Abstract
Rice (Oryza sativa) grain shape, which is controlled by quantitative trait loci (QTL), has a strong effect on yield production and quality. However, the molecular basis for grain development remains largely unknown. In this study, we identified a novel QTL, Slender grain on chromosome 7 (SLG7), that is responsible for grain shape, using backcross introgression lines derived from 9311 and Azucena. The SLG7 allele from Azucena produces longer and thinner grains, although it has no influence on grain weight and yield production. SLG7 encodes a protein homologous to LONGIFOLIA 1 and LONGIFOLIA 2, both of which increase organ length in Arabidopsis. SLG7 is constitutively expressed in various tissues in rice, and the SLG7 protein is located in plasma membrane. Morphological and cellular analyses suggested that SLG7 produces slender grains by longitudinally increasing cell length, while transversely decreasing cell width, which is independent from cell division. Our findings show that the functions of SLG7 family members are conserved across monocots and dicots and that the SLG7 allele could be applied in breeding to modify rice grain appearance.
Keywords: rice, quantitative trait loci, grain shape, cell elongation
RICE (Oryza sativa L.) is a staple food for half of the world’s population (Khush 2001). Three major components, panicle number per plant, grain number per panicle, and grain weight, determine rice yield production. Grain weight is associated with grain size and shape, which are defined as grain length, grain width, and grain thickness (Duan et al. 2014). There is a striking diversity of grain size among the rice species worldwide. The grains of domesticated rice range from 3 to 11 mm in length and from 1.2 to 3.8 mm in width (Fitzgerald et al. 2009). Despite the influence of several environmental factors on plant growth and development, such as water supply and fertilizer level, the final grain size of rice is reasonably constant within a given species.
Rice grain traits are quantitatively inherited. In the past decade, several quantitative trait loci (QTL) controlling grain size and shape have been cloned. GS3, encoding a transmembrane protein containing four putative domains, was the first characterized QTL that regulates grain length (Fan et al. 2006). qGL3 encodes a putative protein phosphatase with a Kelch-like repeat domain, and an aspartate-to-glutamate transition in the second Kelch domain leads to a long-grain phenotype (Zhang et al. 2012). GW6 encodes a GNAT-like protein that harbors intrinsic histone acetyltransferase activity, and an elevated expression enhances grain length and weight by enlarging spikelet hulls and accelerating grain filling (Song et al. 2015). GW2, GW5/qSW5, GS5, and GW8 were identified as regulators of rice grain width. GW2 encodes a previously unknown RING-type protein with E3 ubiquitin ligase, which negatively regulates cell division by degrading its substrate(s) through the ubiquitin-proteasome pathway (Song et al. 2007). GW5/qSW5 encodes a nuclear-located protein that physically interacts with polyubiquitin. Loss of function of GW5/qSW5 leads to a significant increase in sink size, reflecting an increase in cell number in the outer glume of the rice flower (Shomura et al. 2008; Weng et al. 2008). GS5 encodes a putative serine carboxypeptidase (Li et al. 2011), and GW8 encodes squamosa promoter-binding protein-like 16, which belongs to the SBP domain family of transcription factors (Wang et al. 2012). Both are positive regulators of grain width. Higher expression of these two genes contributed to larger grain size through their effects on the cell-cycle machinery. Additionally, the GIF1 gene encodes a cell-wall invertase required for carbon partitioning during early grain filling (Wang et al. 2008), and TGW6 encodes an indole-3-acetic acid-glucose hydrolase affecting the transition from the syncytial to the cellular phase of the endosperm (Ishimaru et al. 2013). GIF1 and TGW6 directly affect the endosperm size and consequently regulate grain size and weight. GS3, qSW5/GW5, GS5, GW8, GIF1, and TGW6 have been artificially selected and widely used in rice-breeding programs. However, the advantageous alleles of GW2, qGL3, and GW6 are present in rare varieties or subspecies and have not been actively exploited.
Most genes involved in rice panicle development, such as SP1, qPE9-1, EP2, DEP2, and DEP3, influence grain size or shape (Li et al. 2009, 2010; Zhou et al. 2009; Zhu et al. 2010; Qiao et al. 2011). Moreover, based on the phenotypic analyses of null mutants, the genes involved in gibberellin and brassinosteroid biosynthesis or signaling also modulate rice grain size (Ashikari et al. 1999; Yamamuro et al. 2000; Hong et al. 2003; Tanabe et al. 2005; Nakagawa et al. 2012). These studies suggested that the control of grain size in rice is complicated. To further reveal the molecular mechanism that determines grain size, genes that affect grain development were identified using mutants and backcross populations as genetic materials. In this study, we isolated and characterized a QTL, Slender grain on chromosome 7 (SLG7), which may produce longer and thinner grains and, if applied to breeding, will significantly improve rice grain yield and appearance quality.
Materials and Methods
Plant materials and growth conditions
A set of backcrossed introgression lines (ILs) derived from a cross between Azucena and 9311 was developed to clone QTL for grain size or shape. Their F1 plants were backcrossed with 9311 for four times, and the BC4F1 seeds were selfed to produce BC4F4 lines (Supporting Information, Figure S1). One of the lines, IL5084, carrying an introgressed segment of chromosome 7 from Azucena, showed extremely slender grains. Rough mapping and fine mapping of SLG7 was carried out using the segregating populations derived from IL5084 and 9311. From the BC5F3 generation, a pair of near-isogenic lines (NILs), NIL-SLG7 and NIL-slg7, was developed to evaluate the genetic effect. NIL-SLG7 carried a small segment including the Azucena SLG7 allele in the 9311 background, whereas its matching line, NIL-slg7, had the homologous segment from 9311. All the plants were grown in the experimental field of Yangzhou University (E 119°25′/N 32°23′). Field management and control of diseases and pests followed the standard procedures to prevent yield loss during the growth period.
Mapping of SLG7
In each generation, short-grain plants (grain length < 10.5 mm) in the derived segregating populations were used to screen recombinants. Fine mapping was performed using a total of 2500 short-grain plants in the BC5F3 and BC5F4 populations. SLG7 was finally narrowed into a 60-kb interval between markers S7-272 and S7-258, and cosegregated with S7-257. The molecular markers used for fine mapping SLG7 are shown in Table S3. The SLG7 candidate genes from Azucena and 9311 genomic DNA were amplified and sequenced. The primers are listed in Table S4.
