ABSTRACT
Cyclic diguanylate (c-di-GMP) is a bacterial second messenger that controls multiple cellular processes. c-di-GMP networks have up to dozens of diguanylate cyclases (DGCs) that synthesize c-di-GMP along with many c-di-GMP-responsive target proteins that can bind and respond to this signal. For such networks to have order, a mechanism(s) likely exists that allow DGCs to specifically signal their targets, and it has been suggested that physical interactions might provide such specificity. Our results show a DGC from Pseudomonas fluorescens physically interacting with its target protein at a conserved interface, and this interface can be predictive of DGC-target protein interactions. Furthermore, we demonstrate that physical interaction is necessary for the DGC to maximally signal its target. If such “local signaling” is a theme for even a fraction of the DGCs used by bacteria, it becomes possible to posit a model whereby physical interaction allows a DGC to directly signal its target protein, which in turn may help curtail undesired cross talk with other members of the network.
IMPORTANCE
An important question in microbiology is how bacteria make decisions using a signaling network made up of proteins that make, break, and bind the second messenger c-di-GMP, which is responsible for controlling many cellular behaviors. Previous work has shown that a given DGC enzyme will signal for specific cellular outputs, despite making the same diffusible molecule as its sibling DGCs in the unpartitioned space of the bacterial cell. Understanding how one DGC differentiates its output from the dozens of other such enzymes in the cell is synonymous with understanding a large component of the bacterial decision-making machinery. We present evidence for a helix on a DGC used to physically associate with its target protein, which is necessary to achieve maximal signaling.
INTRODUCTION
Bacteria are generally thought of as unpartitioned spaces, within which diffusible contents of the cell are free to mix. Paradoxically, many species across the bacterial domain are also known to support complex signaling networks of the small molecule cyclic diguanylate (c-di-GMP) that can regulate a multitude of processes from virulence to biofilm formation to gene transcription (1–3). This leads to an open question: how does a small molecule in a freely diffusible space trigger one cellular output but not another?
Cyclic diguanylate is a near-universal bacterial second messenger that specifies cellular actions by binding to effector proteins or acting on riboswitches (4). This second messenger has come under intense scrutiny, as it regulates virulence in Pseudomonas aeruginosa, Vibrio cholerae, Mycobacterium tuberculosis, and Clostridium difficile, among others (1, 5, 6). Many species of bacteria have dozens of diguanylate cyclases (DGCs) that synthesize c-di-GMP and dozens more phosphodiesterases (PDEs) that degrade this signal. Yet, genetic studies have shown that the absence of a specific DGC or PDE in a network affects one or a few specific processes (5, 7, 8). Given the diversity and number of processes that c-di-GMP regulates, understanding how any given DGC or PDE in the network targets a single cellular process would hold tremendous value for developing novel medical treatments, industrial and civic control of corrosive biofilms, and engineering biological systems responsive to predefined stimuli. In short, understanding network specificity is synonymous with understanding a large component of the bacterial decision-making machinery.
One model to achieve specificity in signaling proposes that DGCs physically interact with effector proteins and PDEs (9), which to date is supported by a domain-level analysis of a c-di-GMP signaling module (10). Importantly, virtually all DGC proteins share a fold of their catalytically active GGDEF domain, as do nearly all PDEs in their EAL domain (11). Additionally, some effector proteins contain an EAL domain that may bind c-di-GMP (12–14). As a result, it is a reasonable question to ask if there are discrete sections of these proteins that may interact with each other.
To approach this topic, we required a bacterial model for c-di-GMP signaling with a known DGC-effector pair. Pseudomonas fluorescens is a Gram-negative bacterium with over two dozen DGCs and at least another dozen PDEs. Within this complex c-di-GMP signaling network is a subnetwork of four DGCs known to affect biofilm formation, including GcbC (8). Recently, a key effector protein of biofilm formation in P. fluorescens was identified; LapD is a c-di-GMP-responsive inner membrane protein that binds c-di-GMP through its defunct EAL domain that can bind, but not degrade, c-di-GMP (15). When bound to c-di-GMP, LapD undergoes a conformational change that sequesters the periplasmic protease LapG (Fig. 1A), thus allowing the large adhesin LapA to accumulate on the cell surface and biofilm formation to commence (12, 16, 17). This simplified subsystem acting against the backdrop of a larger c-di-GMP network provided an ideal model system to test how specificity in signaling is achieved.
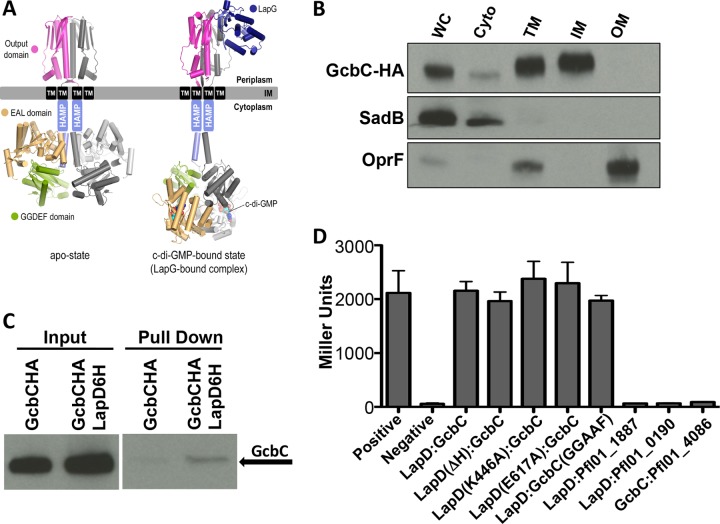
FIG 1 .
GcbC physically interacts with LapD. (A) Dimeric LapD models are based on available crystal structures (L. pneumophila apo-output domain, PDB 4u64; L. pneumophila output domain-P. fluorescens LapG complex, PDB 4u65; P. fluorescens apo-GGDEF-EAL module, PDB 3pjx; P. fluorescens EAL–c-di-GMP, PDB 3pju), as described previously (12, 17). (B) HA-tagged GcbC localizes to the inner membrane fraction of P. fluorescens. Shown are Western blots of whole-cell lysate (WC), cytoplasmic (Cyto), total membrane (TM), inner membrane (IM), and outer membrane (OM) fractions. Control markers for the cytoplasm (SadB) and the outer membrane (OprF) are included. Samples were normalized by protein concentration. (C) Full-length GcbC interacts with full-length LapD in a coprecipitation experiment. From left to right, input of strain tagged with GcbC-HA, input of strain with both GcbC-HA and LapD-6H tagging, coprecipitation of single-tagged GcbC-HA strain, and coprecipitation of double-tagged GcbC-HA and LapD-6H strain. GcbC-HA was detected with antibody to its epitope tag. (D) GcbC and LapD interact via B2H. Shown are the resulting β-galactosidase activities in Miller units, which serve as a measure of protein-protein interactions in vivo. The positive interacting control (Positive), a dimerized leucine zipper protein, and negative control (Negative), the vectors alone, are shown. The x axis shows the proteins, or their mutant variants, used in each experiment. Miller units were assayed after 16 h of incubation from cells scraped from transformation plates. Error bars show standard deviations from 3 biological replicates which were generated from 3 technical replicates per biological replicate. There is no significant difference in the interaction of the wild-type LapD and GcbC protein compared to that of any of the mutant pair combinations.
Based on the findings presented here, we propose a model whereby physical interaction between a DGC and a c-di-GMP binding effector contributes to functional signaling in a larger c-di-GMP network. This model offers one explanation for how a single bacterial cell may have multiple DGCs simultaneously signaling specific cellular processes using the same small molecule while reducing undesired cross talk.
RESULTS
The DGC GcbC physically interacts with its effector LapD.
The DGC GcbC has previously been shown to promote biofilm formation via LapD by controlling LapA localization (8). To test the hypothesis that GcbC physically interacts with LapD to promote c-di-GMP-mediated signaling, we first asked whether the two proteins colocalize to the same region in the cell. LapD is known to be an inner membrane protein (18). To determine if GcbC localizes to the inner membrane, we fractionated whole P. fluorescens cells expressing a hemagglutinin (HA)-tagged version of GcbC from a plasmid into cytoplasmic, inner membrane, and outer membrane components. GcbC was visualized by Western blotting in the inner membrane fraction (Fig. 1B), with SadB and OprF serving as cytosolic and outer membrane fractionation controls, respectively (19, 20).
To test if these inner membrane proteins physically associate with each other or as part of a complex, a 6-histidine-tagged version of LapD and an HA-tagged version of GcbC were coexpressed from a plasmid in P. fluorescens for an in vivo coprecipitation experiment utilizing the full-length proteins from the whole-cell lysate. GcbC was confirmed to associate with LapD, making it part of a new class of catalytically active DGCs demonstrated to physically associate with a c-di-GMP binding effector (Fig. 1C). These tagged versions of GcbC and LapD are used for all subsequent experiments conducted in Pseudomonas fluorescens.
An initial possibility was that this association represents a physical interaction driven by a catalytically activated GcbC that promotes LapD adopting its c-di-GMP-bound form, which is believed to be an extended conformation as mentioned above (Fig. 1A). To investigate this idea, we tested wild-type and mutant constructs of each protein for interaction in a bacterial two-hybrid (B2H) system in Escherichia coli strain BTH101. The B2H system rapidly assesses protein-protein interaction using full-length constructs of these inner membrane proteins. Interaction is measured indirectly via the expression of β-galactosidase using colorimetric reagents. E. coli cells which were cotransformed with LapD and GcbC expressed from the two-hybrid plasmids yielded a positive result for physical interaction compared to the negative control (Fig. 1D), a finding consistent with the pulldown assays. To confirm that this interaction is specific, we also tested each protein for interaction with other c-di-GMP-related proteins. GcbC showed no interaction with Pfl01_4086, a GGDEF-EAL domain-containing, inner membrane protein with a primary structure resembling LapD. Similarly, LapD showed no interaction with the GGDEF domain-containing protein Pfl01_0190 or the GGDEF-EAL domain-containing protein Pfl01_1887 (Fig. 1D).
