Abstract
A Leptinotarsa decemlineata SLC6 NAT gene (LdNAT1) was cloned. LdNAT1 was highly expressed in the larval alimentary canal especially midgut. LdNAT1 mRNA levels were high right after the molt and low just before the molt. JH and a JH analog pyriproxyfen activated LdNAT1 expression. RNAi of an allatostatin gene LdAS-C increased JH and upregulated LdNAT1 transcription. Conversely, silencing of a JH biosynthesis gene LdJHAMT decreased JH and reduced LdNAT1 expression. Moreover, 20E and an ecdysteroid agonist halofenozide repressed LdNAT1 expression, whereas a decrease in 20E by RNAi of an ecdysteroidogenesis gene LdSHD and disruption of 20E signaling by knockdown of LdE75 and LdFTZ-F1 activated LdNAT1 expression. Thus, LdNAT1 responded to both 20E and JH. Moreover, knockdown of LdNAT1 reduced the contents of cysteine, histidine, isoleucine, leucine, methionine, phenylalanine and serine in the larval bodies and increased the contents of these amino acids in the larval feces. Furthermore, RNAi of LdNAT1 inhibited insulin/target of rapamycin pathway, lowered 20E and JH titers, reduced 20E and JH signaling, retarded larval growth and impaired pupation. These data showed that LdNAT1 was involved in the absorption of several neutral amino acids critical for larval growth and metamorphosis.
Insects usually lack the capacity for de novo biosynthesis of ten amino acids (aliphatic: leucine, isoleucine, valine and threonine; aromatic: phenylalanine, tryptophan and histidine; sulfur containing: methionine; basic: arginine and lysine), and must absorb them from food1. Insect midgut cells actively absorb these essential amino acids by transporters2 that mainly belong to the SoLute Carrier (SLC) superfamily3. In total, 9 out of 10 SLC families participate in amino acid transport in insects1. Among the 9 SLC families, SLC6 transporters act in the apical membrane of the alimentary canal, and mediate Na+/K+ ion motive force-coupled transport of amino acids against large chemical gradients4. In contrast, the contributions of SLC7, SLC15, SLC36, SLC38 and SLC43 members, to the essential amino acid absorption in midgut, may be secondary or dispensable1.
SLC6 family comprises two clearly identifiable subfamilies. The basal subfamily consists of Nutrient Amino acid Transporters (NATs) that mainly absorbs large neutral essential amino acids and proline, known as the B0 (“B” and “0” define the Na+-coupled broad substrate spectra for neutral amino acids) and IMINO transporters respectively. The more evolutionary subfamily is the animal-specific neurotransmitter transporters which combine several orthologous clusters of catecholamine (dopamine, norepinephrine and octopamine), indolamine (serotonin) and GABA porters1.
In insects, several SLC6 NATs have been cloned from Lepidopteran species Manduca sexta5,6,7,8,9,10, and Dipteran species Aedes aegypti11, Anopheles gambiae12 and Drosophila melanogaster13. When expressed in Xenopus oocytes, these NATs can absorb a subset of neutral amino acids14. However, in vivo data have not yet been reported.
A high concentration of free amino acids in insects triggers the insulin/target of rapamycin (TOR) signaling pathways15,16. Subsequently, insulin/TOR signaling modifies the titers of 20-hydroxyecdysone (20E) and juvenile hormone (JH) to tune larval growth and metamorphosis17,18,19,20,21,22,23,24. In Leptinotarsa decemlineata (Say), a notorious defoliator of potato, several cytochrome P450 monooxygenases such as Spook, Phantom, Disembodied, Shadow, and Shade (SHD) have been documented to be involved in the biosynthesis of ecdysone in the prothoracic gland, and in the production of 20E in the peripheral tissues25,26. JH has been reported to be produced in the corpora allata. JH acid methyltransferase (JHAMT) participates in JH biosynthesis27,28. During molting of L. decemlineata larvae, either JH or 20E initiates a specific gene expression cascade. Among the activated genes, Krüppel homolog 1 (LdKr-h1) is a JH early-inducible gene28,29, whereas LdE75 and LdFTZ-F1 are 20E-inducible29,30. During mid instar stage, L. decemlineata larvae gnaw a large quantities of potato foliage. At the end of each instar, the larvae stop feeding, and shed their cuticle to allow for further growth. For economical reasons, it appears plausible that, during the molting periods when protein digestion and amino acid absorption are shut down, transcription of relative genes should be downregulated concomitantly, regulated probably by 20E/JH.
In the present paper, we identified a putative NAT1 gene (LdNAT1) in L. decemlineata. For the first time in insect species, we tested the induction of NAT1 by 20E and JH, we knocked down LdNAT1 by RNA interference (RNAi) to investigate the in vivo contributions of LdNAT1 to amino acid absorption in the midgut, as well as to overall beetle biology.
Methods and Materials
Insects rearing and chemicals
L. decemlineata larvae and adults were routinely reared in an insectary according to a previously described method28, and were supplied with potato foliage at vegetative growth or young tuber stages in order to assure sufficient nutrition. At this feeding protocol, L. decemlineata larvae progressed through four distinct instars, with the approximate periods of the first-, second-, third- and fourth-instar stages being 2, 2, 2 and 4 days, respectively. Upon reaching full size, the fourth larval instars spent an additional 4–7 days as non-feeding prepupae. The prepupae then dropped to the soil and burrowed to a depth of 3–5 cm to pupate.
An ecdysteroid agonist halofenozide (Hal) (ChemService, West Chester, USA), 20-hydroxyecdysone (20E) (Sigma-Aldrich, USA), juvenile hormone (JH) (Sigma-Aldrich, USA) and a JH analog pyriproxyfen (Pyr) (Ivy Fine Chemicals Corporation, USA) were purified by reverse phase high performance liquid chromatography before experiments.
Molecular cloning and phylogenetic analysis
Expressed sequence tags of putative LdNAT1, LdInR, Ld4EBP, LdFOXO, LdTOR and LdE75 were obtained from L. decemlineata transcriptome31 and genome data (https://www.hgsc.bcm.edu/arthropods/colorado-potato-beetle-genome-project). The correctness of the sequences was substantiated by polymerase chain reaction (PCR) using primers in Table S1. This was followed by 5′- and 3′-RACE to complete the sequence, with SMARTer RACE cDNA amplification kit (Takara Bio., Dalian, China) and SMARTer RACE kit (Takara Bio.). The antisense/sense gene-specific primers corresponding to the 5′-end and 3′-end of the sequences were listed in Table S1. After obtaining the full-length cDNA, six primer pairs (Table S1) were designed to verify the complete open reading frames. The resulting sequences LdNAT1, LdInR, Ld4EBP, LdFOXO, LdTOR, LdE75A, LdE75B and LdE75C were submitted to GenBank with the accession number of AHH29249, KP331063, KP331062, KR075829, KR075825, KP340510, KP340511 and KT246474 respectively. Transmembrane domains of LdNAT1 were predicted using TMHMM 2.0 (www.cbs.dtu.dk/services/TMHMM).
The representative NAT sequences were retrieved from NCBI, and were aligned with the predicted LdNAT1 using ClustalX (2.1)32. The neighbor-joining (NJ) tree was constructed using MEGA633 under the Poisson correction method. The reliability of NJ tree topology was evaluated by bootstrapping a sample of 1000 replicates.
Preparation of dsRNA
The same method as previously described28 was used to express dsSHD, dsE75, dsFTZ-F1, dsJHAMT, dsAS-C, dsNAT1-1, dsNAT1-2 and dsegfp derived from a 141 bp fragment of LdSHD, a 361 bp common fragment in the three LdE75 isoforms, a 319 bp common fragment in both LdFTZ-F1-1 and LdFTZ-F1-2 cDNAs, a 261 bp fragment of LdJHAMT, a 206 bp fragment of LdAS-C, a 665 bp and a 357 bp fragments of LdNAT1, and a 414 bp fragment of enhanced green fluorescent protein gene. These dsRNAs were individually expressed with specific primers in Table S1, using Escherichia coli HT115 (DE3) competent cells lacking RNase III. Individual colonies were inoculated, and induced to express dsRNA by addition of 0.1 mM isopropyl β-D-1-thiogalactopyranoside. The expressed dsRNA was extracted and confirmed by electrophoresis on 1% agarose gel (data not shown), and quantified using a spectrophotometer (NanoDrop Technologies, Wilmington, DE). Bacteria cells were centrifuged at 5000 ×g for 10 min, and resuspended in 0.05 M phosphate buffered saline (PBS, pH 7.4) at the ratio of 1:1. The bacterial suspensions (at dsRNA concentration of about 0.5 μg/ml) were used for bioassay.