RNA extraction, complementary DNA synthesis, and qRT-PCR
Total RNA from various organs was isolated using an RNA extraction kit following the manufacturer’s instruction (Beijing Tiangen Biotechnology, http://www.tiangen.com/). First-strand complementary DNA (cDNA) was reverse transcribed from ∼1 µg gDNase-treated RNA using a FastQuant RT Kit following the manufacturer’s instruction (Beijing Tiangen Biotechnology). Gene expression analyses were measured using quantitative real-time reverse transcription-PCR (qRT-PCR) with the rice Actin gene as control. The qRT-PCR was performed in a total volume of 25 µl, containing 2 µl of the cDNA, 0.2 mM of each primer, and 12.5 µl of 2× SYBR green PCR master mix (Takara, http://www.takara.com.cn). The qRT-PCR was carried out using an ABI ViiA7 real-time PCR system using the following program: 95° for 3 min and then 40 cycles of 94° for 30 sec, 55° for 30 sec, and 72° for 40 sec. Comparison of the expression level was done with the 2-ΔΔCT method.
Vector construction and plant transformation
To confirm the target gene of SLG7, the full coding regions of ORF1 and ORF2 were amplified from Azucena cDNA and were inserted into the p1300-Actin-3*flag vector to generate two overexpression constructs, pActin::ORF1Azucena and pActin::ORF2Azucena. For the complementary test, a DNA fragment ∼2 kb upstream of the ORF1 translation start site was amplified from Azucena genomic DNA. Both segments were then cloned into the binary pCAMBIA1301 vector to generate a pORF1::ORF1Azucena construct in which the Azucena ORF1 can be driven by its native promoter. All the constructs were transformed by Agrobacterium tumefaciens-mediated transformation (Hiei et al. 1994).
Subcellular localization
For subcellular localization analysis, the coding sequence of SLG7 was fused in-frame with the green fluorescent protein (GFP) coding sequence in the p163-GFP vector to generate CaMV35S::SLG7-GFP. CaMV35S::GFP was used as a control. These two fusion constructs were transfected into rice Nipponbare protoplasts. The transformed protoplasts were examined using a confocal microscope (LSM 700, Leica).
Morphological and cellular analyses
Fresh spikelets from the panicles of NIL-SLG7 and NIL-slg7 at heading time were directly observed with a scanning electron microscope (SEM). For histology, spikelets were fixed in 2% glutaraldehyde, dehydrated in a graded ethanol series, and embedded in Spurr resin. The transverse sections of each spikelet were produced using an ultramicrotome (EM UC7, Leica), then stained with 0.5% toluidine blue, and viewed using a microscope (DM1000, Leica). The areas of the outer parenchyma cell layer of the spikelet palea and lemma were measured using Image J software.
Data availability
Materials and plasmids described here and in the Supporting Information are available upon request.
Results
Identification of a major QTL for slender grains on chromosome 7
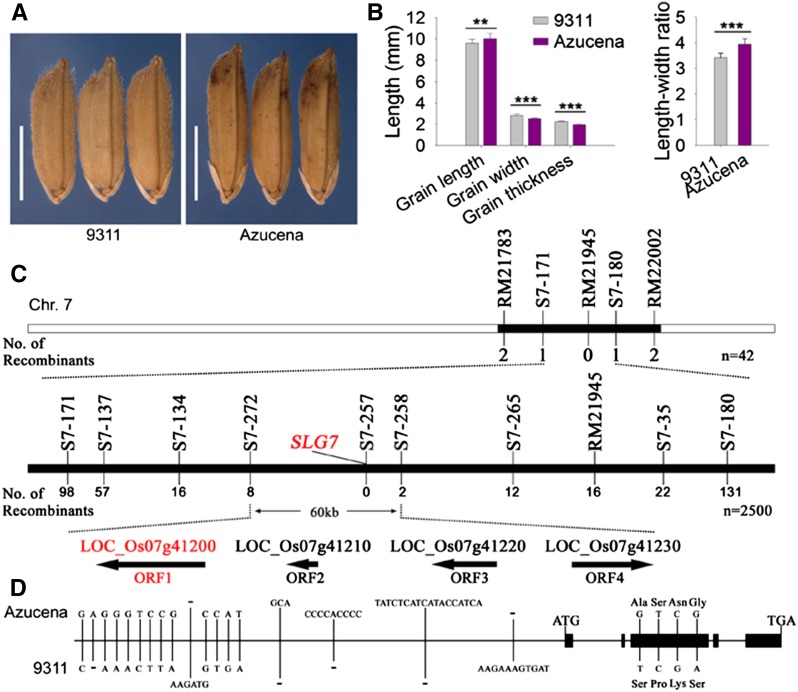
To explore the genetic control of rice grain size and shape, we developed a set of backcrossed introgression lines derived from the cross between Azucena and 9311 (Figure S1). The average length and width of 9311 grains were 9.62 ± 0.36 mm and 2.82 ± 0.14 mm, respectively. By contrast, the average grain length and width of Azucena were 10.03 ± 0.50 mm and 2.54 ± 0.06 mm, respectively (Figure 1, A and B). Using this IL population, several QTL for grain weight and size were mapped (Y. Zhou and G. Liang, unpublished data). One of the BC4F4 lines, IL5084, produced longer but thinner grains than 9311. To reveal the genetic basis of this phenotypic variance, we made a cross between IL5084 and 9311 to generate the derived segregating populations. The distributions of grain traits in a random BC5F3 subpopulation of 400 individuals are illustrated in Figure S2. The ratio of the grain length and length–width ratio fitted well to the expected 3:1 ratio of single locus Mendelian segregation (χ2c = 1.92 < χ20.05,1 = 3.84; χ2c = 2.25 < χ20.05,1 = 3.84, respectively). However, no bimodal distribution was observed in grain width. We also found that grain length was negatively associated with grain width (r = −0.325). Linkage analysis suggested that a major gene, SLG7, located between molecular markers RM21783 and RM22002 on chromosome 7, was responsible for the grain-shape variation (Figure 1C).
Figure 1.
