Abstract
Meiotic chromosome segregation requires pairwise association between homologs, stabilized by the synaptonemal complex (SC). Here, we investigate factors contributing to pairwise synapsis by investigating meiosis in polyploid worms. We devised a strategy, based on transient inhibition of cohesin function, to generate polyploid derivatives of virtually any Caenorhabditis elegans strain. We exploited this strategy to investigate the contribution of recombination to pairwise synapsis in tetraploid and triploid worms. In otherwise wild-type polyploids, chromosomes first sort into homolog groups, then multipartner interactions mature into exclusive pairwise associations. Pairwise synapsis associations still form in recombination-deficient tetraploids, confirming a propensity for synapsis to occur in a strictly pairwise manner. However, the transition from multipartner to pairwise association was perturbed in recombination-deficient triploids, implying a role for recombination in promoting this transition when three partners compete for synapsis. To evaluate the basis of synapsis partner preference, we generated polyploid worms heterozygous for normal sequence and rearranged chromosomes sharing the same pairing center (PC). Tetraploid worms had no detectable preference for identical partners, indicating that PC-adjacent homology drives partner choice in this context. In contrast, triploid worms exhibited a clear preference for identical partners, indicating that homology outside the PC region can influence partner choice. Together, our findings, suggest a two-phase model for C. elegans synapsis: an early phase, in which initial synapsis interactions are driven primarily by recombination-independent assessment of homology near PCs and by a propensity for pairwise SC assembly, and a later phase in which mature synaptic interactions are promoted by recombination.
Keywords: polyploidy, C. elegans, meiosis, synaptonemal complex, recombination
SEGREGATION of homologous chromosomes is a central defining event of meiosis, the specialized cell division program that allows sexually reproducing organisms to reduce their diploid chromosome complement to produce haploid gametes. In order to segregate away from each other, homologous chromosomes must recognize and align with their correct pairing partners during meiotic prophase to allow meiotic recombination to create physical attachments (chiasmata). These physical links between homologs will then promote their correct orientation on the metaphase plate of the meiosis I spindle and ensure their proper partitioning during the first meiotic division. At center stage during the events that prepare homologous chromosomes for segregation is a highly ordered proteinaceous structure called the synaptonemal complex (SC). The SC is composed of two lateral elements that form along the axis of each homolog, linked together by the SC central region. The SC has been shown to be a major regulator of the complex behavior of meiotic chromosomes, acting to stabilize and maintain tight associations between the homologs and playing a role in the maturation of recombination intermediates into fully functional chiasmata.
The nematode Caenorhabditis elegans has emerged as one the premier model systems for investigating key meiotic events, including the mechanisms regulating assembly of the SC (a process known as synapsis). The C. elegans adult hermaphrodite has two gonads, each containing hundreds of germ-cell nuclei that enter and progress through the meiotic prophase program as they travel from the distal tip of the gonad to the uterus. A full gonad therefore represents a developmental time course of nuclei at various stages of meiotic prophase that are organized in a spatiotemporal gradient highly amenable to imaging of meiotic chromosome structures in both live and fixed samples. Moreover, cytological analysis of meiosis can be coupled with genetic screens (Villeneuve 1994; Kelly et al. 2000; Nabeshima et al. 2004) and mutant analyses to discover and characterize the roles of components of the meiotic machinery. This powerful combination of genetics and cytology has enabled discovery of a complex network of factors regulating homolog pairing and synapsis. These include specialized chromosomal sites called “pairing centers” (PCs), located near one end of each chromosome (Rosenbluth and Baillie 1981; Rose et al. 1984; McKim et al. 1988, 1993; Villeneuve 1994), that mediate chromosome movements that are important both for achieving timely homolog pairing and for constraining SC assembly to occur exclusively between correctly paired homologs (MacQueen et al. 2005; Martinez-Perez and Villeneuve 2005; Phillips et al. 2005; Penkner et al. 2007, 2009; Sato et al. 2009).
C. elegans is also amenable to a complementary approach that enables investigation of meiotic mechanisms in the context of a full wild-type complement of meiotic machinery components. This approach involves the use of modified karyotypes, including altered ploidy, as triploid (3n) and tetraploid (4n) worms are viable. Analysis of pairing and synapsis in such challenged situations has provided insights into the principles governing these processes (Mlynarczyk-Evans et al. 2013). For example, this approach revealed that when more than two partners can compete for synapsis, chromosomes are initially sorted into homologous groups regardless of chromosome number and then eventually commit into exclusively pairwise synapsis associations. This study also provided evidence for the operation of “masking mechanisms” that are capable of counterbalancing stringent quality control systems to promote reproductive success. Further, this prior work suggested that experiments integrating the use of altered ploidy with genetic mutants and/or transgenes expressing cytological markers might have potential to generate important new insights into the mechanisms and regulation of meiosis. However, the feasibility of integrating these approaches was limited by the substantial technical difficulty of generating polyploid worms of the desired genotypes.
Here, we have overcome this technical barrier by devising a strategy for generating tetraploid derivatives of virtually any C. elegans strain. Our approach was informed by our finding that impairment of meiotic cohesion function can have very different consequences for male and female gametes, reflecting the very distinct cell division processes associated with spermatocyte and oocyte meiosis. In the current work, we use the ability to manipulate ploidy to investigate how meiotic recombination affects homolog pairing and synapsis. This work reveals a previously unappreciated contribution of meiotic recombination to the maturation of SC-mediated chromosome associations in C. elegans and leads us to propose a two-phase model for the establishment of mature synapsis interactions: in the early phase, synapsis associations are governed by the previously described activities of the PCs and by a propensity of SC to assemble preferentially between pairs of chromosome axes; during the later phase, progression of meiotic recombination solidifies synapsis associations between pairs of homologs.
Materials and Methods
Strains and genetics
Except where noted, all C. elegans strains were cultivated at 20° under standard conditions (Brenner 1974). SP346 (Madl and Herman 1979) was used as our wild-type tetraploid strain, and a mating stock of Bristol N2 provided the wild-type diploid background. Diploid strains for which tetraploid derivatives were generated in this study are referenced in Table 1. Generation of tetraploid derivatives of diploid strains is detailed in the Results section.
Table 1. List of tetraploid strains generated.
| Parental strain | Genotype | Tetraploid derivate | Method |
|---|---|---|---|
| AV776 | spo-11(me44)/nT1 IV; +/nT1[ qIs51[myo-2::gfp Ppes-10::gfp, PF22B7.9::gfp]] V | AV800 | rec-8 RNAi, cross with untreated males |
| AV727 | meIs8 [pie-1p::gfp::cosa-1 + unc-119(+)] II; ltIs37 [pie-1p::mCherry::his-58 + unc-119(+)] IV; ltIs38 [pie-1p::gfp::ph(PLC1delta1) + unc-119(+)] | AV809 | rec-8 RNAi, cross with untreated males |
| AV695 | meIs8 [pie-1p::gfp::cosa-1 + unc-119(+)] II; mnT12 (X; IV) | AV826 | rec-8 RNAi, cross with untreated males |
| DR2078 | mIn1[ dpy-10(e128) mIs14[myo-2::gfp pes-10::gfp]] / bli-2(e768) unc-4(e120) II | AV810 | rec-8 RNAi, cross with untreated males |
| AZ212 | ruIs32 [pie-1::GFP::H2B + unc-119(+)] III | AV822 | rec-8 RNAi, cross with untreated males |
| AZ212 | ruIs32 [pie-1::GFP::H2B + unc-119(+)] III | AV823 | rec-8 RNAi, no cross |
| JU1018 | C. briggsae - mfIs42[Cel-sid-2 + Cel-myo-2::DsRed] | AV824 | rec-8 RNAi, no cross |
Wild-type triploid hermaphrodites were obtained by mating diploid N2 males with SP346 tetraploid hermaphrodites. Similarly, spo-11 mutant triploid hermaphrodites were obtained as GFP− cross-progeny of a cross between AV776 spo-11(me44)/nT1[qIs51] diploid GFP+ males and AV800 tetraploid GFP+ hermaphrodites.
In addition to the strains listed in Table 1, the following strains were used:
FM2: htp-1(gk174) htp-2(tm2543)/nT1 IV; +/nT1[unc-?(n754) let-? qIs50] V
AV276: +/nT1 IV; syp-2(ok307)/nT1[unc-?(n754) let-? qIs50] V
AV393: htp-1(gk174)/nT1 IV; +/nT1[unc-?(n754) let-? qIs50] V
AV739: rec-8(ok978)/nT1[qIs51] IV; +/nT1[let- ?] V; meIs16 [pie-1::mCherry::his-58unc-119 (+)]; ruIs57 [pie-1::GFP::tubulin unc-119 (+)]
AV740: +/nT1[qIs51] IV; +/nT1[let- ?] V; meIs16 [pie-1::mCherry::his-58unc-119 (+)]; ruIs57 [pie-1::GFP::tubulin unc-119 (+)]
meIs16 was obtained by microparticle bombardment of pAA64[pie-1::mCherry::his-58unc-119(+)] (Praitis et al. 2001; Audhya et al. 2005).
rec-8 RNAi
rec-8 RNAi was performed by placing worms on NGM plates containing ampicilin (100 µg/ml) and IPTG (1 mM), seeded with bacteria harboring the W02A2.6/rec-8 clone from the Ahringer library (Kamath and Ahringer 2003). RNAi of C. briggsae rec-8 was performed using the plasmid pBR16 and the C. briggsae strain JU1018. pBR16 was obtained by blunt end cloning of a PCR product obtained by amplification of genomic DNA extracted from the wild-type C. briggsae isolate AF16 using primers oBR242 (ATGCGGGAATTCAGGAAACAT) and oBR243 (CAATCTTCGAAGACTTTCTGG) into EcoRV-digested L4440 (Timmons and Fire 1998). The amplified DNA sequence corresponds to the large exon of CBG12032, the C. briggsae ortholog of rec-8. Observation of univalent chromosomes at diakinesis as well as an extension of the transition zone in gonads of DAPI-stained worms confirmed that pBR16 likely induced RNAi of C. briggsae rec-8.
Cytology
All analyses were performed on 20- to 24-hr post-L4 adults.
For immunofluorescence (IF) experiments in Figure 3, Figure 4, and Figure 5: dissection of gonads, fixation, immunostaining, and DAPI counterstaining were performed as in Martinez-Perez and Villeneuve (2005). The following primary antibodies were used: chicken anti-HTP-3 (1:250, MacQueen et al. 2005), rabbit anti-GFP (1:1000, Yokoo et al. 2012), rabbit anti-SYP-1 (1:250, MacQueen et al. 2002), and guinea pig anti-HIM-8 (1:250, Phillips et al. 2005). Secondary antibodies were Alexa Fluor 488-, 555-, and 647-conjugated goat antibodies directed against the appropriate species (1:400, Life Technologies).
Figure 3.