Bioassay
Our preliminary results revealed that feeding the second-instar larvae with 0.1 μg/mL of 20E-, Hal-, JH- or Pyr-immersed foliage did not affect larvae growth, pupation and adult emergence. In this survey, nine independent bioassays were carried out as previously described25,29, using newly-ecdysed second- and third-instar larvae. The first bioassay was planned to test the effects of 20E and Hal on LdNAT1 expression. For 1 day, ten third-instar larvae were fed leaves which have been immersed in: (1) water (control), (2) 0.1 μg/mL 20E, or (3) 0.1 μg/mL Hal. Each treatment was replicated three times, and was collected to extract total RNA. The second to fourth bioassays were to knock down LdSHD, LdE75 and LdFTZ-F1 and each bioassay had three treatments: (1) PBS-, (2) dsegfp-, (3) dsSHD-, dsE75- or dsFTZ-F1-immerged leaves. Each treatment (ten larvae) was replicated six times and was fed for 3 days. Three replicates were collected to extract total RNA and the other three replicates were used to extract 20E. The fifth bioassay was intended to determine the effects of JH and Pyr on LdNAT1 expression by confining ten third-instar larvae for 1 day in petri dishes with: (1) water (control)-, (2) 0.1 μg/mL JH-, or (3) 0.1 μg/mL Pyr-immersed leaves. Three replicates in each treatment were collected to extract total RNA. The sixth and seventh bioassays were intended to silence LdAS-C and LdJHAMT and had three treatments: (1) PBS-, (2) dsegfp-, (3) dsAS-C- or dsJHAMT-immerged leaves. Six replicates in each treatment (ten larvae) were fed for 3 days to extract total RNA and JH respectively. The eighth and ninth bioassays were planned to knock down LdNAT1 by allowing ten second- and third-instar larvae to ingest: (1) PBS-, (2) dsegfp-, (3) dsNAT1-1-, (4) dsNAT1-2-dipped leaves. Each treatment was repeated 15 times. For extraction of total RNA, free amino acid, 20E and JH, twelve replicates were respectively collected after continuously fed for 3 days. The remaining 3 replicates were used to measure the larval weight, and to observe the larval developing period and the pupation using the methods as described previously28. The surviving larvae were individually weighed 3, 4, 5 and 6 days after treatment. Their development was observed at 4-hr intervals. Larval instars were identified by head capsule width, the appearance of exuviae, the black color of the pronotum, and the anterior beige and posterior black stripe visible on the pronotum of the 3rd and 4th instars respectively. Prepupae were distinctive from larvae by their disappeared black pigmentation, their relative inactivity and their curved body shape. The initiation of pupation was indicated by the soil-digging behavior. The pupation and the adult emergence were recorded during a 4-week trial period. For each bioassay, three biological replicates were carried out.
Real-time quantitative PCR (qPCR)
For tissue expression analysis, RNA samples were extracted from epidermis, foregut, midgut, hindgut, Malpighian tubules, fat body, hemocytes and brain-corpora cardiaca-corpora allata complex of the day 1 fourth-instar larvae. For temporal expression analysis, RNA templates were derived from the first, second, third larval instars at the interval of one day, and from fourth larval instars at the interval of eight hours. For analysis the effects of bioassay, total RNA was extracted from treated larvae. Each sample contained 5–30 individuals and repeated 3 times. The RNA was extracted using SV Total RNA Isolation System Kit (Promega). Purified RNA was subjected to DNase I to remove any residual DNA according to the manufacturer’s instructions. Quantitative mRNA measurements were performed by qRT-PCR, using internal control genes (LdRP4, LdRP18, LdARF1 and LdARF4, the primers listed in Table S1) according to our published results31. An RT negative control (without reverse transcriptase) and a non-template negative control were included for each primer set to confirm the absence of genomic DNA and to check for primer-dimer or contamination in the reactions, respectively. Each sample was repeated three times. Data were analyzed by the 2−ΔΔCT method, using the geometric mean of internal control genes for normalization. All methods and data were confirmed to follow the MIQE (Minimum Information for publication of Quantitative real time PCR Experiments) guidelines34.
Free amino acid analysis
The larvae were collected from 3 replicates, whereas the feces were collected from all 15 replicates after continuously ingested dsNAT1-1 and dsNAT1-2 for 3 days. The foliage samples repeated three times. The collected samples were immediately ground in liquid nitrogen and stored at −80 °C until assayed. Hydrolytic amino acids in the foliage were extracted by acid hydrolysis method. Free amino acids in the bodies and feces were extracted with 80% (v/v) ethanol at 25 °C for 10 min. Samples were centrifuged for 15 min at 10000 ×g and 4 °C. Free amino acid contents in the bodies, feces and foliage were analyzed with a Beckman 6300 Amino Acid Analyzer (Beckman Instruments Inc., Fullerton, CA, USA). The amino acid contents were given as μm per gram.
Quantitative determination of JH and 20E
Hemolymph was collected and JH was extracted following the methods described previously28. A liquid chromatography tandem mass spectrometry was used to quantify JH titers (ng per ml hemolymph)35.
20E was extracted according to an ultrasonic-assisted extraction method29, and its titer (ng per g body weight) was analyzed by a liquid chromatography tandem mass spectrometry-mass spectrometry system using a protocol the same as described36.
Data analysis
The data were given as means ± SE, and were analyzed by analyses of variance (ANOVAs) followed by the Tukey-Kramer test, using SPSS for Windows (SPSS, Chicago, IL, USA). Pupation rate and amino acid composition were subjected to arc-sine transformation before ANOVAs.
Results
Identification of a putative nutrient amino acid transporter transcript
We sequenced a full-length transcript encoding a putative nutrient amino acid transporter in Leptinotarsa decemlineata, and provisionally designated LdNAT1. The correctness of the cDNA was confirmed by end-to-end amplification and sequencing. It was submitted to GenBank with the accession number of AHH29249.
LdNAT1 had a 1920 bp open reading frame encoding a 640-amino acid protein. The LdNAT1 protein was predicted to have 12 transmembrane domains and intracellular C and N termini using TMHMM 2.0 (Figure S1A), which was in agreement with the general structure of transporters in the SLC6 family11. Based on the sequence alignment with selected characterized NATs and a crystallized bacterial NAT13,37, the possible substrate-binding moieties and first and second sodium-binding sites of LdNAT1 were also predicted (Figure S1A).
The evolutionary relationship of NAT-like representatives derived from 6 insect species was evaluated (Figure S1B). As expected, LdNAT1 belongs to the Coleopteran clade. It was first grouped with that from Tribolium castaneum (XP_973741), with 100% of bootstrap value, and then the two and T. castaneum XP_008196787 joined together, supported by 97% of bootstrap value (Figure S1B).
The expression of LdNAT1
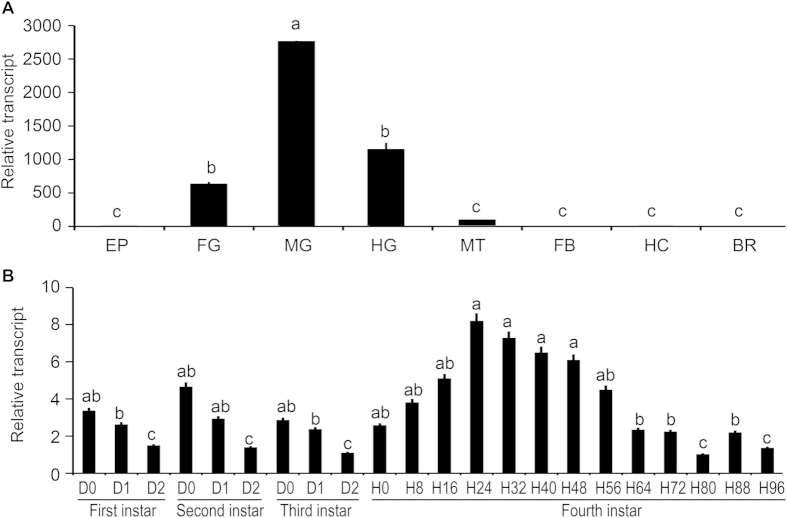
The tissue expression patterns of LdNAT1 were tested by quantitative real-time PCR (qRT-PCR). LdNAT1 was detectable in the epidermis, foregut, midgut, hindgut, Malpighian tubules, fat body, hemocytes and brain-corpora cardiaca-corpora allata complex of the day 1 fourth-instar larvae. LdNAT1 mRNA was high in the midgut and moderate in the foregut and hindgut, whereas it was expressed at low levels in the Malpighian tubules, epidermis, fat body, hemocytes and brain-corpora cardiaca-corpora allata complex (Fig. 1A).
Figure 1. Tissue (A) and temporal (B) expression patterns of LdNAT1.
For tissue expression analysis, cDNA templates are derived from epidermis (EP), foregut (FG), midgut (MG), hindgut (HG), Malpighian tubules (MT), fat body (FB), hemocytes (HE) and brain-corpora cardiaca-corpora allata complex (BR) of the day 1 fourth-instar larvae. For temporal expression analysis, cDNA templates are derived from the first, second, third larval instars at the interval of one day, and from fourth larval instars at the interval of eight hours (D0/H0 indicated newly ecdysed larvae). For each sample, 3 independent pools of 5–30 individuals are measured in technical triplicate using qPCR. The bars represent 2-ΔΔCt method (±SE) normalized to the geometrical mean of housekeeping gene expression. Different letters indicate significant difference at P value < 0.05.