Mapping of SLG7. (A) Grain phenotypes of parents (9311 and Azucena). Bar, 5 mm. (B) Comparison of grain length, grain width, grain thickness, and length–width ratio between 9311 and Azucena. Data are given as mean ± SE (n > 10). Significant at ***0.1% and **1%. (C) Fine mapping of SLG7. SLG7 was primarily mapped between markers S7-171 and S7-180 on chromosome 7 using 42 recessive plants from the BC5F2 segregating population. Then SLG7 was fine-mapped into a 60-kb interval between markers S7-272 and S7-258 and cosegregated with S7-257, using 2500 recessive plants from the BC5F3 and BC5F4 populations. The numbers under each marker indicate the number of recombinants between SLG7 and the molecular markers. Four candidate genes were located within this region, and the target gene is indicated in red. (D) Sequence polymorphisms of SLG7 between 9311 and Azucena.
Fine mapping and cloning of SLG7
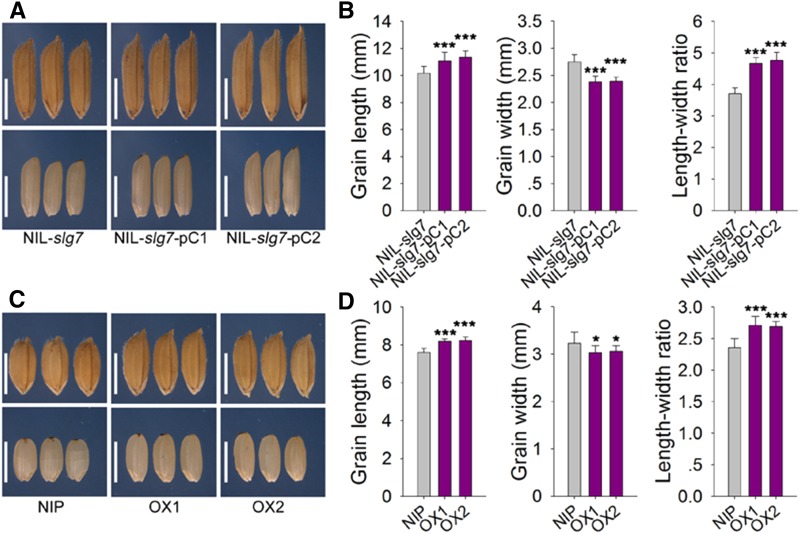
As shown in Figure S2, the grain length showed a bimodal distribution with 10.75 mm as the boundary. Therefore, the plants with short grains (<10.5 mm in length) were considered as recessive individuals, which could be used to screen recombinants. SLG7 was mapped between markers S7-171 and S7-180 using 42 recessive plants from the BC5F2 segregating population (Figure 1C). Fine mapping was further performed using 2500 recessive plants in the BC5F3 and BC5F4 populations. SLG7 was finally narrowed down to a 60-kb interval between markers S7-272 and S7-258, with eight and two recombinants, respectively, and cosegregated with marker S7-257. Within this region, there are four predicted ORFs (Figure 1C). To define the candidate gene of SLG7, the genomic DNA sequences that corresponded to these four ORFs from 9311 and Azucena were sequenced. There was no nucleotide difference between 9311 and Azucena in ORF3 and ORF4, respectively; however, multiple variances were found in ORF1 and ORF2 between the two parents. Thus, ORF1 and ORF2 were considered as good candidates for SLG7. A pair of NILs, NIL-SLG7 and NIL-slg7, was also developed while fine mapping was carried out. NIL-SLG7 carried a small segment including the Azucena SLG7 allele in the 9311 background, whereas its matching line, NIL-slg7, had the homologous segment from 9311. Several constructs were generated and transformed to confirm the candidates. The transgenic NIL-slg7 plants (NIL-slg7-pC1 and NIL-slg7-pC2) produced longer and thinner grains when expressing the Azucena ORF1-coding region under the control of its native promoter (Figure 2, A and B). We also overexpressed the Azucena ORF1-coding region using the Actin promoter in Nipponbare (OX1 and OX2) and found that the grains of the transgenic plants became slender (Figure 2, C and D; Figure S3). However, no visible change in grains was found in the transgenic Nipponbare plants expressing ORF2 using the constitutive Actin promoter (data not shown). Therefore, ORF1 must be the SLG7 gene. Sequence analysis revealed that 12 SNPs and five indels in the promoter region and four SNPs in the coding region were found between 9311 and Azucena (Figure 1D and Table S1). All four SNPs are present in the third exon of SLG7 and cause amino acid residue changes from alanine to serine (Ala462Ser), serine to proline (Ser518Pro), asparagine to lysine (Asn605Lys) and glycine to serine (Gly620Ser) (Figure 1D and Table S1).
Figure 2.
Complementary test and overexpression analysis. (A) Grains (top) and brown rice grains (bottom) of complementary transgenic plants. NIL-slg7-pC1 and NIL-slg7-pC2 are the transgenic lines with pORF1::ORF1Azucena construct under the NIL-slg7 background. Scale bar, 5 mm. (B) Comparison of grain length, grain width, and length–width ratio between NIL-slg7 and the transgenic lines. (C) Grains (top) and brown rice grains (bottom) of overexpression plants. OX1 and OX2 are the transgenic lines with pActin::ORF1Azucena construct under the Nipponbare background. Bar, 5 mm. (D) Comparison of grain length, grain width, and length–width ratio between Nipponbare and the overexpression lines of ORF1Azucena. Data are given as mean ± SE (n > 10). Significant at ***0.1% and *5%.
