Abstract
MicroRNAs (miRNAs) have emerged as key regulators in many pathological processes by suppressing the transcriptional and post-transcriptional expression of target genes. MiR-2008 was previously found to be significantly up-regulated in diseased sea cucumber Apostichopus japonicus by high-through sequencing, whereas the reads of miR-137, a well-documented tumor repressor, displayed no significant change. In the present study, we found that miR-137 expression was slightly attenuated and miR-2008 was significantly enhanced after Vibrio splendidus infection or Lipopolysaccharides application. Further target screening and dual-luciferase reporter assay revealed that the two important miRNAs shared a common target gene of betaine–homocysteine S-methyltransferase (AjBHMT), which exhibited noncorrelated messenger RNA and protein expression patterns after bacterial challenge. In order to fully understand their regulatory mechanisms, we conducted the functional experiments in vitro and in vivo. The overexpression of miR-137 in sea cucumber or primary coelomocytes significantly decreased, whereas the inhibition of miR-137 increased the mRNA and protein expression levels of AjBHMT. In contrast, miR-2008 overexpression and inhibition showed no effect on AjBHMT mRNA levels, but the concentration of AjBHMT protein displayed significant changes both in vitro and in vivo. Consistently, the homocysteine (Hcy) contents were also accordingly altered in the aberrant expression analysis of both miRNAs, consistent with the results of the AjBHMT silencing assay in vitro and in vivo. More importantly, small interfering RNA mediated AjBHMT knockdown and Hcy exposure analyses both significantly increased reactive oxygen species (ROS) production and decreased the number of surviving invasive pathogen in sea cucumber coelomocytes. Taken together, these findings confirmed the differential roles of sea cucumber miR-137 and miR-2008 in regulating the common target AjBHMT to promote ROS production and the clearance of pathogenic microorganisms through Hcy accumulation.
Keywords: Apostichopus japonicus; miR-137; miR-2008; betaine–homocysteine S-methyltransferase; homocysteine; ROS, genetics of immunity
MICRORNAS (miRNAs) are a class of endogenous small noncoding RNAs of ∼22 nucleotides, which typically function as repressors of target genes, resulting in direct messenger RNA (mRNA) degradation or translation inhibition (Bartel 2004). These RNAs play crucial roles in many cellular processes, such as proliferation, differentiation, apoptosis, and survival (Lai 2002; He and Hannon 2004; Miska 2005). The expression of up to 60% of all protein-coding genes is estimated to be under the control of miRNAs (Friedman 2009), and combinatorial regulation has been identified as a prominent feature of miRNA regulation. Thus, a given miRNA might have multiple different mRNA targets, and a given mRNA might similarly be targeted by multiple miRNAs (Krek et al. 2005; Rajewsky 2006). Consistent with the crucial roles of miRNA in the normal functioning of eukaryotic cells, the aberrant expression of miRNAs has also been observed in various diseases (Calin and Croce 2006; Wu et al. 2007). Notably, miRNAs have also emerged as pivotal regulators of the host immune response and key players in host–pathogen interactions (Scaria et al. 2006; Asgari 2011).
Sea cucumber (Apostichopus japonicus) belongs to the phylum Echinodermata, class Holothuroidea. As a representative economic species of Echinoderms, the aquaculture of A. japonicus has become an important sector of the marine industry and has grown rapidly over recent years. The rapid expansion and intensification of farming have also led to a series of problems, including the occurrence of skin ulceration syndrome (SUS), considered as one of the limiting factors in the sustainable development of this industry (Deng et al. 2009). Many reports have demonstrated that the pathogens responsible for the outbreak of SUS include spherical virus (Wang et al. 2005), Vibrio splendidus, and Pseudomonas spp. (Ma et al. 2006; Zhang et al. 2006). Although extensive research concerning the identification of SUS-related pathogens has been conducted, the understanding of the etiology and pathogenesis of this disease remain unclear. In a previous study, we showed abnormal expression level of miR-2008 in diseased A. japonicas by high-throughput sequencing, whereas the reads of miR-137, a well-documented tumor repressor, displayed no change (Li et al. 2012). In human, miR-137 has been extensively investigated and shown to play important roles in various cancers, cell cycle signaling, and embryonic stem cell development. Studies have demonstrated that miR-137 targets cell division control protein 42 (Cdc42), a Rho GTPase family protein up-regulated in many human cancer types, such as colorectal, lung, and breast cancers (Arias-Romero and Chernoff 2013). Through the inhibition of the Cdc42/PAK signaling pathway, miR-137 decreases the proliferation, invasion, and G0/G1 cell cycle progression of colorectal cancer cells (Liu et al. 2011; Zhu et al. 2013). In addition, miR-137 also targets mind bomb-1 (Mib1), a ubiquitin ligase involved in regulating neurogenesis and neurodevelopment (Smrt et al. 2010). Another notable target of miR-137 is micropthalmia-associated transcription factor (MITF), a master regulator of melanocyte development and function, frequently misregulated in human melanomas (Bemis et al. 2008). Recent studies also show that miR-137 is involved in the doxorubicin resistance of neuroblastoma cells by targeting the constitutive androstane receptor (CAR) (Takwi et al. 2013; Chen et al. 2014). Moreover, the epigenetic silencing of miR-137 has been associated with the hypermethylation of the miR-137 promoter (Balaguer et al. 2010; Vrba et al. 2013); however, compared with miR-137, the function of miR-2008 has been rarely studied.
Although the function of several miRNAs in vertebrates and model organisms has been elucidated, scarce miRNA targets have been identified and characterized in nonmodel marine organisms, such as sea cucumber. In the present study, V. splendidus, a well-documented pathogen in sea cucumber SUS, was used to explore the potential roles of miR-137 and miR-2008 in host–pathogen reactions in vitro and in vivo. The decreased expression profiles of miR-137 were detected both in V. splendidus-challenged sea cucumber or LPS-exposed coelomocytes, which slightly differed from the results from diseased samples. More importantly, we observed that both miR-137 and miR-2008 targeted the 3′-UTR of betaine homocysteine S-methyltransferase (AjBHMT) and modulated the transcription and translation of this gene, respectively, Moreover, AjBHMT affects Hcy accumulation in coelomocytes and induces respiratory bursts to kill invasive microorganisms in nonmodel invertebrate pathogen infections.
Materials and Methods
Ethics statement
Sea cucumbers (A. japonicus) and BALB/c mice are commercially cultured animals. All experiments were conducted in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The study protocol was approved by the Experimental Animal Ethics Committee of Ningbo University, China.
Target prediction of miRNAs through RNA-seq
RNA-seq and miRNA target prediction were based on the results of a previous study (Zhang et al. 2013). In brief, the potential targets for miR-137 and miR-2008 were predicted using the miRanda v3.01 toolbox based on the complementary region between miRNAs and mRNAs, as well as the thermodynamic stability of the miRNA–mRNA duplex. All of the mRNAs used for target prediction came from the differentially expressed unigenes obtained above, in which the candidate targets with scores less than the threshold parameter of S > 90 (single-residue pair scores) and a minimum free energy lower than −17 kcal/mol were selected as potential targets.
Experimental animals, bacterial strains, and cell culture
A total of 120 healthy adult sea cucumbers (A. japonicus) of ∼60 ± 10 g were obtained from the Bowang Aquaculture Company (Ningbo, China), randomly assigned to six tanks, and acclimatized in aerated natural seawater [28 practical salinity units at 16° ± 1°] for 3 days before conducting experiments. V. splendidus was initially isolated from the skin ulcerations of diseased sea cucumbers and cultured in liquid 2216E broth (5 g⋅liter−1 tryptone, 1 g⋅liter−1 yeast extract, pH 7.6) at 28°, with shaking at 140 rpm. HEK-293T cells were purchased from ATCC and maintained in DMEM supplemented with 10% FCS at 37° and 5% CO2.
Full-length cDNA clone and analysis of AjBHMT from A. japonicus
Total RNA was extracted using RNAiso plus reagent (Takara), and reverse transcription was performed with 1 μg of RNA using the Primescript First cDNA Synthesis Kit II (Takara). Gene-specific primers (Supporting Information, Table S1) were designed based on the sequences obtained from a previous study (Zhang et al. 2013), and the PCR products were cloned into pMD18-T simple vector (Takara) and bidirectionally sequenced using M13-47 and RV-M primers (Sangon). Subsequently, the cDNA sequence of AjBHMT was analyzed using the BLAST algorithm at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/blast), and the deduced amino acid sequence was analyzed using the Expert Protein Analysis System (http://www.expasy.org/). The percentage of similarity and identity of the full-length amino acid sequences between each gene and its counterparts in other organisms was determined using the Identity and Similarity Analysis program (http://www.biosoft.net/sms/index.html). Multiple alignments of each protein were performed using the ClustalW Multiple Alignment program (http://www.ebi.ac.uk/clustalw2/) and the Multiple Align Show program (http://www.biosoft.net/sms/index.html). Domains in the AjBHMT amino acid sequence were detected using the simple modular architecture research tool (SMART) program (http://www.smart.emblheidelbergde/).