The temporal expression profiles of LdNAT1 were also determined in the larvae. LdNAT1 was expressed throughout all larval stages. Within the first, second and third larval instars, the expression levels of LdNAT1 were high right after the molt than just before the molt. In the fourth larval instar, a peak of LdNAT1 occurred 24 hours after ecdysis. Moreover, LdNAT1 showed two troughs 80 and 96 hours after ecdysis (Fig. 1B).
Juvenile hormone activates the expression of LdNAT1
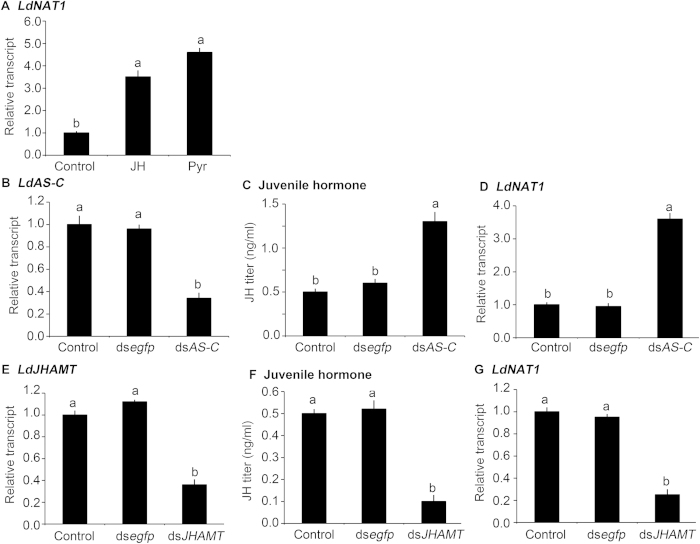
The expression patterns showed that LdNAT1 mRNA levels were correlated with circulating JH. To determine whether JH induces LdNAT1 in vivo, LdNAT1 mRNA level in newly-ecdysed L. decemlineata third-instar larvae were tested after ingestion of water (control)-, JH- and Pyr-immersed foliage for 1 day. Compared with control specimens, LdNAT1 expression levels were significantly increased by 3.4 and 4.0 folds in the larvae that had ingested JH and Pyr (Fig. 2A).
Figure 2. Induction of LdNAT1 expression by juvenile hormone (JH) in Leptinotarsa decemlineata.
The newly-ecdysed third-instar larvae have ingested potato leaves immersed with water (control), 0.1 μg/mL Pyr or JH for 1 day (A). Otherwise, the newly-ecdysed third-instar larvae have ingested PBS-, dsegfp-, or dsAS-C-dipped leaves (B–D); or PBS-, dsegfp-, or dsJHAMT-dipped leaves (E–G) for 3 days. Different letters indicate significant difference at P value < 0.05.
Moreover, LdNAT1 transcription was significantly enhanced by 3.6 fold in the LdAS-C RNAi hypomorphs, in which JH titer was significantly increased (Fig. 2B–D).
Furthermore, JHAMT plays a major role in JH biosynthesis27,28. In this study, we knocked down LdJHAMT by RNAi (Fig. 2E) to lower JH titer (Fig. 2F), and found that LdNAT1 mRNA level was significantly diminished by 61.1% in the LdJHAMT RNAi larvae (Fig. 2G).
20-Hydroxyecdysone inhibits the expression of LdNAT1
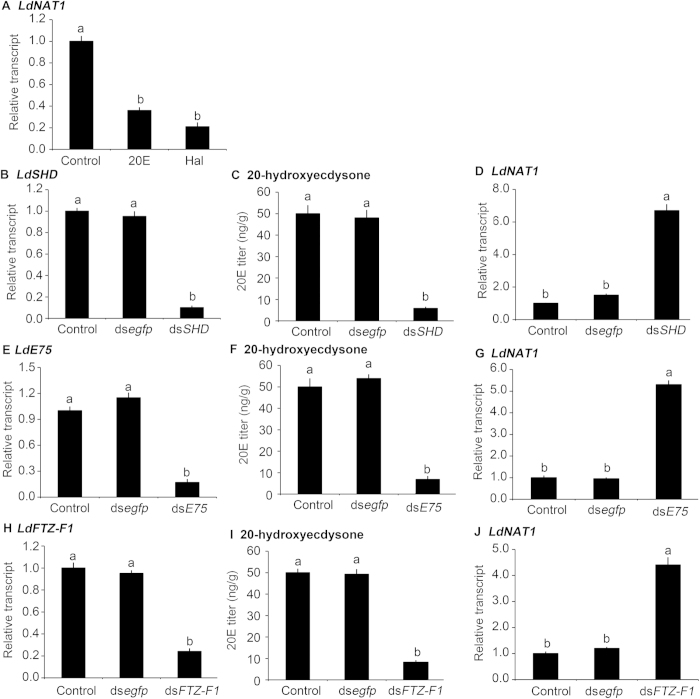
The expression patterns suggest that 20E pulse at the end of each instar inhibits LdNAT1. To verify the suggestion, LdNAT1 mRNA levels in L. decemlineata third-instar larvae were tested after ingestion of water (control)-, Hal- and 20E-immersed foliage for 1 day. Compared with control specimens, LdNAT1 expression levels were significantly decreased by 68.4% and 78.8% in the larvae that had ingested Hal and 20E (Fig. 3A).
Figure 3. Inhibition of LdNAT1 expression by 20-hydroxyecdysone (20E) signaling in Leptinotarsa decemlineata.
The newly-ecdysed third-instar larvae have ingested potato foliage treated with water (control), 0.1 μg/mL halofenozide or 20E for 1 day (A). Otherwise, the newly-ecdysed third-instar larvae have ingested PBS-, dsegfp-, or dsSHD-dipped leaves (B–D); or PBS-, dsegfp-, or dsE75-dipped leaves for 3 days (E–G); or PBS-, dsegfp-, or dsFTZ-F1-dipped leaves (H–J) for 3 days. Different letters indicate significant difference at P value < 0.05.
Since RNAi of LdSHD reduced 20E titer in L. decemlineata25,29, LdNAT1 mRNA level was tested in the third-instar larvae in which LdSHD had been silenced by RNAi (Fig. 3B). As expected, 20E titer had been reduced (Fig. 3C), whereas LdNAT1 mRNA level was significantly increased by 6.1 fold in the LdSHD RNAi hypomorphs, compared with specimens that had ingested dsegfp (Fig. 3D).
LdE75 and LdFTZ-F1 mediated 20E signaling in L. decemlineata29,30. In this study, we found three LdE75 isoforms. We knocked down all these isoforms by dietary introduction of a dsRNA derived from a common fragment in the three LdE75 isoforms (Fig. 3E). 20E titer was significantly decreased (Fig. 3F), whereas LdNAT1 transcription was significantly enhanced in the resulting larvae (Fig. 3G). Moreover, knockdown of LdFTZ-F1 by RNAi (Fig. 3H) also lowered 20E titer (Fig. 3I) but enhanced LdNAT1 transcription by 3.2 fold (Fig. 3J).
Knockdown of LdNAT1 affects the absorption of several neutral amino acids
Hydrolytic amino acids in potato foliage, and free amino acids in bodies and feces of the day 1 third- and fourth-instar larvae were analyzed (Tables S2–S4). The compositions of nineteen proteinogenic amino acids (tryptophan is undetectable) were listed in Table 1. The compositions of cysteine, histidine, lysine, methionine, serine, tyrosine and valine in the feces of the third- and fourth-instar larvae were significantly lower than those in the body and the potato foliage. Similarly, the compositions of glutamate, isoleucine, leucine and phenylalanine in the feces of the fourth-instar larvae were significantly lower than those in the body and the potato foliage (Table 1). These data suggest that a total of 11 amino acids (cysteine, histidine, lysine, methionine, serine, tyrosine, valine, glutamate, isoleucine, leucine, phenylalanine) may be actively absorbed by the larval gut.
