Abstract
The body size of Caenorhabditis elegans is thought to be controlled by sensory inputs because many mutants with sensory cilium structure defects exhibit small body size. The EGL-4 cGMP-dependent protein kinase acts in sensory neurons to reduce body size when animals fail to perceive sensory signals. In addition to body size control, EGL-4 regulates various other behavioral and developmental pathways, including those involved in the regulation of egg laying and chemotaxis behavior. Here we have identified gcy-12, which encodes a receptor-type guanylyl cyclase, as a gene involved in the sensory regulation of body size. Analyses with GFP fusion constructs showed that gcy-12 is expressed in several sensory neurons and localizes to sensory cilia. Genetic analyses indicated that GCY-12 acts upstream of EGL-4 in body size control but does not affect other EGL-4 functions. Our studies indicate that the function of the GCY-12 guanylyl cyclase is to provide cGMP to the EGL-4 cGMP-dependent kinase only for limited tasks including body size regulation. We also found that the PDE-2 cyclic nucleotide phosphodiesterase negatively regulates EGL-4 in controlling body size. Thus, the cGMP level is precisely controlled by GCY-12 and PDE-2 to determine body size through EGL-4, and the defects in the sensory cilium structure may disturb the balanced control of the cGMP level. The large number of guanylyl cyclases encoded in the C. elegans genome suggests that EGL-4 exerts pleiotropic effects by partnering with different guanylyl cyclases for different downstream functions.
Keywords: body size, Caenorhabditis elegans, guanylyl cyclase, cGMP-dependent protein kinase, sensory cilia
THE perception of sensory cues allows an animal to respond appropriately to the environment and to adapt to changing conditions. The Caenorhabditis elegans hermaphrodite has 302 neurons, 60 of which are ciliated sensory neurons. Through the cilia of sensory neurons, C. elegans perceives taste, odor, osmotic change, and pheromones. Similar to the cilia of sensory neurons in other organisms, the sensory cilia in C. elegans are specialized structures where various signaling components, including sensory receptors and G proteins, are localized (Silverman and Leroux 2009).
The importance of cilia-based sensory perception in C. elegans development and behavior has been demonstrated by analyses of a class of mutants that lack a normal cilium structure (Perkins et al. 1986). These mutants, such as che-2, show reduced responses to taste, odor, osmotic change, and pheromones (Lewis and Hodgkin 1977; Bargmann et al. 1993; Fujiwara et al. 1999). Further analyses of these mutants also indicated that normal sensory cilia are required for the regulation of various important aspects of C. elegans development and physiology, including body size, locomotory pattern, life span, and fat storage (Apfeld and Kenyon 1999; Fujiwara et al. 2002; Alcedo and Kenyon 2004; Mak et al. 2006). Interestingly, sensory cues were also shown to regulate life span in Drosophila (Libert et al. 2007), indicating evolutionarily conserved roles of sensory perception through cilia.
Adult body size of che-2 and other cilium defective mutants showed a 20–30% decrease compared with that of the wild-type animal (Fujiwara et al. 2002; Fujiwara et al. 2010). Several other mutants with defects in sensory perception were also reported to exhibit small body size (Kuhara et al. 2002; Lanjuin and Sengupta 2002). A screen for suppressor mutants of small body size due to a che-2 mutation revealed an important role of a cGMP-dependent protein kinase (PKG), EGL-4, in body size regulation (Fujiwara et al. 2002). Mutations in egl-4 result in a large-body phenotype and completely suppress the small-body phenotype of the cilium structure mutant (Fujiwara et al. 2002; Hirose et al. 2003). This suggests that EGL-4 acts to reduce body size, and that cilia-dependent signal(s) keep body size normal by inhibiting EGL-4 activity. egl-4 is expressed in several neurons and other tissues. Although the egl-4 expression in muscle, hypodermis, and intestine has a certain rescuing effect, the expression in sensory neurons is sufficient for normal body size regulation (Fujiwara et al. 2002; Nakano et al. 2004). The mechanism by which EGL-4 PKG activity in sensory neurons affects body size is unknown. Genetic analysis suggests that several components in TGF-β signaling pathways, including the DBL-1/TGF-β ligand and DAF-3/Smad transcription factor, are involved in body-size regulation downstream of EGL-4 (Daniels et al. 2000; Fujiwara et al. 2002; Nakano et al. 2004). EGL-4 also regulates gene expression by antagonizing a histone deacetylase (van der Linden et al. 2008). Another study has shown that EGL-4 affects ploidy in hypodermal cells (Tain et al. 2008). While the precise mechanism is unknown, this accumulating evidence suggests that under the control of sensory inputs, EGL-4 regulates body size by affecting humoral factors through transcriptional regulation and alteration of ploidy in somatic tissues.
