Abstract
CTP synthase (CTPsyn) plays an essential role in DNA, RNA, and lipid synthesis. Recent studies in bacteria, yeast, and Drosophila all reveal a polymeric CTPsyn structure, which dynamically regulates its enzymatic activity. However, the molecular mechanism underlying the formation of CTPsyn polymers is not completely understood. In this study, we found that reversible ubiquitination regulates the dynamic assembly of the filamentous structures of Drosophila CTPsyn. We further determined that the proto-oncogene Cbl, an E3 ubiquitin ligase, controls CTPsyn filament formation in endocycles. While the E3 ligase activity of Cbl is required for CTPsyn filament formation, Cbl does not affect the protein levels of CTPsyn. It remains unclear whether the regulation of CTPsyn filaments by Cbl is through direct ubiquitination of CTPsyn. In the absence of Cbl or with knockdown of CTPsyn, the progression of the endocycle-associated S phase was impaired. Furthermore, overexpression of wild-type, but not enzymatically inactive CTPsyn, rescued the endocycle defect in Cbl mutant cells. Together, these results suggest that Cbl influences the nucleotide pool balance and controls CTPsyn filament formation in endocycles. This study links Cbl-mediated ubiquitination to the polymerization of a metabolic enzyme and reveals a role for Cbl in endocycles during Drosophila development.
Keywords: Cbl, CTP synthase, cytoophidia, endocycle, Drosophila
PROVIDING the raw material for DNA/RNA replication is a key challenge faced by all organisms during rapid growth. One particularly interesting example of this challenge is polyploidy, which arises from endoreplication and is essential for the normal development and specific physiological conditions of many diploid organisms. Organisms often use endoreplication, a type of cell cycle encompassing genomic replication without cell division, to provide nutrients to support the developing egg/embryo and enlarged cell size. Examples of cells and tissues that endocycle include mammalian trophoblast giant cells, plant seeds/roots, and Drosophila egg chambers (Edgar and Orr-Weaver 2001; Lee et al. 2009). In Drosophila, embryogenesis requires the mass production of proteins and high metabolic activity, which are achieved through endoreplication in the terminally differentiated cells of the developing egg chamber.
Drosophila oogenesis provides an excellent system for analyzing developmentally controlled endoreplication. Egg production takes place within 16-cell germline cysts, with the asymmetric and incomplete division of a germline stem cell (Calvi and Spradling 1999). After cyst formation, nurse cells immediately exit the mitotic cycle and begin a series of 10–12 endocycles to reach 512C DNA content to provide proteins and messenger RNAs (mRNAs) for the developing oocyte. Each germline cyst is enveloped by 15–20 somatic follicle cells that divide mitotically to form an epithelial monolayer of ∼1000 cells and then employ three endocycles to reach 16C DNA content during stages 7–10A, the so-called “endocycle stages” (Klusza and Deng 2011). Endoreplication in the follicular epithelium ensures a large amount of eggshell protein production in <24 h (Lilly and Spradling 1996; Calvi et al. 1998). The endocycle in the follicle cells ceases at stage 10B, but some specific genomic foci (e.g., chorion genes) continue to amplify synchronously, an event called gene amplification (Calvi et al. 1998). Notch signaling is responsible for the mitotic cycle–endocycle transition of follicle cells (Deng et al. 2001; Lopez-Schier and St. Johnston 2001), which activates the Cyclin E/Cyclin-Dependent Kinase 2 (CycE/Cdk2) complex to trigger the endocycle transition (Shcherbata et al. 2004).
This rapid series of endoreplication events requires cells to have sufficient stores of the raw materials for DNA synthesis. CTP synthase (CTPsyn) produces CTP to facilitate DNA and RNA synthesis. In both prokaryotes and eukaryotes, CTPsyn is allosterically bound to GTP, activating glutamine hydrolysis to generate ammonia (Long and Pardee 1967; Long et al. 1970; Levitzki and Koshland 1972). Subsequently, CTPsyn catalyzes the ATP-dependent transfer of ammonia from glutamine to the C-4 position of UTP to form CTP (Lieberman 1956; Chakraborty and Hurlbert 1961; Levitzki and Koshland 1971; von der Saal et al. 1985; Endrizzi et al. 2004). Under low concentrations of CTPsyn or in the absence of ATP/UTP/CTP, CTPsyn is present as an inactive monomer. With an increasing concentration of CTPsyn, CTPsyn initially forms inactive dimers and then forms active tetramers in the presence of ATP/UTP/CTP (Anderson 1983; von der Saal et al. 1985). Therefore, CTPsyn monitors cellular nucleotide pools through its four NTP-binding sites, allowing it to match its activity to the concentration of nucleotides (Aronow and Ullman 1987).
Recently, filamentous CTPsyn structures were independently revealed in bacteria, budding yeast, Drosophila, and mammalian cell lines, representing a novel, evolutionarily conserved cellular structure (Ingerson-Mahar et al. 2010; Liu 2010; Noree et al. 2010; Carcamo et al. 2011). In Caulobacter crescentus, the CTPsyn filaments (CtpS filaments) interact with intermediate filaments to regulate bacterial shape (Ingerson-Mahar et al. 2010). In budding yeast, CTP synthase filaments are promoted under the condition of carbon source depletion (Noree et al. 2010). In Drosophila, this structure, termed the “cytoophidia,” is present in a number of tissues, including the salivary gland, testis, mid-gut, and ovary (Buszczak et al. 2007; Liu 2010). Both in yeast and Drosophila, the formation of a filament composed of CTPsyn does not require active tetramers, arguing that the filament is composed of inactive CTPsyn dimers or monomers (Aughey et al. 2014; Noree et al. 2014). However, CTPsyn filaments in bacteria are composed of an inactive form of tetramers (Barry et al. 2014). In mammals, this structure, termed “rods and rings” (RR), appears in both the cytoplasm and the nucleus (Gou et al. 2014) and acts in a cell cycle-independent manner (Carcamo et al. 2011). The RR structure contains not only CTPsyn, but also inosine monophosphase dehydrogenase 2 (IMPDH2), a key enzyme in GTP biosynthesis (Carcamo et al. 2011). Recently, the RR structure was recognized as reflecting the concentration of glutamine, an essential amide nitrogen donor in the nucleotide biosynthesis pathway. The depletion of glutamine forced the formation of the RR structure in mammalian cells (Calise et al. 2014; Gou et al. 2014).
Despite this strikingly broad evolutionary conservation, the function of these filamentous structures and the regulation of their assembly remain elusive. During Drosophila oogenesis, germline cells of the ovary contain two different sizes of CTPsyn filaments in one cell; they can be classified into micro-cytoophidia (1–6 μm) and macro-cytoophidia (10–50 μm) (Liu 2010). While we know that increasing the level of CTPsyn in Drosophila enhances the formation of cytoophidia structures and that CTPsyn activity seems to correlate with the structure in yeast, bacteria, and Drosophila (Chen et al. 2011; Azzam and Liu 2013; Aughey et al. 2014; Barry et al. 2014; Noree et al. 2014; Strochlic et al. 2014), other mechanisms regulating their assembly remain unknown.
We previously analyzed the function of the proto-oncogene Casitas B-lineage lymphoma (Cbl), which is conserved in Caenorhabditis elegans, Drosophila, and mammalian cells. It encodes an E3 ligase that plays a negative role in the receptor tyrosine kinase pathway through its ubiquitin ligase activity (Marmor and Yarden 2004). Ubiquitination of the epidermal growth factor receptor (EGFR) promotes endosomal sorting and lysosomal degradation of this protein (Chang et al. 2008; Wang and Pai 2011). In this role, Cbl determines the polarity of the developing egg by negatively regulating the EGFR in follicle cells (Pai et al. 2000). Cbl can also regulate Notch signaling, an activity related to its short isoform, Cbl-S, during eye and wing development (Wang et al. 2010). Cbl has also been implicated in regulating cell proliferation in the germline, although the mechanism is not known (Epstein et al. 2009). Here, we describe a novel role for Cbl in promoting the endocycle through regulating CTPsyn activity. Moreover, we reveal that reversible modification of ubiquitin regulates the dynamic assembly of CTPsyn structures, the cytoophidia. Together, our data demonstrate a potential role for cytoophidia during Drosophila development and begin to uncover the mechanisms regulating CTPsyn filament assembly.
Materials and Methods
Fly stocks and genetics
The strains used included the following: wild-type (Oregon-R); CblF165/TM3Ser (Pai et al. 2000); CblBB28/TM3Ser (Pai et al. 2006); UAS-λ-top (Queenan et al. 1997); Prosβ61/TM3Sb (Saville and Belote 1993); Hs83-D-Cbl-L-PRDrk-75 (Wang and Pai 2011); Hs83-D-CblS-2 (Pai et al. 2006); UAS-HA-ubiquitin (a gift from C. T. Chien, Institute of Molecular Biology, Academia Sinica, Taiwan); UAS-Dicer2 (Bloomington Stock Center); UAS-CTP synthaseGD12759 (Vienna Drosophila RNAi Center); and GR1, CY2, and 55B-Gal4 (Queenan et al. 1997). For the genetic interaction assay, OreR, Prosβ61/ +, and CblF165/ Prosβ61 were incubated for 3 days at 18° and then at 29° for 2 days. e22cFLP/+; CblF165FRT80B (or CblBB28FRT80B) /hs-mycFRT80B and hs-FLPl22/X; CblF165FRT2A /ovoDFRT2A were used for generating mosaic clones as described previously (Pai et al. 2000). hs-FLPl22/X; CblF165FRT80B/[ubi-nlsRFP]FRT80B was used to generate mosaic clones in the salivary glands. Eggs, 2.5–3.5 hr old, were collected, heat-shocked at 37° for 1.5 hr, and developed to 96-hr third instar larvae.
