Abstract

Protein–carbohydrate interactions play pivotal roles in health and disease. However, defining and manipulating these interactions has been hindered by an incomplete understanding of the underlying fundamental forces. To elucidate common and discriminating features in carbohydrate recognition, we have analyzed quantitatively X-ray crystal structures of proteins with noncovalently bound carbohydrates. Within the carbohydrate-binding pockets, aliphatic hydrophobic residues are disfavored, whereas aromatic side chains are enriched. The greatest preference is for tryptophan with an increased prevalence of 9-fold. Variations in the spatial orientation of amino acids around different monosaccharides indicate specific carbohydrate C–H bonds interact preferentially with aromatic residues. These preferences are consistent with the electronic properties of both the carbohydrate C–H bonds and the aromatic residues. Those carbohydrates that present patches of electropositive saccharide C–H bonds engage more often in CH−π interactions involving electron-rich aromatic partners. These electronic effects are also manifested when carbohydrate–aromatic interactions are monitored in solution: NMR analysis indicates that indole favorably binds to electron-poor C–H bonds of model carbohydrates, and a clear linear free energy relationships with substituted indoles supports the importance of complementary electronic effects in driving protein–carbohydrate interactions. Together, our data indicate that electrostatic and electronic complementarity between carbohydrates and aromatic residues play key roles in driving protein–carbohydrate complexation. Moreover, these weak noncovalent interactions influence which saccharide residues bind to proteins, and how they are positioned within carbohydrate-binding sites.
1. Introduction
There is growing appreciation of the fundamental roles of protein–carbohydrate interactions in biologically and medically important processes. Inhibiting or co-opting these interactions could lead to new classes of therapeutics,1 but despite a few notable successes,2,3 harnessing and controlling these interactions remains challenging. To elucidate and intervene in the biological processes mediated by protein–carbohydrate interactions, an understanding of their molecular basis is critical. Substantial advances are being made in this area.4 Nonetheless, the precise nature and balance of forces that drive the complexation of carbohydrates by proteins are not fully understood.
The importance of hydrogen bonds between the carbohydrate hydroxyl groups and polar moieties of amino acids in the binding of carbohydrates by proteins is well recognized.5−7 However, the role played by hydrophobic aliphatic and aromatic side chains in binding water-soluble carbohydrates is more obscure, with emphasis placed on interactions with carbohydrate C–H groups through the hydrophobic effect.8 Aromatic residues have long been implicated in binding carbohydrates.5,9 Carbohydrate-aromatic interactions are increasingly the subject of study in their own right,10 and an underlying contributer to affinity is the CH−π interaction, i.e., the interaction of an aromatic π-system with a C–H bond.11,12 Indeed, carbohydrate–aromatic interactions have been examined in model systems using a variety of methods, including computational studies; investigation of the folding of synthetic glycopeptides designed to form intramolecular interactions; and the interrogation of small-molecule systems by solution-phase NMR studies.10,13−25
These fundamental studies establish the importance of carbohydrate–aromatic interactions, but some gaps in knowledge remain: The relative propensities of specific monosaccharides and aromatic residues to participate in carbohydrate–aromatic interactions have not been quantified, nor is it known whether certain carbohydrate C–H bonds are prone to engage more than others. Addressing these issues would aid in understanding and predicting the features of protein–carbohydrate complexes, and it would facilitate the design of efficacious inhibitors. Answering these questions depends on understanding the forces underlying carbohydrate–aromatic interactions. CH−π interactions have an agreed dispersion, or van der Waals component. However, additional electrostatic contributions—namely, potentially attractive interactions between partial positive charges on C–H protons and the electronegative π-system—are less certain.17,26 Therefore, the importance of electronic effects in the species—i.e., the factors affecting these charges, such as inductive and stereoelectronic effects—is not established. Theoretical and experimental studies of model carbohydrate–aromatic complexes have found cases both where electronics are important for CH−π interactions,22,24,25 and where they do not play a major role.16,18,21,23
Structural bioinformatics analyses allow protein–carbohydrate interactions to be probed directly at the atomistic level. To date, such analyses have been restricted to specific protein families or carbohydrate residues.17,27 Thus, there is not yet a general understanding of how the structural properties of individual monosaccharides lead to their binding and discrimination through the inherent characteristics and positioning of amino acids within carbohydrate-binding sites in proteins. The increased size of the Protein Data Bank (PDB) over the past decade28 provides a rich source of structural data on protein–carbohydrate complexes.29 We reasoned that quantitative analyses across all protein classes would uncover general and clear principles of protein–carbohydrate interactions, should they exist.
Our analyses reveal that the noncovalently bound carbohydrates make more-numerous and more-specific contacts with protein side chains than do covalently attached carbohydrates (i.e., in glycoproteins) in the PDB. In the binding sites of the former, polar amino acids mostly occur with frequencies expected by chance; aliphatic hydrophobic residues are underrepresented, whereas electron-rich aromatic side chains, particularly tryptophan, are favored. Moreover, there are preferred relative orientations of the aromatic and carbohydrate rings, which depend on the identity of the saccharide residue. CH−π interactions to the electronegative aromatic rings are observed more frequently for more-electropositive C–H bonds, indicating important contributions from both orbital overlap and complementary electronics between the carbohydrate and π-system. This analysis is supported by determination of linear free energy relationships using substituted indoles and methyl glycosides, which highlight a key role for electronic effects in CH−π interactions.
