Abstract
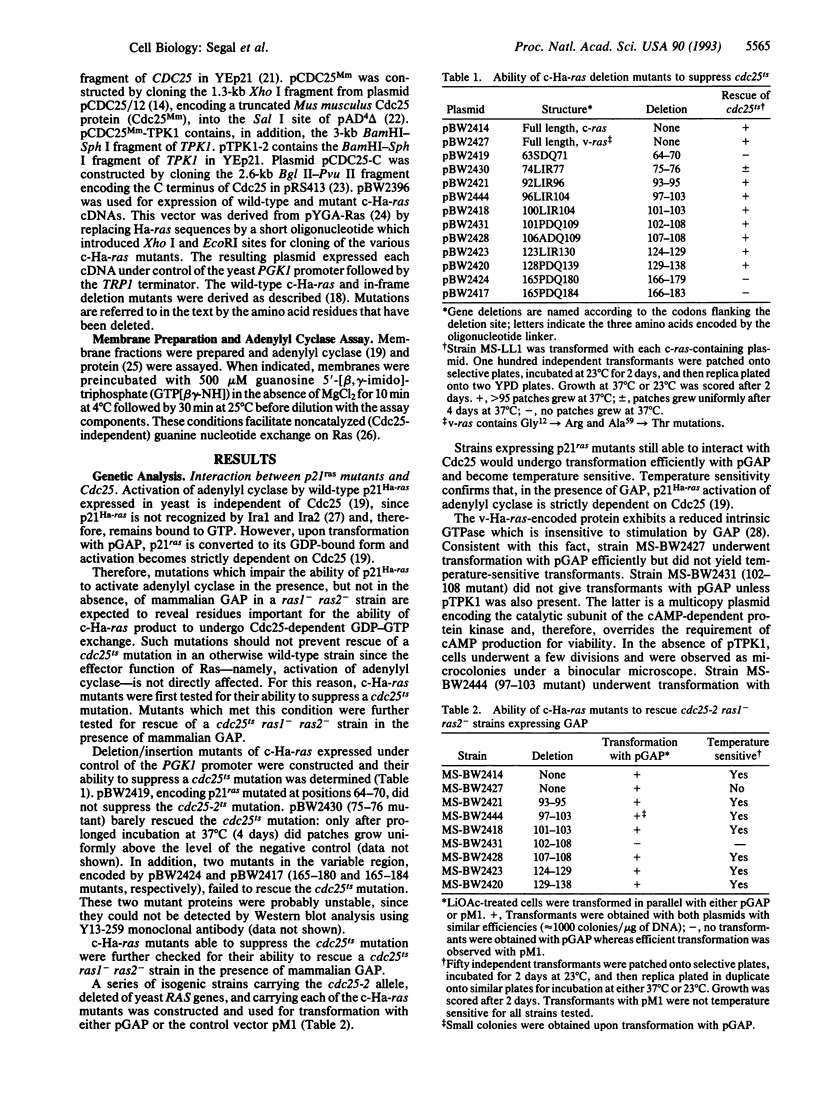
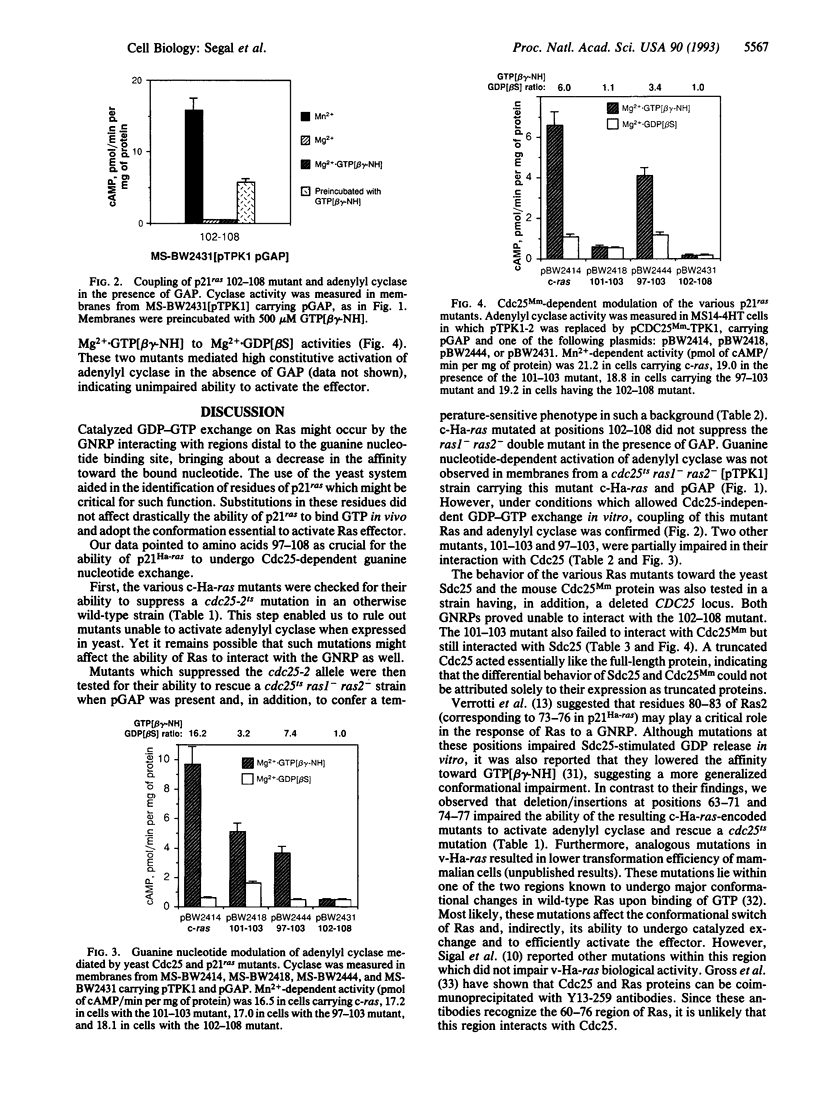
Cdc25 is essential for Ras-mediated activation of adenylyl cyclase in the yeast Saccharomyces cerevisiae. This protein acts by catalyzing GDP-GTP exchange on yeast Ras. Harvey (Ha) ras expressed in S. cerevisiae is also recognized by both Cdc25 and Sdc25, a yeast homolog of Cdc25. Thus it is feasible to examine molecular aspects of mammalian Ras modulation by Cdc25 using the RAS/cAMP pathway in yeast as a model system. Here, we describe mutational analysis of Ha-ras for the identification of residues critical for the ability of Ras to interact with Cdc25 and related guanine nucleotide-release proteins. Mutations within codons 97-108 impaired Ras-mediated activation of adenylyl cyclase in the presence but not in the absence of mammalian GTPase-activating protein. Such mutations, therefore, affected the ability of Ras to undergo GDP-GTP exchange catalyzed by the guanine nucleotide exchanger without preventing Ras activation of the effector. Similar mutations were previously shown to impair the ability of c-ras to transform mammalian cells while having a less drastic effect on v-ras.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adari H., Lowy D. R., Willumsen B. M., Der C. J., McCormick F. Guanosine triphosphatase activating protein (GAP) interacts with the p21 ras effector binding domain. Science. 1988 Apr 22;240(4851):518–521. doi: 10.1126/science.2833817. [DOI] [PubMed] [Google Scholar]
- Ballester R., Michaeli T., Ferguson K., Xu H. P., McCormick F., Wigler M. Genetic analysis of mammalian GAP expressed in yeast. Cell. 1989 Nov 17;59(4):681–686. doi: 10.1016/0092-8674(89)90014-7. [DOI] [PubMed] [Google Scholar]
- Barbacid M. ras genes. Annu Rev Biochem. 1987;56:779–827. doi: 10.1146/annurev.bi.56.070187.004023. [DOI] [PubMed] [Google Scholar]
- Bollag G., McCormick F. Regulators and effectors of ras proteins. Annu Rev Cell Biol. 1991;7:601–632. doi: 10.1146/annurev.cb.07.110191.003125. [DOI] [PubMed] [Google Scholar]
