Abstract
Molecular heterogeneity in human breast cancer has challenged diagnosis, prognosis, and clinical treatment. It is well known that molecular subtypes of breast tumors are associated with significant differences in prognosis and survival. Assuming that the differences are attributed to subtype-specific pathways, we then suspect that there might be gene regulatory mechanisms that modulate the behavior of the pathways and their interactions. In this study, we proposed an integrated methodology, including machine learning and information theory, to explore the mechanisms. Using existing data from three large cohorts of human breast cancer populations, we have identified an ensemble of 16 master regulator genes (or MR16) that can discriminate breast tumor samples into four major subtypes. Evidence from gene expression across the three cohorts has consistently indicated that the MR16 can be divided into two groups that demonstrate subtype-specific gene expression patterns. For example, group 1 MRs, including ESR1, FOXA1, and GATA3, are overexpressed in luminal A and luminal B subtypes, but lowly expressed in HER2-enriched and basal-like subtypes. In contrast, group 2 MRs, including FOXM1, EZH2, MYBL2, and ZNF695, display an opposite pattern. Furthermore, evidence from mutual information modeling has congruently indicated that the two groups of MRs either up- or down-regulate cancer driver-related genes in opposite directions. Furthermore, integration of somatic mutations with pathway changes leads to identification of canonical genomic alternations in a subtype-specific fashion. Taken together, these studies have implicated a gene regulatory program for breast tumor progression.
Keywords: breast cancer, master regulator, regulator–regulon interactions, gene regulatory program
HUMAN breast cancer is the most common malignancy in women, with >200,000 new cases diagnosed each year in the United States (Tolaney and Winer 2007). Breast cancer is a complex disease that is caused by multiple genetic and environmental factors. Molecular heterogeneity in breast tumors has challenged diagnosis, prognosis, and clinical treatment.
Human breast cancer has significant intra- and intertumor molecular heterogeneity. Regarding intratumor heterogeneity, evidence from next-generation sequencing has indicated that different tumor subclones display diversified DNA mutation profiles (Tolaney and Winer 2007; Nik-Zainal et al. 2012; Yates et al. 2015). Furthermore, the mutation profiles are subject to change over time due to the fact that tumor cells can adapt to the selective pressure of therapies (Klein 2013). On the other hand, intertumor variation is manifested by molecular subtypes that represent significant differences in prognosis and survival (Parker et al. 2009). Evidence from gene expression profiling and unsupervised clustering analysis has indicated four major subtypes: luminal A, luminal B, HER2-enriched, and basal-like (Perou et al. 2000). While the majority of the luminal tumors are estrogen receptor (ER)-positive, the other two subtypes, especially the basal-like, are mainly ER-negative tumors. A set of 50 genes (PAM50) has been proposed to classify breast tumor samples into the subtypes (Parker et al. 2009). Six (ESR1, PGR, FOXA1, FOXC1, MYC, and MYBL2) of the 50 genes are transcriptional factor genes (Parker et al. 2009). Recent studies on genomics and transcriptomics of large patient populations have identified additional subtypes (Curtis et al. 2012; Guedj et al. 2012). In the present study, we will focus on the four major subtypes.
High-throughput technologies, including SNP array and next-generation sequencing studies on large cohorts of patient and control populations, have helped identify both common and rare DNA alternations impacting cancer etiology and development (Cancer Genome Atlas Network 2012; Michailidou et al. 2015). Genome-wide association studies have accumulatively identified ∼90 loci that are associated with predisposition to breast cancer risk (Michailidou et al. 2015). The germline mutations collectively account for 16% of the genetic variation in breast cancer (Michailidou et al. 2015). On the other hand, somatic mutations have been identified in tens of breast cancer driver genes, which lead to protein functional changes in tumors (Cancer Genome Atlas Network 2012). However, connections of both the genetic and genomic alternations to the subtypes are largely unknown.
Assuming that there are intrinsic connections between various biological pathways and different tumor subtypes that represent significant differences in prognosis and survival, we suspect that there might be gene regulatory mechanisms that modulate the behavior of the pathways. Thus we hypothesize that identification of master regulators (MRs) and gene regulatory pathways can bridge the gap between genotype and phenotype and shed light on tumor progression.
MRs are transcriptional factor genes that play a pivotal role in modulating downstream pathways or gene networks. Studies have established that ER is such a MR that regulates multiple pathways related to ER-positive breast tumors (Fletcher et al. 2013). However, a complete set of MRs for each tumor subtype, as well as the MR–MR interactions, are little known. Therefore, it is crucial to perform a systematic search for master regulators.
Systems genetics that integrates approaches of genetics, genomics, and multi-level phenotype characterization is powerful in understanding genotype–phenotype dependency (Li et al. 2006; Bjorkegren et al. 2015). Gene–gene interactions are the core of systems genetics (Li et al. 2006; Li and Churchill 2010). But they are challenging to explore in human populations. In this study, using existing data from three large cohorts of human breast cancer populations, including The Cancer Genome Atlas (TCGA) breast tumor cohort (Cancer Genome Atlas Network 2012), the Curtis cohort (Curtis et al. 2012), and the Guedj cohort (Guedj et al. 2012), we systematically scrutinize MR–MR and MR–regulon interactions. Our studies have revealed a gene regulatory program that impacts tumor progression in human breast cancer.
