Telomere erosion and fusion play an important role in the pathology of many common human malignancies including CLL.1,2 Previous studies in CLL have shown that short telomeres defined on the basis of the median value or receiver operating characteristic (ROC) analysis are associated with unmutated IGHV genes, poor risk genomic abnormalities, genomic complexity and high expression of CD38, CD49d, and ZAP70 whereas long telomeres are associated with increasing IGHV mutational load, isolated deletion of 13q and low CD49d expression. In addition, in predominantly diagnostic or mixed patient cohorts, telomere length (TL) predicts time to first treatment and/or overall survival (OS) in multivariate analyses of models incorporating established biomarkers. 3-7 However uncertainties about the most clinically relevant measure of telomere length, the optimal choice of assay, the need for assay standardisation and the lack of published data on the prognostic value of TL in patients entered into randomised trials have hindered the implementation of TL measurement into routine clinical practice. We have attempted to address these issues by measuring telomere length using monochrome multiplex Q-PCR (MMQ-PCR) in 384 patients at randomisation into the UK LRF CLL4 phase 3 chemotherapy trial (Table S1), of whom 111 samples were also screened by single telomere length analysis (STELA). Telomere length and established biomarkers were measured as previously described. 8-13
The mean TL assessed by MMQ-PCR in 384 cases was 3.57 TLU (Telomere Length Units, range 0.61-19.05). For 111 FC patients analysed by both MMQ-PCR and STELA, we showed an excellent correlation between these two TL measurements (Spearman correlation 0.80) (Fig S1), permitting the calibration of the mean TLU to a mean absolute TL of 3.39kb (range 1.93-11.06kb, median 2.92kb [Inter quartile range: 2.58, 3.60]) for our cohort. All subsequent analyses were based on data derived from MMQ-PCR expressed in Kb rather than TLU. TL was found to be significantly associated with IGHV mutation status, ZAP70 and CD38 expression, serum beta-2 microglobulin, TP53 abnormality, 11q deletion, genomic complexity, ATM and SF3B1 mutation but not trisomy 12 or NOTCH1 mutation (Table S2).
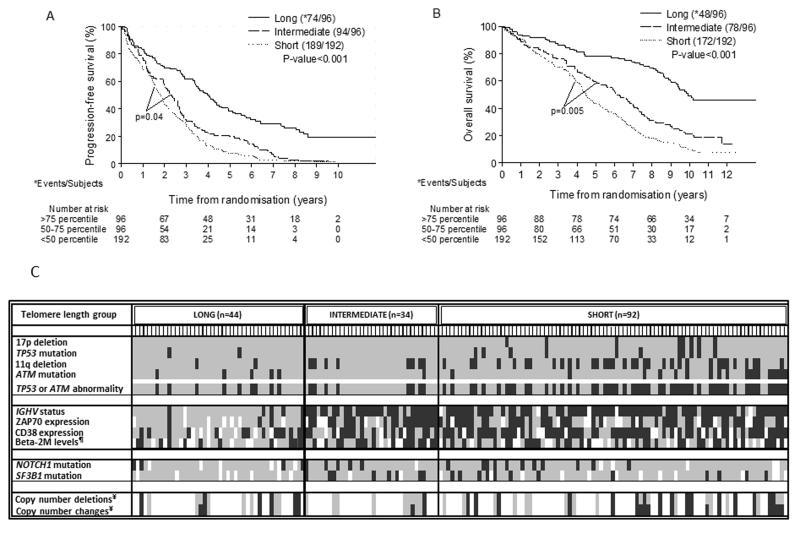
Increasing TL, entered as a continuous variable, was associated with a significant reduction in risk of a PFS event (HR=0.89, 95%CI: 0.85-0.93, p<0.001) and longer OS (HR=0.84, 95%CI: 0.80-0.89, p<0.001) (Table S3). To determine a single TL cut-off value with maximum prognostic power, we employed recursive partitioning and identified the 75th and 80th percentile for PFS (HR: 2.42, p<0.001) and OS (HR: 3.17, p<0.001), respectively (Fig S2), demonstrating that identifying cases with the longest telomeres is key to maximising the prognostic value of these data. We then performed Kaplan-Meier analysis and generated a categorical variable with three groups for telomere length - short (<50 percentile), intermediate (50-75 percentile) and long (>75 percentile) (Fig 1 and S3). The range for the mean TL in the short group was 1.93-2.92 kb (median: 2.58 kb), in the intermediate group, 2.91-3.57 kb (median: 3.14 kb), and in the long group, 3.64-11.06 kb (median: 4.81 kb). Interestingly, we have previously shown that telomere fusions were never detected when the mean telomere length exceeded 3.81kb (as defined by STELA), and the range of telomere lengths in our long and short telomere groups would suggest that telomere fusions would be expected to occur predominantly in the latter group. 