The mechanism by which EGL-4 kinase activity is controlled by sensory inputs is also unclear. Since EGL-4 kinase activity is dependent on cGMP (Stansberry et al. 2001; Hirose et al. 2003), it is likely provided by one or more of the guanylyl cyclases encoded in the C. elegans genome. A total of 34 guanylyl cyclases (GC) have been identified in the C. elegans genome, of which 7 are soluble cyclases and 27 are receptor-type cyclases (Ortiz et al. 2006). Compared with other species, the number of receptor-type GCs encoded in the C. elegans genome is unusually large. For instance, the Drosophila genome contains 6 and mammalian genomes contain 7 (Morton 2004). The GCs in C. elegans are expressed in different subsets of neurons and/or tissues such as muscle and intestine (Ortiz et al. 2006), suggesting a distinct function for each GC. For example, ODR-1 and DAF-11 receptor-type GCs are expressed in many chemosensory neurons and control opening of the TAX-4 cyclic-nucleotide-gated channel, thereby playing indispensable roles in chemosensation (Birnby et al. 2000; L’Etoile and Bargmann 2000). However, the GCs involved in the EGL-4 activation upstream of body size regulation have not been identified.
EGL-4 seems to be a multifunctional regulator with apparently unrelated functions, because egl-4 mutations cause not only the body size phenotype but also a wide variety of phenotypes including defects in the control of egg laying, dauer formation, fat storage, chemosensory behavior, locomotory state, life span, sleep-like state, and even satiety signaling (Trent et al. 1983; Daniels et al. 2000; L’Etoile et al. 2002; Hirose et al. 2003; Raizen et al. 2006; Raizen et al. 2008; You et al. 2008; Hao et al. 2011; O’halloran et al. 2012). Therefore, EGL-4 activity upstream of each of these functions is likely to be controlled separately in C. elegans.
In this article, we report a new suppressor mutation of the small-body phenotype of the che-2 mutant, whose responsible gene encodes a guanylyl cyclase, GCY-12. Analyses of gcy-12 mutants revealed that only some EGL-4 functions require GCY-12, while other functions may require different guanylyl cyclases. We also found that a cyclic nucleotide phosphodiesterase, PDE-2, acts as another EGL-4 regulator of body size control. Our findings point to the importance of cGMP signaling in sensory body size regulation and suggest a complex mechanism underlying the control of cGMP-dependent kinase activity.
Materials and Methods
Strains and genetics
Wild-type animals were the C. elegans strain N2. Worms were grown at 20° with food using standard methods (Brenner 1974).
Strains used in this work included gcy-12(ks99), gcy-12(ks100), gcy-12(nj10), che-2(e1033), egl-4(ky185), egl-4(n478), daf-11(m47), odr-1(n1936), and pde-2(qj6). Double mutants were generated using standard methods and were confirmed by complementation tests or by sequencing. che-2;Ex[che-2/H20::gfp] was provided by Dr I. Katsura, Nationai Institue of Genetics, Japan (Fujiwara et al. 1999).
The pde-2(qj6) mutant was generated by Tc1 transposon insertion and subsequent imprecise excision, which caused deletion of a 1129-bp coding region containing exons 3-5 of pde-2 and the insertion of an extra 132 bp of sequence originating from the Tc1 transposon. Because of a stop codon in the inserted Tc1 sequence, the pde-2(qj6) locus is expected to produce a truncated protein lacking both the GAF domain and the catalytic domain of PDE. A 9-kb genomic DNA fragment containing the pde-2 gene and its 3.0-kb-upstream and 0.5-kb-downstream regions was amplified by PCR and used for a rescue experiment.