Materials and Methods
Three cohorts of human breast cancer populations
In this study, we used existing data generated from three cohorts of human breast cancer populations to identify and cross-validate MRs and gene regulatory pathways. These data include the TCGA breast tumor cohort (Cancer Genome Atlas Network 2012), the Curtis cohort (Curtis et al. 2012), and the Guedj cohort (Guedj et al. 2012). The TCGA breast tumor samples represent a large patient population from the United States, with 90% white women and 10% African American women. In addition to the DNA-Seq data and clinical data, we downloaded RNA-Seq data of 646 primary breast tumor cases (samples) from the TCGA data portal (https://tcga-data.nci.nih.gov/). For cross-validation studies, we also downloaded the microarray-based gene expression data and clinical data from the Curtis cohort (Curtis et al. 2012) and from the Guedj cohort (Guedj et al. 2012). The Curtis cohort encompasses ∼2000 breast tumor samples from the United Kingdom and Canada (Curtis et al. 2012). The Guedj cohort has 537 breast tumor samples from France (Guedj et al. 2012). Among the three cohorts, the breast cancer cases are all females. Regarding age at diagnosis, it is comparable between the TCGA and Curtis cohorts, with a median age of ∼60 (Supporting Information, Figure S1). However, in the Guedj cohort the median age shifts to ∼30. This may partially be explained by the different clinical record systems because age at initial diagnosis was recorded in the Guedj cohort. Regarding pathological phenotypes, such as tumor stage and grade, it is challenging to compare because different pathological systems were used across the cohorts.
Regarding the gene expression data, we processed the data, followed by a data quality check. The TCGA RNA-Seq data downloaded are already processed data (level 3), in which gene expression is quantified as fragments per kilobase transcript per million mapped reads (FPKM). Using the Mbatch tool (http://bioinformatics.mdanderson.org/tcgambatch/), we checked the data for batch effect. The gene expression data from the Curtis and Guedj cohorts are probe-level measurements based on the microarray platforms. Using corresponding human annotation files downloaded from the Bioconductor (http://bioconductor.org), we performed probe-to-gene annotations. Gene-level expression was then quantified as the maximum measurement across the corresponding probes. We also applied the Mbatch tool to check data quality. In addition, the gene expression data were transformed using van der Waerden scores (Lehmann and D’Abrera 1988) to center the mean of each gene to 0 and compare only variance and covariance between genes.
Machine learning to select an assemble of master regulator genes
Supervised machine learning is a powerful method for gene selection and tumor sample classification. Schematic framework of machine leaning is shown in Figure S2. Since RNA-Seq provides a better measurement of gene expression compared to microarray, we used the downloaded TCGA RNA-Seq data for feature (or gene) selection. But findings will be validated in the other two cohorts mentioned above. We split the TCGA primary breast tumor samples (n = 646) into training (3/4) and test (1/4) subsets. Subtype of each sample in the training subset was assigned by use of the PAM50 method (Parker et al. 2009). The subtype information was kindly provided by K. A. Horsley at the University of North Carolina.
Since the first step is to systematically identify MRs for each subtype, we focused on two gene sets for feature selection: (1) a set of ∼1400 well-annotated transcriptional factor (TF) genes (Vaquerizas et al. 2009) and (2) another set of 138 cancer driver genes (Vogelstein et al. 2013). There are only 23 overlapping genes between the two gene sets. As the majority of the cancer driver genes are non-TF genes, the second gene set is used as a control for feature selection. Between the combined gene panel (from gene sets 1 and 2) and the PAM50, only 10 genes overlap, including FOXC1, ESR1, FOXA1, ERBB2, MYC, MDM2, PGR, BCL2, EGFR, and MYBL2.
Regarding feature (or gene) selection, we employed the Random Forest algorithm (Breiman 2001) coupled with recursive gene elimination. Random Forest is one of the state-of-the-art algorithms in supervised machine learning. It is a decision-tree-based method for selection of an ensemble of features that are able to best discriminate the four subtypes of breast tumors. See SI Methods for more details.
Once the feature set is identified by the algorithm, we then train a classifier (or model), using the training data. There are multiple algorithms that can be applied to train a classifier. For example, one algorithm develops a probabilistic model that is based on the Bayesian method (Berger 1985). Application of the classifier to a new tumor sample will generate the posterior probability that the sample belongs to a subtype, given the data and the model. We used this method to train a classifier.
We need a reference for prediction assessment. Using the PAM50 (Parker et al. 2009) method, we called subtypes for the samples in each of the test data sets (Figure S2). Prediction (or classification) accuracy is defined as the percentage of the samples (in each of the test data sets) that have been correctly predicted by the MR16 classifier. This is a measurement of concordance in subtype calling between the PAM50 and the MR16.