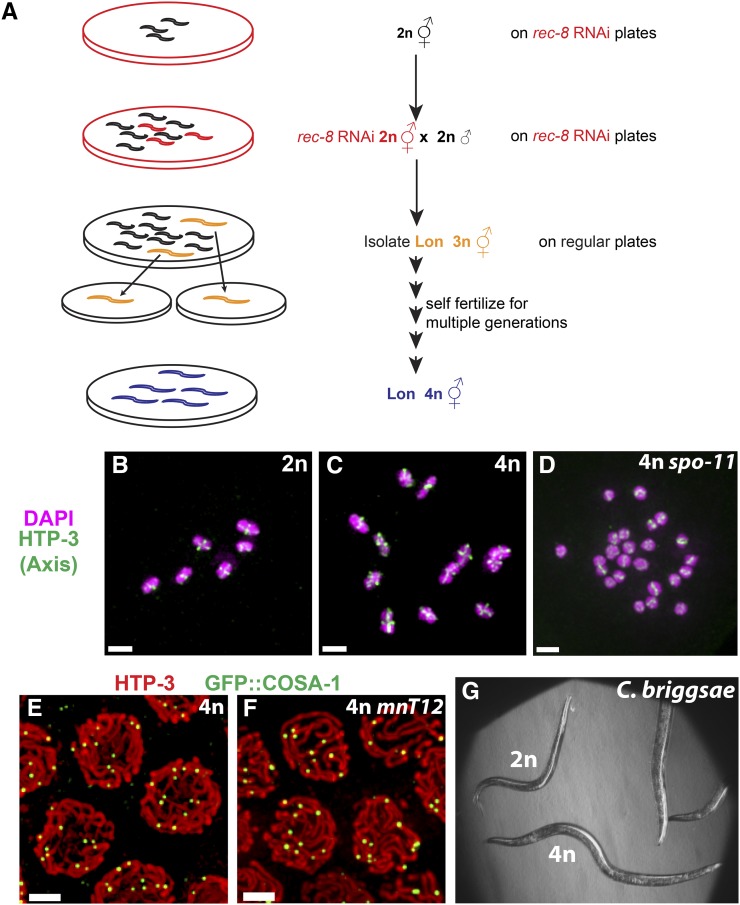
A strategy for generating tetraploid derivatives of any diploid C. elegans strain. (A) Genetic scheme for generating tetraploid strains. Hermaphrodites from a given diploid strain were plated on bacteria expressing dsRNA corresponding to the rec-8 gene to allow RNAi-mediated depletion of REC-8. RNAi-treated F1 worms were transferred to fresh RNAi plates, mated with untreated males of the same strain, and allowed to lay eggs. Each cross plate was analyzed a week later to identify Lon (putative triploid) worms, which were transferred individually to fresh standard OP50 plates at 20° and allowed to produce self-progeny for multiple generations. As triploid worms have very low brood sizes, tetraploid derivatives were readily identified when a plate derived from a putative triploid individual contained large numbers of highly fertile Lon worms. (B–D) Each panel shows the complete complement of chromosomes in a single diakinesis stage oocyte from worms of the indicated genotypes; images are maximum intensity projections. Immunodetection of HTP-3 (green) and DNA counterstaining with DAPI (magenta) allows detection of cruciform chromosome axis structures reflecting the expected presence of 6 and 12 chiasmata, respectively, in wild-type diploid (B) and nT1 heterozygous tetraploid (C) oocytes. In oocytes produced by spo-11 mutant tetraploid hermaphrodites, however, the 24 chromosomes enter the meiotic division as univalents (D) as they failed to initiate meiotic recombination. (E and F) Fields of pachytene nuclei from tetraploid worms carrying a transgene expressing GFP::COSA-1, which marks the sites of meiotic crossovers; the strain in F also contains the fusion chromosome mnT12(X;IV). Chromosome axes and GFP::COSA-1 are visualized using antibodies against HTP-3 (red) and GFP (green), respectively. Images are maximum intensity projections of 3D stacks containing the whole nuclei. In B–F, bar, 2 µm. (G) Bright-field image of a tetraploid (4n) derivative of C. briggsae JU1018 (a strain that was rendered susceptible to feeding RNAi by transgenic expression of C. elegans sid-2), shown with the diploid (2n) for size comparison.
Figure 4.
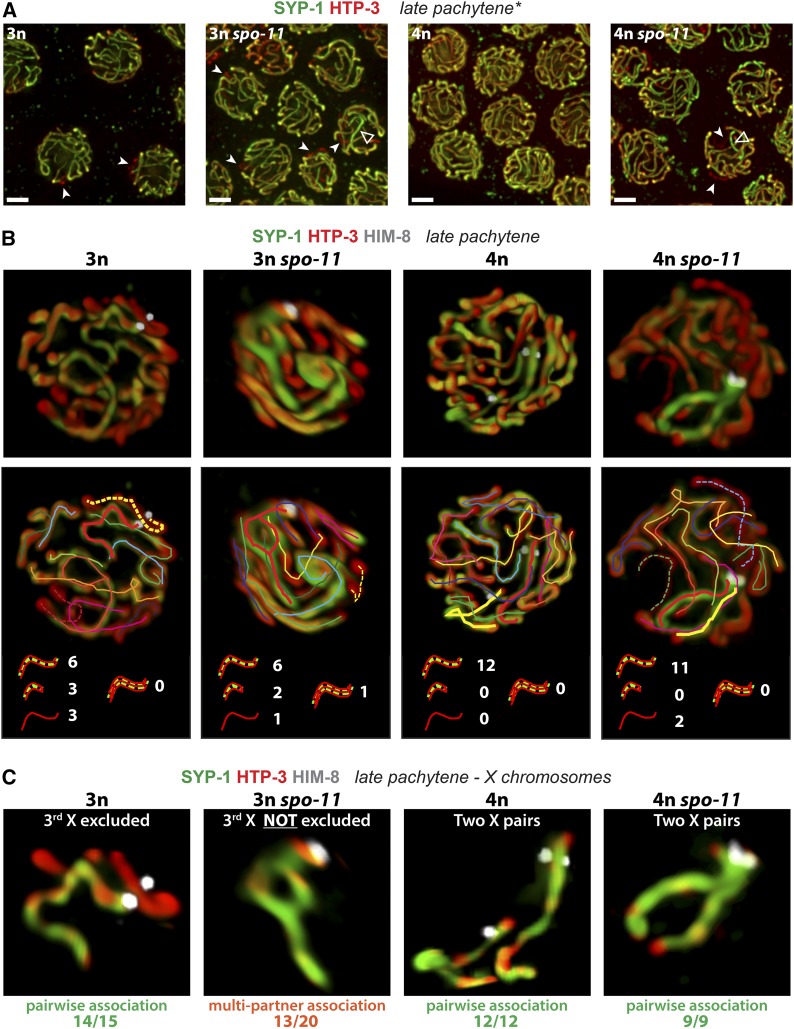
SPO-11 prevents the formation of anomalous multipartner synapsis interactions when three partners compete for synapsis. (A) Synapsis patterns in the indicated genotypes illustrated by immunofluorescence images of nuclei from the late pachytene region of the gonad; images are maximum intensity projections of 3D data stacks encompassing whole nuclei. Previously described differences in meiotic progression between triploid and tetraploid meiocytes were taken into account for definition of the late pachytene region in all analyzed genotypes (see Material and Methods). HTP-3 (red) marks chromosome axes and SYP-1 (green) marks the SC central region; overlap (yellow) indicates synapsed segments. A mixture of nuclei exhibiting apparently complete synapsis and nuclei containing unsynapsed regions (solid arrowheads) was observed in all analyzed genotypes. In addition, some nuclei in both spo-11 mutant triploid and tetraploid strains contained stretches of SC with a higher ratio of central region to axis signal (open arrowheads). Bars, 2 µm. (B) Projected 3D renderings of synapsis configurations in individual nuclei; 3D volume rendering enabled tracing and counting of SCs within each analyzed late pachytene nucleus. Stretches of SC or axes were classified in four categories, illustrated with cartoons below the tracings: (i) long stretches of SYP-1, likely representing pairwise association between two chromosomes; (ii) short stretch of SYP-1, likely representing a single chromosome that had undergone fold-back synapsis; (iii) chromosome axis stretches not colocalizing with any SYP-1 signal, representative of an asynapsed chromosome (dashed lines in tracings); and (iv) SYP-1 associated with more than two partners in the context of multipartner interactions. Classification was assessed on 3D reconstructed nuclei; nuclei from at least two different gonads were analyzed for each genotype. Numbers adjacent to the cartoons indicate the occurrence of the different categories in the example nuclei depicted; frequencies of each category among total nuclei analyzed are reported in the text. (C) Images show synapsis configurations for the X chromosomes from the nuclei depicted in B. X chromosomes were identified by immunofluorescence detection of the X-PC binding protein HIM-8 and were extracted to unambiguously analyze the synapsis configuration of one group of homologs. The most common type of association and its corresponding observed frequency for each genotype is indicated below the sample image. X chromosomes in tetraploids were consistently engaged in pairwise synapsis regardless of spo-11 genotype. In otherwise wild-type triploid nuclei, we regularly observed two of the three X chromosomes engaged in pairwise homologous synapsis, while the third was either asynapsed (as shown here), self-synapsed, or engaged in pairwise nonhomologous synapsis with another partnerless chromosome. In spo-11 mutant triploids, partial or complete multipartner association was observed in 13/20 of the nuclei. Fisher exact test indicates a highly significant difference (P < 10−3) between the observed frequency of multipartner association in spo-11 mutant triploid meiocytes when compared to wild-type triploid.
Figure 5.