Body size measurement
L4 crescent stage animals were transferred to 6-cm seeded NGM plates and maintained in a well-fed condition until body size measurement. For body volume measurement, animals were anesthetized with NaN3 48 hr after the L4 stage and measured using ‘Senchu-gazou-kaiseki-souchi SVK-3A’ (Showa Denki Co., Fukuoka, Japan) as described by Hirose et al. (2003). Each data point represents the average volume of >20 animals. All strains compared in each graph were cultured and measured at the same time/condition.
Screen for che-2 small-body-size suppressors (Chb), mapping, and cloning of gcy-12
che-2(e1033) animals were mutagenized with 50 mM ethyl methanesulfonate (EMS) using the standard protocol (Brenner 1974) and 40,000 F2 progenies (from ∼3000 F1 progenies) were screened for animals that exhibited increased body size but that still retained a ciliary defect. ks100 was mapped using the single nucleotide polymorphism (SNP) method (Wicks et al. 2001), based on a suppressor phenotype of the reduced body size and confined tracking pattern of che-2. The locus was mapped to a ∼120-kb region (between snip markers on the cosmids C54A12 and EEED8) around the center of LGII. Among the cosmid clones covering this area, ZK622 and F08B1 showed rescuing activities; che-2(e1033);ks100 double mutant animals exhibited a decrease in body size when either of the cosmids was introduced. ZK622 and F08B1 are adjoining cosmids and only one gene, gcy-12, exists in the overlapping region. A genomic fragment containing the whole coding region and 3 kb upstream of the predicted start site of the gcy-12 gene also rescued the body size phenotype of ks100.
Dye-filling assay and other behavioral assays
Dye-filling assay: assays were performed as described by Starich et al. (1995) using DiI (10 µg/ml). More than 30 animals per strain were observed.
Chemotaxis: assays were performed as described by Bargmann et al. (1993). Each data point represents the average chemotaxis index of 9–14 chemotaxis plates.
Adaptation to benzaldehyde: assays were performed as described by L’Etoile et al. (2002). To adapt and test animals, benzaldehyde at 1/50000 and 1/300 dilution were used, respectively. Each data point represents the average chemotaxis index of 8–11 chemotaxis plates.
Counting eggs in the uterus: single young adult animal (27 hr after the L4 stage) was bleached and the eggs left behind were counted. Twenty-six to 36 animals per strain were counted.
Dauer formation: eggs were gathered by bleaching well-fed adults and placed on seeded plates (200–300 eggs/plate). After 50 hr at 27°, the numbers of dauers and non-dauers were counted. The criteria for dauer formation were larvae with a long body shape, dark intestine, nonpumping pharynxes, and lethargic movements. Each data point represents the average dauer rate of 11–14 culture plates.
Plasmid construction
genomic gcy-12::gfp:
A genomic gcy-12::gfp construct was made by inserting an 8.1-kb genomic fragment containing a 3-kb promoter region and the entire coding region of gcy-12 into pPD95.77 (GFP expression vector; a gift from Dr A. Fire, Stanford University) in frame using XbaI and MscI sites. The stop codon was substituted to a BalI site by PCR so that GFP was fused to the C terminus of the GCY-12 protein.
gcy-12 expression construct for rescue and overexpression:
A gcy-12 expression construct was made by inserting an 8.1-kb genomic fragment containing a 3-kb promoter region and the entire coding region of gcy-12 into pPD49.26 (expression vector; a gift from Dr A. Fire) using XbaI and KpnI sites.
