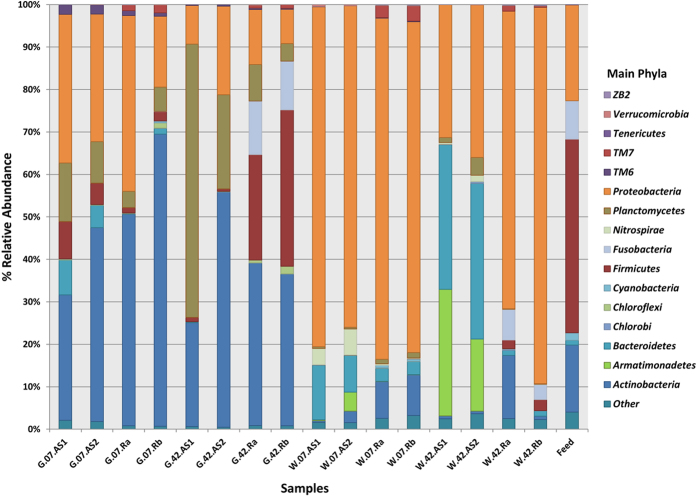
RELATE analysis revealed a strong correlation (Spearman rank correlation Rho 0.807, P < 0.0001) between bacterial communities in gut and water (Table S3). In other words, differences among water and gut samples followed similar patterns, even though the bacterial community composition in gut and water was different (P < 0.001). On day 7, Proteobacteria dominated the water of both systems. In RAS, Proteobacteria was also the dominant phylum on day 42, but Fusobacteria and Firmicutes, which were absent on day 7, appeared. In AS systems, Bacteriodetes and Armatimonadetes replaced a large fraction of the Proteobacteria by day 42. Feed-associated BC was dominated by Firmicutes and Proteobacteria, followed by Actinobacteria and Fusobacteria (Fig. 2).
Figure 2. Cumulative bar charts of the main phyla present in either gut or water samples.
Percentages show the relative abundance of each phylum in the gut or water of each replicate system on either day 7 or 42. Phyla in the feed represent the average values of feed samples from both day 7 and 42. AS1 & AS2: Replicate active suspension system 1 and 2, Ra & Rb: Recirculating aquaculture system a and b.
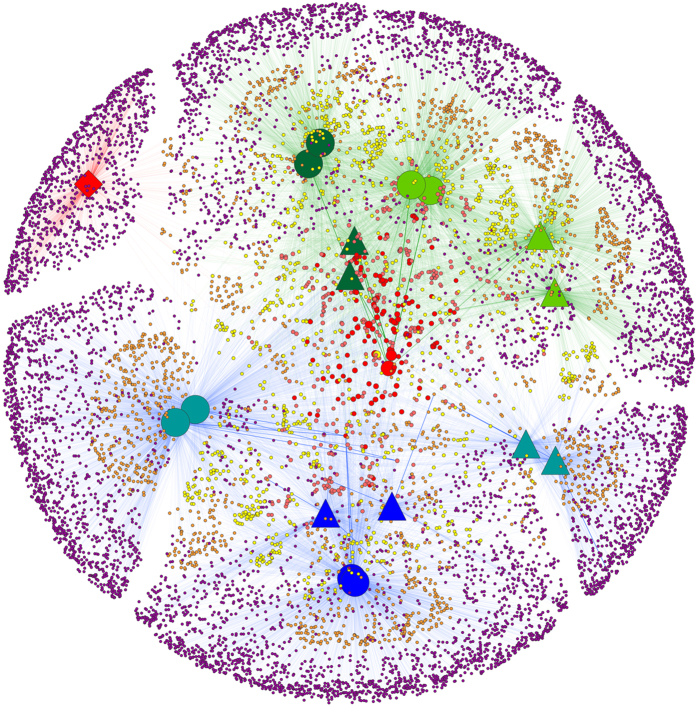
In spite of a high inter-sample correlation, average similarity between gut and water bacterial communities in AS was between 4% and 8% for both days (Table 2). This was mostly due to the low number of subdominant OTUs shared between gut and water (Fig. 3). This low similarity between gut and water on day 7 was attributed to 41 shared OTUs which represented 50% of the cumulative relative abundance in gut and 29% in water. On day 42, 40 shared OTUs between gut and water had a cumulative relative abundance of 90% and 40%, respectively (Table 6).
Figure 3. Network-based analyses of gut, water and feed bacterial communities.
The network diagram is color-coded by: sample type (gut: green, water: blue and feed: red); day 07 (circles), day 42 (triangles) and feed which represents a pooled sample both from day 7 and 42 (square); system (gut AS: Active suspension system with two replicates: dark green; gut RAS: Recirculating aquaculture system with two replicates: light green; water AS: Active suspension system with two replicates: dark blue; water RAS: Recirculating aquaculture system with two replicates: light blue). Sample nodes are oversized to be distinguished from OTU nodes. Colour and size of OTU nodes represent assignments of OTUs to sample nodes based on the amount of samples that were shared with (small nodes are shared with less samples compared to larger ones); purple OTU nodes: OTUs present in only one sample; orange OTU nodes: OTUs shared between two samples; yellow OTU nodes: shared between 3 or 4 samples; pink OTU nodes: shared between 5 to 7 samples; red OTU nodes: shared between 8 to 16 samples). Thickness/brightness of edges connecting two nodes is proportional to the relative abundance of the specific OTU in the physical sample to which it connects.
Table 6. Total number and percentage relative abundance of shared OTUs per system on each sampling day.
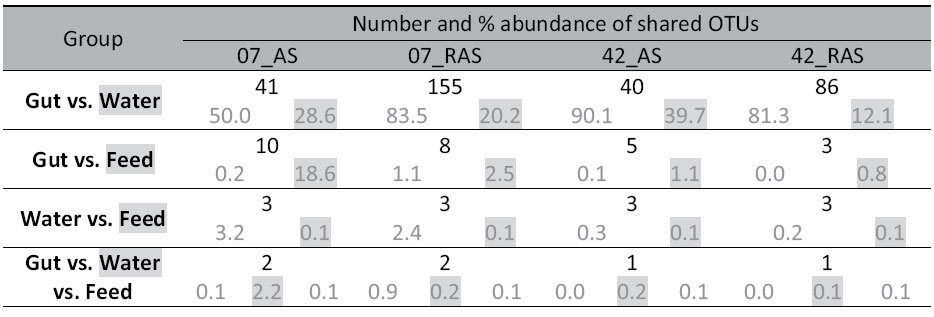
Shaded relative abundance values correspond to shaded sampling group in the first column. Sample types are compared within system for each days. OTU: Operational taxonomic unit; AS & RAS: Active suspension and recirculating aquaculture systems; 07 & 42: Sampling days 7 and 42 respectively.