3′-UTR luciferase reporter assay
The 3′-UTR of AjBHMT mRNA, containing the putative target site of miR-137 and miR-2008, was cloned into the pMIR-REPORT luciferase plasmid (Promega) between the MluI and HindIII restriction sites and subsequently subjected to DNA sequencing for confirmation. The primers used to clone the AjBHMT 3′-UTR are shown in Table S1. The complete 3′-UTR of wild-type AjBHMT was amplified by gene-specific primers with restricted endoenzyme sites of MluI and HindIII. Putative miRNA binding sites were mutated using a PCR approach and served as the mutant type. All purified fragments were digested by double enzymes and ligated into the digested pMIR-REPORT vectors, respectively. These clones were further confirmed by sequencing prior to the luciferase reporter assay. For the transfection experiment, HEK293-T cells were seeded into a 96-well white TC plate with a total volume of 100 μl. Two solutions were prepared in each well as follows: the first solution contained 0.2 μg of pMIR-REPORT containing either the wild-type or mutated AjBHMT 3′-UTR and 0.01 μg of pRL-CMV constructs with 0.25 μl of transfection reagent. The second solution comprised 100 nM miR-137 or miR-2008 mimics and 0.25 μl of transfection reagent. Twenty-five microliters of each solution was mixed together and incubated at room temperature for 20 min. Subsequently the solutions were replaced by 50 μl of medium in each well. At 48-hr post-transfection, the cells were collected for activity determination using the Dual-Luciferase Reporter Assay System (E1910, Promega). The efficiency of each miRNA transfection was confirmed by fluorescence microscope BX53 (Olympus). Luciferase activity was calculated based on the luciferase signal ratio between the two constructs, pMIR-REPORT and pRL-CMV, which could be used to normalize the protein content among different samples. All of the experiments were performed in six replicates.
To explore the effect of combined miR-137 and miR-2008 on AjBHMT, HEK-293T cells were cotransfected with the pMIR-REPORT-BHMT-WT vector, pRL-CMV-Renilla-luciferase and control or serial concentration of miR-137, miR-2008 mimics (50 nM, 100 nM, and 150 nM) or their combinations (50 nM miR-137 + 50 nM miR-2008, 50 nM miR-137 + 100 nM miR-2008, and 100 nM miR-137 + 50 nM miR-2008). The experiments were conducted as mentioned above.
Pathogen microorganism challenge in sea cucumber
An overnight culture of V. splendidus was centrifuged at 5,000 × g for 5 min to harvest the bacteria, and live V. splendidus was subsequently resuspended in filtered seawater (FSW). For the challenge experiments, three tanks were used as controls, and the other three tanks were immersed with a high density of V. splendidus at a final concentration of 107 cfu⋅ml−1. Coelomic fluids were collected from the individuals in each tank and centrifuged at 800 × g for 5 min to harvest the hemocytes at 0, 48, and 96 hr, and the cells were subsequently stored at −80° before RNA extraction.
Expression analysis of mir-137, mir-2008, and AjBHMT in vivo
Total RNA was extracted using RNAiso plus reagent (Takara) according to the manufacturer’s instructions. The SYBR green qRT-PCR assay was used for miRNA and mRNA quantification in the same samples. Briefly, 500 ng of RNA-containing miRNAs was polyadenylated using poly(A) polymerase and converted to cDNA through reverse transcription using the miScript Reverse Transcription Kit (Qiagen). For miRNA analysis, qRT-PCR was performed using the miScript SYBR Green PCR Kit (Qiagen) and the provided miScript Universal primer and miRNA-specific forward primers (for mRNA, the primers were gene-specific forward and reverse primers) on a Rotor-Gene Q 6000 Real-Time PCR cycler (Qiagen).
The miRNA-specific primers were designed based on previously published miRNA sequences (Li et al. 2012), and the gene-specific primers were designed based on previously published mRNA sequences (Zhang et al. 2013) (Table S1). Each reaction was performed in a final volume of 20 μl containing 2 μl of the cDNA, 1 μl of each 10-μM primer, 6 μl of RNase-free water, and 10 μl of SYBR Green PCR Master mix (Qiagen). The amplification profile included initial denaturation at 94° for 15 min, followed by 40 cycles of 94° for 15 sec, 60° for 30 sec, and 70° for 30 sec, in which fluorescence was acquired. Immediately following amplification, a melting curve analysis was performed. Each sample was run in triplicate with three biological replications. The miRNA expression levels were normalized to RNU6B, and the expression levels of mRNAs were normalized to 18S ribosomal RNA.
The 2-∆∆Ct method was used to analyze the expression levels of both miRNA and mRNA, and the value obtained represented the n-fold difference relative to the control (untreated samples). The data were presented as the relative expression levels (means ± SD, n = 9), and all experimental data were subjected to one-way analysis of variance (ANOVA), followed by Duncan’s multiple range test to determine differences in the mean values among the controls. Significant differences between the treated and corresponding control groups at each time point were indicated as *P < 0.05 and **P < 0.01. The error bars in the graphs represent standard deviations.
Primary coelomocyte culture
Sea cucumbers were sterilized in 7% benzalkonium bromide and 75% ethanol for ∼2 min, respectively. Subsequently, the sea cucumbers were dissected using an aseptic surgery technique as previously described (Xing et al. 1998). The coelomic fluids were collected and mixed with an equal volume of anticoagulant solution (0.02 M EGTA, 0.48 M NaCl, 0.019 M KCl, and 0.068 M Tri-HCl, pH 7.6). The cell suspension was filtered through a 100-µm nylon mesh to remove large tissue debris and centrifuged at 800 × g for 10 min at 18°. The cells were washed twice with isotonic buffer (0.001 M EGTA, 0.53 M NaCl, and 0.01 M Tris-HCl, pH 7.6) and resuspended in the Leibovitz’s L-15 cell culture medium (Invitrogen) supplemented with penicillin (100 units⋅ml−1) and streptomycin sulfate (100 µg⋅ml−1), and NaCl (0.39 M) was added to adjust the osmotic pressure. The cell suspension was adjusted to 6 × 105 cells⋅ml−1. Aliquots of 500 µl of cell suspension were dispensed into the wells of 24-well culture microplates. The coelomocytes were incubated for 24 hr at 18° in the darkness prior to use.
Expression analysis of miR-137, miR-2008, and AjBHMT in LPS-exposed primary coelomocytes
For LPS (Sigma) challenge, the primary cultured coelomocytes were stimulated with different concentrations of LPS (0, 0.1, and 1 μg⋅ml−1) for the indicated times. After 3 and 6 hr, the cells were washed once with cold PBS, centrifuged at 800 × g for 5 min at 4°, and then stored at −80° for RNA extraction. The expression levels of miR-137, miR-2008, and AjBHMT were analyzed as described above. Each sample was run in triplicate with three biological replications.
Functional analysis of miRNA in vitro and in vivo
The miRNAs mimics and inhibitors were synthesized at GenePharma and are shown in Table S1. For miR-137 mimics and inhibitor transfections, HiPerFect transfection reagents (Qiagen) were used for the transfection experiment. The miR-137 mimics or inhibitor (2.5 μl of 50 nM), as well as each negative control, were mixed with an equal volume of HiPerFect transfection reagents. The mixture was then transfected into 1000 μl of primary cultured cells. For miR-2008, 100 nM of the mimics or inhibitor were utilized for transfection analysis. At 24 hr post-transfection, the cells were harvested and washed with cold PBS, and centrifuged at 800 × g and 4° for 5 min in preparation for the subsequent expression analysis. The miR-137 and miR-2008 modified mimics specific for in vivo as well as the negative control were designed for the in vivo assay and synthesized by GenePharma (Shanghai). The sequence information is shown in Table S1. These modified mimics were then dissolved into RNase-free water to obtain a working solution of 20 μM. We mixed 10 μl of each modified mimics with 10 μl of transfection reagent and 80 μl of PBS to serve as the transfection solution. Twenty sea cucumbers (60 ± 10 g) were infected with 100 μl of each modified mimic or the negative control. After 24 hr, the treated and control coelomocytes were collected for expression analysis. The assays described above were biologically repeated three times and run in triplicate.
Silencing of AjBHMT using small interfering RNA in vitro and in vivo
Small interfering RNA (siRNA) targeting AjBHMT and a negative control were designed and synthesized by GenePharma. The detailed sequence information is shown in Table S1. For in vitro assay, specific siRNA oligonucleotides (1.0 μl of 20 nM) or the negative control were mixed with 1.0 μl of siRNA-specific transfection reagent (GenePharma). The mixture was added to 1000 μl of primary cultured cells for 24 hr. The treated and control coelomocytes were collected for expression analysis. For in vivo AjBHMT knockdown, 5 μl of AjBHMT-specific siRNA (20 nM) and negative control were mixed with 5 μl of transfection reagent and 90 μl of PBS to serve as the transfection solution. Twenty sea cucumbers (60 ± 10 g) were injected with 100 μl of each AjBHMT siRNA or the negative control. At 24 hr post-transfection, the coelomocytes were collected and stored at −80° until further RNA and protein extraction. AjBHMT expression was analyzed as described above.
