ABSTRACT
Iron, a major protein cofactor, is essential for most organisms. Despite the well-known effects of O2 on the oxidation state and solubility of iron, the impact of O2 on cellular iron homeostasis is not well understood. Here we report that in Escherichia coli K-12, the lack of O2 dramatically changes expression of genes controlled by the global regulators of iron homeostasis, the transcription factor Fur and the small RNA RyhB. Using chromatin immunoprecipitation sequencing (ChIP-seq), we found anaerobic conditions promote Fur binding to more locations across the genome. However, by expression profiling, we discovered that the major effect of anaerobiosis was to increase the magnitude of Fur regulation, leading to increased expression of iron storage proteins and decreased expression of most iron uptake pathways and several Mn-binding proteins. This change in the pattern of gene expression also correlated with an unanticipated decrease in Mn in anaerobic cells. Changes in the genes posttranscriptionally regulated by RyhB under aerobic and anaerobic conditions could be attributed to O2-dependent changes in transcription of the target genes: aerobic RyhB targets were enriched in iron-containing proteins associated with aerobic energy metabolism, whereas anaerobic RyhB targets were enriched in iron-containing anaerobic respiratory functions. Overall, these studies showed that anaerobiosis has a larger impact on iron homeostasis than previously anticipated, both by expanding the number of direct Fur target genes and the magnitude of their regulation and by altering the expression of genes predicted to be posttranscriptionally regulated by the small RNA RyhB under iron-limiting conditions.
IMPORTANCE
Microbes and host cells engage in an “arms race” for iron, an essential nutrient that is often scarce in the environment. Studies of iron homeostasis have been key to understanding the control of iron acquisition and the downstream pathways that enable microbes to compete for this valuable resource. Here we report that O2 availability affects the gene expression programs of two Escherichia coli master regulators that function in iron homeostasis: the transcription factor Fur and the small RNA regulator RyhB. Fur appeared to be more active under anaerobic conditions, suggesting a change in the set point for iron homeostasis. RyhB preferentially targeted iron-containing proteins of respiration-linked pathways, which are differentially expressed under aerobic and anaerobic conditions. Such findings may be relevant to the success of bacteria within their hosts since zones of reduced O2 may actually reduce bacterial iron demands, making it easier to win the arms race for iron.
INTRODUCTION
In nearly all organisms, iron is an essential nutrient that serves as a protein cofactor in pathways ranging from central metabolism to genome maintenance. In single-cell organisms, such as bacteria, maintaining a pool of intracellular iron sufficient to cofactor proteins requires coordination of synthesis of iron-containing proteins and cofactors with iron uptake and iron storage (1, 2). Although this process of iron homeostasis has been well studied in Escherichia coli K-12, most of our understanding comes from analyzing cells grown in the presence of O2 (2–4), conditions known to result in oxidation of Fe2+ to Fe3+, decreased iron solubility, and the formation of reactive oxygen species (5). In contrast, less is known about iron homeostasis during anaerobiosis, conditions in which the soluble form of iron (Fe2+) is more stable and many important iron-requiring activities (e.g., cyclic photosynthesis, N2 fixation, and anaerobic respiration) of bacteria occur (6). We are interested in determining how O2 availability alters the expression of genes needed to maintain cellular pools of iron.
In E. coli K-12, the transcription factor Fur (ferric uptake regulator) (7, 8) and the small RNA RyhB (9, 10) are the major regulators of iron homeostasis. Studies from cells grown under aerobic conditions have led to the prevailing view that Fe2+-Fur binds DNA when iron is sufficient (8, 11, 12), resulting in repression of most of its target genes. Functions repressed by Fur include RyhB (9), the Fe3+-siderophore uptake pathways (e.g., fepA, fhuA, and cirA [7, 13]), one of two Fe-S cluster biogenesis pathways (sufABCDSE [14–16]), the manganese uptake system (mntH [17]), the manganese-containing superoxide dismutase (sodA [18]), and the manganese-containing ribonucleotide reductase complex (nrdHIEF [19]). In contrast, Fur increases the expression of ftnA, encoding an iron storage complex (20). Siderophores are regarded as a major route of iron uptake under aerobic conditions, because their high affinity for Fe3+ compensates for the poor solubility of oxidized iron in the presence of O2 (21, 22). Although repression of iron uptake systems by Fur under iron-sufficient conditions may seem counterintuitive, the low levels of these gene products are apparently adequate to supply iron for protein cofactors and storage.
Upon iron limitation, Fe2+ is not available to bind Fur, and Fur binding to its DNA sites decreases, resulting in decreased expression of Fur-induced genes (i.e., ftnA) and increased expression of most of the Fur regulon, including RyhB (13, 23). The reported changes in Fur-dependent gene transcription under iron-limiting conditions portray a coordinated strategy of reducing iron storage, scavenging limiting iron, and switching to manganese-dependent proteins to replace those requiring iron for function. Although Fur function has not been systematically studied under anaerobic conditions in E. coli, some O2-dependent differences in Fur-regulated genes have been reported. Whereas expression of siderophore-mediated iron transport systems (fepA, fhuA, cirA, tonB, and exbB [7, 24, 25]) is more repressed under anaerobic conditions, expression of Fe2+ transport (feoABC) is increased anaerobically (26, 27).
Expression of the small RNA RyhB under iron-limiting conditions mediates an iron-sparing response to supply iron for critical iron-containing proteins by decreasing translation of certain iron-containing proteins or increasing translation of iron uptake functions (10, 28). Base pairing of RyhB with specific mRNA transcripts results in either enhanced translation through disruption of an inhibitory complex (29) or decreased translation through Hfq recruitment, which is often accompanied by decreased transcript stability (30, 31). Transcription of a few known RyhB targets is repressed under anaerobic conditions (sdhCDAB, acnA, acnB, and fumA) (27, 32, 33), suggesting that these RNAs would not be posttranscriptionally regulated under anaerobic conditions. The fact that transcription of genes encoding other iron-containing respiratory proteins is selectively upregulated under anaerobic conditions (32, 34–37) raises the question of whether these transcripts might be targets of RyhB under anaerobic conditions. Since it is challenging to predict direct targets of small RNAs like RyhB bioinformatically (38, 39), experimental studies are needed to identify RyhB candidates under anaerobic conditions.
Here we address how anaerobiosis affects the Fur and RyhB regulons of E. coli K-12. Chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq) identified in vivo Fur DNA binding sites in the presence and absence of O2. Global gene expression studies of wild-type and Fur− (Δfur) strains cultured under aerobic or anaerobic conditions revealed genes regulated by Fur in an O2-dependent manner. Promoter fusions to lacZ confirmed new Fur targets. Global gene expression studies of strains lacking RyhB (ΔryhB) or RyhB and Fur (Δfur ΔryhB) were used to identify the scope of possible RyhB targets during anaerobiosis. The metallome of cells grown under aerobic and anaerobic conditions was probed to ask if the intracellular availability of iron or other metals changes under anaerobic conditions. Our findings reveal major changes in the Fur and RyhB regulons under anaerobic conditions that tailor these gene expression programs to an O2-free lifestyle.
RESULTS
Fur binds to more genomic regions under anaerobic growth conditions.
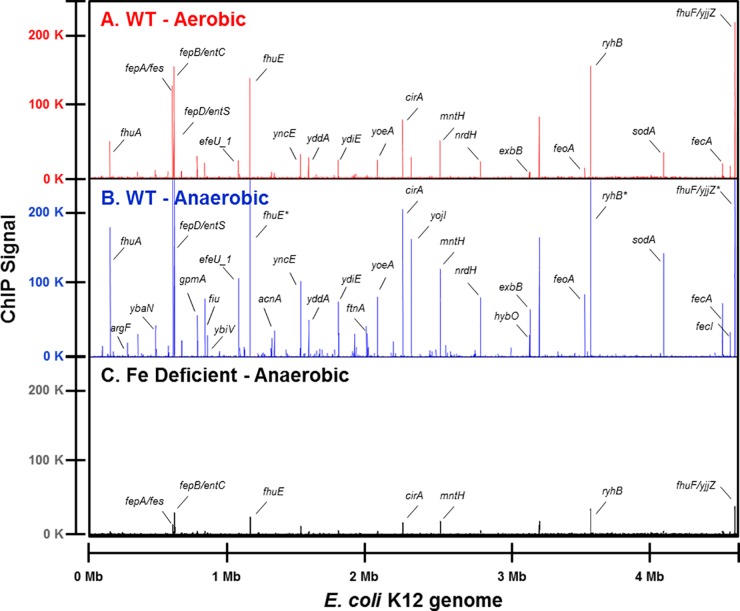
To address how anaerobiosis impacts the Fur regulon, we mapped Fur DNA binding regions genome-wide in E. coli K-12 from cells grown in defined, iron-sufficient medium under aerobic or anaerobic conditions using ChIP-seq (see Table S2 in the supplemental material). Under aerobic conditions, Fur bound to 96 locations, and two-thirds of these binding peaks were found in intergenic regions (Fig. 1A). Under anaerobic conditions, Fur bound all sites identified under aerobic conditions and 157 additional locations (Fig. 1B); only half of these newly identified sites were in intergenic regions. Fur binding under anaerobic conditions was iron dependent because the vast majority of binding locations (247 out of 255) were either eliminated or greatly reduced when assayed under iron-limiting conditions (Fig. 1C). Together, these data show that Fur is bound to more genomic regions under anaerobic conditions, and the iron dependence of its DNA binding suggests that Fur is interacting with its DNA sites in a regulated manner (23).
FIG 1 .
Genome-wide Fur DNA binding. Fur binding across the genome was compared under either aerobic (track A, red) or anaerobic (track B, blue) iron-sufficient (10 µM FeSO4) growth conditions in the wild-type strain, MG1655, or under anaerobic iron-limiting (1.0 µM FeSO4) growth conditions (track C, black) in an iron-uptake-deficient strain (ΔtonB Δfeo ΔzupT) by ChIP-seq. The x axis indicates the genomic position of the ChIP-seq peaks using the MG1655 genome coordinates (version U00096.2), and the y axis indicates the read count after each data set was normalized to 20 million reads; enrichment of Fur DNA binding is indicated by the height of the lines in each track. An asterisk following the gene name indicates the read count extends beyond 250,000. A subset of peaks has been labelled with the corresponding downstream gene for comparison.
Fur binds to less-conserved sequences under anaerobic conditions.
To understand why Fur is bound to more genomic regions under anaerobic conditions, we asked if there were DNA sequence differences between the sites bound only under anaerobic conditions versus those bound under both aerobic and anaerobic growth conditions. The DNA sequences within 100 bp of the summit of the iron-dependent Fur binding peaks were analyzed for overrepresented sequences using the motif-finding algorithm MEME-ChIP (40). The motif derived from the iron-dependent DNA regions bound by Fur under both aerobic and anaerobic growth conditions (Fig. 2, top panel) was similar to the signature inverted repeat 5′-GATAAT-N1-ATTATC-3′ previously described as the Fur dimer binding site (41–44). However, the motif derived from the regions bound by Fur only under anaerobic conditions (Fig. 2, bottom panel) revealed less sequence conservation to this canonical motif. Taken together, this analysis suggests that under anaerobic conditions, Fe2+-Fur is bound to potentially stronger affinity sites represented by the signature Fur motif, as well as potentially weaker affinity DNA sites represented by the less-conserved motif.
FIG 2 .
