Abstract
Recent and rapid advances in genetic and molecular tools have brought spectacular tractability to Caenorhabditis elegans, a model that was initially prized because of its simple design and ease of imaging. C. elegans has long been a powerful model in biomedical research, and tools such as RNAi and the CRISPR/Cas9 system allow facile knockdown of genes and genome editing, respectively. These developments have created an additional opportunity to tackle one of the most debilitating burdens on global health and food security: parasitic nematodes. I review how development of nonparasitic nematodes as genetic models informs efforts to import tools into parasitic nematodes. Current tools in three commonly studied parasites (Strongyloides spp., Brugia malayi, and Ascaris suum) are described, as are tools from C. elegans that are ripe for adaptation and the benefits and barriers to doing so. These tools will enable dissection of a huge array of questions that have been all but completely impenetrable to date, allowing investigation into host–parasite and parasite–vector interactions, and the genetic basis of parasitism.
Keywords: C. elegans, CRISPR, parasitic nematode, tool development
CAENORHABDITIS ELEGANS has been a flagship model organism for over 40 years, and the research community has harnessed it to explore a wide range of biological questions, including but not limited to cell death, developmental timing, genome defense, neurobiology, and aging. The value of C. elegans as a model was not diminished by the lack of genetic tools. However, recent technological advances that enable genome editing, transcriptional and translational manipulation, in vivo labeling, and cell-specific proteomics have opened new avenues of investigation. Thanks to significant investment of time and resources, C. elegans now has an extensive toolkit to facilitate these studies.
In recent years, the global health burden of parasitic nematodes has gained wide attention, as they infect over a billion people (Hotez et al. 2008; Lustigman et al. 2012). Infection is rarely fatal, but those who are infected can suffer from debilitating symptoms. Helminth infection depresses human health, cognition, and productivity, and increases the severity of other common diseases caused by viruses and bacterial pathogens including HIV, malaria, and tuberculosis (Hotez et al. 2008; Lustigman et al. 2012). Furthermore, nematodes also parasitize crops and livestock, causing huge losses in production (Jones et al. 2013; Roeber et al. 2013). The World Health Organization (WHO) has listed certain parasite infections as targets for eradication by the year 2020 (ITFDE 2008; Boatin et al. 2012; Keenan et al. 2013). Efforts to define eradication strategies can be challenging; many parasites have intricate lifecycles with specific host and vector requirements, which in contrast to C. elegans, has limited the types of molecular analyses that can be performed. Recognizing the urgency and scope of these issues, both the C. elegans and the parasitic nematode communities have responded by bringing together the expertise and knowledge of their respective organisms; at two recent international C. elegans meetings, there were workshops to bridge the divide between C. elegans researchers and parasitologists. Furthermore, additional funding opportunities have been created, such as the Faucett Catalyst grant in 2013.
In this “toolbox” review I will: (i) outline the state of technology in several commonly studied parasitic nematodes, which could be employed by C. elegans researchers looking to translate their findings into a biomedically relevant parasite; (ii) highlight tools from C. elegans that could be valuable to parasitologists; and (iii) briefly discuss research areas that can be opened by developing these tools. I will focus on two technologies: fluorescent reporters and the CRISPR/Cas9 system for genome editing.
Lessons Learned from Import into Nonparasites
Beautiful evolution-development and population genetics research has come from using C. elegans, related Caenorhabditis species, and other nonparasitic nematodes such as Pristionchus pacificus. A major impediment to advancing these studies was the lack of genetic tools in these nonparasitic species. It is therefore instructive to examine the barriers faced by researchers seeking to develop C. briggsae and other nonparasitic species as genetic models. Hindsight allows for the identification of particularly successful adaptations, informing fruitful lines of inquiry.
Wei et al. (2014) astutely noted that it typically takes years to build a collection of tools, even those as basic as isolated marker mutations and genetic balancers. Bioinformatic resources such as an annotated genome sequence and single nucleotide polymorphism maps were key in driving development of C. briggsae as a genetic model. The acquisition of a Cbr-unc-119 mutation from a genetic screen provided a selectable marker to permit ballistic transformation and generation of fluorescent reporter strains (Zhao et al. 2010). Similarly, P. pacificus has a sequenced genome and a well-developed toolkit consisting of forward genetic approaches and DNA transformation (Hong and Sommer 2006; Dieterich et al. 2008; Schlager et al. 2009; Cinkornpumin and Hong 2011; Sommer and McGaughran 2013; Witte et al. 2014).
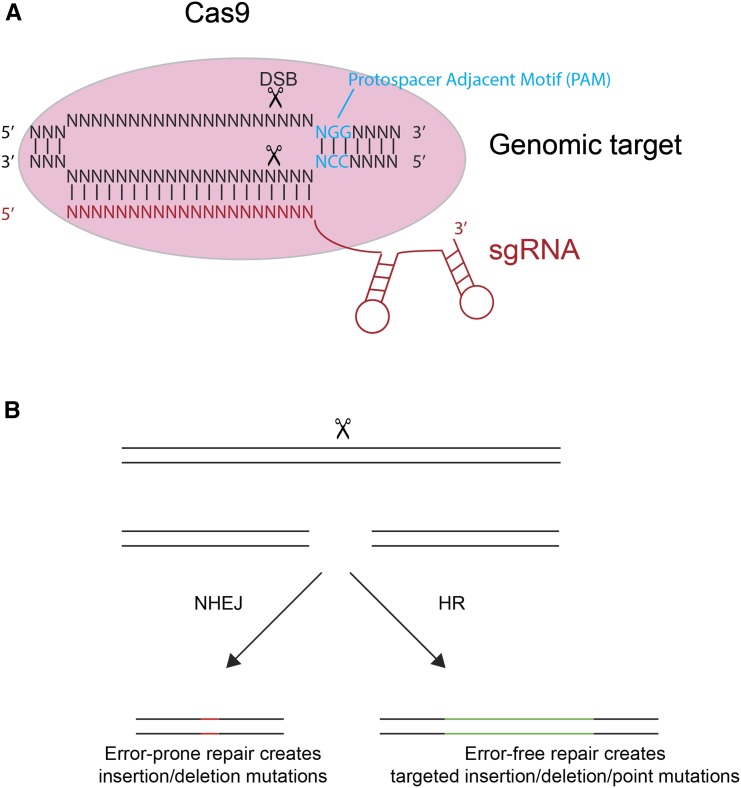
The development of these genetic models has been greatly facilitated by development of site-specific DNA endonucleases to permit genome editing. First, zinc finger nucleases and transcription activator-like effector nucleases (TALENs), and now clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 (Wood et al. 2011; Lo et al. 2013; Wei et al. 2014) provide researchers the ability to inactivate genes or to knockin desired sequences. Zinc finger nucleases were expensive and required complex algorithms to design, while TALENs were cost effective but somewhat laborious to generate. CRISPR/Cas9 editing has largely supplanted these technologies based on its simplicity, modularity, cost effectiveness, and ease of generating new reagents. CRISPR is an adaptive bacterial immune system that uses a DNA endonuclease to destroy incoming foreign DNA (for a more detailed description see Marraffini and Sontheimer 2010). For genome editing approaches, one uses a simplified system consisting of the Cas9 nuclease and a single guide RNA (sgRNA), which performs the combined function of two small RNAs (CRISPR RNA (crRNA) and trans-activating crRNA (trcRNA)) in the Streptococcus pyogenes CRISPR/Cas9 defense (Jinek et al. 2012; Ran et al. 2013). Cas9–sgRNA complexes target 5′-(N20)NGG-3′ motifs in the genome (Marraffini and Sontheimer 2010; Jinek et al. 2012) (Figure 1A). The 20 base pairs at the 5′ end of the sgRNA are homologous to this (N20) sequence and base pair with the opposite strand (Figure 1A). The NGG sequence, known as a protospacer adjacent motif (PAM), is in the genomic target but not the sgRNA and stimulates Cas9 to make a DNA double-stranded break between the third and fourth base 5′ to the PAM (Figure 1A). One can introduce Cas9 and sgRNAs as: (i) ribonucleoprotein (RNP) complexes; (ii) in vitro transcribed RNA; or (iii) encoding double-stranded DNA (dsDNA) molecules such as plasmids or PCR products.
Figure 1.
CRISPR/Cas9 editing. (A) The Cas9:sgRNA complex scans the genome searching for “NGG” PAM sequences (blue text). The 20 bp at the 5′ end of the sgRNA are homologous to the 20 bp 5′ to the PAM in the genomic target site, and the sgRNA pairs with the complementary strand in the genomic target sequence. The PAM sequence instructs Cas9 to make a DNA double-stranded break (DSB) three to four nucleotides 5′ to the PAM. (B) A Cas9-induced DSB can be repaired through error-prone nonhomologous end joining (NHEJ) or error-free homologous recombination (HR) pathways. NHEJ can be used for gene inactivation, as it produces small, random insertion and deletion mutations at the DSB site. By supplying repair templates with homology arms flanking the break site, one can use HR to precisely delete sequences, insert epitopes, or introduce point mutations. To recover HR-mediated knock-ins, one must often either screen large numbers of cells/animals or use positive selection/co-CRISPR approaches; HR is typically less efficient at producing edits compared to NHEJ.
For relatively closely related nematodes, DNA-based CRISPR reagents might be directly imported; Cas9 and sgRNA expression plasmids from C. elegans displayed activity in C. briggsae (Culp et al. 2015). For more distantly related nematodes, an advantage of Cas9–sgRNA in vitro transcription (Chiu et al. 2013; Lo et al. 2013) and RNPs (Cho et al. 2013; Paix et al. 2015) is that they do not require the a priori knowledge of promoters and 3′-UTRs required to express Cas9 or sgRNAs from plasmids or linear dsDNA molecules. Notably, the first genome editing approaches in Caenorhabditis sp. 9 and P. pacificus used in vitro transcribed TALENs as opposed to DNA-based delivery (Wood et al. 2011; Lo et al. 2013). With respect to CRISPR/Cas9, delivery of in vitro transcribed RNA may not be ideal, as Cas9 messenger RNA (mRNA) must be translated while the sgRNA must then complex with the resulting protein product. The window of editing activity is therefore determined by the half-life of the in vitro transcribed sgRNA, the time required to translate Cas9, and the stability of Cas9. A recent report described an RNP-based editing approach in P. pacificus using Cas9 protein and in vitro transcribed sgRNA (Witte et al. 2014). This experimental setup likely represents the highest possible editing efficiency, as a similar approach in C. elegans reported highly efficient genome editing (Paix et al. 2015). However, this approach requires large amounts of Cas9 protein and pure RNA, which can be purchased commercially, though could be cost prohibitive for some groups. Alternatively, labs equipped for biochemistry and planning to do significant amounts of genome editing can make their own Cas9 and T7 RNA polymerase (Fu et al. 2014; Paix et al. 2015), greatly reducing cost.
In the path to developing C. briggsae and P. pacificus as model systems, key steps were the development of DNA transformation approaches, an annotated genome and transcriptome, and the cognate bioinformatic tools required to mine these resources. These investments are essential to realize the true potential of any parasitic nematode as a model.