AjBHMT recombinant protein expression and polyclonal antibody preparation
PCR fragments encoding AjBHMT and Ajβ-actin mature peptides were amplified using gene-specific primers (Table S1) with 5′-end BamHI and NotI, respectively. The PCR products were cloned into pMD18-T simple vector (Takara), completely digested using the restriction enzymes BamHI and NotI (NEB), and subsequently cloned into the pET-28a (+) expression vector (Novagen) between the BamHI and NotI restriction sites. The recombinant plasmids (pET-28a-Ajβ-actin and pET-28a-AjBHMT) were transformed into Escherichia coli BL21 (DE3)-plysS (Novagen) and subjected to DNA sequencing. The positive clones were subsequently incubated in LB medium containing 50 μg⋅ml−1 of kanamycin at 37° and 220 rpm. IPTG, at a final concentration of 1 mM, was added when the culture reached an OD600 of 0.6 and then incubated for an additional 3 hr under the same conditions. The bacteria were harvested through centrifugation at 10,000 × g for 2 min and resuspended in buffer containing 50 mM Tris, 5 mM EDTA, 50 mM NaCl, and 5% glycerol, pH 7.9. After sonication, the recombinant Ajβ-actin and AjBHMT proteins were purified using HiTrap chelating columns (Amersham Biosciences) according to the manufacturer’s instructions. The concentration of the purified proteins was measured using the BCA Protein Assay Kit (Sangon), followed by 15% SDS-PAGE. For polyclonal antibody preparation, 4-week-old rats were intradermally injected with a mixture containing 100 μg of purified His-tagged AjBHMT or Ajβ-actin recombinant protein and an equal volume of complete Freund’s adjuvant (Promega). After 2 weeks, the rats were intramuscularly boosted twice with 75 μg of His-tagged AjBHMT or Ajβ-actin recombinant protein mixed with an equal volume of incomplete Freund’s adjuvant at a 1-week interval. Two weeks after the last immunization, the antiserum was harvested from rat eyes and stored at −80° until further use.
Western blot analysis
For Western blot assay, 48-hr post-transfected cells were washed twice in ice-cold PBS, and the protein was extracted using a Total Protein Extraction Kit (Sangon) according to the manufacturer’s instructions. Subsequently, the protein concentration was measured using the BCA Protein Assay Kit (Sangon). For SDS-PAGE, 25 μg of protein from each sample was used, followed by electrophoretic transfer to a 0.45-mm pore nitrocellulose membrane using an ECL Semi-Dry Blotter (Amersham Biosciences). After blocking with 5% skim milk (in 20 mM Tris-HCl, 150 mM NaCl, and 0.05% Tween-20) at 37° for 1 hr, the membranes were incubated with AjBHMT or Ajβ-actin polyclonal antibodies diluted to 1:500 in 5% skim milk at room temperature for 2 hr. The membranes were washed three times with TBST (20 mM Tris-HCl, 150 mM NaCl, and 0.05% Tween-20) and then incubated with goat-anti-rat IgG (Beyotime) diluted to 1:1000 in 5% skim milk at room temperature for 1.5 hr. After washing three times with TBST for 10 min each, the membrane was incubated in Western Lightning-ECL substrate (Perkin Elmer) prior to exposure onto X-OMAT AR X-ray film (Eastman Kodak, Rochester, NY). The protein bands were quantified using the BioRad Quantity One software package, and the results were derived from the statistical analysis of three independent experiments.
Detection of Hcy concentration
For the in vitro detection of the Hcy concentration at 48 hr post-transfection, the levels of Hcy in the culture medium of cells transfected with miRNA mimics, inhibitor, or siRNA were measured using a cycling enzymatic method. Briefly, the culture medium from each treatment was collected and analyzed using Liquid Stable (LS) 2-Part Homocysteine Reagent (Axis-Shield) on an Olympus AU400 Chemistry Analyzer. Subsequently, the cells from each sample were collected for protein extraction.
For the in vivo detection of the Hcy concentration, 10 μl of miR-137 and miR-2008 modified mimics, 5 μl of AjBHMT siRNA, and the respective negative controls were mixed with transfection reagent and PBS buffer and then injected into sea cucumbers as described above. At 24 hr post-transfection, the coelomocytes were collected and stored at −80° until further use for Hcy detection.
Reactive oxygen species production in vitro
Intracellular reactive oxygen species (ROS) production after Hcy exposure or AjBHMT knockdown was measured as previously described (Wang et al. 2009), with slight modifications. Primary cultured cells were treated with 0, 0.5 or 1 mM Hcy (Sigma). Silencing of AjBHMT expression was conducted as described above. After 24 hr, the cells were incubated with 0.1% nitroblue tetrazolium (NBT) and 0.01 μM phorbol 12-myristate 13-acetate (PMA) for 1 hr. Subsequently, the supernatant was removed through centrifugation at 540 × g for 10 min, and the cells were fixed with 35% methanol. After washing twice with 70% methanol, the cells were resuspended in 0.92 M potassium hydroxide containing 54% dimethyl sulfoxide in a final volume of 1.3 ml to dissolve the reduced NBT (in the form of formazan). The optical density of the sample was measured at 625 nm.
Colony-forming unit assay
Primary cultured cells were treated with 0, 0.5, or 1 mM Hcy and 10 M AjBHMT siRNA and the respective negative controls for 24 hr prior to infection. Subsequently, V. splendidus was added to the cultured cells at a ratio of ∼100 bacteria/cell. After incubation at 18° for 3 and 6 hr, the infected cells were extensively washed with PBS to remove extracellular bacteria, followed by lysing in 1 ml of distilled water. Quantitative culturing was performed using 10-fold serial dilutions. Aliquots of each dilution were inoculated in triplicate onto 2216E agar plates. The plates were incubated overnight at 28°, and subsequently, the colonies were then counted.
ROS production in vivo
In vivo ROS production analysis was performed by flow cytometry (FCM). The FCM analysis was performed using a FACScan (Becton-Dickinson, San Jose, CA). For each sample, a total of 10,000 events were acquired, and the data in forward-scatter (FSC) and side-scatter (SSC) parameters were collected and analyzed using the Cell Quest software.
Before FCM analysis, 0, 10, and 50 mM Hcy were mixed with PBS buffer in a final volume of 100 μl and then injected into 30 sea cucumbers. About 5 μl of AjBHMT siRNA or negative control was mixed with 5 μl of transfection reagent and 90 μl of PBS and injected into 20 sea cucumbers. After treatment for 24 hr, the coelomocytes were collected and adjusted to 6 × 105 cells⋅ml−1 with L15 culture medium. For ROS analysis, ROS was detected using the Reactive Oxygen Species Assay Kit (Beyotime Biotechnology, Jiangsu, China) according to the manufacturer’s instructions. Briefly, 1000 μl of cells was centrifuged at 800 × g and 18° for 5 min and then resuspended in 10 μM DCFH-DA (diluted in L15 culture medium). After incubation at 18° for 20 min in HulaMixer Sample Mixer (Invitrogen), the cells were washed three times by L15 culture medium and then used for FCM test.
Data availability
All list plasmids and strains may be requested from Li Lab. Current versions of protocols will be maintained in our laboratory.
Results
Characterization of miR-137 and miR-2008 expression in response to pathogenic infection in vivo and in vitro
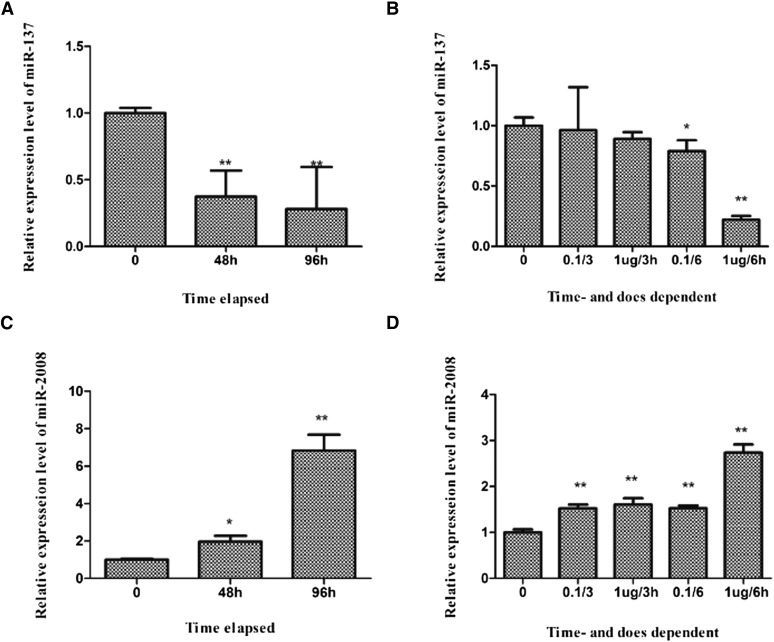
MiR-2008 has been identified as a regulatory factor in sea cucumber SUS outbreaks using deep sequencing (Li et al. 2012). MiR-137 is widely accepted as a tumor suppressor in many humuan cancer developments (Wu et al. 2007; Liu et al. 2011). To fully understand the roles of miR-137 and miR-2008 in the host–pathogen interaction process, we used qPCR to examine the expression profiles of these two miRNAs in response to pathogenic infection or LPS exposure (Figure 1). As shown in Figure 1, miR-137 expression decreased in V. splendidus-challenged sea cucumbers (Figure 1A), with 0.37- and 0.28-fold reductions observed at 48 and 96 hr, respectively. By contrast, miR-2008 expression increased by 1.96-fold and 6.82-fold at 48 hr and 96 hr, respectively (Figure 1C). Further exploration in LPS-exposed primary cultured cells revealed similar patterns of miR-137 (Figure 1B) and miR-2008 (Figure 1D) expression, with a 0.22-fold decrease or 2.74-fold increase at 6 hr, respectively, after exposure to 1 μg⋅ml−1 LPS. However, in our previous study, the expression level of miR-137 displayed no significant change between diseased and healthy juvenile individuals (3–5 g) collected from the indoor ponds of Dalian. The discrepant expression level of miR-137 might result from the different samples or the different states of samples. However, the expression level of miR-2008 exhibited similar expression tendencies between the diseased and pathogen-infected samples. Thus, we suppose that the discrepant results between infected and diseased experiment might indicate a progressively pathogenic process from infection to pathogenesis.