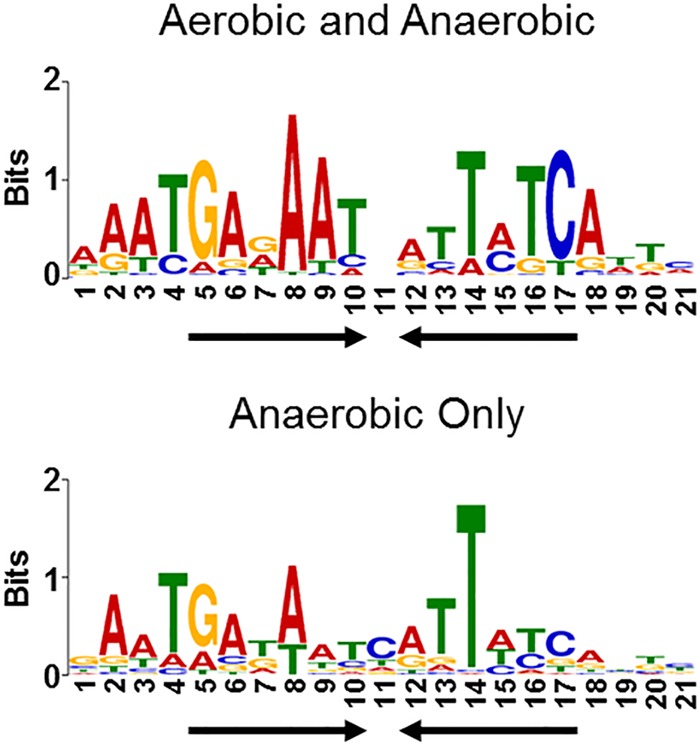
Fur binds less-conserved sequences under anaerobic growth conditions. DNA sequences from iron-dependent Fur ChIP-seq peaks were analyzed by the motif-finding algorithm MEME-ChIP. The height of the letters (in bits on the y axis) represents the degree of conservation at a given position within the aligned sequence set, with perfect conservation being 2 bits. The motif in the top panel was constructed from 90 sequences bound by Fur under both aerobic and anaerobic growth conditions. The motif in the bottom panel was constructed from 157 sequences bound by Fur only under anaerobic growth conditions. Arrows indicate the presence of an inverted repeat.
Despite Fur binding to more locations under anaerobic conditions, the genes regulated by Fur are quite similar between aerobic and anaerobic conditions.
Transcription profiling of Fur+ and Fur− strains was used first to determine whether Fur binding to the sites identified as specific to anaerobic conditions led to transcriptional regulation. Of the 178 operons whose expression we found to be Fur regulated under anaerobic conditions (see Tables S3 to S5 in the supplemental material), only four operons were associated with a Fur ChIP-seq peak that was specific to anaerobic conditions and were not RyhB regulated (see below) (Fig. 3; see Table S3). Expression of three of these operons was increased by Fur under anaerobic conditions, and these encode PreTA, an Fe-S dihydropyrimidine dehydrogenase, Dps, a dual-function nucleoid and iron sequestration protein, and AppY, a transcription activator of anaerobic metabolism. The fourth operon was repressed by Fur under anaerobic conditions and encodes GltA, a citrate synthase, which was reported previously to be regulated by Fur under aerobic conditions (13). Thus, despite the fact that ChIP-seq identified 157 Fur binding sites specific to anaerobic conditions, only four led to detectable transcription regulation under anaerobic conditions.
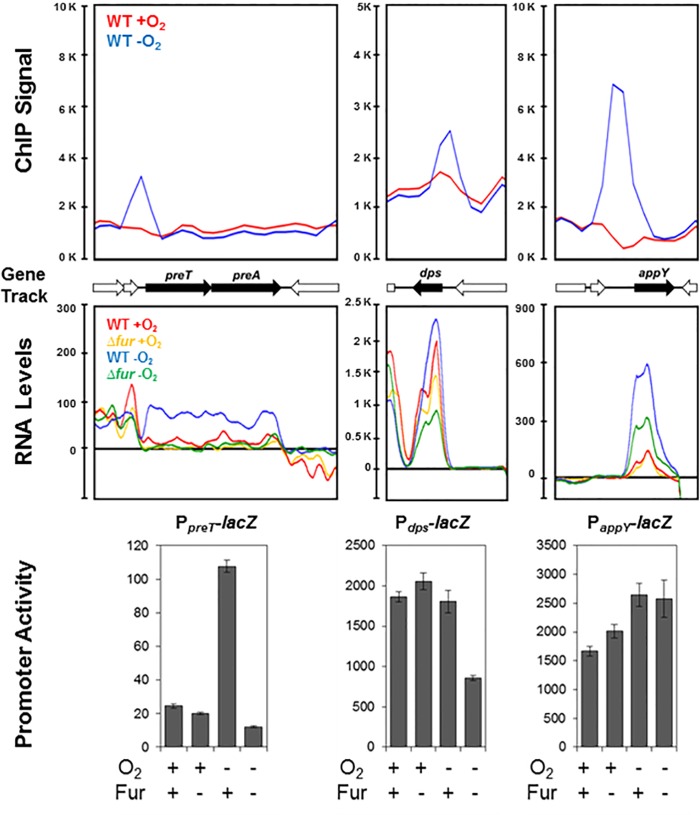
FIG 3 .
Only a few newly identified Fur binding regions lead to transcription regulation. Representative ChIP-seq plots of Fur binding to preT, dps, or appY promoter regions are shown for aerobic (red) or anaerobic (blue) growth conditions (top panel). Plots of preT, dps, or appY RNA levels from transcriptomic data are shown for wild-type (red, blue) and Δfur (yellow, green) strains grown under aerobic (red or yellow) or anaerobic (blue or green) growth conditions (middle panel). Strains bearing promoter-lacZ fusions to the preT, dps, or appY promoters were assayed for β-galactosidase activity in the presence or absence of Fur under aerobic or anaerobic growth conditions and normalized by cell density as a measure of promoter activity (bottom panel). The error bars represent the standard errors from at least three biological replicates.
In contrast, for the DNA regions bound by Fur under both aerobic and anaerobic conditions, more than one-third were associated with operons whose expression was regulated by Fur under anaerobic conditions (see Table S3 in the supplemental material). Indeed, the majority of these 36 operons were already known from studies carried out under aerobic growth conditions to be members of the Fur regulon (7, 13, 23) and include well-known iron homeostasis functions, such as iron acquisition (e.g., fepA-entD and tonB), iron storage (ftnA and bfd), and Fe-S cluster biogenesis (sufABCDSE), as well as the small RNA RyhB (ryhB). Two of the 36 operons, amiA, encoding a peptidoglycan amidohydrolase, and yrbL, a gene of unknown function, were not reported previously to be Fur regulated. In summary, although this genomic approach correctly identifies most of the known Fur regulon and associated binding sites, the majority of Fur binding sites (~200) identified by ChIP-seq from either aerobic or anaerobic conditions do not lead to Fur-dependent changes in transcription under the growth conditions tested here.
Anaerobiosis enhances the magnitude of Fur regulation for most of the Fur regulon.
To further investigate the impact of O2 availability on expression of the four promoter regions bound by Fur only under anaerobic conditions, we compared RNA levels from gene expression profiling of Fur+ and Fur− strains grown under aerobic or anaerobic conditions. For these four operons (preTA, gltA, dps, and appY) (Fig. 3, middle panel; see Table S3 in the supplemental material), we found that Fur-dependent changes in RNA levels were indeed greatest under anaerobic conditions. A similar trend was observed when individual promoter regions of preT and dps were fused to a lacZ reporter gene (Fig. 3, bottom panel) and assayed for Fur-dependent changes in β-galactosidase activity in Fur+ and Fur− strains grown under aerobic or anaerobic conditions. In contrast, we did not observe a comparable effect of Fur on expression of the PappY-lacZ fusion as found with appY transcript levels. Since the activity of the appY promoter is known to be regulated by the nucleoid-associated protein H-NS (45), transplanting this promoter out of its normal genomic context may eliminate the ability of Fur to increase expression, if Fur acts to prevent H-NS repression similar to the mechanism of ftnA induction (20).
The effect of O2 on Fur-dependent regulation of the entire regulon was also examined. By comparing RNA levels from gene expression profiling of Fur+ and Fur− strains, we found that Fur-dependent repression was greater under anaerobic compared to aerobic growth conditions for most of the Fur regulon (Fig. 4, top panel; see Table S3 in the supplemental material). The genomic regions upstream of representative operons were further analyzed by assaying expression from promoter-lacZ fusions in the presence and absence of Fur under both aerobic and anaerobic growth conditions. Expression from promoter-lacZ fusions of three operons (fhuA, bfd, and nrdHIEF) recapitulated the increased anaerobic repression by Fur observed by expression profiling (Fig. 5). In addition, expression from promoter-lacZ fusions also revealed small increases in Fur repression of fepA and fhuE during anaerobiosis that we were unable to detect in genome-wide experiments due to their low expression levels in Fur+ strains (Fig. 5; see Table S3). Although not tested further here, it seems likely that other strongly repressed genes with low expression levels (e.g., fes, fepDGC, entS, and entCEBAH) will also show some degree of Fur-dependent O2 regulation if examined by more sensitive assays. Furthermore, expression of ftnA, which is positively affected by Fur, was also increased under anaerobic conditions (see Table S3). Thus, Fur appears to be more active anaerobically, resulting in O2 regulation of many promoters within its regulon.
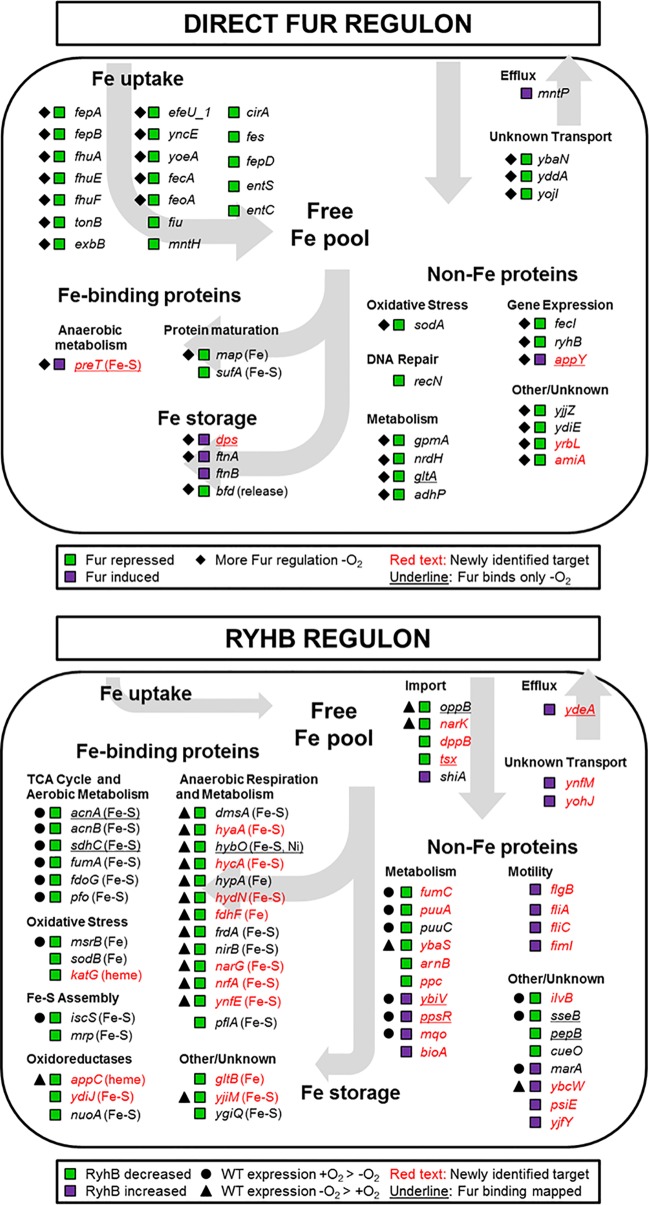
FIG 4 .