odr-1p::gcy-12::gfp, srb-6p::gcy-12::gfp, tax-4p::gcy-12::gfp, ceh-36p::gcy-12::gfp, gpa-9p::gcy-12::gfp, dat-1p::gfp constructs:
First, the GCY-12::GFP expression vector was made by inserting a gcy-12 cDNA (obtained from cDNA clone, yk316d1, provided by Dr Y. Kohara, National Institute of genetics, Japan; the 5′ part was supplemented with a product of 5′ RACE) into pPD95.77 in frame so that GFP was fused to the C terminus of the GCY-12 protein. Then, to generate different promoter-gcy-12 constructs, a 2.4-kb odr-1 promoter region, a 3.3-kb srb-6 promoter region, a 3-kb tax-4 promoter region, a 3.1-kb ceh-36 promoter region, a 2.8-kb gpa-9 promoter region, or a 1.4-kb dat-1 promoter region were inserted into the GCY-12::GFP expression vector.
gcy-12p::gcy-12::gfp and gcy-12p::gcy-12 (ΔECD)::gfp:
gcy-12p::gcy-12::gfp was made by inserting a 3-kb gcy-12 promoter region into the GCY-12::GFP expression vector. For gcy-12p::gcy-12 (ΔECD)::gfp, the coding region for the 442-amino acid (aa) extracellular domain of gcy-12 cDNA was deleted.
Generation of transgenic worms
Transgenic strains were generated by standard micro-injection methods (Mello et al. 1991). Unless otherwise noted, test DNA was injected at 100 ng/µl with myo-3::gfp DNA at 33 ng/µl (a GFP construct expressed in body wall muscle) or lin-44::gfp at 33 ng/µl (a GFP construct expressed in hypodermal cells at the tip of the tail) or ttx-3::mRFP at 33 ng/µl (an mRFP construct expressed in AIY neurons) as a co-injection marker. Generally we isolated multiple (two to five) independent transgenic lines from each injection and confirmed that there were no major differences among them. For gcy-12 overexpression analysis, the plasmid was injected into N2 animals at 150 ng/µl with the co-injection marker, sra-6::gfp (33 ng/µl). To compare the effect of gcy-12 overexpression in different genetic backgrounds, the same extrachromosomal arrays were transferred by mating from N2 to egl-4(ky185) and to egl-4(n478).
Data availability
All strains and plasmids described here and in the Supporting Information are available upon request.
Results
Identification of new Chb mutations, ks99 and ks100
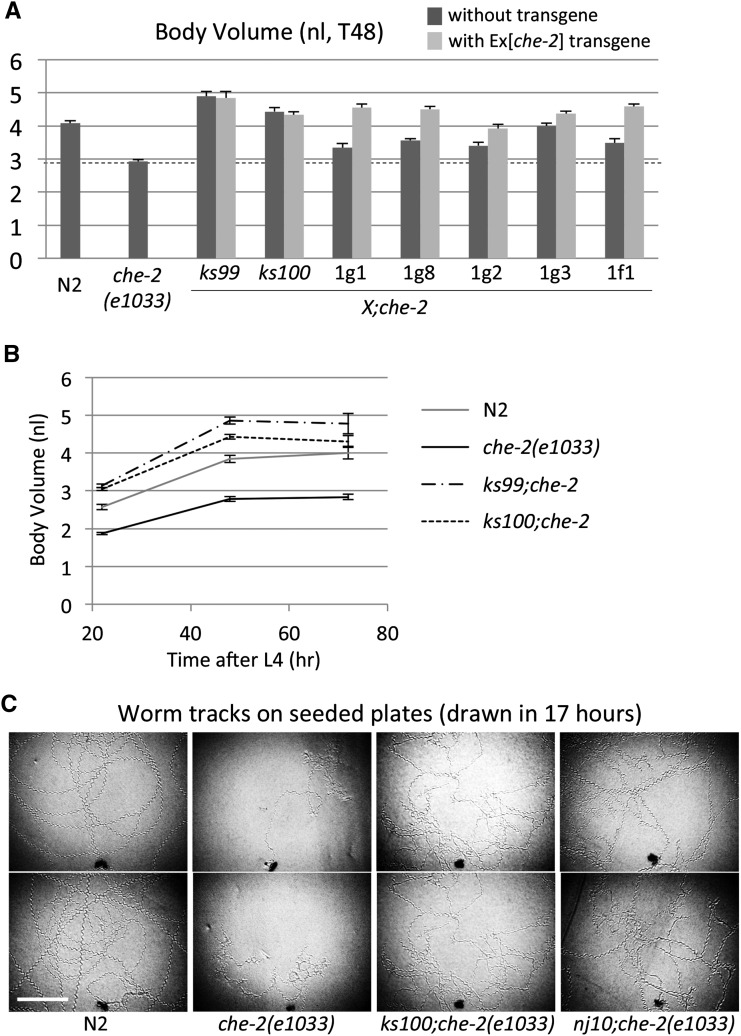
To identify new genes involved in the sensory body size regulation, we performed a Chb (che-2 small-body-size suppressors) screen because a previous screen was apparently not saturated (Fujiwara et al. 2002). We mutagenized che-2(e1033) animals by EMS treatment and looked for animals with an increased body size. In the 6000 haploid-genome scale screening, we isolated six candidate lines (X;che-2) (Figure 1). All six of these lines still demonstrated the dye-filling defect, indicating that they still retained the che-2 mutation.