Table 1. The compositions of amino acid (%) in foliage, the bodies and feces of the day 1 third- and fourth-instar larvae.
| |
The third-instar larvae |
The fourth-instar larvae |
|||
|---|---|---|---|---|---|
| Foliage | Bodies | Feces | Bodies | Feces | |
| A | 8.70 ± 0.41 a | 7.75 ± 0.43 a | 8.58 ± 0.52 a | 7.60 ± 0.37 ab | 6.45 ± 0.54 b |
| R | 4.89 ± 0.27 a | 4.42 ± 0.33 a | 4.44 ± 0.35 a | 4.50 ± 0.29 a | 3.34 ± 0.26 b |
| D | 1.23 ± 0.11 a | 0.91 ± 0.05 a | 0.95 ± 0.07 a | 0.92 ± 0.08 a | 0.71 ± 0.06 a |
| N | 9.79 ± 0.81 a | 7.47 ± 0.54 b | 8.58 ± 0.43 a | 8.25 ± 0.74 ab | 10.81 ± 1.12 a |
| C | 0.25 ± 0.02 b | 1.36 ± 0.12 a | 0.08 ± 0.00 c | 0.88 ± 0.07 ab | 0.06 ± 0.00 c |
| E | 0.96±0.10 ab | 1.38 ± 0.11 a | 1.06 ± 0.13 a | 1.34 ± 0.12 a | 0.80 ± 0.09 b |
| Q | 9.73 ± 0.85 a | 12.33 ± 1.02 a | 9.72 ± 0.81 a | 11.65 ± 1.12 a | 12.79 ± 1.23 a |
| G | 9.62 ± 0.81 c | 9.04 ± 0.75 c | 18.07 ± 1.52 b | 9.16 ± 1.04 c | 24.60 ± 2.33 a |
| H | 1.92 ± 0.15 b | 2.56 ± 0.14 a | 1.58 ± 0.12 b | 2.74 ± 0.16 a | 1.19 ± 0.09 c |
| I | 4.71 ± 0.22 a | 5.09 ± 0.23 a | 5.22 ± 0.36 a | 5.04 ± 0.42 a | 3.92 ± 0.28 b |
| L | 8.87 ± 0.52 a | 7.17 ± 0.48 a | 6.88 ± 0.54 ab | 8.35 ± 0.57 a | 5.94 ± 0.34 b |
| K | 6.49 ± 0.33 a | 7.62 ± 0.35 a | 2.81 ± 0.23 b | 7.52 ± 0.48 a | 2.93 ± 0.23 b |
| M | 1.35 ± 0.12 a | 0.21 ± 0.01 b | 0.01±0.00 c | 0.20 ± 0.03 b | 0.03 ± 0.00 c |
| F | 4.22 ± 0.34 a | 3.38 ± 0.30 ab | 3.91 ± 0.20 a | 3.37 ± 0.18 ab | 2.94 ± 0.18 b |
| P | 6.42 ± 0.53 a | 6.26 ± 0.50 a | 7.39 ± 0.64 a | 5.63 ± 0.59 a | 5.56 ± 0.53 a |
| S | 5.59 ± 0.34 a | 6.81 ± 0.42 a | 4.49 ± 0.35 b | 6.79 ± 0.32 a | 4.15 ± 0.27 b |
| T | 5.32 ± 0.34 bc | 4.65 ± 0.42 c | 8.31 ± 0.72 a | 4.62 ± 0.38 c | 6.26 ± 0.46 b |
| Y | 3.07 ± 0.22 a | 3.19 ± 0.17 a | 2.61 ± 0.14 b | 3.23 ± 0.21 a | 1.96 ± 0.15 c |
| V | 6.79 ± 0.37 b | 8.31 ± 0.42 a | 5.22 ± 0.25 c | 8.12 ± 0.45 a | 5.45 ± 0.30 c |
Note: A, alanine; R, arginine; D, aspartic acid; N, asparagine; C, cysteine; E, glutamate; Q, glutamine; G, glycine; H, histidine; I, isoleucine; L, leucine; K, lysine; M, methionine; F, phenylalanine; P, proline; S, serine; T, threonine; Y, tyrosine; V, valine. Data within each line are subjected to arc-sine transformation and analyzed by ANOVAs followed by the Tukey-Kramer test. Different letters indicate significant difference at P value < 0.05.
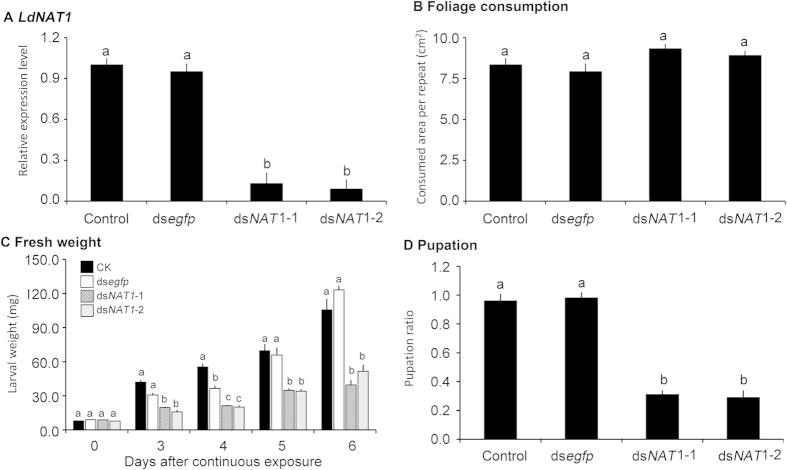
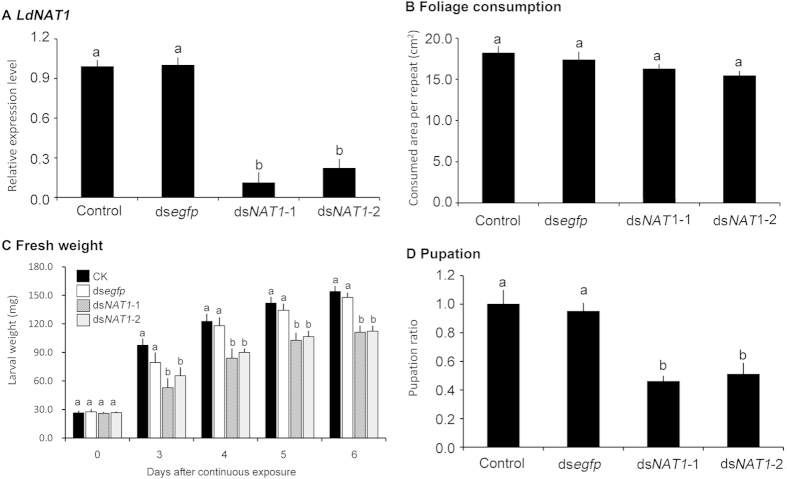
After the day 1 second- and third-instar larvae had ingested foliage treated with PBS-, dsegfp-, dsNAT1-1-, dsNAT1-2-dipped leaves for three days, the larvae reached to the day 1 of third- and fourth-instar stage respectively. The expression levels of LdNAT1 in dsNAT1-1- and dsNAT1-2-fed third-instar larvae were reduced by 87.3% and 91.6% respectively, the transcript levels in dsNAT1-1- and dsNAT1-2-fed fourth-instar larvae were decreased by 88.8% and 77.4% respectively, compared with those in PBS- and dsegfp-fed third- and fourth-instar larvae (Figs 4A and 5A).
Figure 4. Effects of dietary ingestion of dsNAT1 (dsNAT1-1 and dsNAT1-2) by the second-instar L. decemlineata larvae on the expression of the target mRNA (A), foliage consumption (B), larval weight (C) and pupation rate (D).
For relative transcript determination, 3 independent pools of 5–30 individuals are measured in technical triplicate using qRT-PCR. The bars represent 2−ΔΔCt values (±SE), normalized to the geometrical mean of housekeeping gene expression. For larval weight, larval duration and pupation rate, the bars mean averages (±SE). Different letters indicate significant difference at P value < 0.05.
Figure 5. Effects of dietary ingestion of dsNAT1 (dsNAT1-1 and dsNAT1-2) by the third-instar L. decemlineata larvae on the expression of the target mRNA (A), foliage consumption (B), larval weight (C) and pupation rate (D).
For relative transcript determination, 3 independent pools of 5–30 individuals are measured in technical triplicate using qRT-PCR. The bars represent 2−ΔΔCt values (±SE), normalized to the geometrical mean of housekeeping gene expression. For larval weight, larval duration and pupation rate, the bars mean averages (±SE). Different letters indicate significant difference at P value < 0.05.
The consumed foliage areas per day were measured 3 days after the bioassays, the larvae having been fed on PBS-, dsegfp-, dsNAT1-1-, dsNAT1-2-dipped leaves consumed similar amounts of food (Figs 4B, 5B).
The contents of the nineteen proteinogenic amino acids in the bodies and feces of the LdNAT1 RNAi larvae were measured (Table 2, Tables S2, S3). The contents of cysteine, histidine, isoleucine, leucine, methionine, phenylalanine and serine in the bodies of the LdNAT1 RNAi hypomorphs were significantly lower than those in the bodies of PBS- and dsegfp-fed larvae, whereas the contents of these amino acids in the feces of the LdNAT1 RNAi hypomorphs were significantly higher (Table 2, Tables S2, S3). These data indicate that knockdown of LdNAT1 reduces the absorption of the seven amino acids by the larval gut.