In RAS, similarity between gut and water BC was 4 and 8% in the two day respectively (Table 2). Similarity on day 7 was attributed to 155 shared OTUs with a cumulative relative abundance of 83.5% and 20%, in gut and water respectively. On day 42, the 86 shared OTUs had a cumulative relative abundance of 81% and 12% (Table 6).
On the two days, gut shared with feed ten and five OTUs in AS and eight and three in RAS (Table 6). For both systems and days, the similarity between gut and feed was below 1% (Table 2). In AS, OTU 5872 was the only dominant OTU in feed encountered in the gut on day 7. Overall, only two OTUs were shared among gut, water and feed bacterial communities in AS or RAS on day 7, and only one OTU on day 42 (Table 6).
Discussion
The higher levels of nitrate observed in RAS compared to AS could be due to the higher nitrification rates. In RAS, through the water re-use loop, settable solids consisting mainly of organic matter, are removed before aerobic chemoautotrophic nitrifying bacteria convert ammonia via nitrite into nitrate in the biofilter13,14,37. In AS, solids were not removed from the fish rearing unit, but maintained in suspension. The accumulating organic matter in AS was used as substrate for biofloc formation15,38,39,40. Heterotrophic bacteria in bioflocs immobilize NH3-N, reducing the need for nitrification. Nitrification concurs with reduction of alkalinity. This explains why in AS the pH was higher than in RAS. However, the lower pH in RAS did not result in a higher CO2 level, because the amount of free CO2 in a system depends on the total amount of carbon dioxide/bicarbonate/carbonate (i.e. the total alkalinity) present41.
In AS systems, a nitrification-denitrification coupling might have contributed to the observed lower NO3-N concentrations42,43. Nitrification occurs in the aerobic surface layer of bioflocs, whereas denitrification can take place in deeper anoxic layers44,45,46. The high relative abundance of Nitrospira in water bacterial communities of AS on both days, suggests the occurrence of nitrification in those systems. Nitrospira is an aerobic, lithoautotrophic, nitrite-oxidizing bacterium that is commonly encountered in aquaculture systems or in freshwater activated sludge particles47. However, this genus exhibits high substrate affinity and has been mainly reported in filter materials, bio-filters of freshwater, brackish and marine systems37,48,49,50,51.
The second most abundant OTU (14254) in AS water of day 42 belonged to the Comamonadaceae family. Members of this family play a role in nitrate removal as well as in poly-hydroxy-alkanoate (PHA) metabolism. This indicates the metabolic and regulatory relationships between PHA degradation and denitrification by PHA-degrading denitrifiers in anoxic environments52,53,54,55.
The PO4-P concentration in RAS has been higher than in AS. Microbial phosphate assimilation was suggested as the main cause of the lower phosphate levels in biofloc systems when compared to those in RAS42,56,57. The most predominant OTU in RAS water on day 42 was a member of family Rhodocyclaceae. This family contains mainly aerobic or denitrifying bacteria which live in aquatic habitats and exhibit very versatile metabolic capabilities. Rhodocyclaceae species are commonly reported in waste water where they play an important role in bioremediation (e.g. phosphate removal)58,59,60,61,62,63. Information at genus level on its role in phosphorus cycling would be needed to explain differences in abundance between AS and RAS.
DistLM analysis revealed a high correlation between water quality parameters and the bacterial communities present in the water. A high fraction of the total variation (68%) in water BC was explained by pH, NO3-N, PO4-P and conductivity. Ebeling et al. 2006, described under which conditions autotrophic, heterotrophic, nitrifying or denitrifying processes dominate in aquatic systems41. In fact, differences in water quality between RAS and AS were caused by differences in system design and management, which induced differences in composition and functionality of water BC and vice versa. Although the percentage of explained variation in water BC by environmental factors was high, it remains challenging to control bacterial communities composition, species dynamics and functionality because: (a) possibly other factors, presently not measured, are better “predictors” of the bacterial communities and (b) there is a high degree of stochasticity involved in how bacterial communities structures change over time, even in replicated systems17. In addition, interpretation of the DISTLM results should be done with care, especially when sample size is low. Despite the fact that permutation methods allowed us to reach the desirable significance level to test our hypothesis (i.e. correlation between water quality and water microbial communities), the high percentage of explained variation does not directly imply causality between the two datasets, but correlation. Future research, including more systems and data points should be performed in order to verify if the observed correlations are maintained.
The time * system interaction for water BC revealed very interesting trends. First, the similarity of water BC between replicate AS tanks decreased over time, whereas in RAS it remained stable. This suggests that sustainable “replication of water bacterial communities” is more feasible in RAS than in AS. Secondly, the average similarity of water BC between day 7 and 42 in AS was higher than in RAS. Although in RAS more OTUs were shared between days, water bacterial communities in AS remained five times more similar over time compared to that in RAS. This observation could be partly explained by some predominant OTUs in the water, which over time, were more robust in AS than in RAS. For example, Sediminibacterium (OTU 8066) in AS water was among the most predominant OTUs on both days. This genus contains two species exhibiting high versatility in types of used substrates including non-starch polysaccharides (NSPs), oligosaccharides and others. These species were found in lakes, freshwater reservoirs, activated sludge reactors, stream biofilms and nutrient rich environments64,65,66,67,68,69,70.
Water BCs similarity between AS and RAS systems decreased considerably over time, as shown by the noticeable (from 247 to 90) decrease of the number of shared OTUs (Table 4). On day 7, the predominant water OTUs of either AS or RAS were found in both systems though in different relative abundances. On day 42, the predominant OTUs in water of AS systems were not encountered in RAS, and vice versa. Both systems were initially inoculated with water from the same source, and thus with a similar BC, which shows that the system design and function induced the development of a system-specific specialized bacterial community.