2. Experimental Section
To generate the protein–carbohydrate interaction database, context data were obtained from GlyVicinity30,31 for amino acids with any atom within 4.0 Å of any atom of a carbohydrate moiety. In order to deal with any potential mistakes that structures deposited in the PDB28 may contain, which is a problem inherent in any attempt at gaining chemical information from a public structural biology repository,32,33 strict validation criteria were employed. The carbohydrate residues within all of the PDB entries listed by GlyVicinity were validated with the Privateer software,34 according to the following criteria: first, only monosaccharides showing the strongly preferred minimal energy conformation (4C1 for d-sugars, 1C4 for l-sugars) were considered; and second, only models with a good fit to bias-minimized electron density were selected. Only PDB entries deposited along with structure factors—i.e., experimental data—were considered. The selected agreement metric was the real-space correlation coefficient (RSCC), with a minimum cutoff value of 0.8. As the significance of this indicator decreases with decaying resolution, only entries with a reported resolution of 2.0 Å or better were included. Of these, the coordinates of the monosaccharide and amino-acid residues identified were extracted from the parent PDB files, where possible, with examples where the nearby amino acids were identical (as in homooligomeric crystals) discounted. The data set for each examined monosaccharide was obtained using the GlyVicinity assignment of the monosaccharide, with erroneous assignments removed. For each monosaccharide class, structures in which it was found were culled using CD-HIT35 at 95% pairwise protein sequence identity, in order to maximize the data available for each carbohydrate type while minimizing bias from identical protein structures and point mutations.
The relative occurrence of each amino acid in the vicinity of all of the investigated monosaccharides was compared to that in the UniprotKB/Swiss-Prot data bank.36,37 Propensity = (proportion of an amino acid in the data set)/(proportion of that amino acid in UniprotKB); error bars represent 95% confidence assuming a normal approximation of a binomial distribution.
Amino acids interacting with the α-/β-faces were defined as those where the center of the side chain was within 6 Å of the ring atoms or C6 of the carbohydrate.
CH−π interactions were identified using three parameters adapted from those previously used in a study of proteins.38 If multiple C–H bonds fell within these parameters for a single aromatic ring, that with the smallest C-projection distance was taken as the primary interacting C–H bond.
To generate electrostatic surface potentials (ESPs), minimized conformations were generated from Density Functional Theory (B3LYP/6-31+(d)) calculations in the gas phase using Gaussian09.39 ESPs were then generated from Hartree–Fock (B3LYP/6-31(d)) energy calculations of these conformations at isovalue 0.002and visualized using GaussView 5.40
For the NMR experiments, indole, 5-substituted indoles, and deuterium oxide were obtained from Sigma-Aldrich and TCI. 4,4-dimethyl-4-silapentane 1-sulfonic acid (DSS) was obtained from Uvasol. Glycosides (other than methyl-β-d-mannopyranoside, synthesis outlined in Supporting Information) were obtained from Pfanstiem and Sigma-Aldrich. All chemicals were of at least 97% purity. Solutions were prepared on a weight per volume basis. Proton NMR spectra were acquired in D2O on a Bruker Avance-500 500 MHz spectrometer with a DCH cryoprobe. Experiments used a spectral window from 11 to −1 ppm, a 4 s acquisition time, a 2 s relaxation delay, and 64 scans. NMR experiments with a relaxation delay of 15 s were run to verify indole concentration. The shift of the trimethyl peak of DSS was normalized to δDSS = 0 ppm. For the data points shown, three series of experiments were conducted at the same glycoside and indole concentrations: indole only, glycoside only, and mixed samples. The chemical shifts were averaged over three replicates, and the chemical-shift perturbations were reported as Δδ = δindole – δindole-free.
3. Results and Discussion
A Database of Protein–Carbohydrate Interactions
To examine features of protein–carbohydrate interactions, first we used GlyVicinity31 to create a structural database of monosaccharide residues—i.e., free monosaccharides, or separated constituents of larger oligosaccharides—together with proximal amino acids from X-ray crystal structures from the PDB. Strict validation criteria were set to avoid incorporating entries with incorrect nomenclature,32 unlikely conformations, or poorly fitted experimental data.33 For the elucidation of interactions discussed herein, we used the data in its broadest form: We chose 7 of the biologically relevant carbohydrates that occurred most frequently in the data set, as both α- and β-anomers, namely: d-glucose (d-Glc), d-galactose (d-Gal), d-N-acetylglucosamine (d-GlcNAc), d-N-acetylgalactosamine (d-GalNAc), d-mannose (d-Man), d-xylose (d-Xyl), and l-fucose (l-Fuc). We treated each residue as an isolated unit, considering only the pyranose form, and ignoring any modifications of the hydroxyl groups (e.g., O-methylation, O-phosphorylation, etc.). We recognize that substituents on the carbohydrate frameworks may well affect interactions, but our focus on unmodified saccharide residues was simply to maximize the available data and to find general, or first-order, interactions between carbohydrates and their protein hosts. The resulting data set encompassed carbohydrate moieties that could be divided into two groups: covalently bound glycans (from glycoproteins), and ligands bound noncovalently to proteins, Table S1. The overall database provides a means to interrogate many features of protein–carbohydrate complexes in finer detail.
An initial scan of the database indicated that for glycans there were fewer close-contacts between carbohydrate residues and protein side chains in glycosylated proteins than there were for the same monosaccharides from ligands in protein–carbohydrate complexes, Tables 1 and S2. For the four cases with sufficient examples to allow comparisons—α/β-d-Man, α-l-Fuc, and β-d-GlcNAc—the covalently bound carbohydrates made on average approximately one-half to two-thirds the number of contacts with protein side chains, and less than one-fifth of the CH−π interactions, than observed for the corresponding noncovalent complexes. These differences are perhaps not surprising, as the covalent linkage in glycoproteins does not require effective noncovalent interactions to bind the carbohydrate to the protein. An interesting additional possibility, however, is that such interactions may be less likely to occur in glycoproteins, where the glycan can participate in intermolecular protein−carbohydrate interactions as an alternative. Thus, the saccharide’s most-effective binding face is not occluded through an intramolecular interaction, but rather left free to engage in an intermolecular interaction. Without binding partners present in the X-ray crystal structures, whether such trade-offs occur cannot be seen. Whatever the reasons for the lower density of protein–carbohydrate interactions in the glycans, we focused our subsequent analyses on noncovalent protein–carbohydrate complexes, Table 1, as we were interested in the interactions of carbohydrate ligands for this study.