Disease-free survival prediction
Disease-free survival prediction of a classifier is of clinical interest. Since the Guedj cohort has a long follow-up time in metastasis relapse-free survival (Guedj et al. 2012), we focus on this cohort for survival prediction. Using the MR16 from the TCGA training data, we developed a classifier. Application of the classifier to the Guedj cohort can predict the samples into the four major subtypes. We then fit a COX proportional hazards regression model (Cox and Oakes 1984), where the disease-free survival time was used as the response variable, the predicted subtype information as an explanatory variable, plus tumor grade as a covariate. Regarding other cofactors, the effect of age at diagnosis is captured by the subtypes, as the basal-like tumors are frequently detected in young women. Other pathological traits, such as tumor stage and size, are correlated with tumor grade.
In contrast, we used the PAM50 method (Parker et al. 2009) to directly call subtypes for the same samples from the Guedj cohort (Guedj et al. 2012). The PAM50 method often predicts five subtypes, including a normal-like subtype in addition to the four major subtypes. We then fit a similar model for disease-free survival prediction.
Mutual information for identification of target genes
Mutual information is based on the concept of entropy (Shannon 1948). When a population size is large, mutual information is able to capture the nonlinear regulator–regulon interactions (Basso et al. 2005; Carro et al. 2010). See SI Methods for more details. To stringently control false-positive regulator–regulon interactions, we need to take several filtering steps: (1) permutation tests to adjust for multiple hypothesis testing; (2) bootstrap resampling to obtain a consensus network; and (3) data-processing inequality analysis to identify the most likely paths of information flow (Margolin et al. 2006). Detailed methods for the steps were reported previously (Margolin et al. 2006).
Identification of canonical pathway changes in tumors
To associate DNA somatic mutations with gene regulatory pathway changes in tumor subtypes, we focus on the connection between subtype-enriched somatic mutations and subtype-specific pathway amplification, comparing corresponding gene expression between the TCGA tumor samples and the matched normal samples.
Data Available
Data are available at https://tcga-data.nci.nih.gov/.
Results
Identification and validation of master regulator genes
Since genotype–phenotype association is complex in humans, characterization of gene regulatory pathways may bridge the gap between genotype and phenotype.
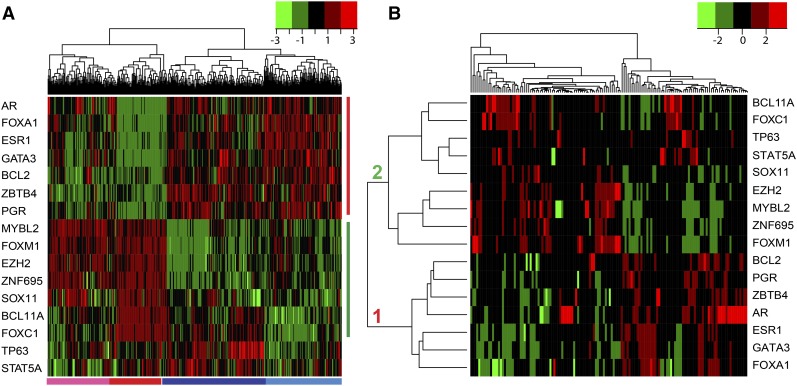
Using the supervised machine learning algorithm as described in Materials and Methods, we have identified an ensemble of 16 genes (Figure 1A). Although the genes are ordered by their ranks in terms of importance (Figure S3), they are an entity that gives rise to the maximum accuracy in classification of the training samples (Figure S4). Furthermore, the 16 genes are among the top ones that are most frequently selected in the repeated computational experiments (Figure S5). Since the gene set plays a significant role in regulating cancer-related genes (will be addressed below), we refer to the 16 genes as master regulators or MR16.
Figure 1.
Patterns of MR gene expression and MR–cancer driver gene interactions in the TCGA breast tumor samples. (A) Heatmap of the MR16 gene expression across the TCGA primary tumor samples. Using the RNA-Seq data and a machine-learning algorithm, we identified the MR16 that can classify the tumor samples into two large clusters: ER-negative and ER-positive. Two subclusters are observed within each of the clusters. The four clusters correspond well to the four tumor subtypes: HER2-enriched (pink bar at the bottom), basal-like (red), luminal A (dark blue), and B (light blue). Vertical red and green bars represent the two groups of the MRs that display distinct expression patterns across the samples. Green and red colors in the heatmap indicate low and high gene expression, respectively. (B) Heatmap of the MR16–cancer driver-related gene interactions. Using the TCGA RNA-Seq data, we computed a mutual information matrix between the MR16 and the cancer driver-related genes. The MR–regulon interaction patterns indicate that two MR groups either up- or down-regulate (in red and green colors, respectively) the cancer driver-related genes in opposite directions.
Using the TCGA data, we then trained a classifier based on the MR16. Application of the classifier to the Curtis (Curtis et al. 2012) and the Guedj (Guedj et al. 2012) data indicated that the luminal A and luminal B samples can be classified with concordance rates of 87 and 85%, respectively, compared to the direct calling by the PAM50 method (Parker et al. 2009). Similarly, HER2-enriched and basal-like tumors can be classified with concordance rates of 90 and 93%, respectively. These intrigued us to predict disease-free survival.