It is important at this point to remember that regarding water BCs in particular, the observations and patterns discovered between the two systems, or over time, are just a useful first starting point. This is firstly because in some cases sample size was low (especially for within system comparisons), though high enough to allow statistical power. Secondly, the environments in this study, although relevant to aquaculture are not natural since they operate under controlled conditions. This means that it is uncertain whether the observed patterns would occur in natural habitats or for example in an extensive outdoor aquaculture system (e.g. a fish pond). Nevertheless, this work can be the basis of future work addressing the implications that rearing systems have on water and fish gut BCs and provide awareness on system microbial management in aquaculture.
The described differences in composition of water BC between the two systems give rise to the question whether these differences had an impact on gut communities, and of course to which extent. The assumption has been that water provides the first microbial colonizers from the moment of mouth opening until first feeding, whereas feed microbiota would dominate during later development9,10,11. Bakke et al. 2013, suggested that relatively small differences in water BC might impose significant differences in gut BC of fish larvae71. In line with this suggestion, our results showed a high and consistent correlation between the bacterial community composition in water and gut (Spearman’s ρ: 0.807).
Although the similarity of water BC between replicate AS decreased over time, the average similarity of gut BC decreased little. Similarly in RAS, a significant decrease in average similarity of water BC over time was not observed in gut which remained eight times higher in gut than in water (Table 2). A considerable number of gut OTUs was shared between day 7 and 42. For example in RAS, OTU 1005 (genus Rhodococcus) was a predominant OTU in the gut on both days. The genus Rhodococcus has been reported in gut microbiota of sole, red rock fish, Norwegian mackerel, USA smelt, rainbow trout and shrimp72,73,74,75.
The relatively larger effect of time on water communities suggests that water BC were more sensitive to environmental changes than bacterial communities residing in the gut of fish larvae. To a certain degree, microbial community stability highly depends upon potential disturbances of the two ecosystems i.e. the gut and the water, as well as the ability of a community to respond resiliently. Preservation of homeostasis at the intestine should be in the gut microbiota’s best interest, in order to provide a convenient long-term habitat. In favor of both, host also contributes to a fairly stable environment leading to a homeostatic interaction between host and gut microbiota. However, during early fish development, rapid anatomical, physiological and immunological changes occur in the gut, which makes it plausible to think that equally substantial changes should have also occurred in the gut communities. Fernandez et al.76 evaluated the behavior of different microbial communities under perturbed conditions and suggested that stability of the functionality does not necessarily imply stability of community structures. This means that in our study, observed changes in the gut community structure over time, could have occurred side-by-side with changes in community functionality. In the same line of thought, observed changes of water physicochemical properties might have been responsible for the relative change of water BCs over time, but whether the latter compromised functional stability of the community, is not known. Further research is required to elucidate the relationship between microbial community structure (i.e. composition, diversity) and its ability to organize in a way that assures the ability of counteracting the stress effects, by maintaining functionality.
Regarding predominant species, an interesting system * day interaction was observed in the gut communities. On day 7, the number of observed gut species in the AS systems was higher than in RAS. This higher richness coincided with higher gut community evenness i.e. higher biodiversity. On day 42 a considerable decrease both in the community richness and evenness was observed in AS. The latter means that a smaller fraction of the different species was present in dominant numbers whereas this was not so evident for fish reared in the RAS systems. Based on our data, it is not possible to clarify with certainty the reason for the observed differences between the two systems. Nevertheless, the fact that similar temporal changes in community biodiversity were also observed in water BCs might point out the role of water as a microbial reservoir available to the fish gut. However, more studies would be necessary to test how changes in community structure of a habitat impact gut microbial biodiversity of the organisms living therein and whether such changes affect the functionality of the gut community.
Similarity of gut bacterial communities between AS and RAS was overall low and further decreased over time. On day 7, OTU 1651 (genus Rhodobacter) was a predominant OTUs in the gut of AS, but absent in RAS. The genus Rhodobacter contains Gram-negative bacteria widely distributed in fresh water as well as marine and hypersaline habitats (Table S4, Figure S3). Rhodobacter species have been reported in the gut of grass carp, rainbow trout, Siberian sturgeon, catfish, Atlantic cod and Pacific white shrimp77,78,79,80,81,82,83, as well as in the water of aquaculture systems84,85.
A similar pattern was observed for the gut BC in RAS. OTU 7966, (family Peptostreptococcaceae) was predominant in the gut on day 42, absent on day 7, and present at very low relative abundance levels in AS. This suggests that both systems and developmental stage of the fish influence the presence and abundance of this OTU. Peptostreptococcaceae is a family of obligate anaerobic bacteria86. In salmon, Peptostreptococcaceae was detected as a member of both the allochthonous and autochthonous microbiota regardless of the fish diet, indicating that Peptostreptococcaceae might be a natural part of the intestinal microbiota of this species87. The family has been also reported in the gut microbiota of yellow catfish88,89 and common carp90.
In this study, out of 11,658 OTUs, two OTUs were detected in all gut and water samples. Of these, Mycobacterium llatzerense, a member of the phylum Actinobacteria, was present at high relative abundance in all gut samples, and less abundant in water samples. The second OTU, a member of the Bradyrhizobiaceae, was among the predominant gut OTUs on day 7 for both systems, less abundant on day 42, and also present in all water samples. Whether its presence in the water was due to fish defecation or whether it originated from water and managed to proliferate in the fish gut is presently not known.
M. llatzerense, has not been previously reported in aquaculture systems but has been reported in hemodialysis water, and drinking water production and distribution systems91. The association of M. llatzerense with groundwater (Table S4, Figure S3) may reflect a more psychrophilic profile of this species compared to other Mycobacteria, and might justify its presence in our systems which rely on well water. Although M. llatzerense has not been previously reported in fish gut microbiota, the high and consistent abundance in all guts suggests the preference of these bacteria for the gut environment of tilapia larvae. M. llatzerense is facultative autotrophic, aerobic, non-fermentative, and involved in hydrogen oxidation and alkaline dephosphorylation92.
