Abstract
Potential molecular alterations based on age and sex are not well defined in diffuse large B-cell lymphoma (DLBCL). We examined global transcriptome DLBCL data from The Cancer Genome Atlas (TCGA) via a systems biology approach to determine the molecular differences associated with age and sex. Collectively, sex and age revealed striking transcriptional differences with older age associated with decreased metabolism and telomere functions and female sex was associated with decreased interferon signaling, transcription, cell cycle, and PD-1 signaling. We discovered that the key genes for most groups strongly regulated immune function activity. Furthermore, older females were predicted to have less DLBCL progression versus older males and young females. Finally, analyses in systems biology revealed that JUN and CYCS signaling were the most critical factors associated with tumor progression in older and male patients. We identified important molecular perturbations in DLBCL that were strongly associated with age and sex and were predicted to strongly influence tumor progression.
Keywords: aging and cancer, non-Hodgkin lymphoma, tumor progression, JUN, sex and cancer, DLBCL, diffuse large B-cell lymphoma, TCGA
Introduction
Older age is an adverse prognostic factor that correlates with inferior survival in diffuse large B-cell lymphoma (DLBCL).1–4 This is likely in part attributable to poorer performance status and inability to tolerate therapy5; however, potential molecular perturbations that may contribute to inferior outcomes are not well defined. In addition, the impact of sex has been shown to be prognostic in DLBCL,6,7 including recent data of improved survival for females receiving rituximab-based therapy.8,9 Molecular perturbations that may be associated with sex-related pathogenesis are largely unexplored.10,11
We performed a comprehensive global transcriptome analyses from The Cancer Genome Atlas (TCGA) on data from pretreatment/baseline DLBCL samples to investigate potential molecular alterations based on age and sex. Furthermore, a novel unbiased method12 of a systems biology approach13–16 was used to identify key genes and the related signaling networks that were most strongly associated with age and sex that predicted tumor progression.
Methods
TCGA transcriptome analysis
Results are based on data from the TCGA research network (cancergenome.nih.gov). Specifically, DLBCL mRNA level 3 data type with a total of 48 data sets from untreated DLBCL patients was available with 33 data sets containing relevant age and sex. Since the median age was 58 years for these data, we defined older patients as ≥59 years (versus ≤58 years as younger patients); data were further segregated based on sex (Supplementary Table 1). Data were imported into MultiExperiment Viewer,17 and statistically significant genes were determined by t-test with a P-value < 0.05 for all comparisons to take forward for pathway analysis. Independent and separate analysis was done for each comparison mentioned in this article using these conditions.
Pathway analysis of the selected genes was performed by using a fold-change ≥1.2 (or ≤−1.2) and then comparing old to young patients and observing pathway relationships using Ingenuity Pathway Analysis (IPA) software (Ingenuity Systems). Upstream regulator analysis from IPA identified any molecule that affected the expression or function of the measured downstream target genes. The activation state of each upstream regulator from the experimental data set is determined by calculating the z-score (≥2, activated or ≤−2, inhibited). Similar analysis through IPA was done predicting biofunctional activity. Gene set enrichment analysis (GSEA)18 was performed using the entire list of genes and with leading edge analysis as defined by Subramanian et al.18 Significant gene sets between age groups were considered with false discovery rate (FDR) <0.05. Network representation of GSEA functions was done using Cytoscape.19 Key genes were determined as previously reported in Beheshti et al.12 and Wage et al.20 by finding the overlapping genes involved in predicting the significant upstream regulators, biofunctions, and GSEA C2 gene sets. More specifically, for each set of genes under analysis, the association with statistically significant pathways and functions were determined through both IPA and GSEA. Common genes were determined to be involved in the analysis of both IPA upstream regulators and biofunctions. These sets of genes were further compared to the GSEA’s leading edge genes (FDR < 0.05). The overlapping genes between these two analyses were considered to be the key genes involved and in control of the majority of predicted functions and activity with the system being analyzed. In previous publications, we have validated key genes determined with this method through experimental approaches involving Western blots, qPCR, and other functional methods.12,20 These experiments proved that the key genes determined with this bioinformatics interrogation are indeed involved in the system being studied. For male-specific analysis, only the overlapping genes for upstream regulators and biofunction predictions were used since no overlap occurred with GSEA. This unbiased method to determine key genes produces molecular factors that are involved in all the significantly regulated functions and pathways and are key regulators involved in age-associated DLBCL. The impact of both the upstream regulators and key genes was associated with tumor progression by determining through the literature how each factor impacts tumor progression (ie, tumor promoter or tumor suppressor). The overall impact was used to determine the global effect on tumor progression based on the specific regulation of each gene. Tumor progression in this manuscript is defined as the predictability of assessing the tumor dynamics based on the biology, rather than clinical data. Specifically, tumor progression describes the outcome of tumor growth predicted from biological factors. This study was performed in accordance with the principles of the Declaration of Helsinki.