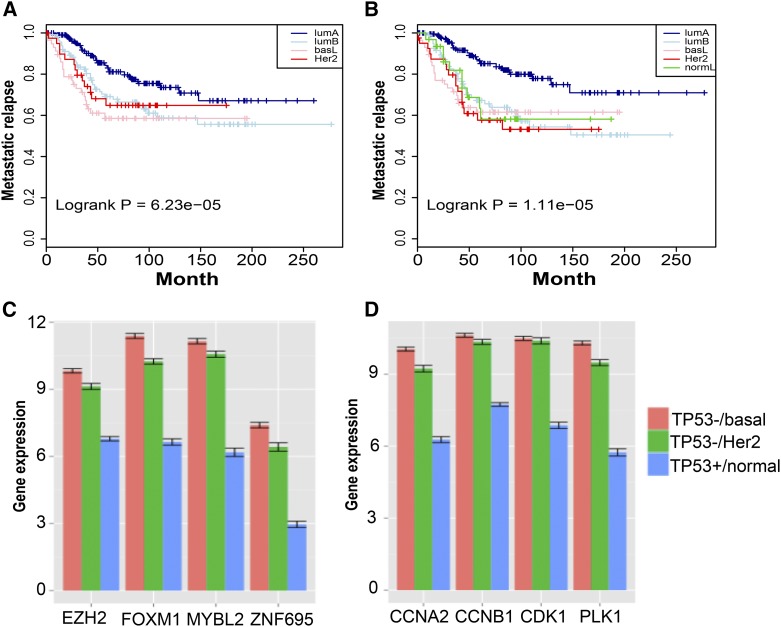
Is the MR16 classifier able to predict disease-free survival in the independent cohorts of breast cancer populations? Since the Guedj cohort has a long follow-up time for metastasis relapse-free survival (Guedj et al. 2012), we focus on this cohort for survival prediction. Corresponding Kaplan–Meier curves are shown in Figure 2A. In contrast, we used the PAM50 method (Parker et al. 2009) to directly call subtypes for the same samples, and corresponding Kaplan–Meier curves are shown in Figure 2B. Comparisons of the two sets of Kaplan–Meier curves indicate that the four major subtypes share similar disease-free survival, indicating that the MR16 has a cross-cohort prediction power similar to the direct prediction by the PAM50. These prediction results are consistent with clinical observations that luminal A tumors display a significantly (log rank P = 6.25 × 10-5) better prognosis, compared to the other subtypes.
Figure 2.
Prediction of disease-free survival by the MR16 and cell cycle pathway changes in ER-negative breast tumors. (A and B) Kaplan–Meier curves of metastatic relapse-free survival. (A) Using the MR16 and TCGA RNA-Seq data, we trained a classifier that was then applied to the Guedj data (Guedj et al. 2012) for subtype classification. We then fit a COX proportional hazards regression model (Cox and Oakes 1984) with tumor grade as a covariate. Kaplan–Meier curves of disease-free survival are shown by subtype. (B) Using the same set of samples from the Guedj data (Guedj et al. 2012), we applied the PAM50 method (Parker et al. 2009) to directly call subtypes, which resulted in five subtypes including a normal-like subtype. Corresponding Kaplan–Meier curves are plotted. Comparisons of the two sets of Kaplan–Meier curves indicate that luminal A tumors are predicted to have better prognosis. (C and D) Association of TP53 somatic mutations with significant overexpression of cell cycle pathways in ER-negative tumors. Using matched tumor samples with TP53 mutations and matched normal samples without TP53 mutations, we compare the expression of representative MRs and cell cycle genes. The y-axis is the averaged gene expression by RNA-Seq in log2_FPKM. Standard errors are plotted on top of the bars. Red, basal-like tumor samples; green, HER2-enriched tumors; blue, normal samples.
Gene expression patterns of the MR16
Gene expression of the MR16 can accurately separate the TCGA primary breast tumor samples into four clusters that correspond to the four major subtypes (Figure 1A). The two large clusters represent luminal (or ER-positive) and nonluminal (or ER-negative) tumors, respectively, indicating that ER status is the major separating factor between the tumor samples. Within each of the main clusters, two subclusters have been classified. In the ER-positive cluster, luminal A and B tumors can be separated by the expression pattern of a combination of the genes, including BCL11A, FOXC1, TP63, and STAT5A (Figure 1A). In the ER-negative cluster, basal-like tumors differ from HER2-enriched tumors mainly by BCL11A and FOXC1 gene expression (Figure 1A). These four clusters correspond well to the four breast tumor subtypes: luminal A, luminal B, HER2-enriched, and basal-like.
Expression of the 16 genes across the TCGA samples has displayed distinct patterns (Figure 1A; Figure S3). In general, the MR16 can be divided into two groups. Group 1 genes, including ESR1, FOXA1, GATA3, PGR, AR, BCL2, and ZBTB4, are highly expressed in luminal tumors, but lowly expressed in nonluminal tumors (Figure 1A). Conversely, group 2 genes, including FOXM1, EZH2, ZNF695, MYBL2, SOX11, BCL11A, and FOXC1, are highly expressed in the nonluminal, especially the basal-like subtype, but lowly expressed in the luminal tumors (Figure 1A).