Table 1. Complete Tables of Statistics by Monosaccharide of All Classes Investigated from Noncovalent Species.
Total examples in data set.
Total proximal amino acids across data set, and composition of these.
Average proximal amino acids per example, and standard deviation.
Average number of amino acids associated with each carbohydrate face, and composition of these.
Facial distribution of CH−π interactions, and average per example.
Aromatic Amino Acids Are Markedly Preferred in Carbohydrate-Binding Sites
The amino acids proximal to carbohydrates were normalized to their occurrence in all protein sequences, Figure 1. Independent of the method of normalization employed (Figure S1), three trends emerged. First, we observed only a small preference for polar, hydrogen-bonding residues within these binding sites; although of these residues, aspartic acid (Asp) and asparagine (Asn) were particularly favored, occurring approximately twice as often as expected by chance. Second, and without exception, aliphatic residues were disfavored in carbohydrate-binding pockets. This exclusion would not be expected if the hydrophobic effect alone played a major role in carbohydrate binding. Third, and most conspicuously, three of the four aromatic residues contacted carbohydrates more frequently than expected by chance, in the order tryptophan (Trp) ≫ tyrosine (Tyr) > histidine (His). These last two observations highlight that carbohydrate–aromatic interactions are a key defining characteristic of carbohydrate-binding sites, whereas, hydrophobic interactions per se are not. They also reveal that not all aromatic residues are equivalent—some are more likely than others to interact with carbohydrates.
Figure 1.

Amino acids proximal to carbohydrates in X-ray crystal structures of protein–carbohydrate complexes. Propensities of amino acids (in order of increasing hydrophobicity41) in carbohydrate-binding sites from the data set compared to the distribution of amino acids across all proteins in Uniprot.37 Alternative methods for normalization are given in Figure S1; however, the overall trends shown here are preserved. Color code: white, hydrogen-bonding side chains; gray, aliphatic hydrophobic side chains, including Gly, Pro, Cys and Met; beige, aromatic side chains.
The Positional Distributions of Aromatic Residues around Carbohydrates Are Biased
We examined the aromatic residues that we identified in detail, postulating that the juxtapositions of carbohydrate and aromatic residues should illuminate the forces that drive protein–carbohydrate interactions. In the following, we illustrate our observations and arguments with comparisons between two well-represented isomers, β-d-Glc and β-d-Gal, that differ in stereochemistry at only the 4-hydroxyl group, Figure 2A,D. The general and discriminating features emerging from this comparison are emblematic of those that we observed more broadly for carbohydrate–protein complexation, Figure S2 and Table 1.
Figure 2.
Distribution of aromatic and aliphatic amino acids around carbohydrates. (A–C) β-d-Glc, and (D–F) β-d-Gal. (A, D) α- and β-faces and ring C–H bonds. (B, E) Centers, represented as spheres, of aromatic and aliphatic side chains interacting with the faces of the carbohydrates (i.e., within 6 Å of any carbohydrate carbon or the ring oxygen). (C, F) Proportions of aromatic and aliphatic side chains interacting with the α- and β-faces reported to the nearest carbon atom of the pyranose ring. See Figure S2 for the analyses for all monosaccharides.
We compared amino-acid distributions around β-d-Glc and β-d-Gal by first focusing on the two distinct surfaces of carbohydrate rings, the α- and β-face, Figure 2A,D. These each present select C–H bonds that differ in stereochemistry and stereoelectronics between monosaccharides configurations. With its completely equatorial arrangement of hydroxyl and alkoxyl groups, β-d-Glc has approximate symmetry, with a polar perimeter in the plane of the saccharide ring bisecting the α- and β-faces consisting of C–H bonds above and below it. These properties have been exploited to design synthetic carbohydrate-binding receptors.16 Consistent with this C–H bond arrangement, we found similar numbers of aliphatic and aromatic contacts on the β-face, and a slight (2.7-fold) preference for aromatic over aliphatic residues on the α-face, Figure 2B, Video S1 and Table 1. We quantified the proportions of side chains nearest each carbon of the carbohydrate to determine how different C–H bonds interacted with the local protein environment, Figure 2C. Our observations largely tracked the direction of the C–H bond, with a higher preference for aromatics and aliphatics on the face toward which the C–H bond was oriented. For example, contacts to both aromatic and aliphatic side chains on the β-face were made by C(2)–H and C(4)–H; those made on the α-face were largely effected by C(1)–H, C(3)–H, and C(5)–H, whereas C6 failed to exhibit a facial preference, presumably because of rotation around the C5–C6 bond.
In contrast, β-d-Gal exhibited marked differences in amino-acid environment between the α- and β-faces, Table 1, Figure 2D–F, Video S2. These findings underscore the importance of the carbohydrate stereochemistry, as the change in configuration at the C4 position has a major effect on interaction with aliphatic and aromatic amino acids. In detail, aliphatic residues were largely excluded from the α-face of β-d-Gal, but aromatic side chains were prevalent, with a 14-fold preference for aromatic moieties. This preference was especially strong at the C(4)–H and C(5)–H positions, Figure 2F, and was much starker than that observed for β-d-Glc C–H protons, indicating more-favorable interactions with aromatics.