We next tested interaction between GcbC and mutants of LapD that have been previously shown to lock this receptor in a functionally active or inactive state (15). Based on these previous studies, we utilized a deletion in the HAMP domain (ΔH) of LapD, which favors the activated conformation of the protein, and the K446A mutation, which promotes the autoinhibited, more compact conformation of LapD (Fig. 1C). We found GcbC able to bind LapD in either state (Fig. 1D).
Since the conformation of LapD depends on whether or not it is c-di-GMP bound, we wanted to test mutant versions of these proteins in which their ability to bind c-di-GMP is impacted. The E617A mutant of LapD (Fig. 1D), which cannot bind c-di-GMP, showed no defect in interacting with GcbC, suggesting that the LapD effector does not need to be in a c-di-GMP-bound state to interact with its partner DGC. Another possibility included interaction that depends on GcbC’s catalytic state. We therefore tested a construct of GcbC with key catalytic residues mutated (GGDEF to GGAAF), as validated previously (8). Interaction between LapD and the catalytically inactive GcbC failed to show a significant decrease from the wild-type interaction.
A C-terminal helix contributes to GcbC interaction with LapD.
Because the B2H system allowed us to quickly test full-length protein-protein interactions, it provided a way to screen for interaction-deficient mutants. Using mutagenic PCR, we created mutant libraries of the gcbC and lapD genes that were cloned into the B2H vectors. Cotransformed E. coli expressing the mutant library of one gene and the wild-type version of the other was screened for mutants that failed to interact, resulting in a white or light blue instead of a blue colony on solid medium amended with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside). The mutations were identified by sequencing, and we created a list of isolates containing a mutation(s) that disrupts the interaction between GcbC and LapD (see Table S1 in the supplemental material).
To identify mutated residues on GcbC that are necessary elements for interaction as opposed to mutations that might simply disrupt overall protein structure or its catalytic activity, the GGDEF domain of GcbC was crystallized and its structure was solved (see Materials and Methods and see also Table S2 in the supplemental material). The 14 mutations found in our screen that mapped to the GGDEF domain clustered into three groups (see Fig. S1A and Table S1). The first group clustered near the domain’s N terminus and was predicted to disrupt the native catalytic regulation of GcbC. In particular, Q355R, S356P, and W361R are located near the inhibitory site of GcbC, with the Q355R and W361R mutations found twice each in the screen. The second group clustered at a pair of helices near the N terminus and was made up of mutations that likely destabilize GcbC’s structure and its ability to actively dimerize, such as D386G. The final two mutations, E477V and A482T, were found at the C terminus of the protein on the α5GGDEF helix and are not obviously responsible for any disruption to GcbC dimerization or catalytic activity. This region of GcbC therefore became our focus for further analysis. Mutations found on LapD were also mapped and are discussed below (see Fig. S1B).
We examined the residues in the vicinity of the two found to disrupt interaction with LapD, and of the nine residues on the N-terminal half of the α5GGDEF helix (residues 477 to 485), five are surface exposed and could conceivably be making contact with another protein: E477, Q478, F481, D484, and K485. The aspartic acid at position 484 is a highly conserved residue that has been reported to be involved in coordinating the product-bound state of a Xanthomonas campestris DGC (21) and is hence analyzed separately to determine any possible contribution to the interaction interface. To test if the remaining four residues played any role in biofilm formation in vivo, we mutated each residue in turn to alanines and tested them separately and in combination for their ability to promote biofilm formation through GcbC. We tested a pGcbC construct as well as the derivative mutations in a background of P. fluorescens lacking the DGC network necessary to make a biofilm, referred to as the Δ4DGC strain. This background lacks the 4 DGCs that have been demonstrated to be the sole DGC contributors to biofilm formation in our minimal medium assay based on a previous analysis of disrupting all DGCs in P. fluorescens (8). All but one of the single alanine substitutions and all of the double and triple mutants showed a significant decrease in biofilm formation (Fig. 2A). The quadruple mutant with all of the residues mutated to alanine showed the smallest amount of biofilm formation, at 44% of that promoted by the wild-type GcbC (P < 5 × 10−5). Several of the mutations appeared additive, including the neighboring residues E477A and Q478A. Intriguingly, the F481A mutation seemed resistant to such additive effects in most mutant pairings (Fig. 2A). Adding the D484A mutant yielded the “Quint-Ala” mutant and resulted in no biofilm detected; recall that D484 has been reported to be involved in coordinating the product-bound state of an X. campestris DGC (21).
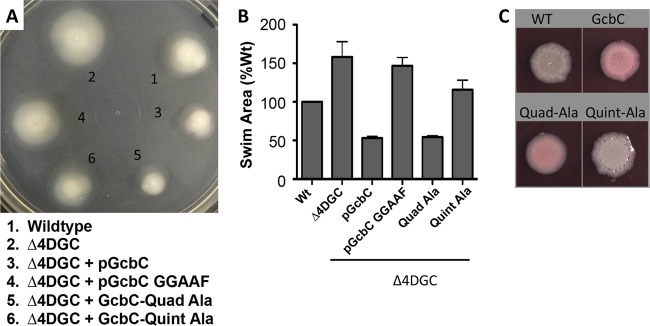
FIG 2 .
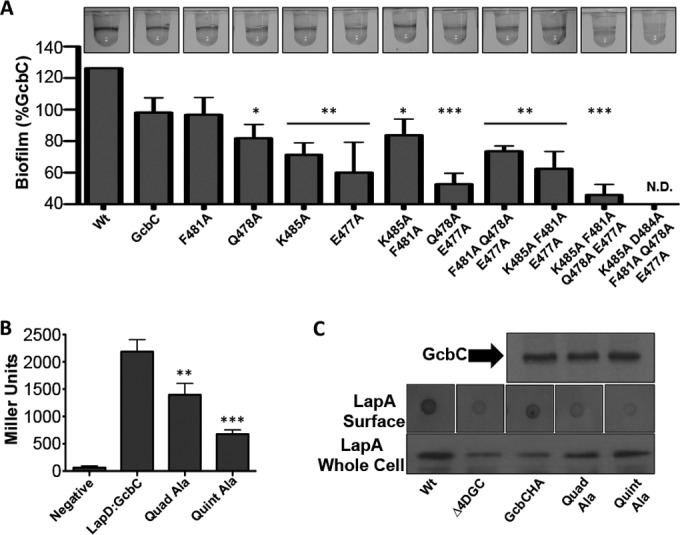
The N terminus of the α5GGDEF helix of GcbC contributes to the interaction with LapD and promotion of biofilm formation. (A) Each amino acid from the N terminus of the α5GGDEF helix was mutated to an alanine alone or in combination to determine accumulative effects on biofilm formation in P. fluorescens. Cells were grown in K10 medium for 6 h at 30°C and stained with 0.1% crystal violet before being dissolved and quantified at OD550. *, P < 0.05; **, P < 0.01; ***, P < 1 × 10−4 (compared to the wild-type GcbC). Error bars show standard deviations from 4 biological replicates which were generated from 8 technical replicates per biological replicate. Biofilm formation was normalized to the wild-type strain, was blanked to the Δ4DGC strain, and is displayed as percent biofilm induction relative to GcbC expressed from a plasmid. Images of biofilm rings formed by each strain are shown at top. (B) The quadruple and quintuple alanine mutants from panel A were assayed for their ability to interact with LapD via B2H. **, P < 0.005; ***, P < 2 × 10−5 (relative to the wild-type GcbC-LapD interaction). Error bars show standard deviations from 4 biological replicates which were generated from 3 technical replicates per biological replicate. (C) (Top) The stability of the HA-tagged quadruple (Quad-Ala) and quintuple (Quint-Ala) alanine GcbC mutants was assayed by Western blotting and compared to wild-type GcbC. (Middle) A dot blot assay was conducted to identify cell surface-associated LapA, which is responsible for biofilm formation. Overnight cultures were normalized to the wild-type strain and blotted onto nitrocellulose paper. When dry, the paper was blotted for LapA. The wild-type and pGcbC strains show accumulation of LapA on the cell surface, while the Δ4DGC, GcbC quadruple alanine (Quad-Ala), and GcbC quintuple alanine (Quint-Ala) mutants show less LapA. (Bottom) Total abundance of LapA was examined by Western blotting on whole-cell lysates of each strain. The Δ4DGC and pGcbC strains show a lower-abundance LapA than did the wild type, while the quadruple (Quad-Ala) and quintuple (Quint-Ala) alanine GcbC mutants show slightly higher levels of total LapA production than does the Δ4DGC strain.
We next wanted to verify that the reduction in biofilm observed in the quadruple mutant was the result of a loss of interaction with LapD as suggested by the original plate-based B2H screen and not due to protein instability or loss of catalytic function. We quantitatively tested the quadruple GcbC mutant for interaction with LapD compared to its wild-type counterpart via B2H using a β-galactosidase assay. We observed a significant decrease in interaction (P < 0.0005), demonstrating that these residues are necessary for full interaction with LapD (Fig. 2B, Quad-Ala). Adding the D484A mutation to the quadruple mutant resulted in a further decrease in interaction (Fig. 2B, Quint-Ala). Because GcbC catalytic activity does not impact interaction with LapD (Fig. 1D), we conclude that this residue is also structurally necessary for interaction with LapD, and its role is analyzed further below. We confirmed that both the quadruple and quintuple mutant protein products are as abundant as wild-type protein via Western blotting (Fig. 2C, top).
We also wanted to confirm that the biofilm defects that we observed were specifically due to loss of signaling to LapD and not due to a pleiotropic effect that GcbC was having elsewhere in the cell. One way to achieve this is to assess LapD function by a dot blot analysis. Our lab has previously demonstrated that LapD functions by sequestering the periplasmic protease LapG only when c-di-GMP levels are high. Under conditions of low c-di-GMP, released LapG may then cleave the large adhesion LapA on the cell surface, releasing it away from the cell (16). Therefore, the functional state of LapD can be assessed by blotting for the presence of absence of LapA on the cell surface, and we have previously shown that an active LapD (e.g., sequestering the LapG protease) results in high surface levels of LapA, while an inactive LapD corresponds to low accumulation of LapA on the surface (16). We would predict that because the mutant variants of GcbC are failing to deliver c-di-GMP to LapD, surface levels of LapA should be low compared to those of a strain expressing wild-type GcbC.