Antibodies
The primary antibodies used for immunohistochemistry included rabbit anti-CTPsyn (1:300; y-88, sc-134457; Santa Cruz), rat anti-HA (1:500; Roche 3F10), mouse anti-MPM2 (1:200; Millipore), mouse anti-Cut (1:50; 2B10); mouse anti-Hnt (1:15 1G9); mouse anti-Flag (1:500; Sigma F1804), and rabbit anti-Myc (1:200; Santa Cruz), while the secondary antibodies included Alexa Fluor 488 or 546 anti-mouse/rabbit/rat antibodies (Molecular Probes). To visualize DNA, 0.5 mg/mL DAPI (Sigma) was added for 5 min at room temperature. To visualize actin, phalloidin (1:100; Molecular Probes) was used. The antibodies used for immunoblotting were anti-D-Cbl 10F1 ascites (Pai et al. 2006), rabbit anti-dCTPsyn, and mouse anti-tubulin (Sigma).
Drug treatment
For MG132 treatment, ovaries were dissected in Schneider’s insect medium (Gibco) and then were incubated in medium containing 60 μM MG132 (20 mM stock dissolved in DMSO) or DMSO (as a control) for 2 hr at room temperature. For Pr619 plus MG132 treatment, ovaries were dissected in Schneider’s medium and then incubated in medium containing 50 μM Pr619 (20 mM stock was dissolved in DMSO) or DMSO for 2 hr at room temperature. After this, the ovaries were transferred to medium containing 60 μM MG132 or DMSO and incubated for another 2 hr at room temperature. After drug treatment, the ovaries were fixed with 4% paraformaldehyde and analyzed by immunostaining.
CTPsyn inhibitor treatment
DON (6-diazo-5-oxo-l-norleucine) (D2141, Sigma) was solubilized at 500 μg/ml in water. Adult females were yeasted for 3–5 days and then fed on wet yeast with or without 500 μg/ml of DON for 24 hr, as previously described (Chen et al. 2011).
Bromodeoxyuridine incorporation assays
Ovaries and salivary glands were collected in Schneider’s medium and then incubated in a medium containing 30 μg/ml bromodeoxyuridine (BrdU) (Sigma) at room temperature for 1 h. The BrdU-incorporated tissues were fixed in 200 μl of 4% formaldehyde containing 600 μl heptane and 0.25% NP-40 for 20 min at room temperature. The samples were treated with 2 N HCl for 30 min, neutralized with 100 mM sodium borate, and washed with PBT three times. For BrdU detection, the samples were incubated with PBT containing 10% horse serum and then with anti-BrdU antibody (1:50, Becton Dickinson) at 4° overnight. Alexa Fluor 488 goat anti-mouse (1:1000; Molecular Probes) was used as the secondary antibody.
Quantification of BrdU-positive cells
For quantification of the percentage of BrdU-positive cells in the Cbl clone, the number of BrdU-positive cells was divided by the total cell number in the Cbl clone or its twin clone. To calculate the percentage of BrdU-positive cells for the genetic interaction experiment, we counted BrdU-positive cells in the first five rows of the anterior and posterior follicle cells per egg chamber in 55B-Gal4::UAS-CTPsynRNAi, 55B-Gal4::UAS-GFP, OreR,CY2-Gal4 in Cbl heterozygous mutants and CY2-Gal4::UAS-CTPsynRNA i with or without Cbl heterozygous mutants and then divided this number by the total number of cells in the five rows.
Transgenic line generation and induction
To generate Flag-CTPsyn, a complementary DNA (cDNA) fragment of CTPsyn isoform C was amplified by PCR from an ovarian cDNA library with the specific primers CTPsyn-C-F 5′-GTCTCGAGATGAAATACATCCTGGTAAC-3′and CTPsyn-C-R 5′-GTCTCGAGTTACTTATGGCCATTGGTAG-3′. The CTPsyn fragment was cloned into pUAST-Flag using XhoI and XbaI restriction enzymes and was in-frame fused to the 3′ end of the Flag-tag. For Flag-CTPsynC399G construction, we employed a mutagenesis technique to generate the mutated-CTPsyn fragments using the following specific primers: dCTPSC t1195g C399G F 5′-CCATTGCTTGGCATCGGCTTGGGTCTGCAAG -3′ and dCTPSC t1195g C399G R 5′-CTTGCAGACCCAAGCCGATGCCAAGCAATGG-3′. The DNA sequence of the product was confirmed before microinjection. For wild-type expression, hs-Gal4::UAS-Flag-CTPsyn transgenic flies were heat-shocked at 37° for 30 min and then incubated for 3 hr at 25°. Subsequently, ovaries were dissected and analyzed by immunostaining. For expression in Cbl mosaic egg chambers, hsFlp/X;hs-Gal4/UAS-Flag-CTPsyn; CblFRT80B/MycFRP80B were heat-shocked for 30 min/day. After 2 days of induction and incubation, the expression level of Flag-CTPsyn and occurrences of the S phase were examined by immunoblotting and by BrdU-incorporation assay, respectively.
Antibody generation
A total of 1–118 amino acids from cg45070 (dCTPsyn in Flybase) was amplified by KOD-plus- DNA polymerase using the paired primers 5′-TAAGCAGAATTCATGAAATACATCCTGGTAACTG-3′ and 5′-ATTGCTCTCGAGAGTGATGTGTGGGACAACTTG-3′ to clone the peptide into a pET32a vector, and 6× His fusion protein was purified by Ni-NTA beads (Qiagen). Antibodies were obtained by immunizing rabbit followed by affinity purification and then were used for Western blotting (1:1000).
Results
Ubiquitination is involved in the regulation of CTPsyn filamentous structure formation
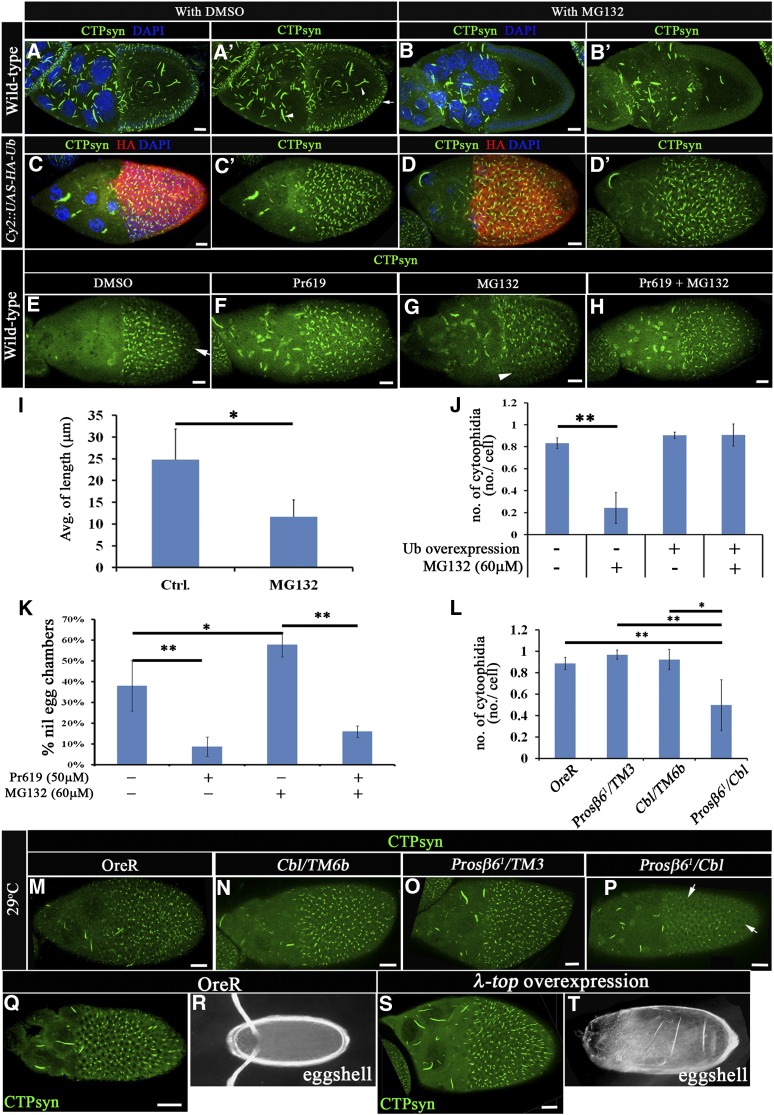
An important insight into the developmental role of CTPsyn filaments can be derived from the identification of the biochemical switch that regulates its assembly. RNA silencing of CTPsyn led to the disassembly of CTPsyn filaments, suggesting that CTPsyn protein levels regulate the CTPsyn filament structure in a dose-dependent manner (Chen et al. 2011). To examine this hypothesis further, we treated Drosophila ovaries with MG132, a proteasome inhibitor that should block the degradation of CTPsyn, leading to an increase in the number of CTPsyn structures. Surprisingly, a reduction in CTPsyn filament formation was found in both the germline cells and the follicle cells of egg chambers treated with MG132, compared to that in the DMSO control (Figure 1, A and B). The length of macro-cytoophidium in control egg chambers was 24.8 ± 7 μm on average, and this length was reduced to 11.6 ± 3.9 μm in egg chambers treated with MG132 (Figure 1I). On average, almost every follicle cell contained one CTPsyn filament (0.83 ± 0.05/per cell, Figure 1J), and a reduction in the number of filaments was observed with MG132 treatment (0.24 ± 0.14/cell, Figure 1J). Despite this observation, not all egg chambers were affected (Supporting Information, Figure S1D). The phenotype, however, did occur in all stages of oogenesis. Moreover, the inhibitory effect of MG132 on CTPsyn filament formation occurred in a dose-dependent manner with a higher dose resulting in a stronger inhibitory response (Figure S1, D and E). A similar, but somewhat less severe phenotype was observed in egg chambers expressing exogenous Flag-CTPsyn induced by hs-Gal4 (Figure S1, A and B). Nevertheless, the CTPsyn protein levels were comparable before and after MG132 treatment (Figure S1C).