Members of the Bradyrhizobiaceae family have been found in sewage treatment plants93, municipal wastewater treatment plants94, active granular activated carbon filters and drinking water distribution systems (Table S4, Figure S3)95. This is in agreement with the presence of Bradyrhizobiaceae in all water samples of this study. In fish, the Bradyrhizobiaceae family was overrepresented in the gut microbiota of sea bass96, and was also reported in the stomach of yellow catfish88. Since the family contains more than ten different genera, it is difficult to speculate about the role and functionality of these microorganisms in the fish gut.
More information and relevant literature regarding major functions and processes as well as environment preference of the encountered OTUs are provided in Table S4 of the supplementary material.
Interestingly, the OTUs shared between gut and water had a higher relative abundance in the gut than in the water. In turn, almost none of the dominant OTUs in water were detected in the gut. This could suggest that host-specificity for particular microbial species is modulated by selective pressures within the host gut attributed to gut habitat (i.e. physiology, anatomy) and host’s genotype97,98,99. On the other hand in water, low abundance of the predominant gut OTUs can be partly explained by fish defecation. Apparently, conditions in water are suboptimal for the growth of defecated bacteria mostly due to the ecological preference of the latter for the gut habitat (i.e. pH, anoxic conditions, etc.), adhesion sites and nutrients availability therein. Despite the significant effort in defining the forces which impact the assembly of gut microbial communities, the underlying mechanisms attributed to the impact of the environment over the host selectivity are still poorly understood.
Feed predominant bacteria were mostly absent in water and gut. Only 1% of gut OTUs was shared with feed at very low relative abundances. It is important at this point to mention that these results do not underestimate the importance of feed per se on gut microbial communities. Feed composition, nutrients, production technology, etc. have been all found to affect gut microbiota in mammals7,100,101,102,103,104 and fish87,105,106,107. This is not the case in our study where the same feed was used in all treatments. We put forward that the low number of subdominant bacteria shared between the feed and the gut indicates the low influence that feed communities had in the gut compared with those present in the water. Bakke et al. 2013, working with cod larvae, suggested that microbiota in water had a stronger influence than microbiota in the feed on the microbial community composition in the gut71. This was clearly pictured in our OTU network (Fig. 3) where only a few OTUs present in feed were also present in water or gut samples. Corynebacterium (OTU 5873) was an exception since it was the predominant OTU in feed but also present in the gut of fish larvae in AS on day 7. Corynebacteria are found in a broad variety of habitats such as soil, plants and food products108.
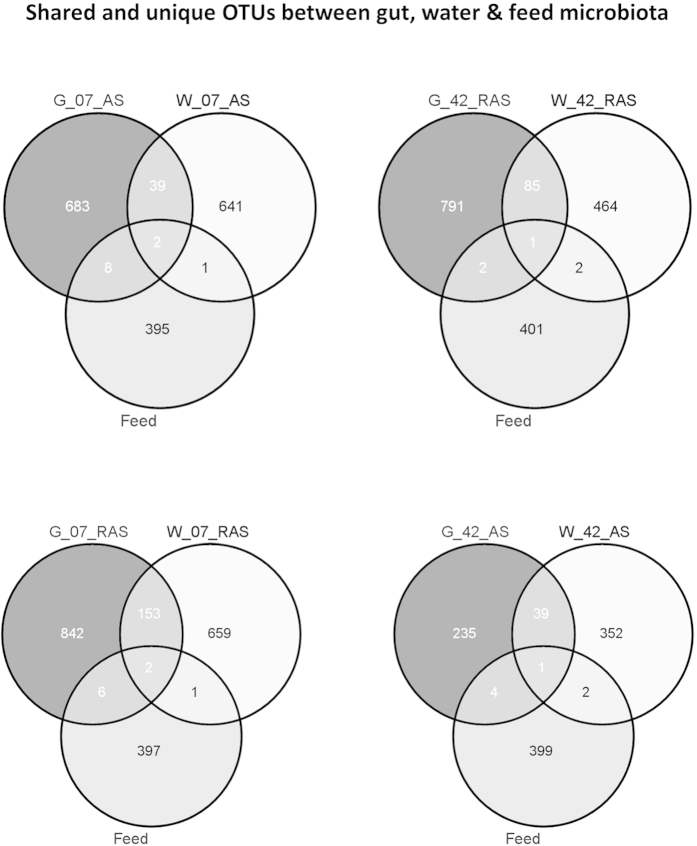
The low average similarity and the small number of OTUs shared between gut, water and feed (Table 2, Fig. 4) suggest that the microbial community in the gut is not a simple reflection of the microbes from the environment. In our study, bacterial communities in rearing water influenced those present in the gut; nevertheless they only had a limited effect on the predominant gut species. Factors affecting community development include: suitability of adhesion sites and substrates availability, host physiology, gut anatomy and abiotic components in different sections of the gut, as well as the host’s non-specific and specific immunity10,109. Thus, the resulting gut microbial community can be very different from the microbiota in water or feed. Sullam et al. (2012) stated that fish hindgut microbial communities resemble much more those of mammals, than those present in water or feed. Our study confirms that fish, like other vertebrates, harbor specialized gastrointestinal communities which are a skewed selection of microbes present in water and feed.
Figure 4. Venn diagrams of bacterial communities between sample types.
Number of reads in all samples was normalized to the minimum number of reads found in one of the samples (1719 reads). The Venn diagram illustrates overlap of bacterial OTUs between gut, water and feed samples for two different systems over time. Numbers inside circles indicate the number of unique OTUs in one of the subsets based on absence/presence data from replicate systems. Numbers of unique OTUs indicate those OTUs that were present in both replicate systems and in at least one of the samples within each replicate system. Numbers in the overlapping regions indicate the number of shared OTUs between subsets. AS & RAS: active suspension and recirculating aquaculture systems; 07 and 42: Sampling day 7 and 42 respectively.