- Botstein D., Falco S. C., Stewart S. E., Brennan M., Scherer S., Stinchcomb D. T., Struhl K., Davis R. W. Sterile host yeasts (SHY): a eukaryotic system of biological containment for recombinant DNA experiments. Gene. 1979 Dec;8(1):17–24. doi: 10.1016/0378-1119(79)90004-0. [DOI] [PubMed] [Google Scholar]
- Bourne H. R., Sanders D. A., McCormick F. The GTPase superfamily: a conserved switch for diverse cell functions. Nature. 1990 Nov 8;348(6297):125–132. doi: 10.1038/348125a0. [DOI] [PubMed] [Google Scholar]
- Bourne H. R., Sanders D. A., McCormick F. The GTPase superfamily: conserved structure and molecular mechanism. Nature. 1991 Jan 10;349(6305):117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- Boy-Marcotte E., Damak F., Camonis J., Garreau H., Jacquet M. The C-terminal part of a gene partially homologous to CDC 25 gene suppresses the cdc25-5 mutation in Saccharomyces cerevisiae. Gene. 1989 Apr 15;77(1):21–30. doi: 10.1016/0378-1119(89)90355-7. [DOI] [PubMed] [Google Scholar]
- Clark S. G., McGrath J. P., Levinson A. D. Expression of normal and activated human Ha-ras cDNAs in Saccharomyces cerevisiae. Mol Cell Biol. 1985 Oct;5(10):2746–2752. doi: 10.1128/mcb.5.10.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Créchet J. B., Poullet P., Mistou M. Y., Parmeggiani A., Camonis J., Boy-Marcotte E., Damak F., Jacquet M. Enhancement of the GDP-GTP exchange of RAS proteins by the carboxyl-terminal domain of SCD25. Science. 1990 May 18;248(4957):866–868. doi: 10.1126/science.2188363. [DOI] [PubMed] [Google Scholar]
- DeFeo-Jones D., Tatchell K., Robinson L. C., Sigal I. S., Vass W. C., Lowy D. R., Scolnick E. M. Mammalian and yeast ras gene products: biological function in their heterologous systems. Science. 1985 Apr 12;228(4696):179–184. doi: 10.1126/science.3883495. [DOI] [PubMed] [Google Scholar]
- Downward J., Riehl R., Wu L., Weinberg R. A. Identification of a nucleotide exchange-promoting activity for p21ras. Proc Natl Acad Sci U S A. 1990 Aug;87(15):5998–6002. doi: 10.1073/pnas.87.15.5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross E., Goldberg D., Levitzki A. Phosphorylation of the S. cerevisiae Cdc25 in response to glucose results in its dissociation from Ras. Nature. 1992 Dec 24;360(6406):762–765. doi: 10.1038/360762a0. [DOI] [PubMed] [Google Scholar]