As expected, similar gene expression patterns have been observed in the Curtis and Guedj cohorts (Figure S6). The same group 1 MRs as defined in the TCGA cohort (Figure 1A) are overexpressed in the luminal tumors, but lowly expressed in the nonluminal tumors. In contrast, the same group 2 MRs are overexpressed in the nonluminal tumors, but lowly expressed in the luminal tumors. The samples in the Curtis validation data display two large clusters that correspond to the luminal and nonluminal tumors (Figure S6A). In each of the two clusters, there are two subclusters. In general, the four clusters correspond to the four major subtypes. The samples in the Guedj cohort also exhibit two large clusters that correspond to the luminal and nonluminal tumors (Figure S6B). In each of the two clusters, the subclusters are more complicated, compared to the TCGA and the Curtis cohorts. This may suggest that there exist more subtypes in the Guedj cohort. The consistent gene expression patterns of the MR16 across different cohorts of breast cancer populations call for further investigation.
Interaction patterns between the MR16 and cancer driver-related genes
What are the relationships between the MR16 and the cancer driver genes? We can approach this question either by performing either simple correlation analysis, which is based on the linearity assumption, or by providing mutual information that is a generalized correlation without such an assumption. Results from Pearson correlation analysis, based on a subset of the MR16 from the TCGA RNA-Seq data, are shown in Figure S7. It appears that there exist moderate positive correlations for the intragroup MRs. Regarding the intergroup MRs, however, there are weak or no correlations between them. In luminal B and basal-like subtypes, the correlation coefficients between the intergroup MRs are negative.
Since the sample sizes are large in the three cohorts of patient populations, we then applied mutual information to model the interactions between the MR16 and their target genes (regulons). The gene panel for mutual information modeling includes (1) the MR16; (2) the 138 cancer driver genes (Vogelstein et al. 2013); and (3) additional cell cycle genes that are known regulons of FOXM1—PLK1, CCNB1, CCNB2, CDK1, CENPF, and UBE2C (Chen et al. 2013). Only four genes (i.e., AR, EZH2, GATA3, and BCL2) overlap between gene sets 1 and 2. The joint gene sets 2 and 3 are referred to as cancer driver-related genes.
Evidence from mutual information modeling indicates that interactions of the MR16 and the cancer driver-related genes display distinct patterns (Figure 1B). The majority of the cancer driver-related genes are found to be either up- or down-regulated by the MR16. This implicates subtype-specific gene regulations in breast tumors, which will be scrutinized below.
Two groups of the MR16 agonistically regulate cell cycle genes
Tumor growth and progression is a hallmark for aggressiveness. The four main breast tumor subtypes have significant differences in tumor prognosis and survival (Parker et al. 2009). Since cell cycle genes play a significant role in regulating tumor growth and progression, we will focus on the interactions of the MR16 and the cell cycle genes.
Examination of the MR–cell cycle gene interactions across the four subtypes gave rise to noting distinct patterns. Many cell cycle genes, including PLK1 and CDK1, are down-regulated by the group 1 MRs, but up-regulated by the group 2 MRs, especially FOXM1, MYBL2, EZH2, and ZNF695 (Figure 3). It is noteworthy that the two group of MRs are clearly defined in the TCGA cohort and validated in both the Curtis and the Guedj cohorts (Figure 1A; Figure S6), based on the gene expression patterns instead of the MR–regulon interaction patterns.
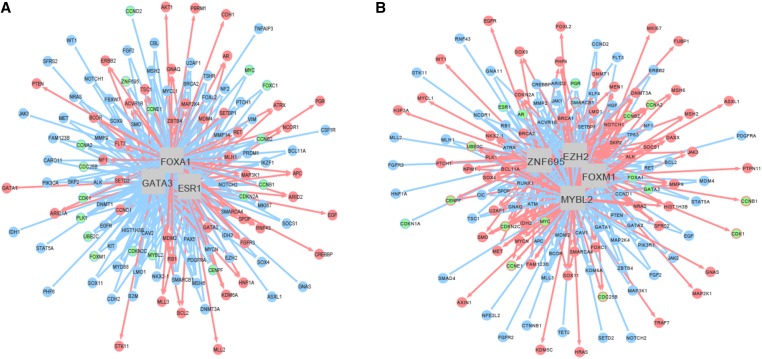
Figure 3.
Cross-subtype gene regulatory networks. The networks are derived from the TCGA RNA-Seq data with 646 primary breast tumor samples across the four major subtypes. The nodes in the circle represent target genes (regulons), and the edges connecting the MRs and the target genes indicate interactions between them. Red and blue colors denote up- and down-regulation, respectively. (A) A gene regulatory network illustrating the interactions between three representative group 1 MRs and regulons. Green nodes are either highlighted cell cycle genes or group 2 MRs. Other group 1 MRs, such as PGR, are up-regulated by the central regulators. However, group 2 MRs, such as FOXM1 and ZNF695, and cell cycle genes, such as CCNB1 and CDK1, are down-regulated by the central MRs. (B) A gene regulatory network illustrating the interactions between four group 2 MRs and regulons. Green nodes are highlighted for either cell cycle genes or group 1 MRs. Other group 2 MRs, such as SOX11 and the cell cycle genes, are up-regulated by the central MRs. However, group 1 MRs, such as ESR1 and AR, are down-regulated by the central MRs.