Results and Discussion
Age- and sex-based differences and interactions
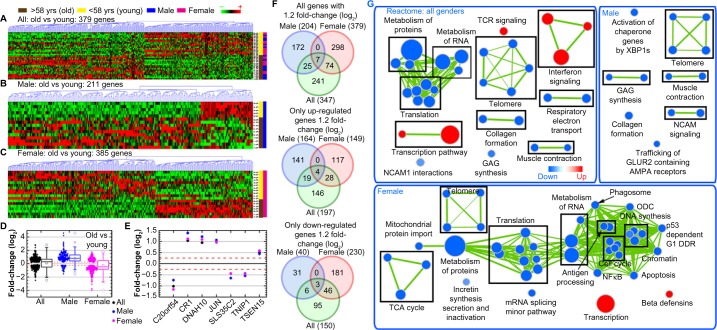
In comparing old versus young DLBCL patients independent of sex (Fig. 1A–C), there was an equal distribution of upregulated and downregulated genes (Fig. 1D). There were, however, several distinct genes associated with older age (Fig. 1E–F). These included JUN (log2 fold-change = 0.967), CR1 (log2 fold-change = 1.036), and DNAH10 (log2 fold-change = 0.946) being most upregulated, while C20orf54 (log2 fold-change = −1.017) was the most downregulated in older DLBCL patients (Fig. 1E). Furthermore, GSEA demonstrated that older age was associated with decreased metabolism and telomere functions21 and also increased immune-related pathways as shown using a network representation of the gene sets associated with these functions (Fig. 1G/Supplementary Tables 2–4).
Figure 1.
Global gene regulation changes as a function of age and sex. Hierarchical clustering of genes by average linkage (UPGMA or Unweighted Pair Group Method with Arithmetic Mean) and Euclidean distance calculation from the DLBCL TCGA mRNA data for (A) older (≥59 years old) versus younger (≤58 years old) DLBCL patient comparisons combining all sexes, (B) male-only comparison in old and young patients, (C) female-only comparison in old and young patients, and (D) average mRNA signal log2 fold-change comparing sex-independent analysis to male and female-specific analyses. Whiskers show the range of the outliers, with max and min values as Ο and the 1 and 99th percentile outliers as X. (E) The average signal log2 fold-change for the seven common genes between the three different sets of analysis. (F) Venn diagrams of the genes with 1.2 fold-change for comparisons between sex-independent analyses and male and female-specific analyses. Separate Venn diagrams for only the up- and downregulated genes are also included. The numbers in the parenthesis indicate the overall number of genes in each group. (G) Network representation of only Reactome-related gene sets from GSEA C2 gene set annotations. Leading edge analysis with an FDR < 0.05 determined significant gene sets enriched for each group. The size of each node reflects the amount of molecules involved in each gene set. The edge (green lines) represents the number of genes associated with the overlap of two gene sets (or nodes) that the edge connects. Clusters were named according to common function in each grouping. Upregulated gene sets were denoted with red color and downregulated gene sets were denoted by blue color.
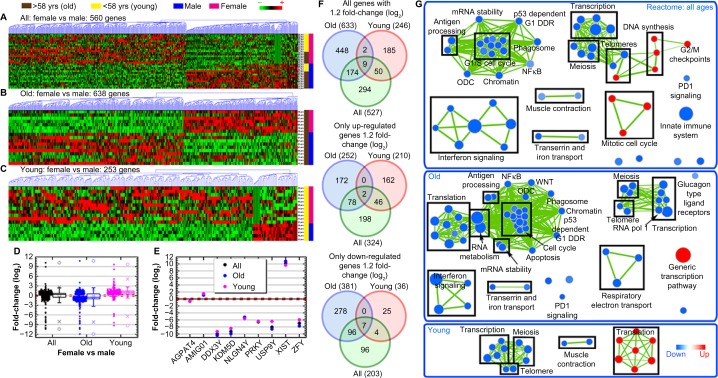
Nine distinct genes were modulated by sex, regardless of age. This included XIST (log2 fold-change = 10.189), which was significantly upregulated in females, and DDX3Y (log2 fold-change = −9.996), KDM5D (log2 fold-change = −9.322), and PRKY (log2 fold-change = −6.554) that were downregulated in females (Fig. 2E–F). GSEA demonstrated that female sex was associated with decreased interferon signaling, transcription, cell cycle, PD-1 signaling, and meiosis with concurrent increases in mitotic cell cycle and DNA synthesis that are illustrated in a network representation of the gene sets associated with each of these functions (Fig. 2G/Supplementary Tables 2–4).
Figure 2.
Global gene regulation changes as a function of age and gender. Hierarchical clustering of genes by average linkage (UPGMA) and Euclidean distance calculation from the DLBCL TCGA mRNA data with a t-test, P-value < 0.05, for (A) female-to-male patient comparisons combining all ages, (B) old patient (>58 years old) only comparisons for female and male patients, and (C) young patient (<58 years old) comparison for female and male patients. (D) The average mRNA signal log2 fold-change comparing age-independent analysis to young and old-specific analyses. Whiskers show the range of the outliers, with max and min values as Ο and the 1 and 99th percentile outliers as X. (E) The average signal log2 fold-change for the nine common genes between the three different sets of analysis. (F) Venn diagrams of the genes with 1.2 fold-change for comparisons between age-independent analysis and old and young-specific analyses with separate Venn diagrams for only the upregulated and downregulated genes. The numbers in the parenthesis indicate the overall number of genes in each group. (G) Network representation of only Reactome-related gene sets from GSEA C2 gene set annotations. Leading edge analysis with an FDR < 0.05 determined significant gene sets enriched for each group. The size of each node reflects the amount of molecules involved in each gene set. The edge (green lines) represents the number of genes associated with the overlap of two gene sets (or nodes) that the edge connects. Clusters were named according to common function in each grouping. Upregulated gene sets are denoted by red color and downregulated gene sets are denoted by blue color.