Table 2. The contents of seven amino acids in the bodies and feces of the Leptinotarsa decemlineata larvae fed on PBS, dsegfp, dsNAT1-1 and dsNAT1-2-immersed foliage for 3 days.
| Amino acid | Origin | Concentration (μmol/g) |
|||
|---|---|---|---|---|---|
| PBS | dsegfp | dsNAT1-1 | dsNAT1-2 | ||
| The third-instar larvae | |||||
| C | Bodies | 11.12 ± 0.51 a | 13.12 ± 0.84 a | 3.21 ± 0.21 b | 2.67 ± 0.14 b |
| Feces | 0.25 ± 0.02 b | 0.33 ± 0.03 b | 2.34 ± 0.14 a | 3.15 ± 0.13 a | |
| H | Bodies | 20.88 ± 1.24 a | 20.02 ± 1.11 a | 10.70 ± 0.52 b | 11.17 ± 0.47 b |
| Feces | 4.69 ± 0.20 b | 4.06 ± 0.28 b | 8.84 ± 0.41 a | 7.87 ± 0.52 a | |
| I | Bodies | 41.49 ± 2.22 a | 42.16 ± 3.04 a | 26.77 ± 1.29 b | 28.13 ± 1.38 b |
| Feces | 15.41 ± 0.84 b | 12.68 ± 0.61 b | 27.22 ± 1.15 a | 29.39 ± 1.74 a | |
| L | Bodies | 58.48 ± 2.41 a | 58.76 ± 1.98 a | 32.45 ± 2.07 b | 34.68 ± 2.15 b |
| Feces | 20.31 ± 1.06 b | 19.75 ± 1.19 b | 37.55 ± 1.24 a | 33.17 ± 2.03 a | |
| M | Bodies | 1.74 ± 0.38 a | 4.26 ± 0.25 a | 0.64 ± 0.04 b | 0.13 ± 0.01 b |
| Feces | 0.02 ± 0.00 b | 0.03 ± 0.00 b | 0.31 ± 0.02 a | 0.25 ± 0.02 a | |
| F | Bodies | 27.55 ± 1.07 a | 28.64 ± 1.13 a | 19.20 ± 1.23 b | 15.73 ± 1.19 b |
| Feces | 11.56 ± 0.81 b | 9.62 ± 0.57 b | 23.69 ± 1.04 a | 18.96 ± 1.07 a | |
| S | Bodies | 55.50 ± 2.11 a | 55.86 ± 4.08 a | 41.88 ± 2.58 b | 34.26 ± 2.14 b |
| Feces | 13.27 ± 1.01 b | 11.67 ± 0.72 b | 31.84 ± 1.22 a | 29.62 ± 1.57 a | |
| The fourth-instar larvae | |||||
| C | Bodies | 6.89 ± 0.51 a | 10.50 ± 0.67 a | 2.10 ± 0.16 b | 0.25 ± 0.02 c |
| Feces | 0.25 ± 0.02 b | 0.25 ± 0.02 b | 4.58 ± 0.37 a | 7.70 ± 0.51 a | |
| H | Bodies | 21.32 ± 0.71 a | 19.86 ± 0.54 a | 9.44 ± 0.35 b | 10.50 ± 0.49 b |
| Feces | 4.69 ± 0.57 b | 4.66 ± 0.41 b | 8.64 ± 0.37 a | 12.06 ± 0.61 a | |
| I | Bodies | 39.21 ± 1.13 a | 45.54 ± 2.02 a | 21.79 ± 1.51 b | 20.38 ± 1.44 b |
| Feces | 15.40 ± 0.87 b | 13.72 ± 0.74 b | 30.73 ± 1.57 a | 27.28 ± 1.63 a | |
| L | Bodies | 65.00 ± 3.54 a | 63.12 ± 4.13 a | 38.40 ± 2.33 b | 37.77 ± 1.79 b |
| Feces | 23.30 ± 1.47 b | 26.91 ± 1.56 b | 46.79 ± 3.64 a | 48.52 ± 4.11 a | |
| M | Bodies | 1.60 ± 0.11 a | 1.54 ± 0.08 a | 0.65 ± 0.02 b | 0.41 ± 0.01 b |
| Feces | 0.15 ± 0.01 b | 0.40 ± 0.02 b | 2.64 ± 0.15 a | 1.51 ± 0.11 a | |
| F | Bodies | 26.25 ± 1.47 a | 29.72 ± 1.55 a | 11.51 ± 0.71 b | 14.34 ± 0.57 b |
| Feces | 11.55 ± 0.57 b | 12.97 ± 0.69 b | 22.34 ± 1.21 a | 24.58 ± 1.54 a | |
| S | Bodies | 52.88 ± 3.74 a | 59.52 ± 3.69 a | 26.64 ± 1.27 b | 28.63 ± 1.34 b |
| Feces | 16.27 ± 0.79 b | 15.34 ± 0.85 b | 31.11 ± 1.45 a | 42.19 ± 1.85 a | |
Free amino acid contents in the bodies and feces are analyzed with a Beckman 6300 Amino Acid Analyzer. Difference of amino acid content within each line is analyzed by ANOVA followed by the Tukey-Kramer test. Data that do not share the same letters are significantly different at P values of 0.05.
Knockdown of LdNAT1 retards larval growth and impairs pupation
When the larvae were weighed after 3, 4, 5 and 6 days of the initiation of the bioassays, the fresh weights were significantly reduced in the LdNAT1 RNAi larvae (Figs 4C and 5C).
Exposure to dsNAT-immersed foliage significantly delayed larval developing stage. The mean periods of those on water-, dsegfp-, dsNAT1-1-, dsNAT1-2-dipped leaves were 14.0, 13.8, 16.0 and 16.3 days in the larvae that had been treated at the second instar stage, and were 12.0, 12.1, 15.0 and 15.0 days in the larvae having been treated at the third instar stage. In addition, the average periods of 2nd, 3rd and 4th instars, and prepupae were further measured. Exposure to dsNAT1-1-, dsNAT1-2-dipped foliage significantly delayed the developing periods of the fourth-instar and prepupae (Table 3).
Table 3. The developing stage of L. decemlineata surviving larvae subjected to dietary dsRNA introduction.
| Larval instar | 2nd | 3rd | 4th | Prepupae | |
|---|---|---|---|---|---|
| Initiation of the bioassay at the early second instar stage | |||||
| CK | 2.0 ± 0.1 a | 2.1 ± 0.2 a | 4.1 ± 0.2 a | 5.8 ± 0.3 a | 14.0 ± 0.4 a |
| dsegfp | 2.2 ± 0.2 a | 2.0 ± 0.1 a | 4.0 ± 0.2 a | 5.6 ± 0.3 a | 13.8 ± 0.5 a |
| dsNAT1-1 | 2.1 ± 0.1 a | 2.1 ± 0.2 a | 5.1 ± 0.2 b | 6.7 ± 0.4 b | 16.0 ± 0.6 b |
| dsNAT1-2 | 2.1 ± 0.1 a | 2.2 ± 0.2 a | 5.2 ± 0.3 b | 6.8 ± 0.4 b | 16.3 ± 0.5 b |
| Initiation of the bioassay at the early third instar stage | |||||
| CK | 2.0 ± 0.1 a | 4.0 ± 0.2 a | 6.0 ± 0.2 a | 12.0 ± 0.3 a | |
| dsegfp | 2.1 ± 0.2 a | 4.2 ± 0.1 a | 5.8 ± 0.4 a | 12.1 ± 0.4 a | |
| dsNAT1-1 | 2.0 ± 0.1 a | 5.4 ± 0.2 b | 7.6 ± 0.3 b | 15.0 ± 0.5 b | |
| dsNAT1-2 | 2.2 ± 0.1 a | 5.3 ± 0.2 b | 7.5 ± 0.3 b | 15.0 ± 0.4 b | |
The larval growth is checked at 4-hr intervals. See text for detail explanation for the identification of instars and prepupae. The data are given as means ± SE, and are subjected one-way ANOVA and followed by the Tukey–Kramer test. Means on the same column followed by the same letters are not significantly different at P < 0.05.
Moreover, the pupation rates were significantly decreased in the LdNAT1 RNAi larvae, compared with those in the PBS- and dsegfp-fed larvae (Figs 4D and 5D).
Knockdown of LdNAT1 disrupts insulin/TOR, JH and 20E signaling pathways
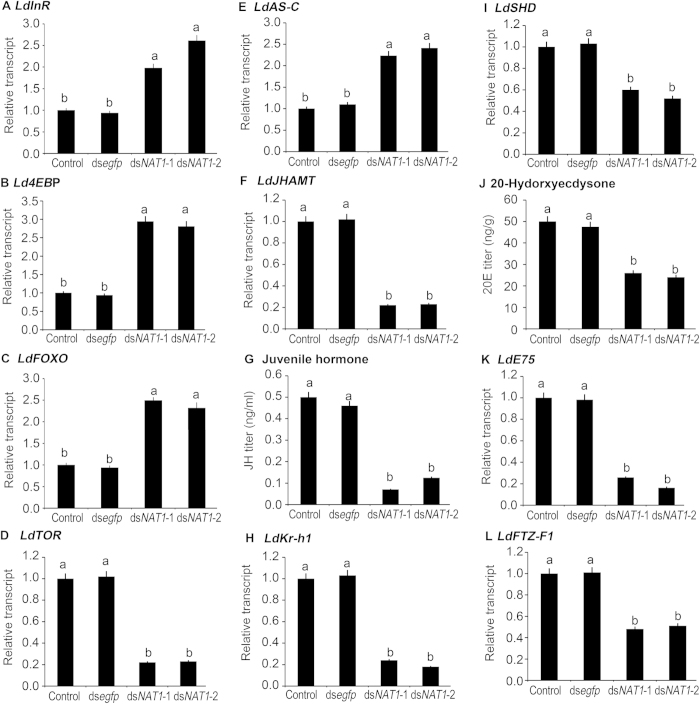
InR, 4EBP, FOXO and TOR are core components in insulin/TOR signaling pathway38. We sequenced cDNAs encoding InR, 4EBP, FOXO and TOR from L. decemlineata. After the initiation of the bioassay for 3 days, the expression levels of the four genes in the fourth-instar alive larvae were tested. LdInR, LdFOXO and Ld4EBP mRNA levels in dsNAT1-1- and dsNAT1-2-fed larvae were significantly upregulated. In contrast, LdTOR mRNA level was significantly downregulated in the LdNAT1 RNAi larvae (Fig. 6A–D).