Figure 1.
Relative expression of miR-137 and miR-2008 in vitro and in vivo. (A) Relative miR-137 expression in V. splendidus-challenged A. japonicus. (B) Relative miR-137 expression in LPS-treated coelomocytes. (C) Relative miR-2008 expression in V. splendidus-challenged A. japonicus. (D) Relative miR-2008 expression in LPS-treated coelomocytes.
Identification of miR-137 and miR-2008 targets
To understand the functional roles of these two miRNAs, candidate targets were predicted using the miRanda software and the available transcriptome RNA-seq data (Zhang et al. 2013) (see Table S2). A total of 22 and 29 genes were predicted as putative targets of miR-137 and miR-2008, respectively. Among these putative targets, AjBHMT was identified as a common target of both miR-137 and miR-2008. More importantly, AjBHMT modulated the Hcy content, which may directly or indirectly lead to cell apoptosis, oxidative stress, or endoplasmic stress. Therefore, AjBHMT was selected as a promising target for functional validation.
Cloning the full-length cDNA of AjBHMT and its response to immune induction in vitro and in vivo
The full-length 1415-bp AjBHMT cDNA was obtained using overlapping ESTs and RACE fragments and deposited in GenBank under the accession no. KJ855191. The full-length cDNA of AjBHMT contained a 5′-UTR of 54 bp, a 3′-UTR of 281 bp, and a putative ORF of 1080 bp encoding a polypeptide of 359 amino acid residues with a predicted molecular mass of 39.72 kDa and theoretical isoelectric point (pI) of 5.49. SMART analysis indicated that AjBHMT had a conserved S-methyl_trans domain (24–323 aa) (see Figure S1).
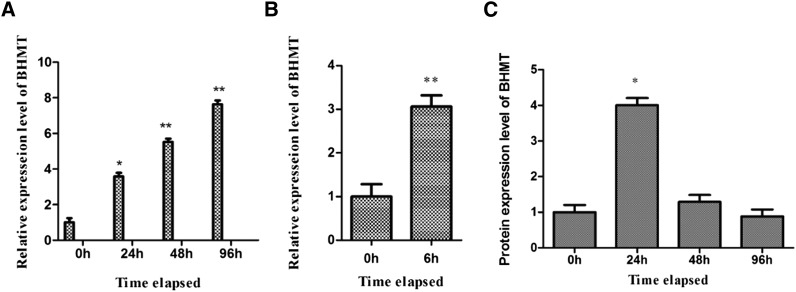
AjBHMT expression was increased not only in the bacteria-challenged individuals, but also in the LPS-treated cells. As shown in Figure 2, a 5.53- and 7.64-fold increase was detected at 48 hr and 96 hr in bacteria-infected samples, respectively, compared with the control group (Figure 2A), and a 3.06-fold increase was detected at 6 hr after exposure to 1 μg⋅ml−1 LPS (Figure 2B). However, the AjBHMT protein level initially increased by 4.01-fold at 24 hr postinfection, but gradually decreased to control levels at 48 hr and continuously declined at 96 hr in V. splendidus-challenged sea cucumbers from our iTRAQ qualification (Zhang et al. 2014) (Figure 2C). These findings indicated that AjBHMT should be tightly regulated at different stages of infection.
Figure 2.
AjBHMT mRNA and protein expression levels in V. splendidus- or LPS-challenged samples. (A) AjBHMT mRNA transcripts in V. splendidus-challenged A. japonicus. (B) AjBHMT mRNA transcripts in LPS-treated coelomocytes. (C) AjBHMT protein expression in V. splendidus-challenged A. japonicus by iTRAQ analysis.
Validation of the interaction between 3′-UTR of AjBHMT and miR-137 or miR-2008 through dual-luciferase reporter assays
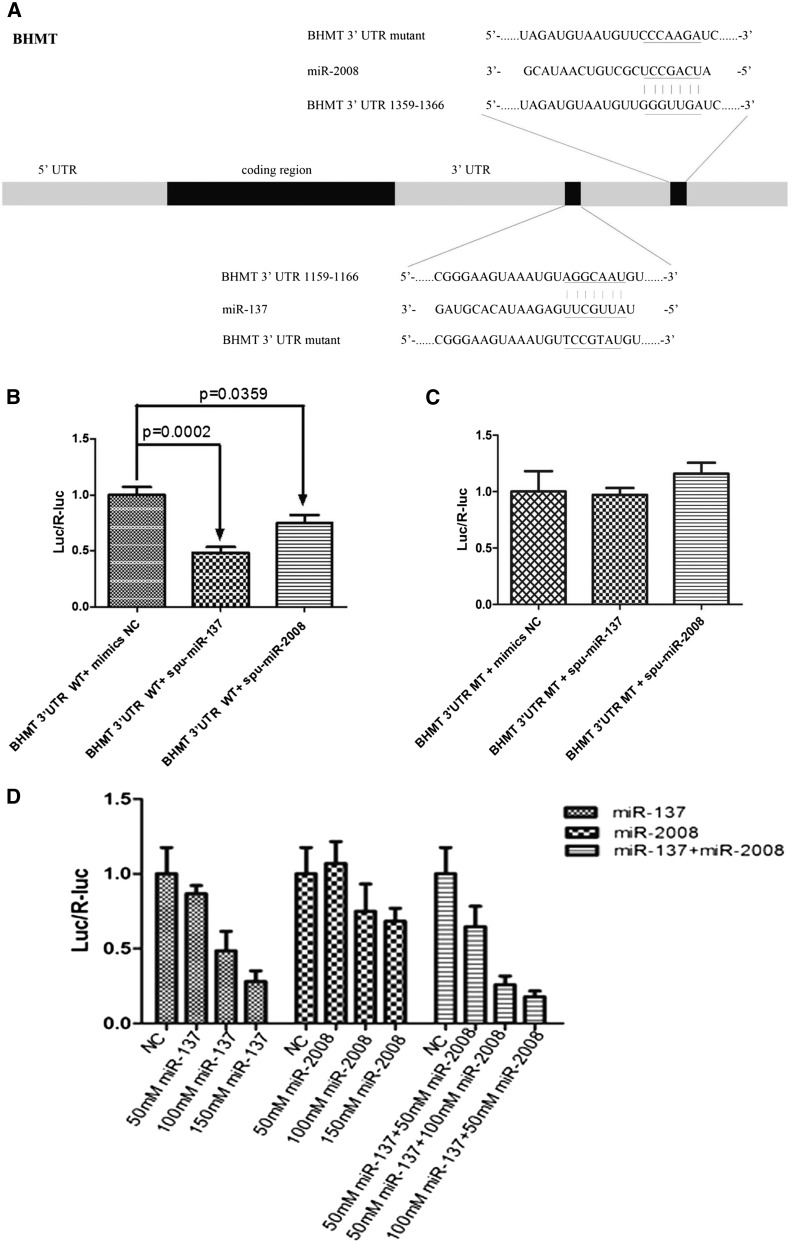
To further validate the predicted regulation of AjBHMT through miR-137 and miR-2008 at the 3′-UTR of this gene, two reporter plasmids containing the putative binding and mutation sites of each miRNA were constructed and used in classical reporter assays in HEK-293T cells. Information concerning the binding and mutant sites of each miRNA in the 3′-UTR of AjBHMT is shown in Figure 3A. These constructs were named pMIR-REPORT-AjBHMT-WT and pMIR-REPORT-AjBHMT-MT. Following the transient cotransfection of HEK-293T cells with pMIR-REPORT-AjBHMT-WT/MT, pRL-CMV-Renilla-luciferase and 100 nM miR-137 or miR-2008 mimics, we observed a 51.39% (P = 0.0002) and 25.07% (P = 0.0359) reduction in luciferase activity in HEK293 cells transfected with pMIR-REPORT-AjBHMT-WT compared with controls for miR-137 and miR-2008, respectively (Figure 3B). No significant variation in luciferase activity in HEK293 cells transfected with pMIR-REPORT-AjBHMT-MT was observed (Figure 3C).