When tested for surface-associated LapA, the wild-type strain and the strain expressing GcbC on a plasmid showed LapA localizing to the cell’s surface (Fig. 2C, middle). Conversely, the control strain lacking biofilm-related DGCs (Δ4DGC), the strain expressing the quadruple alanine GcbC mutant, and the strain expressing the quintuple alanine GcbC mutant showed reduced amounts of LapA at the cell’s surface (Fig. 2C, middle). These results were compared with total levels of LapA obtained from whole-cell lysates to ensure that the mutant variants of GcbC were not resulting in lower levels of LapA production. While the control strain lacking the DGC subnetwork and the strain expressing GcbC on a plasmid showed somewhat lower levels of total LapA than did the wild-type strain, both mutant variants of GcbC showed relatively high levels of LapA, showing that lack of LapA on the surface of the cell is not the result of decreased LapA production (Fig. 2C, bottom).
Next, the GcbC quadruple alanine mutant was expressed in a genetic construct lacking the DGC subnetwork necessary for biofilm formation, with the exception of wild-type GcbC expressed from its native position on the chromosome (Δ3DGC), and tested for biofilm formation. The biofilm made by this strain expressing both the wild-type and mutant versions of GcbC was found to be additive compared to a strain expressing either allele alone, indicating that the mutant allele is not operating in a dominant negative manner (see Fig. S2A in the supplemental material).
Finally, we wanted to verify that the physiological defects in biofilm reduction stemming from the α5GGDEF helix relate specifically to an inability to signal LapD and not a general inability to function as a DGC. Previously, GcbC has been reported to repress swimming motility in P. fluorescens (8); importantly, the impact of GcbC on swimming occurs in a LapD-independent manner. Therefore, to determine in vivo DGC functionality, we tested GcbC and its mutant variants for their ability to repress swimming in a Δ4DGC background as analyzed by swim zone area (Fig. 3A). The Δ4DGC strain swims 158% of the area compared to a wild-type strain in minimal swim medium, while the pGcbC construct in the Δ4DGC background reduced swimming to 53% of that of the wild type (Fig. 3A and B). Further, the pGcbC GGAAF strain lacking the ability to produce c-di-GMP permitted swimming at 147% of the wild-type level, demonstrating that catalytic activity is necessary for GcbC to repress swimming. When the quadruple alanine (Quad-Ala) variant of GcbC is tested, it reduces swimming of the Δ4DGC strain to 54% of the wild-type value (Fig. 3A and B), a value nearly identical to the impact of the wild-type pGcbC plasmid, indicating that the Quad-Ala mutant is capable of functioning normally in vivo in regard to production of c-di-GMP (8). Conversely, when the quintuple alanine (Quint-Ala) variant is tested, the strain swims an area 115% of that of the wild type, demonstrating that the highly conserved aspartic acid at position 484 is likely partially necessary to DGC functionality in addition to its role in interacting with LapD (Fig. 3A and B).
FIG 3 .
The α5GGDEF helix of GcbC contributes to promoting biofilm formation but is not required for GcbC swim motility. (A) Swim phenotypes of wild-type P. fluorescens, the Δ4DGC mutant, and the Δ4DGC mutant expressing GcbC, GcbC GGAAF, or the quadruple (Quad-Ala) and quintuple (Quint-Ala) alanine mutant variants. Strains were grown overnight in LB and normalized to cell density, and 100 µl was dispensed into a 96-well plate. A 200-µl pipette tip was dipped into each well and plunged into an 0.35% agar K10 minimal medium plate supplemented with 0.1% arabinose. Plates were incubated at 30°C for 36 h and photographed. (B) Quantified swim area from experiments performed as in panel A. Error bars show standard deviations from 3 biological replicates which were generated from 3 technical replicates per biological replicate. Swim area was normalized to the wild-type (Wt) strain. (C) The quadruple and quintuple alanine mutants of GcbC were analyzed by Congo red binding by transforming the plasmids into P. aeruginosa. The binding of the red pigment by these P. aeruginosa strains indicates exopolysaccharide production, which serves as a surrogate for the level of c-di-GMP produced. The quadruple mutant (Quad-Ala) shows similar levels of Congo red binding as the wild-type GcbC, while the quintuple mutant (Quint-Ala) shows reduced binding compared to the strain expressing wild-type GcbC. The wild-type (WT) P. aeruginosa PA14 strain served as a control, and strains were grown for 36 h.
These results were mirrored when both the Quad-Ala and Quint-Ala GcbC constructs were tested in P. aeruginosa on Congo red plates for their ability to produce c-di-GMP as assessed indirectly via their ability to promote exopolysaccharide production in P. aeruginosa. The Quad-Ala mutant showed levels of Congo red binding similar to those of the wild-type GcbC construct, whereas the Quint-Ala mutant showed lower levels (Fig. 3C), analogous to the swimming motility findings from P. fluorescens shown in Fig. 3B.
The GcbC α5GGDEF helix is recognized by the α2EAL helix on LapD.
We turned back to the library of B2H mutants to determine if we could identify a LapD-GcbC interaction surface on LapD. We found 14 mutants from LapD’s GGDEF domain and 25 mutants on its EAL domain in our original screen (see Table S1 in the supplemental material). Mutants were found largely in one of two clusters on the EAL domain and were mapped on a previously published crystal structure of LapD (see Fig. S1B and Table S1) (12). The first cluster of mutations is near the c-di-GMP binding pocket, including residues known to be necessary for c-di-GMP binding (see Fig. S1B, cyan residues). These mutations suggest that the residues around the c-di-GMP binding pocket may play a role in the LapD-GcbC interaction, although a LapD that cannot bind c-di-GMP itself can still interact with GcbC (Fig. 1D). The second cluster of mutations is near the N terminus of the domain (see Fig. S1B, red residues) and is discussed below in more detail. The rest of the mutants identified map onto the EAL and GGDEF domains of LapD and showed a largely random distribution (see Fig. S1B, purple residues).
The cluster of mutants identified near the N terminus discussed above pointed to the α2EAL helix (residues 462 to 470; see Fig. S1B in the supplemental material) of the EAL domain of LapD and appeared to form a surface that could feasibly be a compatible interaction interface with the α5GGDEF helix of GcbC (Fig. 4A). To test the α2EAL helix for in vivo function, we mutated these residues in the chromosomal copy of lapD at its native locus. When assayed for biofilm formation in this background, only R463E showed a significant decrease (see Fig. S2B).
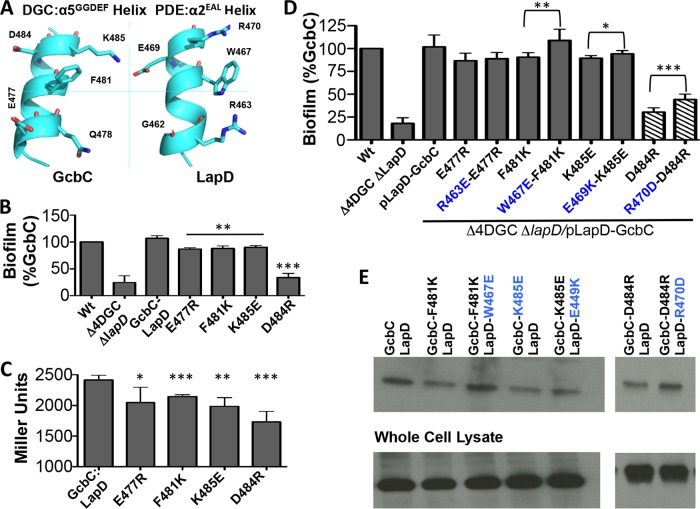
FIG 4 .
The α5GGDEF helix of GcbC interacts with a helix on LapD. (A) The regions of GcbC and LapD that may interact with each other are found on α-helices. Five surface residues that are possible points of contact between the GcbC helix and the LapD helix are shown in this ribbon diagram. (B) Nonconservative mutations in GcbC’s interaction surface were assayed for biofilm formation using the microtiter well assay at 6 h. **, P < 0.01; ***, P < 0.001 (compared to the control strain expressing wild-type GcbC). Error bars show standard deviations from 3 biological replicates which were generated from 8 technical replicates per biological replicate. Biofilm formation was normalized to the wild-type strain and is displayed as percent biofilm induction relative to the strain expressing wild-type GcbC. (C) The same mutations as those shown in panel B were assessed for interaction using a B2H experiment, confirming that GcbC’s α5GGDEF helix contributes to full interaction with LapD. *, P < 0.05; **, P < 0.005; ***, P < 0.001 (compared to interaction between wild-type GcbC and LapD). Error bars show standard deviations from 4 biological replicates which were generated from 3 technical replicates per biological replicate. (D) The disruptive GcbC mutations used in panel B were paired with potential compensatory mutations on LapD and assayed for biofilm formation. Suppression of biofilm disruption was observed for several pairs of mutants. To keep the final pair of mutants in the linear range of the assay, the D484R mutation was induced with 0.1% arabinose as indicated by the hatched bars. *, P < 0.05; **, P < 0.005; ***, P < 0.0005 (for each suppressor mutant relative to its unsuppressed GcbC mutant). Error bars show standard deviations from 7 biological replicates which were generated from 8 technical replicates per biological replicate. Biofilm formation was normalized to the wild-type strain and is displayed as percent biofilm induction relative to GcbC. (E) Compensatory LapD α2EAL helix mutations restore interaction with GcbC α5GGDEF helix mutations. Shown is a Western blot following a coprecipitation assay where cells expressing a 6-histidine-tagged LapD and HA-tagged GcbC were lysed, LapD-6H was immobilized on cobalt resin, and beads were washed before blotting with an anti-HA antibody. The bottom of the panel indicates the level of protein in the whole-cell lysate used for the pulldown assays.