- Kataoka T., Powers S., Cameron S., Fasano O., Goldfarb M., Broach J., Wigler M. Functional homology of mammalian and yeast RAS genes. Cell. 1985 Jan;40(1):19–26. doi: 10.1016/0092-8674(85)90304-6. [DOI] [PubMed] [Google Scholar]
- Kavounis C., Verrotti A. C., De Vendittis E., Bozopoulos A., Di Blasi F., Zahn R., Crechet J. B., Parmeggiani A., Tsernoglou D., Fasano O. Role of glycine-82 as a pivot point during the transition from the inactive to the active form of the yeast Ras2 protein. FEBS Lett. 1991 Apr 9;281(1-2):235–239. doi: 10.1016/0014-5793(91)80401-n. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Marshall M. S., Gibbs J. B., Scolnick E. M., Sigal I. S. Regulatory function of the Saccharomyces cerevisiae RAS C-terminus. Mol Cell Biol. 1987 Jul;7(7):2309–2315. doi: 10.1128/mcb.7.7.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martegani E., Vanoni M., Zippel R., Coccetti P., Brambilla R., Ferrari C., Sturani E., Alberghina L. Cloning by functional complementation of a mouse cDNA encoding a homologue of CDC25, a Saccharomyces cerevisiae RAS activator. EMBO J. 1992 Jun;11(6):2151–2157. doi: 10.1002/j.1460-2075.1992.tb05274.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistou M. Y., Jacquet E., Poullet P., Rensland H., Gideon P., Schlichting I., Wittinghofer A., Parmeggiani A. Mutations of Ha-ras p21 that define important regions for the molecular mechanism of the SDC25 C-domain, a guanine nucleotide dissociation stimulator. EMBO J. 1992 Jul;11(7):2391–2397. doi: 10.1002/j.1460-2075.1992.tb05303.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal M., Marbach I., Engelberg D., Simchen G., Levitzki A. Interaction between the Saccharomyces cerevisiae CDC25 gene product and mammalian ras. J Biol Chem. 1992 Nov 15;267(32):22747–22751. [PubMed] [Google Scholar]