We next examined the impact of sex and age in combination (Fig. 2). There was an overall downregulation of genes for older females (versus older males) with an overall upregulation of genes for younger females (versus younger males) (Fig. 2B–D and F). This provides indication of an overall global difference in the biology signature occurring with age. Older females (versus older males) had downregulation of functions related to cell maintenance (ie, interferon signaling, translation, cell cycle, metabolism, and meiosis), while young females (versus young males) had upregulation in translation (Fig. 2G).
Interestingly, older male DLBCL patients had an overall upregulation of genes, while older female patients had an overall downregulation of genes (Fig. 1B–D); this was verified on sex-specific GSEA analyses (Fig. 1G). As with age differences, this difference indicates an intrinsic biological difference occurring in DLBCL patients with sex. Older male patients had downregulation of structural functions (eg, muscle contraction, telomere, collagen formation), while older female patients had downregulation of cellular maintenance functions (eg, telomere, translation, apoptosis, metabolism, cell cycle) (Fig. 1G/Supplementary Tables 2–4).
Discovery of key genes
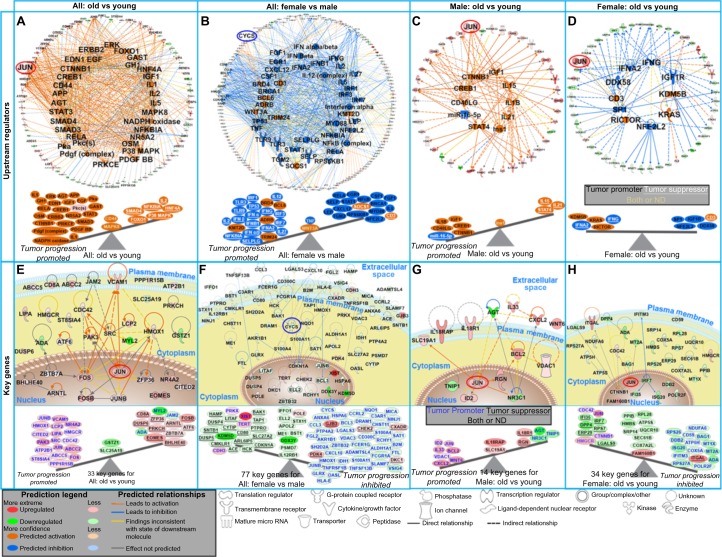
We subsequently determined the most prominent upstream regulators predicted to impact tumor progression. Age-specific molecular analyses (Fig. 3A) predicted that older age was associated with promotion of tumor growth (Supplementary Table 5). Sex-specific analyses (Fig. 3B) predicted females to have increased tumor progression versus males. Analyzing the impact of age within females, however, there was an even balance of both tumor oncogenes and suppressors (Fig. 3D), and in males, older age predicted tumor progression (Fig. 3C). Genes associated with the predictions of the upstream regulators are shown as a network with the key genes discussed below circled (Figs. 3A–D and 4A and B).
Figure 3.
The impact of predicted upstream regulators and key genes from DLBCL patients on tumor progression. (A–D) The top panel shows network representation of the upstream regulators for each type of analysis with the genes involved for each prediction. The second panel shows schematic of the activation states of the upstream regulators from Supplementary Table 2, illustrating the balance between the tumor promoters (black text) and tumor suppressors (white text and underlined) with a predicted activation (orange) or predicted inhibition (blue). The upstream regulators with both promoter and suppressor effects or not determined (ND) effects are shown in the middle of the balance (yellow text). (E–H) The third panel shows gene network analysis for the key genes involved in each type of analysis. The predicted relationships between all genes are also shown. Log2 fold-changes (with a cutoff of 1.2 fold-change) to the gene expression were used to obtain different shades of green for fold-change in downregulated genes, while different shades of red depict fold-change in upregulated genes. The darker the shade of green or red, the greater the fold-change. The bottom panel shows a schematic of the key significant genes (see also Supplementary Table 4) determined to be significant in regulating functions for each comparison illustrating the balance between the tumor promoters (blue text) and tumor suppressors (black text and underlined) with the log2 fold-change color coded as before. Genes with both promoter and suppressor effects or ND effects are shown in the middle of the balance (black text). JUN is circled in red, and CYCS is circled in blue throughout the networks.