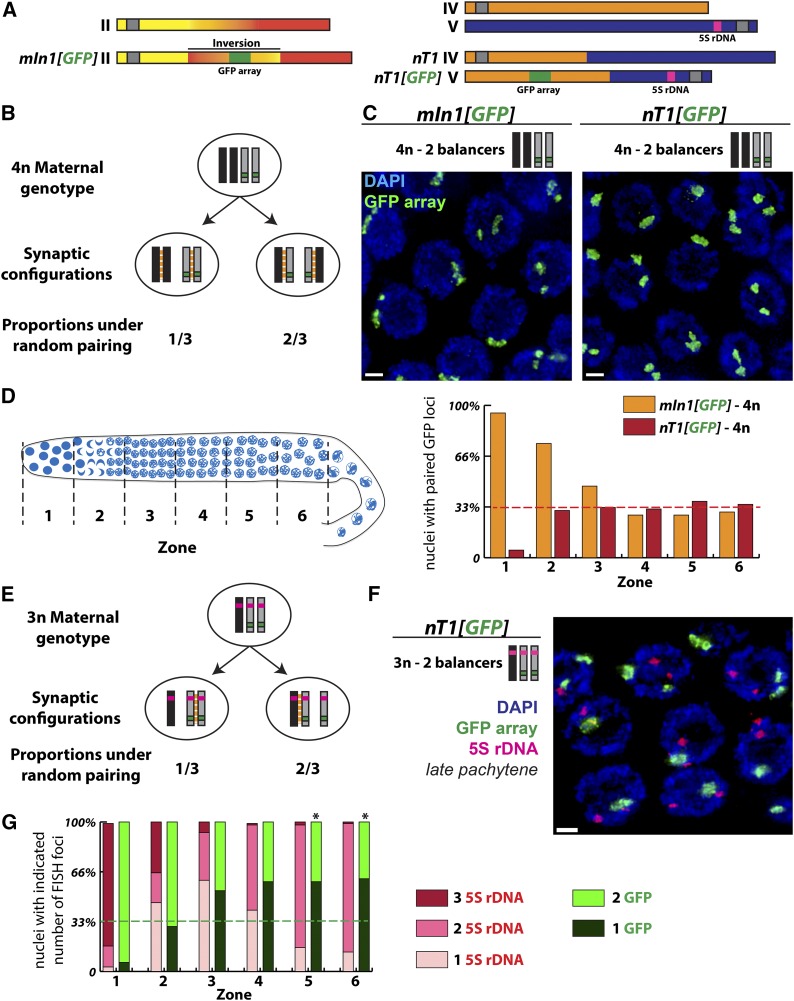
Preference for pairing between identical partners in triploids but not in tetraploids. (A) Schematic representation of chromosomal rearrangements used in this study. The mIn1 rearrangement consists of an inversion of the central third of chromosome II. The nT1 rearrangement is a reciprocal translocation between chromosomes IV and V. GFP transgene arrays have been inserted into these rearrangements to follow their segregation. Of note, the GFP transgene array integrated into nT1 is located on the half-translocation that also carries the 5S rDNA locus and the chromosome V pairing center. (B) Expected proportions of the two possible synaptic configurations in meiocytes carrying two normal sequence (black) and two rearranged chromosomes (gray), under the assumption of random pairing between chromosomes sharing the same pairing center region. (C) Synapsis configurations were analyzed in tetraploid hermaphrodites carrying two normal sequence and two rearranged chromosomes by assessing the pairing status of the GFP transgene array locus using FISH. Images shown are projections of fields of late pachytene meiocytes; bar, 2 µm. (D) Left, schematic representation of germ cells progressing through the meiotic program in a C. elegans gonad; for analysis of pairing by FISH, gonads were divided into six zones of equal size as indicated. Right, histogram representing the frequencies of pairing between the GFP transgene array loci on the rearranged chromosomes in tetraploid (4n) germ cells carrying two copies of mIn1 or nT1. Pairing frequencies are shown for premeiotic nuclei (zone 1), transition zone nuclei (zone 2), and nuclei progressing through the pachytene stage (zones 3–6). The dashed red line indicates the level of pairing at the GFP locus expected under the model that there is no preference for identical synaptic partners. (E) Expected proportions of the two possible pairwise synaptic configurations in triploid (3n) meiocytes carrying one normal sequence chromosome V (black) and two copies of nT1(V) (gray) under the assumption of random pairing between chromosomes sharing the same pairing center region. (F and G) As in C and D, synapsis configurations in triploid meiocytes were inferred using FISH analysis to assess the pairing status of the GFP transgene array located on nT1(V). Image in F shows a projection of a field of late pachytene nuclei, which reveals that the GFP arrays (in green) are detected as paired more frequently than expected based on the hypothesis of random pairing. In these experiments, FISH was also used to detect the 5S rDNA locus (magenta) in the same nuclei. As the 5S locus is present on all three chromosomes competing for pairwise synapsis, the presence of two separate 5S FISH foci serves an indicator that nuclei have progressed to the late pachytene stage in which synapsis associations are predominantly pairwise even in triploids; this further implies that the high level of pairing between the GFP arrays at this stage does not result from persistent associations between all the three partners but instead represents a preference for synapsis to occur between two identical partners in these triploids. Bars, 2 µm. (G) Histogram representing the frequencies of pairing for the two loci analyzed, 5S rDNA in pink and GFP transgene array in green. The dashed green line indicates the level of pairing expected between the GFP arrays under the assumption of random pairing among the three competing partners. Since chromosome movement is prolonged and progression to maximum pairwise synapsis is delayed in triploid germ cells (Mlynarczyk-Evans et al. 2013), the stages of nuclei present in the numbered zones in triploids are not equivalent to those in the tetraploids. Pairing status at the 5S locus indicates that pairwise synapsis configurations predominate in zones 5 and 6. For zones 5 and 6, chi-square analysis indicates a highly significant difference (* P < 10−3) between the observed high incidence of pairing at the GFP array locus on nT1(V) and the level of pairing expected under the assumption of random partner choice, consistent with a strong preference for synapsis between identical partners.
For Figure 2, B, F, and E, immunostaining of dissected male gonads was done as previously described (Gonczy et al. 1999; Oegema et al. 2001) with minor modifications (Schvarzstein et al. 2013). Young adult males were dissected to extrude their gonads, fixed, and permeabilized by freeze cracking in liquid nitrogen followed by soaking in methanol at −20° (Gonczy et al. 1999) for at least 30 min. Gonads were rehydrated in PBS, blocked in AbDil [PBS plus 2% (wt/vol) BSA, 0.1% Triton X-100] as described by Oegema et al. (2001), and incubated at 4° overnight with a combination of different primary antibodies. The following primary antibodies were used at the indicated dilutions: anti-SPD-2 (1:1000) (Kemp et al. 2004), anti-SAS-4 (1:1000) (Delattre et al. 2004), anti-SPE-11 (1:200) (Browning and Strome 1996), and anti-α-tubulin (1:1000; FITC-conjugated DM1A from Sigma). Primary antibodies were detected using Alexa Fluor conjugated secondary antibodies (Life Technologies) at 1:800 dilutions. After washing with PBST (PBS plus 0.1% Triton X-100), gonads were incubated with PBST containing 1 mg/ml Hoechst 33258 (Sigma) and mounted in Molecular Probes ProLong Gold Antifade Mountant (Life Technologies).
Figure 2.
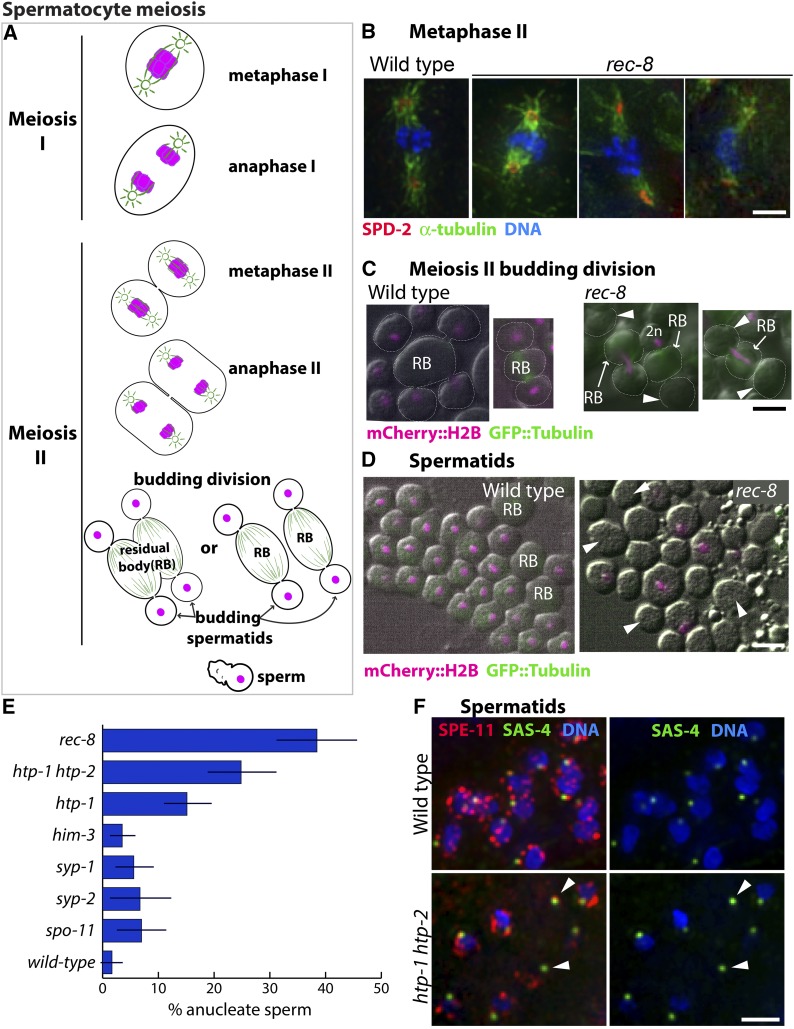
Consequences of impaired meiotic cohesin function during spermatocyte meiotic divisions. (A) Diagram of the C. elegans spermatocyte meiotic divisions; microtubules are indicated in green and chromosomes in magenta. In the first meiotic division, homologs segregate to opposite poles of a spindle that contains centrosomes at its poles. This yields two secondary spermatocytes each of which undergoes meiosis II, giving rise to four haploid spermatids by a specialized budding division. Spermatids bud off and discard ribosomes, microtubules, actin, and other cellular components into the residual body (RB). Spermatids later mature into motile sperm. (B) Images of meiosis II spindles in wild-type and rec-8 mutant secondary spermatocytes, immunostained with anti-SPD-2 and antitubulin antibodies (to highlight centrosomes and spindles) and counterstained with Hoechst to visualize DNA; images are maximum intensity projections of z-stacks encompassing full spindles. rec-8 mutant spermatocytes form bipolar meiosis II spindles but chromosome segregation is impaired. (C) Live images of wild-type (AV740) and rec-8 (AV739) secondary spermatocytes undergoing the budding divisions. Contours of dividing spermatocytes, visualized by DIC, are indicated by white dotted lines; mCherry::histone H2B (shown in magenta) was used to visualize the chromosomes; arrowheads indicate anucleate spermatids. In the wild type, all chromosomes are partitioned into the budding spermatids, with each spermatid receiving a haploid (1n) chromosome complement. In rec-8 mutant spermatocytes, the second meiotic division is abnormal because all sister chromatids are already separated. Three kinds of abnormal budding divisions are shown: right, a division in which all of the DNA was retained in the residual body and two anucleate spermatids were produced; middle, a division yielding one anucleate spermatid and one (presumably 2n) spermatid containing all of the DNA; left, a division in which the chromosomes were partitioned between the residual body and one of the two spermatids, leaving the other spermatid anucleate. (D) Live DIC images of wild-type (AV740) and rec-8 mutant (AV739) spermatids and sperm expressing mCherry::histone H2B (magenta). Arrowheads mark examples of anucleate spermatids and sperm in a rec-8 mutant. (E) Quantification of the proportion of anucleate spermatids/sperm in wild type and several meiosis mutants. Error bars indicate standard deviation. Whereas wild-type sperm rarely have anucleate spermatids or sperm, a quarter or more of the spermatids/sperm produced by htp-1 htp-2 and rec-8 mutants are anucleate. The htp-1 single mutant produces fewer anucleate sperm than the htp-1 htp-2 double mutant, consistent with its less severe defect in meiotic cohesion function (Martinez-Perez et al. 2008; Severson et al. 2009). Fisher’s exact test indicates that htp-1, htp-1 htp-2, and rec-8 mutants have a significantly higher incidence of anucleate sperm (P < 0.0001) when compared either to wild type or to spo-11, him-3, syp-1, and syp-2 mutants (in which sister chromatid cohesin is retained until the second meiotic division). (F) Images of spermatids and sperm from wild type and htp-1 htp-2 mutant males, immunostained for centriolar protein SAS-4 and perinuclear sperm protein SPE-11 and stained with Hoechst to visualize DNA. Arrowheads mark centrioles and SPE-11 staining in anucleate sperm.
For Figure 2, C and D, male germ lines expressing mCherry::H2B and GFP::tubulin were dissected into 3.5 μl of sperm medium (50 mM Hepes, pH 7, 50 mM NaCl, 25 mM KCl, 1 mM MgSO4, 5 mM CaCl2, 1 mg/ml BSA) (Nelson and Ward 1980) and images were acquired at ×100 magnification using a Zeiss Axioimager M2 microscope.
For fluorescence in situ hybridization (FISH) experiments, DNA for 5S ribosomal DNA (rDNA) probes was prepared as in Zhang et al. (2012), and DNA probes for the GFP transgene array were prepared as in Bessler et al. (2010). Probe DNAs were subsequently labeled with either Alexa Fluor 488 or 594 using the Ulysis DNA labeling kit (Life Technologies). Gonad dissection, permeabilization, fixation, hybridization, and DAPI counterstaining were performed as described in Nabeshima et al. (2011).