Figure 1.
The mutants identified in a Chb (che-2 small-body suppressor) screen. (A) The adult stage (48 hr after the L4 stage) body volumes of the che-2(e1033) mutant and of seven suppressor mutants, x;che-2(e1033). The che-2 gene (Ex[che-2]) was introduced into each suppressor mutant to test whether it increased body size further (class II suppressors) or whether body size remained unchanged (class I suppressors). ks99 and ks100 were classified as class I and the other mutants were classified as class II. (B) Time-course analyses of ks99 and ks100 body size. Body volumes were measured at the indicated time points following the L4 stage. (C) Worm tracking patterns on an E. coli lawn. A single worm was left on a seeded plate for 17 hr and allowed to move freely. Two examples of each strain are shown. Scale bar, 5 mm. Error bars indicate SEMs.
Next, we introduced a che-2 transgene (Ex[che-2]) into each candidate line (X;che-2) to rescue the che-2 locus. If the animals with the transgene (X;che-2;Ex[che-2]) exhibited the same body size as those without the transgene (X;che-2), we classified the line into class I. Class I suppressor genes presumably act downstream of che-2 dependent sensory inputs for body size regulation and hence rescue of the che-2 mutation is expected to have no effect on the body size of class I suppressor mutants. In contrast, if the animals with the transgene exhibit an increase in body size relative to control animals without the transgene, we classified the line into class II. Since the effects of che-2 and the suppressor mutation are additive, class II suppressor genes are likely to act in an independent pathway parallel to the che-2 pathway. Out of the six mutations, two (ks99 and ks100) were classified as class I suppressors, while the other four were classified as class II (Figure 1).
Interestingly, we found that ks99 and ks100, but none of the class II mutations, suppressed the confined tracking pattern of che-2 animals (Figure 1 for ks100, data not shown for ks99, see below for nj10). This is a common characteristic of class I mutations, including egl-4 (Fujiwara et al. 2002), indicating that the body size and the locomotory pattern are controlled by the same molecular mechanism.