Figure 3.
Identification and characterization of the miR-137 and miR-2008 binding sites in the 3′-UTR of AjBHMT and cooperative functional effect of miR-137 and miR-2008 on AjBHMT. (A) Schematic representation of the two putative miR-137 and miR-2008 targeting sites in AjBHMT mRNA and the respective mutant sites. (B) HEK-293T cells were cotransfected with the pMIR-REPORT-BHMT-WT vector, carrying the wild-type BHMT 3′-UTR, pRL-CMV-Renilla-luciferase, and control or miR-137 or miR-2008 mimics (final concentration: 100 nM) as indicated. After 24 hr, firefly luciferase activity was measured, and the values were normalized to Renilla luciferase activity. (C) HEK-239T cells were cotransfected with the pMIR-REPORT-BHMT-MT vector, carrying a mutation in the BHMT 3′-UTR, pRL-CMV-Renilla-luciferase, and control or miR-137 or miR-2008 mimics (final concentration: 100 nM) as indicated. (D) HEK-293T cells were transfected with different concentrations of miR-137 and miR-2008, or cotransfected with mixed miR-137 and miR-2008 mimics as indicated.
For the cooperative functional identification of miR-137 and miR-2008 on the AjBHMT, we used serial concentrations of miR-137 or miR-2008 mimics as well as their combination to cotransfect HEK-293T cells with pMIR-REPORT-AjBHMT-WT, pRL-CMV-Renilla-luciferase. Compared with the mimics control group, cells transfected with 50 nM miR-137 mimics or mir-2008 mimics exhibited no significant change in luciferase activity. However, in the miR-137 and miR-2008 cotransfected cells (50 nM miR-137 + 50 nM miR-2008), we found a 35.17% (P = 0.0032) reduction in luciferase activity (Figure 3D). For higher concentration transfection groups (150 nM), miR-137 mimics and mir-2008 mimics displayed 71.92% (P = 0.0005) and 31.66% (P = 0.0049) reduction in luciferase activity, respectively (Figure 3D). Interestingly, two miR-137 and miR-2008 combined groups (50 nM miR-137 + 100 nM miR-2008, 100 nM miR-137 + 50 nM miR-2008) both exhibited higher inhibitory effect by 74.24% (P = 0.0005) and 82.24% (P = 0.0005) reduction compared with 150 nM miR-137 and miR-2008 group, respectively (Figure 3D). Thus we think that these two miRNAs may target to the AjBHMT collaboratively.
miR-137 modulates AjBHMT at the post-transcriptional level, whereas miR-2008 modulates AjBHMT at the translational level in vitro and in vivo
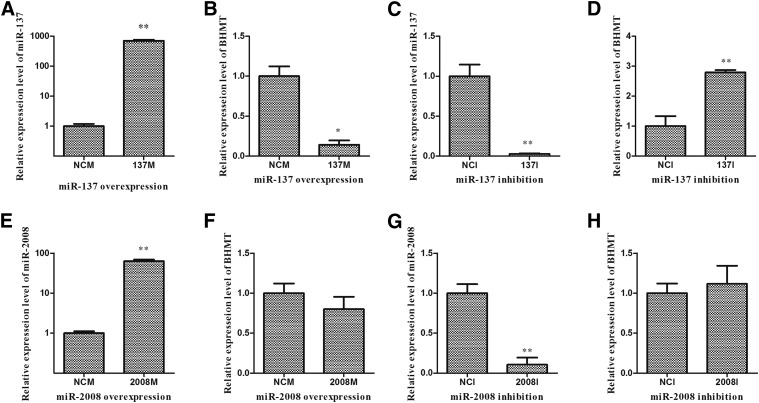
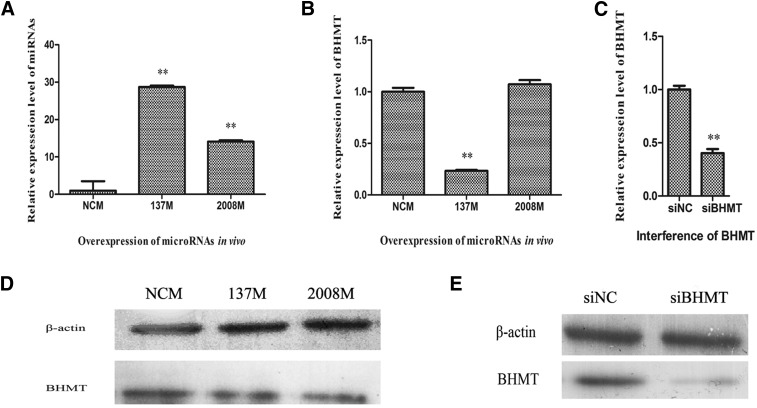
To fully understand the potential roles of miR-137 and miR-2008 in differentially regulating AjBHMT, loss-of-function or gain-of-function assay of miR-137 and miR-2008 was performed in vitro and in vivo. As shown in Figure 4, the qRT-PCR results indicated that the overexpression of miR-137 (Figure 4A) decreased the cellular levels of AjBHMT mRNA (Figure 4B). In contrast, the knockdown of miR-137 (Figure 4C) increased AjBHMT gene expression (Figure 4D). Western blot analysis of miR-137 overexpression and knockdown at 48 hr post-transfection revealed that the protein abundance of AjBHMT was decreased upon miR-137 overexpression and increased upon miR-137 inhibition (Figure 5), consistent with the AjBHMT mRNA expression levels. In vivo analysis further revealed that the overexpression of miR-137 (Figure 6A) decreased the expression of AjBHMT at both the mRNA (Figure 6B) and protein levels (Figure 6D). These results suggested that miR-137 directly targeted AjBHMT by inducing the degradation of AjBHMT mRNA.
Figure 4.
Functional validation of miR-137 and miR-2008 targeting AjBHMT mRNA in vitro. (A) Relative expression of miR-137 in primary cultured cells at 24 hr after miR-137 mimics transfection. (B) AjBHMT mRNA expression in primary cultured cells at 24 hr after transfection with miR-137 mimics. (C) Relative expression of miR-137 in primary cultured cells at 24 hr after miR-137 inhibitor transfection. (D) AjBHMT mRNA expression in primary cultured cells at 24 hr after miR-137 inhibitor transfection. (E) Relative expression of miR-2008 in primary cultured cells at 24 hr after miR-2008 mimics transfection. (F) AjBHMT mRNA expression in primary cultured cells at 24 hr after transfection with the miR-2008 mimics. (G) Relative expression of miR-2008 in primary cultured cells at 24 hr after miR-2008 inhibitor transfection. (H) AjBHMT mRNA expression in primary cultured cells at 24 hr after miR-2008 inhibitor transfection.
Figure 5.
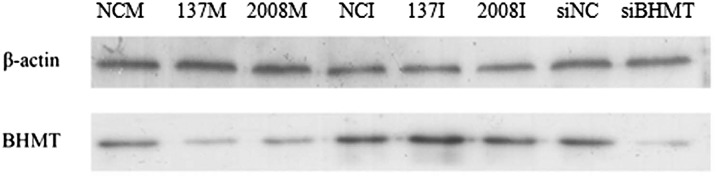
Western blot analysis ofAjBHMT expression in in vitro assays. NCM, negative control of miRNA mimics; 137M, miR-137 mimics; 2008M, miR-2008 mimics; NCI, negative control for the miRNA inhibitor; 137I, miR-137 inhibitor; 2008I, miR-2008 inhibitor; siNC, negative control for the AjBHMT siRNA; and siAjBHMT, AjBHMT siRNA.
Figure 6.
Functional analysis of miR-137 and miR-2008 and AjBHMT in vivo. (A) Relative miR-137 and miR-2008 expression levels after transfection with miRNA modified mimics. (B) Relative AjBHMT expression after transfection with miRNA modified mimics. (C) Relative AjBHMT expression after AjBHMT siRNA transfection. (D) Western blot analysis after transfection with miRNA modified mimics. (E) Western blot analysis after AjBHMT silencing in vivo.
The expression of AjBHMT in primary cultured cells transfected with miR-2008 mimics or the miR-2008 inhibitor is shown in Figure 4. Neither the overexpression nor the inhibition of miR-2008 (Figure 4, E and G) significantly changed AjBHMT mRNA expression (Figure 4, F and H). In contrast, Western blot analysis revealed that the overexpression of miR-2008 decreased AjBHMT protein abundance, and miR-2008 knockdown increased AjBHMT protein abundance (Figure 5). Consistent with the result of in vitro analysis, the results of in vivo assay also showed that the overexpression of miR-2008 (Figure 6A) decreased the expression of AjBHMT protein (Figure 6B), without affecting the levels of AjBHMT mRNA (Figure 6D). Collectively, these observations suggested that miR-2008 decreases AjBHMT protein expression without affecting AjBHMT mRNA expression.