Image collection and analysis
Images in Figure 2B were obtained as stacks of optical sections acquired at 0.1-μm intervals using the DeltaVision deconvolution microscope system and full projections of dividing spermatocytes were generated using SoftWoRx Suite software. For Figure 2, C and D, DIC and epifluorescence images were combined. For Figure 2E, quantification was done using sperm from dissected gonads (three or more animals for each genotype). Sperm chromatin and sperm centrioles were visualized by immunofluorescence imaging using Hoechst DNA stain and antibodies against centriolar protein SAS-4, respectively; antibodies against either sperm-specific perinuclear protein SPE-11 (as in Figure 2F) or α-tubulin were included to aid in distinguishing between spermatids and residual bodies (see below). Numbers of sperm (n) analyzed: wild-type (1496, 10 males), rec-8 (934, 6 males), htp-1htp-2 (1122, 11 males), htp-1 (2078, 11 males), him-3 (598, 8 males), syp-1 (697, 8 males), syp-2 (767, 3 males), spo-11 (619, 10 males). Multiple criteria were used to unambiguously distinguish sperm/spermatids from residual bodies: (1) when viewed using DIC, the cytoplasm of sperm and spermatids has a granular appearance, whereas residual bodies have a smooth, glassy appearance; (2) the centriolar component SAS-4 and perinuclear protein SPE-11 specifically localize to the sperm; and (3) after spermatid budding, the residual body retains most of the tubulin.
Images in Figure 3, Figure 4, and Figure 5 were collected as Z-stacks (at 0.2-µm intervals) using a ×100 N.A. 1.40 objective on a DeltaVison OMX microscopy system, deconvolved, and corrected for registration using SoftWoRx. Final assembly of 2D maximum intensity projections was performed using Fiji (Schindelin et al. 2012), with minor contrast adjustments in Adobe Photoshop. The 3D cropping and rendering of images, as well as contrast adjustment of such 3D rendered surfaces, were performed using the Volocity 5 software package (PerkinElmer). As polyploid strains have an increased tendency for karyotype instability, diakinesis stage oocytes of each analyzed gonad were also imaged and examined to confirm the number the chromosomes as well as some diagnostic features of the studied genotype (e.g., the presence of univalents or bivalents, and/or number of FISH foci).
Identification of late pachytene meiocytes:
As progression through meiotic prophase is altered in germ cells with modified karyotypes (Mlynarczyk-Evans et al. 2013), we used the previously reported position of exit from the early pachytene stage defined by presence of the phosphorylated form of nuclear envelope protein SUN-1 (SUN-1 S8-Pi) as a reference point; for these analyses, the “meiotic zone” of the distal gonad arm was defined as the portion of the gonad extending from entry into the transition zone (indicated by the presence of nuclei with clustered chromosomes) through the last row containing multiple meiotic prophase nuclei. For tetraploid germ cells, “late pachytene” nuclei were defined as being within the last third of the meiotic zone; for triploid germ cells, late pachytene corresponded to the last 10th of the meiotic zone. Multiple overlapping fields covering the whole length of the gonad were acquired for each specimen, and gonads were assembled by iterative use of the “pairwise stitching” plugin (Preibisch et al. 2009) on Fiji to allow identification of nuclei within these defined positions. For analysis of volume-rendered synapsis configurations, nuclei were scored only when staining, resolution and the arrangement of chromosomes within the nucleus permitted unambiguous tracing of SC in 3D rotations; nuclei from at least two different gonads were analyzed for each genotype. The total number of such 3D reconstructed nuclei scored is as follows: 4n, 12; 4n spo-11, 9; 3n, 15; and and 3n spo-11, 20.
FISH quantification:
Whole gonads were reconstructed as described above by iterative pairwise stitching and were then divided into six zones of equal length. FISH signals in nuclei that were fully contained within the stacks and in one zone were analyzed; loci were considered paired when the FISH signals were touching each other in any of the three dimensions and were scored as unpaired otherwise. Two to four gonads were analyzed for each genotype, and the total numbers of nuclei scored in each zone were as follows: from zones 1 to 6, mIn1 heterozygous tetraploids: 43, 55, 68, 64, 40, and 37; nT1 heterozygous tetraploids: 41, 49, 66, 62, 52, and 31; and nT1 heterozygous triploids: 297, 280, 280, 282, 225, and 185.
Statistics
Statistical analyses using Fisher’s exact test were performed using GraphPad Prism software.
Data and reagent availability
All nematode strains generated for this study are available upon request, either from the authors or from the Caenorhabditis Genetics Center.
Results
Impairment of meiotic cohesin function has different consequences for male and female gametes
In C. elegans, as in most metazoa, the cell division processes associated with the production of male and female gametes differ substantially. Oocyte meiosis is characterized by highly asymmetric cell divisions in which chromosomes segregate using acentrosomal barrel-shaped spindles adjacent to the anterior cortex of the newly fertilized zygote. Following anaphase of meiosis I, cytokinesis directed by the central spindle results in one set of homologs being extruded into a polar body, thereby achieving reduction in chromosome number. Only the set of chromosomes retained within the zygote undergoes the equational meiosis II division, which results in extrusion of a second polar body and retention of a single haploid set of chromosomes that will form the maternal pronucleus (Figure 1) (Kim et al. 2013). In contrast, spermatocytes undergo symmetric meiotic divisions using spindles with centrosomes at their poles (Figure 2A). Moreover, both products of spermatocyte meiosis I undergo meiosis II in parallel, and following anaphase II, a specialized budding division process, directed by the centrioles (Peters et al. 2010), packages each of the four meiotic haploid products into individual spermatids, leaving behind microtubules and other cellular components in a structure known as the residual body (Shakes et al. 2009).
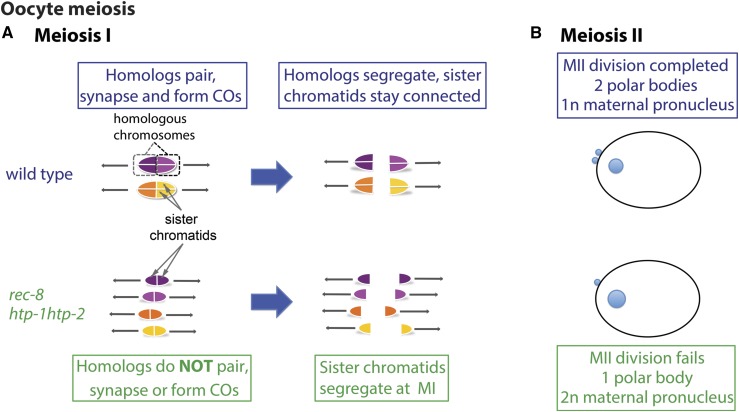
Figure 1.
Consequences of impaired meiotic cohesin function for the oocyte meiotic divisions. Based on Martinez-Perez et al. 2008; Severson et al. 2009, (A) Diagram of chromosome organization and segregation pattern during the first meiotic division in wild-type oocytes and oocytes impaired for meiotic cohesin function (rec-8, htp-1 htp-2). (B) Diagram of the products of oocyte meiosis in newly fertilized zygotes. In wild-type zygotes, two polar bodies (small blue circles at the cortex) and a single haploid (1n) maternal pronucleus reflect the successful execution of two oocyte meiotic divisions. In rec-8 or htp-1 htp-2 mutants, the oocyte meiosis II division fails, resulting in a zygote with a single polar body and a diploid (2n) maternal pronucleus.
In the course of analyzing mutants defective for meiotic sister chromatid cohesion, we discovered that these very different cell division programs can yield very different outcomes when meiotic cohesin function is compromised. Cohesin complexes play multiple roles during the meiotic program, promoting pairing, synapsis, and recombination between homologs during meiotic prophase and mediating regulated segregation during the meiotic divisions (Pasierbek et al. 2001; Chan et al. 2003; Severson et al. 2009; Schvarzstein et al. 2010). During wild-type meiosis, sister chromatid cohesion is released in two steps, enabling reductional segregation of homologs at meiosis I, followed by equational segregation of sister chromatids at meiosis II. Previous studies showed that during oocyte meiosis in C. elegans mutants lacking meiotic cohesin complexes containing the REC-8 subunit (rec-8) or that undergo premature loss of REC-8 cohesin (htp-1htp-2), sister chromatids lose cohesion prematurely and segregate equationally at the meiosis I division (Figure 1A) (Martinez-Perez et al. 2008; Severson et al. 2009); these mutants subsequently fail to complete the meiosis II division and do not extrude a second polar body, resulting in retention of a diploid (or nearly diploid) set of chromosomes in the maternal pronucleus (Figure 1B).
In contrast, we found that loss of REC-8 cohesin function during spermatocyte meiosis frequently results in the formation of gametes that either lack chromosomes entirely or have inherited an abnormal number of chromosomes. Secondary spermatocytes in rec-8 mutant males are able to form metaphase II spindles (Figure 2B), but anaphase II chromosome segregation is severely impaired. Impaired anaphase can result in all chromosomes being packaged into a single spermatid or to some or all chromosomes being left behind in the residual bodies during the budding divisions (shown in combined DIC and epifluorescence live images in Figure 2C). Consequently, such divisions result in a high frequency of anucleate spermatids/sperm (Figure 2, D and E). Anucleate spermatids/sperm are likewise observed at high frequency in the htp-1htp-2 mutant, as depicted in Figure 2F, where anucleate sperm are identified by the presence of both the centriolar component SAS-4 and the sperm-specific protein SPE-11 in cells that lack detectable DNA (see Materials and Methods for details). However, anucleate sperm are much less frequent in other classes of meiotic mutants that lack crossovers (COs) but retain cohesion during meiosis I (Figure 2E). Thus, the same meiotic defect that results in failure to reduce ploidy in the context of female meiosis can lead to the production of gametes devoid of chromosomes in the context of spermatogenesis.
A scheme using transient rec-8 RNAi for efficient generation of stable tetraploid strains
As oocytes produced by rec-8 mutant hermaphrodites often contribute a diploid complement to the developing embryo (Severson et al. 2009), we reasoned that we could efficiently generate polyploid isolates of C. elegans strains by fertilizing diploid oocytes (e.g., produced by a rec-8 mutant hermaphrodite) with haploid sperm produced by a wild-type male to give rise to triploid individuals. Such triploid worms are easily identified as being significantly longer than their diploid counterparts. Further, whereas triploid worms have low fertility owing to the production of aneuploid gametes and offspring, they can eventually give rise to tetraploid descendants, which are much more successful reproductively and can be efficiently propagated. To make this strategy more flexible, we decided to use transient rec-8 RNAi treatment, which can phenocopy many aspects of the mutant phenotype in the female germline (Pasierbek et al. 2001; Severson et al. 2009).
We first tested the strategy, depicted in Figure 3A, by trying to obtain a tetraploid derivative of AV776, in which the spo-11(me44) mutation is balanced by the nT1(IV;V) chromosomal rearrangement. AV776 L4 hermaphrodites were plated at 15° on HT115 bacteria expressing the rec-8 clone from the RNAi feeding library (Kamath and Ahringer 2003). Twenty F1 L4 hermaphrodites from these plates were then transferred to fresh rec-8 RNAi plates and crossed with untreated AV776 males at 15° (four hermaphrodites and 10 males per plate). These plates were screened 7 days later for the presence of long (Lon), likely triploid, individuals. Five potential triploids were identified and transferred individually onto fresh standard plates at 20° and allowed to grow for several generations. Four of the five Lon worms eventually gave rise to stable strains producing Lon worms that were confirmed to be genuine tetraploids by immunostaining experiments. Specifically, we found that diakinesis-stage oocytes from tetraploid nT1 carriers had 12 bivalents (pairs of homologs connected by chiasmata; Figure 3C), whereas oocytes from the recombination-deficient spo-11(me44) homozygotes had 24 univalents (Figure 3D).