Analogous variations in C–H bond interactions were seen for other monosaccharides, Figure S2. For example, for α-d-Glc the only axial hydroxyl is on the α-face, the reverse case to β-d-Gal. Correspondingly, opposite to β-d-Gal, we found a high preference for C–H bonds to interact with aromatic residues on the β-face of α-d-Glc, but little discrimination for those on the α-face, Figure S2A.
Thus, C–H bonds that seem chemically similar, such as the C(4)–H bonds of β-d-Glc and β-d-Gal, have different preferences for interaction with aromatic moieties. Furthermore, preference for aromatics is at the expense of aliphatic amino acids, further discounting the hydrophobic effect as an explanation. Therefore, we sought to elucidate the role of electronics in carbohydrate–aromatic interactions by investigating the electrostatic potentials of the aromatic moieties and carbohydrate C–H bonds.
Role of Electronics in CH−π Interactions
Unlike aliphatic residues, aromatic amino acids present electronegative π-electron systems above and below the planes of the aromatic rings that can interact with carbohydrate C–H bonds through CH−π interactions.10 We posited that if electrostatic contributions are important for CH−π interactions in protein–carbohydrate complexes, differences in the electronics of the aromatic systems and carbohydrate C–H bonds would determine participation in such interactions. We identified CH−π interactions in the data set using a three-parameter operational definition for the interaction27 (Figure 3A), and then we probed for any correlations between the electronics of aromatic and carbohydrate rings, calculated and visualized as electrostatic surface potentials (ESPs), at the sites of the interactions.
Figure 3.

Definition of parameters for CH−π interactions and participating amino acids. (A) Parameters used to identify CH−π interactions:38 CH−π angle (θ, ≤ 40°), CH−π distance (C-X, ≤ 4.5 Å), C-projection distance (Cp–X, ≤ 1.6 Å for His and TrpA; ≤ 2.0 Å for Phe, TrpB, Tyr). (B) Raw-count distribution of aromatic side chains identified making CH−π interactions with carbohydrates. For Trp, CH−π interactions were identified for cases where either the five- or six-membered ring interacts with a CH proton, TrpA and TrpB, respectively, and where the two rings both interact with separate CH protons, TrpA+B. (C) Structure of proteinogenic aromatic amino acids, with corresponding electrostatic surface potentials for the π-systems (highlighted in beige) of the side-chain moieties: indole (Trp); phenol (Tyr); benzene (Phe); imidazole (His). For indole and phenol, the forms as hydrogen-bond donors (H-bonded to water) are shown, as these are predominant in protein X-ray crystal structures.42 To show the differences in the π-systems, the scale is shown from ≥130 kJ mol–1 (electropositive, blue) through neutral (green) to ≤ −130 kJ mol–1 (electronegative, red).
We found that across our database the four aromatic side chains engaged in CH−π interactions with carbohydrate C–H bonds to different extents, with the order Trp > Tyr > phenylalanine (Phe) > His, Figure 3B. This ranking reflects the ESPs of these side chains (Figures 3C and S4A–I) and implies that electron-rich aromatic systems are the most likely to engage in CH−π interactions.
The aforementioned ranking could stem solely from the relative surface areas of the aromatic side chains. When normalized for surface area of the π-systems, however, the most electron-rich Trp remained the most common acceptor of CH−π interactions Figure S5.
The preference for Tyr over Phe also supports the importance of electronics. The aromatic systems of Tyr and Phe both present a similar surface area, comprising 6-carbon-membered rings. Indeed, a study of such interactions between amino acids within protein crystal structures found Phe and Tyr were equally likely to participate as CH−π acceptors,38 possibly highlighting differences for intra- and intermolecular systems. In terms of electronics the two systems are not equivalent. Participation of the Tyr hydroxyl in hydrogen bonding as an H-bond donor—which is the case for almost all examples of Tyr in proteins42—increases the electron-density of the π-system of Tyr, Figure S4. As shown by the ESPs, Figures 3C and S4C–F, this increases the electronegativity of the π-system, hence making it a preferred acceptor over Phe. Trp is almost always involved as an H-bond donor in proteins,42 which increases the electronegativity of the π-system beyond H-bonded Tyr, Figure S4A,B. Interpretation of the data for the side chain of His is complicated by the different hydrogen-bonded and protonation states that it can take; however, its involvement in CH−π interactions in protein–carbohydrate complexes, Figure 3B, and proteins in general,38 is relatively small.
It is striking that the ranking of aromatic amino acids involved in CH−π interactions closely aligns with that observed for cation-π interactions in similar ligand binding systems.43 For many cation-π interactions, such as those of the tetramethylammonium cation, the interaction of the positive charge with electron-rich aromatic rings is mediated by C–H protons,44 and this could be argued to be analogous to a CH−π interaction involving extremely polarized C–H bonds.
Importance of the Electronics of the Carbohydrate C–H Bond
Next, we investigated whether involvement in CH−π interactions also depended on the electronics of the carbohydrate C–H bonds. Such preference could contribute to carbohydrate discrimination: The positivity of the carbohydrate C–H protons results from the overall hydroxyl stereochemistry. Therefore, to compare the C–H protons, we examined the ESPs of the different monosaccharides in more detail.
