Abstract
BACKGROUND
Human papilloma viruses (HPVs) may act early in breast oncogenesis (“hit-and-run” phenomena).
METHODS
The authors used immunohistochemistry for the identification of HPV E7 oncogenic protein expression in 32 sets of benign and subsequent breast cancer specimens from the same Australian patients.
RESULTS
HPV E7 oncoprotein was clearly expressed in the nuclei of 23 (72%) of the 32 benign specimens and 20 (62.5%) of the subsequent 32 breast cancer specimens in the same patients. There was no HPV E7 protein expression in seven (30%) of the 23 breast cancer specimens that had prior HPV E7 protein-positive benign breast biopsies in the same patients.
CONCLUSIONS
This observation suggests that HPV oncogenic influences occur early in some breast cancers. This finding confirms the previous observations. This early influence of HPVs may be the reason why there is no increase in the prevalence of HPV-associated breast cancer in immunocompromised patients as compared to HPV-associated cervical cancer.
Keywords: breast cancer, benign breast, normal breast, human papilloma virus (HPV), HPV E7 protein, PCR, immunohistochemistry, Cervimax
Introduction
High-risk human papilloma viruses (HPVs), which can cause cancer, are controversial causal candidates for breast cancer. The reasons for the controversy are the major conflicts in the evidence. On the one hand, high-risk HPVs have been identified by polymerase chain reaction (PCR) techniques in 25%–100% of breast cancers in more than 30 studies conducted in a wide range of populations.1–3 A meta-analysis showed a significant increase in breast carcinoma risk with HPV positivity; odds ratio = 3.63.1 On the other hand, there is no increase in breast cancer prevalence in immunocompromised patients who have had AIDS or organ transplantations followed by immunosuppression therapies.4 In immunocompromised patients, there is a substantial increase in the prevalence of HPV-associated cancers, such as cervical and head and neck cancers.4 If high-risk HPVs were a major causal factor in breast cancer, Grulich and Vajdic4 argue that there should be an increased prevalence in immunocompromised patients.
Ohba et al3 and Vieira et al5 have independently developed evidence that offers explanations for the conflicting evidence. They have demonstrated that the causal mechanisms for HPVs in breast cancer differ from HPVs in cervical and head and neck cancers. These different mechanisms include the involvement of HPVs in the antiviral and cancer genomic DNA (gDNA) deaminase APOBEC3B. APOBEC3B enzymes influence the human genome and increase the risk of cancer, including breast cancers.6 Vieira et al5 have demonstrated that HPV E6 proteins trigger the upregulation of APOBEC3B. Ohba et al3 have demonstrated that these mechanisms operate early in the many steps of breast oncogenesis. In addition, Moody and Laimins7 have indicated that the induction of genomic instability is an early event in HPV-induced cancers, occurring before the integration of the virus into host chromosomes. These early oncogenic mechanisms may not be influenced at a later stage by the immune system.
We hypothesize that there should be evidence of early HPV oncogenic activity in benign breast cells before the development of malignant breast cancer. As HPV E7 protein is a known oncogenic factor, it should be expressed early in breast oncogenesis. For this reason, using immunohistochemistry (IHC), we conducted a study of HPV E7 in benign breast biopsies from women who subsequently developed breast cancer. We did not investigate the role of HPV E6 because specific antibodies for use in IHC are not available.
Methods
Based on pathology reports, we identified 32 Australian patients who had benign breast biopsies 1–11 years before developing breast cancer. Archival formalin-fixed specimens from these patients were identified and collected from an Australian pathology service (Douglas Hanly Moir Pathology).
We used normal breast specimens from 28 patients who had cosmetic breast surgery as a comparative group. These patients were younger than the patients who developed breast cancer and therefore are not an exact control group. None of these patients had developed breast cancer 10 years after the initial surgery.
We also used breast specimens from 20 patients who had two separate benign biopsies taken at different ages, who had not developed breast cancer.