Conclusion
The culture system strongly affected the composition and development of bacterial communities in water. Despite low similarities between gut and water, observed changes in water bacterial communities were highly correlated with changes in the gut. However, there is no clear evidence that bacteria in the water from one system had a larger impact on gut communities than the other. The potential role of the few shared or discriminant species explaining a fraction of the observed similarity was also evaluated. Most of the predominant or shared OTUs in the fish gut have also been found in other fish species, which suggests that these microorganisms fulfil similar roles in the gut of different species. By studying the roles and functionalities of the shared species, novel insights on their niche and ecological adaptability of microbiota in water and gut, and to a lesser extend feed, will be generated. A better understanding of how microbiota in water, feed and gut interact will help improving the design, microbial management and nutrient cycling of fish rearing systems. Future research should focus on disentangling the key processes governing selection and propagation of water and feed microbiota, which ultimately become important species of the microbial community in the adult fish gut.
Additional Information
How to cite this article: Giatsis, C. et al. The impact of rearing environment on the development of gut microbiota in tilapia larvae. Sci. Rep. 5, 18206; doi: 10.1038/srep18206 (2015).
Supplementary Material
Acknowledgments
This work was funded by The European Community’s Seventh Framework Program (FP7/2007–2013) under grant agreement no. 227197 Promicrobe “Microbes as positive actors for more sustainable aquaculture”.
Footnotes
Author Contributions C.G. and M.V. conceived the experiments, C.G. and H.H. conducted the experiment, C.G., G.B. and D.S. analyzed the data and C.G. wrote the main manuscript text. Authors D.S., H.S., G.B., J.V. and M.V. reviewed the manuscript.
References
- Rawls J. F. Enteric infection and inflammation alter gut microbial ecology. Cell host & microbe 2, 73–74 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekirov I., Russell S. L., Antunes L. C. M. & Finlay B. B. Gut microbiota in health and disease. Physiological reviews 90, 859–904 (2010). [DOI] [PubMed] [Google Scholar]
- Chaucheyras-Durand F. & Durand H. Probiotics in animal nutrition and health. Beneficial microbes 1, 3–9 (2010). [DOI] [PubMed] [Google Scholar]
- McFall-Ngai M. et al. Animals in a bacterial world, a new imperative for the life sciences. Proceedings of the National Academy of Sciences of the United States of America 110, 3229–36 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak S. K. Role of gastrointestinal microbiota in fish. Aquaculture Research 41, 1553–1573 (2010). [Google Scholar]
- Sullam K. E. et al. Environmental and ecological factors that shape the gut bacterial communities of fish: a meta-analysis. Molecular Ecology 21, 3363–3378 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker A. W. et al. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. The ISME journal 5, 220–30 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verschuere L., Rombaut G., Sorgeloos P. & Verstraete W. Probiotic Bacteria as Biological Control Agents in Aquaculture. Microbiology and Molecular Biology Reviews 64, 655–671 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitan K. I., Natvik C. M. & Vadstein O. Drinking rate, uptake of bacteria and microalgae in turbot larvae. Journal of Fish Biology 53, 1145–1154 (1998). [Google Scholar]
- Hansen G. H. H. & Olafsen J. A. A. Bacterial Interactions in Early Life Stages of Marine Cold Water Fish. Microbial Ecology 38, 1–26 (1999). [DOI] [PubMed] [Google Scholar]
- Ringo E. & Birkbeck T. H. Intestinal microflora of fish larvae and fry. Aquaculture research 30, 73–93 (1999). [Google Scholar]
- De Schryver P. & Vadstein O. Ecological theory as a foundation to control pathogenic invasion in aquaculture. The ISME journal 8, 2360–8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crab R., Avnimelech Y., Defoirdt T., Bossier P. & Verstraete W. Nitrogen removal techniques in aquaculture for a sustainable production. Aquaculture 270, 1–14 (2007). [Google Scholar]
- Gutierrez-Wing M. T. & Malone R. F. Biological filters in aquaculture: Trends and research directions for freshwater and marine applications. Aquacultural Engineering 34, 163–171 (2006). [Google Scholar]
- De Schryver P., Crab R., Defoirdt T., Boon N. & Verstraete W. The basics of bio-flocs technology: The added value for aquaculture. Aquaculture 277, 125–137 (2008). [Google Scholar]
- Fujimura K. & Okada N. Development of the embryo, larva and early juvenile of Nile tilapia Oreochromis niloticus (Pisces: Cichlidae). Developmental staging system. Development, growth & differentiation 49, 301–24 (2007). [DOI] [PubMed] [Google Scholar]
- Giatsis C., Sipkema D., Smidt H., Verreth J. & Verdegem M. The Colonization Dynamics of the Gut Microbiota in Tilapia Larvae. PLoS ONE 9, e103641 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lever M. A. et al. A modular method for the extraction of DNA and RNA, and the separation of DNA pools from diverse environmental sample types. Frontiers in Microbiology 6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Bogert B., de Vos W. M., Zoetendal E. G. & Kleerebezem M. Microarray analysis and barcoded pyrosequencing provide consistent microbial profiles depending on the source of human intestinal samples. Applied and environmental microbiology 77, 2071–80 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamady M., Walker J. J., Harris J. K., Gold N. J. & Knight R. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nature methods 5, 235–7 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daims H., Brühl A., Amann R., Schleifer K. H. & Wagner M. The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: development and evaluation of a more comprehensive probe set. Systematic and applied microbiology 22, 434–444 (1999). [DOI] [PubMed] [Google Scholar]
- Guss A. M. et al. Phylogenetic and metabolic diversity of bacteria associated with cystic fibrosis. The ISME journal 5, 20–29 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J. G. et al. QIIME allows analysis of high-throughput community sequencing data. Nature methods 7, 335–6 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis T. Z. et al. NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic acids research 34, W394–9 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Garrity G. M., Tiedje J. M. & Cole J. R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Applied and Environmental Microbiology 73, 5261–5267 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito R. et al. A travel guide to Cytoscape plugins. Nature methods 9, 1069–1076 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson M. J. & Robinson J. Generalized discriminant analysis based on distances. Aust. N. Z. J. Stat. 45, 301–318 (2003). [Google Scholar]
- Legendre P. & Anderson M. J. Distance-based redundancy analysis: testing multispecies responses in multifactorial ecological experiments. Ecological Monographs 69, 1–24 (1999). [Google Scholar]
- Anderson M. J., Ellingsen K. E. & McArdle B. H. Multivariate dispersion as a measure of beta diversity. Ecology Letters 9, 683–693 (2006). [DOI] [PubMed] [Google Scholar]
- Hurlbert S. H. S. The nonconcept of species diversity: a critique and alternative parameters. Ecology 52, 577–586 (1971). [DOI] [PubMed] [Google Scholar]
- Gorley R. N. & Clarke R. K. PRIMER v6: User manual/tutorial. Plymouth, U.K. PRIMER-E Ltd 214 (2006).