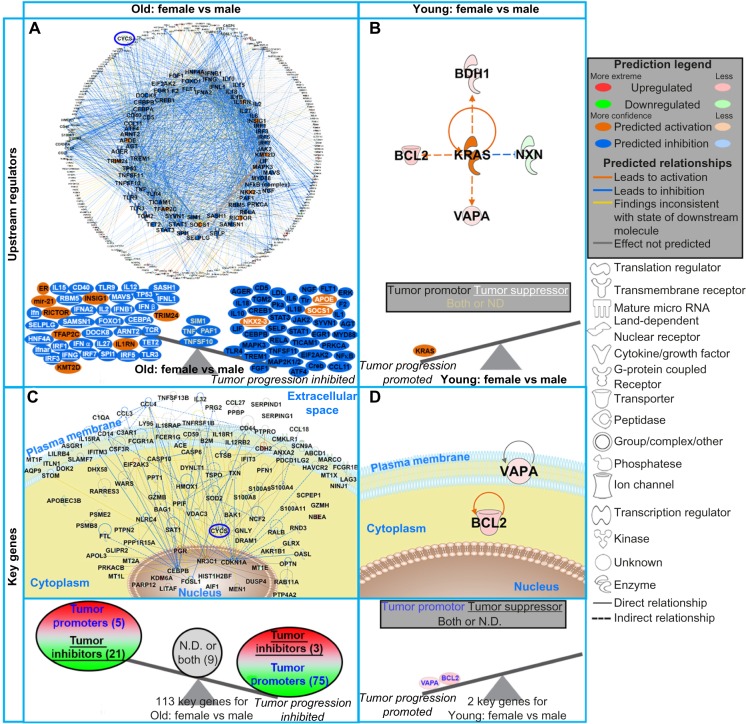
Figure 4.
The impact of predicted upstream regulators and key genes from DLBCL patients on tumor progression. (A and B) The top panel shows network representation of the upstream regulators for each type of analysis with the genes involved for each prediction. The second panel shows schematic of the activation states of the upstream regulators from Supplementary Table 2, illustrating the balance between the tumor promoters (black text) and tumor suppressors (white text and underlined) with a predicted activation (orange) or predicted inhibition (blue). The upstream regulators with both promoter and suppressor effects or ND effects are shown in the middle of the balance (yellow text). (C and D) The third panel shows gene network analysis for the key genes involved in each type of analysis. The pathway analysis was done with IPA software. The predicted relationships between all genes are also shown. Log2 fold-changes (with a cutoff of 1.2 fold-change) to the gene expression were used to obtain different shades of green for fold-change in downregulated genes, while different shades of red depict fold-changes in upregulated genes. The darker the shade of green or red, the greater the fold-change. The bottom panel shows a schematic of the key significant genes (Supplementary Table 4), determined to be significant in regulating many functions for each comparison, illustrating the balance between the tumor promoters (blue text) and tumor suppressors (black text and underlined) with the log2 fold-change color coded as before. Genes with both promoter and suppressor effects or ND effects are shown in the middle of the balance (black text). CYCS is circled in blue throughout the networks. In panel (C), the individual genes are not shown due to the quantity of key genes. For a complete list of key genes, see Supplementary Table 4.
Through an unbiased systems biology approach,12 we determined the key genes predicted to influence tumor progression (Supplementary Table 6). For age (Fig. 3E), JUN, FOS, HMOX, PAK3, and SRC were the key genes involved in the most interactions in tumor progression. For sex, CYCS, JUNB, and FCER1G oncogenes were upregulated in males predicting tumor progression (Fig. 3F), while several genes demonstrated an inhibitory effect on tumor progression in females (Fig. 3H).
This difference can be explained when integrating age into the analysis. Comparing upstream regulators, there was an overall inhibitory effect on tumor progression in older females versus older males (Fig. 4A). For younger females (versus younger males), there was only one upstream regulator predicted to be activated (KRAS) impacting tumor progression (Fig. 4B). In men (Fig. 3G), JUN and BCL2 were centrally involved in tumor progression, while in females (Fig. 3H), JUN, CTNNB1, and IRF7 were the central genes. Collectively, this surprisingly indicated that older DLBCL females were predicted to have a favorable outcome compared with not only older males but also younger females. This demonstrates the importance of not ignoring either age or sex biological differences when studying tumor biology.
The same pattern was observed when studying the influence of key genes on tumor progression. Older females (versus older males) had an overall inhibition of tumor progression with CEBPB, CYCS, CCL4, ANXA2, TXN, and FCER1G being nodes interacting with the key genes having an inhibitory effect (ie, downregulating tumor promoters) (Fig. 4C and Supplementary Table 6).
Through the DAVID Gene Functional Classification Tool22 that measures relationships among different biologically annotated terms based on degrees of co-association of genes, we found that key genes for most groups also strongly regulated immune function activity (Supplementary Tables 7–11). Specifically, comparing old versus young groups, we observed functions related to the immune system (eg, immune system development, T-cell activation, immune response, etc.) appeared in the top 5 clusters; when comparing key genes from females to males, the immune response was observed in the top functional cluster with the greatest functional changes occurring in older populations.