Figure 1.
The proteasome degradation system is involved in the regulation of CTPsyn filaments. (A–B′) Merged-multiple confocal cross sections of middle-stage egg chambers stained with anti-CTPsyn antibody (green) and DAPI (blue). Macro-cytoophidia are presented in germline cells (arrowhead), and CTPsyn filaments are shown in somatic cells (arrow). Wild-type egg chambers were treated with DMSO (as a control, A–A′) or 60 μM of the proteasome inhibitor MG132 (B–B′). The average length of macro-cytoophidium was calculated after DMSO (control, 24.83 ± 7 μm) or 60 μM MG132 (11.67 ± 3.89 μm) treatment, and results are shown in I (n = 10 egg chambers, three biological replicates). (C–H, M–P, Q, and S) Merged multiple confocal surface sections of middle-stage egg chambers were stained with anti-CTPsyn antibody (green). (C–D′) Exogenous HA-ubiquitin-expressing (red) egg chambers were treated with DMSO (C–C′) or 60 μM MG132 (D–D′). The average number of CTPsyn filaments is shown in J (16, 51, 6, and 13 egg chambers for columns 1–4, respectively, with ∼140 follicle cells counted in each egg chamber). On average, the numbers of CTPsyn filaments/cell presented in DMSO- and MG132-treated wild-type egg chambers were 0.8 ± 0.5 and 0.24 ± 0.14, respectively. And the numbers of CTPsyn filaments/cell presented in DMSO- and MG132-treated HA-Ub-expressing egg chambers were 0.9 ± 0.03 and 0.9 ± 0.1, respectively. Wild-type egg chambers were treated with DMSO (E, affected region, arrow), MG132 (G, affected region, arrowhead), Pr619 (F), and MG132 plus Pr619 (H). The percentage of egg chambers showing nil phenotypes was calculated and is shown in K (191, 200, 180, and 187 egg chambers for columns 1–4, respectively, in three biological replicates). (M–P) Wild-type egg chambers (M), CblF165/TM6b (N), Prosβ61/TM3 (O), and Prosβ61/CblF165 (P, affected region, arrow) were incubated at a nonpermissive temperature (29°). The average number of CTPsyn filaments is shown in L (cells/egg chambers: 1255/5, 853/5, 1764/5, and 1021/5 for columns 1–4, respectively.). (Q) Wild-type and (S) λ-top-overexpressing egg chambers. (R and T) Eggs laid by wild-type females (R) or females carrying λ-top-overexpressing egg chambers (T). The results are shown as the mean ± SD; *P < 0.05 and **P < 0.01. Bar, 20 μm.
We reasoned that MG132 treatment could result in two consequences: namely, blockage of protein degradation and depletion of free ubiquitin (Melikova et al. 2006). Given that CTPsyn protein levels remained largely unaffected, we next investigated whether overexpression of ubiquitin could suppress the effect of MG132. Indeed, selective expression of exogenous ubiquitin in follicle cells driven by CY2-Gal4 significantly reduced, from 0.24 ± 0.14/cell to 0.9 ± 0.1/cell, the MG132 effect on CTPsyn filament numbers (Figure 1J), indicating that ubiquitination may play a positive role in the regulation of CTPsyn filaments. In this in vitro-cultured ovary assay, the gradual disassembly of CTPsyn filaments (a “nil” phenotype that completely lacked filament) was observed in ∼20% of egg chambers at 2 hr and in ∼40% of egg chambers at 4 hr in medium containing DMSO (Figure 1K and Figure S1D). To further confirm that ubiquitin modification may play a positive role in filament formation, we next examined the effect of a deubiquitinase-inhibitor, Pr619. Indeed, the CTPsyn filament structures were preserved with Pr619 pretreatment for 2 hr followed by MG132 treatment (Figure 1, E–H and K). Together, these results suggest that ubiquitination is required to maintain CTPsyn filaments.
Since Cbl is an E3 ubiquitin ligase in follicle cells (Pai et al. 2000), we next investigated the role of Cbl in ubiquitin-mediated regulation of CTPsyn filaments using a genetic interaction assay with the Prosβ61 mutant. We used the allele Prosβ61, a temperature-sensitive proteasome subunit mutant (Holden and Suzuki 1973; Saville and Belote 1993), and a Cbl null mutant, CblF165 (Pai et al. 2000). At a nonpermissive temperature (29°), this dominant-negative Prosβ61 allele by itself did not show any significant difference from wild type (Figure 1L). However, the combination of CblF165 and Prosβ61 displayed a synergistic effect on the negative regulation of CTPsyn filament formation (Figure 1, M–P and L), whereas the Cbl heterozygous mutant did not show significant effects. On average, every follicle cell contains 0.9, 1.0, and 0.9 CTPsyn filaments in OreR, CblF165/+, and Prosβ61/+ egg chambers, respectively, but the number of CTPsyn filaments was reduced to 0.5 filaments/cell in CblF165/Prosβ61 egg chambers at a nonpermissive temperature (29°) (Figure 1L). Collectively, these results suggest that ubiquitination and Cbl may regulate the formation of CTPsyn filament structures.
Since disruption of Cbl E3 ligase activity enhances EGFR signaling in follicle cells (Pai et al. 2000; Wang and Pai 2011), we further assessed whether Cbl regulates CTPsyn filament structures through EGFR signaling. The CTPsyn filament was not affected by the overexpression of constitutively activated EGFR (λ-top) (Figure 1, Q–T) nor by activated Ras (RasV12) (Figure S1, F–F′′) in follicle cells. In summary, Cbl regulates the formation of filament structures composed of CTPsyn through its E3 ligase activity and appears to do so independently of EGFR signaling.
Cbl regulates CTPsyn filaments without affecting CTPsyn protein levels
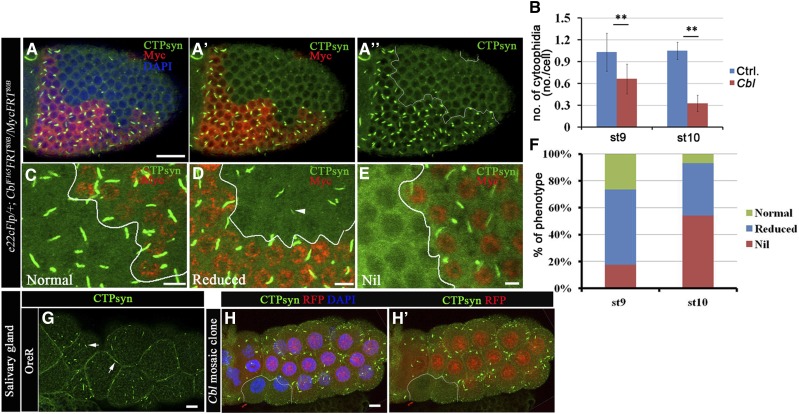
To elucidate the role of Cbl in regulating the formation of CTPsyn filament structure, mosaic Cbl mutant clones were generated in the follicular epithelium. In wild-type egg chambers, at least one CTPsyn filament was detected in every follicle cell (Figure 2, A and B) of stages 9–10 egg chambers. In contrast, few CTPsyn filaments were seen in Cbl mutant follicle cells (Figure 2, A–A′′ and B). We quantified the effects on CTPsyn filament formation by classifying the structures in each cell as (1) normal, (2) reduced in CTPsyn filament length (Figure 2D, “reduced”), or (3) the complete absence of CTPsyn filaments (Figure 2E, “nil”). A reduced phenotype was seen in 57% of Cbl mutant follicle cells, while 18% lacked CTPsyn filaments at stage 9. The defects became more severe at stage 10, when 36% of Cbl mutant cells exhibited a “reduced” phenotype and 58% showed a nil phenotype (Figure 2F). Similar defects in CTPsyn filaments were also observed in follicle cell mutants with another null allele of Cbl (CblBB28) (Figure S2, A–A′) (Pai et al. 2006). However, CTPsyn filaments were maintained as normal in Cbl-mutant cells when these cells were still in the mitotic stages of egg chamber development (before stage 6) (Figure S2, B–B′). To determine if this effect was specific for follicle cells, we examined another cell type that undergoes endocycles and that shows visible CTPsyn filaments, i.e., the salivary glands (Figure 2G). Strikingly, CTPsyn filaments were also absent in the salivary glands of the Cbl mutant (Figure 2, H–H′). In contrast, the structure of the macro-cytoophidia in germline cells (nurse cells and the oocyte) was not obviously affected by the loss of Cbl (Figure S2D). The effects of Cbl on CTPsyn filament were confirmed by rescue experiments in which we ectopically expressed one of the two Cbl isoforms, CblL or CblS, in Cbl mutant follicle cells. Because overexpression of CblL induces lethality during development (Pai et al. 2006), we used D-CblL-PRDrk, a full-length CblL carrying a point mutation in the Drk interaction motif (Wang and Pai 2011). D-CblL-PRDrk and D-CblS each partially restored CTPsyn filaments, reducing the frequency of the nil phenotype of Cbl mutant cells at stage 10 from 79 to 34 and 42%, respectively (Figure S3, B–D). We next investigated whether the E3 ligase activity is required for Cbl-mediated CTPsyn filament maintenance by examining the ability of Cbl-D70Z, a Cbl isoform defective in E3 enzyme activity, to rescue the nil phenotype in Cbl mutant cells. The CTPsyn filament structure in follicle cells, however, could not be restored by D-Cbl-D70Z expression during stage 10 (Figure S3E), suggesting that E3 ligase activity is required for Cbl to maintain CTPsyn filaments. Taken together, these results demonstrate that Cbl regulates the formation of CTPsyn filament structures through its E3 ligase activity.