SLG7 encodes a protein homologous to Arabidopsis LONGIFOLIA1 and LONGIFOLIA2
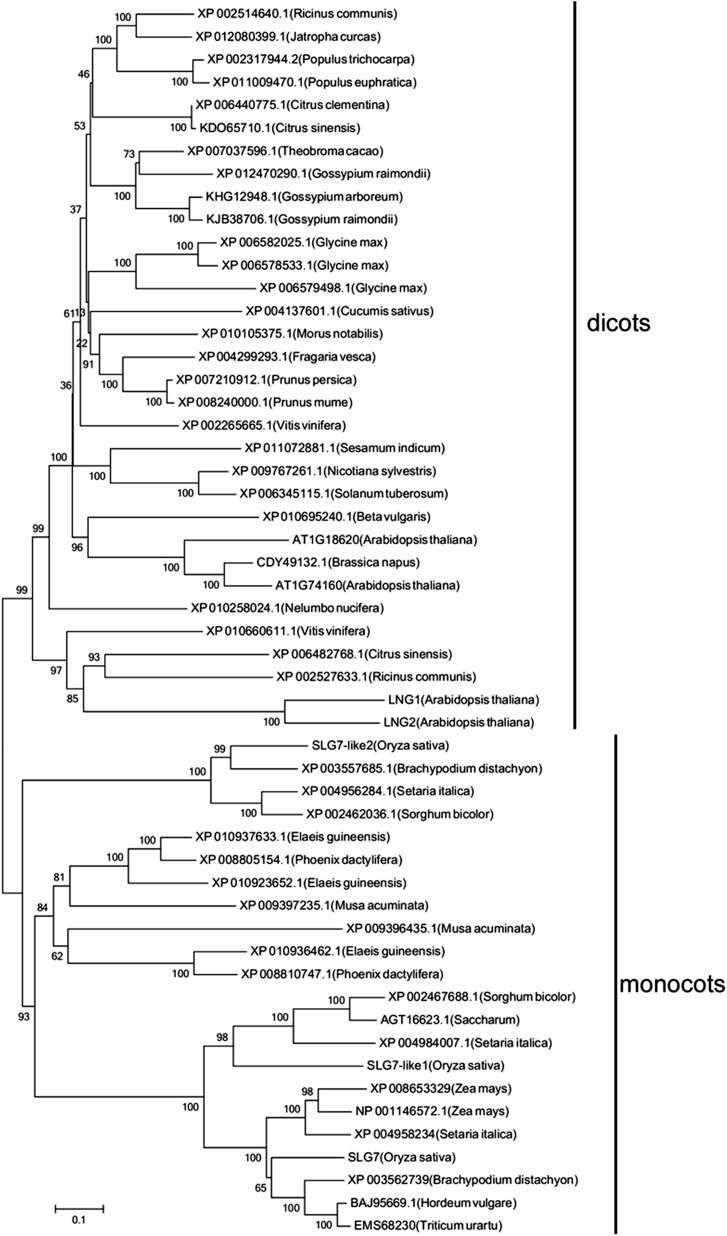
BLASTP searches revealed that the homologs of rice SLG7 are present only in seed plants including gymnosperm and angiosperm. There are two homologs in rice genome, SLG7-like1 (LOC_Os03g30530) and SLG7-like2 (LOC_Os07g01860), having 52% and 30% identities with SLG7, respectively. Both monocot and dicot genomes contained multiple SLG7-like genes (Figure 3). Arabidopsis has four homologs of SLG7, but only two of them, LONGIFOLIA1 (LNG1, At5g15580) and LONGIFOLIA12 (LNG2, At3g02170), were functionally characterized. Both of them activate longitudinal organ expansion (Lee et al. 2006).
Figure 3.
Phylogenetic tree of SLG7-like proteins of the subfamily defined by SLG7. The tree was constructed using MEGA 4.0 based on the neighbor-joining method.
SLG7 inactivates grain and panicle elongation but does not affect yield production
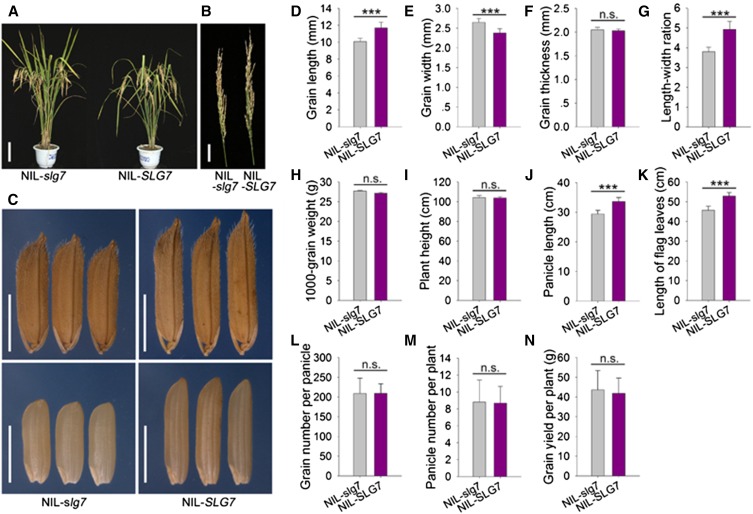
NIL-SLG7 and NIL-slg7 lines were developed to investigate the effect of SLG7 on rice agronomic traits. We confirmed the allele variances of six reported grain-size genes, GS3, GIF1, GW2, GW8, GL3, and GW6, in NIL-SLG7 and NIL-slg7 and found that this pair of NILs shared common alleles at these loci (Table S2). The grain and other agronomic traits of NIL-SLG7 and NIL-slg7 were evaluated under field conditions (Figure 4). The grains of NIL-SLG7 were 19.0% longer and 14.3% narrower than those of NIL-slg7 (Figure 4, C–G). No difference was detected in 1000-grain weight between NIL-SLG7 and NIL-slg7 (Figure 4H), suggesting that the two NILs did not differ from one another with respect to grain weight although their grain shapes were clearly distinct. NIL-SLG7 also produced longer panicles (+14.2%) and flag leaves (+15.8%) compared with NIL-slg7 (Figure 4, A, B, J, and K). No significant differences in plant height and two other rice yield components, grain number per panicle and panicle number per plant, were observed between NIL-SLG7 and NIL-slg7 (Figure 4, A, I, L, and M). As a result, there was no difference in grain yield per plant between the two NILs (Figure 4N). These data indicated that the SLG7 allele from Azucena produces slender grains but has no effect on grain weight and yield production.
Figure 4.
Comparison of the agronomic traits in NIL-slg7 and NIL-SLG7 plants. (A) Plant architecture of NIL-slg7 and NIL-SLG7 plants. Bar, 20 cm. (B) Panicle phenotype of NIL-slg7 and NIL-SLG7 plants. Bar, 5 cm. (C) Phenotypic characterization of grains (top) and brown rice grains (bottom) in NIL-slg7 and NIL-SLG7 plants. Bar, 5 mm. (D–N) Comparison of 11 traits in NIL-slg7 and NIL-SLG7 plants. (D) Grain length. (E) Grain width. (F) Grain thickness. (G) Length–width ratio. (H) 1000-grain weight. (I) Plant height. (J) Panicle length. (K) Length of flag leaves. (L) Grain number per panicle. (M) Panicle number per plant. (N) Grain yield per plant. All data are given as mean ± SE (n > 10). Significant at ***0.1%. n.s., not significant.