- Sigal I. S., Gibbs J. B., D'Alonzo J. S., Scolnick E. M. Identification of effector residues and a neutralizing epitope of Ha-ras-encoded p21. Proc Natl Acad Sci U S A. 1986 Jul;83(13):4725–4729. doi: 10.1073/pnas.83.13.4725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989 May;122(1):19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamanoi F. Yeast RAS genes. Biochim Biophys Acta. 1988 Aug 3;948(1):1–15. doi: 10.1016/0304-419x(88)90002-9. [DOI] [PubMed] [Google Scholar]
- Tanaka K., Nakafuku M., Satoh T., Marshall M. S., Gibbs J. B., Matsumoto K., Kaziro Y., Toh-e A. S. cerevisiae genes IRA1 and IRA2 encode proteins that may be functionally equivalent to mammalian ras GTPase activating protein. Cell. 1990 Mar 9;60(5):803–807. doi: 10.1016/0092-8674(90)90094-u. [DOI] [PubMed] [Google Scholar]
- Tatchell K., Chaleff D. T., DeFeo-Jones D., Scolnick E. M. Requirement of either of a pair of ras-related genes of Saccharomyces cerevisiae for spore viability. Nature. 1984 Jun 7;309(5968):523–527. doi: 10.1038/309523a0. [DOI] [PubMed] [Google Scholar]
- Toda T., Uno I., Ishikawa T., Powers S., Kataoka T., Broek D., Cameron S., Broach J., Matsumoto K., Wigler M. In yeast, RAS proteins are controlling elements of adenylate cyclase. Cell. 1985 Jan;40(1):27–36. doi: 10.1016/0092-8674(85)90305-8. [DOI] [PubMed] [Google Scholar]
- Verrotti A. C., Créchet J. B., Di Blasi F., Seidita G., Mirisola M. G., Kavounis C., Nastopoulos V., Burderi E., De Vendittis E., Parmeggiani A. RAS residues that are distant from the GDP binding site play a critical role in dissociation factor-stimulated release of GDP. EMBO J. 1992 Aug;11(8):2855–2862. doi: 10.1002/j.1460-2075.1992.tb05353.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willumsen B. M., Vass W. C., Velu T. J., Papageorge A. G., Schiller J. T., Lowy D. R. The bovine papillomavirus E5 oncogene can cooperate with ras: identification of p21 amino acids critical for transformation by c-rasH but not v-rasH. Mol Cell Biol. 1991 Dec;11(12):6026–6033. doi: 10.1128/mcb.11.12.6026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittinghofer A., Pai E. F. The structure of Ras protein: a model for a universal molecular switch. Trends Biochem Sci. 1991 Oct;16(10):382–387. doi: 10.1016/0968-0004(91)90156-p. [DOI] [PubMed] [Google Scholar]