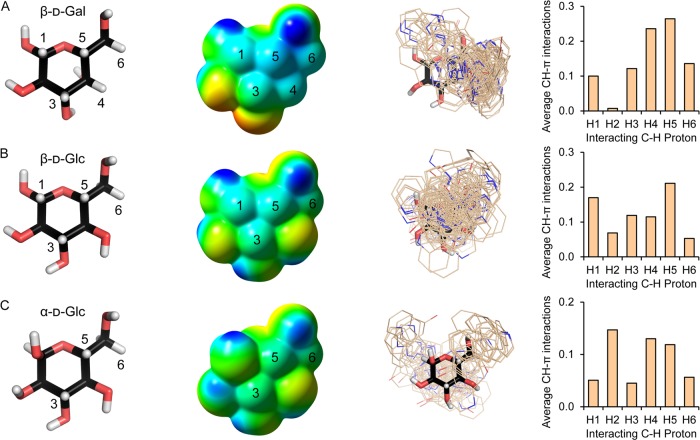
We considered β-d-Gal first, Figure 4A, because carbohydrate–aromatic interactions are already known to play key roles in its binding;9 and indeed, of all the well-represented monosaccharides, our analysis revealed that it made the highest proportions of CH−π interactions, Table 1. While steric hindrance can impact the ability of some C–H bonds (e.g., C(2)–H) to participate in CH−π interactions, the data suggested electronic effects are critical. The configuration of the hydroxyl groups of β-d-Gal give a cluster of C–H bonds on its α-face, formed by C(1)–H, C(3)–H, and C(5)–H and extending to the edge where C(4)–H and one of the C(6)–H atoms are located, Figure 4B. While often described as a “nonpolar patch”,6−8 the ESP indicates that it is in fact partially positive, and this “positive patch” corresponds to the area where interacting side chains are almost exclusively aromatic, Figure 2E,F. One way to rationalize this particularly electropositive patch is through stereoelectronic effects leading to more positive C–H protons: the axial C4-hydroxyl withdraws electron density from C3 and C5 protons via overlap of the C–H σ orbital with the σ* orbital of the C(4)–O bond, and the C4 proton is rendered electron-poor through overlap with σ* orbital of the ring C–O bond.
Figure 4.

Relationship between carbohydrate electrostatic surface potential and formation of CH−π interactions. (A) Orthogonal views of a minimized conformation of β-d-Gal, representative of the majority of those found in the database, which has the ω-angle favored by Gal in solution and in protein crystal structures,45 in stick-model representation with C–H protons numbered systematically. (B) ESP calculated for the minimized conformation. To show the differences in the C–H bonds, the scale is shown from ≥260 kJ mol–1 (electropositive, blue) through neutral (green) to ≤ −260 kJ mol–1 (electronegative, red). This is double that used for the aromatic systems; i.e., similar changes in color here signify bigger differences than in Figure 3C. (C) Juxtaposed aromatic moieties of amino acids engaged in CH−π interactions with β-d-Gal.
Superposition of the subset of aromatic side chains engaged in CH−π interactions revealed them located predominantly over the most electropositive C–H bonds of C4 and C5, Figure 4C and Video S3. Very few examples interacted with the C(2)–H of the β-face, for which the electrostatic potential is more neutral. That the more-positive protons of the carbohydrate interact more frequently with the electron-rich aromatic systems is consistent with a contribution from electrostatics to CH−π interactions.
To test the importance of electronics more generally, we compared the ESPs of further carbohydrates and assessed their engagement in CH−π interactions, Figures 5, S6 and S7. In all cases, our findings support a role for an electrostatic contribution to the CH−π interactions. As the electronics of the carbohydrate C–H bonds are determined by the identity of the monosaccharide and the anomer, this leads to distinct modes of interaction for the different classes. For example, β-d-Gal and β-d-Glc more often than not engaged in CH−π interactions with proximal aromatic residues; however, such contacts were less common in binding sites of α-d-Man, α-l-Fuc, α-d-Xyl, and α- and β-d-GlcNAc, which do not present such electropositive C–H bonds, Table 1 and Figure S6.
Figure 5.
Hydroxyl group stereochemistry influences carbohydrate electrostatics and CH−π interactions. (A) β-d-Gal, (B) β-d-Glc, and (C) α-d-Glc. Column 1: Stick models for representative minimized conformations viewed from the α-faces with C–H protons numbered. Column 2: Normalized calculated ESPs for the same orientation of the minimized conformation. The scale is shown from ≥260 kJ mol–1 (electropositive, blue) through neutral (green) to ≤ −260 kJ mol–1 (electronegative, red); as with Figure 4B this is double that used for the aromatic systems in Figure 3C. Column 3: The distributions of aromatic side chains that form CH−π interactions with the monosaccharides. Column 4: Average frequency of involvement of the monosaccharide C–H protons in the CH−π interactions. For complete analyses for all monosaccharides see Figures S6 and S7.
The α-faces of β-d-Glc and β-d-Gal isomers are sterically similar, Figure 5A,B, and yet the propensity for the two carbohydrates to engage in CH−π interactions on this face differed. This is because the α-face C–H protons are comparatively more electropositive for β-d-Gal, which should promote CH−π interactions, particularly those involving the C4 and C5 protons, Figure 5A. 97% of CH−π interactions occurred on the α-face for β-d-Gal, at an average of almost one interaction per example, Table 1. The corresponding α-face protons of β-d-Glc are less electropositive, and, as a result, CH−π interactions were less frequent, Figure 5B. 68% of interactions occurred on the α-face for β-d-Glc, just over 0.5 per example on average.
Examination of other, albeit less-well represented, monosaccharides in our database provided further support for electronic effects, Figures S6 and S7. For example, for both α-d-Gal and α-d-Glc the axial hydroxyl on the α-face reduces the electropositivity, and correspondingly, there are CH−π interactions, of the α-face C–H bonds compared to the β-anomers, Figures S6A,C and S7A,C. For α-d-Glc the most positive C–H bonds are on the β-face, and this is where most CH−π interactions occurred, Figure 5C. Disruption or reduction of the electropositive patches led to lesser involvement in CH−π interactions. For α-d-Man, the 1,2-diaxial arrangement of hydroxyl groups prevent there being any very electropositive C–H protons, Figures S6E and S7E.