Figure 6. Effects of dietary ingestion of dsNAT1 (dsNAT1-1, dsNAT1-2) by the L. decemlineata third-instar larvae on insulin/TOR (the left column, A–D), juvenile hormone (the middle column, E–H) and 20-hydroxyecdysone (the right column, I–L) signaling pathways.
Relative transcripts are 2−ΔΔCt values (±SE), normalized to the geometrical mean of housekeeping gene expression. Juvenile hormone and 20-hydroxyecdysone titers are tested by a liquid chromatography tandem mass spectrometry system and a liquid chromatography tandem mass spectrometry-mass spectrometry system. Different letters indicate significant difference at P value < 0.05.
In the LdNAT1 RNAi hypomorphs, LdAS-C expression levels were significantly increased (Fig. 6E), whereas LdJHAMT mRNA levels were dramatically reduced (Fig. 6F) and JH titers were significantly decreased (Fig. 6G). As a result, LdKr-h1 transcript levels were diminished (Fig. 6H).
Finally, ingestion of dsNAT1-1 and dsNAT1-2 by the third-instar larvae significantly reduced LdSHD expression levels (Fig. 6I), decreased 20E titers (Fig. 6J), and diminished the transcript levels of LdE75 and LdFTZ-F1-1 (Fig. 6K,L).
Discussion
In this survey, a putative LdNAT1 were cloned from L. decemlineata. LdNAT1 had a high amino acid similarity to homologs from other insect species. The phylogenetic result indicated that LdNAT1 was distantly related to other insect NAT1-like proteins. Moreover, LdNAT1 was predicted to have 12 transmembrane domains, which was in agreement with the general structure of transporters in the SLC6 family11. It appears that LdNAT1 may be among insect NAT-SLC6 members, and may play a principal role in active absorption and distribution of essential amino acids in L. decemlineata.
Tissue expression of LdNAT1
In insects, amino acids taken up from midgut need to traverse at least three membranes to reach their intracellular site of use: (1) uptake into an epithelial gut cell, (2) basolateral efflux from the gut cell into the hemolymph, and (3) uptake into somatic cell. In general, SLC6 transporters potentially mediate the uptake steps (1) and (3), whereas other SLC members are involved in the efflux step (2)1,39.
In this survey, tissue expression profiles revealed that LdNAT1 mRNA levels were high or moderate in the larval gut, and lower in other surveyed tissues. Similarly, NAT genes in mammalians, other insect species and nematodes are highly expressed in the apical membranes of the alimentary canal, as well as in other organs with elevated requirements for essential amino acids1,11,12,13,37,39. For example, MsKAAT1 in M. sexta7, AeAAT111 and AeNAT514 in A. aegypti, AgNAT637 and AgNAT812 in A. gambiae were highly expressed in the larval guts. Moreover, insect NATs showed unique expression patterns in neurons of the central ganglia and sensory system, which suggest their role as substrate providers for the synthesis of monoamine neurotransmitters12,37.
Therefore, the tissue expression patterns of LdNAT1 are compatible with the common idea that NATs function in: (1) active epithelial uptake of amino acids from the lumen of the gut to support systemic amino acid requirements; (2) active uptake of amino acids into specific cells to support their specialized metabolism or growth; and (3) control of the extracellular concentration of neurotransmitter amino acids in the context of synaptic transmission39.
JH triggers whereas 20E represses the expression of LdNAT1
Essential amino acids are absorbed by midgut in insects11,12,13. In L. decemlineata midgut, several cysteine proteases such as intestain A through E have been reported to digest foliage protein40,41. Vacuolar H+-ATPases energize Na+ and/or K+/H+ antiport42 to actively transport Na+/K+ from the hemolymph into the midgut lumen. Na+/K+ ions are expected to subsequently trigger Na+/K+-dependent NATs to absorb amino acids in L. decemlineata, similar to the NAT-SLC6 members reported in other insect species43.
In some insect species, genes encoding proteases, ATPase subunits and some NAT members are regulated by JH/20E. In Spodoptera litura larvae, for example, a putative serine protease gene Slctlp2 was induced by JH III but not 20E44. In Helicoverpa armigera, the expression of a trypsin-like serine protease gene HaTLP was upregulated by a JH analog methoprene and downregulated by 20E in vivo45. In A. aegypti, the transcription of a serine-type protease gene JHA15 is activated by JH in the newly emerged female adults46. Moreover, the expression of V-ATPase subunit genes were repressed by 20E in M. sexta47. Furthermore, two SLC7 genes, JhI-21 and minidiscs (mnd), were JH-inducible in D. melanogaster48. However, whether NAT-SLC6 genes in insects respond to JH/20E remains unproven.
In several insect species, JH peaks and troughs are observed at each ecdysis. In a termite Cryptotermes secundus, for example, within each instar the JH titer rose shortly before or right after the molt, and then dropped sharply49. Similar phenomenon was observed in L. decemlineata27 and M. sexta47. In this study, temporal expression patterns indicate that LdNAT1 transcript level appears to be positively correlated with the titer of JH. Thus, we determined whether the correlation had any biological significance. As expected, we discovered that either JH or Pyr induced the expression of LdNAT1 in an in vivo bioassay. Moreover, we found that knockdown of LdAS-C to increase JH titer27 activated LdNAT1 expression. Conversely, silencing LdJHAMT to inhibit JH biosynthesis reduced LdNAT1 transcript in L. decemlineata final instar larvae.
Temporal expression patterns also imply that 20E inhibits LdNAT1 transcription. As expected, we found that LdNAT1 expression was dramatically decreased in L. decemlineata specimens having ingested 20E or an ecdysteroid agonist Hal. Conversely, a decrease in 20E in the LdSHD RNAi hypomorphs activated the expression of LdNAT1. Thus, 20E represses the transcription of LdNAT1 in L. decemlineata. In holometabolous insects such as D. melanogaster, 20E signal directly induces transcription of early 20E-response genes such as DmBR-C, DmE74A and DmE75A, and upregulates an early-late gene DmHR3 during larval-pupal transition50. DmHR3 then induces DmβFTZ-F1 expression in mid-prepupae51. In this survey, knockdown of either LdE75 or LdFTZ-F1 reduced 20E titer, as our previously reported results29, the expression levels of LdNAT1 in both LdE75 and LdFTZ-F1 RNAi larvae were significantly increased. Therefore, our results suggested that LdNAT1 repression required complete 20E signaling pathway.
In response to JH and 20E, LdNAT1 transcription is activated right after the molt, and is repressed just before the molt. Thus, its protein may function in absorption of amino acids at the mid instar stage when L. decemlineata larvae are actively feeding.
Involvement of LdNAT1 in uptake of neutral amino acids
Xenopus oocyte-expressed insect transporters, such as M. sexta MsKAAT15 and MsCATCH110, A. aegypti AaNAT111 and D. melanogaster DmNAT113, shared relatively broad substrate spectra. MsKAAT1 absorbed phenylalanine, tryptophan, isoleucine, leucine, valine, methionine and alanine5,7,8,9, whereas MsCAATCH1 preferred threonine in the presence of K+ but preferred proline in the presence of Na+ 10. AeAAT1 from A. aegypti had notable apparent affinities and transport velocities for phenylalanine, cysteine, histidine, alanine, serine and methionine11. DmNAT1 in D. melanogaster transported threonine, isoleucine, leucine, valine, histidine, phenylalanine, tyrosine, tryptophan, methionine, cysteine, alanine, proline, serine, asparagine and glycine, with virtually equal apparent affinities and transport velocities13. In contrast, several Dipteran NAT-SLC6 members showed narrow specialization for absorption of essential amino acids12,14,37. However, their orphan orthologs are absent in D. melanogaster or outside Diptera species14.
In this survey, by comparing the compositions of nineteen proteinogenic amino acids (tryptophan is undetectable) in potato foliage, and in bodies and feces of the day 1 third- and fourth-instar larvae, we found that a total of 11 amino acids (cysteine, histidine, lysine, methionine, serine, tyrosine, valine, glutamate, isoleucine, leucine, phenylalanine) may be actively absorbed by L. decemlineata larval gut. In the LdNAT1 RNAi larvae, the contents of cysteine, histidine, isoleucine, leucine, methionine, phenylalanine and serine in the bodies were significantly lower, whereas the contents of these amino acids in the feces were significantly higher, compared with those in the bodies and feces of PBS- and dsegfp-fed larvae. Therefore, LdNAT1 in L. decemlineata appears an SLC6-NAT transporter belonging to the B(0) system, like most of its homologs in Dipteran and Lepidopteran insect species5,10,11,13.