14 The risk of progression was increased 2-fold for the intermediate group (HR: 2.07, 95% CI: 1.52-2.82, p<0.001) and by 2.7 times for the short group (HR: 2.67, 95% CI: 2.03-3.53, p<0.001) when compared to the long group while the risk for OS was increased 2.3 and 3.5 times for the intermediate and short groups respectively (Table S3 and Fig S4). The median PFS and OS for the 96 patients in the long TL group was 4.0 and 9.9 years respectively, and these patients were 63% (HR: 0.37, 95% CI: 0.28-0.49, p<0.001) and 72% (HR: 0.28, 95% CI: 0.21-0.39, p<0.001) less likely to progress or die compared to patients within the short TL group (Table S4).
Fig 1. Kaplan-Meier plots of mean telomere length (from MMQPCR) divided into three groups for PFS (A) and OS (B), and distribution of CLL biomarkers within the telomere length groups for 170 patients with complete data for TP53 and ATM status (C).
For (A) and (B), Log-rank P-value for each of the lower quartiles when compared to the Long group is <0.001. Long: >75 percentile; Intermediate: 50-75 percentile; Short: <50 percentile. The median PFS and OS for the 96 patients in the long TL group was 4.0 and 9.9 years respectively. This longer median survival was sustained when the analyses were performed with similarly categorised TL groups using measurements from both MMQPCR and STELA in the 111 patients with data for both (PFS: 8.1 and 5.1 years; OS: not reached and 9.8 years, Fig S5 and 6). For (C) data are presented in decreasing telomere length as measured by MMQPCR and divided into the three groups as described in the text. LONG: >75 percentile, INTERMEDIATE: 50-75 percentile, SHORT: <50 percentile. Each short vertical line (below the TL group name) corresponds to a patient. The presence and absence of each of the biomarker status listed on the left are represented by black and grey boxes respectively whereas white boxes indicate missing data on the biomarker status. *ZAP70 is expressed if >10%-positive cells by flow-cytometry, #CD38 is expressed if >7%-positive cells by flow-cytometry, ¶Beta- M: beta-2 microglobulin (present: >4 mg/L), ¥Present: if: >3 deletions/changes per patient.
Further investigations into the genomic and immunogenetic context of the three TL groups in 170 CLL4 patients with complete TP53 and ATM deletion and mutational data showed that poor-risk features, such as unmutated IGHV genes, TP53 abnormalities, biallelic ATM inactivation, and genomic complexity were found at a higher frequencies in the intermediate and especially in the short TL groups (Fig 1, Table S5). The short TL group also included occasional patients that lacked poor risk biomarkers thus identifying additional patients with poor outcome after first-line chemotherapy. Conversely, the long TL group captured patients with extended survival despite having poor-risk features. Interestingly in the long TL group, patients with TP53 abnormalities or biallelic ATM inactivation predominantly had mutated IGHV genes whereas they were unmutated in the short TL group.