Anaerobiosis impacts the expression of the Fur and RyhB regulons. The expression pattern of operons regulated directly by Fur (top panel [data from Table S3 in the supplemental material]) and operons that are candidates for RyhB regulation (bottom panel [data from Table S4 in the supplemental material]) were compared and grouped by cellular iron functions. In the top panel, green squares indicate operons repressed by Fur, whereas purple squares indicate operons induced by Fur. Diamonds indicate operons exhibiting more Fur regulation under anaerobic conditions, and underlined operons are bound by Fur only under anaerobic conditions. In the bottom panel, green squares indicate operons whose transcript levels are decreased by RyhB, whereas purple squares indicate operons that are increased by RyhB. Operons exhibiting ≥2-fold-increased wild-type expression under aerobic or anaerobic conditions are marked with a circle or triangle, respectively, and operons where upstream Fur DNA binding was mapped are underlined. Newly identified Fur- or RyhB-regulated genes are in red text. Proteins known to contain an Fe cofactor are indicated.
FIG 5 .
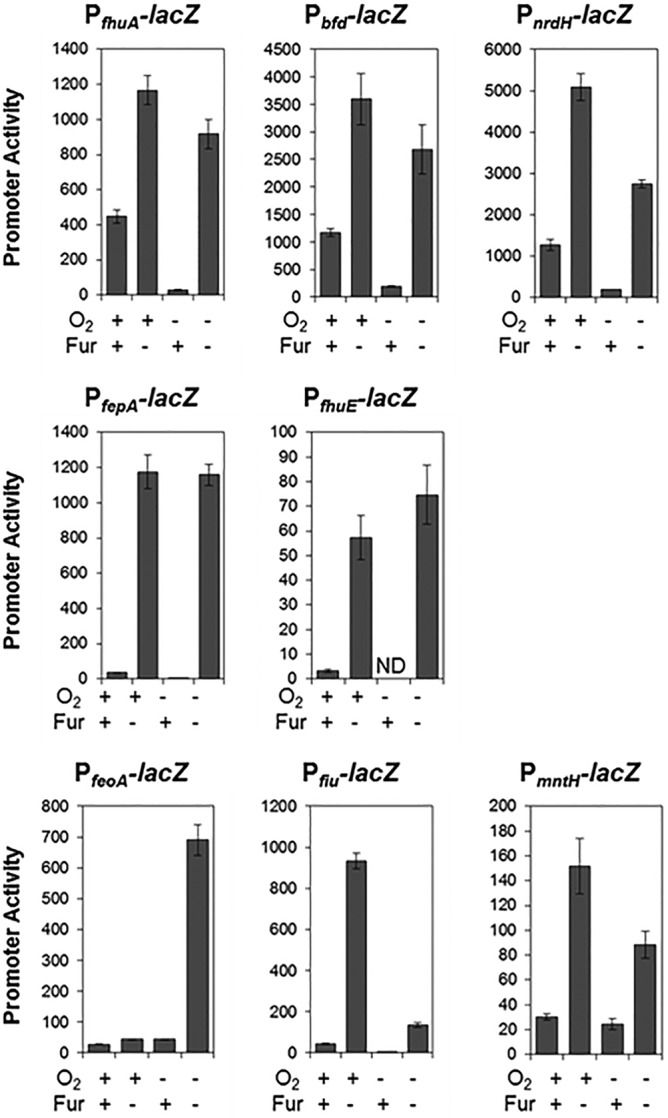
Effect of O2 on expression of select Fur regulon members. Strains bearing promoter-reporter gene (lacZ) fusions to several Fur-regulated promoters (fhuA, bfd, nrdH, fepA, fhuE, feoA, fiu, and mntH) were assayed for β-galactosidase activity in the presence or absence of Fur under aerobic or anaerobic growth conditions. Promoter activity was normalized to cell density. ND indicates that the promoter activity was below our detection limit. The error bars represent the standard errors from at least three biological replicates.
However, for some Fur-repressed operons, O2 regulation was not solely mediated by Fur. For example, transcript levels from several operons known to be Fur targets (fiu-ybiX, feoABC, yddAB-pqqL, sufABCDSE, yoeA, cirA, mntH, nrdHIEF, yrbL, fecABCDE, and sodA) showed O2-dependent differences in Fur− strains (see Table S3). The genomic regions upstream of three such Fur-regulated operons (feoA, fiu, and mntH) were further analyzed by assaying expression from promoter-lacZ fusions in the presence and absence of Fur under both aerobic and anaerobic conditions (Fig. 5). In the case of feoABC, Fur repression was limited to anaerobic conditions, likely due to the known activation of this operon by the anaerobic transcription factors ArcA and FNR (26, 27). In contrast, Fur repression of fiu and mntH was greater under aerobic than anaerobic conditions. Although the mechanism is not known, MntR also represses mntH (46), and ArcA binds to the fiu promoter region (27). Finally, although not tested here, expression of the sufABCDSE, nrdHIEF, and sodA operons is known to also be controlled by the transcription factor IscR, whose activity is regulated by O2 availability (19, 47, 48). Therefore, it is probable that, in addition to Fur, transcription factors such as IscR, ArcA, or FNR further modulate the expression of operons within this group under anaerobic conditions. Thus, anaerobiosis appears to have a major effect on expression of the Fur regulon.
Many genes indirectly regulated by Fur appear to be novel RyhB targets.
The small regulatory RNA RyhB is known to posttranscriptionally decrease expression of select iron-containing proteins and increase expression of certain iron uptake functions in an effort to spare iron for critical functions during iron-limiting, aerobic growth conditions (10, 28). Because RyhB is elevated in strains lacking Fur, we reasoned that some of the operons indirectly regulated by Fur (i.e., those that lacked a ChIP-seq peak) might represent novel RyhB targets. Using the criteria that transcripts regulated by RyhB should return to wild-type levels when strains lacking Fur are also deleted for ryhB, we found that nearly one-third of the operons indirectly controlled by Fur under anaerobic growth conditions are candidates for direct RyhB regulation (see Table S4 in the supplemental material). The effect of RyhB on RNA levels was generally small (~2-fold), in agreement with previous reports (49), although a few showed >5-fold changes. Control experiments comparing expression of wild-type (Fur+ RyhB+) to Fur+ RyhB− strains revealed very little differential gene expression as expected (see Table S7 in the supplemental material). The remaining genes, whose expression was not RyhB regulated but which were indirectly influenced by Fur, are reported in Table S5 in the supplemental material.
Of the 46 operons regulated by RyhB under anaerobic conditions, most are new candidates for RyhB regulation (Fig. 4, bottom panel; see Table S4 in the supplemental material). For example, 13 of the 15 operons whose RNA levels were increased by RyhB expression are new potential targets. In addition to the known RyhB-dependent increase of shiA transcripts (29), we observed increases in RNA levels for proteins involved in cellular motility (flgBCDEFG, fliAZY, fliC, and fimICDFGH), metabolism (e.g., ybiV, ppsR, and bioA), and transport (e.g., ydeA, ynfM, and yohJ) (Fig. 4, bottom panel; see Table S4). If these increases are due to direct effects of RyhB, then these data suggest that the positive effect of RyhB on transcript stability may be broader than just iron homeostasis. In contrast, the RNA levels of the 31 operons decreased by RyhB expression (Fig. 4, bottom panel; see Table S4) under anaerobic conditions encode mostly iron-containing proteins, consistent with the paradigm of iron sparing (28). Some of these operons were previously known or predicted to be regulated by RyhB (e.g., sodB, frdABCD, pflA, nuoABCEFGHIJKLMN, dmsABC, hypABCDE, and nirBDC) (39, 49). However, most operons are new candidates for RyhB regulation and encode protein complexes involved in anaerobic respiration and metabolism (hyaABCDEF, hycABCDEFGHI, hydN-hypF, fdhF, narGHJI, nrfABCDEF, appCB-yccB-appA, ynfEFGH, narK, and dppBCDF).
The suite of RyhB downregulated genes differs between aerobic and anaerobic conditions.
Many of the newly identified downregulated RyhB candidates are expressed preferentially under anaerobic conditions (27, 32, 33, 36), providing a plausible explanation for why they were not previously detected when cells grown under aerobic conditions were analyzed. Conversely, some known RyhB targets were not found to be regulated by RyhB under anaerobic conditions—presumably because they were not sufficiently expressed. To test how extensively O2 influences the transcription of RyhB targets, we compared RNA levels from Fur− versus Fur− RyhB− strains grown under both aerobic and anaerobic growth conditions. Indeed, of the 44 transcripts negatively regulated by RyhB, 13 are expressed at higher levels in the presence of O2, 14 are similarly expressed whether O2 is present, and 17 are expressed at higher levels under anaerobic conditions (Fig. 4, bottom panel; see Table S4 in the supplemental material).
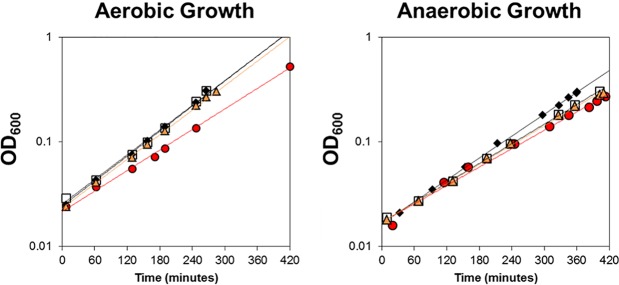
We also found that Fur− strains grew slower than Fur+ strains under aerobic but not anaerobic conditions (Fig. 6). However, wild-type growth rates were reestablished in aerobic Fur− strains upon deletion of RyhB. The fact that this growth rate phenotype is only observed under aerobic growth conditions suggests the genes whose transcription is limited to aerobic conditions and are targeted by RyhB may be responsible for the observed growth defect. Together, these analyses highlight a major role of O2-dependent transcriptional changes in determining which mRNAs are targeted by RyhB to promote an iron-sparing response.
FIG 6 .
Growth of strains lacking Fur and RyhB. Cell density was measured over time for the wild-type (black diamonds), Δfur (red circles), ΔryhB (open squares), and Δfur ΔryhB (orange triangles) strains under aerobic or anaerobic growth conditions in MOPS glucose minimal medium with 10 µM FeSO4. Trend lines were added for ease of visualization. This representative growth curve was replicated several times.
Iron-containing proteins that are not regulated by RyhB.
By analyzing the expression levels of all annotated iron-binding proteins (according to the EcoCyc database [50]) in our data set, we could separate out the iron-binding proteins that appear to evade RyhB regulation. This group was enriched in heme biosynthetic enzymes, cytochrome maturation functions, and heme proteins (see Table S6 in the supplemental material). In addition, genes coding for iron-containing proteins that function in cofactor biosynthesis (e.g., bioB, lipA, nadA, ispG, and ispH), DNA repair (nth and mutY), RNA modification (rlmC, rlmD, rlmN, queG, ttcA, tsaD, and miaB), and transcriptional regulation (soxR, nsrR, and fnr) also do not appear to be subject to RyhB regulation (see Table S6). Perhaps some of these processes escape RyhB regulation because they are more critical to cellular function (e.g., RNA modification) or too costly to abandon (e.g., synthesis of cofactors such as heme, thiamine, biotin, ubiquinone, or NAD+) upon iron deprivation.
Possible dual regulation of genes by Fur and RyhB.
Our data suggest that several operons are potentially coregulated by Fur and RyhB because we found an upstream Fur DNA binding site and differential gene expression in Fur− compared to Fur− RyhB− strains. For example, cirA, fecABCDE, yddAB-pqqL, yncE, ybiV, ydeA, and ppsR are bound upstream by Fur, and their RNA levels are increased by RyhB (see Tables S3 and S4 in the supplemental material). In fact, cirA is known to be regulated by Fur (7) and RyhB (51). Maximizing expression of known (cirA and fecABCDE) and predicted (yddAB-pqqL and yncE) iron uptake systems during iron-limiting conditions could be advantageous. Yet for a second group of genes (including acnA, sdhCDAB, hybOAB, oppBCDF, tsx, sseB, and pepB), the potential opposing effects of Fur and RyhB and additional regulation by transcription factors such as ArcA and IscR preclude any conclusions without additional data (see Table S4).