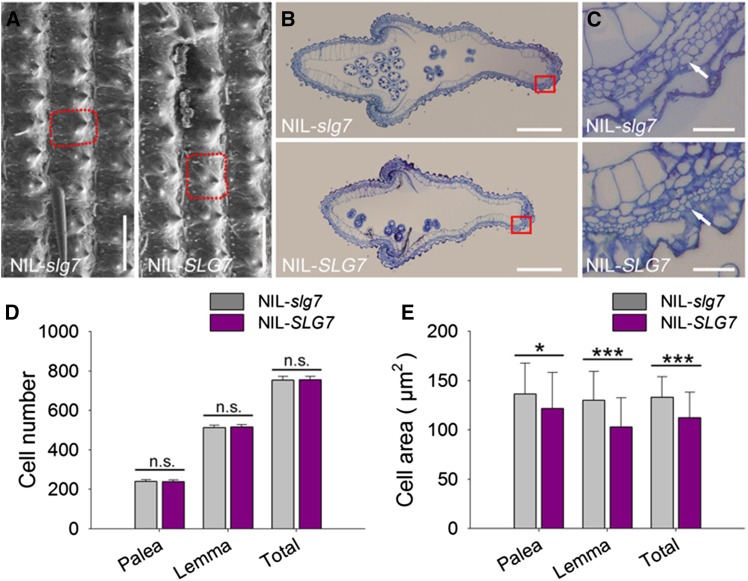
SLG7 promotes cell expansion but has no effect on cell division
To investigate whether cell number or cell size contributes to the development and patterning of grain shape, we compared the length and width of the outer glume epidermal cells using SEM. The outer epidermal cells of NIL-SLG7 were longer but narrower than those of NIL-slg7 (Figure 5A). We further examined the cross sections of spikelet hull central parts between NIL-SLG7 and NIL-slg7 (Figure 5, B and C). The number of outer parenchyma cell layers in NIL-SLG7 did not differ from that in NIL-SLG7 (Figure 5D). However, the cell sizes of outer parenchyma cell layers of the palea and lemma in NIL-SLG7 were 10.8 and 20.8% smaller, respectively, than those in NIL-slg7 (Figure 5E). These results suggested that SLG7 produces slender grains by longitudinally increasing cell length while transversely decreasing cell width, which is independent from cell division.
Figure 5.
The effect of SLG7 on cell number and area in lemma/palea. (A) Scanning electron microscope photos of outer glume surfaces. One of the outer glume cells in NIL-slg7 and NIL-SLG7 hulls, respectively, is boxed by the red dotted line. Bars, 100 μm. (B) Cross section of middle spikelet hulls in NIL-slg7 and NIL-SLG7 plants. Bars, 500 μm. (C) Magnified photos of spikelet hull cross section boxed in B. White arrow shows the outer parenchyma cell layers. Bars, 50 μm. (D) Comparisons of cell number in the cross sections of the outer parenchyma cell layer of NIL-slg7 and NIL-SLG7 hulls. Data are given as mean ± SE (n = 4). n.s., not significant. (E) Comparisons of cell area in the cross sections of the outer parenchyma cell layer of NIL-slg7 and NIL-SLG7 hulls. Data are given as mean ± SE (n = 80). Significant at ***0.1% and *5%.
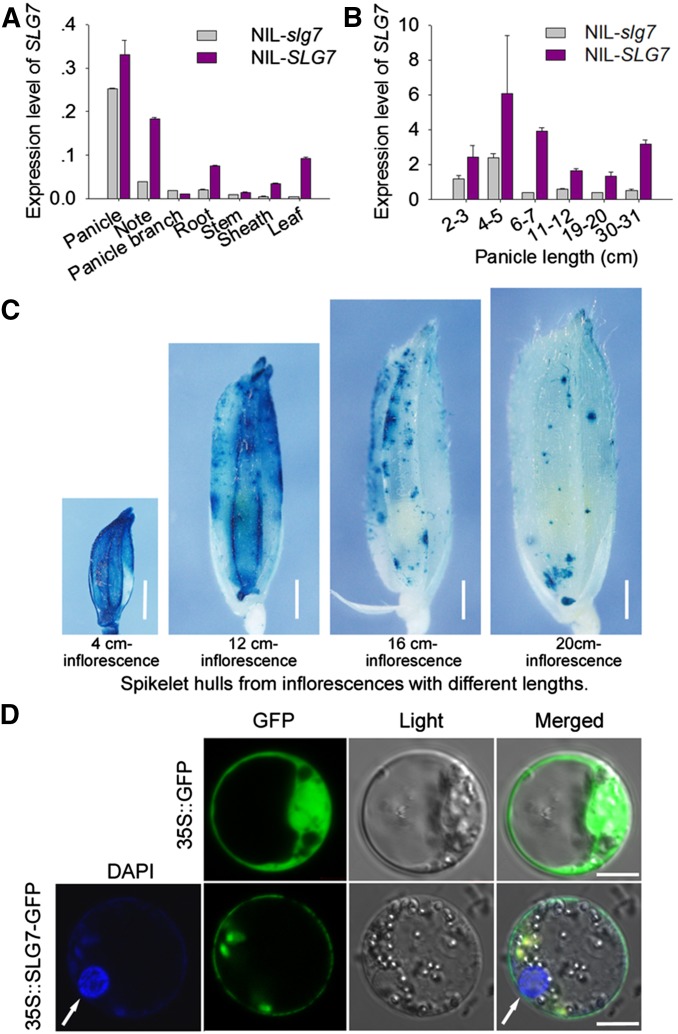
Expression pattern of SLG7 and subcellular localization of the SLG7 protein
qRT-PCR was used to investigate the expression pattern of SLG7. As shown in Figure 6A, SLG7 was constitutively expressed in leaf, panicle, stem, sheath, node, and root. The transcripts in panicle accumulated to a higher level than those in other tissues, which was consistent with the biological function of SLG7 in the regulation of rice inflorescences and grain development. Additionally, the difference of expression level was observed throughout the whole panicle developmental process (Figure 6B). These results implied that the sequence variations in promoter may affect the expression level of SLG7. The expression pattern of SLG7 was further analyzed using transgenic plants expressing the β-glucuronidase (GUS) reporter gene under the control of SLG7 promoter. A strong GUS staining was detected in the spikelet hulls of transgenic lines, and the GUS activities in the spikelet hulls from young inflorescences were much higher than those from developed inflorescences (Figure 6C). These data suggested that the expression of SLG7 decreased with the development of inflorescences and grains. To investigate the subcellular localization of SLG7, we constructed an SLG7-GFP fusion the expression of which was driven by the CaMV 35S promoter. The expression of this fusion protein in rice protoplasts clearly suggested that SLG7 is a plasma membrane protein (Figure 6D).