Key steps in developing a nematode as a model
Annotated genome and transcriptome
Method(s) of transgenesis
Genome editing to enable rapid genetic analyses
Existing Technologies in Parasites
The number of nematodes with sequenced genomes is ever expanding, due to efforts of many groups such as the 959 Nematode Genomes Project (Kumar et al. 2014), the 50 Helminth Genomes Initiative, and the Filarial Worms Genomes Project. Nematode.net (Wylie et al. 2004; Martin et al. 2015), and WormBase ParaSite (Li et al. 2015) provide useful tools for navigating these sequences. Importantly, the plummeting cost and increasing sensitivity of high throughput sequencing allow analyses of the genome, transcriptome, or variome of a given parasite, provided clean DNA or RNA can be obtained. For example, these tools have led to insight in RNA interference (RNAi) and microRNA (miRNA) biology in a range of nematodes (Dalzell et al. 2011; Wang et al. 2011; Winter et al. 2012; Ahmed et al. 2013; Poole et al. 2014; Sarkies et al. 2015), discovery of parasite-secreted RNAs that can modulate host biology (Buck et al. 2014; Zamanian et al. 2015), and led to revelation that germline-expressed genomic DNA is lost in Ascaris suum somatic cells (Wang et al. 2012).
In contrast to these genomic/transcriptomic approaches, powerful genetic tools, such as transgenesis, RNAi, and CRISPR editing, rely on parasite culture. In this section, I will highlight the most useful genetic tools of several species of parasites (Strongyloid spp., Brugia malayi, and A. suum) noting that Strongyloid species provide the most advanced toolkit; tools in other parasites currently lag far behind.
Strongyloides spp.: the most sophisticated toolkit
Strongyloides spp. are soil-dwelling helminths with complex lifecycles involving alternation of free-living and parasitic stages (heterogony). Infectious L3 (iL3) stage larvae residing in the soil burrow through the skin of their host, migrate to alveolar macrophages, are coughed up, and swallowed. During this migration, they molt twice so that adult nematodes reside in the gut where the parthenogenic parasite females produce eggs (Table 1). These eggs are either shed in feces or hatch within the intestine. The shed eggs hatch outside the host and either directly develop to iL3 larvae, the parasitic stage, or enter an alternate, free-living stage (Viney and Lok 2015). For most species of Strongyloides, there is a single free-living generation, and all progeny of the males and females develop to iL3 (Viney and Lok 2015). Interestingly, the felid parasite Strongyloides planiceps can undergo several sequential free-living generations, with the proportion of iL3 progeny increasing in each generation until no more free-living animals are produced (Yamada et al. 1991).
Table 1. Strongyloides spp. overview.
| Lifecycle | Health burden | Transgenesis | Genetic tools |
|---|---|---|---|
| • iL3 larvae penetrate skin, make it to small intestine | • Estimated 30–100 million infected | • Gonadal microinjection of adults (Li et al. 2006; 2011) | • Promoter:fluorescent protein reporters (Li et al. 2006; 2011; Junio et al. 2008; Stoltzfus et al. 2014) |
| • Adults produce larvae that are shed in stool | • Asymptomatic to mild manifestations in skin, lung, and intestines | • Transposons for insertional mutagenesis and stable transformation (Shao et al. 2012) | |
| • Larvae excreted into the environment can either develop directly into iL3 or enter a free-living stage, developing to adults outside the host | • Infection can be life threatening in immunocompromised individuals | ||
| • S. stercoralis larvae in large intestine can autoinfect by penetrating the intestine wall and migrating back to small intestine |
The free-living larvae develop to adulthood and mate to produce eggs, which hatch and develop to i L3 larvae. In the case of Strongyloides stercoralis, autoinfection cycles are possible. Eggs can hatch inside the intestine, develop to iL3 larvae, penetrate the intestinal mucosa or perianal skin, and migrate randomly to other organs. If these larvae migrate to the intestine via the lungs, the infection cycle can begin anew (Lok 2007; Viney and Lok 2015). Importantly, there is a free-living cycle in soil that is exploited in the lab to culture the parasites for one generation outside of a host (Viney and Lok 2015). S. stercoralis (dog or gerbil host) and S. ratti (rat host), two species commonly studied in the lab, both can be transformed by gonadal microinjection (Li et al. 2006, 2011). A gene expression “toolkit” has been constructed for S. stercoralis using a gfpS65C gene from a C. elegans expression system and the S. stercoralis era-1 3′-UTR (Li et al. 2006, 2011); cloning S. stercoralis promoter sequences into this vector allows for cell- and tissue-specific expression of GFP. This system was further refined by making it Gateway (Invitrogen) cloning compatible, allowing researchers to combine any promoter, coding sequence, and 3′-UTR to create expression vectors (Stoltzfus et al. 2014). In S. ratti, adaptation of the piggyBAC transposon system, commonly used in Drosophila melanogaster, allows for insertional mutagenesis and stable integration of transgenes (Shao et al. 2012). Notably, toggling between C. elegans and Strongyloides spp. has provided insight into genetic control of developmental transition to the iL3 stage (Wang et al. 2009, 2015; Stoltzfus et al. 2012a,b; 2014). Parastrongyloides trichosuri, a parasite of Australian possums of the genus Trichosurus, has great promise as a model for Strongyloides (Grant et al. 2006b). Unlike S. ratti and S. stercoralis, P. trichosuri can be maintained in a free-living state in the absence of a host for over 90 generations (Grant et al. 2006b), and gonadal microinjection of DNA can produce heritable transgenesis (Grant et al. 2006a).
Brugia: National Institutes of Health core funding driving tool development
Brugia species belong to the filarial nematode family, members of which cause lymphatic filariasis (B. malayi, B. timori, and Wuchereria bancrofti) and subcutaneous filariasis (Loa loa, Mansonella streptocerca, Onchocerca volvulus) in humans. Dog heartworms (Dirofilaria immitis), an important parasite from a veterinary perspective, are also filarial nematodes. Interestingly, many members of this family carry Wolbachia pipientis as an obligate endosymbiote (Paily et al. 2009). I will focus on Brugia malayi, as it has the most sophisticated toolkit. Disease caused by this parasite can range from asymptomatic to acute episodes of inflammation involving skin, lymph nodes, and lymphatic vessels (Chandy et al. 2011) (Table 2). The parasite is transmitted by a mosquito vector; next, a specialized L1 stage (microfilariae) is taken up by mosquitoes in a bloodmeal (Paily et al. 2009). The microfilariae shed an extracuticular sheath to become proper L1s and migrate to the thoracic muscle where they molt twice to become L3 larvae (Paily et al. 2009). The L3 larvae migrate to the mosquito’s head and enter the human host when taking a bloodmeal. The L3s molt twice in unknown locations in the human host to become adults, which reside in the lymphatic system and produce an average of 10,000 microfilariae a day (Paily et al. 2009).
Table 2. B. malayi overview.
| Lifecycle | Health burden | Transgenesis | Genetic tools |
|---|---|---|---|
| • L1–L3 in mosquito, L4–adult in mammalian host | • 120 million with lymphatic filariasis | • Bombardment of embryos and adult females (Higazi et al. 2002) | • Luciferase reporters in embryos and L3s (Xu et al. 2011) |
| • Adults live in lymphatic vessels | • B. malayi causes 20 million of these cases | • Gonadal microinjection of adult females (Higazi et al. 2002) | • Immunofluorescence (Landmann et al. 2012) |
| • Adult females can release up to 10,000 of specialized L1 larvae (microfilariae) per day | • Disfiguring lymphodema, elephantiasis, and scrotal swelling | • Calcium phosphate precipitate-mediated transformation of L3s in vivo or ex vivo (Xu et al. 2011) | • RNAi: on adult females to knock down embryo gene expression (Aboobaker and Blaxter 2003; Landmann et al. 2012); on L3 larvae ex vivo (Kushwaha et al. 2012); on L2 and L3 larvae in vivo (in mosquito vector; Song et al. 2010); Note RNAi may be variable in efficiency |
In 1969, Dr. Paul Thompson was awarded a National Institutes of Health (NIH) contract to establish the Filariasis Research Reagent Resource Center (FR3) to supply worms for his studies and those of other researchers (Michalski et al. 2011). Currently funded by the National Institute of Allergy and Infectious Diseases (NIAID) of the NIH, the FR3 maintains the lifecycle of B. malayi, B. pahangi, D. immitis, and the rodent parasite, Acanthocheilonema vitae. The role of distributing these reagents has recently been passed onto Biodefense and Emerging Infections Research Resources Repository (BEI Resources), which the NIAID established to provide reagents and tools for the study of a range of pathogens, emerging infectious disease agents, and nonpathogenic microbes. BEI ships parasites, vectors, and molecular and serological reagents at no cost. The FR3 and BEI have provided the single greatest value to lymphatic filariasis infection research, providing reagents to researchers around the globe. These resources have driven progress in these filarial nematodes, as otherwise facilities for rearing both the mosquito vector and mammalian host, typically jirds, would be required.
Tool development in B. malayi has been enabled by the range of DNA transformation techniques available: microinjection in adult females (Higazi et al. 2002), bombardment of adult females and embryos (Higazi et al. 2002), and calcium phosphate precipitate-mediated transformation of L3 larvae in cell culture media ex vivo or in jird peritonea in vivo (Xu et al. 2011). Available tools include: (i) immunofluorescence (Landmann et al. 2012); (ii) secreted Gaussia luciferase (Xu et al. 2011) and nonsecreted firefly and Renilla luciferase reporters (Liu et al. 2010), which can be used to study various aspects of gene expression, including miRNA control (Liu et al. 2015); and (iii) RNAi ex vivo on adult females to interrogate embryo gene function (Aboobaker and Blaxter 2003; Landmann et al. 2012), on L3 larvae ex vivo (Kushwaha et al. 2012), and in vivo in the mosquito vector to target L2 and L3 larvae (Song et al. 2010). It is important to note that in many parasitic nematodes, including Brugia spp., RNAi efficacy is extremely variable, with robust knockdown only observed for a limited number of genes (Geldhof et al. 2006; Maule et al. 2011; Dalzell et al. 2012). A recent RNAi protocol using heterogenous short interfering RNAs (hsiRNAs) has shown promise in knocking down a number of genes in the hands of two different groups (Landmann et al. 2012; Winter et al. 2013); however, more positive results are required to confirm the broad applicability of this tool.
B. pahangi, a canid parasite very similar to B. malayi, is often used to study Brugia biology, as: (i) it is easier to get large amounts of the parasite, (ii) the tools listed above should also work in B. pahangi, (iii) it has a sequenced genome (Lau et al. 2015), and (iv) it does not typically infect humans. Many researchers prefer working with B. pahangi, as the microfilariae can be harvested from dogs, unlike B. malayi, in which microfilariae are harvested from cats, which can be difficult to handle (J. Sakanari, personal communication).
Ascaris: a big target
Ascaris are large nematodes, up to 35 cm long, that infect pigs (A. suum) and humans (A. lumbricoides); notably, cross-infection is possible (Dold and Holland 2011). Most work has been done with A. suum, primarily owing to the availability of samples from the pork industry. A. suum can infect its host directly through contaminated food or water or via a paratenic host (Dold and Holland 2011) (Table 3). The pig ingests eggs with L2 larvae, which then hatch and then undergo hepatic migration (Dold and Holland 2011). These larvae molt to L3, migrate to the lungs where they enter alveoli, are coughed up, and swallowed (Dold and Holland 2011). Finally in the gut, they molt twice to become adults and mate; adults produce upwards of 200,000 eggs per day (Dold and Holland 2011).