O2-dependent regulation of the metallome.
Since we found that anaerobic conditions led to increased Fur-dependent repression of ferric uptake pathways and genes encoding other divalent cation-binding proteins (Mn-binding SodA and NrdHIEF), we assayed whether cellular metal levels changed between aerobic and anaerobic conditions. The cellular levels of 11 elements (Mn, Co, Ni, Zn, Mg, P, S, Fe, K, Cu, and Ca) were measured by inductively coupled plasma mass spectrometry (ICP-MS) (Table 1). Fe was present at 19.1 ng/mg cell pellet during aerobic growth, corresponding to ~0.0063% of the cellular dry weight—~3-fold less than previous reports (2)—and showed a small 1.2-fold increase during anaerobic growth. In contrast, Mn, Co, and Ca levels showed large O2-dependent differences in abundance: anaerobic cellular Mn and Co levels decreased 37-fold and 5.5-fold, respectively, whereas anaerobic Ca levels increased 2.9-fold compared to those in aerobic cells (Table 1). These data show that O2 availability has a broad effect on cellular metal homeostasis.
TABLE 1 .
Results from whole-cell elemental analysis
| Element | Elemental analysis result, ng/mg cell pellet (SE)a |
Aerobic/anaerobic ratio | |
|---|---|---|---|
| Aerobic growth | Anaerobic growth | ||
| Mn | 3.23 (0.253) | 0.0882 (0.00427) | 37 |
| Co | 0.213 (0.0229) | 0.0389 (0.0000995) | 5.5 |
| Ni | 0.0369 (0.00354) | 0.0223 (0.00567) | 1.7 |
| Zn | 6.49 (0.150) | 4.06 (0.0727) | 1.6 |
| Mg | 422 (5.30) | 321 (3.16) | 1.3 |
| P | 3,190 (31.7) | 2,670 (9.54) | 1.2 |
| S | 1,680 (27.8) | 1,550 (16.8) | 1.1 |
| Fe | 19.1 (1.12) | 22.8 (0.265) | 0.84 |
| K | 1,000 (38.9) | 1,560 (9.14) | 0.64 |
| Cu | 0.494 (0.0302) | 0.818 (0.139) | 0.60 |
| Ca | 24.7 (0.594) | 70.5 (4.23) | 0.35 |
Nanograms of element per milligram of packed cell pellet as determined by ICP-MS. Assuming 70% of cell pellet is H2O, dividing by 0.3 and multiplying by 100 should approximate the percentage of dry weight. SE, standard error from three biological replicates of cells grown in MOPS glucose minimal medium.
DISCUSSION
The findings reported here show that the lack of O2 produces large and previously unreported effects on metal ion homeostasis in the enteric bacterium E. coli. Specifically we found that O2 availability impacts the expression of genes regulated by the two iron global regulators, Fur and RyhB, but for different reasons. Under anaerobic conditions, the positive or negative effects of the transcription factor Fur on expression of many genes were enhanced. In contrast, the O2-dependent changes in the genes posttranscriptionally regulated by RyhB could be attributed to differential transcription of these target mRNAs. This regulatory hierarchy suggests that the iron proteome may be differentially remodeled under iron starvation conditions, depending on O2 availability. Finally, enhanced Fur-dependent repression of manganese-cofactored enzymes was accompanied by a dramatic decrease in cellular manganese levels, suggesting previously unknown rewiring of the metallome under anaerobic growth conditions.
Adaptation of the Fur regulon to anaerobic conditions.
While our analysis of anaerobic cells allowed us to identify some new genes regulated by Fur, our major finding was the enhancement in Fur regulation in response to anaerobiosis. Thus, our results provide new insight into the sensitivity of the control of iron homeostasis in E. coli to O2 availability (summarized in Fig. 7). For example, expression of genes encoding several Fe3+-siderophore uptake systems was decreased under anaerobic conditions, consistent with decreased demand for ferric uptake. Fur also increased expression of the genes encoding two iron storage proteins, ferritin A and Dps, and decreased expression of bfd, encoding a protein that would facilitate iron release from bacterioferritin (52), suggesting that iron storage may be increased anaerobically. However, the process by which iron is stored under anaerobic conditions in E. coli is unclear because the best-studied mechanisms for iron storage require O2, mineralizing ~1,000 to 3,000 iron atoms/ferritin (1, 3) and ~20 to 500 irons/Dps (2, 53). Thus, in the absence of an O2-dependent mineralization mechanism, less iron may actually be stored anaerobically, despite the increase in dps and ftnA expression. Nevertheless, we observed a small increase in total cellular iron levels under anaerobic conditions, raising the possibility that iron storage could be increased. Although the bulk of cellular iron is assumed to be in a bound state, allocated between iron-bound proteins and storage forms (2, 54), the overall distribution of iron in anaerobically grown cells is not known. Determining whether this increased iron is present in iron stores and protein cofactors or is unbound will be critical in addressing if Fur activity is enhanced under anaerobic conditions because of an increase in the “labile” iron pool.
FIG 7 .
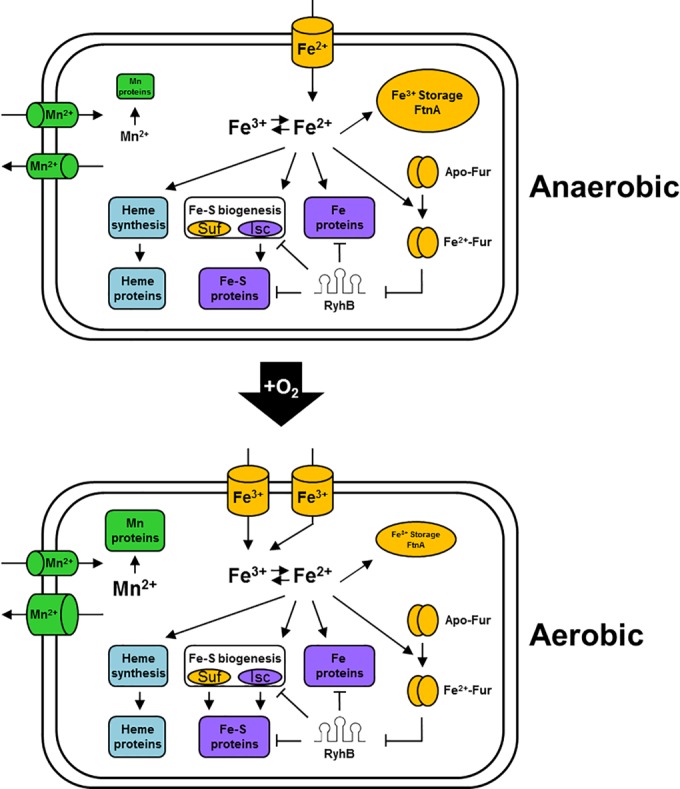
Iron homeostasis pathways are reprogrammed in response to O2 availability: a model. Under both aerobic and anaerobic conditions, we propose that iron is distributed to iron storage complexes, Fur, iron-binding proteins, or Fe-S biogenesis and heme synthesis pathways. However, under anaerobic growth conditions, iron uptake is shifted to ferrous transport systems, expression of iron storage proteins is increased, and Fe-S biogenesis occurs primarily by the housekeeping Isc pathway. When expressed under anaerobic iron-limiting conditions, we propose that RyhB targets anaerobically induced Fe-S-binding and iron-binding proteins of anaerobic respiratory pathways in addition to other constitutively expressed RyhB targets. Whereas under aerobic growth conditions, iron uptake is shifted to ferric transport systems, iron storage gene expression is decreased, and Fe-S biogenesis is still mainly by the housekeeping Isc pathway, but expression of the stress-induced Suf pathway is increased. When expressed under aerobic iron-limiting conditions, RyhB switches from targeting Fe-S-binding and iron-binding proteins of anaerobic respiratory pathways to those of aerobic respiratory pathways and the TCA cycle. The abundance of Mn, Mn proteins, and Mn efflux systems also increases under aerobic growth conditions.
Our results also reinforce previous studies that Fe-S cofactor biosynthesis is regulated by O2 and iron availability (Fig. 7). First, expression of both the housekeeping Isc pathway and the stress-induced Suf Fe-S cluster biogenesis pathways is decreased under anaerobic conditions, due to the regulators IscR (47, 55) and Fur (14), respectively. Since some Fe-S clusters are known to be labile to O2 or reactive oxygen species (4), the decrease in expression of Fe-S biogenesis pathways under anaerobic conditions might reflect a decreased demand for Fe-S clusters under conditions where clusters are more stable. However, when iron is limiting, this coordinate control of Fe-S biogenesis pathways should be disrupted under anaerobic conditions because RyhB downregulates expression of the Isc pathway (56), whereas the loss of Fur repression promotes an increase in expression of the Suf pathway (14–16). Surprisingly, the enzymes required for heme biosynthesis or many heme-containing proteins were not found to be part of the Fur or RyhB regulon. How the flux of iron into this pathway is controlled remains to be determined.
RyhB connects iron status to O2-dependent transcriptional networks.
This study also reveals extensive integration of the RyhB network with those that respond to O2 limitation, ensuring that cells produce the most appropriate suite of iron-containing proteins, depending on environmental conditions. For example, transcription of several genes encoding iron-containing proteins is regulated by O2 to tailor protein production to the appropriate mode of energy conservation (e.g., tricarboxylic acid [TCA] cycle and aerobic and anaerobic respiratory pathways) (6, 32, 37). When external iron is sufficient, intracellular iron is available to synthesize the appropriate complement of iron-containing proteins, and transcriptional regulation by O2 is the primary point of control. However, when external iron is not sufficient, RyhB is expressed and a second level of control is added (Fig. 7), which decreases mRNA levels of a subset of iron-containing proteins, making iron available for more “essential” iron proteins in the so-called “iron-sparing response” (28, 57).
The observed downregulation of components of the TCA cycle and respiratory pathways by RyhB under both aerobic and anaerobic growth conditions suggests that this small RNA selectively targets respiration-linked energy conservation pathways to maintain the function of other iron-binding proteins under iron-limiting conditions. Surprisingly, targeting these mRNAs only affected the growth rate under aerobic conditions. Although the downregulation of pathways that generate NADH (i.e., TCA cycle) and its oxidation (i.e., NADH dehydrogenase I) by RyhB in E. coli may be sufficient to explain this decreased growth rate, we cannot exclude contributions from the downregulation of other RyhB targets, such as superoxide dismutase B (SodB), which functions in reducing oxidative stress (58). It is noteworthy that Bacillus subtilis appears to exert a similar strategy in that Fur− strains cannot grow on the respiratory substrate succinate unless these strains also lack the small RNA FsrA, the B. subtilis equivalent of RyhB (59).
Anaerobiosis may impact the metallation state of other divalent cation-containing proteins.
Comparison of the metallomes of E. coli K-12 between aerobic and anaerobic conditions revealed a large decrease in cellular manganese levels during anaerobiosis. This decrease in manganese was accompanied by enhanced anaerobic repression by Fur of two major Mn-containing enzymes, the Mn-superoxide dismutase encoded by sodA (18, 60) and the Mn-ribonucleotide reductase encoded by nrdHIEF (19). Both of these enzymes have iron-containing isozymes, encoded by sodB and nrdAB or nrdDG, which substitute for their Mn counterparts under anaerobic conditions (61–64). Thus, decreasing synthesis of the Mn isozymes when they are not needed under anaerobic conditions serves both to avoid wasting energy in synthesizing unnecessary polypeptides and also to possibly avoid their mismetallation when Mn is decreased; the importance of maintaining the cellular Fe/Mn ratio for protein metal ion selectivity and cellular physiology has recently been reviewed (54).