Immunohistochemistry
We used standard manual IHC methods for the identification of HPV E7 protein expression in both the benign and subsequent breast cancer specimens from the same patients. The antibodies were anti-HPV E7 monoclonal “Cervimax”—Valdospan GmbH. These antibodies have recently been developed. The specificity of these antibodies has been demonstrated experimentally and by epidemiological studies.8,9 The HPV E7 antibody reacts with a wide range of HPV types including high risk for cancer HPV 16 and 18. The experiments using Cervimax HPV E7 antibodies for IHC analyses were repeated several times using (i) different concentrations of the antibodies and (ii) without antigen retrieval. The best results were achieved without antigen retrieval. Good outcomes for HPV E7 (antibody dilution 1/500) were achieved with clear staining of both cytoplasm and nuclei of breast cancer cells. The use of this dilution is different from the manufacturer’s recommendation.
An HPV type 18-positive cervical intraepithelial neoplasia specimen was used as a positive control for Cervimax antibodies (See Fig. 1).
Figure 1.
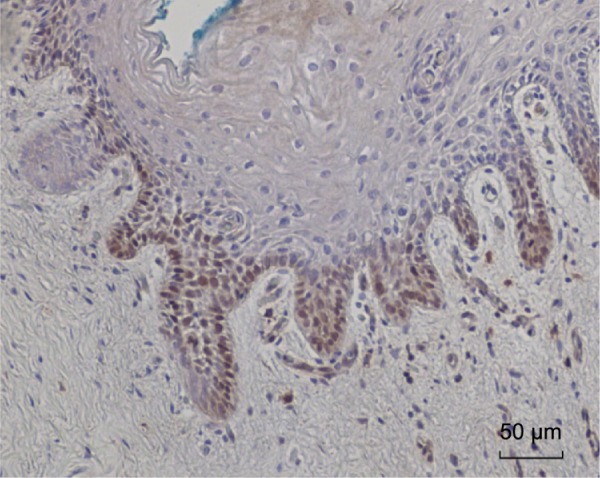
HPV E7 protein expression identified in HPV type 18-positive cervical intraepithelial neoplasia (CIN 1). This is a positive control for Cervimax antibodies used in the immunohistochemistry analyses.
PCR analyses
HPV gene sequences were identified by PCR. Standard PCR, semi-nested PCR, and real-time PCR were used for the detection of HPV. All PCR products were sequenced.
Preparation of gDNA
For standard and real-time PCR, the methods were as described in Steinau et al.10 The primers used were β-actin 5′ forward (5′-CTTCT-GCCGTTTTCCGTAGG-3′) and β-actin 3′ reverse (5′-TGGGATGGGGAGTCTGTTCA-3′) at the final concentration of 1 μM. Thermal cycles were 94°C for 15 minutes, 94°C for 30 seconds, 55°C for 30 seconds, and 72°C for 45 seconds for 30 cycles. HotStarTaq Master Mix Kit (QIAGEN) was used for the PCR reaction master mix. gDNA samples, which were β-actin positive, were selected for the detection of HPV genomes.
Standard PCR
The primers used for semi-nested PCR were My11 (5′-GCMCAGGGWCATAAYAATGG-3′)11 to modified GP6+ (5′-AAATCATATTCCTCMMCAT-GTC-3′).12 The second round was GP5+ (5′-TTTGTTACT-GTGGTAGATACTAC-3′)11 to GP6+. These primers were degenerated for HPV 16 and 18 but were also capable of bringing up types 3, 11, 12, 45, 58, 73, and 75.
Stringent negative controls were used in parallel with all PCR analyses. These negative controls were no DNA (water) and a reagent blank, as well as sequencing of the products of these controls in case the bands could not be seen on a gel. Negative outcomes of PCR analyses of selected breast cancer specimens provided negative controls based on breast tissues. The positive control for HPV was an HPV 18-positive HeLa cell line.