The Hcy concentration is directly modulated through changes in AjBHMT in vitro and in vivo
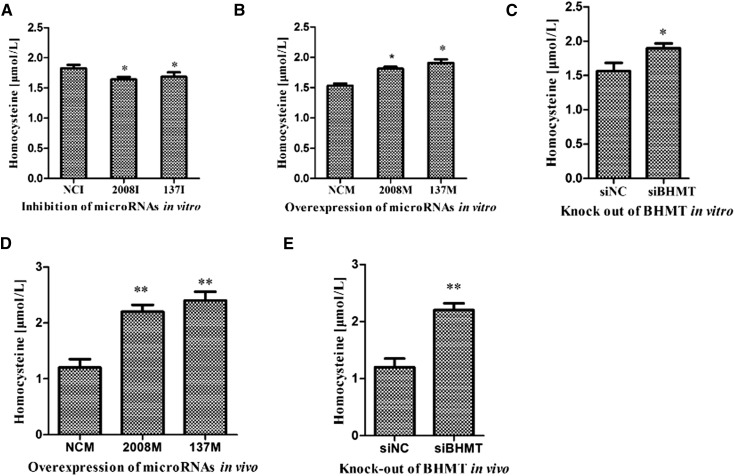
As a substrate of BHMT metabolism, the Hcy content was considered as an important index for changes in BHMT expression. Therefore, we explored alterations in the Hcy concentration in the culture medium of cells transfected with miRNA mimics or inhibitors at 48 hr post-transfection. The inhibition of miR-137 and miR-2008 decreased the levels of Hcy in the culture medium compared with the negative control (Figure 7A), whereas the overexpression of miR-137 and miR-2008 increased the levels of Hcy in the culture medium (Figure 7B). The abundance of Hcy was also increased upon siRNA-mediated AjBHMT silencing (Figure 7C), further suggesting that miR-137 and miR-2008 modulated the Hcy concentration by targeting AjBHMT in vitro. In vivo, the overexpression of miR-137 and miR-2008 (Figure 7D), as well as the siRNA-mediated AjBHMT silencing (Figure 7E) also causes the accumulation of the Hcy concentration, consistent with the result of in vitro analysis.
Figure 7.
Correlation analysis of Hcy contents with miR-137, miR-2008, and AjBHMT in vitro and in vivo. (A) miR-137 or miR-2008 inhibitor transfection in vitro. (B) miR-137 or miR-2008 mimics transfection in vitro. (C) AjBHMT silencing in vitro. (D) miR-137 or miR-2008 modified mimics transfection in vivo. (E) AjBHMT silencing in vivo.
siRNA-mediated AjBHMT silencing decreases the bacterial survival rate through the promotion of Hcy expression and ROS production in vitro and in vivo
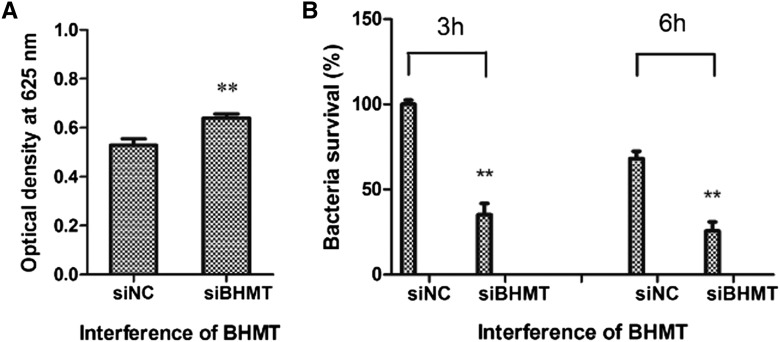
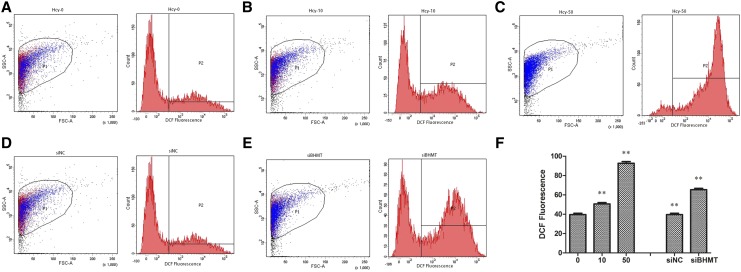
The injection of AjBHMT siRNA specifically inhibited AjBHMT mRNA expression (Figure 6C) and protein synthesis in vivo (Figure 6E). A dramatic increase in Hcy expression was observed after AjBHMT deletion (Figure 7E). Previous studies have indicated that Hcy elevates respiratory burst in immune-competent cells (Vladychenskaya and Boldyrev 2009). Therefore, we investigated the ROS production and bacterial survival rate in cells transfected with AjBHMT siRNA. As shown in Figure 8, AjBHMT silencing significantly accelerated intracellular ROS production (Figure 8A) and decreased the bacterial survival rate by 35.3% and 25.7% at 3 and 6 hr postinfection, respectively (Figure 8B). We used the 2′,7′-dichlorofluorescin diacetate (DCFH-DA) method to directly evaluate the effects of AjBHMT-mediated respiratory burst on sea cucumbers and assayed the results using ECM. As shown in Figure 9, AjBHMT silencing increased intracellular ROS production BY 65.4% (Figure 9, E and F) compared with the control group (Figure 9, D and F).
Figure 8.
Determination of ROS production and bacterial survival rate after AjBHMT knockdown in vivo. (A) ROS production assay. (B) Bacterial survival rate assay.
Figure 9.
ROS production assay after AjBHMT knockdown in vivo by flow cytometry. (A) Hcy control; (B) 10 mM Hcy injection; (C) 50 mM Hcy injection; (D) negative control for siBHMT; (E) AjBHMT silencing; and (F) bar chart representing ROS production as described in A–E.
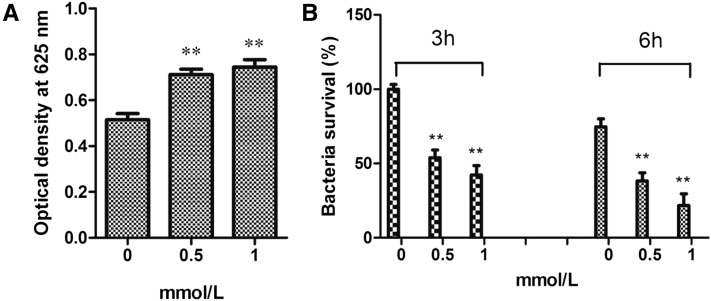
Hcy exposure also minimizes the bacterial survival rate through an increase in ROS production in vitro and in vivo
To evaluate the effect of Hcy on ROS production and the bacterial survival rate, we stimulated primary cultured cells with different concentrations of Hcy. As shown in Figure 10, exposure to 0.5 and 1 mM Hcy significantly increased intracellular ROS production (Figure 10A) and decreased the bacterial survival rate (Figure 10B). In response to stimulation with 0.5 mM Hcy, 55.1% and 41.0% bacteria survived at 3 and 6 hr postinfection, respectively, and 1 mM Hcy exposure decreased the bacterial survival rate to 37.0% and 21.1% at 3 and 6 hr postinfection (Figure 10B). To further confirm Hcy-mediated ROS production in vivo, we injected sea cucumbers with different concentrations of Hcy and assayed ROS production in the coelomocytes using ECM. The results shown in Figure 10 demonstrated that 10 mM Hcy exposure increased ROS production to 50.6% (Figure 9, B and F) and 50 mM Hcy stimulation increased ROS production to 92.7% (Figure 9, C and F) compared with the respective control groups showing 40% ROS production (Figure 9, A and F).
Figure 10.
Effect of Hcy on ROS production and the bacterial survival rate in primary cultured cells. (A) ROS production after Hcy stimulation. (B) Bacterial survival rate after Hcy stimulation.
Discussion
MicroRNA-mediated gene silencing is a widespread regulatory mechanism implicated in host–pathogen interactions and improvements in cell-specific targeting for gene and cell therapy (Brown and Naldini 2009). Accumulative evidence indicated that miRNAs modulate target gene expression at the mRNA or protein level, and a given mRNA can be targeted by several miRNAs. However, scarce information is available concerning the modulation of a specific target through different miRNAs at different levels, particularly in nonmodel invertebrates. In the present study, we explored the expression patterns of miR-137, miR-2008, and AjBHMT in pathogen-challenged individuals and LPS-exposed cells using qRT-PCR. The results suggested an inverse relationship between miR-137 and AjBHMT expression, as well as a parallel relationship between miR-2008 and AjBHMT expression. In a previous study, we performed a iTRAQ-based proteomics analysis of pathogen-challenged sea cucumbers, showing that the abundance of AjBHMT protein increased at 24 hr postinfection but decreased at 48 and 96 hr postinfection. Thus, AjBHMT could be regulated through miR-137 at the post-transcriptional level via direct mRNA degradation and through miR-2008 at the translational level via translation repression. To confirm this hypothesis, we applied the overexpression and inhibition of miR-137 and miR-2008 to explore the gene or protein levels of AjBHMT using qRT-PCR and Western blotting in vitro and in vivo. The results indicated that miR-137 and miR-2008 differentially regulated AjBHMT expression. This differential regulation might represent a highly efficient mechanism for the tight regulation of target proteins. Given that a single transcript can produce hundreds of proteins, the different mechanisms of miR-137 and miR-2008 act in cooperation to post-transcriptionally control AjBHMT through a “fine-tuning” model (Sevignani et al. 2006). Meanwhile the miR-137-mediated degradation of AjBHMT mRNA may lead to a sharp decline in the abundance of AjBHMT protein abundance, the translation repression of AjBHMT induced by miR-2008 only reduced the AjBHMT protein levels, which could be quickly recovered as this kind of repression often is relatively mild (Baek et al. 2008; Selbach et al. 2008). The discrepancy between the mRNA expression level and the protein abundance of AjBHMT may also result from some other regulatory factors. However, this topic is beyond the scope of the present study.