We then scrutinized subtype-specific networks in the TCGA cohort. The MR–cell cycle gene interaction patterns are consistent with that observed across the subtypes (Figure S8). Furthermore, the group 2 MRs, including FOXM1, MYBL2, EZH2, and ZNF695, cooperatively up-regulate the cell cycle genes in the basal-like tumors. Similar results are observed in the HER2-enriched tumors (data not shown). On the contrary, the group 1 MRs, such as ESR1, FOXA1, and GATA3, cooperatively down-regulate the cell cycle genes in the luminal tumors. These examples clearly demonstrate that MR–cell cycle gene interactions behave in a subtype-specific fashion.
Although the cell cycle genes are known target genes for FOXM1 and MYBL2 (Chen et al. 2013), the emergence of at least two other partners (EZH2 and ZNF695) in regulating the cell cycle genes is new.
Validation of the regulator–regulon interactions
We then used the Curtis and the Guedj cohorts to validate the MR–cell cycle gene interactions. We focus on validation of the regulator–regulon interaction patterns, for example, the interactions between the two groups of MRs and the cell cycle genes. We are not testing a specific regulator–regulon interaction, since the different population sizes can affect the filtering steps, which may result in a discrepant report of the interaction.
Using the same methods, we modeled the MR–regulon interactions in both the Curtis and the Guedj cohorts. As expected, the MR–cell cycle gene interaction patterns are congruent; that is, the group 1 MRs down-regulate expression of the cell cycle genes in ER-positive tumors; in contrast, the group 2 MRs up-regulate expression of the genes in ER-negative tumors (Figure S9 and Figure S10). The subtype-specific interactions imply that the cell cycle pathways play a critical role in promoting progression of ER-negative tumors.
Association of DNA somatic mutations with gene regulatory pathways
Since breast cancer is a heterogeneous disease in which various subclones from a tumor can exhibit different mutation profiles (Cancer Genome Atlas Network 2012), identification of canonical pathway changes in each subtype has therapeutic implications.
It is noteworthy that DNA somatic mutations in cancer driver genes are unevenly distributed across tumor subtypes (Cancer Genome Atlas Network 2012). For example, TP53 is one of the top three genes (TP53, PIK3CA, and GATA3) that are most frequently mutated in breast cancer (Cancer Genome Atlas Network 2012). Regarding mutation types, 60% of the TP53 mutations are missense mutations that lead to dysfunctional proteins (Table S1). Interestingly, the TP53 mutations are overwhelmingly enriched in HER2-enriched and basal-like subtypes (Cancer Genome Atlas Network 2012).
Since the TP53 gene is known to play a key role in regulating cell cycle arrest and apoptosis (Amundson et al. 1998), we then managed to associate the subtype-specific TP53 somatic mutations with the overexpression of the cell cycle pathways in the ER-negative breast tumors. Using breast tumor samples with TP53 mutations and matched normal samples without TP53 mutations, we examined expression of representative group 2 MRs and the cell cycle genes as well. Results from TCGA RNA-Seq data indicate that the four group 2 MRs (FOXM1, MYBL2, EZH2, and ZNF695) are significantly (P = 0.002) overexpressed in both HER2-enriched and basal-like subtypes, compared to the normal samples (Figure 2C). Similarly, representative cell cycle genes are significantly overexpressed in the two subtypes (Figure 2D). In contrast, these are not observed in luminal A and B subtypes (data not shown). These clearly indicate the association of TP53 somatic mutations with up-regulated cell cycle pathways in ER-negative tumors. Furthermore, the association reflects canonical genomic alternations in the ER-negative breast tumors.
Discussion
We have identified an ensemble of 16 master regulators as an entity that has the maximum prediction power in the training data. In terms of subtype classification and survival prediction, the MR16 is comparable to the PAM50 (Parker et al. 2009). Compared to the PAM50, the MR16 has only six overlapping genes (ESR1, PGR, FOXA1, FOXC1, BCL2, and MYBL2). Among the top four genes (i.e., ESR1, GATA3, FOXM1, and EZH2) in the MR16, three (GATA3, FOXM1, and EZH2) are nonoverlapping genes. The GATA3 gene is the key regulator of luminal progenitor cell differentiation (Carr et al. 2012). Previous studies have indicated that FOXM1 is a key regulator of cell proliferation and is overexpressed in many cancer types including breast cancer (Kwok et al. 2010). In the present study, FOXM1 and EZH2 are critical genes that play a significant role in regulation of the cell cycle gene expression in ER-negative tumors. However, the three genes are missed in the PAM50. Furthermore, results from mutual information modeling indicate that the majority of the PAM50 genes are predicted to be target genes of the MR16 (Figure S11).