We next tested these mutations with regard to disruption of LapD-GcbC interactions using the B2H assay. We were unable to show statistically significant reductions in interactions for individual residues on the helix of LapD (see Fig. S2C in the supplemental material). Given the lack of apparent defects in LapD-GcbC interactions arising from single mutations on LapD, we could not be sure that the LapD mutants identified in the screen and the biofilm defects arising from the LapD mutants were specifically due to disruption of interaction with GcbC. We therefore wanted to employ an alternative strategy to determine if the LapD α-helix spanning residues 462 to 470 (the α2EAL helix) might serve as an interface with GcbC. To accomplish this, we set out to make nonconservative substitutions (i.e., charge reversals) to members of GcbC’s α5GGDEF helix in order to disrupt interaction and biofilm formation and to then make potentially compensatory mutations on LapD’s α2EAL helix in an attempt to restore interaction and biofilm formation.
We first created an expression construct with tagged versions of LapD and GcbC to test these mutant variants for biofilm formation. These four mutants also showed defects in biofilm formation compared to the wild-type GcbC, again with the D484R mutant showing the largest effect (Fig. 4B). We then made nonconservative substitutions on GcbC’s α5GGDEF helix and tested the mutant proteins for interaction with LapD via B2H. Four of the mutant variants showed a significant decrease in interaction with LapD, with the D484R mutant showing the largest effect (Fig. 4C).
We then constructed several compensatory mutants of LapD that we predicted might restore the functional interaction. The constructs were tested for biofilm formation in a Δ4DGC genetic background in which the lapD gene had also been deleted. We observed three examples of a LapD compensatory mutation suppressing the biofilm-deficient phenotype of a GcbC mutant (Fig. 4D). The LapD(R463E) mutation showed a very modest and not significant suppression of the GcbC(E477R) allele. The LapD(W467E) mutation showed significant suppression of the GcbC(F481K) allele, and the LapD(E469K) mutation showed a small but significant suppression of the GcbC(K485E) allele. Finally, the LapD(R470D) mutation also showed significant suppression of the GcbC(D484R) allele.
To show that the compensatory mutations discussed in this section were acting in an allele-specific manner, we matched several LapD mutants with GcbC mutants in a manner in which we would not expect restoration to occur (i.e., negatively charged residues paired with negatively charged residues or residues which we would expect to be too far apart to interact). We found that these mismatched mutants failed to restore biofilm formation and that the LapD helix must be mutated in an allele-specific manner to restore biofilm to mutants on the α5GGDEF helix of GcbC (see Fig. S2D in the supplemental material).
We next confirmed that the biofilm restoration seen in Fig. 4D was in fact due to a restoration of interaction between GcbC and LapD. To this end, we used the same constructs from Fig. 4B and conducted coprecipitation experiments. We observed interaction defects with the point mutations F481K, K485E, and D484R in the GcbC helix (Fig. 4E) and demonstrated partial to full rescue of interaction using the corresponding point mutations W467E, E469K, and R470D, respectively, in the LapD helix (Fig. 4E). From these results, we conclude that the LapD α2EAL helix can recognize the α5GGDEF helix of GcbC, and together these α-helices form part of the interaction interface of GcbC and LapD (Fig. 4A).
The DGC α5GGDEF helix is conserved in other species and can be used to predict LapD interaction partners.
We next explored whether this interaction model might have predictive power; that is, could we use the α5GGDEF helix sequences from DGCs to determine possible partners for LapD proteins in other species? To test this idea in a different species, we turned to P. aeruginosa, which has a LapD homolog with 62% identity and 75% similarity to P. fluorescens LapD. Importantly, the α2EAL helix in the P. aeruginosa LapD is nearly identical to the P. fluorescens LapD (Fig. 5A; see also Fig. S3 and Table S3 in the supplemental material). We originally searched for a GcbC homolog in P. aeruginosa, but no homolog was found. We reasoned that if this helical interaction is a general feature of DGC signaling in the Lap system and if the α5GGDEF helix is conserved as a secondary structure in most DGCs, then it may be possible to identify the DGC interacting with the P. aeruginosa LapD by examining each of the α5GGDEF helices of every DGC in P. aeruginosa to find one which matches the GcbC α5GGDEF helix from P. fluorescens.
FIG 5 .
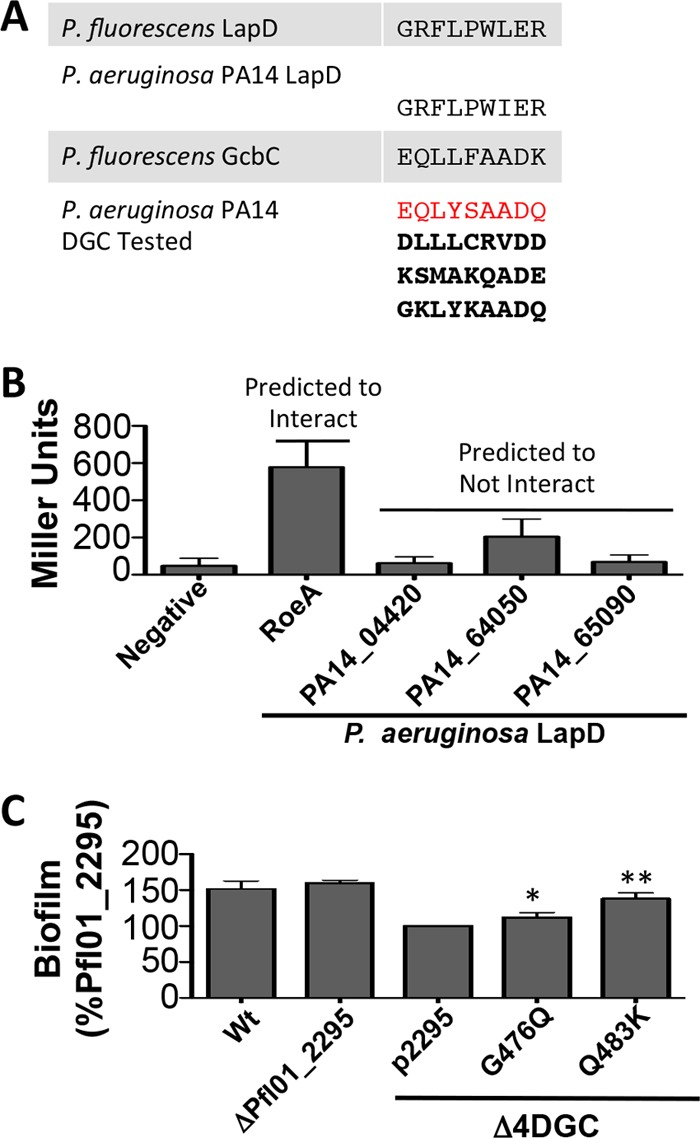
The N-terminal residues of the α5GGDEF helix of GGDEF domains can be predictive of DGC function. (A) The residues of the P. fluorescens and P. aeruginosa PA14 LapD α2EAL helix are shown, along with the N-terminal α5GGDEF residues of GcbC and potential P. aeruginosa PA14 DGC enzymes predicted to interact and not interact with P. aeruginosa PA14 LapD. The sequence highlighted in red showed the highest match out of the 28 potential sequences of P. aeruginosa examined, while the three remaining boldface sequences are among the lowest predicted matches. See Table S3 in the supplemental material for the full list of DGCs and sequences examined. (B) The P. aeruginosa PA14 DGCs from panel A were tested by B2H for interaction with P. aeruginosa PA14 LapD. BTH101 E. coli cells were grown on selective plates for 24 h before being collected by scraping. β-Galactosidase activity is expressed as Miller units. The data show the standard deviations from 4 biological replicates that were generated from 3 technical replicates per biological replicate. (C) The DGC Pfl01_2295 is partially “rewired” with residues from GcbC’s α5GGDEF helix, allowing an increased ability to promote biofilm formation. Biofilm formation was assayed after 6 h in a microtiter dish. Protein expression was induced with 0.05% l-arabinose. From left to right, wild-type (Wt) P. fluorescens; Pfl01_2295 deletion strain, the Δ4DGC strain lacking the DGCs needed to make a biofilm; Pfl01_2295 expressed on a plasmid; GcbC-like mutation G476Q; and GcbC-like mutation Q483K. All Pfl01_2295 constructs were expressed in the Δ4DGC background. *, P < 0.05; **, P < 0.005 (compared to the strain carrying the wild-type Pfl01_2295 on a plasmid). Error bars show standard deviations from 3 biological replicates which were generated from 8 technical replicates per biological replicate. Biofilm formation was normalized to the Pfl01_2295-expressing strain and blanked to the Δ4DGC strain and is displayed as percent biofilm induction relative to Pfl01_2295. See also Fig. S4 in the supplemental material.
We next surveyed all such DGC helices in the c-di-GMP network of P. aeruginosa. Table S3 in the supplemental material shows the residues from the N-terminal portion of the α5GGDEF helix in GcbC from P. fluorescens and each of the matching residues from the 28 GGDEF domain-containing proteins in P. aeruginosa. Among these DGCs in P. aeruginosa, PA14_50060, also known as RoeA, appeared to be the closest match to P. fluorescens GcbC (Fig. 5A). RoeA is a predicted inner membrane DGC with a GGDEF cyclase domain, and deleting RoeA has previously been shown to reduce biofilm formation and increase swarming in P. aeruginosa via its impact on the production of the Pel exopolysaccharide (22).
To test if RoeA interacts with P. aeruginosa LapD, we cloned both genes into the B2H vectors. Additionally, we selected three more DGCs that we would predict not to interact with LapD based on their α5GGDEF helices—PA14_04420, PA14_64050, and PA14_65090. These proteins include one predicted transmembrane DGC and two predicted cytoplasmic DGCs of various domain architectures. When tested by B2H, RoeA interacted with the P. aeruginosa LapD, while the three negative-control DGCs failed to interact (Fig. 5B). We conclude that the P. aeruginosa LapD utilizes its α2EAL helix in the same manner as does the P. fluorescens LapD as one interface surface to contact the α5GGDEF helix of RoeA.