Figure 6.
Expression pattern of SLG7 and subcellular localization of SLG7 protein. (A) Relative expression levels of SLG7 in different tissues analyzed by qRT-PCR. (B) Quantitative expression analysis of SLG7 in 2- to 3-, 4- to 5-, 6- to 7-, 11- to 12-, 19- to 20-, and 30- to 31-cm stages of NIL-slg7 and NIL-SLG7 panicles. The expression level of the rice Actin gene was amplified as a control. Values are means ± SE of three independent experiments. (C) GUS activities in the spikelet hulls from inflorescences with different lengths. Bars, 1 mm. (D) Subcellular localization of SLG7 protein. Localization of 35S::GFP and 35S::SLG7-GFP in rice protoplast cells is shown. The photos were taken in an optic field to examine cell morphology (light), in a dark field to localize green fluorescence (GFP), and in combination (merged). White arrow shows the nucleus stained by DAPI. Bars, 5 μm.
Sequence diversities of SLG7 in germplasms
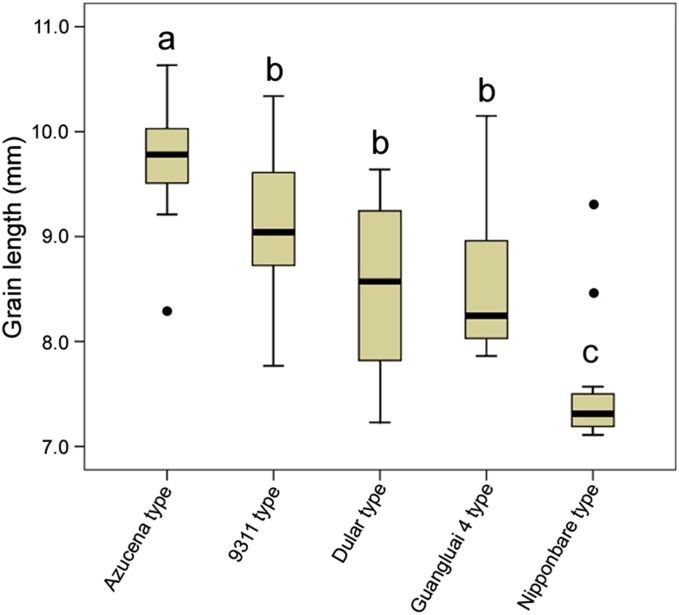
It was detected that allelic variances of SLG7 among 50 rice germplasms from a wide geographic range in Asia and America. The SLG7 alleles were classified into five types, namely Azucena, 9311, Nipponbare, Guangluai 4, and Dular (Table S1). The Nipponbare and Guangluai four haplotypes were exclusively found in japonica and indica subspecies, respectively, while the other three haplotypes were detected in both subspecies. The Azucena SLG7 alleles were identified in 13 germplasms from the United States, the Philippines, Indonesia, and southern China, suggesting that SLG7 is not a rare allele and has been collected during rice breeding. In addition, the varieties carrying the Azucena-type SLG7 allele are significantly higher than those having the other four SLG7 alleles in grain length (Figure 7). This result indicated that SLG7 is a positive factor for grain length and plays a dominant role in shaping extremely long rice grains.
Figure 7.
Comparison of the grain length in the five SLG7 allelic groups. The box plot was generated by the SPSS 10.0 software.
Discussion
Rice grain shape is a determinant of evolutionary fitness and is also an important agronomic trait for yield production and appearance quality. Several genes for this trait have been characterized recently (Zuo and Li 2014). However, the molecular components and regulatory network have not been fully illustrated.
To reveal more genes for rice grain size or shape, we developed a set of backcrossed introgression lines using 9311 as the recipient and Azucena as the donor. The first indica rice to have its whole-genome sequenced is 9311; it forms long grains because of a nonsense mutation in the second exon of the GS3 gene in comparison with short-grain varieties including Nipponbare (Fan et al. 2006; Takano-Kai et al. 2009; Wang et al. 2011). Azucena produces longer and thinner grains than 9311. A map-based cloning strategy was employed to isolate the target gene, SLG7, responsible for the slender grains of Azucena. The data from NILs and transgenic lines showed that the SLG7 allele from Azucena promotes spikelet elongation and produces slender grains under both 9311 and Nipponbare backgrounds. Surprisingly, SLG7 has no effect on grain weight, probably because of the reduced grain width that accompanies the increased grain length (Figure 4). These results imply that SLG7 is different from the other grain size-related QTL that directly modify grain weight and yield. Varieties with long and slender grains are preferred by consumers and cultivars in the United States, China, and most Asian countries. By contrast, short and round grain varieties are popular in Japan, South Korea, and Sri Lanka (Juliano and Villareal 1993). Therefore, SLG7 can be used to develop varieties with diverse grain shapes to meet different market demands.
The BLASTP analysis revealed that SLG7 shares numerous putative homologs (Figure 3), including two functionally characterized genes, LNG1 (At5g15580) and LNG2 (At3g02170), both of which function in the elongation of leaves, siliques, and seeds in Arabidopsis (Lee et al. 2006). LNG2 (also named TON1 Recruiting Motif 1) is identified as a microtubule-associated protein, which is able to target TON1 to cortical microtubules (Drevensek et al. 2012). We found that SLG7 homologs are restricted to the land plants and are found within several important crops including barley (Hordeum vulgare), millet (Setaria italica), maize (Zea mays), and sorghum (Sorghum bicolor) (Figure 3). In view of the results in rice and Arabidopsis, SLG7 homologs represent key components controlling grain and organ size and could be employed to improve agronomic traits in crop breeding.