Identification of gene regulatory pathways is critical for mechanistic understanding of genotype–phenotype dependency. The molecular subtypes in human breast cancer are associated with significant differences in prognosis and survival (Parker et al. 2009). Our studies have indicated that there exist intrinsic connections between specific molecular pathways and the subtypes. Furthermore, the pathways are regulated by the two groups of the MR16. Therefore, gene regulatory mechanisms play a significant role in shaping the subtypes.
Statistical evidence from information theory, based on three large cohorts of breast cancer populations, has implicated a gene regulatory program related to breast tumor progression. The program is characteristic of two types of gene–gene interactions: MR–MR and MR–regulon interactions.
Regarding MR–MR interactions, the intragroup MRs interact in a hierarchical and synergistic fashion. Recent studies using co-immunoprecipitation and ChIP-Seq technologies have indicated that group 1 MRs, including FOXA1, ESR1, and GATA3, are involved in the same protein complex (Theodorou et al. 2013). However, interactions of the four main group 2 MRs (FOXM1, MYBL2, EZH2, and ZNF695) are less known, although FOXM1 and MYBL2 are known to cooperatively bind to the promoter regions of the cell cycle genes (Wang and Gartel 2011; Chen et al. 2013). EZH2 is a member of the polycomb repressive complex 2 (PRC2) that methylates lysine 27 of histone H3 (H3K27). The canonical function of EZH2 is repression of tumor suppressor genes through H3K27me3 (Yamaguchi and Hung 2014). On the other hand, accumulating evidence has indicated that EZH2 is able to activate target genes by directly binding to the regulatory regions (Lee et al. 2011; Gonzalez et al. 2014). ZNF695 is a gene with little function known to date. Interestingly, the MR16 are all TF genes, although 115 non-TF cancer driver genes (Vogelstein et al. 2013), including HER2 (or ERBB2) and PTEN, were incorporated in the gene panel for feature selection.
Contrary to the synergistic intragroup MR–MR interactions, the intergroup MR–MR interactions operate in an agnostic fashion. An interesting question is, “What is the biological mechanism underlying the inhibition between the two groups of MRs?” A recent study by Carr et al. (2012) indicated that FOXM1 represses GATA3 gene expression in the mammary gland by recruiting the methyltransferase DNMT3b to the binding sites within the GATA3 promoter, thereby leading to methylation-induced gene silencing and the undifferentiated state of luminal progenitors. More in-depth studies are needed to elucidate the intra- and intergroup MR–MR interactions.
Regarding MR–regulon interactions, the two groups of MRs either up- or down-regulate different subsets of cancer driver-related genes in a subtype-specific fashion. This may help us understand pathway interactions in each tumor subtype. For example, the group 2 MRs are highly expressed in HER2-enriched and basal-like tumors. Correspondingly, the cell cycle genes are cooperatively up-regulated by the four major MRs in group 2. These two lines of evidence indicate that cell cycle pathways are overexpressed in the two subtypes, but not in the luminal A and B subtypes. Furthermore, the subtype-specific amplification of the cell cycle pathways coincides with the subtype-specific TP53 mutations. Therefore, it is suggested that TP53 mutations and corresponding cell cycle pathway amplification represent canonical genomic alternations in HER2-enriched and basal-like tumors.
Since tumor heterogeneity on the DNA level has challenged clinical breast cancer care (Yates et al. 2015), the present study has illustrated the benefit of combining both DNA mutations and gene expression in the identification of canonical pathway changes in human breast cancer. Identification of canonical genomic alternations is critical for precision medicine.
Supplementary Material
Acknowledgments
We thank The Cancer Genome Atlas network; C. Curtis and the collaborative consortium; and M. Guedj and colleagues for generating the data.
Author contributions: R.L. provided study design, data analysis, and paper writing; J.C. executed data process and management; and J.I. collaborated in cancer biology research.
Footnotes
Communicating editor: S. K. Sharan
Supporting information is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.180125/-/DC1.