However, among 11 amino acids being suggested active absorption by the larval gut in L. decemlineata in this survey, lysine, tyrosine, valine and glutamate can not be absorbed by LdNAT1. This indicates that there are other functional NATs in L. decemlineata. Consistent with our results, a total of 9 SLC families are present in insects out of 10 SLC families participating in mammalian amino acid transport 1. Work is in progress to identify these transporters.
Knockdown of LdNAT1 impaired larval development
Silencing LdNAT1 caused obvious negative effects: larval growth was retarded; development period was lengthened, and pupation was impaired. Similarly, RNAi of AeNAT5 increased larval mortality during ecdysis and dramatically suppressed adult emergence in A. aegypti14. Moreover, SLC6 was prominent in bacteria and archaea, and often served in environmental absorption of tryptophan, phenylalanine, tyrosine and methionine. Depriving the organism of such amino acids, or knocking out bacterial SLC6 transporters, limited the exponential growth of bacterial populations52,53,54,55,56. In Caenorhabditis elegans, snf-5 encodes a SLC6 family NAT. A loss-of-function mutation in snf-5 increased dauer formation and reduced dauer maintenance upon starvation39.
In insect, nutritional deprivation inhibited insulin/TOR pathway38 to modulate insect growth and metamorphosis22,23,24. Thus, we measured the expression levels of LdInR, Ld4EBP, LdFOXO and LdTOR. As expected, the transcription of LdInR, Ld4EBP and LdFOXO was upregulated in L. decemlineata. In contrast, the mRNA level of LdTOR was reduced.
Our results further revealed that exposure to dsNAT1-1-, dsNAT1-2-dipped foliage significantly delayed the developing periods of the fourth-instar and prepupae, whenever we treated the larvae at the second-instar or at the third-instar stages. Thus, NAT is required for dietary intake of amino acids supporting growth and development to critical weight at the fourth (final) instar stage. Moreover, we further discovered that knockdown of LdNAT1 reduced the mRNA level of an ecdysteroidogenesis gene LdSHD, decreased 20E titer, and lowered the expression of two 20E-response gene LdE75 and LdFTZ-F129. Furthermore, silencing LdNAT1 increased the mRNA level of LdAS-C, reduced the expression of a JH biosynthesis gene LdJHAMT, decreased JH titer, and diminished the transcription of a JH early-inducible gene LdKr-h1 in L. decemlineata larvae.
There are at least two alternative, but not mutually exclusive, explanations for the results. Firstly, delay of the onset of critical weight and reduced insulin/TOR signaling change the timing of 20E pulse and the level of circulating JH in the LdNAT1 RNAi hypomorphs. Alternatively, the reduced insulin/TOR signaling decreases circulating 20E and JH22,23,24. Disturbed 20E and JH signals subsequently delay larval development and impaired pupation. Accordingly, a model for the influence of knockdown of LdNAT1 on the larval-pupal metamorphosis is proposed in L. decemlineata (Fig. 7).
Figure 7. A model for LdNAT1 knockdown on Leptinotarsa larval-pupal metamorphosis.
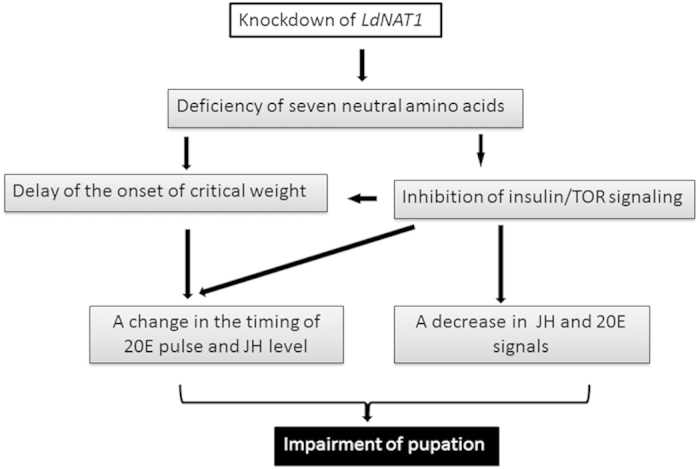
Knockdown of LdNAT1 results in the deficiency of seven neutral amino acids, i.e., cysteine, histidine, isoleucine, leucine, methionine, phenylalanine and serine. This delays the onset of critical weight and inhibits insulin/TOR signaling. The onset of critical weight and the inhibited insulin/TOR signaling may change the timing of the 20E pulse and JH level in the final instar larvae. Moreover, the reduced insulin/TOR signaling may also decrease 20E and JH signals. Thus, the growth is retarded and the pupation is impaired in the LdNAT1 RNAi hypomorphs.
Additional Information
How to cite this article: Fu, K.-Y. et al. Knockdown of a nutrient amino acid transporter gene LdNAT1 reduces free neutral amino acid contents and impairs Leptinotarsa decemlineata pupation. Sci. Rep. 5, 18124; doi: 10.1038/srep18124 (2015).
Supplementary Material
Acknowledgments
This research was supported by the National Natural Science Foundation of China (31272047 and 31360442), and the Fundamental Research Funds for the Central Universities (KYTZ201403).
Footnotes
Author Contributions K.Y.F. performed all experiments, analyzed the data. W.C.G. and T.A. participated in the experiments. G.Q.L. conducted the study and wrote the manuscript. All authors approved the final manuscript.
References
- Boudko D. Y. Molecular basis of essential amino acid transport from studies of insect nutrient amino acid transporters of the SLC6 family (NAT-SLC6). J. Insect Physiol. 58, 433–449 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfersberger M. G. Uniporters, symporters and antiporters. J. Exp. Physiol. 196, 5–6 (1994). [DOI] [PubMed] [Google Scholar]
- Hediger M. A. et al. The ABCs of solute carriers: physiological, pathological and therapeutic implications of human membrane transport proteins introduction. Pflügers Archiv. 447, 465–468 (2004). [DOI] [PubMed] [Google Scholar]
- Broer S. The SLC6 orphans are forming a family of amino acid transporters. Neurochem. Int. 48, 559–567 (2006). [DOI] [PubMed] [Google Scholar]
- Castagna M. et al. Cloning and characterization of a potassium-coupled amino acid transporter. Proc. Natl. Acad. Sci. USA. 95, 5395–5400 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quick M. & Stevens B. R. Amino acid transporter CAATCH1 is also an amino acid-gated cation channel. J. Biol. Chem. 276, 33413–33418 (2001). [DOI] [PubMed] [Google Scholar]
- Vincenti S., Castagna M. Peres A. & Sacchi V. Substrate selectivity and pH dependence of KAAT1 expressed in Xenopus laevis oocytes. J. Membr. Biol. 174, 213–224 (2000). [DOI] [PubMed] [Google Scholar]
- Castagna M. et al. Molecular characteristics of mammalian and insect amino acid transporters: implications for amino acid homeostasis. J. Exp. Physiol. 200, 269–286 (1997). [DOI] [PubMed] [Google Scholar]
- Peres A. & Bossi E. Effects of pH on the uncoupled, coupled and pre-steady-state currents at the amino acid transporter KAAT1 expressed in Xenopus oocytes. J. Physiol. 525, 83–89 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman D., Harvey W. & Stevens B. A novel electrogenic amino acid transporter is activated by K+ or Na+, is alkaline pH-dependent, and is Cl-independent. J. Biol. Chem. 275, 24518–24526 (2000). [DOI] [PubMed] [Google Scholar]
- Boudko D. et al. Ancestry and progeny of nutrient amino acid transporters. Proc. Natl. Acad. Sci. USA. 102, 1360–1365 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meleshkevitch E. A. et al. Molecular characterization of the first aromatic nutrient transporter from the sodium neurotransmitter symporter family. J. Exp. Physiol. 209, 3183–3198 (2006). [DOI] [PubMed] [Google Scholar]
- Miller M. M., Popova L. B., Meleshkevitch E. A., Tran P. V. & Boudko D. Y. The invertebrate B(0) system transporter, D. melanogaster NAT1, has unique D-amino acid affinity and mediates gut and brain functions. Insect Biochem. Mol. Biol. 38, 923–931 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meleshkevitch E. A. et al. A novel eukaryotic Na+ methionine selective symporter is essential for mosquito development. Insect Biochem. Mol. Biol. 43, 755–767 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewell J. L. et al. Differential regulation of mTORC1 by leucine and glutamine. Science 347, 194–198 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chantranupong L., Wolfson R. L. & Sabatini D. M. Nutrient-sensing mechanisms across evolution. Cell 161, 67–83 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q. & Brown M. R. Signaling and function of insulin-like peptides in insects. Ann. Rev. Entomol. 51, 1–24 (2006). [DOI] [PubMed] [Google Scholar]