We subsequently used this strategy to generate tetraploid derivatives of strains harboring other chromosome rearrangements and/or transgenes expressing a variety of different germ cell markers (Table 1). These include a strain expressing GFP::COSA-1, which marks the sites of crossover recombination events in late pachytene and diplotene meiocytes (Yokoo et al. 2012) (Figure 3E), in combination with mCherry::histone H2B and a plasma membrane marker (Audhya et al. 2005; McNally et al. 2006); a strain carrying the mnT12(IV:X) fusion chromosome in combination with the GFP::COSA-1-expressing transgene (Yokoo et al. 2012) (Figure 3F); and a strain heterozygous for the mIn1 chromosomal inversion and recessive morphological markers (see below). Thus, our strategy provides a reliable and generalizable means to generate tetraploid strains with complex genotypes that would have been nearly impossible to construct by other methods.
As some strains are defective at mating, we sought to test whether the crossing step in our original strategy was essential for generation of tetraploid strains. We therefore sought to generate tetraploid derivatives of AZ212, a transgenic strain with germline expression of a fusion between GFP and histone H2B (Praitis et al. 2001), both by crossing rec-8 RNAi-treated hermaphrodites with untreated males (original scheme described in Figure 3A, step 2) or by simply transferring the rec-8 RNAi treated F1 self-progeny onto fresh RNAi plates. Lon worms were isolated using both strategies (3 for the cross plates, 18 for the “noncross” plates). Although most Lon worm obtained by the selfing strategy proved to be sterile, we did successfully generate one tetraploid strain using this approach; for comparison, each of the 3 Lon putative triploids obtained through the original crossing scheme gave rise to tetraploid strains. We also successfully used the selfing strategy to generate a tetraploid derivative of C. briggsae JU1018 (Figure 3G), a transgenic strain that is sensitive to RNAi by feeding (Nuez and Felix 2012).
Role of meiotic recombination in establishing pairwise synapsis
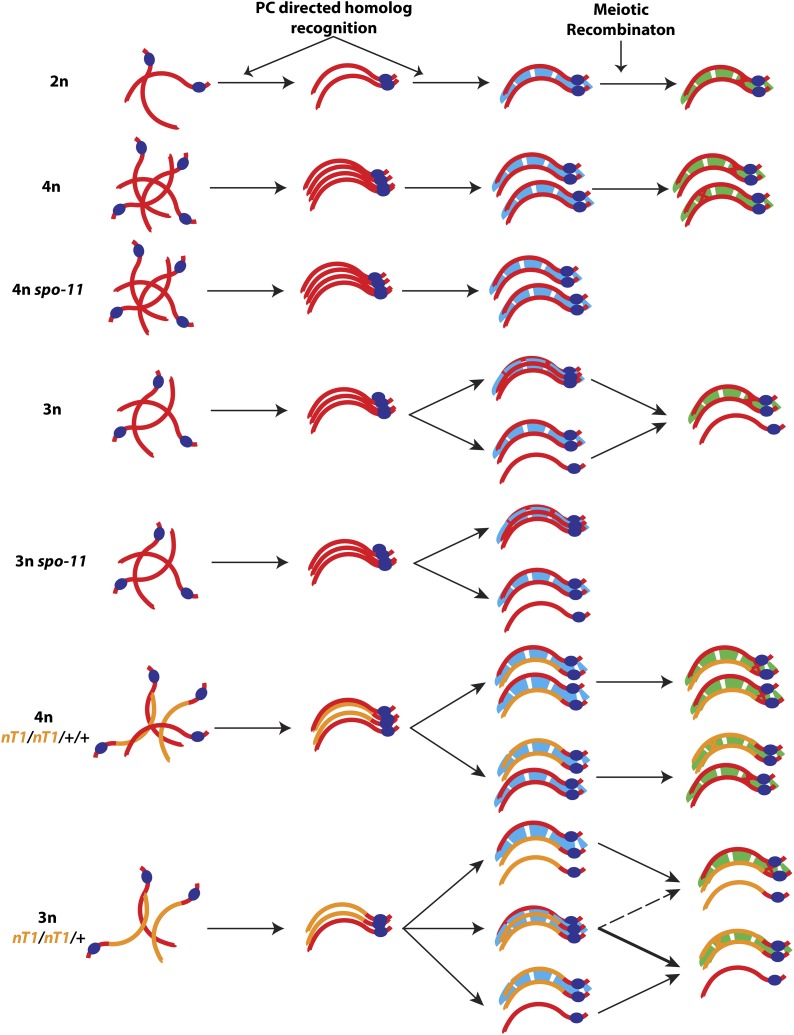
We had previously analyzed the processes of pairing and synapsis in worms with altered ploidy (Mlynarczyk-Evans et al. 2013) and observed that chromosomes were first sorted into groups of homologs regardless of their number. In the context of a wild-type meiotic machinery, chromosomes then preferentially achieved pairwise associations even in situation where more than two partners could compete for synapsis. However, by analyzing the synapsis configurations in trisomic (triplo-X) meiocytes mutant for spo-11, which are deficient in forming the double-strand DNA breaks (DSBs) that initiate meiotic recombination, we observed a persistence of association between all three X chromosomes at stages where their wild-type counterparts had excluded the third X chromosome. This observation suggested that meiotic recombination may play a role in restricting synapsis associations to a strictly pairwise state, a possibility that we tested further in the current work.
We investigated the contribution of recombination to achieving pairwise synapsis in C. elegans by comparing synapsis configurations in both triploid and tetraploid meiocytes competent or deficient for initiating meiotic recombination. To evaluate synapsis, we stained for the meiotic chromosome axis protein HTP-3 to identify all the chromosomes, and for the central region protein SYP-1 to identify synapsed regions (MacQueen et al. 2002; Goodyer et al. 2008) (Figure 4). As we aimed to focus on mature synapsis interactions, we specifically analyzed late pachytene meiocytes, where synapsis is maximal in both diploids and polyploids. As our previous analysis had revealed that triploid germ cell nuclei are significantly delayed in meiotic progression (Mlynarczyk-Evans et al. 2013), we used criteria from that prior study to identify nuclei at the appropriate stage (see Material and Methods for details). Images of late pachytene nuclei in recombination-proficient polyploids were consistent with our previous report showing that most tetraploid meiocytes display apparent full pairwise synapsis, while triploid nuclei, which carry one additional copy of each of the six chromosomes, display zero to two unsynapsed chromosomes (Figure 4A).
At first glance, nuclei from spo-11 polyploids appeared largely similar to their wild-type counterparts: only a few unsynapsed axes were detected in each triploid spo-11 nucleus, and apparent full synapsis was observed in most tetraploid spo-11 nuclei (Figure 4A). We did note that a subset of late pachytene nuclei in the spo-11 mutants exhibited an uneven distribution of SYP-1 staining among the axes (Figure 4A, empty arrowheads). This feature is also present in spo-11 diploids and suggests that the molecular architecture of the SCs formed in the absence of DSBs may be somewhat different from that present in wild-type meiocytes.
To better determine the specific synapsis configurations achieved in nuclei of all analyzed genotypes, we used 3D rendering of IF samples to trace and count every single SC and/or chromosome axis within a given nucleus (Figure 4B). As expected, in recombination-proficient tetraploids, we could typically identify 12 SC tracks, presumably representing full normal synapsis between each of the 12 pairs of homologs (11/12 nuclei). Analysis of spo-11 tetraploid meiocytes similarly revealed that most chromosomes in most nuclei achieved pairwise synapsis interactions (7/9 nuclei with 12 SC tracks). Further, the occasional synapsis defects detected (i.e., two unsynapsed axes or one unsynapsed axis and one short SC track likely representing fold-back self-synapsis of a single chromosome) do not represent deviations from the general rule that synapsis interactions occur preferentially between pairs of chromosome axes. We could therefore conclude that, in the presence of an even number of chromosomes competing for synapsis, meiotic recombination is largely dispensable for achieving pairwise synapsis associations. This finding corroborates and extends our previous observation of a strong drive for synapsis to occur in a strictly pairwise manner (Mlynarczyk-Evans et al. 2013).
As expected by the presence of an odd number of competing partners, synapsis defects were much more abundant in triploid nuclei. In addition to long SC tracks similar to those observed in tetraploids, which likely represent full-length pairwise synapsis associations (most between homologous but some between nonhomologous chromosomes), we detected a few unsynapsed chromosome axes and/or self-synapsed chromosomes (identified as short SC tracks) in every single triploid nucleus. Moreover, genuine multipartner associations persisting into late pachytene were observed unambiguously in the spo-11 triploids. These were readily identified in cases where a multipartner association was incomplete and gave rise to configurations where a bubble could be observed in the middle of a SYP-1 stretch (red track, Figure 4B, 3n spo-11) or when a single SC track forked into two (Supporting Information, Figure S1A, blue tracks).
To further evaluate the effect of recombination on pairwise synapsis, we estimated and compared the incidence of nuclei with multipartner associations in spo-11 mutant and recombination-proficient triploids. Specifically, we counted the numbers of SC tracks and unsynapsed axes to try to account for the presence of every single chromosome within each scored nucleus. If all SC tracks were explainable by strictly pairwise association between chromosome axes, a given SC track would account for the presence of either two chromosomes or one self-synapsed chromosome. In wild-type trip loids, the pairwise hypothesis can fully explain the distribution of SC tracks and unsynapsed chromosome axes in 16 of the 17 nuclei analyzed. In contrast, the pairwise hypothesis can explain only 5 of the 21 spo-11 triploid nuclei analyzed, and the presence of at least one multipartner association is necessary to account for all chromosomes in the remaining 16 nuclei (see Figure S1B for an example). This difference between recombination-proficient and recombination-deficient triploids was highly significant (P < 0.0001, two-tailed Fisher’s exact test).
Our immunofluorescence analysis also included antibodies detecting HIM-8, a zinc finger protein that specifically binds the X chromosome pairing center (Phillips et al. 2005), enabling unambiguous identification of the X chromosomes and analysis of their synapsis configurations (Figure 4C). We found that the X chromosomes were engaged in pairwise synapsis associations exclusively in tetraploids (both wild-type and spo-11) and predominantly in wild-type triploids, whereas multipartner associations involving the X chromosomes were detected in more than half of the spo-11 triploid nuclei.
Altogether, this analysis demonstrates that although initiation of meiotic recombination is not strictly required for the establishment of exclusive pairwise interactions, some aspect(s) of the meiotic recombination program contribute to the maturation of synapsis associations into a strictly pairwise state when an odd number of partners are competing for synapsis.
Homology in the vicinity of the PC dictates partner choice in tetraploids
We had previously reported that in polyploid meiocytes, chromosomes were first sorted in groups of homologs, sharing the same PC, and then achieved pairwise synapsis (Mlynarczyk-Evans et al. 2013). This implies that from each group of homologs, interactions between two homologous partners must somehow be selected to mature into fully pairwise synapsis associations. We aimed to investigate the extent to which homology in regions of the chromosomes besides the PCs might contribute to this homolog selection process. To address this issue, we used polyploid worms heterozygous for rearranged chromosomes to create “competitive pairing” situations in which identical and nonidentical chromosomes sharing the same PC compete to establish pairwise relationships.