The mechanism by which Mn levels decrease under anaerobic conditions is not known. Perhaps decreased Mn transport via the MntH Mn2+/Fe2+:H+ symporter, which is driven by the proton motive force (65), could be limiting under the fermentative conditions of growth used in these experiments. We found that expression of the Mn exporter MntP (66) is reduced anaerobically (see Table S3 in the supplemental material), so it would seem unlikely to play a role in this response. Interestingly, expression of the Mn-dependent isozyme of phosphoglycerate mutase, GpmM, is induced during anaerobiosis (67). If this is the major form of the enzyme under anaerobic conditions, it is possible that cells have a system to prioritize manganese loading of GpmM under conditions of decreased cellular Mn. The MntS/RybA protein has been suggested to be an Mn chaperone (46), which could perhaps carry out this activity.
Fur binds the canonical Fur motif under either aerobic or anaerobic conditions.
Prior to this study, the in vivo DNA binding sites of Fur had been mapped only under aerobic growth conditions (23). We found good agreement between the locations of 59 high-signal-to-noise, iron-dependent Fur binding regions in our study and the binding regions mapped with the higher-resolution “ChIP-exo” approach (23), which combines ChIP with λ exonuclease digestion and high-throughput sequencing (see Table S2 in the supplemental material). Less overlap between our studies and ChIP-exo experiments was observed for peaks with low signal-to-noise ratio (ChIP signal of <3,000), which may be attributed to the complexity in resolving the background of ChIP experiments. Nevertheless, a large number of Fur binding regions mapped under either aerobic or anaerobic conditions did not result in transcriptional regulation. Some of these sites could function under different physiological conditions, as was proposed for similar “transcriptionally inactive” binding regions found with the transcription factor FNR (36), or some may contribute to the overall nucleoid structure of the genome (68). Perhaps the necessity of global regulators to bind more degenerate DNA sites to achieve regulation within many promoter regions might lead to binding at other regions of the genome as an unintended consequence. We also did not observe Fur binding in vivo to some sites predicted by an information theory model (44), indicating that not all high-quality DNA sites are accessible to transcription factor binding. This property was also previously observed for the transcription factor FNR (36), providing additional support to the notion that global regulators compete for occupancy in vivo with other DNA binding proteins.
Why is Fur binding increased anaerobically?
Bioinformatic analysis of the regions bound by Fur under both aerobic and anaerobic conditions revealed DNA sites similar to the Fur motif defined from previous studies (44). In contrast, the Fur binding sites specific to anaerobic conditions were not predicted by this weight matrix model, consistent with the notion that these represent weaker affinity sites. Because Fur occupancy of these less-conserved sequences increased under anaerobic conditions, we propose that there must be more active Fur under anaerobic conditions to bind to these putative weaker affinity sites. Increased Fur DNA occupancy of previously known Fur sites could also explain increased repression and induction of many Fur-regulated genes during anaerobiosis. While increased Fur DNA binding could result from greater Fur protein abundance in the cell during anaerobiosis, we did not observe an increase in fur expression under anaerobic conditions. Thus, the mechanism to explain increased Fur activity and whether it is connected to the small increases in cellular iron observed under anaerobic conditions requires further study.
In summary, our in vivo DNA binding and expression data suggest that Fur activity increases during anaerobiosis. Our findings reveal that in the absence of O2, the Fur regulon is modified such that transcription of iron uptake genes is geared toward Fe2+ and expression of iron storage proteins is increased. Furthermore, Mn levels and expression of Mn-cofactored enzymes that have iron counterparts are decreased under anaerobic conditions. We also found that many potential targets of RyhB are also transcriptionally regulated by O2, implying that the iron proteome is likely to be differentially remodeled in response to iron deprivation under anaerobic conditions compared to aerobic conditions.
MATERIALS AND METHODS
Strain construction for global analyses and promoter activity assays.
Relevant strains are listed in Table S1 in the supplemental material. Sequences of primers are available upon request. E. coli K-12 MG1655 (F− λ− rph-1) served as the wild-type strain. To construct the Fur− strain, PK9427, Δfur::kan from the Keio collection (69) was moved into MG1655 by transduction with P1 vir, selecting for kanamycin resistance (Kmr). The kan cassette was removed by transforming strains with pCP20, encoding the FLP recombinase (70). To construct the RyhB− (PK10474) and Fur− RyhB− (PK10475) strains, ΔryhB::cat from PK7875 was moved into MG1655 and PK9427, respectively, by transduction with P1 vir, selecting for chloramphenicol resistance (Cmr). PK7875 was made by Pl vir transduction of ΔryhB::cat from EM1238 (9) into MG1655 and selection for Cmr.
Strains bearing chromosomal promoter-lacZ fusions were constructed as previously described (47). Briefly, promoter regions of interest were amplified from MG1655 and cloned into pPK7035. A lacI-kan-promoter-lacZ fragment, amplified from pPK7035 derivatives, was then electroporated into either BW25993 containing pKD46 (70) or PK9012 (MG1655 λc1857 ΔmutS::Tn10 Δcro-bioA). PK9012 was constructed in a manner previously described (55). Chromosomal promoter-lacZ fusions were moved into MG1655, PK9427, and JEM609 by transduction with P1 vir, selecting for Kmr. All constructs were confirmed by colony PCR and/or DNA sequencing.
Growth of strains for global analyses.
Strains were grown in gas-sparged Roux bottles at 37°C in MOPS (morpholinepropanesulfonic acid) minimal medium with 0.2% glucose and the indicated amount of FeSO4 (36). For ChIP-seq or ChIP-chip (ChIP with microarray technology) analysis of MG1655 or PK9427 and for transcriptomic analysis of MG1655, PK9427, PK10474, and PK10475 the medium contained 10 µM FeSO4. For ChIP-seq analysis of JEM609 (MG1655 ΔlacZ, ΔtonB, ΔfeoABC, ΔzupT [19]) the medium contained 1.0 µM FeSO4 to promote iron deficiency. A gas mixture of 70% N2, 5% CO2, and 25% O2 was used for aerobic experiments, and a gas mixture of 95% N2 and 5% CO2 was used for anaerobic experiments (36). Cells were harvested at an optical density at 600 nm (OD600) of 0.3 to 0.35, measured using a Perkin Elmer Lambda 25 UV/visible spectrophotometer.
Chromatin immunoprecipitation followed by high-throughput sequencing or hybridization to a microarray chip.
ChIP assays were performed as previously described (36) using antibodies specific to Fur that were purified over a His6-Fur-bound HiTrap N-hydroxysuccinimide (NHS)-activated high-performance (HP) column (GE Healthcare) (71). Western blot analysis, performed as previously described (72), showed that the purified antibody was specific for Fur (see Fig. S1 in the supplemental material).
For ChIP-seq experiments, DNA enriched from three replicates of aerobic Fur+ cultures (MG1655 or MG1655 PfepA-lacZ), three replicates of anaerobic Fur+ cultures (MG1655 or MG1655 PfepA-lacZ), two replicates of anaerobic iron-deficient cultures (JEM609 or JEM609 PfepA-lacZ), or one combined-input sample was submitted to the University of Wisconsin—Madison DNA Sequencing Facility for library construction and Illumina sequencing (Illumina Genome Analyzer IIx or Illumina HiSeq2000 [all single-end, 1 × 50 bp]) per the manufacturer’s recommendations. Strains bearing PfepA-lacZ allowed for readout of cellular iron status (see Fig. S2 in the supplemental material). Illumina sequencing FASTQ files were reformatted to the Sanger format using the FASTQ Groomer script (73) and reads (aerobic Fur+ read counts of 21,140,380, 25,480,433, and 25,181,847; anaerobic Fur+ read counts of 17,617,567, 16,084,736, and 18,255,201; anaerobic iron-deficient read counts of 18,361,478 and 20,432,024; and an input read count of 23,481,977) were mapped to the E. coli K-12 MG1655 genome (version U00096.2) using the Bowtie 2 algorithm (default settings) (74). Greater than 90% of reads mapped to the genome for all samples. Enriched regions were identified using the peak-calling algorithm MOSAiCS (75, 76) using a false discovery rate (FDR) of 0.1. The dPeak algorithm (77) was used to deconvolute close-proximity peaks. A total of 517 unique peaks, having been found in at least two replicates, were identified across all strains and growth conditions. Data sets were normalized to 20 million reads, and 262 peaks of low read count (<2,500 reads at the peak summit) were removed because they were present in both Fur+ and iron-deficient cultures (DBChIP [78]; P < 0.05) or did not visually conform to a peak above the local background. Normalized ChIP-seq data files were visualized with MochiView (79). The final peak list is given in Table S2 in the supplemental material. Fur binding site motifs were constructed by analyzing the 100 bp upstream and downstream of the dPeak-identified peak summits, submitting the sequences to MEME-ChIP (40), and using the overrepresented sequences to construct the position weight matrix (PWM).
For ChIP-chip experiments, DNA enriched from one replicate of an anaerobic Fur− culture (PK9472) and input DNA was amplified, labeled, and hybridized to a custom-made E. coli K-12 MG1655 tiled-genome microarray (Roche NimbleGen, Inc., Madison, WI [80]); hybridized microarrays were scanned using a GenePix 4000B (Axon Instruments) microarray scanner as previously described (36). ChIP and input data were quantile normalized using “normalize.Quantiles” from the preprocessCore package (81) in R (82), and enriched binding regions were identified using the CMARRT peak-calling algorithm (P < 0.1) (83). Enriched regions were removed from the final Chip-seq peak list if they were also present in the Fur− strain data.
RNA isolation and whole-genome transcriptomic microarray analysis.
RNA was isolated from two biological replicates of MG1655, PK9472, PK10474, and PK10475 under aerobic or anaerobic growth conditions as previously described (36). Ten micrograms of RNA was reverse transcribed and labeled with Amersham Cy3 monoreactive dye (GE Healthcare) as previously described (80). The purified, labeled cDNAs were fragmented with DNase I (0.1 U per µg of cDNA) for 10 min at 37°C. DNase I was then inactivated at 95°C for 10 min; samples from RyhB− and Fur− RyhB− strains also included 0.85 mM EDTA in this reaction. Approximately 0.6 to 1.5 µg of precipitated Cy3-labeled cDNA was hybridized to a custom-made E. coli K-12 MG1655 tiled-genome microarray (Roche NimbleGen, Inc., Madison, WI [80]) as previously described (36). Hybridized microarrays were scanned using a GenePix 4000B (Axon Instruments) microarray scanner, and the photomultiplier tube (PMT) was adjusted so that the median fluorescence was just below 100.
Raw probe intensities were normalized across all samples using the Robust Multichip Average (RMA) algorithm in the NimbleScan software package (version 2.5 [84]). After probes were matched to gene coordinates (“IRanges” package [85] in R [82]), differential expression of genes between experiments was determined using an analysis of variance (ANOVA) test (“anova.test” in R [82]), and P values were adjusted using the Benjamini-Hochberg false discovery rate control procedure (86) (“p.adjust” with method = “BH” in R [82]; FDR, <0.01) to address the multiple testing issue. For differentially expressed genes, the experiments in which gene expression was significantly different from those of other experiments were identified by a post hoc test using the Tukey’s honestly significant difference method (87) (“TukeyHSD” in R [82]; P < 0.01). Genes were further required to show at least a 1.5-fold change in expression between experiments. A cutoff of 1.5-fold change in expression between experiments was chosen based on known regulation of the Isc pathway by RyhB (56). Regulation of an operon was reported to be RyhB dependent only if expression changed under the condition in which the transcript was most expressed. The expression value of a given gene is the sum of the intensity of the probes that overlap that gene, from both biological replicates, divided by the length of that gene, log2 transformed.