- Vafopoulou X. The coming of age of insulin-signaling in insects. Front. Physiol. 5, 216 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith W. A., Lamattina A. & Collins M. Insulin signaling pathways in lepidopteran ecdysone secretion. Front. Physiol. 5, 19 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen I. A., Attardo G. M., Rodriguez S. D. & Drake L. L. Four-way regulation of mosquito yolk protein precursor genes by juvenile hormone-, ecdysone-, nutrient-, and insulin-like peptide signaling pathways. Front. Physiol. 5, 103 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H.-J. et al. Two insulin receptors determine alternative wing morphs in planthoppers. Nature 519, 464–467 (2015). [DOI] [PubMed] [Google Scholar]
- Nässel D. R., Liu Y. & Luo J. Insulin/IGF signaling and its regulation in Drosophila. Gen. Comp. Endocr. 220, doi: 10.1016/j.ygcen.2014.11.021 (2015). [DOI] [PubMed] [Google Scholar]
- Mizoguchi A. & Okamoto N. Insulin-like and IGF-like peptides in the silkmoth Bombyx mori: discovery, structure, secretion, and function. Front. Physiol. 4, 217 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim C. & Denlinger D. L. Insulin signaling and the regulation of insect diapause. Front. Physiol. 4, 189 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Y. et al. The P450 enzyme Shade mediates the hydroxylation of ecdysone to 20-hydroxyecdysone in the Colorado potato beetle, Leptinotarsa decemlineata. Insect Mol. Biol. 23, 632–643 (2014). [DOI] [PubMed] [Google Scholar]
- Wan P.-J. et al. Identification of cytochrome P450 monooxygenase genes and their expression profiles in cyhalothrin-treated Colorado potato beetle, Leptinotarsa decemlineata. Pest. Biochem. Physiol. 107, 360–368 (2013). [DOI] [PubMed] [Google Scholar]
- Meng Q.-W. et al. Involvement of a putative allatostatin in regulation of juvenile hormone titer and the larval development in Leptinotarsa decemlineata (Say). Gene 554, 105–113 (2015). [DOI] [PubMed] [Google Scholar]
- Zhou L.-T. et al. RNA interference of a putative S-adenosyl-L-homocysteine hydrolase gene affects larval performance in Leptinotarsa decemlineata (Say). J. Insect Physiol. 59, 1049–1056 (2013). [DOI] [PubMed] [Google Scholar]
- Liu X.-P. et al. Involvement of FTZ-F1 in the regulation of pupation in Leptinotarsa decemlineata (Say). Insect Biochem. Mol. Biol. 55, 51–60 (2014). [DOI] [PubMed] [Google Scholar]
- Guo W.-C. et al. Functions of nuclear receptor HR3 during larval-pupal molting in Leptinotarsa decemlineata (Say) revealed by in vivo RNA interference. Insect Biochem. Mol. Biol. 63, 23–33 (2015). [DOI] [PubMed] [Google Scholar]
- Shi X.-Q. et al. Validation of reference genes for expression analysis by quantitative real-time PCR in Leptinotarsa decemlineata (Say). BMC Res. Notes 6, 93 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M. A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007). [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A. & Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin S. A. et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622 (2009). [DOI] [PubMed] [Google Scholar]
- Cornette R., Gotoh H., Koshikawa S. & Miura T. Juvenile hormone titers and caste differentiation in the damp-wood termite Hodotermopsis sjostedti (Isoptera, Termopsidae). J. Insect Physiol. 54, 922–930 (2008). [DOI] [PubMed] [Google Scholar]
- Zhou J. et al. Quantitative determination of juvenile hormone III and 20-hydroxyecdysone in queen larvae and drone pupae of Apis mellifera by ultrasonic-assisted extraction and liquid chromatography with electrospray ionization tandem mass spectrometry. J. Chromatogr. B 879, 2533–2541 (2011). [DOI] [PubMed] [Google Scholar]
- Meleshkevitch E. et al. Cloning and functional expression of the first eukaryotic Na+-tryptophan symporter, AgNAT6. J. Exp. Biol. 212, 1559–1567 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh A. L. & Smith W. A. Nutritional sensitivity of fifth instar prothoracic glands in the tobacco hornworm, Manduca sexta. Journal of Insect Physiology 57, 809–818 (2011). [DOI] [PubMed] [Google Scholar]
- Metzler R., Meleshkevitch E. A., Fox J., Kim H. & Boudko D. Y. An SLC6 transporter of the novel B(0,)- system aids in absorption and detection of nutrient amino acids in Caenorhabditis elegans. J. Exp. Biol. 216, 2843–2857 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petek M. et al. A complex of genes involved in adaptation of Leptinotarsa decemlineata larvae to induced potato defense. Arch. Insect Biochem. Physiol. 79, 153–181 (2012). [DOI] [PubMed] [Google Scholar]
- Gruden K. et al. Molecular basis of Colorado potato beetle adaptation to potato plant defence at the level of digestive cysteine proteinases. Insect Biochem. Mol. Biol. 34, 365–375 (2004). [DOI] [PubMed] [Google Scholar]
- Fu K.-Y., Guo W.-C., Lü F.-G., Liu X.-P & Li G.-Q. Response of the vacuolar ATPase subunit E to RNA interference and four chemical pesticides in Leptinotarsa decemlineata. Pest. Biochem. Physiol. 114,16–23 (2014). [DOI] [PubMed] [Google Scholar]
- Castagna M., Bossi E. & Sacchi V. Molecular physiology of the insect K-activated amino acid transporter 1 (KAAT1) and cation-anion activated amino acid transporter/channel 1 (CAATCH1) in the light of the structure of the homologous protein LeuT. Insect Mol. Biol. 18, 265–279 (2009). [DOI] [PubMed] [Google Scholar]
- Zhang C. et al. A chymotrypsin-like serine protease cDNA involved in food protein digestion in the common cutworm, Spodoptera litura: Cloning, characterization, developmental and induced expression patterns, and localization. J. Insect Physiol. 56, 788–799 (2010). [DOI] [PubMed] [Google Scholar]
- Sui Y. P., Wang J. X. & Zhao X. F. The impacts of classical insect hormones on the expression profiles of a new digestive trypsin-like protease (TLP) from the cotton bollworm, Helicoverpa armigera. Insect Mol. Biol. 18, 443–452 (2009). [DOI] [PubMed] [Google Scholar]
- Bian G., Raikhel A. S. & Zhu J. Characterization of a juvenile hormone-regulated chymotrypsin-like serine protease gene in Aedes aegypti mosquito. Insect Biochem. Mol. Biol. 38, 190–200 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reineke S., Wieczorek H. & Merzendorfer H. Expression of Manduca sexta V-ATPase genes mvB, mvG and mvd is regulated by ecdysteroids. J. Exp. Biol. 205, 1059–1067 (2002). [DOI] [PubMed] [Google Scholar]
- Dubrovsky E. B., Dubrovskaya V. A. & Berger E. M Juvenile hormone signaling during oogenesis in Drosophila melanogaster. Insect Biochem. Mol. Biol. 32, 1555–1565 (2002). [DOI] [PubMed] [Google Scholar]
- Korb J., Hoffmann K. & Hartfelder K. Molting dynamics and juvenile hormone titer profiles in the nymphal stages of a lower termite, Cryptotermes secundus (Kalotermitidae)–signatures of developmental plasticity. J. Insect Physiol. 58, 376–383 (2012). [DOI] [PubMed] [Google Scholar]
- Horner M. A., Chen T. & Thummel C. S. Ecdysteroid regulation and DNA binding properties of Drosophila nuclear hormone receptor superfamily members. Dev. Biol. 168, 490–502 (1995). [DOI] [PubMed] [Google Scholar]
- Ruaud A. F., Lam G. & Thummel C. S. The Drosophila nuclear receptors DHR3 and betaFTZ-F1 control overlaping developmental responses in late embryos. Development 137, 123–131 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Androutsellis-Theotokis A. et al. Characterization of a functional bacterial homologue of sodium-dependent neurotransmitter transporters. J. Biol. Chem. 278, 12703–12709 (2003). [DOI] [PubMed] [Google Scholar]
- Chye M. L., Guest J. R. & Pittard J. Cloning of the aroP gene and identification of its product in Escherichia coli K-12. J. Bacteriol. 167, 749–753 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quick M. et al. State-dependent conformations of the translocation pathway in the tyrosine transporter Tyt1, a novel neurotransmitter:sodium symporter from Fusobacterium nucleatum. J. Biol. Chem. 281, 26444–26454 (2006). [DOI] [PubMed] [Google Scholar]
- Trotschel C. et al. Methionine uptake in Corynebacterium glutamicum by MetQNI and by MetPS, a novel methionine and alanine importer of the NSS neurotransmitter transporter family. Biochemistry 47, 12698–12709 (2008). [DOI] [PubMed] [Google Scholar]
- Burkovski A. & Kramer R. Bacterial amino acid transport proteins: occurrence, functions, and significance for biotechnological applications. Appl. Microbiol. Biot. 58, 265–274 (2002). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.