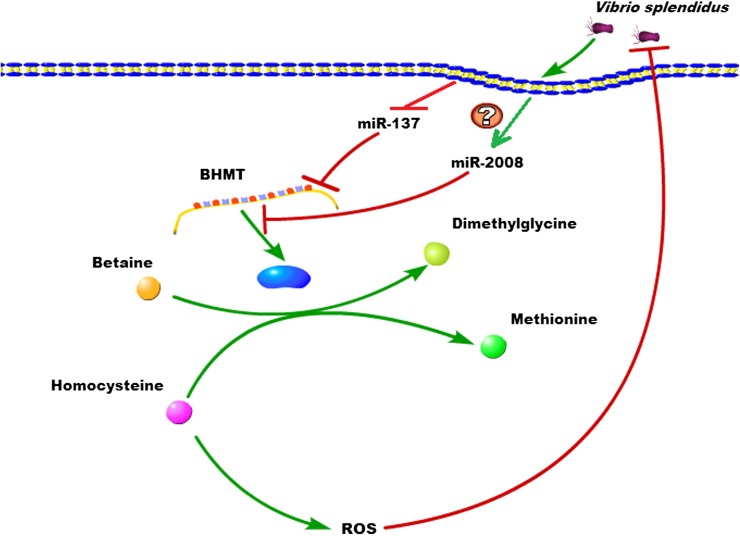
BHMT was first identified as a facilitator of methyl group donation for the remethylation of Hcy into methionine (Pajares and Pérez-Sala 2006), and reduced BHMT functions can elevate Hcy levels (Ji et al. 2007). Consistently, in the present study, the overexpression of miR-137 and miR-2008 and silencing of AjBHMT elevated the Hcy content, whereas the inhibition of miR-137 and miR-2008 decreased the Hcy levels. In the past decade, Hcy has been identified as a risk factor for numerous diseases. Many studies have demonstrated that a high Hcy concentration increases intracellular ROS generation, and the imbalance of Hcy metabolism results in cellular immune dysfunction in metabolic syndromes. ROS are important intracellular second messengers that mediate the production of inflammatory cytokines (Gorowara et al. 1998). Abundant evidence indicated that excessive Hcy in endothelial cells induces endothelial dysfunction through the inhibition of arginine transport (Jin et al. 2007), elevates ROS levels through autooxidation, and diminishes the activities of antioxidative enzymes, such as glutathione peroxidase-1 (Handy et al. 2005) and superoxide dismutase (Yamamoto et al. 2000). Furthermore, recent studies indicate that Hcy induces the production of superoxide anion (Alvarez-Maqueda et al. 2004; Au-Yeung et al. 2006), exerts important effects on cellular NO production (Tsen et al. 2003; Zhang et al. 2000), and interferes with de novo GSH synthesis (Toroser and Sohal 2007) to improve intracellular ROS levels. In a previous study (Li et al. 2012), we demonstrated that the extremely high cellular abundance of miR-2008 can reduce the activity of AjBHMT significantly in SUS-diseased samples, whereas the normal abundance of miR-137 maintains low abundance of AjBHMT mRNA. Both of these events resulted in difficult recovered AjBHMT activity and continued high Hcy concentration. As described above, elevated Hcy concentrations induce inflammatory responses and increase ROS production, which play a critical role in cellular pathophysiology and induce damage to organisms (Lee et al. 2013). This kind of excess accumulation of Hcy and ROS production may contribute to the SUS outbreak of A. japonicus. Numerous studies have implicated ROS in oxidative defense mechanisms responsible for bacterial death (Slauch 2011). Thus, we evaluated the effect of Hcy on intracellular ROS generation and bacterial clearance, and we confirmed that Hcy induced ROS generation and observably decreased the bacterial survival rate. A schematic representation of the involvement of miR-137 and miR-2008 in the host–pathogen interaction by regulating intracellular ROS generation through cotargeting of AjBHMT to modulate cellular Hcy concentration is shown in Figure 11.
Figure 11.
Schematic representation of the involvement of miR-137 and miR-2008 in the host–pathogen interaction by cotargeting AjBHMT.
Intriguingly, in the early phase of V. splendidus infection, the down-regulation of miR-137 expression could increase the level of AjBHMT expression, resulting from an increasing demand for methionine in protein synthesis or methyl donation. Previous studies have confirmed that the aberrant methylation of the miR-137 promoter contributes to miR-137 silencing (Langevin et al. 2010, 2011; Dang et al. 2013). In the present study, we speculated that the continuous down-regulation of miR-137 might reflect an increasing level of methylation, forming a feedback loop that increases AjBHMT levels and decreases in miR-137 levels to further increase the concentration of AjBHMT. Elevated AjBHMT activity might promote the expression of miR-2008 to maintain appropriate cellular AjBHMT levels. Thus, our future studies should focus on the upstream regulation of these two miRNAs.
In the present study, we confirmed that miR-137 and miR-2008 played a pivotal role in host–pathogen interactions through cotargeting of the AjBHMT gene to regulate Hcy metabolism, thereby affecting cellular oxidative-redox stress and improving bacterial elimination. Immunometabolism is an emerging frontier that demonstrates the interplay between immunological and metabolic processes. The multilevel interactions between metabolic and immune systems suggest the importance of understanding the pathogenic mechanisms (Mathis and Shoelson 2011). The results of the present study suggested a new horizon for dissecting the pathogenesis of infectious diseases from the perspective of miRNA-mediated cross-talk between immunology and metabolism.
Supplementary Material
Acknowledgments
We thank Dr. Sebastian Shimeld from Oxford University for proofreading. This work was financially supported through the National Natural Science Foundation of China (31522059, 41576139), the Zhejiang Provincial Natural Science Foundation of China (LR14C190001), Collaborative Innovation Center for Zhejiang Marine High-Efficiency and Healthy Aquaculture, and the K. C. Wong Magna Fund at Ningbo University.
Footnotes
Communicating editor: B. Lazzaro
Supporting information is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.178871/-/DC1.