Promoter activity measurements by β-galactosidase assay.
Strains bearing promoter-lacZ fusions were grown at 37°C in MOPS minimal medium containing 10 µM FeSO4 and 0.2% glucose to an OD600 of ~0.2 to 0.4 under either aerobic or anaerobic conditions, and promoter activity was measured as previously described (88). Differences in aerobic and anaerobic cell counts for cells grown in minimal medium were corrected by multiplying aerobic activity by 1.5 (72). Assays were repeated at least three times, and error bars represent the standard error from these biological replicates.
Cellular element analysis.
Triplicate cultures of MG1655 were grown in MOPS minimal medium containing 10 µM FeSO4 and 0.2% glucose under either aerobic or anaerobic growth conditions in gas-sparged Roux bottles as in global analyses. At an OD600 of ~0.4, equal numbers of cells were centrifuged, resuspended in MOPS minimal medium containing 10 µM FeSO4, 0.2% glucose, and 10 mM diethylenetriaminepentaacetic acid (DTPA [Sigma Aldrich]), and incubated for 15 min at 37°C to remove contaminating surface metals (89). Cells were centrifuged and resuspended twice in 20 mM Tris-HCl (pH 7.4) and then transferred to preweighed 10% HCl-treated tubes. Cells were centrifuged, aspirated of the remaining liquid, and resuspended in H2O to 0.33 mg cell pellet/µl. Cells were lysed on ice by sonication with a cup horn-equipped Misonix S-4000 sonicator at 10-s on-off intervals of 60% output for 60 min. At the Wisconsin State Laboratory of Hygiene, 200 µl of this cell lysate was digested with 100 µl of tetramethylammonium hydroxide (TMAH) for 1 h at 70°C prior to dilution with 4% HNO3 (90) and element analysis by magnetic sector inductively coupled plasma mass spectrometry using a Thermo-Finnigan Element 2 plasma mass spectrometer (91).
Microarray data accession number.
ChIP-seq, ChIP-chip, and tiling array data sets have been deposited in the Gene Expression Omnibus (GEO) under accession no. GSE74933.
SUPPLEMENTAL MATERIAL
Western blots of cellular proteins to determine Fur antibody specificity. The reactivity of purified Fur antibodies to Fur (~17 kDa) in cells was examined by Western blot analysis of strains lacking or containing Fur. Cells were grown in MOPS glucose minimal medium with 10 µM FeSO4 to an OD600 of 0.1, and cell proteins were separated by SDS-PAGE and analyzed by Western blotting with affinity-purified Fur antibodies. Lanes: A, molecular mass standards; B, Fur+ strain, 3.2 × 107 cells; C, Fur+ strain, 6.5 × 107 cells; D, Δfur strain, 3.2 × 107 cells; E, Δfur strain, 6.5 × 107 cells. Download
Analysis of iron-uptake-deficient strain. To determine if Fur DNA binding was iron-dependent under anaerobic conditions, we performed ChIP-seq analyses of an iron-uptake-deficient strain (ΔtonB Δfeo ΔzupT) grown under iron-limiting conditions. We showed that the cultures used for ChIP-seq analysis were indeed iron deficient by comparing promoter activities of a known iron-dependent, Fur-repressed promoter, PfepA, in wild-type (WT), Fur-lacking (Δfur), and iron-uptake-deficient (ΔtonB Δfeo ΔzupT) strains. Promoter activity was determined from the β-galactosidase activity of a PfepA-lacZ gene fusion strain normalized to cell density. The concentration of FeSO4 in the MOPS glucose minimal medium was 10 µM for the iron-sufficient growth conditions and 1.0 µM for the iron-deficient conditions. Download
Strains and plasmids. This table details the strains and plasmids used to perform the experiments described in this study.
Fur binding regions mapped across the E. coli K-12 genome under aerobic or anaerobic growth conditions using ChIP-seq. In vivo Fur DNA binding sites identified through ChIP-Seq experiments are listed according to whether Fur DNA binding was observed in the presence or absence of O2 and whether Fur DNA binding was iron-dependent or iron-independent. Additional information includes the gene predicted to be associated with the Fur DNA binding site, the location of the ChIP signal relative to that gene, if the gene associated with the Fur DNA binding site is Fur or RyhB regulated, and if the Fur DNA binding site was previously known.
Operons directly regulated by Fur during aerobic and/or anaerobic growth. Transcription profiling results are reported as the log2-transformed mRNA levels for the first gene in each operon determined to be directly regulated by Fur (i.e., contained a ChIP-seq peak [see Table S2 in the supplemental material]). The data are presented for wild-type, Fur−, and Fur−‑ RyhB− strains grown in the presence or absence of O2 and are separated by whether Fur DNA binding occurs under only anaerobic growth conditions or under both aerobic and anaerobic growth conditions. A reference is provided if Fur regulation was previously known.
Operons regulated by RyhB during aerobic and/or anaerobic growth. Transcription profiling results are reported as the log2-transformed mRNA levels for each gene in operons predicted to be regulated by RyhB. The data are presented for wild-type, Fur−, and Fur−RyhB− strains grown in the presence or absence of O2 and are separated by whether expression of the operon is decreased or increased by RyhB and whether expression of the operon is greater under aerobic or anaerobic growth conditions. A reference is provided if Fur and/or RyhB regulation was previously known.
Genes indirectly regulated by Fur and not regulated by RyhB. Transcription profiling results are reported as the log2-transformed mRNA levels for the first gene in each operon determined to be indirectly regulated by Fur and not regulated by RyhB. The data are presented for wild-type and Fur− strains grown in the presence or absence of O2 and separated by whether the Fur-dependent change in expression occurs under primarily aerobic or anaerobic growth conditions. A reference is provided if Fur regulation was previously known. It is also indicated whether a Fur binding site was previously predicted using bioinformatics despite the absence of an in vivo Fur DNA binding site in this study.
Iron-containing proteins whose transcript levels do not appear to be affected by RyhB expression. Transcription profiling results are reported as the log2-transformed mRNA levels for genes encoding iron-binding proteins, which do not appear to be regulated by RyhB. The data are presented for wild-type, Fur−, and Fur−RyhB− strains grown in the presence or absence of O2. Genes are separated into parts a and b based on wild-type expression levels.
Log2 expression for all genes in the E. coli K-12 genome under all experimental conditions. Transcription profiling results are reported as the log2-transformed mRNA levels for all genes in the E. coli genome. The data are presented for the wild-type, Fur−, Fur− RyhB−, and RyhB− strains grown in the presence or absence of O2 and are organized by the gene accession number known as the b-number.
ACKNOWLEDGMENTS
We thank Marie Adams at the University of Wisconsin—Madison DNA Sequencing Facility for help with ChIP library construction and high-throughput sequencing. We acknowledge Martin Shafer of the Wisconsin State Lab of Hygiene for help with whole-cell element analyses. We thank Jason M. Peters for advice on tiling microarray experiments. We thank Jim A. Imlay for advice and strain JEM609.
This work was funded by a grant from the NIH to P.J.K. (R01GM045844). N.A.B. was supported by University of Wisconsin—Madison NIH Chemistry Biology Interface training grant T32GM008505.
Footnotes
Citation Beauchene NA, Myers KS, Chung D, Park DM, Weisnicht AM, Keleş S, Kiley PJ. 2015. Impact of anaerobiosis on expression of the iron-responsive Fur and RyhB regulons. mBio 6(6):e01947-15. doi:10.1128/mBio.01947-15.
REFERENCES
- 1.Andrews SC, Robinson AK, Rodríguez-Quiñones F. 2003. Bacterial iron homeostasis. FEMS Microbiol Rev 27:215–237. doi: 10.1016/S0168-6445(03)00055-X. [DOI] [PubMed] [Google Scholar]
- 2.Andrews S, Norton I, Salunkhe AS, Goodluck H, Aly WS, Mourad-Agha H, Cornelis P. 2013. Control of iron metabolism in bacteria. Met Ions Life Sci 12:203–239. [DOI] [PubMed] [Google Scholar]
- 3.Bradley JM, Moore GR, Le Brun NE. 2014. Mechanisms of iron mineralization in ferritins: one size does not fit all. J Inorg Biochem 19:775–785. doi: 10.1007/s00775-014-1136-3. [DOI] [PubMed] [Google Scholar]
- 4.Imlay JA. 2006. Iron-sulphur clusters and the problem with oxygen. Mol Microbiol 59:1073–1082. doi: 10.1111/j.1365-2958.2006.05028.x. [DOI] [PubMed] [Google Scholar]
- 5.Imlay JA. 2003. Pathways of oxidative damage. Annu Rev Microbiol 57:395–418. doi: 10.1146/annurev.micro.57.030502.090938. [DOI] [PubMed] [Google Scholar]
- 6.Kiley PJ, Donohue TJ. 2010. Global responses of bacteria to O2 deprivation. ASM Press, Washington, DC. [Google Scholar]
- 7.Hantke K. 1981. Regulation of ferric iron transport in Escherichia coli K12: isolation of a constitutive mutant. Mol Gen Genet 182:288–292. doi: 10.1007/BF00269672. [DOI] [PubMed] [Google Scholar]
- 8.Bagg A, Neilands JB. 1987. Ferric uptake regulation protein acts as a repressor, employing iron (II) as a cofactor to bind the operator of an iron transport operon in Escherichia coli. Biochemistry 26:5471–5477. doi: 10.1021/bi00391a039. [DOI] [PubMed] [Google Scholar]
- 9.Masse E, Gottesman S. 2002. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc Natl Acad Sci U S A 99:4620–4625. doi: 10.1073/pnas.032066599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salvail H, Massé E. 2012. Regulating iron storage and metabolism with RNA: an overview of posttranscriptional controls of intracellular iron homeostasis. Wiley Interdiscip Rev RNA 3:26–36. doi: 10.1002/wrna.102. [DOI] [PubMed] [Google Scholar]
- 11.Escolar L, de Lorenzo V, Pérez-Martíín J. 1997. Metalloregulation in vitro of the aerobactin promoter of Escherichia coli by the Fur (ferric uptake regulation) protein. Mol Microbiol 26:799–808. doi: 10.1046/j.1365-2958.1997.6211987.x. [DOI] [PubMed] [Google Scholar]
- 12.Escolar L, Perez-Martin J, de Lorenzo V. 1998. Coordinated repression in vitro of the divergent fepA-fes promoters of Escherichia coli by the iron uptake regulation (Fur) protein. J Bacteriol 180:2579–2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McHugh JP, Rodriguez-Quinones F, Abdul-Tehrani H, Svistunenko DA, Poole RK, Cooper CE, Andrews SC. 2003. Global iron-dependent gene regulation in Escherichia coli—a new mechanism for iron homeostasis. J Biol Chem 278:29478–29486. doi: 10.1074/jbc.M303381200. [DOI] [PubMed] [Google Scholar]
- 14.Mettert EL, Kiley PJ. 2014. Coordinate regulation of the Suf and Isc Fe-S cluster biogenesis pathways by IscR is essential for viability of Escherichia coli. J Bacteriol 196:4315–4323. doi: 10.1128/JB.01975-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee K, Yeo W, Roe J. 2008. Oxidant-responsive induction of the suf operon, encoding a Fe-S assembly system, through Fur and IscR in Escherichia coli. J Bacteriol 190:8244–8247. doi: 10.1128/JB.01161-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Outten FW, Djaman O, Storz G. 2004. A suf operon requirement for Fe-S cluster assembly during iron starvation in Escherichia coli. Mol Microbiol 52:861–872. doi: 10.1111/j.1365-2958.2004.04025.x. [DOI] [PubMed] [Google Scholar]