Table 3. Ascaris spp. overview.
| Lifecycle | Health burden | Transgenesis | Genetic tools |
|---|---|---|---|
| • After ingestion of infective eggs, larvae hatch, penetrate intestine wall, and migrate to lungs by way of liver | • Estimated 800–1000 million infected with A. lumbricoides | • Bombardment in A. suum (Davis et al. 1999) | • Whole A. suum embryo extracts for biochemistry (Hannon et al. 1990; Denker et al. 2002; Lall et al. 2004) |
| • Larvae develop further, are coughed up, swallowed, and develop to adults in the small intestine | • Asymptomatic to light abdominal discomfort | • RNAi by microinjection into A. suum pseudocoelom (McCoy et al. 2015) | |
| • Adult females can release up to 200,000 eggs per day | • Heavy infections can cause intestinal blockage and stunt child development and growth | ||
The large size of Ascaris makes it amenable for biochemical studies. Whole cell embryo extracts have been used for in vitro studies of transcription, RNA processing, and translation, and have provided much initial insight into nematode splicing (Hannon et al. 1990; Davis et al. 1999; Denker et al. 2002; Lall et al. 2004). Fertilized zygotes arrest before pronuclear fusion, allowing storage of large numbers of embyos for years at 4°, which can then be synchronously released by exposure to an acidic environment that mimics the host stomach (R. E. Davis, personal communication). Postfertilization cell cycles are slow (13 hr, compared to 30 min in C. elegans) (Azzaria and McGhee 1992), allowing for excellent temporal resolution of early development. Although pigs are needed to maintain the lifecycle, embryonic and early larval development can be studied in vitro, and migration from the intestine to the liver and lungs can be studied in rabbits, mice, or rats (Campbell and Timinski 1965; Galvin 1968; Enobe et al. 2006). From a toolkit perspective, transient transformation is possible by bombardment, usually at the 32- to 64-cell stage, thereby introducing DNA or RNA into embryos (Davis et al. 1999). Excitingly, a recent report described a protocol in A. suum in which pseudocoelemic microinjection of dsRNA produced robust knockdown of multiple target transcripts in a range of tissues throughout the animal (McCoy et al. 2015). Exposure to Ascaris pseudocoelomic fluid can cause acute, severe allergic reactions in some laboratory workers (Kennedy and Qureshi 1986; Kennedy et al. 1987; Kennedy 2013) and has resulted in some researchers moving to other systems (A. Fire, personal communication).
Other Nematodes of Significant Interest and Importance
While I have focused on three parasitic nematode species with the best genetic toolkits (Strongyloides spp., Brugia spp., and A. suum), I stress that there are numerous other important parasitic nematodes not discussed in this review (Hotez et al. 2008; Jones et al. 2013; Roeber et al. 2013); many of these have complete, annotated genomes, or sequencing of those genomes is in progress. In this section, I will highlight several clinically or economically important species of nematodes and provide references for the reader to find information about these parasites.
Soil-dwelling helminths such as hookworm (Ancylostoma duodenale and Necator americanus) or whipworm (Trichuris trichiura) are important human pathogens that each infect hundreds of millions (Lustigman et al. 2012). A relative of T. trichiura that infects mice (T. muris) is a useful laboratory model for study of whipworms and used as a model gastrointestinal nematode for immunological studies (Foth et al. 2014). Parasites of livestock and crops pose an equally important problem to human well-being. Haemonchus contortus is the central model species for strongyloid parasitic nematodes of livestock, and provides a paradigm to study emergence of anthelmintic resistance (Gilleard 2013). Notably, C. elegans has been successfully used as a heterologous system to functionally characterize H. contortus genes (for example, see Glendinning et al. 2011). Ostertagia ostertagi is a gastrointestinal parasite of cattle and arguably the most important livestock parasite globally (Rinaldi and Geldhof 2012). Toxocara canis is emerging as an important zoonosis and has good genomic resources (Gasser 2013; Zhu et al. 2015).
Plant parasitic nematodes compromise global food security, causing an estimated US$80 billion in losses per year (Nicol et al. 2011). The rootknot nematodes (Meloidogyne spp.) attack a wide range of plants, including many common vegetables. The second juvenile stage (J2) of this parasite creates a permanent feeding site on the affected plant root, leading to formation of several giant cells on which the sedentary J2 animal extracts nutrients. Two rootknot nematodes, Meloidogyne hapla and M. incognita, have sequenced genomes that provided insight into the adaptations and genes required for their obligate parasitic lifecycle (Abad et al. 2008; Opperman et al. 2008). Cyst nematodes (Heterodera and Globodera spp.) are important pests for a range of crops, such as members of the Solanaceae family (i.e., potatoes and tomatoes), sugar beets, soy beans, and cereals; these parasites cause significant agroeconomic loss (Jones et al. 2013). Of the cyst nematodes, the potato cyst nematode (Globodera pallida), has a sequenced genome (Cotton et al. 2014); the soybean cyst nematode (Heterodera glycines) and sugar beet cyst nematode (Heterodera schachtii) genomes are in the process of being sequenced. There is a range of other important plant parasitic nematodes, nicely reviewed by Jones et al. (2013). Notably, unlike animal and human parasitic nematodes, RNAi appears to work very well in a range of plant parasitic nematodes, offering the possibility of genetic study (Maule et al. 2011). Nematodes can also be beneficial to agriculture (Lacey et al. 2015); entomopathogenic nematodes such as Steinernema spp. (Dillman et al. 2015) or Heterorhabditis bacteriophora (Bai et al. 2013) target a wide range of insect pests.
C. elegans Tools: Low Hanging Fruit Ripe for Picking
For all tools that I will describe, a system for transgenesis is a prerequisite; the system does not need to be elegant, one just needs to get DNA, RNA, or protein into the organism (Figure 2). Gonadal microinjection, bombardment, electroporation, or transformation are all possibilities. The observation that B. malayi L3 larvae can be transformed in culture may allow transformation/transfection methods commonly used in mammalian cells to be used in particular nematodes at select stages (i.e., cationic lipids, recently described iTOP for protein delivery) (D’Astolfo et al. 2015). Nematodes that can be coaxed to molt in culture may be more susceptible to these transfection approaches (Rajan et al. 2003; Xu et al. 2011). Notably, many important nematodes such as hookworms, whipworms, and plant parasitic nematodes currently do not have established DNA transformation protocols. As such, a significant investment is required to overcome this crucial barrier to further genetic and molecular study. However, the reward for doing so is great: these particular parasites pose a significant burden to human health and agriculture, the barrier is surmountable, and there is enormous potential for advancing the field using the tools that follow. In the following sections, I will outline several technologies that should be transferable to any nematode for which transformation is possible: reporters for gene expression and protein localization, methods to enrich transgenic animals, and last but certainly not least, genome editing with CRISPR/Cas9, which could greatly accelerate research into these diverse and biomedically and economically relevant organisms. Importantly, for those interested in developing these tools in a parasitic nematode, many of the C. elegans reagents have been deposited in AddGene, a nonprofit plasmid repository.
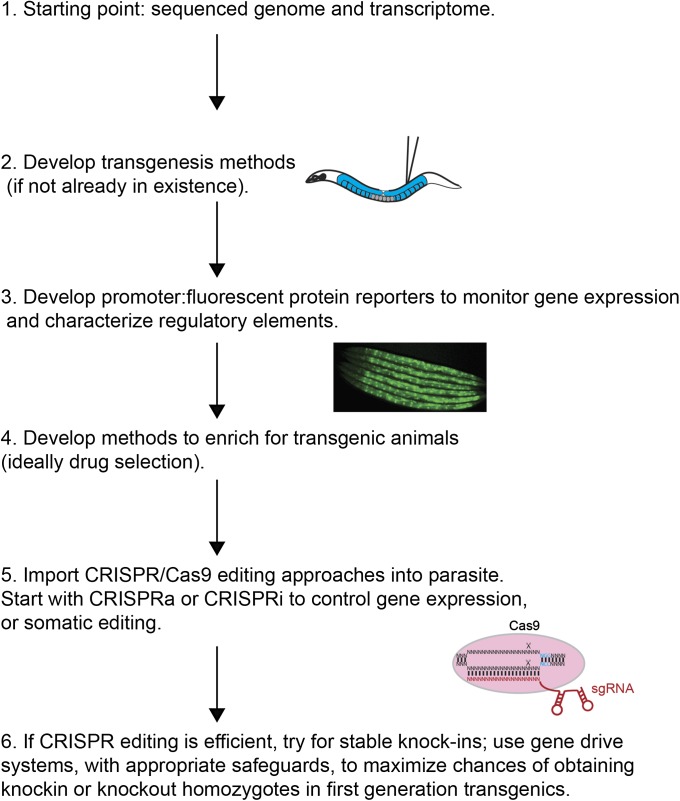
Figure 2.
Recommended pipeline for technology import into parasitic nematodes. One should start with a parasitic nematode species that has an annotated genome or transcriptome or generate these datasets. For the described genetic tools, a method of transgenesis is a prerequisite. Promoter:FP reporters should be used to characterize regulatory elements, and monitor gene expression, temporally and spatially. The regulatory elements characterized through the FP reporters are useful for development of selection systems and CRISPR editing approaches. Positive selection systems, such as drug resistance or FP reporter expression, help identify transgenic animals. Drug selection cassettes are preferable, as they could allow for in vivo selection of transgenic parasites in host animals. For CRISPR approaches, it is best to start with activation or repression of gene expression using CRISPRa or CRISPRi, respectively, or generation of indel mutations using somatic CRISPR editing. These approaches should work in first generation transgenic animals, circumventing issues with requisite host passage. If these methods are robust, the lifecycle can be maintained, and once a positive selection system is in place, one can attempt to generate stable knockouts or knockins. A gene drive system is recommended, with appropriate safeguards, to create homozygotes in first generation transgenics, and subsequently in any genetic background to which the transgene is introduced.
Look to your neighbors
Before starting, a phylogenetic perspective (Blaxter et al. 1998; Holterman et al. 2006) may be instructive for gauging how likely a given methodology from C. elegans is to work in a given parasite. Clade V nematodes such as hookworms may be more successful recipients than clade III nematodes, such as Brugia spp., or clade IV nematodes, such as Strongyloides spp. Indeed, C. elegans has been a useful system to study aspects of H. contortus (Clade V) gene function (for examples, see Glendinning et al. 2011; Law et al. 2015). Another important consideration is to learn from other free-living models (Lok 2009). The modifications required for development of a transgenesis method in P. pacificus were useful to improve methodologies in Strongyloides spp. (Lok 2009). As nicely observed by Lok (2009), the necromenic association of P. pacificus with the scarab beetle requires many of the “same crucial functions and life-cycle transitions that medically important parasitic nematodes execute in their vertebrate hosts.” Looking to these free-living models may help identify optimal conditions for generating lines of nematodes that stably express and inherit transgenes.
Reporter systems: an engine to drive other technologies
Developing reporter systems such as luciferase, beta-galactosidase (LacZ), or fluorescent proteins (FPs) are useful for two reasons: (i) they provide experimental information on putative regulatory DNA sequences, and (ii) they provide information on tissue- and stage specificity of gene expression. Obtaining a collection of validated promoters and 3′-UTRs is essential for developing DNA-based expression systems to drive cotransformation markers and express genes of interest. To date luciferase reporters have been used in B. malayi and fluorescent reporters such as GFP (Li et al. 2006; Li et al. 2011; Xu et al. 2011; Stoltzfus et al. 2014) and mRFPmars (Junio et al. 2008) have been employed in Strongyloides spp. The use of luciferase reporters in B. malayi has provided valuable information about splicing, gene regulation by the ecdysone receptor, and study of select promoters (Shu et al. 2003; Higazi and Unnasch 2004; Tzertzinis et al. 2010). Firefly luciferase was initially used to identify regions of the HSP70 promoter required for transgene expression (Shu et al. 2003), though a caveat of this approach is that animals must be homogenized for the assay. Gaussia luciferase, a secreted variant, can be measured in live animals thus allowing for monitoring of promoter activity in vivo (Xu et al. 2011). However, luciferase reporters cannot be used to examine spatial expression patterns.