Real-time PCR
The HPV L1 gene in gDNA samples was amplified using a real-time PCR machine (Rotor Gene Q, QIAGEN). Thermal cycles used were 95°C for five minutes, 95°C for 10 seconds, and 60°C for 30 seconds for 60 cycles. The PCR reaction mix was the QuantiFast SYBR Green kit (QIAGEN) and was used according to the manufacturer’s recommendation. The primers used for the detection of HPV were GP5+ forward (5′-TTTGTTACTGTGGTAGATAC-TAC-3′) and GP6+ reverse (5′-AAATCATATTCCTC-MMCATGTC-3′) at the final concentration of 1 μM.13 The quantity of gDNA used for PCR was determined by estimating theoretical gene copies of HPV 16 in breast and breast cancer tissues. A positive control for HPV was a purified PCR product containing the HPV L1 region. It was amplified with MY11 forward and MY09 reverse (5′-CGTCCMAR-RGGAWACTGATC-3′)12 primers. Negative controls were (i) ultrapure H2O (no DNA template) and (ii) extraction blank (a DNA extraction control without formalin-fixed paraffin-embedded tissues).
The PCR products were analyzed and selected for sequencing based on amplification profiles and the achievement of the known melting point. The analysis was performed using Rotor Gene Q software (QIAGEN).
Sequencing the PCR products and identification of HPV types
The HPV PCR products from GP5 to GP6 were sequenced to determine the HPV type. The HPV genotypes were identified by BLAST via the US National Center for Biotechnology Information.
Confirmatory PCR studies
DNA extracts from six selected (for positive HPV) benign and breast cancer specimens were independently analyzed by the Antonsson group at the QIMR Berghofer Medical Research Institute, Brisbane, for the presence of HPV DNA using their previously published methods.14
Results
HPV sequences
HPV types 12, 16, 18, 45, 58, 73, 124 plus af019978 were identified in the benign and subsequent breast cancers as shown in Supplementary Table 1. af019978 is an untyped HPV that had previously been identified in a Sydney patient.
Confirmation of data based on PCR
DNA extracts from six HPV-positive benign and breast cancers (extracted and analyzed at the University of New South Wales laboratories) were independently analyzed at the Antonsson laboratories in Brisbane, Australia. HPV types 16 and 18 were identified in two of six specimens. The identification of different HPV types indicates that contamination is unlikely.
HPV E7 oncoprotein expression in benign and subsequent breast cancers
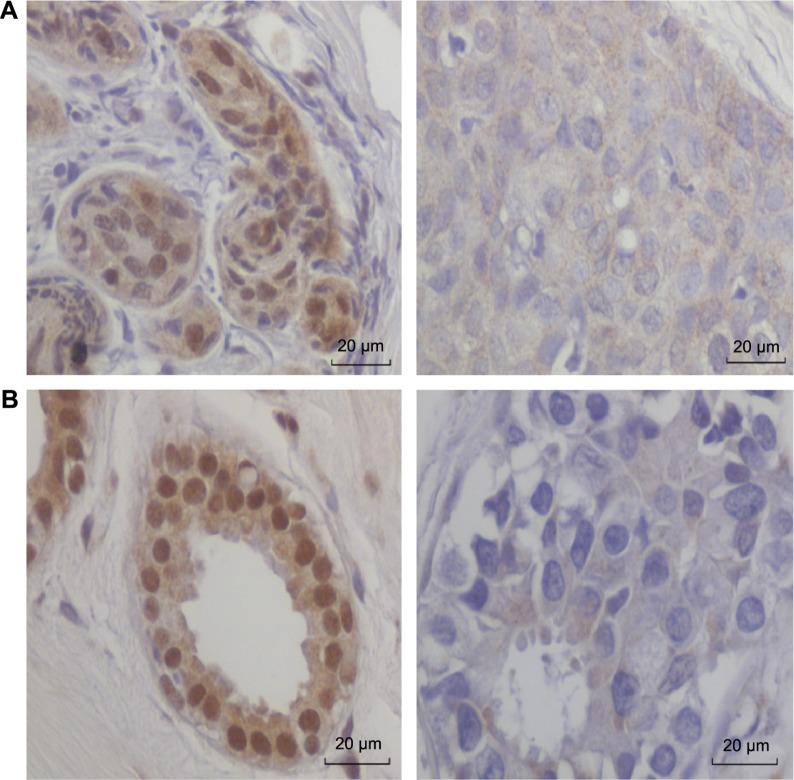
Thirty-two sets of benign breast and subsequent breast cancer specimens from the same patients were used for assessing HPV E7 protein expression. HPV E7 oncoprotein was clearly expressed in the nuclei of 23 (72%) of the 32 benign specimens and 20 (62.5%) of the subsequent 32 breast cancer specimens in the same patients. There was no HPV E7 protein expression in eight (30%) of the 23 subsequent breast cancers that had prior HPV E7 protein-positive benign breast biopsies in the same patients. These data are shown in Supplementary Table 1, Table 1, and illustrated in Figure 2. It is noted in Figure 2 and Supplementary Table 1 that HPV E7 protein was frequently detected in both the nuclei and the cytoplasm of the same cells.