- Oliveros J. C. VENNY. An interactive tool for comparing lists with Venn Diagrams. BioinfoGP of CNB-CSIC (2007). at < http://bioinfogp.cnnb.csic.es/tools/venny/index.html>
- Pruesse E., Peplies J. & Glöckner F. O. SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics (Oxford, England) 28, 1823–1829 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C. et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic acids research 41, D590–6 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Molecular biology and evolution 17, 540–552 (2000). [DOI] [PubMed] [Google Scholar]
- Ronquist F. et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic biology 61, 539–542 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreier H. J., Mirzoyan N. & Saito K. Microbial diversity of biological filters in recirculating aquaculture systems. Current opinion in biotechnology 21, 318–25 (2010). [DOI] [PubMed] [Google Scholar]
- Avnimelech Y. Feeding with microbial flocs by tilapia in minimal discharge bio-flocs technology ponds. Aquaculture 264, 140–147 (2007). [Google Scholar]
- Azim M. E. & Little D. C. The biofloc technology (BFT) in indoor tanks: Water quality, biofloc composition, and growth and welfare of Nile tilapia (Oreochromis niloticus). Aquaculture 283, 29–35 (2008). [Google Scholar]
- Hargreaves J. a. Photosynthetic suspended-growth systems in aquaculture. Aquacultural Engineering 34, 344–363 (2006). [Google Scholar]
- Ebeling J. M., Timmons M. B. & Bisogni J. J. Engineering analysis of the stoichiometry of photoautotrophic, autotrophic, and heterotrophic removal of ammonia–nitrogen in aquaculture systems. Aquaculture 257, 346–358 (2006). [Google Scholar]
- Da Silva K. R., Wasielesky W. & Abreu P. C. Nitrogen and Phosphorus Dynamics in the Biofloc Production of the Pacific White Shrimp, Litopenaeus vannamei. Journal of the World Aquaculture Society 44, 30–41 (2013). [Google Scholar]
- Ray A. J., Dillon K. S. & Lotz J. M. Water quality dynamics and shrimp (Litopenaeus vannamei) production in intensive, mesohaline culture systems with two levels of biofloc management. Aquacultural Engineering 45, 127–136 (2011). [Google Scholar]
- De Kreuk M. K., Heijnen J. J. & van Loosdrecht M. C. M. Simultaneous COD, nitrogen, and phosphate removal by aerobic granular sludge. Biotechnology and bioengineering 90, 761–9 (2005). [DOI] [PubMed] [Google Scholar]
- Satoh H., Nakamura Y., Ono H. & Okabe S. Effect of oxygen concentration on nitrification and denitrification in single activated sludge flocs. Biotechnology and bioengineering 83, 604–7 (2003). [DOI] [PubMed] [Google Scholar]
- Third K. a, Burnett N. & Cord-Ruwisch R. Simultaneous nitrification and denitrification using stored substrate (PHB) as the electron donor in an SBR. Biotechnology and bioengineering 83, 706–20 (2003). [DOI] [PubMed] [Google Scholar]
- Mußmann M. et al. Colonization of freshwater biofilms by nitrifying bacteria from activated sludge. FEMS microbiology ecology 85, 104–15 (2013). [DOI] [PubMed] [Google Scholar]
- Sugita H., Nakamura H. & Shimada T. Microbial communities associated with filter materials in recirculating aquaculture systems of freshwater fish. Aquaculture 243, 403–409 (2005). [Google Scholar]
- Kruse M. et al. Relevance and diversity of Nitrospira populations in biofilters of brackish RAS. PloS one 8, e64737 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoi S., Ebihara N., Washio S. & Sugita H. Nitrite-oxidizing bacteria, Nitrospira, distribution in the outer layer of the biofilm from filter materials of a recirculating water system for the goldfish Carassius auratus. Aquaculture 264, 297–308 (2007). [Google Scholar]
- Brown M. N., Briones A., Diana J. & Raskin L. Ammonia-oxidizing archaea and nitrite-oxidizing nitrospiras in the biofilter of a shrimp recirculating aquaculture system. FEMS microbiology ecology 83, 17–25 (2013). [DOI] [PubMed] [Google Scholar]
- Khan S. T., Horiba Y., Yamamoto M. & Hiraishi A. Members of the Family Comamonadaceae as Primary Poly(3-Hydroxybutyrate-co-3-Hydroxyvalerate)-Degrading Denitrifiers in Activated Sludge as Revealed by a Polyphasic Approach. Applied and environmental microbiology 68, 3206–3214 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginige M. P., Keller J. & Blackall L. L. Investigation of an Acetate-Fed Denitrifying Microbial Community by Stable Isotope Probing, Full-Cycle rRNA Analysis, and Fluorescent In Situ Hybridization-Microutoradiography. Applied and environmental microbiology 71, 8683 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadaie T. et al. Reducing Sludge Production and the Domination of Comamonadaceae by Reducing the Oxygen Supply in the Wastewater Treatment Procedure of a Food-Processing Factory. Bioscience, Biotechnology, and Biochemistry 71, 791–799 (2007). [DOI] [PubMed] [Google Scholar]
- Adav S. S., Lee D.-J. & Lai J. Y. Microbial community of acetate utilizing denitrifiers in aerobic granules. Applied microbiology and biotechnology 85, 753–62 (2010). [DOI] [PubMed] [Google Scholar]
- Brito L. O. et al. Water quality and growth of Pacific white shrimp Litopenaeus vannamei (Boone) in co-culture with green seaweed Ulva lactuca (Linaeus) in intensive system. Aquaculture International 22, 497–508 (2013). [Google Scholar]
- Luo G. et al. Growth, digestive activity, welfare, and partial cost-effectiveness of genetically improved farmed tilapia (Oreochromis niloticus) cultured in a recirculating aquaculture system and an indoor biofloc system. Aquaculture 422–423, 1–7 (2014). [Google Scholar]
- Quan Z.-X., Im W.-T. & Lee S.-T. Azonexus caeni sp. nov., a denitrifying bacterium isolated from sludge of a wastewater treatment plant. International journal of systematic and evolutionary microbiology 56, 1043–6 (2006). [DOI] [PubMed] [Google Scholar]