LacZ is a very sensitive reporter gene, and it can be used to gauge expression patterns in vivo, but the animals must be fixed (Fire et al. 1990). Of note, LacZ reporters have been validated in transgenic Parastrongyloides trichuris (Grant et al. 2006a). Beware that there may be issues with endogenous beta-galactosidase expression, which can result in spurious background signal, particularly if the endogenous beta-galactosidase is activated at the slightly alkaline pH resulting from the fixation process (Weiss et al. 1999).
The reporters of choice in C. elegans are fluorescent proteins. FPs can be scored in living animals, enabling the collection of temporal and spatial expression data for both genes and proteins. A catalog of regulatory elements has been exploited to drive gene expression constitutively, inducibly, or in a temporally and spatially defined pattern. Autofluorescence may interfere with imaging in parasites, but the palette of FPs used in C. elegans continues to expand, virtually guaranteeing a solution to meet the needs of the model. Directed selection of mutant variants of the original GFP from Aequorea victoria and FPs from other organisms, such as Discosoma sp. (Shaner et al. 2004), led to the creation of red, yellow, and green FPs, as well as ultrabright FPs (3xVenus, mNeonGreen, tdTomato (Nagai et al. 2002; Shaner et al. 2004; Heisler et al. 2005; Shaner et al. 2013) and photostable FPs that cover a range of wavelengths (far red, near IR, blue, cyan, orange). In selecting promoters for expression of FPs in desired spatial patterns, one should make educated decisions based on multispecies alignments to identify conserved regulatory elements, and/or make use of existing RNA-seq data. For example, the promoter and 3′-UTR of the BmHsp70 gene drive luciferase reporter activity (Shu et al. 2003), and RNA-seq data confirm high expression of this transcript (Choi et al. 2011). Adding two copies of a nuclear localization sequence to drive nuclear expression of the FP, or three copies to drive nucleolar expression, can concentrate the FP to facilitate cell identification and visualization. Fusion of FPs to proteins of interest allows for live in vivo monitoring of protein localization, though the large size of the FP tag can potentially interfere with protein function or interaction with endogenous partners; occasionally one will need to try several different fusion locations of the FP within the protein of interest.
There are important caveats to be aware of, gleaned from the extensive use of FP reporters in C. elegans. While promoters provide temporal and spatial control of gene expression, FPs do not always require a promoter for expression (A. Fire, personal communication). They do, however, require a C. elegans 3′-UTR. The unc-54 3′-UTR was used initially, and is still frequently used (Fire et al. 1990). More recently, the let-858 3′-UTR (Kelly et al. 1997), which results in uniform expression across cell types, and the tbb-2 3′-UTR (Merritt et al. 2008), which permits germline expression of transgenes, have also been used. Introns are important for expression in C. elegans, and often short, synthetic introns are added to genes to facilitate expression (Fire et al. 1990). More recently developed LacZ reporters containing multiple synthetic introns are much more active in C. elegans than the original single intron LacZ reporter (A. Fire, personal communication). The same trend of increased expression with more introns is also true for GFP, but less dramatic, likely due to the shorter size of the GFP mRNA (A. Fire, personal communication). It is unclear whether these C. elegans synthetic introns will work in all parasites, but the C. elegans gfp65C cassette, which contains three synthetic introns, expresses in Strongyloides spp., a nematode highly diverged from C. elegans (Blaxter et al. 1998; Li et al. 2006, 2011). As introns could differ extensively, whether C. elegans FP cassettes can be directly used in a given parasite needs to be assessed on a case-by-case basis.
Enriching for transgenics
Once transgenesis and FP reporters are established in a model nematode, the next step is to develop methods to maintain and enrich for transgenic animals. In C. elegans, transgenic animals were initially selected by using phenotypic markers, such as rol-6(su1006) (Kramer et al. 1990), which produced visible changes in transgenic animals (i.e., rolling in a circle) (Mello et al. 1991). These phenotypic markers are identified through laborious genetic screens. Moreover, phenotypic markers could impair processes such as invasion, migration, and establishment in the host that are critical for propagation as transgenics. Markers in C. elegans and P. pacificus such as dominant rol-6 could impair parasite motility to the point where the worms are innocuous (Kramer and Johnson 1993; Schlager et al. 2009; Arribere et al. 2014). Promoter:FP reporters were used next, in which transgenic animals had all cells labeled with the FP (Gu et al. 1998), or FP expression in specific cell types (i.e., pharyngeal muscle or body-wall muscle; Okkema et al. 1993). Maintenance of lines carrying either phenotypic or FP cotransformation markers requires manual picking of marker positive animals, which can be labor intensive for experiments that require large amounts of transgenic animals. Instruments such as the COPAS Biosort (Union Biometrica) may allow for enrichment of transgenic animals through automated separation of fluorescent and nonfluorescent animals; transgenic iL3 S. stercoralis larvae were selected from a mixed stage population using this instrument (Junio et al. 2008). Historically, to select for transgenic animals with minimal effort, rescue of mutants that displayed phenotypes such as temperature-sensitive embryonic and larval lethality (pha-1(e2123); Granato et al. 1994), or defective movement and inability to enter a stress-resistant dauer stage (unc-119(ed3); Maduro and Pilgrim 1995; Praitis et al. 2001), were used as transformation markers. Again, these markers were the product of laborious genetic screens. Recently, a number of antibiotic resistance cassettes have been developed for C. elegans. Use of hygromycin (Radman et al. 2013), puromycin (Semple et al. 2010), and G-418 (geneticin) (Giordano-Santini et al. 2010) selection identifies and maintains animals with no manual manipulation required. Though these systems require constitutively active promoters to drive expression of the resistance cassette, the C. elegans systems use ribosomal protein promoters, which are well conserved in parasites, and standard C. elegans 3′-UTRs (unc-54 or let-858). Through a combination of multispecies genomic DNA alignments and expression data, candidate regulatory elements in a parasite of choice can be identified to drive expression of drug resistance cassettes.
CRISPR/Cas9 editing: bringing genetics to the intractable
CRISPR/Cas9 editing has brought genetics to an ever-increasing number of organisms. The development of CRISPR/Cas9 editing in the protozoan parasite, Cryptosporidium parvum highlights many of the challenges that will be involved in developing CRISPR/Cas9 editing in parasitic nematodes. C. parvum sporozoites, a motile, spore-like state of Apicomplexan parasites, were transfected with plasmids encoding Cas9 and the sgRNA template, and a luciferase–neomycin resistance fusion gene (Vinayak et al. 2015). Mice were infected with the transfected sporozoites and subsequently treated with the neomycin analog paromomycin (Vinayak et al. 2015). Antibiotic-resistant parasites expressing a stably integrated luciferase–neomycin resistance fusion gene were then recovered in the feces of the infected mouse (Vinayak et al. 2015). These experiments highlight the need for well-characterized regulatory elements, development of antibiotic selection systems to enrich for transgenics, and the challenges implicit to working with an organism that requires passage through a host.
Depending on the parasitic nematode and transgenesis methods available, one can choose between the different approaches previously discussed (RNPs, RNA, DNA). The latter is very cost effective, but requires knowledge of constitutive RNA polymerase II and III promoters for Cas9 and sgRNA expression, respectively. RNPs would be the recommended starting point, as they produce higher editing rates than DNA-based approaches (Paix et al. 2015), and require no knowledge of regulatory elements for a given parasite. Moreover, as a biochemically savvy lab can purify Cas9 and T7 RNA polymerase, this approach could be very cost effective.
An emerging theme in CRISPR/Cas9 editing is that not all sgRNAs are created equal; half of all sgRNAs in C. elegans tested have had very low efficiencies (Arribere et al. 2014; Kim et al. 2014; Paix et al. 2014). Large-scale genomic screens in mammalian cells have led to algorithms to predict sgRNA efficiency (Doench et al. 2014; Xu et al. 2015). At least one of these algorithms (Doench et al. 2014) has predictive power in C. elegans (Katic et al. 2015). The most consistent predictor of highly active sgRNAs is two guanines at the 3′ end of the 20-bp targeting sequence (Farboud and Meyer 2015). With active sgRNAs one occasionally gets homozygous knockout alleles in a single generation, an important point for parasitic nematodes in which mating to homozygose a mutation may be challenging.
Insertion–deletion (indel) mutations, caused by repair of Cas9-induced DNA double-stranded breaks through error-prone nonhomologous end joining (NHEJ) (Figure 1B), should be the first edits attempted since these occur at high frequency. Once NHEJ-mediated indels are possible in a parasite, one can then try to import homologous recombination-based knockin approaches (Figure 1B). Using oligonucleotides as repair templates to knock in point mutations and small epitopes has been successful in a range of organisms, including C. elegans (Arribere et al. 2014; Paix et al. 2014; Zhao et al. 2014; Ward 2015) and require no cloning, as they are commercially available. Though PAGE purification boosts editing efficiency, this cost can be avoided if one screens more animals (Ward 2015).
Finding rare knockin animals can pose a “needle in the haystack” type problem; several direct screening approaches have required screening dozens to hundreds of animals (Paix et al. 2014; Zhao et al. 2014). To circumvent this issue, C. elegans researchers have developed efficient methods to enrich for knockins. Co-conversion/co-CRISPR approaches select for a Cas9-dependent editing event that produces a visible phenotype. Selecting for this marker phenotype enriches for edits at other loci. Markers include mutations that produce abnormal movement [dpy-10(cn64), rol-6(su1006), unc-22 (loss-of-function)] or repair of a conditional lethal mutation [pha-1(e2123) (Arribere et al. 2014; Kim et al. 2014; Ward 2015]. Again, these markers were the product of laborious genetic screens and may not be desirable to use in parasites, as they could compromise processes required for propagation. One approach that may work in parasites would be the use of a frame-shifted or mutated FP or drug resistance cassette as a co-CRISPR marker. Selecting for Cas9-mediated edits that restore FP or drug resistance gene expression should enrich for the desired edit. CRISPR/Cas9-mediated restoration of frame-shifted GFP expression has been described in C. elegans, but not tested for use as a co-CRISPR marker (Waaijers et al. 2013). Using RNPs is compatible with co-CRISPR (Paix et al. 2015), which further boosts editing efficiency and minimizes screening.
Alternatively, selection cassettes can be introduced into the knockin construct to select for insertion. Selection markers have included phenotypic markers [unc-119 (+) or sqt-1(e1350); Dickinson et al. 2013; 2015], GFP transgenes (Norris et al. 2015), and drug resistance cassettes (hygromycin or G-418) (Chen et al. 2013; Dickinson et al. 2015; Norris et al. 2015). By flanking the markers with LoxP sites, Cre-mediated excision can be used to remove the cassette. Although this excision leaves a 34-bp insertion, it is often placed within intronic sequences to minimize any deleterious effect to coding regions (Dickinson et al. 2015; Norris et al. 2015). Cleverly, Dickinson et al. (2015) included the Cre gene under a heat-shock inducible promoter in their “self-excising cassette,” which allowed researchers to use hygromycin selection for the knockin, followed by heat shock to remove the cassette. These selection-based schemes are powerful, but require more information about promoters and UTRs, and would take significantly more investment to develop for a given parasite.