The CH−π interactions of α-l-Fuc also suggested a contribution of electrostatics over hydrophobic or simple steric effects particularly well: The lack of oxygen at C6 relative to α-d-Gal reduces the electropositivity of the C–H protons at C5 and C6, and correspondingly fewer CH−π interactions, despite Fuc being the more hydrophobic overall, Figures S6M and S7M.
Electronic Effects Promote Carbohydrate–Aromatic Interactions in Solution
Finally, and as an experimental test, we probed how our two exemplar carbohydrate residues, β-d-Glc and β-d-Gal, interacted with aromatic residues in aqueous solution. We used 1H NMR spectroscopy to follow the association of indole (as a Trp surrogate) and the two β-methyl-glycosides. In both cases, there were small but measurable and reproducible upfield shifts (negative Δδ) indicative of CH−π interactions13 of some, but not all, C–H protons of the carbohydrates, Figures 6A, S8, and S9. Moreover, the magnitudes of the changes differed between protons, with the NMR data, Figures 6A and S9, in good agreement with the database-derived propensities, Figure 5. As predicted, carbohydrate–aromatic interactions were stronger for β-methyl-d-Gal than for β-methyl-d-Glc. For the former, larger chemical-shift changes were observed for the C1, C3, C4, and C5 protons, i.e., all on the electropositive α-face of the monosaccharide. The interactions with β-methyl-d-Glc were weaker, consistent with a less-electropositive α-face and our database analysis, Figures 6A and S9. Indole gave stronger CH−π interactions than previously reported for phenol or benzene,13 in accord with the observed preference for Trp in carbohydrate-binding sites, Table S3. Our findings are in accord with those of others on model peptides,14 and between methyl glycosides with the free amino acids l-Phe, l-Trp, and l-Tyr.15 Again, these data suggest that the favorable CH−π interactions make critical contributions to the binding of some but not all saccharides.
Figure 6.

1H NMR chemical shift perturbations in carbohydrate–aromatic interactions in solution. (A) Interactions between methyl glycosides and 7.5 mM indole in D2O. The circle color and size is scaled to represent the chemical-shift change relative to indole-free solutions (Δδ = δindole – δindole-free). From left to right: β-d-Gal, β-d-Glc, and β-d-Man. (B) Δδ shift for H5 and methyl C–H protons of methyl-β-d-Gal versus the Hammett σp parameter of the 5-substituent in a series of substituted indoles. To allow for solubility limitations, all perturbations were normalized to 7.5 mM indole using the linear dependence of chemical-shift perturbation on indole concentration, Figure S9. Linear fits of the data are shown for H5 (gradient = 5.7, R2 = 0.86) and Me (gradient = 2.1, R2 = 0.63). Δδ values were independent of glycoside concentration. ppb = parts per billion.
Our analyses of the ESPs suggested that other saccharides, less-well represented in our bioinformatics study, also present clusters of electropositive C–H bonds that might facilitate favorable CH−π interactions. One such carbohydrate epitope is β-d-Man. Because of the axial C(2)–OH, the α-face C–H bonds of β-d-Man (at C1, C2, C3 and C5) form an electropositive patch analogous to that of β-d-Gal, Figure S6F. Therefore, we postulated that β-d-Man should engage in CH−π interactions at these positions. This hypothesis was supported by the relatively small number of examples in our structural database, Table 1. By 1H NMR we detected similar CH−π interaction strengths as those observed for β-methyl-d-Gal. As predicted, the indole interacted with the most-electropositive C–H protons on the α-face of β-d-Man, Figures 6A and S9.
To examine further electronic effects in the associations in solution, we carried out a linear free energy (Hammett) analysis of the binding of methyl-β-d-Gal to different 5-substituted indoles, Figures S10 and S4. We monitored changes in chemical shift for the most perturbed Gal ring proton, C(5)–H, Figure 6B. Electron-rich indoles gave larger changes in chemical shift than did indole itself, indicating that the former engaged in stronger CH−π interactions. In contrast, electron-poor indoles afforded weaker interactions, and the strongly electron-withdrawing nitro-substituent appeared to abolish the interactions entirely. The linear trend observed, Figure 6B, indicates that electronic effects are critical in CH−π interactions.
4. Conclusions
In summary, we provide a quantitative assessment of the interactions made between protein side chains and the pyranose forms of the most-common monosaccharides found across all high-resolution structures of protein–carbohydrate complexes in the Protein Data Bank. We have quantified biases in the amino-acid occurrence in the immediate vicinities of the carbohydrates, with a preponderance of aromatic residues, and particularly the electron-rich side chain of tryptophan, above and/or below the plane of the carbohydrate rings. This preference for aromatics is at the expense of aliphatic hydrophobic residues. Thus, it is not simply the case that the faces of the carbohydrate are sequestered through the hydrophobic effect. Our data indicate that two effects are at play. As a first-order effect, the electronegative faces of the aromatic rings engage in favorable electrostatic interactions with certain electropositive faces of the carbohydrates. In addition, a more-specific and more-intimate second-order effect operates. Specifically, polarized, electropositive C–H bonds of the carbohydrate engaging in CH−π interactions with a contacting aromatic ring. This model is supported by calculation of the electrostatic surface potentials of both the carbohydrate and arene rings, examination of the proximity of individual carbohydrate carbon atoms to the aromatic groups, and the linear free energy relationship analysis. Moreover, because the electrostatic surfaces, and, importantly, the electropositive characters of C–H bonds differ between carbohydrate isomers, the aromatic side chains engage with different regions of the carbohydrate. This not only provides a mechanism contributing to the binding of carbohydrates by proteins, but also for discriminating between one monosaccharide and other closely similar structures within their binding sites.