In addition, the somatic form of cytochrome C (CYCS) was shown to be a critical inhibitory factor in older females versus older males (Fig. 4C), independent of age (Fig. 3F). CYCS plays a central role in the mitochondrial electron-transport chain and is a factor released rapidly in apoptotic stimulus when a cell undergoes DNA damage or metabolic stress.23 Prior data have shown that increased CYCS expression is associated with inferior survival in DLBCL.24
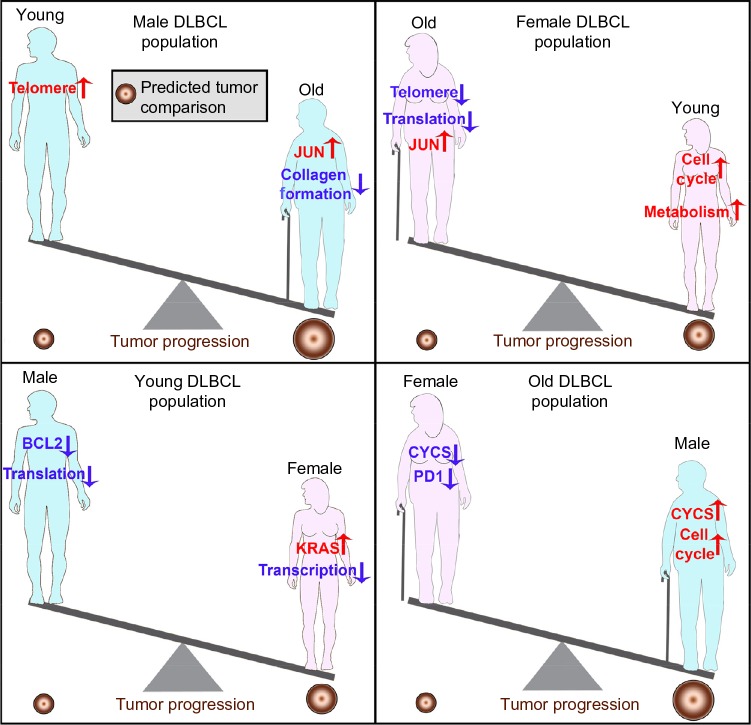
The overall goal for the treatment of DLBCL is to provide improved individualized treatment strategies and/or to discover key conserved biomarkers for DLBCL. The global impact from these factors predicted that tumor progression should be promoted for older DLBCL patients (Fig. 5), which is in agreement with the epidemiological data.25 Our data also demonstrated that the older female population with DLBCL should have a better outcome based on the biology (Fig. 5). Previous gender-based population studies have demonstrated that older female populations tend to have better outcome and lower incidence of DLBCL compared to male populations, but the biological reasons have not been previously investigated.10,11 Additionally, JUN was the only specific key gene that was commonly upregulated for both older male and female DLBCL patients when compared with younger patients and is also part of the seven common genes (Figs. 1E and 3). Blonska et al recently showed that JUN signaling is key in promoting DLBCL growth through the interaction with the microenvironment.26 The current data expand on this finding and further demonstrate that JUN is a critical factor that is upregulated as a function of age, independent of gender. An effective treatment strategy for older DLBCL patients may be via use of specific JUN small molecule inhibitors.27–29 A schematic of the overall predictions of tumor growth based on the biology is presented to summarize the results (Fig. 5).
Figure 5.
Schematic of DLBCL growth differences associated with age and sex, based on the biological predictions from TCGA data. The tumor size varies depending on age and sex. The regulation of key factors and functions are shown within the schematic.
Conclusions
Collectively, findings herein reinforce the importance of biological understanding in DLBCL. JUN was a predicted upstream regulator, and it was the only commonly upregulated key gene in older DLBCL patients, independent of sex. Treatment strategies targeting JUN should be investigated for older DLBCL patients.27–29 For sex, CYCS was shown to be a critically connected factor. Additionally, an extensive literature search reveals that all differences and key factors reported between age and sex for DLBCL patients were not found to occur in normal sex and aged populations. Although this study is limited by a small sample size potentially increasing the false-positive rate, there is significant association with additional molecular and epidemiological data found in independent publications. Continued examination of the impact of sex-based molecular changes should be explored not only for DLBCL but also for all types of cancer. Altogether, understanding how molecular factors interact and change as a function of age and sex, and how this impacts tumor progression, may improve our understanding of carcinogenesis as well as lead to enhanced therapeutic strategies.
Supplementary Materials
Supplementary Table 1. Number of patients from the TCGA dataset used for each grouping.
Supplementary Table 2. Complete list of Gene Set Enrichment Analysis (GSEA) for Reactome gene sets for old DLBCL patients (>58 years old) compared to young DLBCL patients (<58 years old) with gender independent and gender specific analysis and Female vs Male DLBCL patients age independent analysis. Leading edge analysis with a FDR < 0.05 determined significant gene sets enriched for each group. The normalized enrichment score (NES) indicates whether the gene set is up- (>0) or down-regulated (<0) for each group determined with the # of leading edge genes (appears in parenthesis).
Supplementary Table 3. Complete list of Gene Set Enrichment Analysis (GSEA) for Reactome gene sets for old DLBCL patients (>58 years old) compared to young DLBCL patients (<58 years old) with gender independent and gender specific analysis and Female vs Male DLBCL patients age independent analysis. Leading edge analysis with a FDR < 0.05 determined significant gene sets enriched for each group. The FDR q-values are shown for each gene set.