CRISPR/Cas9 editing: a critical assessment
Following the development of RNAi in C. elegans, much hope was raised for using this tool for genetic studies in parasitic nematodes. However, recent genomics work has revealed that there is extensive diversity in RNAi and Piwi-interacting RNA (piRNA) biology between C. elegans and different nematode species (Dalzell et al. 2011; Sarkies et al. 2015). While RNAi has been effective in plant parasitic nematodes, the results in human and animal parasitic nematodes have been variable (Maule et al. 2011). It is therefore important to critically evaluate the chances of CRISPR working in a given parasitic nematode. CRISPR/Cas9 editing efficiency in C. elegans depends on the type of edit, location of the DNA double-stranded break, and whether a selection scheme is used; Table 4 denotes the current range of editing efficiencies with current approaches in C. elegans.
Table 4. Current range of CRISPR/Cas9 editing efficiencies in C. elegans.
| Edit type | Selection | No. injected | Success rate |
|---|---|---|---|
| Indels | No selection, 3′-GG sgRNAs | 7–33 | 10–72% of screened transgenic marker positive F1 (Farboud and Meyer 2015) |
| Indels | co-CRISPR | 20–40 | 36–88% of screened co-CRISPR marker positive F1 (Kim et al. 2014) |
| Small knockins (point mutations, stop codons, epitopes) | co-CRISPR | 20–40 | 8–50% of screened co-CRISPR marker positive F1 (Arribere et al. 2014; Kim et al. 2014; Ward 2015) |
| Small knockins (point mutations, stop codons, epitopes) | co-CRISPR, 3′-GG sgRNAs | 20–40 | 30–63% of screened co-CRISPR marker positive F1 (Farboud and Meyer 2015; J. D. Ward, unpublished data) |
| GFP-sized knockin | co-CRISPR | 17–40 | 50% of screened co-CRISPR marker positive F1 (Paix et al. 2015) |
| GFP-sized knockin | co-CRISPR, Cas9 RNP | 10–20 | 2–80% of screened co-CRISPR marker positive F1 (Paix et al. 2015) |
| GFP-sized knockin | Drug selection | 6–90 | 0–25% of injected parental animal (due to selection, do not know how many F1 are screened (Dickinson et al. 2015; Norris et al. 2015) |
Based on these efficiencies, in order to generate small indels or knockins in a given parasite, one would require dozens of transgenics, while larger knockins would require hundreds of transgenics, if not more. Since C. elegans hermaphrodites self-fertilize, knockins can be recovered by selecting candidates, allowing them to lay eggs, and genotyping the parental animal. The requirement of host passage and lack of in vitro culture for most parasitic nematodes means that one will most likely need to identify knockins through scoring a selectable cointegration marker (i.e., drug resistance or fluorescent markers). Here, I critically review the likelihood of implementing CRISPR/Cas9 editing in the three highlighted parasitic nematodes (Strongyloides spp., Brugia spp., and A. suum).
The best opportunity for generating stable knockins is S. ratti. Transgenesis rates of 3.7–7.4% transgenics per F1 screened have been reported for S. ratti, yielding 6–17 transgenic animals from 40–60 injected parental animals (Li et al. 2011). For S. stercoralis, the transgenesis rates were higher (5.6–33.5%), and more transgenic F1’s were obtained (2–204) from a similar number of injected parents when using the same plasmids as in the S. ratti experiments (Li et al. 2011). While S. ratti does not tolerate microinjection as well as S. stercoralis, 50–100% of infectious S. ratti iL3s develop to reproductively active females in five days (Li et al. 2011); this is in contrast to S. stercoralis in which on average only 6.5% of iL3s develop to adult females in a gerbil host (Nolan et al. 1993; Junio et al. 2008). Based on transformation efficiency, survival rate, and assuming only 10% of transgenics are germline transformants, Li et al. (2011) estimated that generation of a single germline transformed line would require 167 transgenic iL3s in S. stercoralis and 20 transgenic iL3s in S. ratti. Based on the number of transgenics typically obtained through gonadal microinjection, it has not yet been possible to establish stable lines in S. stercoralis (J. Lok, personal communication).
Shao et al. (2012) developed an insertional mutagenesis protocol in S. ratti which is instructive about how one would generate stable transgenics using CRISPR/Cas9 editing. Using a GFP-marked cassette, they observed 7.9% GFP-positive animals in the first generation, which went to 0.5% of 6014 animals in the second generation. Through selecting and reinfecting with GFP-positive iL3 animals, the percentage of GFP-positive progeny went to 15.6% in the third generation, 99.0% in the fourth, 82.4% in the fifth, and 98.7% in the sixth (Shao et al. 2012). The lines obtained had stable transgene expression over 10 generations and a number of discrete insertions were obtained (Shao et al. 2012). It would take ∼31 days from initiating a transgenesis experiment to establishment of fourth generation mating pairs, and an additional week to obtain the fifth generation iL3 progeny from this cross (J. Lok, personal communication; Lok and Unnasch 2013). Each additional passage through a rat would take seven days from inoculation of iL3 to recovery of next generation larvae in the feces (J. Lok, personal communication).
For Brugia spp., injection of females to transform developing embryos is difficult and can irreversibly damage the animal (Higazi et al. 2002), while biolistic transformation of embryos renders them developmentally incompetent (Shu et al. 2003). Calcium phosphate precipitate-mediated transformation of L3 larvae is the remaining avenue for stable transformation. L3 larvae transformed with a Gaussia luciferase reporter and subsequently induced to molt ex vivo were not able to develop further when inoculated into a jird host (Xu et al. 2011). In contrast, L3s incubated in a calcium phosphate precipitate mix containing the Gaussia luciferase reporter and directly inoculated into jirds were able to develop to adulthood, with 60–67% of adults secreting detectable luciferase (Xu et al. 2011). In a typical experiment, 20 adult females and 20 adult males are recovered in the peritoneal cavity from 300-400 injected L3s, (∼10% of L3s develop to adulthood; T. Unnasch, personal communication). A fecund adult female can then produce millions of progeny. Importantly, microfilariae in the next generation expressed the luciferase reporter, with an estimate of 0.47% transgenic animals in the F1 population (Xu et al. 2011). This population was fed to the mosquito vector, and L3 larvae expressing the luciferase reporter were obtained, suggesting that the transgene had been maintained through the entire lifecycle (Xu et al. 2011). This generation of transgenics is a significant undertaking: it takes 120–150 days postinoculation to recover adults and microfilariae, and 14 additional days for microfilariae fed to mosquitos to develop to L3s (Xu et al. 2011). Depending on the efficiency of CRISPR/Cas9 editing, these studies suggest that stable transformation may by feasible but will depend on transformation efficiency. It would also be critical to develop selectable markers, such as fluorescent reporters or drug selection cassettes, to identify transgenic animals.
A strength of A. suum as a model is its extreme fecundity; females can produce 200,000 eggs per day, and one can biolistically transform 700,000 embryos. Though the lack of fluorescent markers makes it difficult to determine transformation efficiency, 30–40% of embryos are hit with gold particles in a typical bombardment experiment (R. E. Davis, personal communication). These experiments are typically performed on 32- to 64-cell embryos, which will make any transgenics mosaic and decrease the chance of obtaining a stable line (Davis et al. 1999). Ideally, one would bombard 1-cell stage embryos, but these would fail to develop, and the incredibly robust permeability barrier prevents other approaches such as electroporation (R. E. Davis, personal communication). The development of RNAi protocols by injection may pave the way for new methods of germline transformation, which would increase the chances of obtaining a stable knockin (McCoy et al. 2015). It is important to note a major barrier to stable transgenesis in Ascaris is that most labs obtain adult worms from local slaughterhouses; there are likely only a handful of labs that maintain the lifecycle in the pig. If stable transgenesis is attempted, it would take 3–4 weeks for the embryos to develop to L2 larvae and 2–3 months for the development of fertile adults.
The longer lifecycles and potential requirement for multiple host passages to obtain stable knockins poses a significant challenge to implementing these tools, which may be insurmountable for many parasites. There are several approaches through which one could circumvent some of these issues. Generating homozygotes in F1 transgenics using a “mutagenic chain reaction” (Gantz and Bier 2015), would limit the need for multiple passages and mating to establish a line. This system consists of a cassette containing a Cas9 transgene, an sgRNA targeting a specific genomic locus of interest, and homology flanking the sgRNA target site. The cassette can be used as a template for repair of the Cas9-induced DSB resulting in insertion of the cassette at the break site. Cas9 and the sgRNA can then be expressed from the cassette and result in a DSB in the homologous chromosome, and insertion of the cassette into this chromosome through homologous recombination, resulting in a homozygote for the cassette insertion. Gene drive systems are controversial due to the risk of inadvertently altering wild population genetic diversity, and their use requires ethical oversight and best practice guidelines to prevent an inappropriate biological release (Akbari et al. 2015).
An important consideration in importing CRISPR/Cas9 editing into parasites is that many genes of interest for vaccine or drug targets will be essential for critical aspects of host–parasite interaction such as invasion, migration through tissues, and adult longevity. Mutations in these types of genes would be difficult, if not impossible to propagate as they could render the parasite innocuous. Another option would be to implement CRISPR/Cas9 methodology variations that allow assays to be performed in first generation transgenic animals. Somatic editing using CRISPR/Cas9 in C. elegans appears much more robust than germline editing: for five genes tested, 86–97% penetrance of a known recessive phenotype were observed, with targeting of one gene producing a less penetrant phenotype (20% of animals) (Shen et al. 2014). Alternatively, one could activate or repress target gene expression through CRISPR activation (CRISPRa) or CRISPR interference (CRISPRi), respectively (Qi et al. 2013; Gilbert et al. 2014). These methods use the fusion of a transcriptional activator or repressor to a nuclease-dead Cas9 protein. With somatic editing or CRISPRa/i, one could score a phenotype of interest relative to a control and then sacrifice the animals to experimentally confirm CRISPR efficiency. Somatic mutations can be detected by PCR followed by digestion with mismatch cutting nucleases such as CEL-1 or T7E1, while gene expression changes can be detected by qRT-PCR. These approaches will rely on expression of Cas9 and the sgRNAs from promoters of interest. One could add an additional level of control through use of conditional systems such as the drug-inducible, split-Cas9 system (Zetsche et al. 2015). In this approach, Cas9 is split into two fragments that are each fused to rapamycin-binding dimerization domains (Zetsche et al. 2015). Cas9 is inactive until rapamycin is added, bringing the two fragments into proximity. This system could be expressed under either ubiquitous or tissue-specific promoter to allow genetic manipulation with temporal control. Such an approach could be used to study specific aspects of parasitism, such as tissue invasion or migration in first generation transgenic animals.
Future Perspectives
The import of these approaches combined with other technologies such as deep sequencing and mass spectrometry will synergize and allow extremely rapid progress on a range of biomedically and agriculturally important parasites. In Figure 2, I suggest a workflow for stepwise development of tools in a given parasitic nematode, starting with basic transgenesis methods and FP reporters, and building to sophisticated genome editing tools. This workflow represents a significant investment in time and money and would be best undertaken by groups of researchers. The payoff from such a development scheme would be worthwhile as it could aid discovery of novel interventions. For example, one could generate fluorescent reporters to readout pathway activity, which could allow for more targeted drug screening. Developing genetics in these parasites also presents a rich opportunity for evo–devo studies; studying both C. elegans and a parasitic nematode is a win–win, as conserved pathways can be dissected in C. elegans and a researcher can toggle back and forth between the model and the parasite. Nonconserved pathways are interesting from an evolutionary perspective, as one can explore how selective pressure may have shaped these differences. There are incredible open questions waiting to be explored spanning the biology and evolution of host–parasite and parasite–vector interactions. Parasites often have unique sets of genes, some of which are likely central to the unique lifecycle and biology of a given parasite, and we need robust new tools to explore their function. Finally, the intimate interaction of parasites with their cognate host offers potential insight into our own biology. The study of phage allowed us to probe bacterial transcription and gene regulation, and work on Listeria revealed mechanisms underlying mammalian cytoskeletal dynamics. The range of parasites with specific lifecycles, target tissues, and unique gene products to modulate host pathways is an incredibly rich experimental vein to tap. We just need to develop the right tools to realize this potential.