- 17.Patzer SI, Hantke K. 2001. Dual repression by Fe2+-Fur and Mn2+-MntR of the mntH gene, encoding an NRAMP-like Mn2+ transporter in Escherichia coli. J Bacteriol 183:4806–4813. doi: 10.1128/JB.183.16.4806-4813.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tardat B, Touati D. 1993. Iron and oxygen regulation of Escherichia coli MnSOD expression: competition between the global regulators Fur and ArcA for binding to DNA. Mol Microbiol 9:53–63. doi: 10.1111/j.1365-2958.1993.tb01668.x. [DOI] [PubMed] [Google Scholar]
- 19.Martin JE, Imlay JA. 2011. The alternative aerobic ribonucleotide reductase of Escherichia coli, NrdEF, is a manganese-dependent enzyme that enables cell replication during periods of iron starvation. Mol Microbiol 80:319–334. doi: 10.1111/j.1365-2958.2011.07593.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nandal A, Huggins CCO, Woodhall MR, McHugh J, Rodríguez-Quiñones F, Quail MA, Guest JR, Andrews SC. 2010. Induction of the ferritin gene (ftnA) of Escherichia coli by Fe2+-Fur is mediated by reversal of H-NS silencing and is RyhB independent. Mol Microbiol 75:637–657. doi: 10.1111/j.1365-2958.2009.06977.x. [DOI] [PubMed] [Google Scholar]
- 21.Chu BC, Garcia-Herrero A, Johanson TH, Krewulak KD, Lau CK, Peacock RS, Slavinskaya Z, Vogel HJ. 2010. Siderophore uptake in bacteria and the battle for iron with the host; a bird’s eye view. Biometals 23:601–611. doi: 10.1007/s10534-010-9361-x. [DOI] [PubMed] [Google Scholar]
- 22.Raymond KN, Dertz EA, Kim SS. 2003. Enterobactin: an archetype for microbial iron transport. Proc Natl Acad Sci U S A 100:3584–3588. doi: 10.1073/pnas.0630018100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seo SW, Kim D, Latif H, O’Brien EJ, Szubin R, Palsson BO. 2014. Deciphering Fur transcriptional regulatory network highlights its complex role beyond iron metabolism in Escherichia coli. Nat Commun 5:4910. doi: 10.1038/ncomms5910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hantke K, Zimmermann L. 1981. The importance of the exbB gene for vitamin-B12 and ferric iron transport. FEMS Microbiol Lett 12:31–35. doi: 10.1111/j.1574-6968.1981.tb07606.x. [DOI] [Google Scholar]
- 25.Young GM, Postle K. 1994. Repression of tonB transcription during anaerobic growth requires Fur binding at the promoter and a second factor binding upstream. Mol Microbiol 11:943–954. doi: 10.1111/j.1365-2958.1994.tb00373.x. [DOI] [PubMed] [Google Scholar]
- 26.Kammler M, Schon C, Hantke K. 1993. Characterization of the ferrous iron uptake system of Escherichia coli. J Bacteriol 175:6212–6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park DM, Akhtar MS, Ansari AZ, Landick R, Kiley PJ. 2013. The bacterial response regulator ArcA uses a diverse binding site architecture to regulate carbon oxidation globally. PLoS Genet 9:e1003839. doi: 10.1371/journal.pgen.1003839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jacques J, Jang S, Prévost K, Desnoyers G, Desmarais M, Imlay J, Massé E. 2006. RyhB small RNA modulates the free intracellular iron pool and is essential for normal growth during iron limitation in Escherichia coli. Mol Microbiol 62:1181–1190. doi: 10.1111/j.1365-2958.2006.05439.x. [DOI] [PubMed] [Google Scholar]
- 29.Prévost K, Salvail H, Desnoyers G, Jacques J, Phaneuf É, Massé E. 2007. The small RNA RyhB activates the translation of shiA mRNA encoding a permease of shikimate, a compound involved in siderophore synthesis. Mol Microbiol 64:1260–1273. doi: 10.1111/j.1365-2958.2007.05733.x. [DOI] [PubMed] [Google Scholar]
- 30.Masse E, Escorcia FE, Gottesman S. 2003. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev 17:2374–2383. doi: 10.1101/gad.1127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prevost K, Desnoyers G, Jacques J, Lavoie F, Masse E. 2011. Small RNA-induced mRNA degradation achieved through both translation block and activated cleavage. Genes Dev 25:385–396. doi: 10.1101/gad.2001711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spiro S, Guest JR. 1991. Adaptive responses to oxygen limitation in Escherichia coli. Trends Biochem Sci 16:310–314. doi: 10.1016/0968-0004(91)90125-F. [DOI] [PubMed] [Google Scholar]
- 33.Gunsalus RP, Park S. 1994. Aerobic-anaerobic gene regulation in Escherichia coli: control by the ArcAB and Fnr regulons. Res Microbiol 145:437–450. doi: 10.1016/0923-2508(94)90092-2. [DOI] [PubMed] [Google Scholar]
- 34.Constantinidou C, Hobman JL, Griffiths L, Patel MD, Penn CW, Cole JA, Overton TW. 2006. A reassessment of the FNR regulon and transcriptomic analysis of the effects of nitrate, nitrite, NarXL, and NarQP as Escherichia coli K12 adapts from aerobic to anaerobic growth. J Biol Chem 281:4802–4815. doi: 10.1074/jbc.M512312200. [DOI] [PubMed] [Google Scholar]
- 35.Kang Y, Weber KD, Qiu Y, Kiley PJ, Blattner FR. 2005. Genome-wide expression analysis indicates that FNR of Escherichia coli K-12 regulates a large number of genes of unknown function. J Bacteriol 187:1135–1160. doi: 10.1128/JB.187.3.1135-1160.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Myers KS, Yan H, Ong IM, Chung D, Liang K, Tran F, Keleş S, Landick R, Kiley PJ. 2013. Genome-scale analysis of Escherichia coli FNR reveals complex features of transcription factor binding. PLoS Genet 9:e1003565. doi: 10.1371/journal.pgen.1003565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Unden G, Bongaerts J. 1997. Alternative respiratory pathways of Escherichia coli: energetics and transcriptional regulation in response to electron acceptors. Biochim Biophys Acta 1320:217–234. doi: 10.1016/S0005-2728(97)00034-0. [DOI] [PubMed] [Google Scholar]
- 38.Tjaden B, Goodwin SS, Opdyke JA, Guillier M, Fu DX, Gottesman S, Storz G. 2006. Target prediction for small, noncoding RNAs in bacteria. Nucleic Acids Res 34:2791–2802. doi: 10.1093/nar/gkl356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wright PR, Richter AS, Papenfort K, Mann M, Vogel J, Hess WR, Backofen R, Georg J. 2013. Comparative genomics boosts target prediction for bacterial small RNAs. Proc Natl Acad Sci U S A 110:E3487–E3496. doi: 10.1073/pnas.1303248110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Machanick P, Bailey TL. 2011. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics 27:1696–1697. doi: 10.1093/bioinformatics/btr189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Lorenzo V, Wee S, Herrero M, Neilands JB. 1987. Operator sequences of the aerobactin operon of plasmid-ColV-K30 binding the ferric uptake regulation (Fur) repressor. J Bacteriol 169:2624–2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lavrrar JL, Christoffersen CA, McIntosh MA. 2002. Fur-DNA interactions at the bidirectional fepDGC-entS promoter region in Escherichia coli. J Mol Biol 322:983–995. doi: 10.1016/S0022-2836(02)00849-5. [DOI] [PubMed] [Google Scholar]
- 43.Lavrrar JL, McIntosh MA. 2003. Architecture of a Fur binding site: a comparative analysis. J Bacteriol 185:2194–2202. doi: 10.1128/JB.185.7.2194-2202.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen Z, Lewis KA, Shultzaberger RK, Lyakhov IG, Zheng M, Doan B, Storz G, Schneider TD. 2007. Discovery of Fur binding site clusters in Escherichia coli by information theory models. Nucleic Acids Res 35:6762–6777. doi: 10.1093/nar/gkm631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Atlung T, Sund S, Olesen K, Brondsted L. 1996. The histone-like protein H-NS acts as a transcriptional repressor for expression of the anaerobic and growth phase activator AppY of Escherichia coli. J Bacteriol 178:3418–3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Waters LS, Sandoval M, Storz G. 2011. The Escherichia coli MntR miniregulon includes genes encoding a small protein and an efflux pump required for manganese homeostasis. J Bacteriol 193:5887–5897. doi: 10.1128/JB.05872-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Giel JL, Rodionov D, Liu M, Blattner FR, Kiley PJ. 2006. IscR-dependent gene expression links iron-sulphur cluster assembly to the control of O2-regulated genes in Escherichia coli. Mol Microbiol 60:1058–1075. doi: 10.1111/j.1365-2958.2006.05160.x. [DOI] [PubMed] [Google Scholar]
- 48.Yeo W, Lee J, Lee K, Roe J. 2006. IscR acts as an activator in response to oxidative stress for the suf operon encoding Fe-S assembly proteins. Mol Microbiol 61:206–218. doi: 10.1111/j.1365-2958.2006.05220.x. [DOI] [PubMed] [Google Scholar]
- 49.Masse E, Vanderpool CK, Gottesman S. 2005. Effect of RyhB small RNA on global iron use in Escherichia coli. J Bacteriol 187:6962–6971. doi: 10.1128/JB.187.20.6962-6971.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Keseler IM, Collado-Vides J, Santos-Zavaleta A, Peralta-Gil M, Gama-Castro S, Muniz-Rascado L, Bonavides-Martinez C, Paley S, Krummenacker M, Altman T, Kaipa P, Spaulding A, Pacheco J, Latendresse M, Fulcher C, Sarker M, Shearer AG, Mackie A, Paulsen I, Gunsalus RP, Karp PD. 2011. EcoCyc: a comprehensive database of Escherichia coli biology. Nucleic Acids Res 39:D583–D590. doi: 10.1093/nar/gkq1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Salvail H, Caron M, Bélanger J, Massé E. 2013. Antagonistic functions between the RNA chaperone Hfq and an sRNA regulate sensitivity to the antibiotic colicin. EMBO J 32:2764–2778. doi: 10.1038/emboj.2013.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Quail MA, Jordan P, Grogan JM, Butt JN, Lutz M, Thomson AJ, Andrews SC, Guest JR. 1996. Spectroscopic and voltammetric characterisation of the bacterioferritin-associated ferredoxin of Escherichia coli. Biochem Biophys Res Commun 229:635–642. doi: 10.1006/bbrc.1996.1856. [DOI] [PubMed] [Google Scholar]
- 53.Zeth K. 2012. Dps biomineralizing proteins: multifunctional architects of nature. Biochem J 445:297–311. doi: 10.1042/BJ20120514. [DOI] [PubMed] [Google Scholar]
- 54.Helmann JD. 2014. Specificity of metal sensing: iron and manganese homeostasis in Bacillus subtilis. J Biol Chem 289:28112–28120. doi: 10.1074/jbc.R114.587071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Giel JL, Nesbit AD, Mettert EL, Fleischhacker AS, Wanta BT, Kiley PJ. 2013. Regulation of iron-sulphur cluster homeostasis through transcriptional control of the Isc pathway by [2Fe-2S]-IscR in Escherichia coli. Mol Microbiol 87:478–492. doi: 10.1111/mmi.12052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Desnoyers G, Morissette A, Prévost K, Massé E. 2009. Small RNA-induced differential degradation of the polycistronic mRNA iscRSUA. EMBO J 28:1551–1561. doi: 10.1038/emboj.2009.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Massé E, Salvail H, Desnoyers G, Arguin M. 2007. Small RNAs controlling iron metabolism. Curr Opin Microbiol 10:140–145. doi: 10.1016/j.mib.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 58.Imlay JA. 2008. Cellular defenses against superoxide and hydrogen peroxide. Annu Rev Biochem 77:755–776. doi: 10.1146/annurev.biochem.77.061606.161055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smaldone GT, Revelles O, Gaballa A, Sauer U, Antelmann H, Helmann JD. 2012. A global investigation of the Bacillus subtilis iron-sparing response identifies major changes in metabolism. J Bacteriol 194:2594–2605. doi: 10.1128/JB.05990-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tardat B, Touati D. 1991. Two global regulators repress the anaerobic expression of MnSOD in Escherichia coli::Fur (ferric uptake regulation) and Arc (aerobic respiration control). Mol Microbiol 5:455–465. doi: 10.1111/j.1365-2958.1991.tb02129.x. [DOI] [PubMed] [Google Scholar]