Table 1.
HPV E7 protein expression in the nuclei of benign breast cells and nuclei of subsequent breast cancer cells in the same patients. HPV E7 protein expression in benign breast tissues is significantly higher than in subsequent breast cancer in the same patients. This suggests that HPV E7 oncogenic activity occurs in the early benign breast stages of breast oncogenesis.
| HPV E7 BENIGN BREAST IMMUNOHISTOCHEMISTRY | HPV E7 BREAST CANCER IMMUNOHISTOCHEMISTRY | NUMBER OF PATIENTS n = 32 |
|---|---|---|
| Pos | Neg or slight | 7 (22%) |
| Pos | Pos | 10 (31%) |
| Neg | Pos | 2 (6%) |
| Neg | Neg | 13 (41%) |
Figure 2.
HPV E7 protein expression (based on immunohistochemistry) in benign breast and subsequent breast cancer in the same patients. HPV E7 protein is strongly expressed in the cell nuclei of the benign breast tissue. There is no HPV E7 protein expression in the nuclei of the breast cancer cells. Patient A. Benign breast at the age of 33 and subsequent breast cancer at the age of 44 years. Patient B. Benign breast at age 44 and subsequent breast cancer at age 48 years.
This is the same pattern of HPV E7 distribution observed in prostate cancer by Pascale et al.9 This pattern of HPV E7 distribution has now been observed in two different cancer types (breast cancer and prostate cancer), indicating that it may be a general HPV E7 distribution pattern in cancer cells.
Benign and later repeated benign breast biopsies
As shown in Table 2 and Supplementary Table 2, HPV E7 protein was expressed in two (10%) of 20 of the first benign breast biopsy and eight (40%) of 20 of the subsequent benign breast specimens. The biopsies of the benign breast tissues were taken several years apart from the same patients. The pattern of HPV E7 protein expression is very different from the pattern shown in Table 1. There was a negative HPV E7 protein expression in both prior and later biopsies in a significant majority of patients—12 (60%) of 20. None of these patients are known to have developed breast cancer.
Table 2.
HPV E7 protein expression in the nuclei of benign breast cells and nuclei of subsequent benign breast cells in the same patients. The biopsies of the benign breast tissues were taken several years apart from the same patients. None of these patients are known to have developed breast cancer. The pattern of HPV E7 protein expression is very different from the pattern shown in Table 1. There was a negative HPV E7 protein expression in both prior and later biopsies in a significant majority of patients.
| HPV E7 BENIGN BREAST INITIAL BIOPSY IMMUNOHISTOCHEMISTRY | HPV E7 BENIGN BREAST FOLLOW UP BIOPSY IMMUNOHISTOCHEMISTRY | NUMBER OF PATIENTS n = 20 |
|---|---|---|
| Pos | Neg | 0 |
| Pos | Pos | 2 (10%) |
| Neg | Pos | 6 (30%) |
| Neg | Neg | 12 (60%) |
HPV E7 protein was expressed in 10 (36%) of the 28 normal breast specimens. In both these comparative groups (normal and benign), the fact that none of these women developed cancer that parallels the cervical infection, where >90% of new infections are cleared.
Discussion
We have demonstrated that HPV E7 oncoprotein was clearly expressed in the nuclei of 23 (72%) of the 32 benign specimens and 20 (62.5%) of the subsequent 32 breast cancer specimens that developed 1–11 years later in the same patients. In seven (30%) sets of specimens, there was a high HPV E7 protein expression in the benign breast specimen but no HPV E7 protein expression in the subsequent breast cancer specimen in the same patient.