Figure 2.
Cbl regulates the CTPsyn filamentous structure. (A–F) Merged confocal sections of stage 9–10 egg chambers stained with anti-CTPsyn (green) and anti-Myc (red) antibodies (A–A′′). Myc-positive signals marked wild-type and Cbl heterozygous cells, whereas Cbl homozygous mutant cells were marked by the absence of Myc staining. (B) Average numbers of CTPsyn filaments/cell were calculated from stage 9 and 10 egg chambers of the genotype e22cFlp/+; Cbl F165FRT80B/MycFRT80B. (C) Normal CTPsyn filament structures are shown in green and are represented by the green columns in F. The “reduced” phenotype (F, blue column) indicates a shortening (arrowhead in D) of CTPsyn filaments in Cbl mutant cells. The nil phenotype (red column) represents the lack of CTPsyn filaments (34 and 72 egg chambers at stages 9 and 10, respectively). (G) Larval salivary glands of OreR were stained with anti-CTPsyn antibody. CTPsyn filamentous structures are shown in green (arrow). (H–H′) Larval salivary glands containing a clone of Cbl mutant cells are marked by the absence of RFP (outlined in white) and stained with anti-CTPsyn (green) antibody. The results are shown as the mean ± SD; **P < 0.01. Bar, 20 μm.
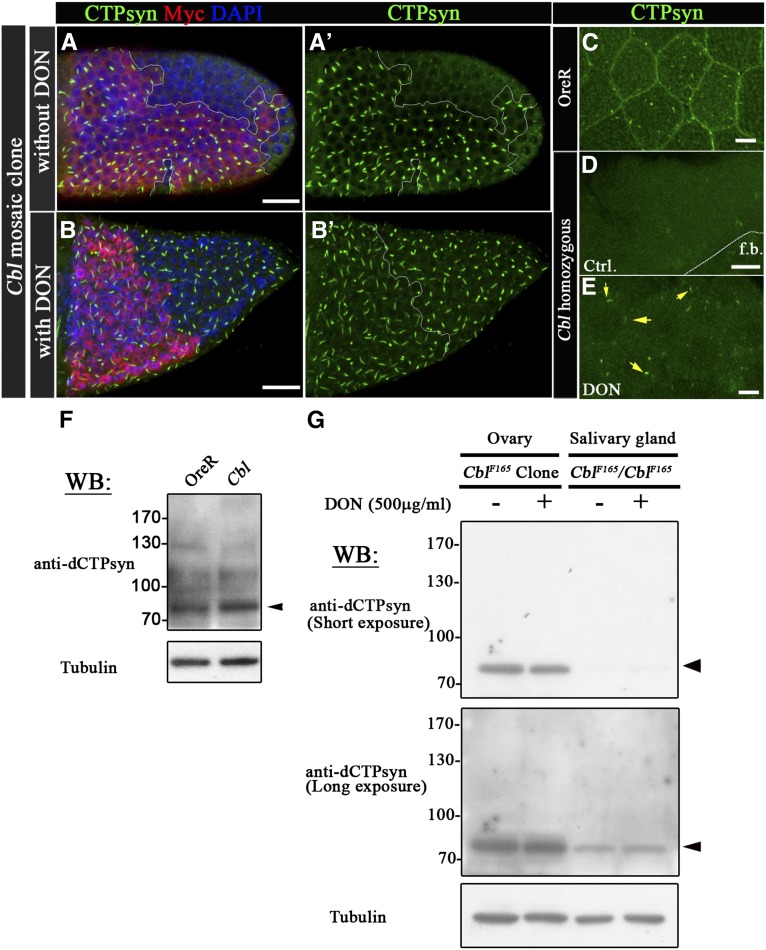
To investigate whether disruption of CTPsyn filament structures in Cbl mutant cells is caused by protein degradation, we first treated egg chambers with the CTPsyn inhibitor DON, an analog of glutamine, which blocks glutamine hydrolysis and promotes the formation of CTPsyn filaments in wild-type flies (Chen et al. 2011). Strikingly, the CTPsyn filament structure was restored by DON treatment in Cbl mutant follicle cells (Figure 3, B–B′) and salivary glands (Figure 3E). In contrast, the CTPsyn filament structures could not be rescued in CTPsyn-depleted follicle cells that were produced by RNA interference (RNAi) knockdown (Figure S4, D and E). To directly test whether loss of Cbl affected the levels of CTPsyn, we examined Cbl mutant salivary glands, which, as we demonstrated above, lack normal CTPsyn filaments. Immunoblotting revealed that the protein levels of CTPsyn were not reduced in the absence of Cbl (Figure 3F). This was consistent with results from a previous study and suggests that DON may induce structural alteration in CTPsyn, leading to CTPsyn polymer formation (Barry et al. 2014). DON treatment did not show effects on CTPsyn filament in follicle cells expressing CTPsyn RNAi (Figure S4) and did not elevate CTPsyn protein levels in Cbl mutant salivary glands or mosaic egg chambers (Figure 3G). These results suggest that Cbl contributes to stabilization of the CTPsyn filament structure rather than maintenance of CTPsyn protein levels.
Figure 3.
Cbl maintains CTPsyn filamentous structures without effects on CTPsyn protein levels. (A–B′) The labeling of cells is the same as described in Figure 2; i.e., homozygous mutant cells are indicated by the absence of red Myc staining. (B–B′) CTPsyn filaments were restored partially in Cbl mutant cells through a 500-μg/ml DON treatment. (C–E) CTPsyn filaments were labeled with anti-CTPsyn antibody (green) in OreR and Cbl homozygous salivary glands with or without DON treatment. The CTPsyn filamentous structures were observed in Cbl homozygous salivary glands with DON treatment (arrow in E). (F–G) CTPsyn expression levels were analyzed by Western blots with anti-dCTPsyn antibody. Tubulin served as the loading control. Cell lysates were extracted from OreR, Cbl homozygous salivary glands (F), or Cbl-mosaic ovaries and Cbl homozygous salivary glands with or without DON treatment (G). f.b. indicates fat bodies. Bar, 20 μm.
Cbl is involved in endoreplication
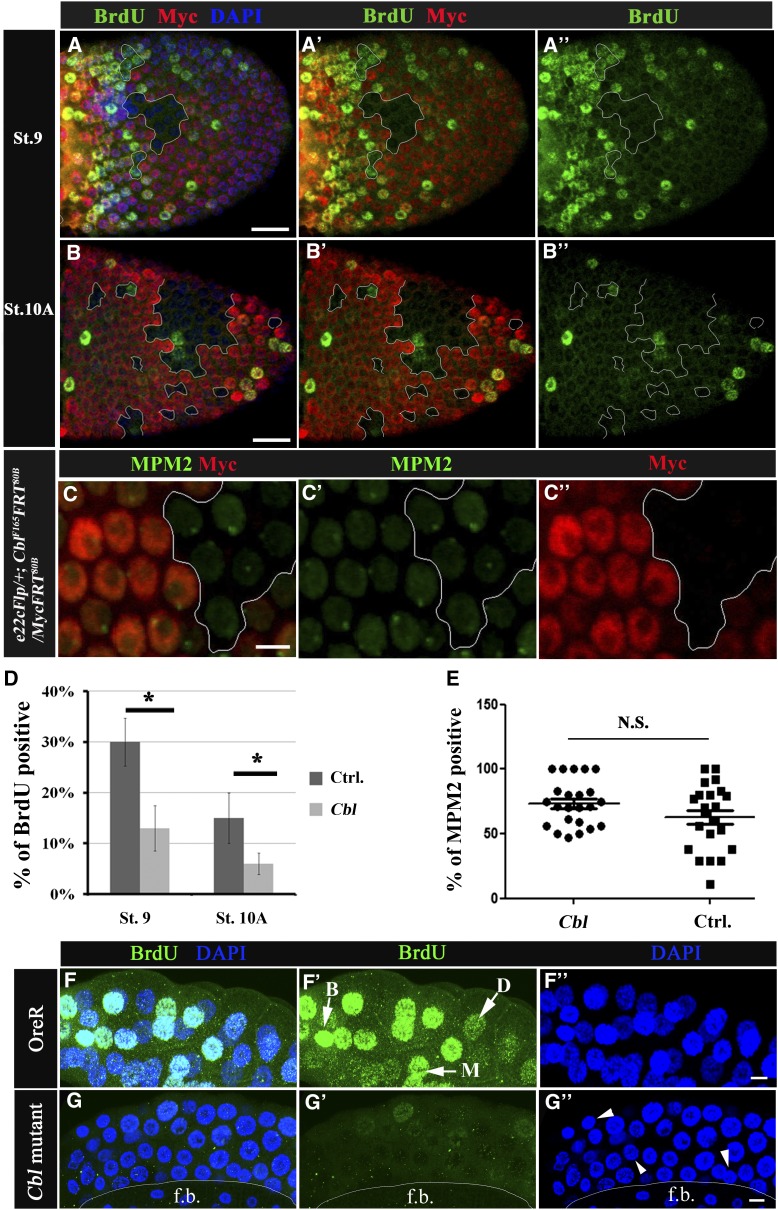
Nucleotide pools are important for DNA replication. In follicle cells, the mass production of eggshell proteins, including vitelline membrane and chorion proteins, are promoted by amplifying the DNA templates for mRNA synthesis via a series of endocycles, followed by gene amplification. This requires cells to ramp up DNA synthesis in a process that involves altering the cell cycle and DNA replication program (Calvi et al. 1998; Tower 2004; Lee et al. 2009). Since Cbl regulates the CTPsyn-composed filamentous structures, which may influence the balance of the nucleotide pool, we next examined whether Cbl plays a role in endocycles, using a BrdU incorporation assay in stages 9–10B of egg chambers. Different degrees of endocycle defect were found in Cbl mutant follicle cells: 30 and 13% of wild-type (Myc-positive) cells were BrdU-positive during stages 9 and 10A, respectively, compared to 13% (Figure 4, A–A′′ and D) and 6% (Figure 4, B–B′′ and D) in Cbl mutant cells, respectively. This defect was not observed in stage-10B egg chambers (Figure S5, A–A′′) in which gene amplification took over endocycles in follicle cells.