The GcbC α5GGDEF helix contributes to target protein specificity.
Our final analysis of the α5GGDEF helix on GcbC was to determine its possible sufficiency for specifying a target protein. We hypothesized that if this helix was sufficient to specify a given effector protein, transplanting residues from one DGC to another might alter its interaction partners or function. To this end, we transplanted the entire GcbC N-terminal portion of the α5GGDEF helix examined in this study onto several unrelated P. fluorescens DGCs that do not interact with LapD or contribute toward biofilm formation. In no case did the hybrid protein perform better at interacting with LapD or forming a biofilm than the unmodified protein (data not shown).
Alternatively, to determine if the GcbC α5GGDEF helix can have any impact on rewiring another DGC, we decided to utilize a DGC that already shared substantial domain-level architecture with GcbC and substitute the GcbC residues one at a time onto this DGC to assess any impact on its ability to govern biofilm formation. That is, we hypothesized that under a model where the α5GGDEF helix is one of several important factors for interaction, residues on this helix would need to be paired with additional structural elements to function properly.
We selected the inner membrane DGC Pfl01_2295 due to its nearly identical domain architecture compared to GcbC but unrelated α5GGDEF helix (see Fig. S4A in the supplemental material). Under our standard assay conditions, deletion of the Pfl01_2295 gene has no impact on biofilm formation, and overexpression from a plasmid only partially restores biofilm formation to a strain lacking its native biofilm subnetwork (Fig. 5C). Since transplantations of the entire α5GGDEF helix region did not yield DGCs that could better signal to LapD, we attempted to rewire Pfl01_2295 by replacing amino acids on its α5GGDEF helix one at a time with the analogous residue from GcbC and looked for substitutions that showed biofilm restoration. Of the four surface-facing residues on the N-terminal portion of the GcbC α5GGDEF helix not shared by Pfl01_2295, two substitutions (G476Q and Q483K) resulted in a more “GcbC-like” α5GGDEF helix and significantly increased biofilm formation when transplanted onto Pfl01_2295 (Fig. 5C). To be sure that an unexpected increase of c-di-GMP was not responsible for the increased biofilm formation, we assessed c-di-GMP production via Congo red binding in P. aeruginosa, as well as by mass spectrometry in P. fluorescens, and we found no difference among the mutants and no pattern or trend that could explain the increased biofilm of the two α5GGDEF helix substitutions (see Fig. S4B).
DISCUSSION
How bacterial cells with many DGCs curate the outputs of these signaling proteins has been an open question in microbiology. Beyond biofilm formation in P. fluorescens and P. aeruginosa, specificity in DGC signaling has been observed in many other organisms. In Caulobacter crescentus, specificity in c-di-GMP signaling beyond what can be achieved with the global c-di-GMP pool has been linked to cell fate determination (23). Further evidence from C. crescentus suggests that the DGC PleD can contribute to this process by being activated only once it is sequestered at the poles, providing one possible mechanism that can drive spatial and temporal signaling specificity (24). Spatial sequestration upon c-di-GMP binding can also play a role in effector output, as in the example of SgmT in Myxococcus xanthus (25). In Vibrio cholerae, several DGCs have been found which promote biofilm formation where specificity is partially achieved in response to low temperature (26). In a particularly exquisite example of DGC specificity, the DGC YdaM in Escherichia coli specifically activates the transcription factor MlrA but only after it is itself derepressed by c-di-GMP signaling from another DGC (10). Notably, physical interaction was explicitly invoked in this example to help explain specificity of signaling.
We present a model of DGC-effector signaling that relies on protein-protein interaction to achieve localized c-di-GMP effects by minimizing cross talk to the rest of the network. We demonstrate that GcbC localizes to the inner membrane where LapD is found and that they physically interact with one another. We further demonstrate that this physical interaction is necessary for full signaling between GcbC and LapD, providing an example of the concept that c-di-GMP networks are ordered and regulated by physical proximity between c-di-GMP reading effectors and their DGC and PDE partners. Surprisingly, we find that this interaction endures nearly unperturbed regardless of the DGC’s ability to make c-di-GMP, the effector’s ability to bind the dinucleotide, or the regulatory state of the effector. This finding suggests that LapD and GcbC may often be associated with each other and that a change in environment that promotes biofilm formation may be rapidly communicated from GcbC to LapD via a preformed complex.
We put forward the GcbC α5GGDEF helix as an element contributing to the interaction between GcbC and LapD. We further postulate that the α2EAL helix of LapD is necessary for full signaling and that this helix forms part of the interaction surface with GcbC (Fig. 6). The interaction between GcbC and LapD in P. fluorescens described here provides a framework to begin to understand specific DGC outputs with EAL domain-containing proteins. As defects for a given mutation in GcbC are often modest, and as mutating all residues in this region does not result in total loss of interaction, we conclude that the α5GGDEF helix likely acts as a fine-tuning mechanism for the GcbC-LapD interaction. We also note that residue D484 may play a role in the product-bound state of GcbC, as this widely conserved residue was argued to be involved in a novel DGC-inhibited state in X. campestris in a recent publication (21), possibly explaining the residue’s requirement for the general function of GcbC in both its biofilm and swim regulatory roles. However, because D484R can be partially compensated for in biofilm formation and interaction by a LapD R470D mutant, we conclude that D484R is also involved in the GcbC-LapD interaction. We also conclude that the remaining residues from the GcbC α5GGDEF helix impact only the DGC’s signaling specificity to LapD, as GcbC’s regulation of swim motility was not affected when the remaining four surface residues were replaced with alanines.
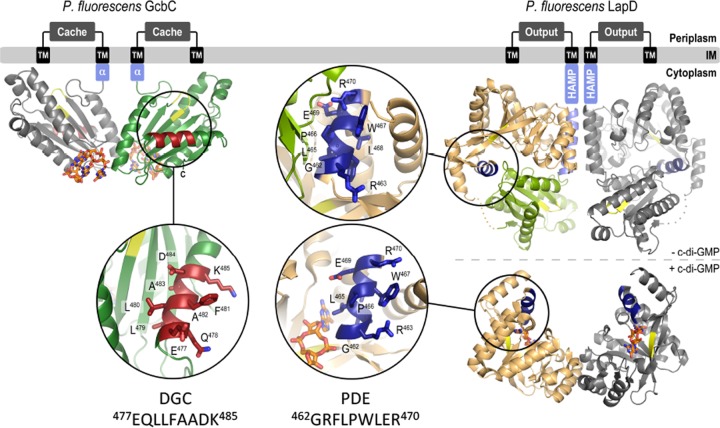
FIG 6 .
Model for GcbC-LapD interactions. (Left) GcbC bound to c-di-GMP in inactive dimer. The inset shows the α5GGDEF helix (residues 477 to 485) in red. (Right) LapD in its unbound state (above) and c-di-GMP-bound, active state (below). Insets show the α2EAL helix of LapD in dark blue. The helix is exposed in the c-di-GMP-bound state of LapD.
We further provide evidence that this helical interaction between LapD and a DGC is not limited to a single species of bacteria but can also be found in the LapD-RoeA interaction of P. aeruginosa, demonstrating that these helices may be predictive of interaction in some cases and could help identify interaction nodes in c-di-GMP networks that make use of the Lap system. Interestingly, we recently demonstrated that the LapD-LapG system of P. aeruginosa is required to regulate the proteolytic processing of the cell surface protein CdrA (27). CdrA was shown previously to bind an exopolysaccharide produced by P. aeruginosa (28); it is therefore intriguing that in this microbe RoeA, which has been shown to be required for exopolysaccharide production (22), interacts with LapD.
It is also interesting to consider the possibilities of what α5GGDEF helices from other GGDEF domain-containing proteins and α2EAL helices from other EAL domain-containing proteins may be responsible for beyond signaling of LapD homologs. P. fluorescens harbors an additional 36 GGDEF domain-containing proteins besides GcbC. It will merit analysis as to whether the α5GGDEF helix in these other DGCs may also be involved in DGC-effector interactions. In addition, this model may aid in understanding specificity between and among DGC and PDE enzymes, which can also include interactions between GGDEF and EAL domains. Additionally, we note that many PilZ domains contain an α-helix near their c-di-GMP binding site that could be involved in interaction with GGDEF or EAL domains. Intriguingly, a recent publication (29) describes a crystal structure of a PilZ domain interacting with the α2EAL helix of an EAL domain (see Fig. S4C in the supplemental material), suggesting that the helices described here may be used broadly as interaction elements among components of c-di-GMP networks.
It bears noting that many DGCs, PDEs, and effectors are dual-domain proteins, containing both a GGDEF and an EAL domain. Dual-domain proteins would be expected to contain both an α5GGDEF helix and an α2EAL helix, and depending on the state of the protein, each could be masked or available. As previously mentioned, LapD is thought to undergo a large conformational shift upon activation by c-di-GMP binding, allowing for differential exposure of such regulatory sites (Fig. 6). Whether these helices could help modulate intramolecular interactions or whether they could allow for multiple DGCs, PDEs, or effectors to form multiprotein complexes remains an open question.