This observation suggests that HPV oncogenic influences, such as the expression of HPV E7 oncoproteins, occur early in the step-by-step oncogenic progression in breast cancer. This finding parallels the observations by Ohba et al3 that high-risk HPVs exert their oncogenic influences at an early stage of breast oncogenesis by influencing the biology of APOBEC3B enzymes, which in turn influence genomic stability and lead to an increased risk of breast cancer.3,6 The influence of HPVs on APOBEC3B has been confirmed by Vieira et al.5
These mechanisms differ from HPV mechanisms in cervical cancer that include the influences of HPV E6 and E7 oncoproteins, which work in concert to disrupt cell cycle regulation, inhibit apoptosis, and stimulate cell cycle progression by binding and inhibiting the p53 and p110RB tumor suppressor genes.15 In most instances, persistent infection and continued presence of HPV are required for cervical oncogenesis.
Limitations of this investigation
There are limitations to this investigation because the number of patients in this study is less. Therefore, formal statistical analyses are not justified. However, the outcomes are striking as shown in Figure 2. It is possible that as oncogenesis progresses, the biology of the breast cells could become damaged and unable to express proteins such as HPV E7. However, this is an unlikely explanation as estrogen and progesterone receptor proteins were strongly expressed.
The findings that HPVs probably act early in breast oncogenesis also offer an explanation why there is no increase in the prevalence of HPV-associated breast cancer in immunocompromised patients.4
Conclusions
These observations suggest that HPV oncogenic influences occur early in the step-by-step oncogenic progression in some breast cancers. This finding parallels and confirms previous observations that high-risk HPVs exert their oncogenic influences at an early stage of breast oncogenesis. This early HPV oncogenic influence in breast cancer differs from HPV-associated cervical cancer in which persistent HPV infections are usually required for oncogenesis.
Supplementary Materials
Supplementary table 1. Benign breast and subsequent breast cancer in the same patients. Human papilloma virus (HPV) sequences and HPV E7 protein expression.
Supplementary table 2. HPV E7 protein expression in repeated biopsies of the same patients.
Acknowledgments
The contribution of Annika Antonsson of the Berghofer Queensland Institute of Medical Research and the donation of archival specimens by Douglass Hanly Moir Pathology are gratefully acknowledged.
Footnotes
ACADEMIC EDITOR: Goberdhan P. Dimri, Editor in Chief
PEER REVIEW: Six peer reviewers contributed to the peer review report. Reviewers’ reports totaled 1,546 words, excluding any confidential comments to the academic editor.
FUNDING: Authors disclose no external funding sources. This study was financed by the authors.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
Paper subject to independent expert blind peer review. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE).
Author Contributions
Conceived and designed the study: WKG, JSL, CN, NJW. Analyzed the data: WKG, JSL, CN, NJW. Wrote the first draft of the manuscript: JSL, WKG. Agree with the manuscript results and conclusions: WKG, JSL, CN, NJW, RC, WD. Jointly developed the structure and arguments for the paper: WKG, JSL, CN, NJW. Made critical revisions and approved final version: WKG, JSL, CN, NJW. Identified the patients, reviewed pathology reports, collected the archival specimens, JL, RC, WD. All authors reviewed and approved the final manuscript.
Ethical Considerations
This study has formal ethical approval by the University of New South Wales Human Research Ethics Committee— number HREC HC11421. Participants gave their written, informed consent to participate in this study. The research was conducted in accordance with the principles of the Declaration of Helsinki.
Ethical approval for the follow-up of women who donated normal breast tissues was given by the New South Wales Population and Health Services Research Ethics Committee—number 2009/12/203. This Ethics Committee waived the need for consent. The reasons for waiving consent were (i) the specimens were archival having been collected in 1999, 2000, and 2001, (ii) all specimens were “de-identified” to the research group, and (iii) retrospective approaches to donors, all of whom had cosmetic surgery, may have caused unnecessary anxiety.