Supplementary Table 4. Complete list of Gene Set Enrichment Analysis (GSEA) for Reactome gene sets for old DLBCL patients (>58 years old) compared to young DLBCL patients (<58 years old) with gender independent and gender specific analysis and Female vs Male DLBCL patients age independent analysis. Leading edge analysis with a FDR < 0.05 determined significant gene sets enriched for each group. The p-values are shown for each gene set.
Supplementary Table 5. Upstream regulators predicted to be activated or inhibited (as indicated by the activation z-score) for old DLBCL patients (>58 years old) compared to young DLBCL patients (<58 years old) or Female to Male DLBCL patients through the use of Ingenuity Pathway Analysis (IPA) software. Regulation z-score indicates the degree of inhibition (for negative values) or activation (for positive values). The third column denotes the effects the upstream regulators have on tumor progression based on the literature (reference in superscript). N.D. represents upstream regulators which are Not Determined to have any effects on tumor progression in the literature.
Supplementary Table 6. Key genes involved in age-dependent changes for DLBCL patients comparing age/gender independent analysis with age/gender dependent analysis. These key genes were determined by finding the common genes that overlap with the significant upstream regulators, the biofunction analysis and Gene Set Enrichment Analysis (GSEA) from C2 annotations with FDR < 0.05. Male data set was only done with common genes between the significant upstream regulators and the biofunction analysis, since no overlap was found with GSEA data. The second column denotes the effects these genes have on tumor progression based on the literature. Genes with no reported effects on tumors were identified as Not Determined (ND).
Supplementary Table 7. All: Old vs Young. The top 15 functional annotation clusters determined from key genes for All: Old vs Young DLBCL patients. This was determined through DAVID Gene Functional Classification Tool. The enrichment scores were determined by DAVID through the geometric mean of the EASE scores (modified Fisher Exact).
Supplementary Table 8. Female: Old vs Young. The top 15 functional annotation clusters determined from key genes for Female: Old vs Young DLBCL patients. This was determined through DAVID Gene Functional Classification Tool. The enrichment scores were determined by DAVID through the geometric mean of the EASE scores (modified Fisher Exact).
Supplementary Table 9. Male: Old vs Young. All functional annotation clusters determined from key genes for Male: Old vs Young DLBCL patients. This was determined through DAVID Gene Functional Classification Tool. The enrichment scores were determined by DAVID through the geometric mean of the EASE scores (modified Fisher Exact).
Supplementary Table 10. All: Female vs Male. The top 15 functional annotation clusters determined from key genes for All: Female vs Male DLBCL patients. This was determined through DAVID Gene Functional Classification Tool. The enrichment scores were determined by DAVID through the geometric mean of the EASE scores (modified Fisher Exact).
Supplementary Table 11. Old: Female vs Male. The top 15 functional annotation clusters determined from key genes for Old: Female vs Male DLBCL patients. This was determined through DAVID Gene Functional Classification Tool. The enrichment scores were determined by DAVID through the geometric mean of the EASE scores (modified Fisher Exact).
Acknowledgments
The results published here are in whole or part based upon data generated by the TCGA Research Network: http://cancergenome.nih.gov.
Footnotes
ACADEMIC EDITOR: J. T. Efird, Editor in Chief
PEER REVIEW: Five peer reviewers contributed to the peer review report. Reviewers’ reports totaled 1109 words, excluding any confidential comments to the academic editor.
FUNDING: This research was supported in part by R01 CA164311 (AME). The authors confirm that the funder had no influence over the study design, content of the article, or selection of this journal.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
Paper subject to independent expert blind peer review. All editorial decisions madeby independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE).
Author Contributions
Designed the study, performed research and all data analysis, and wrote the manuscript: AB, AME. Provided exchange of ideas and assisted with editing the manuscript: DN, JTM, CRV. All authors reviewed and approved of the final manuscript.