- 61.Kargalioglu Y, Imlay JA. 1994. Importance of anaerobic superoxide dismutase synthesis in facilitating outgrowth of Escherichia coli upon entry into an aerobic habitat. J Bacteriol 176:7653–7658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hopkin KA, Papazian MA, Steinman HM. 1992. Functional differences between manganese and iron superoxide dismutases in Escherichia coli K-12. J Biol Chem 267:24253–24258. [PubMed] [Google Scholar]
- 63.Jordan A, Aragall E, Gibert I, Barbé J. 1996. Promoter identification and expression analysis of Salmonella typhimurium and Escherichia coli nrdEF operons encoding one of two class I ribonucleotide reductases present in both bacteria. Mol Microbiol 19:777–790. doi: 10.1046/j.1365-2958.1996.424950.x. [DOI] [PubMed] [Google Scholar]
- 64.Garriga X, Eliasson R, Torrents E, Jordan A, Barbé J, Gibert I, Reichard P. 1996. nrdD and nrdG genes are essential for strict anaerobic growth of Escherichia coli. Biochem Biophys Res Commun 229:189–192. doi: 10.1006/bbrc.1996.1778. [DOI] [PubMed] [Google Scholar]
- 65.Makui H, Roig E, Cole ST, Helmann JD, Gros P, Cellier MFM. 2000. Identification of the Escherichia coli K-12 Nramp orthologue (MntH) as a selective divalent metal ion transporter. Mol Microbiol 35:1065–1078. doi: 10.1046/j.1365-2958.2000.01774.x. [DOI] [PubMed] [Google Scholar]
- 66.Martin JE, Waters LS, Storz G, Imlay JA. 2015. The Escherichia coli small protein MntS and exporter MntP optimize the intracellular concentration of manganese. PLoS Genet 11:e1004977. doi: 10.1371/journal.pgen.1004977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fraser HI, Kvaratskhelia M, White MF. 1999. The two analogous phosphoglycerate mutases of Escherichia coli. FEBS Lett 455:344–348. doi: 10.1016/S0014-5793(99)00910-2. [DOI] [PubMed] [Google Scholar]
- 68.Browning DF, Grainger DC, Busby SJ. 2010. Effects of nucleoid-associated proteins on bacterial chromosome structure and gene expression. Curr Opin Microbiol 13:773–780. doi: 10.1016/j.mib.2010.09.013. [DOI] [PubMed] [Google Scholar]
- 69.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Witte K, Schuh AL, Hegermann J, Sarkeshik A, Mayers JR, Schwarze K, Yates JR III, Eimer S, Audhya A. 2011. TFG-1 function in protein secretion and oncogenesis. Nat Cell Biol 13:550–558. doi: 10.1038/ncb2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sutton VR, Mettert EL, Beinert H, Kiley PJ. 2004. Kinetic analysis of the oxidative conversion of the [4Fe-4S]2+ cluster of FNR to a [2Fe-2S]2+ cluster. J Bacteriol 186:8018–8025. doi: 10.1128/JB.186.23.8018-8025.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Blankenberg D, Gordon A, Von Kuster G, Coraor N, Taylor J, Nekrutenko A. 2010. Manipulation of FASTQ data with Galaxy. Bioinformatics 26:1783–1785. doi: 10.1093/bioinformatics/btq281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sun G, Chung D, Liang K, Keleş S. 2013. Statistical analysis of ChIP-seq data with MOSAiCS. Methods Mol Biol 1038:193–212. doi: 10.1007/978-1-62703-514-9_12. [DOI] [PubMed] [Google Scholar]
- 76.Kuan PF, Chung D, Pan G, Thomson JA, Stewart R, Keleş S. 2011. A statistical framework for the analysis of ChIP-Seq data. J Am Stat Assoc 106:891–903. doi: 10.1198/jasa.2011.ap09706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chung D, Park D, Myers K, Grass J, Kiley P, Landick R, Keles S. 2013. dPeak: high resolution identification of transcription factor binding sites from PET and SET ChIP-Seq data. PLOS Comput Biol 9:e1003246. doi: 10.1371/journal.pcbi.1003246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liang K, Keles S. 2012. Detecting differential binding of transcription factors with ChIP-seq. Bioinformatics 28:121–122. doi: 10.1093/bioinformatics/btr605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Homann OR, Johnson AD. 2010. MochiView: versatile software for genome browsing and DNA motif analysis. BMC Biol 8:49. doi: 10.1186/1741-7007-8-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Peters JM, Mooney RA, Grass JA, Jessen ED, Tran F, Landick R. 2012. Rho and NusG suppress pervasive antisense transcription in Escherichia coli. Genes Dev 26:2621–2633. doi: 10.1101/gad.196741.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bolstad BM. preprocessCore: a collection of pre-processing functions. R package version 1.20.0. www.bioconductor.org.
- 82.R Development Core Team 2008. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 83.Kuan PF, Chun H, Keles S. 2008. CMARRT: a tool for the analysis of ChIP-chip data from tiling arrays by incorporating the correlation structure. Pac Symp Biocomput 208:515–526. [PMC free article] [PubMed] [Google Scholar]
- 84.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. 2003. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res 31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lawrence M, Huber W, Pagès H, Aboyoun P, Carlson M, Gentleman R, Morgan MT, Carey VJ. 2013. Software for computing and annotating genomic ranges. PLOS Comput Biol 9:e1003118. doi: 10.1371/journal.pcbi.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B Stat Methodol 57:289–300. [Google Scholar]
- 87.Montgomery DC. 2000. Design and analysis of experiments, 5 ed. John Wiley & Sons, New York, NY. [Google Scholar]
- 88.Miller J. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 89.Keyer K, Imlay JA. 1996. Superoxide accelerates DNA damage by elevating free-iron levels. Proc Natl Acad Sci U S A 93:13635–13640. doi: 10.1073/pnas.93.24.13635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.McShane WJ, Pappas RS, Wilson-McElprang V, Paschal D. 2008. A rugged and transferable method for determining blood cadmium, mercury, and lead with inductively coupled plasma-mass spectrometry. Spectrochim Acta B At Spectrosc 63:638–644. doi: 10.1016/j.sab.2008.03.016. [DOI] [Google Scholar]
- 91.Langer EK, Johnson KJ, Shafer MM, Gorski P, Overdier J, Musselman J, Ross JA. 2011. Characterization of the elemental composition of newborn blood spots using sector-field inductively coupled plasma-mass spectrometry. J Expo Sci Environ Epidemiol 21:355–364. doi: 10.1038/jes.2010.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Western blots of cellular proteins to determine Fur antibody specificity. The reactivity of purified Fur antibodies to Fur (~17 kDa) in cells was examined by Western blot analysis of strains lacking or containing Fur. Cells were grown in MOPS glucose minimal medium with 10 µM FeSO4 to an OD600 of 0.1, and cell proteins were separated by SDS-PAGE and analyzed by Western blotting with affinity-purified Fur antibodies. Lanes: A, molecular mass standards; B, Fur+ strain, 3.2 × 107 cells; C, Fur+ strain, 6.5 × 107 cells; D, Δfur strain, 3.2 × 107 cells; E, Δfur strain, 6.5 × 107 cells. Download
Analysis of iron-uptake-deficient strain. To determine if Fur DNA binding was iron-dependent under anaerobic conditions, we performed ChIP-seq analyses of an iron-uptake-deficient strain (ΔtonB Δfeo ΔzupT) grown under iron-limiting conditions. We showed that the cultures used for ChIP-seq analysis were indeed iron deficient by comparing promoter activities of a known iron-dependent, Fur-repressed promoter, PfepA, in wild-type (WT), Fur-lacking (Δfur), and iron-uptake-deficient (ΔtonB Δfeo ΔzupT) strains. Promoter activity was determined from the β-galactosidase activity of a PfepA-lacZ gene fusion strain normalized to cell density. The concentration of FeSO4 in the MOPS glucose minimal medium was 10 µM for the iron-sufficient growth conditions and 1.0 µM for the iron-deficient conditions. Download
Strains and plasmids. This table details the strains and plasmids used to perform the experiments described in this study.
Fur binding regions mapped across the E. coli K-12 genome under aerobic or anaerobic growth conditions using ChIP-seq. In vivo Fur DNA binding sites identified through ChIP-Seq experiments are listed according to whether Fur DNA binding was observed in the presence or absence of O2 and whether Fur DNA binding was iron-dependent or iron-independent. Additional information includes the gene predicted to be associated with the Fur DNA binding site, the location of the ChIP signal relative to that gene, if the gene associated with the Fur DNA binding site is Fur or RyhB regulated, and if the Fur DNA binding site was previously known.
Operons directly regulated by Fur during aerobic and/or anaerobic growth. Transcription profiling results are reported as the log2-transformed mRNA levels for the first gene in each operon determined to be directly regulated by Fur (i.e., contained a ChIP-seq peak [see Table S2 in the supplemental material]). The data are presented for wild-type, Fur−, and Fur−‑ RyhB− strains grown in the presence or absence of O2 and are separated by whether Fur DNA binding occurs under only anaerobic growth conditions or under both aerobic and anaerobic growth conditions. A reference is provided if Fur regulation was previously known.
Operons regulated by RyhB during aerobic and/or anaerobic growth. Transcription profiling results are reported as the log2-transformed mRNA levels for each gene in operons predicted to be regulated by RyhB. The data are presented for wild-type, Fur−, and Fur−RyhB− strains grown in the presence or absence of O2 and are separated by whether expression of the operon is decreased or increased by RyhB and whether expression of the operon is greater under aerobic or anaerobic growth conditions. A reference is provided if Fur and/or RyhB regulation was previously known.
Genes indirectly regulated by Fur and not regulated by RyhB. Transcription profiling results are reported as the log2-transformed mRNA levels for the first gene in each operon determined to be indirectly regulated by Fur and not regulated by RyhB. The data are presented for wild-type and Fur− strains grown in the presence or absence of O2 and separated by whether the Fur-dependent change in expression occurs under primarily aerobic or anaerobic growth conditions. A reference is provided if Fur regulation was previously known. It is also indicated whether a Fur binding site was previously predicted using bioinformatics despite the absence of an in vivo Fur DNA binding site in this study.
Iron-containing proteins whose transcript levels do not appear to be affected by RyhB expression. Transcription profiling results are reported as the log2-transformed mRNA levels for genes encoding iron-binding proteins, which do not appear to be regulated by RyhB. The data are presented for wild-type, Fur−, and Fur−RyhB− strains grown in the presence or absence of O2. Genes are separated into parts a and b based on wild-type expression levels.
Log2 expression for all genes in the E. coli K-12 genome under all experimental conditions. Transcription profiling results are reported as the log2-transformed mRNA levels for all genes in the E. coli genome. The data are presented for the wild-type, Fur−, Fur− RyhB−, and RyhB− strains grown in the presence or absence of O2 and are organized by the gene accession number known as the b-number.