Our final observation regarding the residues found on the N-terminal portion of the α5GGDEF helix is that they can be predictive of function in some cases. By examining the said residues of GGDEF domains in P. aeruginosa, we were able to identify a DGC that interacts with the P. aeruginosa LapD and is known to help regulate biofilm formation. Further, we could similarly identify several DGCs of various architectures and cellular locations that do not interact with P. aeruginosa LapD. Whether or not this structure can generally be used to predict interaction partners will have to be verified using effectors beyond LapD homologs. Additionally, the predictive value of the primary sequence of a given α5GGDEF helix may be limited when the configuration of these surface-exposed side chains is unrestricted in three dimensions, making it difficult to guess among many possible composite surfaces. Our experiments with Pfl01_2295 further highlighted how these residues can be predictive of function, as an unrelated DGC was able to partially promote biofilm formation when rewiring its α5GGDEF helix to look more GcbC-like. There is a precedent for rewiring bacterial proteins to change their interaction and hence function (30), though in the case of these DGCs it appears that physical association may depend on more than just a single helix-helix interaction and may require other, unidentified structural elements. Furthermore, not every residue substitution that we initially tried resulted in increased biofilm formation. This finding may indicate that some substitutions increase this DGC’s affinity for certain PDEs that normally regulate GcbC, or it may mean that all interaction surfaces on a given DGC work synergistically and the substitution of one interaction surface cannot make up for another. Further, the finding that Pfl01_2295 could not fully replace GcbC may indicate that this DGC has an activation signal different from GcbC or that there are other surfaces on Pfl01_2295 that disallow undesired interaction partners.
While physical interaction between DGCs and effectors helps explain a fundamental gap in the field, our model has several implications and limitations that must be addressed. One difficulty that we encountered in our study was identifying phenotypes for LapD mutations. While the location of the LapD interaction surface was originally inferred from a clustering of mutations in our B2H screen, no single mutation showed a dramatic defect for interaction of LapD with GcbC as assessed by B2H. There are several possible explanations for this finding. Our previous work has shown that LapD is dynamic in its physical state (Fig. 6), and its GGDEF and EAL domains come into contact when LapD is inactive and move apart when LapD is activated (12). It is possible that α2EAL helix mutations disrupt the inactive state of LapD, causing this receptor protein to readily adopt its active conformation, which could promote biofilm formation and/or interaction with GcbC. Alternatively, some of the mutations found in the B2H screen of LapD may make unexpectedly large disruptions to the entire α2EAL helix and will require crystallographic studies to precisely identify how the region’s structure is impacted. Finally, some DGC and PDE proteins from the large c-di-GMP network in P. aeruginosa have been demonstrated to be involved in global signaling, where cell-wide levels of c-di-GMP are impacted by the function of specific enzymes (31). It is unknown how a global c-di-GMP signal might be integrated with the local signaling that we put forth here, nor is it immediately apparent how DGC/PDE-effector complexes might insulate themselves from or be influenced by global c-di-GMP levels.
While physical interaction between DGCs and effectors provides a framework to understand specificity in c-di-GMP signaling, these findings leave open the question of how proximity can tame a network of dozens of DGCs, which may simultaneously produce the same small, diffusible molecule. Further study is merited to determine how often physical interaction is required for a DGC to signal its effector, the totality of interaction surfaces that DGCs use in such cases, and the frequency with which the α5GGDEF helix is utilized in other DGCs or the α2EAL helix is utilized by PDEs and effectors to contact other members of a bacterium’s c-di-GMP network.
MATERIALS AND METHODS
Strains and media.
P. fluorescens Pfl01, P. aeruginosa PA14, and E. coli S17-1 and BTH101 were used throughout the study (see Table S4 in the supplemental material). Bacteria were grown in liquid LB medium or on 1.5% agar LB plates. P. fluorescens was grown at 30°C, while E. coli was grown at 37°C. The vector pMQ72 (see Table S4) was used for expression in P. fluorescens and induced at 0.1% arabinose. See Text S1 in the supplemental material for additional details and Table S5 for the primers used to build all constructs and mutants used in this study.
Bacterial two-hybrid assay.
BTH101 E. coli cells were cotransformed by electroporation with 50 ng of each vector and allowed to recover in 1 ml of LB for 1 h at 37°C based on a previously described system (32). The culture was then diluted 1:200 in fresh LB, and 50 µl of the diluent was plated onto LB plates containing kanamycin (Kn), carbenicillin (Cb), and IPTG (isopropyl-β-d-thiogalactopyranoside). Cells were scraped from the plates after 24 h and assayed for β-galactosidase activity as described in Text S1 in the supplemental material.
Coprecipitations.
Subcultured cells were lysed with a French press. Whole-cell lysate was normalized to protein concentration, and 1.8 mg of the lysate was added to 40 ml of cobalt resin. The coprecipitation buffer contained 10 mM MgCl2, 20 mM Tris buffer (pH 8), 0.8% Thesit, and 5 mM imidazole. The lysate and resin mixtures were rotated at 4°C for 25 min, followed by centrifugation. The supernatant was removed, and samples were washed with the coprecipitation buffer once for 15 min at 4°C, followed by 3 washes for 5 min at room temperature. Samples were treated with 60 ml of 10% 2-mercaptoethanol in SDS and boiled for 5 min prior to analysis via gel electrophoresis and blotting as described in Text S1 in the supplemental material.
Biofilm assay.
The biofilm assay was conducted as described previously (33). Briefly, overnight cultures were grown from single colonies. Cultures were normalized to optical density, and 1.5 ml was added to 100 µl of K10T-1 in a 96-well polystyrene plate (Costar). Plates were incubated at 30°C for 6 h in a humid chamber. The liquid from the wells was discarded, and the wells were stained with 125 µl of 0.1% crystal violet for 20 min at room temperature. The plate was subsequently subjected to two rinses with water and then was dried overnight. Biofilms were quantified by destaining the wells with 150 µl of a solution of 30% methanol and 10% acetic acid for 20 min. One hundred twenty-five microliters of the destained biofilm suspension from each well was transferred to a flat-bottom 96-well plate and quantified as optical density at 550 nm (OD550).
Swim assay.
P. fluorescens Pfl01 strains were grown up in selective LB overnight. One-hundred-microliter aliquots were pipetted into a 96-well dish, and a 200-µl pipette tip was dipped into each well and then plunged into 0.35% K10 swim agar plates supplemented with 0.1% arabinose. Plates were photographed at 36 h, and swim area was analyzed with ImageJ software.
Congo red assessment of diguanylate cyclase activity.
Variants of DGCs were expressed from the arabinose-inducible promoter carried on plasmid pMQ72 to assess their cyclase activity. These variants were transformed into P. aeruginosa PA14, and 2.5 µl of overnight cultures was plated on Congo red plates with 0.1% arabinose to induce expression of the DGC.
Quantification of c-di-GMP levels.
Strains of P. fluorescens were grown overnight in LB selective medium. Seventy-five microliters of overnight culture was added to 5 ml of K10 medium supplemented with arabinose as indicated in the text. Strains were grown for 6 h, pelleted, and resuspended in 250 µl of extraction buffer (40% methanol, 40% acetonitrile, 20% double-distilled water [ddH2O] plus 1.M formic acid) and incubated at −20°C for 1 h. Samples were repelleted, and 200 µl of the extraction was added to 8 µl of 15% NH4HCO3. Samples were analyzed by liquid chromatography-mass spectrometry (LC-MS) and compared to a standard curve of known c-di-GMP concentration, and results were normalized to the dry weight of the bacterial pellets that generated the extractions.
Crystallization, data collection, and structure solution.
Protein crystals were obtained for GcbCGGDEF bound to c-di-GMP by hanging-drop vapor diffusion, mixing equal volumes (1 µl) of protein at concentrations of 10 to 30 mg/ml and reservoir solution, followed by incubation at 20°C. The reservoir solution consisted of 1.0 M ammonium sulfate, 0.1 M HEPES (pH 7.0), and 0.5% (wt/vol) polyethylene glycol 8000 (PEG8000). Crystals were cryoprotected by soaking them in their respective reservoir solutions supplemented with 25% xylitol, flash frozen, and stored in liquid nitrogen. Data were collected on frozen crystals at 100 K on beamline A1 at the Cornell High-Energy Synchrotron Source (CHESS; Cornell University, Ithaca, NY). See Text S1 in the supplemental material for analysis details.
Protein structure accession numbers.
Coordinates and structure factors for GcbC have been deposited in the Protein Data Bank (PDB) and assigned accession number 5EUH.