Next, we estimated the adjusted impact of the telomere length categories on PFS and OS after controlling for confounding variables using multivariate Cox regression. The short TL category, treatment allocation (CHL, FDR, FC), unmutated IGHV genes, the presence of an 11q deletion and TP53 abnormality emerged as strong predictors of shorter PFS when the stepwise backward and forward selection method was employed to arrive at a final model (Table 1). The highest increase in risk of progression was for patients with a TP53 abnormality (HR: 2.51, 95% CI: 1.66 to 3.81, p<0.001) followed by the short group for telomere length (short vs long HR: 2.10, 95% CI: 1.37 to 3.21). It is interesting to note that short telomere length had a more detrimental effect on progression than unmutated IGHV genes or 11q deletion. Shorter telomere length, unmutated IGHV genes and a TP53 abnormality remained as strong predictors of shorter overall survival as well.
Table 1. Predictors of overall and progression-free survival.
| Factor | HR (95% CI) | P-value |
|---|---|---|
|
| ||
| PFS | ||
|
|
|
|
| Male vs female | 1.34 (0.999 to 1.80) | 0.05 |
| Treatment | <0.001 | |
| Fludarabine vs. chlorambucil | 0.72 (0.52 to 0.996) | 0.047 |
| (Fludarabine + cyclophosphamide) vs. chlorambucil | 0.38 (0.28 to 0.50) | <0.001 |
| Telomere length | <0.001 | |
| Intermediate vs. Long | 1.31 (0.83 to 2.05) | 0.246 |
| Short vs. Long | 2.10 (1.37 to 3.21) | 0.001 |
| IGHV unmutated vs. mutated | 1.59 (1.12 to 2.25) | 0.01 |
| TP53 abnormal vs. normal | 2.51 (1.66 to 3.81) | <0.001 |
| 11q deleted vs. not deleted | 1.46 (1.07 to 1.98) | 0.02 |
| OS | ||
| Telomere length | 0.003 | |
| Intermediate vs. Long | 1.34 (0.72 to 2.46) | 0.355 |
| Short vs. Long | 2.21 (1.27 to 3.87) | 0.005 |
| IGHV unmutated vs. mutated | 2.08 (1.22 to 3.57 | 0.01 |
| TP53 abnormal vs. normal | 2.11 (1.26 to 3.53) | 0.004 |
| ZAP70 expressed vs. not expressed§ | 0.65 (0.45 to 0.96) | 0.03 |
| 13q deleted vs. not deleted | 0.67 (0.47 to 0.95) | 0.02 |
Telomere length groups: Long: >75 percentile; Intermediate: 50-75 percentile; Short: <50 percentile. Candidates entered in the iterative backward-forward selection method were factors with P-values ≤0.05 in the univariable analysis (see Figure S4). Age was entered as a continuous variable for the multivariable analysis.There was a small but significant negative association between age and shorter TL (data not included). The final models were based on 292 subjects and 269 events for PFS and 201 subjects and 153 deaths for OS.
expressed if expression levels >10%
We then used sensitivity-specificity analysis to derive likelihood ratios (LR+ and LR− and the LR+/LR− ratio) to judge the relative discriminatory power of each of the four biomarkers (TL, IGHV, TP53 and 11q deletion) that were strong predictors of PFS and/or OS in our whole cohort, to predict both the presence and absence of PFS and OS events at last follow-up in the 292 patients with complete TL, IGHV, 11q deletion and TP53 abnormality data (Tables S6-S9). One caveat was that LR+/LR− for PFS events could not be estimated for TP53 alone as the false negative rates were zero.