Acknowledgments
I am indebted to Elizabeth Silva for many helpful discussions and editorial comments on this manuscript; Andy Fire for comments on the manuscript and a very helpful discussion on regulatory elements and development of FP reporters; Christian Frøkjaer-Jensen for his thoughts on importing FPs from C. elegans into parasites; John Gilleard and Tom Maier for advice on parasitic nematodes of livestock and plants, respectively; Brenda Beerntsen, Richard Davis, James Lok, and Thomas Unnasch for helpful discussions on nematode lifecycles and transgenesis; Sara Lustigman, Gabriela Monsalve, Lindsey Pack, Judy Sakanari, and Keith Yamamoto for advice and comments on the manuscript; and the reviewers for their many excellent comments and suggestions. J.D.W. was supported by the National Institute Of General Medical Sciences of the National Institutes of Health under award no. K99GM107345.
Footnotes
Communicating editor: O. Hobert
Literature Cited
- Abad P., Gouzy J., Aury J.-M., Castagnone-Sereno P., Danchin E. G. J., et al. , 2008. Genome sequence of the metazoan plant-parasitic nematode Meloidogyne incognita. Nat. Biotechnol. 26: 909–915. [DOI] [PubMed] [Google Scholar]
- Aboobaker A. A., Blaxter M. L., 2003. Use of RNA interference to investigate gene function in the human filarial nematode parasite Brugia malayi. Mol. Biochem. Parasitol. 129: 41–51. [DOI] [PubMed] [Google Scholar]
- Ahmed R., Chang Z., Younis A. E., Langnick C., Li N., et al. , 2013. Conserved miRNAs are candidate post-transcriptional regulators of developmental arrest in free-living and parasitic nematodes. Genome Biol. Evol. 5: 1246–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbari O. S., Bellen H. J., Bier E., Bullock S. L., Burt A., et al. , 2015. Safeguarding gene drive experiments in the laboratory. Science 349: 927–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arribere J. A., Bell R. T., Fu B. X. H., Artiles K. L., Hartman P. S., et al. , 2014. Efficient Marker-Free Recovery of Custom Genetic Modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics 198: 837–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzaria M., McGhee J. D., 1992. DNA synthesis in the early embryo of the nematode Ascaris suum. Dev. Biol. 152: 89–93. [DOI] [PubMed] [Google Scholar]
- Bai X., Adams B. J., Ciche T. A., Clifton S., Gaugler R., et al. , 2013. A Lover and a Fighter: The Genome Sequence of an Entomopathogenic Nematode Heterorhabditis bacteriophora. PLoS One 8: e69618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaxter M. L., De Ley P., Garey J. R., Liu L. X., Scheldeman P., et al. , 1998. A molecular evolutionary framework for the phylum Nematoda. Nature 392: 71–75. [DOI] [PubMed] [Google Scholar]
- Boatin B. A., Basáñez M.-G., Prichard R. K., Awadzi K., Barakat R. M. et al., 2012 A research agenda for helminth diseases of humans: towards control and elimination. PLoS Negl. Trop. Dis. 6: e1547. [DOI] [PMC free article] [PubMed]
- Buck A. H., Coakley G., Simbari F., McSorley H. J., Quintana J. F., et al. , 2014. Exosomes secreted by nematode parasites transfer small RNAs to mammalian cells and modulate innate immunity. Nat. Commun. 5: 5488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell W. C., Timinski S. F., 1965. Immunization of rats against Ascaris suum by means of nonpulmonary larval infections. J. Parasitol. 51: 712–716. [PubMed] [Google Scholar]
- Chandy A., Thakur A. S., Singh M. P., Manigauha A., 2011. A review of neglected tropical diseases: filariasis. Asian Pac. J. Trop. Med. 4: 581–586. [DOI] [PubMed] [Google Scholar]
- Chen C., Fenk L. A., de Bono M., 2013. Efficient genome editing in Caenorhabditis elegans by CRISPR-targeted homologous recombination. Nucleic Acids Res. 41: e193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu H., Schwartz H. T., Antoshechkin I., Sternberg P. W., 2013. Transgene-free genome editing in Caenorhabditis elegans using CRISPR-Cas. Genetics 195: 1167–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S. W., Lee J., Carroll D., Kim J.-S., Lee J., 2013. Heritable gene knockout in Caenorhabditis elegans by direct injection of Cas9-sgRNA ribonucleoproteins. Genetics 195: 1177–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y.-J., Ghedin E., Berriman M., McQuillan J., Holroyd N., et al. , 2011. A Deep Sequencing Approach to Comparatively Analyze the Transcriptome of Lifecycle Stages of the Filarial Worm, Brugia malayi. PLoS Negl. Trop. Dis. 5: e1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinkornpumin, J. K., and R. L. Hong, 2011 RNAi mediated gene knockdown and transgenesis by microinjection in the necromenic nematode Pristionchus pacificus. JoVE. 16: e3270. [DOI] [PMC free article] [PubMed]
- Cotton, J. A., C. J. Lilley, L. M. Jones, T. Kikuchi, A. J. Reid et al, 2014 The genome and life-stage specific transcriptomes of Globodera pallida elucidate key aspects of plantparasitism by a cyst nematode. 15: 1–17. [DOI] [PMC free article] [PubMed]
- Culp, E., C. Richman, D. Sharanya, B. Gupta, 2015 Genome Editing in Caenorhabditis briggsae using the CRISPR/Cas9 System. bioRxiv. 10.1101/021121. [DOI] [PMC free article] [PubMed]
- Dalzell J. J., Mcveigh P., Warnock N. D., Mitreva M., Bird D. M., et al. , 2011. RNAi Effector Diversity in Nematodes. PLoS Negl. Trop. Dis. 5: e1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalzell J. J., Warnock N. D., McVeigh P., Marks N. J., Mousley A., et al. , 2012. Considering RNAi experimental design in parasitic helminths. Parasitology 139: 589–604. [DOI] [PubMed] [Google Scholar]
- D’Astolfo D. S., Pagliero R. J., Pras A., Karthaus W. R., Clevers H., et al. , 2015. Efficient Intracellular Delivery of Native Proteins. Cell 161: 674–690. [DOI] [PubMed] [Google Scholar]
- Davis R. E., Parra A., LoVerde P. T., Ribeiro E., Glorioso G., et al. , 1999. Transient expression of DNA and RNA in parasitic helminths by using particle bombardment. Proc. Natl. Acad. Sci. USA 96: 8687–8692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denker J. A., Zuckerman D. M., Maroney P. A., Nilsen T. W., 2002. New components of the spliced leader RNP required for nematode trans-splicing. Nature 417: 667–670. [DOI] [PubMed] [Google Scholar]
- Dickinson D. J., Pani A. M., Heppert J. K., Higgins C. D., Goldstein B., 2015. Streamlined Genome Engineering with a Self-Excising Drug Selection Cassette. Genetics 200: 1035–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson D. J., Ward J. D., Reiner D. J., Goldstein B., 2013. Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat. Methods 10: 1028–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterich C., Clifton S. W., Schuster L. N., Chinwalla A., Delehaunty K., et al. , 2008. The Pristionchus pacificus genome provides a unique perspective on nematode lifestyle and parasitism. Nat. Genet. 40: 1193–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillman A. R., Macchietto M., Porter C. F., Rogers A., Williams B., et al. , 2015. Comparative genomics of Steinernema reveals deeply conserved gene regulatory networks. Genome Biol. 16: 200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench J. G., Hartenian E., Graham D. B., Tothova Z., Hegde M., et al. , 2014. Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat. Biotechnol. 32: 1262–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dold C., Holland C. V., 2011. Ascaris and ascariasis. Microbes Infect. 13: 632–637. [DOI] [PubMed] [Google Scholar]
- Enobe C. S., Araujo C. A., Perini A., Martins M. A., Macedo M. S., et al. , 2006. Early stages of Ascaris suum induce airway inflammation and hyperreactivity in a mouse model. Parasite Immunol. 28: 453–461. [DOI] [PubMed] [Google Scholar]
- Farboud B., Meyer B. J., 2015. Dramatic Enhancement of Genome Editing by CRISPR/Cas9 Through Improved Guide RNA Design. Genetics 199: 959–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A., Harrison S. W., Dixon D., 1990. A modular set of lacZ fusion vectors for studying gene expression in Caenorhabditis elegans. Gene 93: 189–198. [DOI] [PubMed] [Google Scholar]
- Foth B. J., Tsai I. J., Reid A. J., Bancroft A. J., Nichol S., et al. , 2014. Whipworm genome and dual-species transcriptome analyses provide molecular insights into an intimate host-parasite interaction. Nat. Genet. 46: 693–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedland A. E., Tzur Y. B., Esvelt K. M., Colaiácovo M. P., Church G. M., et al. , 2013. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat. Methods 10: 741–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu B. X. H., Hansen L. L., Artiles K. L., Nonet M. L., Fire A. Z., 2014. Landscape of target:guide homology effects on Cas9-mediated cleavage. Nucleic Acids Res. 42: 13778–13787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvin T. J., 1968. Development of human and pig ascaris in the pig and rabbit. J. Parasitol. 54: 1085–1091. [PubMed] [Google Scholar]
- Gantz V. M., Bier E., 2015. The mutagenic chain reaction: a method for converting heterozygous to homozygous mutations. Science 348: 442–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser R. B., 2013. A perfect time to harness advanced molecular technologies to explore the fundamental biology of Toxocara species. Vet. Parasitol. 193: 353–364. [DOI] [PubMed] [Google Scholar]
- Geldhof P., Murray L., Couthier A., Gilleard J. S., McLauchlan G., et al. , 2006. Testing the efficacy of RNA interference in Haemonchus contortus. Int. J. Parasitol. 36: 801–810. [DOI] [PubMed] [Google Scholar]
- Gilbert L. A., Horlbeck M. A., Adamson B., Villalta J. E., Chen Y., et al. , 2014. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell 159: 647–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilleard J. S., 2013. Haemonchus contortus as a paradigm and model to study anthelmintic drug resistance. Parasitology 140: 1506–1522. [DOI] [PubMed] [Google Scholar]
- Giordano-Santini R., Milstein S., Svrzikapa N., Tu D., Johnsen R., et al. , 2010. An antibiotic selection marker for nematode transgenesis. Nat. Methods 7: 721–723. [DOI] [PubMed] [Google Scholar]
- Glendinning S. K., Buckingham S. D., Sattelle D. B., Wonnacott S., Wolstenholme A. J., 2011. Glutamate-Gated Chloride Channels of Haemonchus contortus Restore Drug Sensitivity to Ivermectin Resistant Caenorhabditis elegans. PLoS One 6: e22390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granato M., Schnabel H., Schnabel R., 1994. pha-1, a selectable marker for gene transfer in C. elegans. Nucleic Acids Res. 22: 1762–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant W. N., Skinner S. J. M., Newton-Howes J., Grant K., Shuttleworth G., et al. , 2006a Heritable transgenesis of Parastrongyloides trichosuri: A nematode parasite of mammals. Int. J. Parasitol. 36: 475–483. [DOI] [PubMed] [Google Scholar]
- Grant W. N., Stasiuk S., Newton-Howes J., Ralston M., Bisset S. A., et al. , 2006b Parastrongyloides trichosuri, a nematode parasite of mammals that is uniquely suited to genetic analysis. Int. J. Parasitol. 36: 453–466. [DOI] [PubMed] [Google Scholar]