Figure 4.
Cbl is involved in the endocycle. (A–C′′) Mosaic egg chambers containing Cbl mutant follicle cell clones. Cells are labeled as described in Figure 2. (A–B′′) BrdU incorporation is shown in the follicular epithelium containing mosaic clones for Cbl (Myc-negative cells, outlined in white) of stage 9 (A–A′′) and stage 10A (B–B′′) egg chambers. The BrdU signal is in green; nuclei were stained with DAPI (blue). (C–C′′) The MPM2 signal (in green) was detected in Cbl mutant cells and wild-type follicle cells in stage-9 egg chambers. (D) Percentages of BrdU-positive vs. Myc-negative cells (Cbl, light gray column) and Myc-positive twin clone cells with wild-type Cbl genes (Ctrl, dark gray column). Results are shown as the mean ± SEM; *P < 0.05. There were 16 and 18 egg chambers at stages 9 and 10, respectively. N = 3. (E) MPM2 positivity was assessed as described above; N.S. indicates not significant. (F–G′′) BrdU incorporation (shown in green) in wild-type (F–F′′) and in Cbl homozygous (G–G′′) salivary glands. In F′, BrdU signals were classified as bright (B), medium (M), or dark (D) (arrow). (F′′ and G′′) DNA was stained with DAPI (blue), and a reduction in nuclear size in Cbl mutant (arrowhead in G′′) was observed. f.b. indicates fat bodies. Bar, 20 μm.
We further confirmed the effect of Cbl on endoreplication in another polyploid tissue, the salivary gland. While the Cbl homozygous larvae were of normal size, the size of the Cbl mutant salivary glands were decreased. Simultaneously, both the reduced DAPI nuclear signals and small nucleus size (Figure 4G′′, arrowheads) were found in Cbl mutants, which is consistent with previous reports that endoreplication was found to be positively correlated with cell size (Edgar and Orr-Weaver 2001). Therefore, we further measured the nuclear size and DAPI fluorescence intensity of cells in Cbl-mosaic egg chambers and salivary glands. On average, the nuclear sizes were not significantly different between cells in twin clones and Cbl mutant clones, either in egg chambers or salivary glands (Figure S6, A and C). While the DAPI nuclear signal of Cbl mutant follicle cells showed no difference (Figure S6B), the DAPI signal was reduced in Cbl mutant salivary gland cells (Figure S6D).
Significantly reduced BrdU signals were found in Cbl homozygote mutant salivary glands (Figure 4G), indicating interference with the S phase of the endocycle. The density of the BrdU signal could be classified into three categories: bright, medium, and dark (Figure 4F′). The number of nuclei with a bright BrdU signal decreased by 18% in Cbl mutant cells, whereas the number of nuclei with dark BrdU signal increased by 21% (Figure S6F). The defect in endocycle occurrence might result from the loss of the license for cell cycle entry or from a defect in upstream Notch signaling. To determine whether the endocycle defect in Cbl mutant cells resulted from a CycE/Cdk2 activation defect, the number of cells recognized by an MPM2 monoclonal antibody was examined in Cbl mutant follicle cell clones. This antibody stains a subnuclear sphere of endocycling follicle cells and shows a positive correlation with CycE/Cdk2 activity (Calvi et al. 1998; White et al. 2007, 2011). The number of MPM2-positive cells did not change significantly in Cbl mutant clones (Figure 4, C–C′′ and E). This phenomenon was also observed in the salivary glands of Cbl mutants (Figure S5C). Similarly, by examining positive and negative effectors downstream of Notch in follicle cells, we demonstrated that Notch signaling was not altered either (Figure S7). These results indicate that Cbl is involved in endocycle regulation and may act downstream of Notch signaling.
Depletion of CTPsyn disrupts endocycles in follicle cells
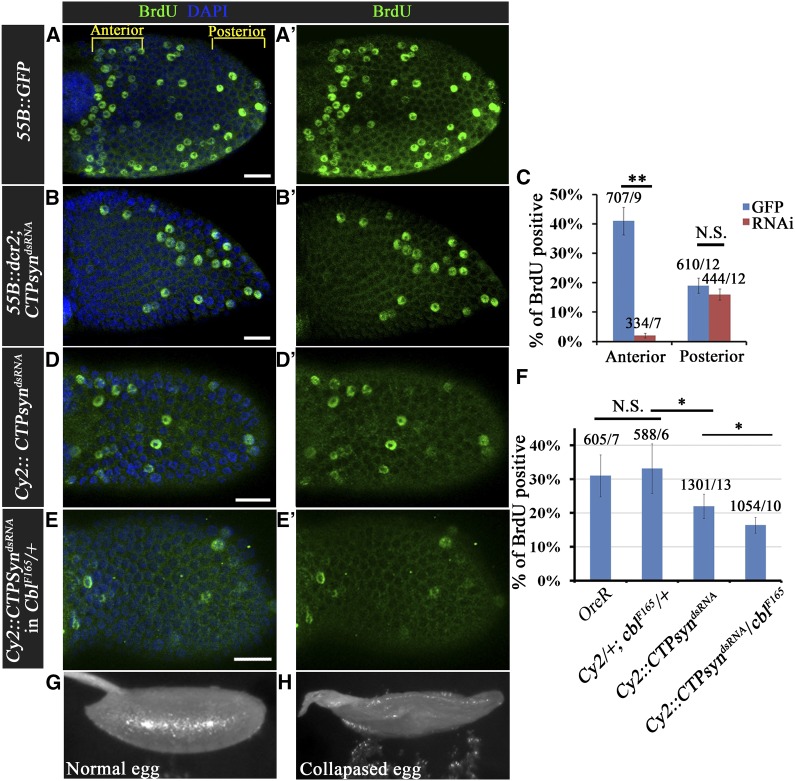
To further investigate whether CTPsyn activity is involved in the endocycle process, we examined DNA replication under CTPsyn depletion conditions. Dicer 2 (Dcr2) CTPsyn RNAi or GFP were driven by 55B-Gal4, which was expressed only in anterior follicle cells, allowing posterior follicle cells to serve as an internal control. Of the GFP-expressing control cells, 41% were BrdU-positive (Figure 5, A–A′ and C), while in CTPsyn-depleted cells the percentage of BrdU-positive cells was decreased significantly to 2% (Figure 5, B–B′ and C). The number of MPM2-positive cells did not decrease under CTPsyn-depleted conditions (Figure S5E), suggesting that CTPsyn depletion could reduce DNA replication, but not entry into the S phase. The depletion of CTPsyn could lead to an imbalance in nucleotide pools, which could disrupt the endocycle event in Drosophila follicle cells. We further examined the relationship between Cbl and CTPsyn in endocycle regulation by investigating the endocycle in CTPsyn-depleted follicle cells in Cbl +/null mutants. The Cbl heterozygous and wild-type egg chambers showed similar percentages of BrdU-positive cells situated in the anterior follicular epithelium (Figure 5F). Silencing of CTPsyn without Dcr2 led to a reduction in BrdU-positive cells from 33 to 22% (Figure 5, D–D′ and F). The percentage of BrdU-positive cells decreased further to 16% (Figure 5, E–E′ and F) in a Cbl heterozygote background. In addition, the nuclear size of follicle cells was reduced from 31.2 to 24.0 and 19.9 μm in CY2-Gal4/+;CTPsynRNAi/+ and CY2-Gal4/+;CTPsynRNAi /CblF165 stage-10A egg chambers, respectively (Figure S6E). Whereas Cbl heterozygous females generally produced normal eggs (Figure 5G), 21% of collapsed eggs were laid by females carrying a Cbl heterozygous mutation with CTPsyn depletion (Figure 5H). Therefore, Cbl interacts genetically with CTPsyn to regulate the endocycle. Taken together, Cbl may control the endocycle by maintaining the nucleotide pool balance in Drosophila follicle cells.
Figure 5.
Cbl genetically interacts with CTPsyn. (A–B′) BrdU incorporation (green) was seen in stage-9 egg chambers expressing GFP (A–A′) or CTPsyndsRNA (B–B′) in their anterior follicle cells. (C) Quantification of BrdU-positive cells in the anterior and posterior regions of GFP-expressing (blue bars) or CTPsyndsRNA-expressing (red bars) egg chambers. (D–E′) BrdU incorporation was seen in follicle cells with CTP depletion in wild-type (D and D′) and Cbl heterozygous (E and E′) backgrounds. (F) Statistics for the percentage of BrdU-positive cells in the first five rows of anterior follicle cells/egg chamber with and without CTPsyndsRNA expression in wild-type and Cbl heterozygotes. (G) Wild-type eggshells and (H) collapsed eggshells laid by females with CTPsyn depletion in a Cbl heterozygous background. DNA was stained with DAPI (blue). Results are shown as the mean ± SEM; *P < 0.05 and **P < 0.01; N.S. indicates not significant. The numbers in C and F indicate the numbers of cells/egg chamber. Bar, 20 μm.