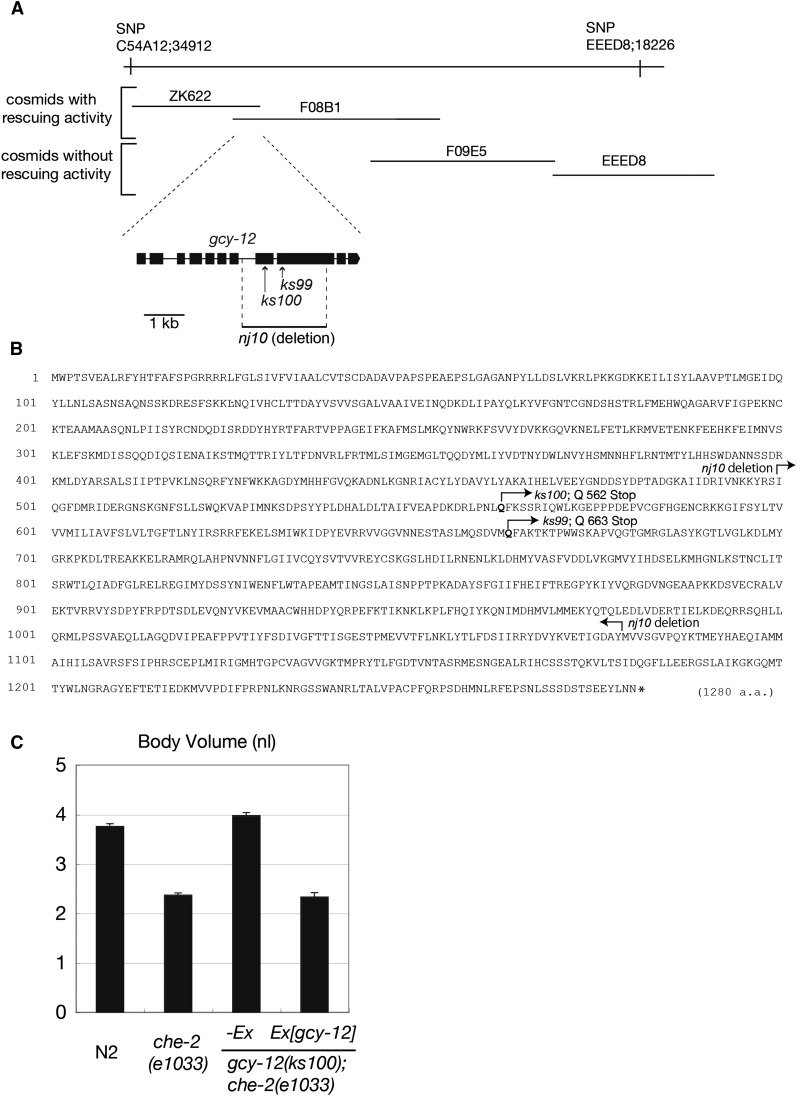
Both ks99 and ks100 are mutations in gcy-12
ks99 and ks100 mutations were linked to linkage group II (LGII); therefore, we performed a complementation test and found that ks99 and ks100 could not complement each other’s Chb phenotype (data not shown), suggesting that they are mutations in the same gene. Subsequently, ks100 was used for further mapping to identify the responsible gene. By SNP mapping, we have narrowed the mutation site to a ∼120-kb region (between snip markers on cosmids C54A12 and EEED8) that is near the center of LGII. We introduced several cosmids covering the area into che-2(e1033);ks100 double mutant animals and examined the rescuing activity of each cosmid, the rescue phenotype being a decrease in body size. Two adjoining cosmids, ZK622 and F08B1, showed a rescue, and only one gene, gcy-12, exists in the overlapping region. Because general toxicity of introduced DNA can reduce body size, we first sequenced the gcy-12 locus of ks100 and ks99 animals for mutations. As expected, both ks100 and ks99 had mutations in the coding region of gcy-12, which are predicted to result in early termination of translation (Figure 2, A and B). The genomic fragment containing the whole coding region and 3 kb upstream of the predicted start site showed rescuing activity of the Chb phenotype (Figure 2C).
Figure 2.
gcy-12 is a new Chb mutant. (A) Genetic and physical maps of ks100. SNP mapping placed ks100 in a ∼120-kb region around the center of LGII. Two cosmids, ZK622 and F08B1, rescued ks100. Only one gene, gcy-12 was encoded in the overlap region of the two cosmids. (B) gcy-12 encodes a 1280-aa receptor-type guanylyl cyclase. The amino acid sequence was predicted from a cDNA obtained by RT–PCR and an EST clone, yk316d1. Mutation sites in ks99 and ks100 and the amino acid sequence deleted in nj10 are shown. (C) The transgene containing the entire gcy-12 coding region (Ex[gcy-12]) reduced the body size of the gcy-12(ks100);che-2(e1033) mutant to that of the che-2(e1033) mutant. Body volumes at the adult stage (48 hr after the L4 stage) are shown. Error bars indicate SEMs.