We first evaluated meiosis in tetraploid worms carrying two copies of a normal-sequence chromosome and two copies of a rearranged chromosome that shares the same PC (Figure 5A). In diploids, such rearranged chromosomes pair and synapse efficiently with the normal-sequence chromosome sharing the homologous PC region (MacQueen et al. 2005). We reasoned that tetraploid worms with two copies of each type of chromosome could allow us to test whether, given the opportunity, chromosomes would exhibit a preference for pairing and synapsing with identical partners.
We evaluated two different tetraploid strains heterozygous for two different rearrangements: mIn1, which harbors an inversion of the central portion of chromosome II, and nT1 (IV;V), a reciprocal translocation between chromosomes IV and V. In diploids, both of these rearrangements serve as balancer chromosomes, as pairwise synapsis initiated at chromosome segments that share PCs will juxtapose nonhomologous chromosome segments (MacQueen et al. 2005). Both rearrangements also harbor large transgene arrays containing many copies of a transgene that drives expression of GFP in the pharynx, enabling identification of worms carrying such chromosomes.
For mIn1, we first assessed the frequencies of GFP(−) worms among the self-progeny of mIn1 heterozygous tetraploids (Figure S2). Interestingly, we found that the frequencies of GFP(−) self-progeny produced by mIn1 heterozygous tetraploids matched expectations for random pairing among the competing chromosomes, followed by independent assortment (Figure S2). Thus, this analysis implied a lack of preference for identical pairing partners in tetraploids carrying two copies of mIn1 and two copies of a normal-sequence chromosome II.
Cytological analysis of homolog pairing in heterozygous tetraploids provided a direct demonstration of this lack of preference for identical pairing partners (Figure 5, B–D). For this analysis, the GFP transgene arrays on the rearranged chromosomes served as chromosome-specific FISH targets, enabling us to distinguish between synapsis configurations in which the identical partners are paired (one FISH signal) and configurations where synapsis occurs between nonidentical partners (two separated FISH signals). For both mIn1 and nT1, the frequencies of paired FISH signals in pachytene nuclei matched expectations for random partner choice among normal-sequence and rearranged chromosomes, indicating lack of a preference for identical synapsis partners in tetraploid worms undergoing meiosis. These data suggest that in tetraploids, homology in the region of the chromosome contiguous with the PC is the primary driver of synapsis partner choice.
Surprisingly, the GFP transgene arrays used as FISH targets for these experiments behaved very differently from each other in the premeiotic region of the germ line. Whereas no pairing between the arrays was detected in premeiotic germ cell nuclei of nT1/nT1/+/+ tetraploids, consistent with the known behavior of endogenous loci in diploids (MacQueen et al. 2002; Nabeshima et al. 2011), the transgene arrays were very consistently paired in premeiotic nuclei in mIn1/mIn1/+/+ tetraploids (Figure 5D, zone 1). This premeiotic pairing may reflect heterochromatin-like associations, as histone modifications characteristic of heterochromatin are known to be enriched on repetitive transgene arrays in C. elegans germ cells (Bessler et al. 2010). However, this premeiotic association of the transgene array on mIn1 was lost upon entry into meiosis, indicating that the meiotic pairing and synapsis machinery has the capacity to dissolve preexisting interchromosomal interactions.
Homology distant from the PC contributes to partner choice in nT1/nT1/+ triploids
We also tested whether homology outside of the PC region contributed to partner choice in triploids. For this analysis, we conducted FISH experiments analyzing the pairing of both the GFP transgene array locus and the 5S rDNA locus in triploid animals carrying two copies of nT1 V and one normal-sequence chromosome V (Figure 5F). The 5S rDNA locus is located to the right of the nT1 breakpoint on chromosome V and is therefore present on the same half-translocation (nT1[GFP] V) that also harbors both the chromosome V PC and the transgene array (Figure 5A). Analysis of the dynamics of pairing at the 5S rDNA locus was consistent with the previously described dynamics of homolog pairing and synapsis in triploid germ cells (Mlynarczyk-Evans et al. 2013): No significant pairing was detected at the 5S locus in premeiotic germ cells (Figure 5G, zone 1), and a strong association between all three homologs was evident upon entry into meiosis (Figure 5G, zones 2 and 3). Upon progression through meiotic prophase toward the late pachytene zone, the single 5S FISH signal resolved into two separate FISH signals (Figure 5G, zones 4–6), reflecting two of the three partners eventually achieving a pairwise association and the third being excluded (and either engaging in nonhomologous synapsis or remained asynapsed). This analysis of the 5S rDNA locus thus allowed us to infer that in nuclei within zones 5 and 6 of the gonad, the chromosomes being assayed for partner preference had achieved pairwise synapsis.
In contrast to the tetraploids, analysis of pairing at the GFP transgene array locus revealed that nT1/nT1/+ triploids exhibit a strong preference for synapsis to occur between identical partners. We observed a single GFP FISH signal in about two-thirds of pachytene nuclei (Figure 5, zones 5 and 6), characteristic of pairing occurring preferentially between the two nT1 chromosomes. This represents a highly significant departure from the one-third frequency expected under the hypothesis of random pairing between partners sharing the same PC (Figure 5E; P < 10−3). Thus, when three chromosomes sharing the same PC compete for exclusive pairwise association in nT1/nT1/+ triploids, there is a significant bias favoring association between the two identical chromosomes, suggesting that homology outside the pairing center contributes to synapsis partner choice in this context.
Discussion
Efficient generation of polyploid strains for investigating diverse biological processes
Tetraploid derivatives of both C. elegans and C. briggsae were previously obtained by crossing heat-shock-treated males and hermaphrodites followed by screening for animals with longer body size, reduced fertility, unusually low incidence of progeny displaying phenotypes of recessive markers, and/or a high incidence of male self-progeny, all of which are potential indicators of hyperploidy (Madl and Herman 1979; Woodruff et al. 2010). This approach likely relies on the temperature sensitivity of meiotic processes such as synaptonemal complex assembly (Bilgir et al. 2013) that makes heat-shocked animals more prone to producing highly aneuploid gametes and therefore to produce an occasional triploid animal. Unfortunately, the method is limited in its applicability as it also relies on the use of genetic markers to help to identify the triploid intermediates and has proved to have low efficiency.
We have described here a highly generalizable scheme that can generate tetraploid derivatives of virtually any C. elegans strain by using the previously reported potential of some meiotic mutants to produce diploid gametes. This flexible strategy provides a means to efficiently generate and analyze tetraploid strains carrying mutations, chromosome rearrangements and/or transgenes, which we exploited to gain further insights into the processes of pairing and synapsis during meiotic prophase. The strategy described here could also prove to be a valuable tool to interrogate processes relevant to cell and genome biology, physiology, or development in a model animal. For example, analyzing how the cell and its components, such as the nucleus or mitotic spindle, scale in response to an increase in DNA content may enable interrogation of the genetic circuits responsible for such scaling (Levy and Heald 2012). Further, polyploid strains may be useful for investigating the mechanisms that regulate endoreplication in tissues in which cells are normally polyploid (Hedgecock and White 1985; Lozano et al. 2006). We also noted during our study that tetraploid strains can give rise to aneuploid derivatives and may potentially be more permissive for aneuploidy than diploid strains, suggesting that our strategy may provide tools for analyzing how differences in dosage of both sex-linked and autosomal genes are tolerated or mitigated to ensure developmental success.
Multiple inputs contribute to the pairwise nature of synapsis interactions: a two-phase model for synapsis during C. elegans meiosis
In many organisms, including S. cerevisiae, M. musculus, and A. thaliana, initiation and progression of meiotic recombination are required to establish SC-mediated pairwise association between homologous chromosomes (Giroux et al. 1989; Edelmann et al. 1999; Baudat et al. 2000; Romanienko and Camerini-Otero 2000; Grelon et al. 2001). This strict dependence is not universally conserved, however, since in organisms such as C. elegans and D. melanogaster, mutants that fail to initiate meiotic recombination can efficiently pair and synapse their homologs (Dernburg et al. 1998; McKim et al. 1998). Nonetheless, identification of defects in establishing exclusive pairwise synapsis interactions among X chromosomes in trisomic triplo-X worms deficient in initiating meiotic recombination (Mlynarczyk-Evans et al. 2013) led us to reinterrogate the relationship between these processes in C. elegans.
Taking advantage of our ability to generate polyploid derivatives of diploid strains, we analyzed how the ability to initiate meiotic recombination affected synapsis interactions in a context where synapsis was challenged by the presence of supernumerary chromosomes. We found that in triploid germ cells, the ability to exclude multipartner synapsis interactions was impaired in the absence of recombination initiation, whereas failure to initiate recombination did not impair the pairwise nature of synapsis association in tetraploid meiocytes. Efficient establishment of exclusively pairwise synapsis interactions therefore relies on meiotic recombination only when an odd number of partners compete for synapsis.
Our observations reveal a previously hidden contribution of meiotic recombination to synapsis and support a model (Figure 6) in which mature synapsis interactions are the product of a two-phase process involving: (1) an early phase, in which early interactions are both (a) promoted by activities of the PCs and (b) constrained by a propensity of the SC to assemble in a strictly pairwise manner between two and only two partner axes; and (2) a later phase during which recombination-based interactions solidify the associations between one pair of homologs while simultaneously excluding any additional partner.
Figure 6.
A two-phase model for establishment of mature synapsis interactions. We propose that mature synapsis interactions are established as part of a two-phase process. During the early phase of synapsis, homologs are sorted and aligned based on the activity of their PCs (dark blue ovals), PC-associated factors, and PC-adjacent homology, culminating in assembly of an early SC (light blue). This early SC preferentially assembles in a pairwise manner, but in situations where three homologs compete for synapsis, the environment can permit synapsis-like associations among more than two partners. During the later phase of synapsis, recombination-based interactions solidify associations between two partners to form a mature SC (green); this process can drive resolution of multipartner associations into mature pairwise configurations. See Discussion for details.
In this model, early synapsis interactions can temporarily accommodate multipartner associations, but a strong drive leads to preferential assembly of SCs between pairs of chromosomes. As a consequence, any transient multipartner associations that might occur in tetraploids would be strongly outcompeted by a more stable configuration in which the four homologs are distributed in two synapsed pairs. In contrast, in triploids and trisomic triplo-X germ cells, an odd number of homologs can be maintained in close alignment for a prolonged period by the activity of the PCs (Mlynarczyk-Evans et al. 2013), making the formation or persistence of unfavorable multipartner synapsis associations possible. We note that our analysis did not distinguish whether the observed three-partner associations reflected bona fide three-way synapsis in which SC central region components simultaneously link three different chromosome axes or whether they reflected local pairwise synapsis associations combined with partner switches. The observed tendency for the SC central region to bridge pairs of axes would tend to argue for the latter hypothesis, although the apparently processive nature of SC assembly does not seem compatible with partner switching (MacQueen et al. 2005; Rog and Dernburg 2015).
Our analysis of partner choice in polyploid worms heterozygous for chromosomal rearrangements provides further support for this two-phase model. We suggest that when early synapsis associations can be driven to occur in a strictly pairwise manner, as is the case in tetraploid germ cells, homology assessment occurs only (or primarily) in the vicinity of the PCs, resulting in random association between normal sequence and rearranged chromosomes sharing the same PC. In contrast, persistence of multipartner interactions until later stages would offer an opportunity for recombinational interactions to assess homology along the whole length of the chromosomes and consequently to promote associations preferentially between fully identical chromosomes. Although this was not directly tested due to technical limitations, we believe that such a scenario likely occurs in triploids and can explain the observed bias favoring association between the two rearranged chromosomes.