Cbl regulates endocycle by modulating CTPsyn activity
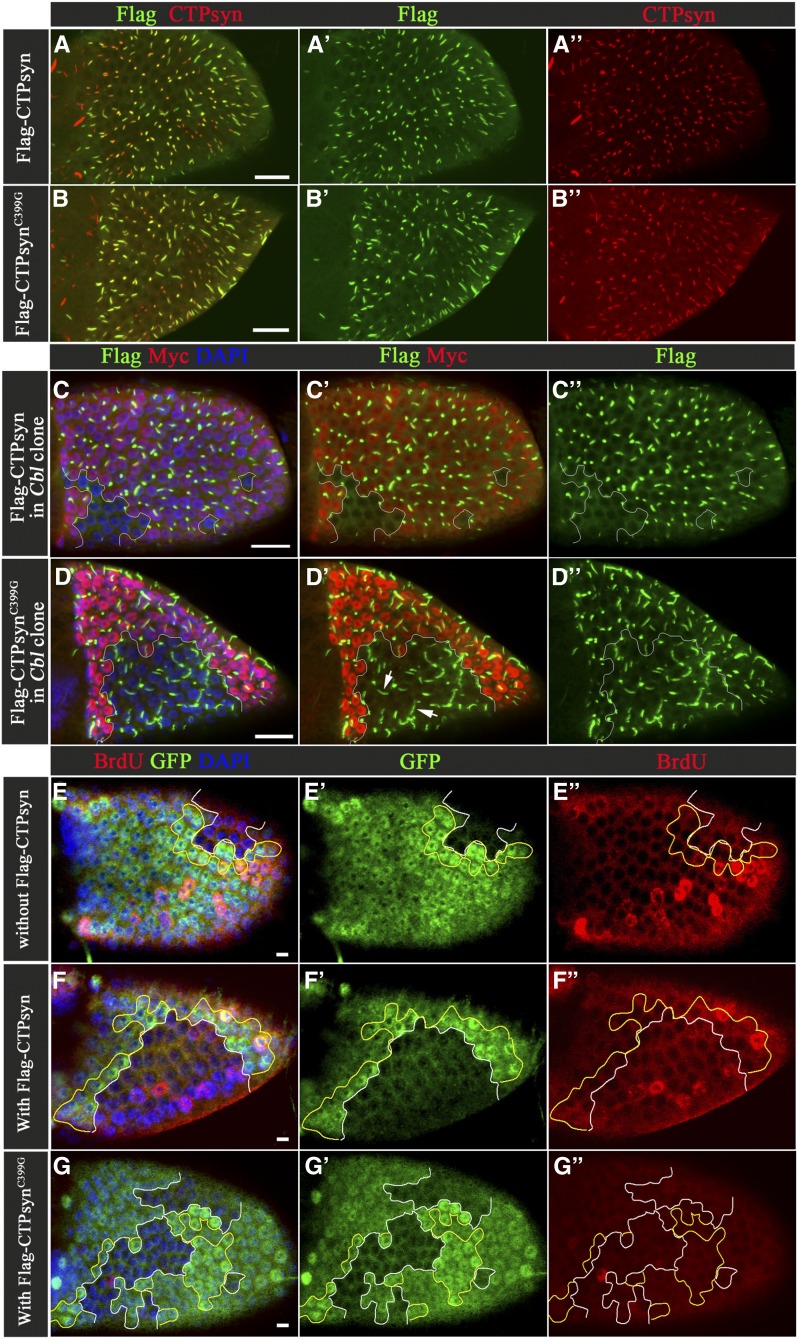
If the endoreplication defect in the Cbl mutant cell was due to an imbalance in nucleotides during the S phase, overexpression of CTPsyn might increase the activity of CTPsyn, rescuing the endoreplication defect. We found that exogenously expressed Flag-CTPsyn driven by hS-Gal4 could be incorporated into the CTPsyn filament structure in wild-type cells (Figure 6A), but that a CTPsyn filament structure was not seen in Cbl mutant cells despite overexpression of CTPsyn (Figure 6C). Nevertheless, the BrdU assay showed that overexpression of Flag-CTPsyn restored DNA replication from 16 to 25% in Cbl mutant cells (Figure 6, E–F′′ and Figure S8F). These results suggest that in Cbl mutant cells CTPsyn activity might not be adequate and that overexpression of CTPsyn could rescue this defect. If true, the catalytically defective mutant should not overcome the defect in Cbl mutant. Therefore, we generated a UAS-Flag-CTPsynC399G mutant, a mutation homologous to the C404G Ura7p mutant in yeast, which cannot bind its glutamine substrate (Noree et al. 2014). In agreement with our prediction, we found that this mutant could not restore the replication defect in Cbl mutant. Interestingly, this mutant also played a dominant-negative role in wild-type follicle cells (Figure 6G and Figure S8G), and in wild-type follicle cells, this Flag-CTPsynC399G mutant protein could be incorporated into a filament structure that contains endogenous wild-type CTPsyn (Figure 6B). Moreover, the Flag- CTPsynC399G mutant still formed filament structures in Cbl mutant follicle cells at similar expression levels to the wild-type CTPsyn (Figure 6D and Figure S8A), suggesting that the Flag-CTPsynC399G mutant protein escapes Cbl-mediated regulation in the process of CTPsyn filament formation. Glutamine depletion has been shown to induce the formation of CTPsyn polymers in a cell culture system (Calise et al. 2014). A similar effect on CTPsyn filaments was found in Drosophila S2 cells (Figure S8B′). To our surprise, this mutant could form filament structure in S2 cells in complete Schneider’s Drosophila medium (Figure S8B′), indicating that the polymerization process of this mutant may differ from that of wild-type CTPsyn. In sum, Cbl influences endocycles by affecting the nucleotide pool balance and is required for wild-type CTPsyn to form filaments.
Figure 6.
Cbl regulates endocycle through CTPsyn. (A–B′′) Merged surface confocal sections of Flag-CTPsyn-overexpressing egg chambers were stained with anti-Flag (A′ and B′, green) and anti-CTPsyn (A′′ and B′′, red) antibodies. (C–D′′) Multiple merged-surface confocal sections of Flag-CTPsyn-overexpressing egg chambers were stained with anti-Flag (green) antibody. The Flag-CTPsyn signal is shown in Cbl+/+or Cbl-/+cells (Myc-positive) and Cbl−/− follicular cells (Myc-negative). (E–G′′) BrdU incorporation is shown in Cbl mosaic stage-9 egg chambers with or without Flag-CTPsyn transgene expression. Cbl−/− clone cells are marked by the absence of GFP (outlined in white), while Cbl+/+ twin clone cells are marked by strong GFP-positive staining (outlined in yellow). BrdU signal is shown in red. DAPI-stained nuclei (blue). Bar: 20 μm (A–D) and 10 μm (E–G).
Next, we investigated whether Cbl-mediated regulation of filament formation is through direct ubiquitination of CTPsyn. The lysates of Drosophila S2 cells expressing HA-ubiquitin and Flag-CTPsyn were subjected to immunoprecipitation using anti-Flag antibody, and HA signals were detected in the immunoprecipitation products (Figure S8C). Furthermore, lysates of ovarian and salivary gland cells expressing HA-ubiquitin and Flag-CTPsyn were analyzed under similar conditions. However, no ubiquitination signal on CTPsyn was detected in either ovaries or salivary glands (Figure S8, D and E). Collectively, these results suggest that Cbl-mediated regulation of CTPsyn filament formation in follicle cells or salivary glands is not through direct ubiquitination of the CTPsyn protein itself.
Discussion
In this study, we revealed ubiquitin modification as a regulatory mechanism for the assembly of CTPsyn filaments throughout all stages of oogenesis (Figure 1 and Figure S1). Moreover, we identified Cbl as a specific E3 ubiquitin ligase that maintains these structures during endocycles (Figure 2) and showed that Cbl mutation resulted in a endoreplication-associated defect in the endocycle S phase (Figure 4). We also demonstrated that CTPsyn activity is essential for endoreplication (Figure 5) and that overexpression of wild-type CTPsyn, but not the catalytically defective mutant, is capable of rescuing the endocycle defect in Cbl mutant cells (Figure 6, E–G, and Figure S8). These results suggest that the activity of CTPsyn is insufficient to provide nucleotides for rapid endoreplication in Cbl mutant cells. Disruption of the CTPsyn filamentous structure in the absence of Cbl (Figure 2) may account for the defects in endocycles.
The ubiquitin modification of proteins by polyubiquitination or multi-monoubiquitination serves as a posttranslational regulatory mechanism to control protein stability or protein–protein interactions, respectively (Sadowski et al. 2012). In the salivary glands of Cbl null larvae, the CTPsyn protein level was not diminished (Figure 3F), suggesting that Cbl and CTPsyn filament structures are not required for stabilization of CTPsyn. However, the fact that D-Cbl-D70Z could not restore CTPsyn filament structures in Cbl mutant cells indicates that the E3 ligase activity of Cbl is required for the maintenance of the CTPsyn filament structure. In addition, we found that reversible ubiquitination regulates the dynamic assembly of both macro-and micro-cytoophidia in egg chambers. These results suggest that ubiquitination may provide a signal for polymerization of CTPsyn. While the activities of several enzymes have been reported to be controlled by ubiquitination-mediated protein degradation (Mattiroli and Sixma 2014) and by enzymatic inactivation through conformational changes (Sagar et al. 2007), the CTPsyn filament represents the first case in which an enzyme polymeric structure is regulated by ubiquitination.