SUPPLEMENTAL MATERIAL
LapD mutants identified by B2H. (A) The structure of the GGDEF domain of GcbC is shown with three clusters of mutations found in a screen to limit interaction with LapD. The residues shown in purple likely impact active dimerization or protein stability, residues in cyan likely impact the catalytic activity of GcbC, and residues in red map to the N terminus of the α5GGDEF helix of GcbC. See also Table S1. (B) The LapD EAL domain shows clustering of mutants that impact interaction. Mutations in red likely impact the ability of the α2EAL helix to recognize GcbC, mutations in cyan likely impact the ability of LapD to bind c-di-GMP, and mutations in purple may affect the structural stability of LapD but do not appear to be part of the interaction surface. Mutations that were found on the same plasmid as a mutation from one of the clusters have been removed for simplicity of displaying the data, but all mutations identified are listed in Table S1. Mutations found in the GGDEF domain of LapD in the B2H screen map randomly throughout the domain. The most common mutation found was introduction of proline residues, likely destabilizing large sections of the domain; such mutations likely do not directly impact recognition of GcbC by LapD. Download
Characterization of GcbC and LapD mutations. (A) The GcbC Quad-Ala mutation is not dominant negative. The plasmid containing GcbC Quad-Ala was electroporated into a strain of P. fluorescens that retains its native copy of GcbC on the chromosome but lacks the other three DGCs necessary for biofilm formation (Δ3DGC). Biofilm formation was quantified at 550 nm after 6 h of incubation in a microtiter plate assay. The strain was compared to the Δ3DGC strain alone and the Δ4DGC + GcbC Quad-Ala strain. Error bars show standard deviations from 3 biological replicates which were generated from 8 technical replicates per biological replicate. Strains were normalized to the wild-type strain, and data are displayed as percent biofilm induction relative to the wild-type strain. (B) A LapD α2EAL helix mutation is defective for biofilm formation. Mutations in the LapD helix were introduced into the chromosomal copy of the lapD gene. Biofilm formation was quantified at 550 nm after 6 h of incubation in a microtiter plate assay. **, P < 0.01 compared to the control strain lacking the 4DGC subnetwork and expressing GcbC on a plasmid. Error bars show standard deviations from 3 biological replicates which were generated from 8 technical replicates per biological replicate. Strains were normalized to the wild-type strain, and data are displayed as percent biofilm induction relative to the wild-type strain. (C) Single α2EAL helix mutations demonstrate no significant reduction in interaction between LapD and GcbC. One mutation caused an increase in interaction but led to a decrease in biofilm formation as seen in panel A. *, P < 0.05 compared to the wild-type GcbC-LapD interaction. Error bars show standard deviations from 3 biological replicates which were generated from 3 technical replicates per biological replicate. (D) Matching GcbC α5GGDEF helix defects with the incorrect compensatory mutant on LapD’s α2EAL helix does not result in biofilm suppression, demonstrating that the compensatory mutants in Fig. 3C are likely allele specific in their suppression of biofilm defects. Hatched bars on the right represent addition of 0.1% arabinose to keep the strain carrying the GcbC D484R mutation in the linear range of the assay. Error bars show standard deviations from 3 biological replicates which were generated from 8 technical replicates per biological replicate. Download
The N terminus of the α5GGDEF helix can be used to predict interaction between DGCs and related effectors. The domain architecture and α2EAL helices for P. fluorescens and P. aeruginosa LapD are shown along with the domain architecture and α5GGDEF helices for GcbC and RoeA. Due to the resemblance of the RoeA helix to the GcbC helix, RoeA was predicted to interact with P. aeruginosa PA14 LapD. Transmembrane domains (TM) are depicted in light blue, and the other domains are labeled in the figure. Download
The α5GGDEF helix of a DGC can partially impact function. (A) The domain architectures for GcbC and Pfl01_2295 are shown. Transmembrane domains (TM) are depicted in light blue, and the other domains are labeled in the figure. The N terminus of the α5GGDEF helix for each is shown on the right. The α5GGDEF helix of Pfl01_2295 was modified to become more GcbC-like and used to test biofilm formation in Fig. 5C. (B) (Top) Pfl01_2295 and its mutant variants were transformed on a plasmid into Pseudomonas aeruginosa PA14 and analyzed on Congo red plates with 0.1% arabinose as described in Materials and Methods. (Bottom) The same constructs as in panel B along with control strains were induced with 0.1% arabinose in Pseudomonas fluorescens, and extracts were analyzed by LC-MS to quantify c-di-GMP production as described in Materials and Methods. Error bars represent standard deviation and data were obtained from three subcultured replicates. (C) Xanthomonas axonopodis EAL domain of FimX interacting with PilZ. The interaction is partially mediated by the α2EAL helix, which is the same helix used by LapD to contact GcbC (dark blue, PDB 4FOU) (29). Download
GcbC and LapD mutants isolated in this study using the B2H screen. Shown is a complete list of the amino acid changes in GcbC and LapD associated with the mutants isolated as a consequence of the bacterial 2-hybrid (B2H) screen described in the text.
Crystallographic data collection and refinement for GcbC. Shown is a list of the relevant crystallographic data for the structure of GcbC.
Candidate α5GGDEF helices from PA14 for interaction with LapD. Shown is a complete list of the sequences of the α5 of all GGDEF motif-containing proteins in P. aeruginosa PA14.
Strains and plasmids used in this study. Shown is a complete list of strains used, their genotypes, and whether they were commercially available (with company name), generated during previous studies (with the appropriate reference) or built during the course of this work (listed as “This Study”). All plasmids are also listed, as is their source as outlined above for strains.
Primers used in this study. Shown is a complete list of primers used in this work, the primer sequences, and related information.
This document contains additional details of the methods used in this work, as well as the appropriate citations describing previously published methods. Download
ACKNOWLEDGMENTS
This work was supported by grant R01-AI097307 (H.S./G.A.O.), by a Fleming Fellowship (K.M.G.), and by grant T32-GM08704 (K.M.D.) and by the NSF grant MCB-9984521 (G.A.O.). Part of this work is based upon research conducted at the Cornell High Energy Synchrotron Source (CHESS), which is supported by the National Science Foundation and the National Institutes of Health/National Institute of General Medical Sciences under NSF award DMR-1332208, using the Macromolecular Diffraction at CHESS (MacCHESS) facility, which is supported by award GM-103485 from the National Institute of General Medical Sciences, National Institutes of Health.
We declare no conflicts of interest.
K.M.D., K.M.G., A.J.C., H.S., and G.A.O. conceived and designed the experiments. K.M.D., K.M.G., and A.J.C. performed the experiments. K.M.D. and K.M.G. analyzed the data. H.S. contributed reagents, materials, and analysis tools. K.M.D., K.M.G., H.S., and G.A.O. wrote the paper.
Footnotes
Citation Dahlstrom KM, Giglio KM, Collins AJ, Sondermann H, O’Toole GA. 2015. Contribution of physical interactions to signaling specificity between a diguanylate cyclase and its effector. mBio 6(6):e01978-15. doi:10.1128/mBio.01978-15.
REFERENCES
- 1.Tamayo R, Pratt JT, Camilli A. 2007. Roles of cyclic diguanylate in the regulation of bacterial pathogenesis. Annu Rev Microbiol 61:131–148. doi: 10.1146/annurev.micro.61.080706.093426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Srivastava D, Hsieh M, Khataokar A, Neiditch MB, Waters CM. 2013. Cyclic di-GMP inhibits Vibrio cholerae motility by repressing induction of transcription and inducing extracellular polysaccharide production. Mol Microbiol 90:1262–1276. doi: 10.1111/mmi.12432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martinez-Gil M, Ramos-Gonzalez MI, Espinosa-Urgel M. 2014. Roles of cyclic di-GMP and the Gac system in transcriptional control of the genes coding for the Pseudomonas putida adhesins LapA and LapF. J Bacteriol 196:1484–1495. doi: 10.1128/JB.01287-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Romling U, Galperin MY, Gomelsky M. 2013. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev 77:1–52. doi: 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kulasakara H, Lee V, Brencic A, Liberati N, Urbach J, Miyata S, Lee DG, Neely AN, Hyodo M, Hayakawa Y, Ausubel FM, Lory S. 2006. Analysis of Pseudomonas aeruginosa diguanylate cyclases and phosphodiesterases reveals a role for bis-(3′-5′)-cyclic-GMP in virulence. Proc Natl Acad Sci U S A 103:2839–2844. doi: 10.1073/pnas.0511090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tischler AD, Camilli A. 2004. Cyclic diguanylate (c-di-GMP) regulates Vibrio cholerae biofilm formation. Mol Microbiol 53:857–869. doi: 10.1111/j.1365-2958.2004.04155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ha D, Richman ME, O’Toole GA. 2014. Deletion mutant library for investigation of functional outputs of cyclic diguanylate metabolism in Pseudomonas aeruginosa PA14. Appl Environ Microbiol 80:3384–3393. doi: 10.1128/AEM.00299-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Newell PD, Yoshioka S, Hvorecny KL, Monds RD, O’Toole GA. 2011. Systematic analysis of diguanylate cyclases that promote biofilm formation by Pseudomonas fluorescens Pf0-1. J Bacteriol 193:4685–4698. doi: 10.1128/JB.05483-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bobrov AG, Kirillina O, Forman S, Mack D, Perry RD. 2008. Insights into Yersinia pestis biofilm development: topology and co-interaction of Hms inner membrane proteins involved in exopolysaccharide production. Environ Microbiol 10:1419–1432. doi: 10.1111/j.1462-2920.2007.01554.x. [DOI] [PubMed] [Google Scholar]
- 10.Lindenberg S, Klauck G, Pesavento C, Klauck E, Hengge R. 2013. The EAL domain protein YciR acts as a trigger enzyme in a c-di-GMP signalling cascade in E. coli biofilm control. EMBO J 32:2001–2014. doi: 10.1038/emboj.2013.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schirmer T, Jenal U. 2009. Structural and mechanistic determinants of c-di-GMP signalling. Nat Rev Microbiol 7:724–735. doi: 10.1038/nrmicro2203. [DOI] [PubMed] [Google Scholar]