Organ size or shape is determined by cell proliferation and cell expansion, two processes that are coordinated and compensated (Orozco-Arroyo et al. 2015). Morphological and cellular analyses indicated that SLG7 has a function in regulating cell elongation in spikelets (Figure 5). Similar to our data, LNG1 and LNG2 were also reported to play a positive role in increasing leaf and silique length through longitudinal cell elongation in Arabidopsis (Lee et al. 2006). These results suggested that the functions of SLG7 family members are probably conserved across monocots and dicots.
Very recently, two novel alleles of SLG7, GL7 and GW7, were identified (S. Wang et al. 2015; Y. Wang et al. 2015). GL7 was characterized as a positive regulator for grain length, and the functional allele produced longer seeds resulting from elongated longitudinal cells (Y. Wang et al. 2015). By contrast, GW7 was reported to influence cell proliferation, rather than cell expansion (S. Wang et al. 2015). We compared the expression levels of 25 genes, including 14 putatively involved in the G1/S and 11 in the G2/M phase, as shown in a previous report (Li et al. 2011). None of them showed more than a twofold change in transcript levels between NIL-SLG7 and NIL-slg7 inflorescences (Figure S4A). A similar result was also observed between Nipponbare and the SLG7 overexpression transgenic line (Figure S4B). It seems that our results are similar to the data from GL7. Genes regulating rice grain size or shape on chromosome 7, such as qGL7, qGL7-2, and GS7, have been mapped using several populations (Bai et al. 2010; Shao et al. 2010, 2012; Qiu et al. 2012). These four genes are close to SLG7, but none of them have been cloned. Perhaps there is a gene cluster associated with rice grain size or shape on the long arm of chromosome 7.
Nucleotide diversities of SLG7 were analyzed in 50 accessions (Table S1). The Azucena SLG7 allele was found in 13 germplasms from the United States, the Philippines, Indonesia, and China (Table S1), suggesting that this allele has been selected in breeding in these areas. It was noted that one variety with the Azucena SLG7 allele, Zenith, did not produce longer and thinner grains, which is different from the other materials within this group. Thus, the genetic basis for the grain size in this variety was of interest. GS3 and GW8 are two widely distributed major determinants for rice grain length, and the mutation gs3 and gw8 alleles promote the elongation of spikelets (Fan et al. 2006; Takano-Kai et al. 2009; Wang et al. 2012). Sequence analysis indicated that Zenith has negative alleles at the GS3 and GW8 loci, respectively (Table S5). This can be explained by the fact that Zenith lacks the gs3 and gw8 alleles, which are able to increase grain length, although it has a functional SLG7 gene. These data also suggested that SLG7 could be combined with other genes for grain size and shape to regulate rice grain appearance in breeding.
Supplementary Material
Acknowledgments
This work was supported by grants from the Key Program of Basic Research of China (2013CBA01405); the National Natural Science Foundation of China (31471458 and 31271679); the Key Science and Technology Support Program of Jiangsu Province (BE2015341); the Natural Science Foundation of Jiangsu Province (BK20150010 and BK20130725); and the Priority Academic Program Development of Jiangsu Higher Education Institutions.
Footnotes
Communicating editor: J. A. Birchler
Supporting information is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.181115/-/DC1.
Literature Cited
- Ashikari M., Wu J., Yano M., Sasaki T., Yoshimura A., 1999. Rice gibberellin-insensitive dwarf mutant gene Dwarf 1 encodes the alpha-subunit of GTP-binding protein. Proc. Natl. Acad. Sci. USA 96: 10284–10289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai X., Luo L., Yan W., Kovi M. R., Zhan W., et al. , 2010. Genetic dissection of rice grain shape using a recombinant inbred line population derived from two contrasting parents and fine mapping a pleiotropic quantitative trait locus qGL7. BMC Genet. 11: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drevensek S., Goussot M., Duroc Y., Christodoulidou A., Steyaert S., et al. , 2012. The Arabidopsis TRM1–TON1 interaction reveals a recruitment network common to plant cortical microtubule arrays and eukaryotic centrosomes. Plant Cell 24: 178–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan P., Rao Y., Zeng D., Yang Y., Xu R., et al. , 2014. SMALL GRAIN 1, which encodes a mitogen-activated protein kinase kinase 4, influences grain size in rice. Plant J. 77: 547–557. [DOI] [PubMed] [Google Scholar]
- Fan C., Xing Y., Mao H., Lu T., Han B., et al. , 2006. GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor. Appl. Genet. 112: 1164–1171. [DOI] [PubMed] [Google Scholar]
- Fitzgerald M. A., McCouch S. R., Hall R. D., 2009. Not just a grain of rice: the quest for quality. Trends Plant Sci. 14: 133–139. [DOI] [PubMed] [Google Scholar]
- Hiei Y., Ohta S., Komari T., Kumashiro T., 1994. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium tumeficience and sequence analysis of the boundaries of the T-DNA. Plant J. 6: 271–282. [DOI] [PubMed] [Google Scholar]
- Hong Z., Ueguchi-Tanaka M., Umemura K., Uozu S., Fujioka S., et al. , 2003. A rice brassinosteroid-deficient mutant, ebisu dwarf (d2), is caused by a loss of function of a new member of cytochrome P450. Plant Cell 15: 2900–2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimaru K., Hirotsu N., Madoka Y., Murakami N., Hara N., et al. , 2013. Loss of function of the IAA-glucose hydrolase gene TGW6 enhances rice grain weight and increases yield. Nat. Genet. 45: 707–711. [DOI] [PubMed] [Google Scholar]
- Juliano B. O., Villareal C. P., 1993. Grain quality evaluation of world rices, pp, International Rice Research Institute, Manila, Philippines. [Google Scholar]