REFERENCES
- 1.Li N, Bi X, Zhang Y, Zhao P, Zheng T, Dai M. Human papillomavirus infection and sporadic breast carcinoma risk: a meta-analysis. Breast Cancer Res Treat. 2011;126:515–520. doi: 10.1007/s10549-010-1128-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Simões PW, Medeiros LR, Simões Pires PD, et al. Prevalence of human papillomavirus in breast cancer: a systematic review. Int J Gynecol Cancer. 2012;22:343–347. doi: 10.1097/IGC.0b013e31823c712e. [DOI] [PubMed] [Google Scholar]
- 3.Ohba K, Ichiyama K, Yajima M, et al. In vivo and in vitro studies suggest a possible involvement of HPV infection in the early stage of breast carcinogenesis via APOBEC3B induction. PLoS One. 2014;9:e97787. doi: 10.1371/journal.pone.0097787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grulich AE, Vajdic CM. The epidemiology of cancers in human immunodeficiency virus infection and after organ transplantation. Semin Oncol. 2015;42:247–257. doi: 10.1053/j.seminoncol.2014.12.029. [DOI] [PubMed] [Google Scholar]
- 5.Vieira VC, Leonard B, White EA, et al. Human papillomavirus E6 triggers upregulation of the antiviral and cancer genomic DNA deaminase APOBEC3B. MBio. 2014;5(6):e2234–e2214. doi: 10.1128/mBio.02234-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burns MB, Lackey L, Carpenter MA, et al. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature. 2013;21:366–370. doi: 10.1038/nature11881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moody CA, Laimins LA. Human papillomavirus oncoproteins: pathways to transformation. Nat Rev Cancer. 2010;10:550–560. doi: 10.1038/nrc2886. [DOI] [PubMed] [Google Scholar]
- 8.Faoro V, Barbazza R, Bonin S, Brunetti D, Sulfaro S, Stanta G. Detection of HPV E7 oncoviral protein in cervical lesions by a new antibody. Appl Immunohistochem Mol Morphol. 2013;21:341–350. doi: 10.1097/PAI.0b013e318269bf5d. [DOI] [PubMed] [Google Scholar]
- 9.Pascale M, Pracella D, Barbazza R, et al. Is human papillomavirus associated with prostate cancer survival? Dis Markers. 2013;35:607–613. doi: 10.1155/2013/735843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Steinau M, Patel SS, Unger ER. Efficient DNA extraction for HPV genotyping in formalin-fixed, paraffin-embedded tissues. J Mol Diagn. 2011;13:377–381. doi: 10.1016/j.jmoldx.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jacobs MV, de Roda Husman AM, van den Brule AJ, Snijders PJ, Meijer CJ, Walboomers JM. Group-specific differentiation between high- and low-risk human papillomavirus genotypes by general primer-mediated PCR and two cocktails of oligonucleotide probes. J Clin Microbiol. 1995;33:901–905. doi: 10.1128/jcm.33.4.901-905.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gravitt PE, Peyton CL, Alessi TQ, et al. Improved amplification of genital human papillomaviruses. J Clin Microbiol. 2000;38:357–361. doi: 10.1128/jcm.38.1.357-361.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lawson JS, Glenn WK, Tran DD, Ngan CC, Duflou JA, Whitaker NJ. Identification of human papilloma viruses in atheromatous coronary artery disease. Front Cardiovasc Med. 2015(2):17. doi: 10.3389/fcvm.2015.00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Antonsson A, Nancarrow DJ, Brown IS, et al. Australian Cancer Study High-risk human papillomavirus in esophageal squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev. 2010;19:2080–2087. doi: 10.1158/1055-9965.EPI-10-0033. [DOI] [PubMed] [Google Scholar]
- 15.Zur Hausen H. Papillomaviruses and cancer: from basic studies to clinical application. Nat Rev Cancer. 2002;2:342–350. doi: 10.1038/nrc798. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary table 1. Benign breast and subsequent breast cancer in the same patients. Human papilloma virus (HPV) sequences and HPV E7 protein expression.
Supplementary table 2. HPV E7 protein expression in repeated biopsies of the same patients.