- Gu T., Orita S., Han M., 1998. Caenorhabditis elegans SUR-5, a novel but conserved protein, negatively regulates LET-60 Ras activity during vulval induction. Mol. Cell. Biol. 18: 4556–4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon G. J., Maroney P. A., Ayers D. G., Shambaugh J. D., Nilsen T. W., 1990. Transcription of a nematode trans-spliced leader RNA requires internal elements for both initiation and 3′ end-formation. EMBO J. 9: 1915–1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisler M. G., Ohno C., Das P., Sieber P., Reddy G. V., et al. , 2005. Patterns of Auxin Transport and Gene Expression during Primordium Development Revealed by Live Imaging of the Arabidopsis Inflorescence Meristem. Curr. Biol. 15: 1899–1911. [DOI] [PubMed] [Google Scholar]
- Higazi T. B., Unnasch T. R., 2004. Intron encoded sequences necessary for trans splicing in transiently transfected Brugia malayi. Mol. Biochem. Parasitol. 137: 181–184. [DOI] [PubMed] [Google Scholar]
- Higazi T. B., Merriweather A., Shu L., Davis R., Unnasch T. R., 2002. Brugia malayi: transient transfection by microinjection and particle bombardment. Exp. Parasitol. 100: 95–102. [DOI] [PubMed] [Google Scholar]
- Holterman M., van der Wurff A., van den Elsen S., van Megen H., Bongers T., et al. , 2006. Phylum-wide analysis of SSU rDNA reveals deep phylogenetic relationships among nematodes and accelerated evolution toward crown Clades. Mol. Biol. Evol. 23: 1792–1800. [DOI] [PubMed] [Google Scholar]
- Hong R. L., Sommer R. J., 2006. Pristionchus pacificus: a well-rounded nematode. BioEssays 28: 651–659. [DOI] [PubMed] [Google Scholar]
- Hotez P. J., Brindley P. J., Bethony J. M., King C. H., Pearce E. J., et al. , 2008. Helminth infections: the great neglected tropical diseases. J. Clin. Invest. 118: 1311–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ITFDE, 2008 Summary of the Twelfth Meeting of the International Task Force for Disease Eradication. The Carter Center, Atlanta. [Google Scholar]
- Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J. A., et al. , 2012. A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science 337: 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J. T., Haegeman A., Danchin E. G. J., Gaur H. S., Helder J., et al. , 2013. Top 10 plant-parasitic nematodes in molecular plant pathology. Mol. Plant Pathol. 14: 946–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junio A. B., Li X., Massey H. C., Jr, Nolan T. J., Todd Lamitina S., et al. , 2008. Strongyloides stercoralis: cell- and tissue-specific transgene expression and co-transformation with vector constructs incorporating a common multifunctional 3′ UTR. Exp. Parasitol. 118: 253–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katic I., Xu L., Ciosk R., 2015. CRISPR/Cas9 genome editing in Caenorhabditis elegans: evaluation of templates for homology-mediated repair and knock-ins by homology-independent DNA repair. G3 (Bethesda) 5: 1649–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keenan J. D., Hotez P. J., Amza A., Stoller N. E., Gaynor B. D., et al. , 2013. Elimination and eradication of neglected tropical diseases with mass drug administrations: a survey of experts. PLoS Negl. Trop. Dis. 7: e2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly W. G., Xu S., Montgomery M. K., Fire A., 1997. Distinct requirements for somatic and germline expression of a generally expressed Caernorhabditis elegans gene. Genetics 146: 227–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy, M. W., 2013 Ascaris-Antigens, Allergens, Immunogenetics, Protein Structures, pp. 51–79 in Ascaris: The Neglected Parasite, edited by C. Holland. Academic Press.
- Kennedy M. W., Qureshi F., 1986. Stage-specific secreted antigens of the parasitic larval stages of the nematode Ascaris. Immunology 58: 515–522. [PMC free article] [PubMed] [Google Scholar]
- Kennedy M. W., Qureshi F., Haswell-Elkins M., Elkins D. B., 1987. Homology and heterology between the secreted antigens of the parasitic larval stages of Ascaris lumbricoides and Ascaris suum. Clin. Exp. Immunol. 67: 20–30. [PMC free article] [PubMed] [Google Scholar]
- Kim H., Ishidate T., Ghanta K. S., Seth M., Conte D., et al. , 2014. A co-CRISPR strategy for efficient genome editing in Caenorhabditis elegans. Genetics 197: 1069–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer J. M., Johnson J. J., 1993. Analysis of mutations in the sqt-1 and rol-6 collagen genes of Caenorhabditis elegans. Genetics 135: 1035–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer J. M., French R. P., Park E. C., Johnson J. J., 1990. The Caenorhabditis elegans rol-6 gene, which interacts with the sqt-1 collagen gene to determine organismal morphology, encodes a collagen. Mol. Cell. Biol. 10: 2081–2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Koutsovoulos G., Kaur G., Blaxter M., 2014. Toward 959 nematode genomes. Worm 1: 42–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushwaha S., P. K. Singh, M. Shahab, M. Pathak, and S. M. Bhattacharya, 2012 In vitro silencing of Brugia malayi trehalose-6-phosphate phosphatase impairs embryogenesis and in vivo development of infective larvae in jirds. PLoS Negl. Trop. Dis. 6: e1770. [DOI] [PMC free article] [PubMed]
- Lacey L. A., Grzywacz D., Shapiro-Ilan D. I., Frutos R., Brownbridge M., et al. , 2015. Insect pathogens as biological control agents: back to the future. J. Invertebr. Pathol. 132: 1–41. [DOI] [PubMed] [Google Scholar]
- Lall S., Friedman C. C., Jankowska-Anyszka M., Stepinski J., Darzynkiewicz E., et al. , 2004. Contribution of trans-splicing, 5′ -leader length, cap-poly(A) synergism, and initiation factors to nematode translation in an Ascaris suum embryo cell-free system. J. Biol. Chem. 279: 45573–45585. [DOI] [PubMed] [Google Scholar]
- Landmann F., Foster J. M., Slatko B. E., Sullivan W., 2012. Efficient in vitro RNA interference and immunofluorescence-based phenotype analysis in a human parasitic nematode, Brugia malayi. Parasit. Vectors 5: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau Y.-L., Lee W.-C., Xia J., Zhang G., Razali R., et al. , 2015. Draft genome of Brugia pahangi: high similarity between B. pahangi and B. malayi. Parasit. Vectors 8: 451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law W., Wuescher L. M., Ortega A., Hapiak V. M., Komuniecki P. R., et al. , 2015. Heterologous expression in remodeled C. elegans: a platform for monoaminergic agonist identification and anthelmintic screening. PLoS Pathog. 11: e1004794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Cowley A., Uludag M., Gur T., McWilliam H., et al. , 2015. The EMBL-EBI bioinformatics web and programmatic tools framework. Nucleic Acids Res. 43: W580–W584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Massey H. C., Jr, Nolan T. J., Schad G. A., Kraus K., et al. , 2006. Successful transgenesis of the parasitic nematode Strongyloides stercoralis requires endogenous non-coding control elements. Int. J. Parasitol. 36: 671–679. [DOI] [PubMed] [Google Scholar]
- Li X., Shao H., Junio A., Nolan T. J., Massey H. C., et al. , 2011. Transgenesis in the parasitic nematode Strongyloides ratti. Mol. Biochem. Parasitol. 179: 114–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Oliveira A., Chauhan C., Ghedin E., Unnasch T. R., 2010. Functional analysis of putative operons in Brugia malayi. Int. J. Parasitol. 40: 63–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Voronin D., Poole C. B., Bachu S., Rogers M. B., et al. , 2015. Functional analysis of microRNA activity in Brugia malayi. Int. J. Parasitol. 45: 579–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo T.-W., Pickle C. S., Lin S., Ralston E. J., Gurling M., et al. , 2013. Precise and heritable genome editing in evolutionarily diverse nematodes using TALENs and CRISPR/Cas9 to engineer insertions and deletions. Genetics 195: 331–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lok, J. B., 2007 Strongyloides stercoralis: a model for translational research on parasitic nematode biology. WormBook: the online review of C. elegans biology: 1–18. [DOI] [PMC free article] [PubMed]
- Lok J. B., 2009. Transgenesis in parasitic nematodes: building a better array. Trends Parasitol. 25: 345–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lok, J. B., and Unnasch T. R., 2013 Transgenesis in animal parasitic nematodes: Strongyloides spp and Brugia spp WormBook: the online review of C. elegans biology: 1–18. [DOI] [PMC free article] [PubMed]
- Lustigman, S., R. K. Prichard, A. Gazzinelli, W. N. Grant, B. A. Boatin et al., 2012 A research agenda for helminth diseases of humans: the problem of helminthiases. PLoS Negl. Trop. Dis. 6: e1582. [DOI] [PMC free article] [PubMed]
- Maduro M., Pilgrim D., 1995. Identification and cloning of unc-119, a gene expressed in the Caenorhabditis elegans nervous system. Genetics 141: 977–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini L. A., Sontheimer E. J., 2010. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat. Rev. Genet. 11: 181–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin J., Rosa B. A., Ozersky P., Hallsworth-Pepin K., Zhang X., et al. , 2015. Helminth.net: expansions to Nematode.net and an introduction to Trematode.net. Nucleic Acids Res. 43: D698–D706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maule A. G., Mcveigh P., Dalzell J. J., Atkinson L., Mousley A., et al. , 2011. An eye on RNAi in nematode parasites. Trends Parasitol. 27: 505–513. [DOI] [PubMed] [Google Scholar]
- McCoy C. J., Warnock N. D., Atkinson L. E., Atcheson E., Martin R. J., et al. , 2015. RNA interference in adult Ascaris suum - an opportunity for the development of a functional genomics platform that supports organism-, tissue- and cell-based biology in a nematode parasite. Int. J. Parasitol. 45: 673–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello C. C., Kramer J. M., Stinchcomb D., Ambros V., 1991. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10: 3959–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt C., Rasoloson D., Ko D., Seydoux G., 2008. 3′ UTRs are the primary regulators of gene expression in the C. elegans germline. Curr. Biol. 18: 1476–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalski M. L., Griffiths K. G., Williams S. A., Kaplan R. M., Moorhead A. R., 2011. The NIH-NIAID Filariasis Research Reagent Resource Center. PLoS Negl. Trop. Dis. 5: e1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai T., Ibata K., Park E. S., Kubota M., Mikoshiba K., et al. , 2002. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 20: 87–90. [DOI] [PubMed] [Google Scholar]
- Nicol J. M., Turner S. J., Coyne D. L., Nijs L. D., Hockland S., et al. , 2011, pp. 21–44 in Genomics and Molecular Genetics of Plant–Nematode Interactions, edited by Jones J. T., Gheysen G., Fenoll C. Springer-Verlag, Heidelberg. [Google Scholar]
- Nolan T. J., Megyeri Z., Bhopale V. M., Schad G. A., 1993. Strongyloides stercoralis: the first rodent model for uncomplicated and hyperinfective strongyloidiasis, the Mongolian gerbil (Meriones unguiculatus). J. Infect. Dis. 168: 1479–1484. [DOI] [PubMed] [Google Scholar]