- Khush G. S., 2001. Green revolution: the way forward. Nat. Rev. Genet. 2: 815–822. [DOI] [PubMed] [Google Scholar]
- Lee Y. K., Kim G. T., Kim I. J., Park J., Kwak S. S., et al. , 2006. LONGIFOLIA1 and LONGIFOLIA2, two homologous genes, regulate longitudinal cell elongation in Arabidopsis. Development 133: 4305–4314. [DOI] [PubMed] [Google Scholar]
- Li F., Liu W., Tang J., Chen J., Tong H., et al. , 2010. Rice DENSE AND ERECT PANICLE 2 is essential for determining panicle outgrowth and elongation. Cell Res. 20: 838–849. [DOI] [PubMed] [Google Scholar]
- Li S., Qian Q., Fu Z., Zeng D., Meng X., et al. , 2009. Short panicle1 encodes a putative PTR family transporter and determines rice panicle size. Plant J. 58: 592–605. [DOI] [PubMed] [Google Scholar]
- Li Y., Fan C., Xing Y., Jiang Y., Luo L., et al. , 2011. Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat. Genet. 43: 1266–1269. [DOI] [PubMed] [Google Scholar]
- Nakagawa H., Tanaka A., Tanabata T., Ohtake M., Fujioka S., et al. , 2012. Short grain1 decreases organ elongation and brassinosteroid response in rice. Plant Physiol. 158: 1208–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco-Arroyo G., Paolo D., Ezquer I., Colombo L., 2015. Networks controlling seed size in Arabidopsis. Plant Reprod. 28: 17–32. [DOI] [PubMed] [Google Scholar]
- Qiao Y., Piao R., Shi J., Lee S. I., Jiang W., et al. , 2011. Fine mapping and candidate gene analysis of dense and erect panicle 3, DEP3, which confers high grain yield in rice (Oryza sativa L.). Theor. Appl. Genet. 122: 1439–1449. [DOI] [PubMed] [Google Scholar]
- Qiu X., Gong R., Tan Y., Yu S., 2012. Mapping and characterization of the major quantitative trait locus qSS7 associated with increased length and decreased width of rice seeds. Theor. Appl. Genet. 125: 1717–1726. [DOI] [PubMed] [Google Scholar]
- Shao G., Tang S., Luo J., Jiao G., Wei X., et al. , 2010. Mapping of qGL7–2, a grain length QTL on chromosome 7 of rice. J. Genet. Genomics 37: 523–531. [DOI] [PubMed] [Google Scholar]
- Shao G., Wei X., Chen M., Tang S., Luo J., et al. , 2012. Allelic variation for a candidate gene for GS7, responsible for grain shape in rice. Theor. Appl. Genet. 125: 1303–1312. [DOI] [PubMed] [Google Scholar]
- Shomura A., Izawa T., Ebana K., Ebitani T., Kanegae H., et al. , 2008. Deletion in a gene associated with grain size increased yields during rice domestication. Nat. Genet. 40: 1023–1028. [DOI] [PubMed] [Google Scholar]
- Song X. J., Huang W., Shi M., Zhu M. Z., Lin H. X., 2007. A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat. Genet. 39: 623–630. [DOI] [PubMed] [Google Scholar]
- Song X. J., Kuroha T., Ayano M., Furuta T., Nagai K., et al. , 2015. Rare allele of a previously unidentified histone H4 acetyltransferase enhances grain weight, yield, and plant biomass in rice. Proc. Natl. Acad. Sci. USA 112: 76–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano-Kai N., Jiang H., Kubo T., Sweeney M., Matsumoto T., et al. , 2009. Evolutionary history of GS3, a gene conferring grain length in rice. Genetics 182: 1323–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe S., Ashikari M., Fujioka S., Takatsuto S., Yoshida S., et al. , 2005. A novel cytochrome P450 is implicated in brassinosteroid biosynthesis via the characterization of a rice dwarf mutant, dwarf11, with reduced seed length. Plant Cell 17: 776–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Chen S., Yu S., 2011. Functional markers developed from multiple loci in GS3 for fine marker-assisted selection of grain length in rice. Theor. Appl. Genet. 122: 905–913. [DOI] [PubMed] [Google Scholar]
- Wang E., Wang J., Zhu X., Hao W., Wang L., et al. , 2008. Control of rice grain-filling and yield by a gene with a potential signature of domestication. Nat. Genet. 40: 1370–1374. [DOI] [PubMed] [Google Scholar]
- Wang S., Wu K., Yuan Q., Liu X., Liu Z., et al. , 2012. Control of grain size, shape and quality by OsSPL16 in rice. Nat. Genet. 44: 950–954. [DOI] [PubMed] [Google Scholar]
- Wang S., Li S., Liu Q., Wu K., Zhang J., et al. , 2015. The OsSPL16–GW7 regulatory module determines grain shape and simultaneously improves rice yield and grain quality. Nat. Genet. 47: 949–954. [DOI] [PubMed] [Google Scholar]
- Wang Y., Xiong G., Hu J., Jiang L., Yu H., et al. , 2015. Copy number variation at the GL7 locus contributes to grain size diversity in rice. Nat. Genet. 47: 944–948. [DOI] [PubMed] [Google Scholar]
- Weng J., Gu S., Wan X., Gao H., Guo T., et al. , 2008. Isolation and initial characterization of GW5, a major QTL associated with rice grain width and weight. Cell Res. 18: 1199–1209. [DOI] [PubMed] [Google Scholar]
- Yamamuro C., Ihara Y., Wu X., Noguchi T., Fujioka S., et al. , 2000. Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell 12: 1591–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Wang J., Huang J., Lan H., Wang C., et al. , 2012. Rare allele of OsPPKL1 associated with grain length causes extra-large grain and a significant yield increase in rice. Proc. Natl. Acad. Sci. USA 109: 21534–21539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Zhu J., Li Z., Yi C., Liu J., et al. , 2009. Deletion in a quantitative trait gene qPE9–1 associated with panicle erectness improves plant architecture during rice domestication. Genetics 183: 315–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu K., Tang D., Yan C., Chi Z., Yu H., et al. , 2010. Erect panicle2 encodes a novel protein that regulates panicle erectness in indica rice. Genetics 184: 343–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J., Li J., 2014. Molecular genetic dissection of quantitative trait loci regulating rice grain size. Annu. Rev. Genet. 48: 99–118. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Materials and plasmids described here and in the Supporting Information are available upon request.