These bioinformatics and experimental findings provide a strong construct for understanding the fundamental forces underpinning protein–carbohydrate interactions, and they have implications for studies of their molecular recognition. For instance, by increasing the electropositivity of C–H bonds, carbohydrate binding should be facilitated via improved carbohydrate–aromatic interactions. In this way, carbohydrates with electron-withdrawing O-acylated or O-sulfated groups could form stronger CH−π interactions. Similarly, hydrogen bonding or calcium-ion coordination to key carbohydrate hydroxyl groups could increase the strength of CH−π interactions. Given the vital role that carbohydrate–protein interactions play in biology, one strategy for designing glycomimetic drugs would be to exploit specific CH−π interactions, or the general presence of electron-rich aromatic rings to complement electropositive faces of carbohydrates in binding sites. While the importance of CH−π interactions in carbohydrate-based environments is apparent from our studies, this class of interaction plays roles within wider ligand binding, the structure of macromolecules and proteins, and in the mechanisms of chemical reactions.12 Therefore, appreciation of the impact of stereoelectronic effects on these and similar noncovalent interactions has potential for application within many contexts.
Acknowledgments
KLH thanks the EPSRC-funded Bristol Chemical Synthesis Centre for Doctoral Training (EP/G036764/1) and the University of Bristol, for a PhD studentship. GJB and DNW are grateful to the EPSRC/NSF for funding (EP/J001430). JA was supported by the BBSRC (BB/K008153/1). DNW holds a Royal Society Wolfson Research Merit Award. LLK acknowledges support from the NIH (GM049975), and RCD thanks the Molecular Biosciences Training Grant (T32GM007215) for support. The data reported in this paper are available in the Supporting Information, and the coordinates of carbohydrates with proximal amino acids used have been made available on the Internet (http://coiledcoils.chm.bris.ac.uk/protein-carbohydrate-ints), in the form of PyMOL46 sessions. We thank Robert Brown for assistance with the synthesis of β-methylmannopyranoside, and Robert Newberry and members of the TG, LLK, and DNW laboratories for helpful discussions.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.5b08424.
The authors declare no competing financial interest.
Supplementary Material
References
- Transforming Glycoscience: A Roadmap for the Future; National Academies Press: Washington, D.C., 2012. [PubMed] [Google Scholar]
- Ernst B.; Magnani J. L. Nat. Rev. Drug Discovery 2009, 8, 661. 10.1038/nrd2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiessling L. L.; Splain R. A. Annu. Rev. Biochem. 2010, 79, 619. 10.1146/annurev.biochem.77.070606.100917. [DOI] [PubMed] [Google Scholar]
- Solís D.; Bovin N. V.; Davis A. P.; Jiménez-Barbero J.; Romero A.; Roy R.; Smetana K.; Gabius H.-J. Biochim. Biophys. Acta, Gen. Subj. 2015, 18501186. 10.1016/j.bbagen.2014.03.016. [DOI] [PubMed] [Google Scholar]
- Quiocho F. Annu. Rev. Biochem. 1986, 55, 287. 10.1146/annurev.bi.55.070186.001443. [DOI] [PubMed] [Google Scholar]
- Weis W. I.; Drickamer K. Annu. Rev. Biochem. 1996, 65, 441. 10.1146/annurev.bi.65.070196.002301. [DOI] [PubMed] [Google Scholar]
- Gabius H.-J.; André S.; Jiménez-Barbero J.; Romero A.; Solís D. Trends Biochem. Sci. 2011, 366298. 10.1016/j.tibs.2011.01.005. [DOI] [PubMed] [Google Scholar]
- Lemieux R. U. Acc. Chem. Res. 1996, 298373. 10.1021/ar9600087. [DOI] [Google Scholar]
- Vyas N. K. Curr. Opin. Struct. Biol. 1991, 15732. 10.1016/0959-440X(91)90172-P. [DOI] [Google Scholar]
- Asensio J. L.; Ardá A.; Cañada F. J.; Jiménez-Barbero J. Acc. Chem. Res. 2013, 464946. 10.1021/ar300024d. [DOI] [PubMed] [Google Scholar]
- Salonen L. M.; Ellermann M.; Diederich F. Angew. Chem., Int. Ed. 2011, 50214808. 10.1002/anie.201007560. [DOI] [PubMed] [Google Scholar]
- Nishio M.; Umezawa Y.; Fantini J.; Weiss M. S.; Chakrabarti P. Phys. Chem. Chem. Phys. 2014, 16, 12648. 10.1039/c4cp00099d. [DOI] [PubMed] [Google Scholar]
- Fernández-Alonso M. C.; Cañada F. J.; Jiménez-Barbero J.; Cuevas G. J. Am. Chem. Soc. 2005, 127207379. 10.1021/ja051020+. [DOI] [PubMed] [Google Scholar]
- Laughrey Z. R.; Kiehna S. E.; Riemen A. J.; Waters M. L. J. Am. Chem. Soc. 2008, 1304414625. 10.1021/ja803960x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbussche S.; Díaz D.; Fernández-Alonso M. C.; Pan W.; Vincent S. P.; Cuevas G.; Cañada F. J.; Jiménez-Barbero J.; Bartik K. Chem. - Eur. J. 2008, 14, 7570. 10.1002/chem.200800247. [DOI] [PubMed] [Google Scholar]