Literature Cited
- Alvarez-Maqueda M., El Bekay R., Monteseirín J., Alba G., Chacón P., et al. , 2004. Homocysteine enhances superoxide anion release and NADPH oxidase assembly by human neutrophils. Effects on MAPK activation and neutrophil migration. Atherosclerosis 172(2): 229–238. [DOI] [PubMed] [Google Scholar]
- Arias-Romero L. E., Chernoff J., 2013. Targeting Cdc42 in cancer. Expert Opin. Ther. Targets 17(11): 1263–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asgari S., 2011. Role of microRNAs in insect host-microorganism interactions. Front. Physiol. 2: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au-Yeung K. K., Yip J. C., Siow Y. L., 2006. Folic acid inhibits homocysteine-induced superoxide anion production and nuclear factor kappa B activation in macrophages. Can. J. Physiol. Pharmacol. 84(1): 141–147. [DOI] [PubMed] [Google Scholar]
- Baek D., Villén J., Shin C., Camargo F. D., Gygi S. P., et al. , 2008. The impact of microRNAs on protein output. Nature 455(7209): 64–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaguer F., Link A., Lozano J. J., Cuatrecasas M., Nagasaka T., et al. , 2010. Epigenetic silencing of miR-137 is an early event in colorectal carcinogenesis. Cancer Res. 70(16): 6609–6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D. P., 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116(2): 281–297. [DOI] [PubMed] [Google Scholar]
- Bemis L. T., Chen R., Amato C. M., Classen E. H., Robinson S. E., et al. , 2008. MicroRNA-137 targets microphthalmia-associated transcription factor in melanoma cell lines. Cancer Res. 68(5): 1362–1368. [DOI] [PubMed] [Google Scholar]
- Brown B. D., Naldini L., 2009. Exploiting and antagonizing microRNA regulation for therapeutic and experimental applications. Nat. Rev. Genet. 10(8): 578–585. [DOI] [PubMed] [Google Scholar]
- Calin G. A., Croce C. M., 2006. MicroRNA signatures in human cancers. Nat. Rev. Cancer 6(11): 857–866. [DOI] [PubMed] [Google Scholar]
- Chen, S., N. He, J. Yu, L. Li, Y. Hu et al., 2014 Post-transcriptional regulation by miR-137 underlies the low abundance of CAR and low rate of bilirubin clearance in neonatal mice. Life Sci. pii: S0024–3205(14): 00444–00445. [DOI] [PubMed]
- Dang J., Bian Y. Q., Sun J. Y., Chen F., Dong G., et al. , 2013. MicroRNA-137 promoter methylation in oral lichen planus and oral squamous cell carcinoma. J. Oral Pathol. Med. 42(4): 315–321. [DOI] [PubMed] [Google Scholar]
- Deng H., He C., Zhou Z., Liu C., Tan K., 2009. Isolation and pathogenicity of pathogens from skin ulceration disease and viscera ejection syndrome of the sea cucumber Apostichopus japonicus. Aquaculture 287(1): 18–27. [Google Scholar]
- Friedman R. C., Farh K. K., Burge C. B., Bartel D. P., 2009. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 19(1): 92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorowara S., Sapru S., Ganguly N. K., 1998. Role of intracellular second messengers and reactive oxygen species in the pathophysiology of V. cholera O139 treated rabbit ileum. Biochim. Biophys. Acta 1407(1): 21–30. [DOI] [PubMed] [Google Scholar]
- Handy D. E., Zhang Y., Loscalzo J., 2005. Homocysteine down-regulates cellular glutathione peroxidase (GPx1) by decreasing translation. J. Biol. Chem. 280(16): 15518–15525. [DOI] [PubMed] [Google Scholar]
- He L., Hannon G. J., 2004. MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 5(7): 522–531. [DOI] [PubMed] [Google Scholar]
- Ji C., Shinohara M., Kuhlenkamp J., Chan C., Kaplowitz N., 2007. Mechanisms of protection by the betaine-homocysteine methyltransferase/betaine system in HepG2 cells and primary mouse hepatocytes. Hepatology 46(5): 1586–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L., Caldwell R. B., Li-Masters T., Caldwell R. W., 2007. Homocysteine induces endothelial dysfunction via inhibition of arginine transport. J. Physiol. Pharmacol. 58(2): 191–206. [PubMed] [Google Scholar]
- Krek A., Grün D., Poy M. N., Wolf R., Rosenberg L., et al. , 2005. Combinatorial microRNA target predictions. Nat. Genet. 37(5): 495–500. [DOI] [PubMed] [Google Scholar]
- Lai E. C., 2002. MicroRNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat. Genet. 30(4): 363–364. [DOI] [PubMed] [Google Scholar]
- Langevin S. M., Stone R. A., Bunker C. H., Grandis J. R., Sobol R. W., et al. , 2010. MicroRNA-137 promoter methylation in oral rinses from patients with squamous cell carcinoma of the head andneck is associated with gender and body mass index. Carcinogenesis 31(5): 864–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langevin S. M., Stone R. A., Bunker C. H., Lyons-Weiler M. A., LaFramboise W. A., 2011. MicroRNA-137 promoter methylation is associated with poorer overall survival in patients with squamous cell carcinoma of the head and neck. Cancer 117(7): 1454–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. S., Lee S. J., Seo K. W., Bae J. U., Park S. Y., et al. , 2013. Homocysteine induces COX-2 expression in macrophages through ROS generated by NMDA receptor-calcium signaling pathways. Free Radic. Res. 47(5): 422–431. [DOI] [PubMed] [Google Scholar]
- Li C., Feng W., Qiu L., Xia C., Su X., et al. , 2012. Characterization of skin ulceration syndrome associated microRNAs in sea cucumber Apostichopus japonicus by deep sequencing. Fish Shellfish Immunol. 33: 436–441. [DOI] [PubMed] [Google Scholar]
- Liu M., Lang N., Qiu M., Xu F., Li Q., et al. , 2011. miR-137 targets Cdc42 expression, induces cell cycle G1 arrest and inhibits invasion in colorectal cancer cells. Int. J. Cancer 128(6): 1269–1279. [DOI] [PubMed] [Google Scholar]
- Ma Y., Xu G., Chang Y., Chen J., Shi C., 2006. Bacterial pathogens of skin ulceration disease in cultured sea cucumber Apostichopus japonicus (Selenka) juveniles. J. Dalian Ocean Univer. 21(1): 13–18. [Google Scholar]
- Mathis D., Shoelson S. E., 2011. Immunometabolism: an emerging frontier. Nat. Rev. Immunol. 11(2): 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miska E. A., 2005. How microRNAs control cell division, differentiation and death. Curr. Opin. Genet. Dev. 15(5): 563–568. [DOI] [PubMed] [Google Scholar]
- Pajares M. A., Pérez-Sala D., 2006. Betaine homocysteine S-methyltransferase: just a regulator of homocysteine metabolism? Cell. Mol. Life Sci. 63(23): 2792–2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajewsky N., 2006. microRNA target predictions in animals. Nat. Genet. 38: S8–S13. [DOI] [PubMed] [Google Scholar]
- Scaria V., Hariharan M., Maiti S., Pillai B., Brahmachari S. K., 2006. Host-virus interaction: a new role for microRNAs. Retrovirology 3: 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selbach M., Schwanhäusser B., Thierfelder N., Fang Z., Khanin R., et al. , 2008. Widespread changes in protein synthesis induced by microRNAs. Nature 455(7209): 58–63. [DOI] [PubMed] [Google Scholar]
- Sevignani C., Calin G. A., Siracusa L. D., Croce C. M., 2006. Mammalian microRNAs: a small world for fine-tuning gene expression. Genome 17(3): 189–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slauch J. M., 2011. How does the oxidative burst of macrophages kill bacteria? Still an open question. Mol. Microbiol. 80(3): 580–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smrt R. D., Szulwach K. E., Pfeiffer R. L., Li X., Guo W., et al. , 2010. MicroRNA miR-137 regulates neuronal maturation by targeting ubiquitin ligase mind bomb-1. Stem Cells 28(6): 1060–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takwi A. A., Wang Y., Wu J., Michaelis M., Cinatl J., et al. , 2013. miR-137 regulates the constitutive androstane receptor and modulates doxorubicin sensitivity in parental and doxorubicin-resistant neuroblastoma cells. Oncogene 128(6): 1269–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toroser D., Sohal R. S., 2007. Age-associated perturbations in glutathione synthesis in mouse liver. Biochem. J. 405(3): 583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsen C. M., Hsieh C. C., Yen C. H., Lau Y. T., 2003. Homocysteine altered ROS generation and NO accumulation in endothelial cells. Chin. J. Physiol. 46(3): 129–136. [PubMed] [Google Scholar]
- Vladychenskaya E. A., Boldyrev A. A., 2009. Effect of homocysteine on respiratory burst in neutrophils induced by chemotaxis factor fMLP. Neurochem. J. 3(1): 64–69. [Google Scholar]
- Vrba L., Muñoz-Rodríguez J. L., Stampfer M. R., Futscher B. W., 2013. miRNA gene promoters are frequent targets of aberrant DNA methylation in human breast cancer. PLoS One 8(1): e54398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Chang Y., Xu G., Song L., 2005. Isolation and ultrastructure of an enveloped virus in cultured sea cucumber Apostichopus japonicus (Selenka). J. Fish. Sci. China 12(6): 766–770. [Google Scholar]
- Wang T., Sun Y., Jin L., Xu Y., Wang L., et al. , 2009. Enhancement of non-specific immune response in sea cucumber (Apostichopus japonicus) by Astragalus membranaceus and it polysaccharides. Fish Shellfish Immunol. 27(6): 757–762. [DOI] [PubMed] [Google Scholar]
- Wu W., Sun M., Zou G. M., Chen J., 2007. MicroRNA and cancer: Current status and prospective. Int. J. Cancer 120(5): 953–960. [DOI] [PubMed] [Google Scholar]
- Xing J., Leung M. F., Chia F. S., 1998. Quantitative analysis of phagocytosis by amebocytes of a sea cucumber, Holothuria leucospilota. Invertebr. Biol. 117(1): 13–22. [Google Scholar]
- Yamamoto M., Hara H., Adachi T., 2000. Effects of homocysteine on the binding of extracellular-superoxide dismutase to the endothelial cell surface. FEBS Lett. 486(2): 159–162. [DOI] [PubMed] [Google Scholar]
- Zhang C., Wang Y., Rong X., 2006. Isolation and identification of causative pathogen for skin ulcerative syndrome in Apostichopus japonicus. J. Fish. China 30: 118–123. [Google Scholar]
- Zhang P., Li C., Zhu L., Su X., Li Y., et al. , 2013. De novo assembly of the sea cucumber Apostichopus japonicus hemocytes transcriptome to identify miRNA targets associated with skin ulceration syndrome. PLoS One 8(9): e73506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Li C., Zhang P., Jin C., Pan D., et al. , 2014. iTRAQ-based proteomics reveals novel members involved in pathogen challenge in sea cucumber Apostichopus japonicus. PLoS One 9(6): e100492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Li H., Jin H., Ebin Z., Brodsky S., et al. , 2000. Effects of homocysteine on endothelial nitric oxide production. Am. J. Physiol. Renal Physiol. 279(4): F671–F678. [DOI] [PubMed] [Google Scholar]
- Zhu X., Li Y., Shen H., Li H., Long L., et al. , 2013. miR-137 inhibits the proliferation of lung cancer cells by targeting Cdc42 and Cdk6. FEBS Lett. 587(1): 73–81. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All list plasmids and strains may be requested from Li Lab. Current versions of protocols will be maintained in our laboratory.