The best predictor of PFS events was the combined short and intermediate TL groups (cut-off <75th percentile), alone (LR+/LR−: 15.54) or in combination with a TP53 abnormality (LR+/LR−: 16.35) (Table S6). 81% of patients who progressed or died had short or intermediate length telomeres. Consistent with the recursive partitioning data, the best predictor of long PFS (absence of PFS events) was the long TL group alone (LR+/LR− : 15.26) or in combination with wild type TP53 (LR+/LR−: 17.20) (Table S7). Long TL correctly predicted the 18/23 patients who did not have a PFS event (sensitivity of 78.3%), while 218/269 patients without long TL did have a PFS event during follow-up (specificity of 81%). As with the presence of PFS events, the inclusion of IGHV mutational status alone or combined with TL and TP53 data did not increase the LR+/LR− ratio further (Table S7). For OS, the LR+/LR− for the short TL sub-group was 4.70, with the highest ratio for TP53 abnormality (LR+/LR−: 8.83), with sensitivity, specificity and accuracy rates of 84.5%, 51.5% and 77.1%, respectively (Table S8). The long TL category provided a LR+/LR− ratio of 5.82 to predict the absence of an OS event, second to TP53 (LR+/LR−: 8.61) but with much higher specificity (Table S9). In each case LR ratios were improved for TL in combination with IGHV mutational status, TP53 data, or both.
In summary, our data confirm that both MMQ-PCR and STELA can provide clinically relevant information in the range of telomere lengths found in CLL. Importantly we demonstrate for the first time that TL is superior both to established and to recently discovered genomic biomarkers for predicting prolonged PFS following chemotherapy. A study of telomere length using MMQ-PCR in 620 patients entered into the CLL8 trial currently presented in abstract form only, identified TL as an independent marker of PFS but not OS in models which incorporated a similar range of biomarkers to those employed in our study, suggesting that TL also has prognostic significance in patients treated with chemo-immunotherapy. 15 It remains to be seen whether TL will retain prognostic value in patients treated solely with non-genotoxic therapies. Whilst these novel therapies show very significant promise, long-term data on side effects and drug resistance are awaited and they may be unsuitable or unaffordable for some patients. In contrast, immunochemotherapy can offer good long-term outcomes with acceptable toxicity to subgroups of patients with CLL 14. In conclusion, TL, which usually remains stable on sequential analyses 5 warrants further evaluation in CLL patients receiving therapy. Factors such as cost, reproducibility and availability will determine the assay method of choice.
Supplementary Material
Footnotes
Conflict of Interest
All authors declare no conflict of interest.
References
- 1.Lin TT, Letsolo BT, Jones RE, Rowson J, Pratt G, Hewamana S, et al. Telomere dysfunction and fusion during the progression of chronic lymphocytic leukemia: evidence for a telomere crisis. Blood. 2010;116:1899–907. doi: 10.1182/blood-2010-02-272104. [DOI] [PubMed] [Google Scholar]
- 2.Jones CH, Pepper C, Baird DM. Telomere dysfunction and its role in haematological cancer. Br J Haematol. 2012;156:573–87. doi: 10.1111/j.1365-2141.2011.09022.x. [DOI] [PubMed] [Google Scholar]
- 3.Rossi D, Lobetti Bodoni C, Genuardi E, Monitillo L, Drandi D, Cerri M, et al. Telomere length is an independent predictor of survival, treatment requirement and Richter’s syndrome transformation in chronic lymphocytic leukemia. Leukemia. 2009;23:1062–72. doi: 10.1038/leu.2008.399. [DOI] [PubMed] [Google Scholar]