Our model is based on the premise that the abnormal synapsis configurations observed in spo-11 mutant triploids reflect a role for recombination in promoting pairwise synapsis. Based on considerations addressed below, we favor this interpretation over an alternative hypothesis, i.e., that such synapsis configurations instead reflect persistence of nuclei that would have been culled by DNA damage checkpoint-induced apoptosis in the context of a wild-type meiotic machinery. This alternative view might be suggested by the finding that aneuploid meiocytes generated in the context a diploid germ line (by experimentally induced mitotic errors) are eliminated by triggering apoptosis in a spo-11-dependent manner (Stevens et al. 2013). However, loss of spo-11 function in the context of systemic aneuploidy, i.e., in triplo-X worms, rather than reducing the frequency of apoptosis, instead results in elevated apoptosis (Mlynarczyk-Evans et al. 2013). Further, elevated apoptosis in spo-11 triplo-X worms is correlated with persistence of multipartner associations among the three X chromosomes (Mlynarczyk-Evans et al. 2013), indicating that such configurations are detected despite elevated apoptosis. Moreover, this elevated apoptosis is suppressed in pch-2; spo-11 triplo-X worms (B.R., unpublished results), consistent with the idea that loss of spo-11 in the context of systemic aneuploidy increases the occurrence of synaptic defects that trigger the synapsis checkpoint (Bhalla and Dernburg 2005).
The observed preference for identical pairing partners detected in nT1/nT1/+ triploids is likewise not readily explained by selective culling of nuclei in which rearranged and normal sequence chromosomes are heterosynapsed, since all nuclei in such triploid germ cells have extensive heterosynapsis and/or asynapsis. Further, we did not observe an increase in the frequency of paired GFP FISH signals during progression through the pachytene stage in mIn1/mIn1/+/+ tetraploid germ lines, contrary to what would have been expected if persisting DSBs were sufficient to trigger the selective elimination of meiocytes with heterosynapsed chromosomes. Thus, we infer that apoptosis is not a major factor affecting the frequencies of pairing detected in our experiments.
A key feature of our model is that some properties of the SC are changing in response to engagement of the meiotic recombination program. We have invoked a recombination-dependent change in the state of the SC in order to explain two features: (1) the requirement for SPO-11 to eliminate multipartner associations in triploids, and (2) the observed preference for identical synapsis partners in nT1/nT1/+ triploids. The latter highlights a unique opportunity created by our approach, i.e., a competitive pairing scenario that enabled us to provide evidence for a recombination-dependent phase of synapsis in the context of a full wild-type inventory of SC and recombination machinery components. Additional support for the idea that recombination can influence the behavior of SC subunits comes from the prior observation that recombination is required for the delayed and aberrant association of SYP proteins with chromosome axes that occurs in a cra-1 mutant (which lacks the regulatory subunit of an enzyme responsible for N-terminal protein acetylation) (Smolikov et al. 2008). Further, the variation in the ratio of axis to SC central region components that we observe in a subset of late pachytene nuclei in the spo-11 mutant background suggests that even in the context of pairwise associations, a mature wild-type SC may be structurally distinct from the SCs present at later stages in recombination-deficient mutants, which we propose are blocked in the earlier synapsis phase. Interestingly, several factors have been shown previously to associate transiently with the SCs during pachytene progression with a dynamics dependent on meiotic recombination. These include the predicted SUMO-ligase ZHP-3, which first associates along the lengths of SCs and then redistributes to concentrate on smaller domains defined by the crossover sites (Jantsch et al. 2004; Bhalla et al. 2008) and the AAA+-ATPase PCH-2, which associates with the SCs during early-mid pachytene but is lost from the chromosomes at late pachytene in a crossover-dependent manner (Deshong et al. 2014). The dynamic association of these factors with the SC may represent another reflection of the transition from one synapsis phase to another that we propose in our model.
Implications of a two-state view of the SC
Recent work has demonstrated that the C. elegans SC central region proteins are important not only for promoting the formation of meiotic COs, but also for limiting their numbers by contributing to CO interference (Hayashi et al. 2010; Libuda et al. 2013). Inherent in the concept of CO interference is the ability of a nascent CO to change the environment in which it occurs in a manner that inhibits other recombination events in its vicinity from maturing into COs. Accordingly, the demonstrated involvement of the SC proteins in both promoting and antagonizing CO formation strongly suggested that progression of CO recombination must trigger a change in state of the SC. The current work supports this scenario by providing evidence that the ability of synapsed chromosomes to engage in recombination does indeed affect the properties and behavior of SCs during C. elegans meiosis.
The two-state view of the SC proposed here may also be relevant to the interrelationships between recombination and meiotic prophase progression. Several recent studies have converged on the conclusion that formation and repair of DSBs during meiosis are governed by a checkpoint-like negative feedback network in which germ cells monitor the formation of recombination intermediates that have the capacity to become interhomolog crossovers (“CO-eligible intermediates”) and couple detection of such intermediates to progression through meiotic prophase (Rosu et al. 2011, 2013; Stamper et al. 2013; Woglar et al. 2013). If sufficient intermediates have formed to guarantee a CO for each homolog pair, the cell will be permitted to undergo a major transition affecting multiple distinct aspects of the meiotic program, including cessation of competence for DSB formation and shutting down of access to the homolog as a template for DSB repair; if the condition is not met, this transition will be delayed. This model explains a substantial body of data and provides a strong conceptual framework for thinking about how germ cells ensure CO formation while minimizing the danger to genome integrity posed by DSBs. However, it has been challenging to envision how germ cells might be able to detect chromosomes that lack CO-eligible intermediates. The two-state SC model proposed here suggests a possible solution: germ cells may in fact be monitoring the status of the SCs rather than recombination intermediates per se. By provoking a change in SC state, a nascent recombination intermediate could extinguish a “wait progression” signal emanating from an early-state SC, thereby enabling progression. Thus, a transition between two SC states can be envisaged as a way for a homolog pair to signal that it has successfully acquired a CO-eligible intermediate and is ready to proceed to the later stages. In this context, the SC would not only function in promoting and limiting CO formation, but would also coordinate the proper execution of these events with meiotic progression to ensure reproductive success.
Sexual dimorphism of reproductive cell divisions as a driver of cohesin evolution?
The jumping off point for this work was our discovery that the very different cell division programs associated with female and male gametogenesis can yield very different outcomes in terms of chromosome inheritance when meiotic cohesin function is compromised. We found that the exact same impairment of meiotic cohesion function that results in retention of an extra set of chromosomes during female meiosis frequently leads to loss of some or all chromosomes during spermatogenesis. Thus, mutations that disrupt meiotic cohesion function can favor inheritance of maternal chromosomes on the one hand while at the same time disfavoring inheritance of paternal chromosomes. This observation suggests that sexual dimorphism in the reproductive cell division programs represents an inherent source of conflict, distinct from meiotic drive, that may contribute to evolution of the meiotic machinery, particularly for meiosis-specific cohesin components and their regulators
Supplementary Material
Acknowledgments
We thank the Caenorhabditis Genetics Center (funded by National Institutes of Health (NIH) Office of Research Infrastructure Programs P40 OD010440) for strains and A. Dernburg, K. Oegema, and K. O’Connell for antibodies. We thank D. Pattabiraman, D. Libuda, S. Ramakrishnan, C. Girard, and A. Woglar for discussions and comments on the manuscript; K. Nabeshima for technical advice for FISH experiments; and B. X. Fu for help with bright-field imaging of whole worms. This work was supported by a Canadian Institutes of Health Research postdoctoral fellowship to M.S., by NIH grant 1S10OD01227601 to the Stanford Cell Sciences Imaging Facility, by the Stanford Discovery Science Innovation Fund, and by NIH grants R01GM53804 and R01GM67268 to A.M.V.
Footnotes
Communicating editor: M. Colaiácovo
Supporting information is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.182279/-/DC1.
Literature Cited
- Audhya A., Hyndman F., McLeod I. X., Maddox A. S., Yates J. R., 3rd, et al. , 2005. A complex containing the Sm protein CAR-1 and the RNA helicase CGH-1 is required for embryonic cytokinesis in Caenorhabditis elegans. J. Cell Biol. 171: 267–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudat F., Manova K., Yuen J. P., Jasin M., Keeney S., 2000. Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Mol. Cell 6: 989–998. [DOI] [PubMed] [Google Scholar]
- Bessler J. B., Andersen E. C., Villeneuve A. M., 2010. Differential localization and independent acquisition of the H3K9me2 and H3K9me3 chromatin modifications in the Caenorhabditis elegans adult germ line. PLoS Genet. 6: e1000830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalla N., Dernburg A. F., 2005. A conserved checkpoint monitors meiotic chromosome synapsis in Caenorhabditis elegans. Science 310: 1683–1686. [DOI] [PubMed] [Google Scholar]
- Bhalla N., Wynne D. J., Jantsch V., Dernburg A. F., 2008. ZHP-3 acts at crossovers to couple meiotic recombination with synaptonemal complex disassembly and bivalent formation in C. elegans. PLoS Genet. 4: e1000235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilgir C., Dombecki C. R., Chen P. F., Villeneuve A. M., Nabeshima K., 2013. Assembly of the synaptonemal complex is a highly temperature-sensitive process that is supported by PGL-1 during Caenorhabditis elegans meiosis. G3 (Bethesda) 3: 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning H., Strome S., 1996. A sperm-supplied factor required for embryogenesis in C. elegans. Development 122: 391–404. [DOI] [PubMed] [Google Scholar]
- Chan R. C., Chan A., Jeon M., Wu T. F., Pasqualone D., et al. , 2003. Chromosome cohesion is regulated by a clock gene paralogue TIM-1. Nature 423: 1002–1009. [DOI] [PubMed] [Google Scholar]
- Delattre M., Leidel S., Wani K., Baumer K., Bamat J., et al. , 2004. Centriolar SAS-5 is required for centrosome duplication in C. elegans. Nat. Cell Biol. 6: 656–664. [DOI] [PubMed] [Google Scholar]
- Dernburg A. F., McDonald K., Moulder G., Barstead R., Dresser M., et al. , 1998. Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell 94: 387–398. [DOI] [PubMed] [Google Scholar]
- Deshong A. J., Ye A. L., Lamelza P., Bhalla N., 2014. A quality control mechanism coordinates meiotic prophase events to promote crossover assurance. PLoS Genet. 10: e1004291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelmann W., Cohen P. E., Kneitz B., Winand N., Lia M., et al. , 1999. Mammalian MutS homologue 5 is required for chromosome pairing in meiosis. Nat. Genet. 21: 123–127. [DOI] [PubMed] [Google Scholar]
- Giroux C. N., Dresser M. E., Tiano H. F., 1989. Genetic control of chromosome synapsis in yeast meiosis. Genome 31: 88–94. [DOI] [PubMed] [Google Scholar]