Ubiquitin modification has been found in human CTPS1 using a proteomic approach (Udeshi et al. 2013). Our data indicate that Cbl-related ubiquitination of the CTPsyn protein itself might not occur in Drosophila follicle cells or salivary glands, suggesting that Cbl might act on an unknown protein that coordinates the formation of CTPsyn filament structures in an ubiquitination-dependent manner (Figure S8, D and E). We initially detected a CTPsyn filament-like structure using an antibody that was raised to recognize human phosphorylated PKD peptide. This structure was also recognized by anti-CTPsyn antibodies. Furthermore, even though Drosophila PKD has been excluded from the CTPsyn complex, we believe that CTPsyn is a complex that includes many types of enzymes, such as inosine-5′-monophosphate dehydrogenase and other proteins. This notion is supported by the recent finding that DAck colocalizes with CTPsyn filaments in germline cells (Strochlic et al. 2014).
Even though Cbl regulates CTPsyn filament structures only in follicle cells and not in germline cells, our inhibitor treatment assays and other genetic approaches in ovaries and other tissues demonstrated that CTPsyn filaments in germline cells, follicle cells, and salivary glands are sensitive to ubiquitin modification, suggesting that ubiquitination may be a general mechanism for the regulation of CTPsyn filament structures. Therefore, one possible explanation for the different effects of Cbl is that different cells may control CTPsyn filaments by different types of E3-mediated ubiquitination. Furthermore, the role of Cbl in CTPsyn filament formation seems to be crucial only in polyploid cells (Figure 2), such as follicle cells during the late endocycle stages (i.e., stages 9–10A) and the salivary glands. In contrast, in diploid cells, such as follicle cells at the mitotic stage, CTPsyn filaments are normally maintained in Cbl mutant cells (Figure S2, B–B′). One possibility is that the endocycle places additional regulations on CTPsyn filament formation in follicle cells, such as increased dynamics of assembly/disassembly. We anticipated that, if Cbl-mediated ubiquitination is required for assembly, then the polymerization of CTPsyn filaments would be reduced in the absence of Cbl, while depolymerization would be unaffected, resulting in a net effect of filament shortening. Indeed, a range of effects on CTPsyn filament structures were seen in Cbl mutant cells, suggesting that further levels of regulation might affect the role of Cbl in the formation of CTPsyn filament structures. Further studies on the mechanisms of polymerization and depolymerization of CTPsyn filaments may provide greater insight into ubiquitination-mediated regulation.
It is conceivable that to adapt quickly for growth under various physiological conditions, cells must provide sufficient materials to synthesize DNA, RNA, and lipids. Consistent with this interpretation, a special requirement for human CTP synthase1 (CTPS1) activity in DNA replication was revealed recently in receptor-mediated activation of T cells by anti-CD3 and anti-CD28 antibodies (Martin et al. 2014). The synthesis of phospholipids for membrane expansion is controlled by the levels of CTP, and the research group of Jeffrey R. Peterson at Fox Chase Cancer Center, Philadelphia, has demonstrated a role for CTPsyn in regulating the phospholipid composition of Drosophila germline cells (Chang and Carman 2008; Strochlic et al. 2014). Our results demonstrated that supplying the enzymatic activity of CTPsyn can restore endoreplication defects in Cbl mutant cells, indicating that Cbl may be involved in maintaining the nucleotide balance. During endocycles DNA replication reaches 16C within 24 hr in follicle cells and 1024C within 120 hr in the salivary glands. Loss of Cbl regulation resulted in a 50% reduction in DNA replication function (Figure 4D and Figure S6F). This defect could not be detected by measurement of nuclear size (Figure S6, A and C); however, it was reflected in the DAPI fluorescence signal in Cbl mutant salivary gland cells (Figure S6D). Given that in Cbl mutant cells CTPsyn filament structures are reduced, one possible explanation for these results is that CTPsyn filament structures are required to allow CTPsyn activity to meet the demand for nucleotide pools during endocycle. However, we still cannot exclude the possibility that Cbl regulates CTPsyn filament structures and CTPsyn activity independently.
Interestingly, CTPsyn filaments were observed under many specific physiological conditions. Nutrition depletion seems to be a common factor in the induction of this structure in yeast, brains of Drosophila larva, and mammalian cells (Aughey et al. 2014; Barry et al. 2014; Noree et al. 2010, 2014). In Drosophila S2 cells, we found that glutamine depletion induced filament formation, whereas normal Schneider’s medium allowed very little filament formation. Interestingly, also, the glutamine-binding defect mutant C399G, an enzymatically inactive CTPsyn in vivo, seemed to mimic the wild-type CTPsyn in glutamine-depleted conditions so that filaments were formed in normal medium (Figure S8, B′ and B′′). Indeed, this mutant was not regulated by Cbl and formed filament structures in Cbl mutant cells. Given that this inactive mutant mingled with endogenous wild-type CTPsyn to form filaments, we hypothesize that CTPsyn filaments may be composed of different types of CTPsyn, including active, inactive, or a mixture of both forms.
CTP biosynthesis is one of several biosynthesis pathways implicated in liver malignancy and kidney carcinoma, and CTPsyn activity is increased in hepatomas (Williams et al. 1978). As the transition from tetraploidy to aneuploidy is considered to be an important step in cancer development (Davoli and de Lange 2011), the role of the proto-oncogene Cbl in the regulation of CTPsyn activity during endocycles implies a new mechanism through which Cbl promotes cancer formation. In this study, we defined a new role for the proto-oncogene Cbl in endocycles. We demonstrated that Cbl-mediated ubiquitination controls the assembly of CTPsyn filaments and may regulate the nucleotide supply for rapid endoreplication in Drosophila.
Supplementary Material
Acknowledgments
We thank Hai-Wei Pi, Fly Core Taiwan, the Bloomington Stock Center, and the Vienna Stock Center for providing the reagents and fly stocks; Trudi Schüpbach, Ji-Long Liu, Mark Peifer, Jörg Grosshans, Cheng-Ting Chien, Hai-Wei Pi, Bertrand Tan, Scott C. Schuyler, Tzu-Chien Van Wang, Chien-Kuo Lee, and Shu-Yuan Yang for commenting on the manuscript; and members of the Pai laboratory for their comments and helpful discussions. This work was supported by grants from the National Science Council of the Republic of China (NSC 101-2811-B-182-017 to P.-Y.W. and MOST103-2311-B-182-004-MY3 to L.-M.P.); the Chang Gung Memorial Hospital (CMRPD-2B0061-3 to L.-M.P.); and the Ministry of Education, Taiwan (EMRPD1E1421 to L.-M.P.).
Footnotes
Communicating editor: R. J. Duronio
Supporting information is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.180737/-/DC1.