- 4.Roos G, Kröber A, Grabowski P, Kienle D, Bühler A, Döhner H, et al. Short telomeres are associated with genetic complexity, high-risk genomic aberrations, and short survival in chronic lymphocytic leukemia. Blood. 2008;111:2246–52. doi: 10.1182/blood-2007-05-092759. [DOI] [PubMed] [Google Scholar]
- 5.Mansouri L, Grabowski P, Degerman S, Svenson U, Gunnarsson R, Cahill N, et al. Short telomere length is associated with NOTCH1/SF3B1/TP53 aberrations and poor outcome in newly diagnosed chronic lymphocytic leukemia patients. Am J Hematol. 2013;88:647–51. doi: 10.1002/ajh.23466. [DOI] [PubMed] [Google Scholar]
- 6.Grabowski P, Hultdin M, Karlsson K, Tobin G, Aleskog A, Thunberg U, et al. Telomere length as a prognostic parameter in chronic lymphocytic leukemia with special reference to VH gene mutation status. Blood. 2005;105:4807–12. doi: 10.1182/blood-2004-11-4394. [DOI] [PubMed] [Google Scholar]
- 7.Sellmann L, de Beer D, Bartels M, Opalka B, Nückel H, Dührsen U, et al. Telomeres and prognosis in patients with chronic lymphocytic leukaemia. Int J Hematol. 2011;93:74–82. doi: 10.1007/s12185-010-0750-2. [DOI] [PubMed] [Google Scholar]
- 8.Skowronska A, Parker A, Ahmed G, Oldreive C, Davis Z, Richards S, et al. Biallelic ATM Inactivation Significantly Reduces Survival in Patients Treated on UK CLL4 Trial. J Clin Oncol. 2012;30:4524–32. doi: 10.1200/JCO.2011.41.0852. [DOI] [PubMed] [Google Scholar]
- 9.Rose-Zerilli M, Forster J, Parker H, Parker A, Rodriguez A, Chaplin T, et al. ATM mutation rather than BIRC3 deletion and/or mutation predicts reduced survival in 11q-deleted chronic lymphocytic leukemia, data from the UK LRF CLL4 trial. Haematologica. 2014;99:736–42. doi: 10.3324/haematol.2013.098574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oscier DG, Rose-Zerilli MJJ, Winkelmann N, Gonzalez de Castro D, Gomez B, Forster J, et al. The clinical significance of NOTCH1 and SF3B1 mutations in the UK LRF CLL4 trial. Blood. 2013;120:4441–3. doi: 10.1182/blood-2012-05-429282. [DOI] [PubMed] [Google Scholar]
- 11.Oscier D, Wade R, Davis Z, Morilla A, Best G, Richards S, et al. Prognostic factors identified three risk groups in the LRF CLL4 trial, independent of treatment allocation. Haematologica. 2010;95:1705–12. doi: 10.3324/haematol.2010.025338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gonzalez D, Martinez P, Wade R, Hockley S, Oscier D, Matutes E, et al. Mutational status of the TP53 gene as a predictor of response and survival in patients with chronic lymphocytic leukemia: results from the LRF CLL4 trial. J Clin Oncol. 2011;29:2223–9. doi: 10.1200/JCO.2010.32.0838. [DOI] [PubMed] [Google Scholar]
- 13.Catovsky D, Richards S, Matutes E, Oscier D, Dyer MJ, Bezares RF, et al. Assessment of fludarabine plus cyclophosphamide for patients with chronic lymphocytic leukaemia (the LRF CLL4 Trial): a randomised controlled trial. Lancet. 2007;370:230–9. doi: 10.1016/S0140-6736(07)61125-8. [DOI] [PubMed] [Google Scholar]
- 14.Lin TT, Norris K, Heppel NH, Pratt G, Allan JM, Allsup DJ, et al. Telomere dysfunction accurately predicts clinical outcome in chronic lymphocytic leukaemia, even in patients with early stage disease. Br J Haematol. 2014;167:214–23. doi: 10.1111/bjh.13023. [DOI] [PubMed] [Google Scholar]
- 15.Jebaraj BM, Busch R, Zenz T, Bühler A, Winkler D, Schnaiter A, et al. Telomere Length and Treatment Outcome In Chronic Lymphocytic Leukemia: Results From The CLL8 Trial, Abstracts from the 55th American Society of Hematology Annual Meeting and Exposition; New Orleans, LA. 2013.p. 671. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.