- Fernández N., Sierra-Alvarez R., Amils R., Field J. a. & Sanz J. L. Compared microbiology of granular sludge under autotrophic, mixotrophic and heterotrophic denitrification conditions. Water science and technology 59, 1227–36 (2009). [DOI] [PubMed] [Google Scholar]
- Tsai Y.-P., You S.-J., Pai T.-Y. & Chen K.-W. Effect of cadmium on composition and diversity of bacterial communities in activated sludges. International Biodeterioration & Biodegradation 55, 285–291 (2005). [Google Scholar]
- Ebrahimi S. et al. Performance and microbial community composition dynamics of aerobic granular sludge from sequencing batch bubble column reactors operated at 20 degrees C, 30 degrees C, and 35 degrees C. Applied microbiology and biotechnology 87, 1555–68 (2010). [DOI] [PubMed] [Google Scholar]
- Adav S. S., Lee D.-J. & Lai J.-Y. Potential cause of aerobic granular sludge breakdown at high organic loading rates. Applied microbiology and biotechnology 85, 1601–10 (2010). [DOI] [PubMed] [Google Scholar]
- Guo F., Zhang S.-H., Yu X. & Wei B. Variations of both bacterial community and extracellular polymers: the inducements of increase of cell hydrophobicity from biofloc to aerobic granule sludge. Bioresource technology 102, 6421–8 (2011). [DOI] [PubMed] [Google Scholar]
- Kim Y.-J., Nguyen N.-L., Weon H.-Y. & Yang D.-C. Sediminibacterium ginsengisoli sp. nov., isolated from soil of a ginseng field, and emended descriptions of the genus Sediminibacterium and of Sediminibacterium salmoneum. International journal of systematic and evolutionary microbiology 63, 905–12 (2013). [DOI] [PubMed] [Google Scholar]
- Kahlisch L., Henne K., Gröbe L., Brettar I. & Höfle M. G. Assessing the viability of bacterial species in drinking water by combined cellular and molecular analyses. Microbial ecology 63, 383–97 (2012). [DOI] [PubMed] [Google Scholar]
- Torrentó C. et al. Enhanced denitrification in groundwater and sediments from a nitrate-contaminated aquifer after addition of pyrite. Chemical Geology 287, 90–101 (2011). [Google Scholar]
- Wang H., Hu C., Hu X., Yang M. & Qu J. Effects of disinfectant and biofilm on the corrosion of cast iron pipes in a reclaimed water distribution system. Water research 46, 1070–8 (2012). [DOI] [PubMed] [Google Scholar]
- Singleton D. R., Richardson S. D. & Aitken M. D. Pyrosequence analysis of bacterial communities in aerobic bioreactors treating polycyclic aromatic hydrocarbon-contaminated soil. Biodegradation 22, 1061–73 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khemkhao M., Nuntakumjorn B., Techkarnjanaruk S. & Phalakornkule C. Effect of chitosan on UASB treating POME during a transition from mesophilic to thermophilic conditions. Bioresource technology 102, 4674–81 (2011). [DOI] [PubMed] [Google Scholar]
- Besemer K. et al. Unraveling assembly of stream biofilm communities. The ISME journal 6, 1459–68 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakke I., Skjermo J., Vo T. A. & Vadstein O. Live feed is not a major determinant of the microbiota associated with cod larvae (Gadus morhua). Environmental Microbiology Reports 5, 537–548 (2013). [DOI] [PubMed] [Google Scholar]
- Kim D. H., Brunt J. & Austin B. Microbial diversity of intestinal contents and mucus in rainbow trout (Oncorhynchus mykiss). Journal of Applied Microbiology 102, 1654–1664 (2007). [DOI] [PubMed] [Google Scholar]
- Sanchez L. M., Wong W. R., Riener R. M., Schulze C. J. & Linington R. G. Examining the fish microbiome: vertebrate-derived bacteria as an environmental niche for the discovery of unique marine natural products. PloS one 7, e35398 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanggaard B. et al. The microflora of rainbow trout intestine: a comparison of traditional and molecular identification. Aquaculture 182, 1–15 (2000). [Google Scholar]
- Huber I. et al. Phylogenetic analysis and in situ identification of the intestinal microbial community of rainbow trout (Oncorhynchus mykiss, Walbaum). Journal of Applied Microbiology 96, 117–132 (2004). [DOI] [PubMed] [Google Scholar]
- Fernandez A. S. et al. Flexible community structure correlates with stable community function in methanogenic bioreactor communities perturbed by glucose. Applied and environmental microbiology 66, 4058–67 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S. et al. Composition, diversity, and origin of the bacterial community in grass carp intestine. PloS one 7, e30440 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong S. et al. Aquacultured Rainbow Trout (Oncorhynchus mykiss) Possess a Large Core Intestinal Microbiota That Is Resistant to Variation in Diet and Rearing Density. Applied and Environmental Microbiology 79, 4974–4984 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield G. S. et al. Characterization of rainbow trout (Oncorhynchus mykiss) intestinal microbiota and inflammatory marker gene expression in a recirculating aquaculture system. Aquaculture 307, 95–104 (2010). [Google Scholar]
- Geraylou Z. et al. Prebiotic effects of arabinoxylan oligosaccharides on juvenile Siberian sturgeon (Acipenser baerii) with emphasis on the modulation of the gut microbiota using 454 pyrosequencing. FEMS Microbiology Ecology 86, 357–371 (2013). [DOI] [PubMed] [Google Scholar]
- Di Maiuta N., Schwarzentruber P., Schenker M. & Schoelkopf J. Microbial population dynamics in the faeces of wood-eating loricariid catfishes. Letters in Applied Microbiology 56, 401–407 (2013). [DOI] [PubMed] [Google Scholar]
- Fjellheim A. J., Playfoot K. J., Skjermo J. & Vadstein O. Inter-individual variation in the dominant intestinal microbiota of reared Atlantic cod (Gadus morhua L.) larvae. Aquaculture Research 1–10 (2011). [Google Scholar]
- Zhang M. et al. Characterization of the intestinal microbiota in Pacific white shrimp, Litopenaeus vannamei, fed diets with different lipid sources. Aquaculture 434, 449–455 (2014). [Google Scholar]