REFERENCES
- 1.Shankland KR, Armitage JO, Hancock BW. Non-Hodgkin lymphoma. Lancet. 2012;380(9844):848–57. doi: 10.1016/S0140-6736(12)60605-9. [DOI] [PubMed] [Google Scholar]
- 2.Sarkozy C, Coiffier B. Diffuse large B-cell lymphoma in the elderly: a review of potential difficulties. Clin Cancer Res. 2013;19(7):1660–9. doi: 10.1158/1078-0432.CCR-12-2837. [DOI] [PubMed] [Google Scholar]
- 3.Dixon DO, Neilan B, Jones SE, et al. Effect of age on therapeutic outcome in advanced diffuse histiocytic lymphoma: the Southwest Oncology Group experience. J Clin Oncol. 1986;4(3):295–305. doi: 10.1200/JCO.1986.4.3.295. [DOI] [PubMed] [Google Scholar]
- 4.Klapper W, Kreuz M, Kohler CW, et al. Molecular Mechanisms in Malignant Lymphomas Network Project of the Deutsche Krebshilfe. Patient age at diagnosis is associated with the molecular characteristics of diffuse large B-cell lymphoma. Blood. 2012;119(8):1882–7. doi: 10.1182/blood-2011-10-388470. [DOI] [PubMed] [Google Scholar]
- 5.Nabhan C, Smith SM, Helenowski I, et al. Analysis of very elderly (>/=80 years) non-Hodgkin lymphoma: impact of functional status and co-morbidities on outcome. Br J Haematol. 2012;156(2):196–204. doi: 10.1111/j.1365-2141.2011.08934.x. [DOI] [PubMed] [Google Scholar]
- 6.Sarkozy C, Mounier N, Delmer A, et al. Impact of BMI and gender on outcomes in DLBCL patients treated with R-CHOP: a pooled study from the LYSA. Lymphoma. 2014;2014:12. [Google Scholar]
- 7.Hasselblom S, Ridell B, Nilsson-Ehle H, Andersson PO. The impact of gender, age and patient selection on prognosis and outcome in diffuse large B-cell lymphoma – a population-based study. Leuk Lymphoma. 2007;48(4):736–45. doi: 10.1080/10428190601187703. [DOI] [PubMed] [Google Scholar]
- 8.Pfreundschuh M, Poeschel V, Zeynalova S, et al. Optimization of rituximab for the treatment of diffuse large B-cell lymphoma (II): extended rituximab exposure time in the SMARTE-R-CHOP-14 trial of the German high-grade non-Hodgkin lymphoma study group. J Clin Oncol. 2014;32(36):4127–33. doi: 10.1200/JCO.2013.54.6861. [DOI] [PubMed] [Google Scholar]
- 9.Pfreundschuh M, Müller C, Zeynalova S, et al. Suboptimal dosing of rituximab in male and female patients with DLBCL. Blood. 2014;123(5):640–6. doi: 10.1182/blood-2013-07-517037. [DOI] [PubMed] [Google Scholar]
- 10.Ansell P, Simpson J, Lightfoot T, et al. Non-Hodgkin lymphoma and autoimmunity: does gender matter? Int J Cancer. 2011;129(2):460–6. doi: 10.1002/ijc.25680. [DOI] [PubMed] [Google Scholar]
- 11.Szekely E, Hagberg O, Arnljots K, Jerkeman M. Improvement in survival of diffuse large B-cell lymphoma in relation to age, gender, International Prognostic Index and extranodal presentation: a population based Swedish Lymphoma Registry study. Leuk Lymphoma. 2014;55(8):1838–43. doi: 10.3109/10428194.2013.853297. [DOI] [PubMed] [Google Scholar]
- 12.Beheshti A, Benzekry S, McDonald JT, et al. Host age is a systemic regulator of gene expression impacting cancer progression. Cancer Res. 2015;75(6):1134–43. doi: 10.1158/0008-5472.CAN-14-1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuchenbaecker KB, Ramus SJ, Tyrer J, et al. GEMO Study Collaborators. Breast Cancer Family Registry. HEBON. KConFab Investigators. Australian Cancer Study (Ovarian Cancer Investigators) Australian Ovarian Cancer Study Group; Consortium of Investigators of Modifiers of BRCA1 and BRCA2 Identification of six new susceptibility loci for invasive epithelial ovarian cancer. Nat Genet. 2015;47(2):164–71. doi: 10.1038/ng.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang J, Liu B, Jiang X, et al. A systems biology-based gene expression classifier of glioblastoma predicts survival with solid tumors. PLoS One. 2009;4(7):e6274. doi: 10.1371/journal.pone.0006274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anguiano A, Tuchman SA, Acharya C, et al. Gene expression profiles of tumor biology provide a novel approach to prognosis and may guide the selection of therapeutic targets in multiple myeloma. J Clin Oncol. 2009;27(25):4197–203. doi: 10.1200/JCO.2008.19.1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang Y, Han L, Yuan Y, Li J, Hei N, Liang H. Gene co-expression network analysis reveals common system-level properties of prognostic genes across cancer types. Nat Commun. 2014;5:3231. doi: 10.1038/ncomms4231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saeed AI, Bhagabati NK, Braisted JC, et al. TM4 microarray software suite. Methods Enzymol. 2006;411:134–93. doi: 10.1016/S0076-6879(06)11009-5. [DOI] [PubMed] [Google Scholar]
- 18.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Merico D, Isserlin R, Stueker O, Emili A, Bader GD. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One. 2010;5(11):e13984. doi: 10.1371/journal.pone.0013984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wage J, Ma L, Peluso M, et al. Proton irradiation impacts age-driven modulations of cancer progression influenced by immune system transcriptome modifications from splenic tissue. J Radiat Res. 2015;56(5):792–803. doi: 10.1093/jrr/rrv043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153(6):1194–217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 23.Ow YP, Green DR, Hao Z, Mak TW. Cytochrome c: functions beyond respiration. Nat Rev Mol Cell Biol. 2008;9(7):532–42. doi: 10.1038/nrm2434. [DOI] [PubMed] [Google Scholar]
- 24.Wilkinson ST, Johnson DB, Tardif HL, Tome ME, Briehl MM. Increased cytochrome c correlates with poor survival in aggressive lymphoma. Oncol Lett. 2010;1(2):227–30. doi: 10.3892/ol_00000040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Howlader N, Noone AM, Krapcho M, et al. SEER Cancer Statistics Review, 1975–2011. Bethesda, MD: 2014. Available at: http://seer.cancer.gov/csr/1975_2011/. based on November 2013 SEER data submission. [Google Scholar]
- 26.Blonska M, Zhu Y, Chuang HH, et al. Jun-regulated genes promote interaction of diffuse large B-cell lymphoma with the microenvironment. Blood. 2015;125(6):981–91. doi: 10.1182/blood-2014-04-568188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bogoyevitch MA, Arthur PG. Inhibitors of c-Jun N-terminal kinases: JuNK no more? Biochim Biophys Acta. 2008;1784(1):76–93. doi: 10.1016/j.bbapap.2007.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koch P, Gehringer M, Laufer SA. Inhibitors of c-Jun N-terminal kinases: an update. J Med Chem. 2015;58(1):72–95. doi: 10.1021/jm501212r. [DOI] [PubMed] [Google Scholar]
- 29.LoGrasso P Kamenecka T. Inhibitors of c-jun-N-terminal kinase (JNK) Mini Rev Med Chem. 2008;8(8):755–66. doi: 10.2174/138955708784912120. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. Number of patients from the TCGA dataset used for each grouping.