Literature Cited
- Anderson P. M., 1983. CTP synthetase from Escherichia coli: an improved purification procedure and characterization of hysteretic and enzyme concentration effects on kinetic properties. Biochemistry 22: 3285–3292. [DOI] [PubMed] [Google Scholar]
- Aronow B., Ullman B., 1987. In situ regulation of mammalian CTP synthetase by allosteric inhibition. J. Biol. Chem. 262: 5106–5112. [PubMed] [Google Scholar]
- Aughey G. N., Grice S. J., Shen Q. J., Xu Y., Chang C. C., et al. , 2014. Nucleotide synthesis is regulated by cytoophidium formation during neurodevelopment and adaptive metabolism. Biol. Open 3: 1045–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzam G., Liu J. L., 2013. Only one isoform of Drosophila melanogaster CTP synthase forms the cytoophidium. PLoS Genet. 9: e1003256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry R. M., Bitbol A. F., Lorestani A., Charles E. J., Habrian C. H., et al. , 2014. Large-scale filament formation inhibits the activity of CTP synthetase. eLife 3: e03638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buszczak M., Paterno S., Lighthouse D., Bachman J., Planck J., et al. , 2007. The carnegie protein trap library: a versatile tool for Drosophila developmental studies. Genetics 175: 1505–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calise S. J., Carcamo W. C., Krueger C., Yin J. D., Purich D. L., et al. , 2014. Glutamine deprivation initiates reversible assembly of mammalian rods and rings. Cell. Mol. Life Sci. 71: 2963–2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvi B. R., Spradling A. C., 1999. Chorion gene amplification in Drosophila: a model for metazoan origins of DNA replication and S-phase control. Methods 18: 407–417. [DOI] [PubMed] [Google Scholar]
- Calvi B. R., Lilly M. A., Spradling A. C., 1998. Cell cycle control of chorion gene amplification. Genes Dev. 12: 734–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carcamo W. C., Satoh M., Kasahara H., Terada N., Hamazaki T., et al. , 2011. Induction of cytoplasmic rods and rings structures by inhibition of the CTP and GTP synthetic pathway in mammalian cells. PLoS One 6: e29690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty K. P., Hurlbert R. B., 1961. Role of glutamine in the biosynthesis of cytidine nucleotides in Escherichia coli. Biochim. Biophys. Acta 47: 607–609. [DOI] [PubMed] [Google Scholar]
- Chang W. L., Liou W., Pen H. C., Chou H. Y., Chang Y. W., et al. , 2008. The gradient of Gurken, a long-range morphogen, is directly regulated by Cbl-mediated endocytosis. Development 135: 1923–1933. [DOI] [PubMed] [Google Scholar]
- Chang Y. F., Carman G. M., 2008. CTP synthetase and its role in phospholipid synthesis in the yeast Saccharomyces cerevisiae. Prog. Lipid Res. 47: 333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K., Zhang J., Tastan O. Y., Deussen Z. A., Siswick M. Y., et al. , 2011. Glutamine analogs promote cytoophidium assembly in human and Drosophila cells. J. Genet. Genomics 38: 391–402. [DOI] [PubMed] [Google Scholar]
- Davoli T., de Lange T., 2011. The causes and consequences of polyploidy in normal development and cancer. Annu. Rev. Cell Dev. Biol. 27: 585–610. [DOI] [PubMed] [Google Scholar]
- Deng W. M., Althauser C., Ruohola-Baker H., 2001. Notch-Delta signaling induces a transition from mitotic cell cycle to endocycle in Drosophila follicle cells. Development 128: 4737–4746. [DOI] [PubMed] [Google Scholar]
- Edgar B. A., Orr-Weaver T. L., 2001. Endoreplication cell cycles: more for less. Cell 105: 297–306. [DOI] [PubMed] [Google Scholar]
- Endrizzi J. A., Kim H., Anderson P. M., Baldwin E. P., 2004. Crystal structure of Escherichia coli cytidine triphosphate synthetase, a nucleotide-regulated glutamine amidotransferase/ATP-dependent amidoligase fusion protein and homologue of anticancer and antiparasitic drug targets. Biochemistry 43: 6447–6463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein A. M., Bauer C. R., Ho A., Bosco G., Zarnescu D. C., 2009. Drosophila Fragile X protein controls cellular proliferation by regulating cbl levels in the ovary. Dev. Biol. 330: 83–92. [DOI] [PubMed] [Google Scholar]
- Gou K. M., Chang C. C., Shen Q. J., Sung L. Y., Liu J. L., 2014. CTP synthase forms cytoophidia in the cytoplasm and nucleus. Exp. Cell Res. 323: 242–253. [DOI] [PubMed] [Google Scholar]
- Holden J. J., Suzuki D. T., 1973. Temperature-sensitive mutations in Drosophila melanogaster. XII. The genetic and developmental effects of dominant lethals on chromosome 3. Genetics 73: 445–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingerson-Mahar M., Briegel A., Werner J. N., Jensen G. J., Gitai Z., 2010. The metabolic enzyme CTP synthase forms cytoskeletal filaments. Nat. Cell Biol. 12: 739–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klusza S., Deng W. M., 2011. At the crossroads of differentiation and proliferation: precise control of cell-cycle changes by multiple signaling pathways in Drosophila follicle cells. BioEssays 33: 124–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. O., Davidson J. M., Duronio R. J., 2009. Endoreplication: polyploidy with purpose. Genes Dev. 23: 2461–2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitzki A., Koshland D. E., Jr, 1971. Cytidine triphosphate synthetase. Covalent intermediates and mechanisms of action. Biochemistry 10: 3365–3371. [DOI] [PubMed] [Google Scholar]
- Levitzki A., Koshland D. E., Jr, 1972. Role of an allosteric effector. Guanosine triphosphate activation in cytosine triphosphate synthetase. Biochemistry 11: 241–246. [DOI] [PubMed] [Google Scholar]
- Lieberman I., 1956. Enzymatic amination of uridine triphosphate to cytidine triphosphate. J. Biol. Chem. 222: 765–775. [PubMed] [Google Scholar]
- Lilly M. A., Spradling A. C., 1996. The Drosophila endocycle is controlled by Cyclin E and lacks a checkpoint ensuring S-phase completion. Genes Dev. 10: 2514–2526. [DOI] [PubMed] [Google Scholar]
- Liu J. L., 2010. Intracellular compartmentation of CTP synthase in Drosophila. J. Genet. Genomics 37: 281–296. [DOI] [PubMed] [Google Scholar]
- Long C. W., Pardee A. B., 1967. Cytidine triphosphate synthetase of Escherichia coli B. I. Purification and kinetics. J. Biol. Chem. 242: 4715–4721. [PubMed] [Google Scholar]
- Long C. W., Levitzki A., Koshland D. E., Jr, 1970. The subunit structure and subunit interactions of cytidine triphosphate synthetase. J. Biol. Chem. 245: 80–87. [PubMed] [Google Scholar]
- Lopez-Schier H., St. Johnston D., 2001. Delta signaling from the germ line controls the proliferation and differentiation of the somatic follicle cells during Drosophila oogenesis. Genes Dev. 15: 1393–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmor M. D., Yarden Y., 2004. Role of protein ubiquitylation in regulating endocytosis of receptor tyrosine kinases. Oncogene 23: 2057–2070. [DOI] [PubMed] [Google Scholar]
- Martin E., Palmic N., Sanquer S., Lenoir C., Hauck F., et al. , 2014. CTP synthase 1 deficiency in humans reveals its central role in lymphocyte proliferation. Nature 510: 288–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattiroli F., Sixma T. K., 2014. Lysine-targeting specificity in ubiquitin and ubiquitin-like modification pathways. Nat. Struct. Mol. Biol. 21: 308–316. [DOI] [PubMed] [Google Scholar]
- Melikova M. S., Kondratov K. A., Kornilova E. S., 2006. Two different stages of epidermal growth factor (EGF) receptor endocytosis are sensitive to free ubiquitin depletion produced by proteasome inhibitor MG132. Cell Biol. Int. 30: 31–43. [DOI] [PubMed] [Google Scholar]
- Noree C., Sato B. K., Broyer R. M., Wilhelm J. E., 2010. Identification of novel filament-forming proteins in Saccharomyces cerevisiae and Drosophila melanogaster. J. Cell Biol. 190: 541–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noree C., Monfort E., Shiau A. K., Wilhelm J. E., 2014. Common regulatory control of CTP synthase enzyme activity and filament formation. Mol. Biol. Cell 25: 2282–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai L. M., Barcelo G., Schupbach T., 2000. D-cbl, a negative regulator of the Egfr pathway, is required for dorsoventral patterning in Drosophila oogenesis. Cell 103: 51–61. [DOI] [PubMed] [Google Scholar]
- Pai L. M., Wang P. Y., Chen S. R., Barcelo G., Chang W. L., et al. , 2006. Differential effects of Cbl isoforms on Egfr signaling in Drosophila. Mech. Dev. 123: 450–462. [DOI] [PubMed] [Google Scholar]
- Queenan A. M., Ghabrial A., Schupbach T., 1997. Ectopic activation of torpedo/Egfr, a Drosophila receptor tyrosine kinase, dorsalizes both the eggshell and the embryo. Development 124: 3871–3880. [DOI] [PubMed] [Google Scholar]
- Sadowski M., Suryadinata R., Tan A. R., Roesley S. N., Sarcevic B., 2012. Protein monoubiquitination and polyubiquitination generate structural diversity to control distinct biological processes. IUBMB Life 64: 136–142. [DOI] [PubMed] [Google Scholar]
- Sagar G. D., Gereben B., Callebaut I., Mornon J. P., Zeold A., et al. , 2007. Ubiquitination-induced conformational change within the deiodinase dimer is a switch regulating enzyme activity. Mol. Cell. Biol. 27: 4774–4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saville K. J., Belote J. M., 1993. Identification of an essential gene, l(3)73Ai, with a dominant temperature-sensitive lethal allele, encoding a Drosophila proteasome subunit. Proc. Natl. Acad. Sci. USA 90: 8842–8846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherbata H. R., Althauser C., Findley S. D., Ruohola-Baker H., 2004. The mitotic-to-endocycle switch in Drosophila follicle cells is executed by Notch-dependent regulation of G1/S, G2/M and M/G1 cell-cycle transitions. Development 131: 3169–3181. [DOI] [PubMed] [Google Scholar]
- Strochlic T. I., Stavrides K. P., Thomas S. V., Nicolas E., O’Reilly A. M., et al. , 2014. Ack kinase regulates CTP synthase filaments during Drosophila oogenesis. EMBO Rep. 15: 1184–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tower J., 2004. Developmental gene amplification and origin regulation. Annu. Rev. Genet. 38: 273–304. [DOI] [PubMed] [Google Scholar]
- Udeshi N. D., Svinkina T., Mertins P., Kuhn E., Mani D. R., et al. , 2013. Refined preparation and use of anti-diglycine remnant (K-epsilon-GG) antibody enables routine quantification of 10,000s of ubiquitination sites in single proteomics experiments. Mol. Cell. Proteomics 12: 825–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von der Saal W., Anderson P. M., Villafranca J. J., 1985. Mechanistic investigations of Escherichia coli cytidine-5′-triphosphate synthetase. Detection of an intermediate by positional isotope exchange experiments. J. Biol. Chem. 260: 14993–14997. [PubMed] [Google Scholar]
- Wang P. Y., Pai L. M., 2011. D-Cbl binding to Drk leads to dose-dependent down-regulation of EGFR signaling and increases receptor-ligand endocytosis. PLoS One 6: e17097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Chen Z., Bergmann A., 2010. Regulation of EGFR and Notch signaling by distinct isoforms of D-cbl during Drosophila development. Dev. Biol. 342: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White A. E., Leslie M. E., Calvi B. R., Marzluff W. F., Duronio R. J., 2007. Developmental and cell cycle regulation of the Drosophila histone locus body. Mol. Biol. Cell 18: 2491–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White A. E., Burch B. D., Yang X. C., Gasdaska P. Y., Dominski Z., et al. , 2011. Drosophila histone locus bodies form by hierarchical recruitment of components. J. Cell Biol. 193: 677–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J. C., Kizaki H., Weber G., Morris H. P., 1978. Increased CTP synthetase activity in cancer cells. Nature 271: 71–73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.