- Gonczy P., Schnabel H., Kaletta T., Amores A. D., Hyman T., et al. , 1999. Dissection of cell division processes in the one cell stage Caenorhabditis elegans embryo by mutational analysis. J. Cell Biol. 144: 927–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodyer W., Kaitna S., Couteau F., Ward J. D., Boulton S. J., et al. , 2008. HTP-3 links DSB formation with homolog pairing and crossing over during C. elegans meiosis. Dev. Cell 14: 263–274. [DOI] [PubMed] [Google Scholar]
- Grelon M., Vezon D., Gendrot G., Pelletier G., 2001. AtSPO11–1 is necessary for efficient meiotic recombination in plants. EMBO J. 20: 589–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M., Mlynarczyk-Evans S., Villeneuve A. M., 2010. The synaptonemal complex shapes the crossover landscape through cooperative assembly, crossover promotion and crossover inhibition during Caenorhabditis elegans meiosis. Genetics 186: 45–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedgecock E. M., White J. G., 1985. Polyploid tissues in the nematode Caenorhabditis elegans. Dev. Biol. 107: 128–133. [DOI] [PubMed] [Google Scholar]
- Jantsch V., Pasierbek P., Mueller M. M., Schweizer D., Jantsch M., et al. , 2004. Targeted gene knockout reveals a role in meiotic recombination for ZHP-3, a Zip3-related protein in Caenorhabditis elegans. Mol. Cell. Biol. 24: 7998–8006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath R. S., Ahringer J., 2003. Genome-wide RNAi screening in Caenorhabditis elegans. Methods 30: 313–321. [DOI] [PubMed] [Google Scholar]
- Kelly K. O., Dernburg A. F., Stanfield G. M., Villeneuve A. M., 2000. Caenorhabditis elegans msh-5 is required for both normal and radiation-induced meiotic crossing over but not for completion of meiosis. Genetics 156: 617–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp C. A., Kopish K. R., Zipperlen P., Ahringer J., O’Connell K. F., 2004. Centrosome maturation and duplication in C. elegans require the coiled-coil protein SPD-2. Dev. Cell 6: 511–523. [DOI] [PubMed] [Google Scholar]
- Kim S., Spike C., Greenstein D., 2013. Control of oocyte growth and meiotic maturation in Caenorhabditis elegans. Adv. Exp. Med. Biol. 757: 277–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy D. L., Heald R., 2012. Mechanisms of intracellular scaling. Annu. Rev. Cell Dev. Biol. 28: 113–135. [DOI] [PubMed] [Google Scholar]
- Libuda D. E., Uzawa S., Meyer B. J., Villeneuve A. M., 2013. Meiotic chromosome structures constrain and respond to designation of crossover sites. Nature 502: 703–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano E., Saez A. G., Flemming A. J., Cunha A., Leroi A. M., 2006. Regulation of growth by ploidy in Caenorhabditis elegans. Curr. Biol. 16: 493–498. [DOI] [PubMed] [Google Scholar]
- MacQueen A. J., Colaiacovo M. P., McDonald K., Villeneuve A. M., 2002. Synapsis-dependent and -independent mechanisms stabilize homolog pairing during meiotic prophase in C. elegans. Genes Dev. 16: 2428–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacQueen A. J., Phillips C. M., Bhalla N., Weiser P., Villeneuve A. M., et al. , 2005. Chromosome sites play dual roles to establish homologous synapsis during meiosis in C. elegans. Cell 123: 1037–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madl J. E., Herman R. K., 1979. Polyploids and sex determination in Caenorhabditis elegans. Genetics 93: 393–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Perez E., Villeneuve A. M., 2005. HTP-1-dependent constraints coordinate homolog pairing and synapsis and promote chiasma formation during C. elegans meiosis. Genes Dev. 19: 2727–2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Perez E., Schvarzstein M., Barroso C., Lightfoot J., Dernburg A. F., et al. , 2008. Crossovers trigger a remodeling of meiotic chromosome axis composition that is linked to two-step loss of sister chromatid cohesion. Genes Dev. 22: 2886–2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKim K. S., Howell A. M., Rose A. M., 1988. The effects of translocations on recombination frequency in Caenorhabditis elegans. Genetics 120: 987–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKim K. S., Peters K., Rose A. M., 1993. Two types of sites required for meiotic chromosome pairing in Caenorhabditis elegans. Genetics 134: 749–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKim K. S., Green-Marroquin B. L., Sekelsky J. J., Chin G., Steinberg C., et al. , 1998. Meiotic synapsis in the absence of recombination. Science 279: 876–878. [DOI] [PubMed] [Google Scholar]
- McNally K., Audhya A., Oegema K., McNally F. J., 2006. Katanin controls mitotic and meiotic spindle length. J. Cell Biol. 175: 881–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlynarczyk-Evans S., Roelens B., Villeneuve A. M., 2013. Evidence that masking of synapsis imperfections counterbalances quality control to promote efficient meiosis. PLoS Genet. 9: e1003963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabeshima K., Villeneuve A. M., Hillers K. J., 2004. Chromosome-wide regulation of meiotic crossover formation in Caenorhabditis elegans requires properly assembled chromosome axes. Genetics 168: 1275–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabeshima K., Mlynarczyk-Evans S., Villeneuve A. M., 2011. Chromosome painting reveals asynaptic full alignment of homologs and HIM-8-dependent remodeling of X chromosome territories during Caenorhabditis elegans meiosis. PLoS Genet. 7: e1002231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson G. A., Ward S., 1980. Vesicle fusion, pseudopod extension and amoeboid motility are induced in nematode spermatids by the ionophore monensin. Cell 19: 457–464. [DOI] [PubMed] [Google Scholar]
- Nuez I., Felix M. A., 2012. Evolution of susceptibility to ingested double-stranded RNAs in Caenorhabditis nematodes. PLoS One 7: e29811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oegema K., Desai A., Rybina S., Kirkham M., Hyman A. A., 2001. Functional analysis of kinetochore assembly in Caenorhabditis elegans. J. Cell Biol. 153: 1209–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasierbek P., Jantsch M., Melcher M., Schleiffer A., Schweizer D., et al. , 2001. A Caenorhabditis elegans cohesion protein with functions in meiotic chromosome pairing and disjunction. Genes Dev. 15: 1349–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penkner A., Tang L., Novatchkova M., Ladurner M., Fridkin A., et al. , 2007. The nuclear envelope protein Matefin/SUN-1 is required for homologous pairing in C. elegans meiosis. Dev. Cell 12: 873–885. [DOI] [PubMed] [Google Scholar]
- Penkner A. M., Fridkin A., Gloggnitzer J., Baudrimont A., Machacek T., et al. , 2009. Meiotic chromosome homology search involves modifications of the nuclear envelope protein Matefin/SUN-1. Cell 139: 920–933. [DOI] [PubMed] [Google Scholar]
- Peters N., Perez D. E., Song M. H., Liu Y., Muller-Reichert T., et al. , 2010. Control of mitotic and meiotic centriole duplication by the Plk4-related kinase ZYG-1. J. Cell Sci. 123: 795–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips C. M., Wong C., Bhalla N., Carlton P. M., Weiser P., et al. , 2005. HIM-8 binds to the X chromosome pairing center and mediates chromosome-specific meiotic synapsis. Cell 123: 1051–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praitis V., Casey E., Collar D., Austin J., 2001. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics 157: 1217–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preibisch S., Saalfeld S., Tomancak P., 2009. Globally optimal stitching of tiled 3D microscopic image acquisitions. Bioinformatics 25: 1463–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rog O., Dernburg A. F., 2015. Direct visualization reveals kinetics of meiotic chromosome synapsis. Cell Rep. 10: 1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanienko P. J., Camerini-Otero R. D., 2000. The mouse Spo11 gene is required for meiotic chromosome synapsis. Mol. Cell 6: 975–987. [DOI] [PubMed] [Google Scholar]
- Rose A. M., Baillie D. L., Curran J., 1984. Meiotic pairing behavior of two free duplications of linkage group I in Caenorhabditis elegans. Mol. Gen. Genet. 195: 52–56. [DOI] [PubMed] [Google Scholar]
- Rosenbluth R. E., Baillie D. L., 1981. The genetic analysis of a reciprocal translocation, eT1(III; V), in Caenorhabditis elegans. Genetics 99: 415–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosu S., Libuda D. E., Villeneuve A. M., 2011. Robust crossover assurance and regulated interhomolog access maintain meiotic crossover number. Science 334: 1286–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosu S., Zawadzki K. A., Stamper E. L., Libuda D. E., Reese A. L., et al. , 2013. The C. elegans DSB-2 protein reveals a regulatory network that controls competence for meiotic DSB formation and promotes crossover assurance. PLoS Genet. 9: e1003674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato A., Isaac B., Phillips C. M., Rillo R., Carlton P. M., et al. , 2009. Cytoskeletal forces span the nuclear envelope to coordinate meiotic chromosome pairing and synapsis. Cell 139: 907–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., et al. , 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9: 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schvarzstein M., Wignall S. M., Villeneuve A. M., 2010. Coordinating cohesion, co-orientation, and congression during meiosis: lessons from holocentric chromosomes. Genes Dev. 24: 219–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schvarzstein M., Pattabiraman D., Bembenek J. N., Villeneuve A. M., 2013. Meiotic HORMA domain proteins prevent untimely centriole disengagement during Caenorhabditis elegans spermatocyte meiosis. Proc. Natl. Acad. Sci. USA 110: E898–E907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severson A. F., Ling L., van Zuylen V., Meyer B. J., 2009. The axial element protein HTP-3 promotes cohesin loading and meiotic axis assembly in C. elegans to implement the meiotic program of chromosome segregation. Genes Dev. 23: 1763–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakes D. C., Wu J. C., Sadler P. L., Laprade K., Moore L. L., et al. , 2009. Spermatogenesis-specific features of the meiotic program in Caenorhabditis elegans. PLoS Genet. 5: e1000611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolikov S., Schild-Prufert K., Colaiacovo M. P., 2008. CRA-1 uncovers a double-strand break-dependent pathway promoting the assembly of central region proteins on chromosome axes during C. elegans meiosis. PLoS Genet. 4: e1000088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamper E. L., Rodenbusch S. E., Rosu S., Ahringer J., Villeneuve A. M., et al. , 2013. Identification of DSB-1, a protein required for initiation of meiotic recombination in Caenorhabditis elegans, illuminates a crossover assurance checkpoint. PLoS Genet. 9: e1003679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens D., Oegema K., Desai A., 2013. Meiotic double-strand breaks uncover and protect against mitotic errors in the C. elegans germline. Curr. Biol. 23: 2400–2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons L., Fire A., 1998. Specific interference by ingested dsRNA. Nature 395: 854. [DOI] [PubMed] [Google Scholar]
- Villeneuve A. M., 1994. A cis-acting locus that promotes crossing over between X chromosomes in Caenorhabditis elegans. Genetics 136: 887–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woglar A., Daryabeigi A., Adamo A., Habacher C., Machacek T., et al. , 2013. Matefin/SUN-1 phosphorylation is part of a surveillance mechanism to coordinate chromosome synapsis and recombination with meiotic progression and chromosome movement. PLoS Genet. 9: e1003335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff G. C., Eke O., Baird S. E., Felix M. A., Haag E. S., 2010. Insights into species divergence and the evolution of hermaphroditism from fertile interspecies hybrids of Caenorhabditis nematodes. Genetics 186: 997–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoo R., Zawadzki K. A., Nabeshima K., Drake M., Arur S., et al. , 2012. COSA-1 reveals robust homeostasis and separable licensing and reinforcement steps governing meiotic crossovers. Cell 149: 75–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Miley N., Zastrow M. S., MacQueen A. J., Sato A., et al. , 2012. HAL-2 promotes homologous pairing during Caenorhabditis elegans meiosis by antagonizing inhibitory effects of synaptonemal complex precursors. PLoS Genet. 8: e1002880. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.