- Barwell N. P.; Davis A. P. J. Org. Chem. 2011, 76166548. 10.1021/jo200755z. [DOI] [PubMed] [Google Scholar]
- Nishio M. Phys. Chem. Chem. Phys. 2011, 13, 13873. 10.1039/c1cp20404a. [DOI] [PubMed] [Google Scholar]
- Tsuzuki S.; Uchimaru T.; Mikami M. J. Phys. Chem. A 2011, 1154111256. 10.1021/jp2045756. [DOI] [PubMed] [Google Scholar]
- Fernández-Alonso M. C.; Díaz D.; Berbis M. Á.; Marcelo F.; Cañada J.; Jiménez-Barbero J. Curr. Protein Pept. Sci. 2012, 138816. 10.2174/138920312804871175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimmerová M.; Kozmon S.; Nečasová I.; Mishra S. K.; Komárek J.; Koča J. PLoS One 2012, 710e46032. 10.1371/journal.pone.0046032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W.; Enck S.; Price J. L.; Powers D. L.; Powers E. T.; Wong C.-H.; Dyson H. J.; Kelly J. W. J. Am. Chem. Soc. 2013, 135269877. 10.1021/ja4040472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santana A. G.; Jiménez-Moreno E.; Gómez A. M.; Corzana F.; González C.; Jiménez-Oses G.; Jiménez-Barbero J.; Asensio J. L. J. Am. Chem. Soc. 2013, 13593347. 10.1021/ja3120218. [DOI] [PubMed] [Google Scholar]
- Lucas R.; Peñalver P.; Gómez-Pinto I.; Vengut-Climent E.; Mtashobya L.; Cousin J.; Maldonado O. S.; Perez V.; Reynes V.; Aviñó A.; Eritja R.; González C.; Linclau B.; Morales J. C. J. Org. Chem. 2014, 7962419. 10.1021/jo402700y. [DOI] [PubMed] [Google Scholar]
- Jiménez-Moreno E.; Gómez A. M.; Bastida A.; Corzana F.; Jiménez-Oses G.; Jiménez-Barbero J.; Asensio J. L. Angew. Chem., Int. Ed. 2015, 54144344. 10.1002/anie.201411733. [DOI] [PubMed] [Google Scholar]
- Jiménez-Moreno E.; Jiménez-Osés G.; Gómez A. M.; Santana A. G.; Corzana F.; Bastida A.; Jiménez-Barbero J.; Asensio J. L. Chem. Sci. 2015, 6, 6076. 10.1039/C5SC02108A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuzuki S.; Fujii A. Phys. Chem. Chem. Phys. 2008, 10, 2584. 10.1039/b718656h. [DOI] [PubMed] [Google Scholar]
- Abbott D. W.; van Bueren A. L. Curr. Opin. Struct. Biol. 2014, 28, 32. 10.1016/j.sbi.2014.07.004. [DOI] [PubMed] [Google Scholar]
- Berman H. M.; Coimbatore Narayanan B.; Di Costanzo L.; Dutta S.; Ghosh S.; Hudson B. P.; Lawson C. L.; Peisach E.; Prlić A.; Rose P. W.; Shao C.; Yang H.; Young J.; Zardecki C. FEBS Lett. 2013, 58781036. 10.1016/j.febslet.2012.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo S.; Song K. C.; Desaire H.; MacKerell A. D.; Im W. J. Comput. Chem. 2011, 32143135. 10.1002/jcc.21886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GlyVicinity. http://glycosciences.de/tools/glyvicinity (accessed Nov 6, 2013).
- Lütteke T.; Frank M.; von der Lieth C.-W. Nucleic Acids Res. 2004, 33, D242. 10.1093/nar/gki013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lütteke T.; Frank M.; von der Lieth C.-W. Carbohydr. Res. 2004, 33951015. 10.1016/j.carres.2003.09.038. [DOI] [PubMed] [Google Scholar]
- Agirre J.; Davies G.; Wilson K.; Cowtan K. Nat. Chem. Biol. 2015, 11, 303. 10.1038/nchembio.1798. [DOI] [PubMed] [Google Scholar]
- Agirre J.; Iglesias-Fernández J.; Rovira C.; Davies G. J.; Wilson K. S.; Cowtan K. D. Nat. Struct. Mol. Biol. 2015, 22, 833. 10.1038/nsmb.3115. [DOI] [PubMed] [Google Scholar]
- Li W.; Godzik A. Bioinformatics 2006, 22131658. 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- ProtScale Tool. http://web.expasy.org/protscale/pscale/A.A.Swiss-Prot.html (accessed Nov 10, 2013).
- Bairoch A.; Apweiler R.; Wu C. H.; Barker W. C.; Boeckmann B.; Ferro S.; Gasteiger E.; Huang H.; Lopez R.; Magrane M.; Martin M. J.; Natale D. A.; O’Donovan C.; Redaschi N.; Yeh L.-S. L. Nucleic Acids Res. 2004, 33, D154. 10.1093/nar/gki070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandl M.; Weiss M. S.; Jabs A.; Sühnel J.; Hilgenfeld R. J. Mol. Biol. 2001, 307, 357. 10.1006/jmbi.2000.4473. [DOI] [PubMed] [Google Scholar]
- Gaussian 09, Revision D.01; Gaussian, Inc.: Wallingford, CT, 2009.
- GaussView, Version 5; Gaussian, Inc.: Wallingford, CT, 2009.
- Fauchere J.-L.; Pliska V. E. Eur. J. Med. Chem. 1983, 18, 369. [Google Scholar]
- McDonald I. K.; Thornton J. M. J. Mol. Biol. 1994, 2385777. 10.1006/jmbi.1994.1334. [DOI] [PubMed] [Google Scholar]
- Mecozzi S.; West A. P.; Dougherty D. A. Proc. Natl. Acad. Sci. U. S. A. 1996, 932010566. 10.1073/pnas.93.20.10566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougherty D. A. Acc. Chem. Res. 2013, 464885. 10.1021/ar300265y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner K. N.; Woods R. J. Proc. Natl. Acad. Sci. U. S. A. 2001, 981910541. 10.1073/pnas.191362798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The PyMOL Molecular Graphics System, Version 1.7.0.3; Schrödinger, LLC: New York, NY, 2014.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.