- Tang Y. et al. Identification of bacterial community composition in freshwater aquaculture system farming of Litopenaeus vannamei reveals distinct temperature-driven patterns. International journal of molecular sciences 15, 13663–80 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rurangwa E. & Verdegem M. C. J. J. Microorganisms in recirculating aquaculture systems and their management. Reviews in Aquaculture 5, 1–14 (2014). [Google Scholar]
- Ringø E., Strøm E. & Tabachek J. Intestinal microflora of salmonids: a review. Aquaculture Research 26, 773–789 (1995). [Google Scholar]
- Hartviksen M. et al. Alternative dietary protein sources for Atlantic salmon (Salmo salar L.) effect on intestinal microbiota, intestinal and liver histology and growth. Aquaculture Nutrition 20, 381–398 (2014). [Google Scholar]
- Wu S., Tian J., Wang G., Li W. & Zou H. Characterization of bacterial community in the stomach of yellow catfish (Pelteobagrus fulvidraco). World journal of microbiology & biotechnology 28, 2165–74 (2012). [DOI] [PubMed] [Google Scholar]
- Wu S. et al. Microbial diversity of intestinal contents and mucus in yellow catfish (Pelteobagrus fulvidraco). Aquaculture 303, 1–7 (2010). [Google Scholar]
- Van Kessel M. A. et al. Pyrosequencing of 16S rRNA gene amplicons to study the microbiota in the gastrointestinal tract of carp (Cyprinus carpio L.). AMB Express 1, 41 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrou S. et al. Diversity, community composition, and dynamics of nonpigmented and late-pigmenting rapidly growing mycobacteria in an urban tap water production and distribution system. Applied and environmental microbiology 79, 5498–508 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomila M., Ramirez A., Gascó J. & Lalucat J. Mycobacterium llatzerense sp. nov., a facultatively autotrophic, hydrogen-oxidizing bacterium isolated from haemodialysis water. International journal of systematic and evolutionary microbiology 58, 2769–73 (2008). [DOI] [PubMed] [Google Scholar]
- Zhang T., Shao M.-F. & Ye L. 454 Pyrosequencing Reveals Bacterial Diversity of Activated Sludge From 14 Sewage Treatment Plants. The ISME journal 6, 1137–47 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye L. & Zhang T. Bacterial communities in different sections of a municipal wastewater treatment plant revealed by 16S rDNA 454 pyrosequencing. Applied microbiology and biotechnology 97, 2681–90 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R. et al. Diversity of bacteria and mycobacteria in biofilms of two urban drinking water distribution systems. 270, 261–270 (2012). [DOI] [PubMed] [Google Scholar]
- Carda-Diéguez M., Mira A. & Fouz B. Pyrosequencing survey of intestinal microbiota diversity in cultured sea bass (Dicentrarchus labrax) fed functional diets. FEMS Microbiology Ecology 87, 451–459 (2014). [DOI] [PubMed] [Google Scholar]
- Yan Q., van der Gast C. J. & Yu Y. Bacterial community assembly and turnover within the intestines of developing zebrafish. PLoS ONE 7, e30603 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawls J. F., Mahowald M. A., Ley R. E. & Gordon J. I. Reciprocal Gut Microbiota Transplants from Zebrafish and Mice to Germ-free Recipients Reveal Host Habitat Selection. Cell 127, 423–433 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarrete P. et al. PCR-TTGE analysis of 16S rRNA from rainbow trout (Oncorhynchus mykiss) gut microbiota reveals host-specific communities of active bacteria. PLoS ONE 7, e31335–e31335 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott K. P., Duncan S. H., Louis P. & Flint H. J. Nutritional influences on the gut microbiota and the consequences for gastrointestinal health. Biochemical Society transactions 39, 1073–8 (2011). [DOI] [PubMed] [Google Scholar]
- Flint H. J., Duncan S. H., Scott K. P. & Louis P. Interactions and competition within the microbial community of the human colon: links between diet and health. Environmental microbiology 9, 1101–1111 (2007). [DOI] [PubMed] [Google Scholar]
- Turnbaugh P. J. et al. The effect of diet on the human gut microbiome: A metagenomic analysis in humanized gnotobiotic mice. Science translational medicine 1, 6ra14 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muegge B. D. et al. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science 332, 970–4 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delsuc F. et al. Convergence of gut microbiomes in myrmecophagous mammals. Molecular ecology 23, 1301–17 (2014). [DOI] [PubMed] [Google Scholar]
- Merrifield D. L., Olsen R. E., Myklebust R. & Ringo E. Dietary Effect of Soybean (Glycine max) Products on Gut Histology and Microbiota of Fish, Soybean and Nutrition, Prof. El-Shemy Hany (Ed.), ISBN: 978-953-307-536-5, InTech, 10.5772/20101. Available from: http://www.intechopen.com/books/soybean-and-nutrition/dietary-effect-of-soybean-glycine-max-products-on-gut-histology-and-microbiota-of-fish [DOI] [Google Scholar]
- Ingerslev H. C. et al. The development of the gut microbiota in rainbow trout (Oncorhynchus mykiss) is affected by first feeding and diet type. Aquaculture 424–425, 24–34 (2014). [Google Scholar]
- Meziti A., Mente E. & Kormas K. A. Gut bacteria associated with different diets in reared Nephrops norvegicus. Systematic and applied microbiology 35, 473–82 (2012). [DOI] [PubMed] [Google Scholar]
- Yassin a. F., Kroppenstedt R. M. & Ludwig W. Corynebacterium glaucum sp. nov. International Journal of Systematic and Evolutionary Microbiology 53, 705–709 (2003). [DOI] [PubMed] [Google Scholar]
- Clements K. D., Angert E. R., Montgomery W. L. & Choat J. H. Intestinal microbiota in fishes: what’s known and what’s not. Molecular ecology 23, 1891–8 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.