Literature Cited
- Amundson S. A., Myers T. G., Fornace A. J., Jr, 1998. Roles for p53 in growth arrest and apoptosis: putting on the brakes after genotoxic stress. Oncogene 17: 3287–3299. [DOI] [PubMed] [Google Scholar]
- Basso K., Margolin A. A., Stolovitzky G., Klein U., Dalla-Favera R., et al. , 2005. Reverse engineering of regulatory networks in human B cells. Nat. Genet. 37: 382–390. [DOI] [PubMed] [Google Scholar]
- Berger, J., 1985 Satistical Decision Theory and Bayesian Analysis. Springer Series in Statistics, Ed. 2. Springer-Verlag, Berlin; Heidelberg, Germany; New York. [Google Scholar]
- Bjorkegren J. L., Kovacic J. C., Dudley J. T., Schadt E. E., 2015. Genome-wide significant loci: How important are they? Systems genetics to understand heritability of coronary artery disease and other common complex disorders. J. Am. Coll. Cardiol. 65: 830–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiman L., 2001. Random Forest. Mach. Learn. 45: 5–32. [Google Scholar]
- Cancer Genome Atlas Network, 2012. Comprehensive molecular portraits of human breast tumours. Nature 490: 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr J. R., Kiefer M. M., Park H. J., Li J., Wang Z., et al. , 2012. FoxM1 regulates mammary luminal cell fate. Cell Reports 1: 715–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carro M. S., Lim W. K., Alvarez M. J., Bollo R. J., Zhao X., et al. , 2010. The transcriptional network for mesenchymal transformation of brain tumours. Nature 463: 318–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Muller G. A., Quaas M., Fischer M., Han N., et al. , 2013. The forkhead transcription factor FOXM1 controls cell cycle-dependent gene expression through an atypical chromatin binding mechanism. Mol. Cell. Biol. 33: 227–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox D., Oakes D., 1984. Analysis of Survival Data. Chapman & Hall, New York. [Google Scholar]
- Curtis C., Shah S. P., Chin S. F., Turashvili G., Rueda O. M., et al. , 2012. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486: 346–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher M. N., Castro M. A., Wang X., de Santiago I., O’Reilly M., et al. , 2013. Master regulators of FGFR2 signalling and breast cancer risk. Nat. Commun. 4: 2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez M., Moore H., Li X., Toy K., Huang W., et al. , 2014. EZH2 expands breast stem cells through activation of NOTCH1 signaling. Proc. Natl. Acad. Sci. USA 111: 3098–3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guedj M., Marisa L., de Reynies A., Orsetti B., Schiappa R., et al. , 2012. A refined molecular taxonomy of breast cancer. Oncogene 31: 1196–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein C., 2013. Selection and adaptation during metastatic cancer progression. Nature 501.: 365–372. [DOI] [PubMed] [Google Scholar]
- Kwok J., Peck B., Monteiro L., Schwenen H. D., Millour J., et al. , 2010. FOXM1 confers acquired cisplatin resistance in breast cancer cells. Mol. Cancer Res. 8: 24–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Li Z., Wu Z., Aau M., Guan P., et al. , 2011. Context-specific regulation of NF-κB target gene expression by EZH2 in breast cancers. Mol. Cell 43: 798–810. [DOI] [PubMed] [Google Scholar]
- Lehmann, E., and H. J. M. D’Abrera, 1988 Nonparametrics: Statistical Methods Based on Ranks. New York: McGraw-Hill.
- Li R., Churchill G., 2010. Epistasis contributes to the genetic buffering of plasma HDL cholesterol in mice. Physiol. Genomics 42A: 228–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Tsaih S. W., Shockley K., Stylianou I. M., Wergedal J., et al. , 2006. Structural model analysis of multiple quantitative traits. PLoS Genet. 2: e114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin A. A., Nemenman I., Basso K., Wiggins C., Stolovitzky G., et al. , 2006. ARACNE: an algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinformatics 7(Suppl 1): S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michailidou K., Beesley J., Lindstrom S., Canisius S., Dennis J., et al. , 2015. Genome-wide association analysis of more than 120,000 individuals identifies 15 new susceptibility loci for breast cancer. Nat. Genet. 47: 373–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nik-Zainal S., Van Loo P., Wedge D. C., Alexandrov L. B., Greenman C. D., et al. , 2012. The life history of 21 breast cancers. Cell 149: 994–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker J. S., Mullins M., Cheang M. C., Leung S., Voduc D., et al. , 2009. Supervised risk predictor of breast cancer based on intrinsic subtypes. J. Clin. Oncol. 27: 1160–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perou C. M., Sorlie T., Eisen M. B., van de Rijn M., Jeffrey S. S., et al. , 2000. Molecular portraits of human breast tumours. Nature 406: 747–752. [DOI] [PubMed] [Google Scholar]
- Shannon C., 1948. A mathemetical theory for communication. Bell Syst. Tech. J. 27: 379–423. [Google Scholar]
- Theodorou V., Stark R., Menon S., Carroll J. S., 2013. GATA3 acts upstream of FOXA1 in mediating ESR1 binding by shaping enhancer accessibility. Genome Res. 23: 12–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolaney S.M., Winer E. P., 2007. Follow-up care of patients with breast cancer. Breast 16(Suppl 2): S45–S50. [DOI] [PubMed] [Google Scholar]
- Vaquerizas J. M., Kummerfeld S. K., Teichmann S. A., Luscombe N. M., 2009. A census of human transcription factors: function, expression and evolution. Nat. Rev. Genet. 10: 252–263. [DOI] [PubMed] [Google Scholar]
- Vogelstein B., Papadopoulos N., Velculescu V. E., Zhou S., Diaz L. A., Jr, et al. , 2013. Cancer genome landscapes. Science 339: 1546–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Gartel A. L., 2011. The suppression of FOXM1 and its targets in breast cancer xenograft tumors by siRNA. Oncotarget 2: 1218–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi H., Hung M. C., 2014. Regulation and role of EZH2 in cancer. Cancer Res. Treat. 46: 209–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates L. R., Gerstung M., Knappskog S., Desmedt C., Gundem G., et al. , 2015. Subclonal diversification of primary breast cancer revealed by multiregion sequencing. Nat. Med. 21: 751–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.