- Norris A. D., Kim H.-M., Colaiácovo M. P., Calarco J. A., 2015. Efficient genome editing in Caenorhabditis elegans with a toolkit of dual-marker selection cassettes. Genetics 201: 449–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okkema P. G., Harrison S. W., Plunger V., Aryana A., Fire A., 1993. Sequence requirements for myosin gene expression and regulation in Caenorhabditis elegans. Genetics 135: 385–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opperman C. H., Bird D. M., Williamson V. M., Rokhsar D. S., Burke M., et al. , 2008. Sequence and genetic map of Meloidogyne hapla: A compact nematode genome for plant parasitism. Proc. Natl. Acad. Sci. USA 105: 14802–14807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paily K. P., Hoti S. L., Das P. K., 2009. A review of the complexity of biology of lymphatic filarial parasites. J. Parasit. Dis. 33: 3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paix A., Wang Y., Smith H. E., Lee C.-Y. S., Calidas D., et al. , 2014. Scalable and versatile genome editing using linear DNAs with microhomology to Cas9 Sites in Caenorhabditis elegans. Genetics 198: 1347–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paix A., Folkmann A., Rasoloson D., Seydoux G., 2015. High efficiency, homology-directed genome editing in Caenorhabditis elegans using CRISPR-Cas9 ribonucleoprotein complexes. Genetics 201: 47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole C. B., Gu W., Kumar S., Jin J., Davis P. J., et al. , 2014. Diversity and expression of MicroRNAs in the filarial parasite, Brugia malayi. PLoS One 9: e96498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praitis V., Casey E., Collar D., Austin J., 2001. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics 157: 1217–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi L. S., Larson M. H., Gilbert L. A., Doudna J. A., Weissman J. S., et al. , 2013. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152: 1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radman I., Greiss S., Chin J. W., 2013. Efficient and rapid C. elegans transgenesis by bombardment and hygromycin B selection. PLoS One 8: e76019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajan T. V., Paciorkowski N., Kalajzic I., McGuiness C., 2003. Ascorbic acid is a requirement for the morphogenesis of the human filarial parasite Brugia malayi. J. Parasitol. 89: 868–870. [DOI] [PubMed] [Google Scholar]
- Ran F. A., Hsu P. D., Wright J., Agarwala V., Scott D. A., et al. , 2013. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8: 2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaldi M., Geldhof P., 2012. Immunologically based control strategies for ostertagiosis in cattle: Where do we stand? Parasite Immunol. 34: 254–264. [DOI] [PubMed] [Google Scholar]
- Roeber F., Jex A. R., Gasser R. B., 2013. Impact of gastrointestinal parasitic nematodes of sheep, and the role of advanced molecular tools for exploring epidemiology and drug resistance: an Australian perspective. Parasit. Vectors 6: 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkies P., Selkirk M. E., Jones J. T., Blok V., Boothby T., et al. , 2015. Ancient and novel small RNA pathways compensate for the loss of piRNAs in multiple independent nematode lineages. PLoS Biol. 13: e1002061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlager B., Wang X., Braach G., Sommer R. J., 2009. Molecular cloning of a dominant roller mutant and establishment of DNA-mediated transformation in the nematode Pristionchus pacificus. Genesis 47: 300–304. [DOI] [PubMed] [Google Scholar]
- Semple J. I., Garcia-Verdugo R., Lehner B., 2010. Rapid selection of transgenic C. elegans using antibiotic resistance. Nat. Methods 7: 725–727. [DOI] [PubMed] [Google Scholar]
- Shaner N. C., Campbell R. E., Steinbach P. A., Giepmans B. N. G., Palmer A. E., et al. , 2004. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22: 1567–1572. [DOI] [PubMed] [Google Scholar]
- Shaner N. C., Lambert G. G., Chammas A., Ni Y., Cranfill P. J., et al. , 2013. A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nat. Methods 10: 407–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao H., Li X., Nolan T. J., Massey H. C., Pearce E. J., et al. , 2012. Transposon-mediated chromosomal integration of transgenes in the parasitic nematode Strongyloides ratti and establishment of stable transgenic lines. PLoS Pathog. 8: e1002871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z., Zhang X., Chai Y., Zhu Z., Yi P., et al. , 2014. Conditional knockouts generated by engineered CRISPR-Cas9 endonuclease reveal the roles of coronin in C. elegans neural development. Dev. Cell 30: 625–636. [DOI] [PubMed] [Google Scholar]
- Shu L., Katholi C. R., Higazi T., Unnasch T. R., 2003. Analysis of the Brugia malayi HSP70 promoter using a homologous transient transfection system. Mol. Biochem. Parasitol. 128: 67–75. [DOI] [PubMed] [Google Scholar]
- Sommer R. J., McGaughran A., 2013. The nematode Pristionchus pacificus as a model system for integrative studies in evolutionary biology. Mol. Ecol. 22: 2380–2393. [DOI] [PubMed] [Google Scholar]
- Song C., Gallup J. M., Day T. A., Bartholomay L. C., Kimber M. J., 2010. Development of an in vivo RNAi protocol to investigate gene function in the filarial nematode, Brugia malayi. PLoS Pathog. 6: e1001239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoltzfus J. D., Massey H. C., Nolan T. J., Griffith S. D., Lok J. B., 2012a Strongyloides stercoralis age-1: a potential regulator of infective larval development in a parasitic nematode. PLoS One 7: e38587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoltzfus J. D., Minot S., Berriman M., Nolan T. J., Lok J. B., 2012b RNAseq analysis of the parasitic nematode Strongyloides stercoralis reveals divergent regulation of canonical dauer pathways. PLoS Negl. Trop. Dis. 6: e1854. [DOI] [PMC free article] [PubMed]
- Stoltzfus J. D., Bart S. M., Lok J. B., 2014. cGMP and NHR signaling co-regulate expression of insulin-like peptides and developmental activation of infective larvae in Strongyloides stercoralis. PLoS Pathog. 10: e1004235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzertzinis G., Egaña A. L., Palli S. R., Robinson-Rechavi M., Gissendanner C. R., et al. , 2010. Molecular evidence for a functional ecdysone signaling system in Brugia malayi. PLoS Negl. Trop. Dis. 4: e625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinayak S., Pawlowic M. C., Sateriale A., Brooks C. F., Studstill C. J., et al. , 2015. Genetic modification of the diarrhoeal pathogen Cryptosporidium parvum. Nature 523: 477–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viney, M. E., J. B. Lok, 2015 The Biology of Strongyloides Spp WormBook: the online review of C. elegans biology: 1–17. [DOI] [PMC free article] [PubMed]
- Waaijers S., Portegijs V., Kerver J., Lemmens B. B. L. G., Tijsterman M., et al. , 2013. CRISPR/Cas9-targeted mutagenesis in Caenorhabditis elegans. Genetics 195: 1187–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Czech B., Crunk A., Wallace A., Mitreva M., et al. , 2011. Deep small RNA sequencing from the nematode Ascaris reveals conservation, functional diversification, and novel developmental profiles. Genome Res. 21: 1462–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Mitreva M., Berriman M., Thorne A., Magrini V., et al. , 2012. Silencing of Germline-Expressed Genes by DNA Elimination in Somatic Cells. Dev. Cell 23: 1072–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Zhou X. E., Motola D. L., Gao X., Suino-Powell K., et al. , 2009. Identification of the nuclear receptor DAF-12 as a therapeutic target in parasitic nematodes. Proc. Natl. Acad. Sci. USA 106: 9138–9143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Stoltzfus J., You Y.-J., Ranjit N., Tang H., et al. , 2015. The nuclear receptor DAF-12 regulates nutrient metabolism and reproductive growth in nematodes. PLoS Genet. 11: e1005027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward J. D., 2015. Rapid and precise engineering of the Caenorhabditis elegans genome with lethal mutation co-conversion and inactivation of NHEJ repair. Genetics 199: 363–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Q., Shen Y., Chen X., Shifman Y., Ellis R. E., 2014. Rapid creation of forward-genetics tools for C. briggsae using TALENs: lessons for nonmodel organisms. Mol. Biol. Evol. 31: 468–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss D. J., Liggitt D., Clark J. G., 1999. Histochemical discrimination of endogenous mammalian beta-galactosidase activity from that resulting from lac-Z gene expression. Histochem. J. 31: 231–236. [DOI] [PubMed] [Google Scholar]
- Winter A. D., Weir W., Hunt M., Berriman M., Gilleard J. S., et al. , 2012. Diversity in parasitic nematode genomes: the microRNAs of Brugia pahangi and Haemonchus contortus are largely novel. BMC Genomics 13: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter A. D., McCormack G., Myllyharju J., Page A. P., 2013. Prolyl 4-hydroxlase activity is essential for development and cuticle formation in the human infective parasitic nematode Brugia malayi. J. Biol. Chem. 288: 1750–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte H., Moreno E., Rödelsperger C., Kim J., Kim J.-S., et al. , 2014. Gene inactivation using the CRISPR/Cas9 system in the nematode Pristionchus pacificus. Dev. Genes Evol. 225: 55–62. [DOI] [PubMed] [Google Scholar]
- Wood A. J., Lo T.-W., Zeitler B., Pickle C. S., Ralston E. J., et al. , 2011. Targeted genome editing across species using ZFNs and TALENs. Science 333: 307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wylie T., Martin J. C., Dante M., Mitreva M. D., Clifton S. W., et al. , 2004. Nematode.net: a tool for navigating sequences from parasitic and free-living nematodes. Nucleic Acids Res. 32: D423–D426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Xiao T., Chen C.-H., Li W., Meyer C. A., et al. , 2015. Sequence determinants of improved CRISPR sgRNA design. Genome Res. 25: 1147–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S., Liu C., Tzertzinis G., Ghedin E., Evans C. C., et al. , 2011. In vivo transfection of developmentally competent Brugia malayi infective larvae. Int. J. Parasitol. 41: 355–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada M., Matsuda S., Nakazawa M., Arizono N., 1991. Species-specific differences in heterogonic development of serially transferred free-living generations of Strongyloides planiceps and Strongyloides stercoralis. J. Parasitol. 77: 592–594. [PubMed] [Google Scholar]
- Zamanian M., Fraser L. M., Agbedanu P. N., Harischandra H., Moorhead A. R., et al. , 2015. Release of small RNA-containing exosome-like vesicles from the human filarial parasite Brugia malayi. PLoS Negl. Trop. Dis. 9: e0004069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B., Volz S. E., Zhang F., 2015. A split-Cas9 architecture for inducible genome editing and transcription modulation. Nat. Biotechnol. 33: 139–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P., Zhang Z., Ke H., Yue Y., Xue D., 2014. Oligonucleotide-based targeted gene editing in C. elegans via the CRISPR/Cas9 system. Cell Res. 24: 247–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z., Flibotte S., Murray J. I., Blick D., Boyle T. J., et al. , 2010. New tools for investigating the comparative biology of Caenorhabditis briggsae and C. elegans. Genetics 184: 853–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X.-Q., Korhonen P. K., Cai H., Young N. D., Nejsum P. et al, 2015. Genetic blueprint of the zoonotic pathogen Toxocara canis. Nat. Commun. 6: 6145. [DOI] [PMC free article] [PubMed] [Google Scholar]