Supplementary Table 2. Complete list of Gene Set Enrichment Analysis (GSEA) for Reactome gene sets for old DLBCL patients (>58 years old) compared to young DLBCL patients (<58 years old) with gender independent and gender specific analysis and Female vs Male DLBCL patients age independent analysis. Leading edge analysis with a FDR < 0.05 determined significant gene sets enriched for each group. The normalized enrichment score (NES) indicates whether the gene set is up- (>0) or down-regulated (<0) for each group determined with the # of leading edge genes (appears in parenthesis).
Supplementary Table 3. Complete list of Gene Set Enrichment Analysis (GSEA) for Reactome gene sets for old DLBCL patients (>58 years old) compared to young DLBCL patients (<58 years old) with gender independent and gender specific analysis and Female vs Male DLBCL patients age independent analysis. Leading edge analysis with a FDR < 0.05 determined significant gene sets enriched for each group. The FDR q-values are shown for each gene set.
Supplementary Table 4. Complete list of Gene Set Enrichment Analysis (GSEA) for Reactome gene sets for old DLBCL patients (>58 years old) compared to young DLBCL patients (<58 years old) with gender independent and gender specific analysis and Female vs Male DLBCL patients age independent analysis. Leading edge analysis with a FDR < 0.05 determined significant gene sets enriched for each group. The p-values are shown for each gene set.
Supplementary Table 5. Upstream regulators predicted to be activated or inhibited (as indicated by the activation z-score) for old DLBCL patients (>58 years old) compared to young DLBCL patients (<58 years old) or Female to Male DLBCL patients through the use of Ingenuity Pathway Analysis (IPA) software. Regulation z-score indicates the degree of inhibition (for negative values) or activation (for positive values). The third column denotes the effects the upstream regulators have on tumor progression based on the literature (reference in superscript). N.D. represents upstream regulators which are Not Determined to have any effects on tumor progression in the literature.
Supplementary Table 6. Key genes involved in age-dependent changes for DLBCL patients comparing age/gender independent analysis with age/gender dependent analysis. These key genes were determined by finding the common genes that overlap with the significant upstream regulators, the biofunction analysis and Gene Set Enrichment Analysis (GSEA) from C2 annotations with FDR < 0.05. Male data set was only done with common genes between the significant upstream regulators and the biofunction analysis, since no overlap was found with GSEA data. The second column denotes the effects these genes have on tumor progression based on the literature. Genes with no reported effects on tumors were identified as Not Determined (ND).
Supplementary Table 7. All: Old vs Young. The top 15 functional annotation clusters determined from key genes for All: Old vs Young DLBCL patients. This was determined through DAVID Gene Functional Classification Tool. The enrichment scores were determined by DAVID through the geometric mean of the EASE scores (modified Fisher Exact).
Supplementary Table 8. Female: Old vs Young. The top 15 functional annotation clusters determined from key genes for Female: Old vs Young DLBCL patients. This was determined through DAVID Gene Functional Classification Tool. The enrichment scores were determined by DAVID through the geometric mean of the EASE scores (modified Fisher Exact).
Supplementary Table 9. Male: Old vs Young. All functional annotation clusters determined from key genes for Male: Old vs Young DLBCL patients. This was determined through DAVID Gene Functional Classification Tool. The enrichment scores were determined by DAVID through the geometric mean of the EASE scores (modified Fisher Exact).
Supplementary Table 10. All: Female vs Male. The top 15 functional annotation clusters determined from key genes for All: Female vs Male DLBCL patients. This was determined through DAVID Gene Functional Classification Tool. The enrichment scores were determined by DAVID through the geometric mean of the EASE scores (modified Fisher Exact).
Supplementary Table 11. Old: Female vs Male. The top 15 functional annotation clusters determined from key genes for Old: Female vs Male DLBCL patients. This was determined through DAVID Gene Functional Classification Tool. The enrichment scores were determined by DAVID through the geometric mean of the EASE scores (modified Fisher Exact).