- 12.Navarro MVAS, Newell PD, Krasteva PV, Chatterjee D, Madden DR, O’Toole GA, Sondermann H. 2011. Structural basis for c-di-GMP-mediated inside-out signaling controlling periplasmic proteolysis. PLoS Biol 9:e1000588. doi: 10.1371/journal.pbio.1000588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qi Y, Chuah MLC, Dong X, Xie K, Luo Z, Tang K, Liang Z. 2011. Binding of cyclic diguanylate in the non-catalytic EAL domain of FimX induces a long-range conformational change. J Biol Chem 286:2910–2917. doi: 10.1074/jbc.M110.196220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davis NJ, Cohen Y, Sanselicio S, Fumeaux C, Ozaki S, Luciano J, Guerrero-Ferreira RC, Wright ER, Jenal U, Viollier PH. 2013. De- and repolarization mechanism of flagellar morphogenesis during a bacterial cell cycle. Genes Dev 27:2049–2062. doi: 10.1101/gad.222679.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Newell PD, Monds RD, O’Toole GA. 2009. LapD is a bis-(3′,5′)-cyclic dimeric GMP-binding protein that regulates surface attachment by Pseudomonas fluorescens Pf0-1. Proc Natl Acad Sci U S A 106:3461–3466. doi: 10.1073/pnas.0808933106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Newell PD, Boyd CD, Sondermann H, O’Toole GA. 2011. A c-di-GMP effector system controls cell adhesion by inside-out signaling and surface protein cleavage. PLoS Biol 9:e1000587. doi: 10.1371/journal.pbio.1000587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chatterjee D, Cooley RB, Boyd CD, Mehl RA, O’Toole GA, Sondermann H. 2014. Mechanistic insight into the conserved allosteric regulation of periplasmic proteolysis by the signaling molecule cyclic-di-GMP. Elife 3:e03650. doi: 10.7554/eLife.03650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hinsa SM, O’Toole GA. 2006. Biofilm formation by Pseudomonas fluorescens WCS365: a role for LapD. Microbiology 152:1375–1383. doi: 10.1099/mic.0.28696-0. [DOI] [PubMed] [Google Scholar]
- 19.Ballok AE, Bahl CD, Dolben EL, Lindsay AK, St. Laurent JD, Hogan DA, Madden DR, O’Toole GA. 2012. Epoxide-mediated CifR repression of cif gene expression utilizes two binding sites in Pseudomonas aeruginosa. J Bacteriol 194:5315–5324. doi: 10.1128/JB.00984-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caiazza NC, O’Toole GA. 2004. SadB is required for the transition from reversible to irreversible attachment during biofilm formation by Pseudomonas aeruginosa PA14. J Bacteriol 186:4476–4485. doi: 10.1128/JB.186.14.4476-4485.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang C, Chin K, Chuah ML, Liang Z, Wang AH, Chou S. 2011. The structure and inhibition of a GGDEF diguanylate cyclase complexed with (c-di-GMP)(2) at the active site. Acta Crystallogr D Biol Crystallogr 67:997–1008. doi: 10.1107/S090744491104039X. [DOI] [PubMed] [Google Scholar]
- 22.Merritt JH, Ha D, Cowles KN, Lu W, Morales DK, Rabinowitz J, Gitai Z, O’Toole GA. 2010. Specific control of Pseudomonas aeruginosa surface-associated behaviors by two c-di-GMP diguanylate cyclases. mBio 1:e00183-10. doi: 10.1128/mBio.00183-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abel S, Bucher T, Nicollier M, Hug I, Kaever V, Abel Zur Wiesch P, Jenal U. 2013. Bi-modal distribution of the second messenger c-di-GMP controls cell fate and asymmetry during the Caulobacter cell cycle. PLoS Genet 9:e1003744. doi: 10.1371/journal.pgen.1003744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paul R, Weiser S, Amiot NC, Chan C, Schirmer T, Giese B, Jenal U. 2004. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev 18:715–727. doi: 10.1101/gad.289504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petters T, Zhang X, Nesper J, Treuner-Lange A, Gomez-Santos N, Hoppert M, Jenal U, Søgaard-Andersen L. 2012. The orphan histidine protein kinase SgmT is a c-di-GMP receptor and regulates composition of the extracellular matrix together with the orphan DNA binding response regulator DigR in Myxococcus xanthus. Mol Microbiol 84:147–165. doi: 10.1111/j.1365-2958.2012.08015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Townsley L, Yildiz FH. 2015. Temperature affects c-di-GMP signalling and biofilm formation in Vibrio cholerae. Environ Microbiol doi: 10.1111/1462-2920.12799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cooley RB, Smith TJ, Leung W, Tierney V, Borlee BR, O’Toole GA, Sondermann H. 2015. Cyclic di-GMP-regulated periplasmic proteolysis of a Pseudomonas aeruginosa type Vb secretion system substrate. J Bacteriol doi: 10.1128/JB.00369-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Borlee BR, Goldman AD, Murakami K, Samudrala R, Wozniak DJ, Parsek MR. 2010. Pseudomonas aeruginosa uses a cyclic-di-GMP-regulated adhesion to reinforce the biofilm extracellular matrix. Mol Microbiol 75:827–842. doi: 10.1111/j.1365-2958.2009.06991.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guzzo CR, Dunger G, Salinas RK, Farah CS. 2013. Structure of the PilZ-FimXEAL-c-di-GMP complex responsible for the regulation of bacterial type IV pilus biogenesis. J Mol Biol 425:2174–2197. doi: 10.1016/j.jmb.2013.03.021. [DOI] [PubMed] [Google Scholar]
- 30.Skerker JM, Perchuk BS, Siryaporn A, Lubin EA, Ashenberg O, Goulian M, Laub MT. 2008. Rewiring the specificity of two-component signal transduction systems. Cell 133:1043–1054. doi: 10.1016/j.cell.2008.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuchma SL, Brothers KM, Merritt JH, Liberati NT, Ausubel FM, O’Toole GA. 2007. BifA, a cyclic-di-GMP phosphodiesterase, inversely regulates biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J Bacteriol 189:8165–8178. doi: 10.1128/JB.00586-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karimova G, Pidoux J, Ullmann A, Ladant D. 1998. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc Natl Acad Sci U S A 95:5752–5756. doi: 10.1073/pnas.95.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O’Toole GA, Kolter R. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol Microbiol 28:449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
LapD mutants identified by B2H. (A) The structure of the GGDEF domain of GcbC is shown with three clusters of mutations found in a screen to limit interaction with LapD. The residues shown in purple likely impact active dimerization or protein stability, residues in cyan likely impact the catalytic activity of GcbC, and residues in red map to the N terminus of the α5GGDEF helix of GcbC. See also Table S1. (B) The LapD EAL domain shows clustering of mutants that impact interaction. Mutations in red likely impact the ability of the α2EAL helix to recognize GcbC, mutations in cyan likely impact the ability of LapD to bind c-di-GMP, and mutations in purple may affect the structural stability of LapD but do not appear to be part of the interaction surface. Mutations that were found on the same plasmid as a mutation from one of the clusters have been removed for simplicity of displaying the data, but all mutations identified are listed in Table S1. Mutations found in the GGDEF domain of LapD in the B2H screen map randomly throughout the domain. The most common mutation found was introduction of proline residues, likely destabilizing large sections of the domain; such mutations likely do not directly impact recognition of GcbC by LapD. Download
Characterization of GcbC and LapD mutations. (A) The GcbC Quad-Ala mutation is not dominant negative. The plasmid containing GcbC Quad-Ala was electroporated into a strain of P. fluorescens that retains its native copy of GcbC on the chromosome but lacks the other three DGCs necessary for biofilm formation (Δ3DGC). Biofilm formation was quantified at 550 nm after 6 h of incubation in a microtiter plate assay. The strain was compared to the Δ3DGC strain alone and the Δ4DGC + GcbC Quad-Ala strain. Error bars show standard deviations from 3 biological replicates which were generated from 8 technical replicates per biological replicate. Strains were normalized to the wild-type strain, and data are displayed as percent biofilm induction relative to the wild-type strain. (B) A LapD α2EAL helix mutation is defective for biofilm formation. Mutations in the LapD helix were introduced into the chromosomal copy of the lapD gene. Biofilm formation was quantified at 550 nm after 6 h of incubation in a microtiter plate assay. **, P < 0.01 compared to the control strain lacking the 4DGC subnetwork and expressing GcbC on a plasmid. Error bars show standard deviations from 3 biological replicates which were generated from 8 technical replicates per biological replicate. Strains were normalized to the wild-type strain, and data are displayed as percent biofilm induction relative to the wild-type strain. (C) Single α2EAL helix mutations demonstrate no significant reduction in interaction between LapD and GcbC. One mutation caused an increase in interaction but led to a decrease in biofilm formation as seen in panel A. *, P < 0.05 compared to the wild-type GcbC-LapD interaction. Error bars show standard deviations from 3 biological replicates which were generated from 3 technical replicates per biological replicate. (D) Matching GcbC α5GGDEF helix defects with the incorrect compensatory mutant on LapD’s α2EAL helix does not result in biofilm suppression, demonstrating that the compensatory mutants in Fig. 3C are likely allele specific in their suppression of biofilm defects. Hatched bars on the right represent addition of 0.1% arabinose to keep the strain carrying the GcbC D484R mutation in the linear range of the assay. Error bars show standard deviations from 3 biological replicates which were generated from 8 technical replicates per biological replicate. Download
The N terminus of the α5GGDEF helix can be used to predict interaction between DGCs and related effectors. The domain architecture and α2EAL helices for P. fluorescens and P. aeruginosa LapD are shown along with the domain architecture and α5GGDEF helices for GcbC and RoeA. Due to the resemblance of the RoeA helix to the GcbC helix, RoeA was predicted to interact with P. aeruginosa PA14 LapD. Transmembrane domains (TM) are depicted in light blue, and the other domains are labeled in the figure. Download
The α5GGDEF helix of a DGC can partially impact function. (A) The domain architectures for GcbC and Pfl01_2295 are shown. Transmembrane domains (TM) are depicted in light blue, and the other domains are labeled in the figure. The N terminus of the α5GGDEF helix for each is shown on the right. The α5GGDEF helix of Pfl01_2295 was modified to become more GcbC-like and used to test biofilm formation in Fig. 5C. (B) (Top) Pfl01_2295 and its mutant variants were transformed on a plasmid into Pseudomonas aeruginosa PA14 and analyzed on Congo red plates with 0.1% arabinose as described in Materials and Methods. (Bottom) The same constructs as in panel B along with control strains were induced with 0.1% arabinose in Pseudomonas fluorescens, and extracts were analyzed by LC-MS to quantify c-di-GMP production as described in Materials and Methods. Error bars represent standard deviation and data were obtained from three subcultured replicates. (C) Xanthomonas axonopodis EAL domain of FimX interacting with PilZ. The interaction is partially mediated by the α2EAL helix, which is the same helix used by LapD to contact GcbC (dark blue, PDB 4FOU) (29). Download
GcbC and LapD mutants isolated in this study using the B2H screen. Shown is a complete list of the amino acid changes in GcbC and LapD associated with the mutants isolated as a consequence of the bacterial 2-hybrid (B2H) screen described in the text.
Crystallographic data collection and refinement for GcbC. Shown is a list of the relevant crystallographic data for the structure of GcbC.
Candidate α5GGDEF helices from PA14 for interaction with LapD. Shown is a complete list of the sequences of the α5 of all GGDEF motif-containing proteins in P. aeruginosa PA14.
Strains and plasmids used in this study. Shown is a complete list of strains used, their genotypes, and whether they were commercially available (with company name), generated during previous studies (with the appropriate reference) or built during the course of this work (listed as “This Study”). All plasmids are also listed, as is their source as outlined above for strains.
Primers used in this study. Shown is a complete list of primers used in this work, the primer sequences, and related information.
This document contains additional details of the methods used in this work, as well as the appropriate citations describing previously published methods. Download