gcy-12 encodes a receptor-type guanylyl cyclase. It consists of an extracellular ligand-binding domain, a transmembrane domain, a kinase-like domain, and a guanylyl cyclase catalytic domain. The nonsense mutations in ks99 and ks100 are predicted to result in GCY-12 proteins that lack all intracellular domains, including the catalytic domain, indicating that both are likely to be null alleles. To further characterize gcy-12 mutants, we also analyzed a deletion allele, nj10, obtained by the TMP/UV method (Gengyo-Ando and Mitani 2000). nj10 has a 2197-bp deletion removing exon 8 and part of exon 9 (Figure 2, A and B). The deleted region corresponds to the transmembrane domain, the kinase-like domain, and about half of the guanylyl cyclase domain; therefore, the nj10 allele represents a null allele. gcy-12(nj10) suppressed the small body size of the che-2 mutant in the same manner as ks100 and ks99 (Supporting Information, Figure S1).
Genetic interaction between gcy-12 and egl-4
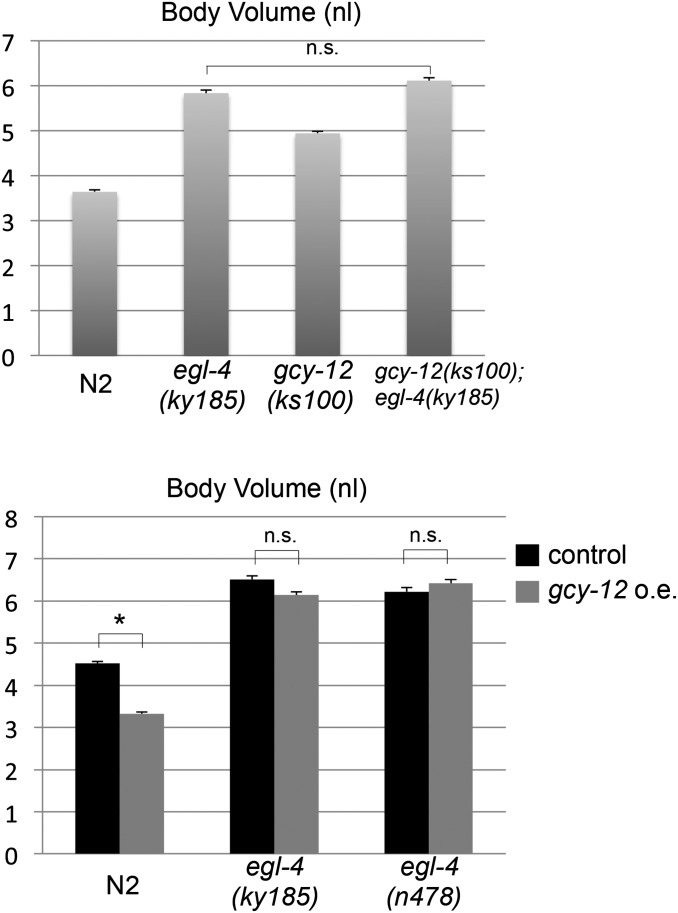
A key regulator of the sensory body-size regulation is the cGMP-dependent protein kinase EGL-4. Mutations in egl-4 result in the class I Chb phenotype and the expression of egl-4 cDNA in several sensory neurons can rescue this phenotype (Fujiwara et al. 2002). The EGL-4 kinase is activated by cGMP (Stansberry et al. 2001; Hirose et al. 2003); therefore, it is conceivable that cGMP produced by the GCY-12 guanylyl cyclase activates EGL-4 in vivo. To examine this possibility by genetic analyses, we first made a gcy-12;egl-4 double mutant. For the egl-4 mutant, we mainly used egl-4(ky185) because it has a 777-bp deletion that removes the coding sequence corresponding to a major part of the kinase domain and is therefore likely a null allele (Fujiwara et al. 2002). The gcy-12, as well as the egl-4 mutant, shows an increased body size compared with wild-type animals (N2) (Figure 3 and Figure S2). However, the gcy-12;egl-4 double mutant did not become larger than the egl-4 single mutant, indicating that gcy-12 and egl-4 act in the same genetic pathway. However, we noted that the body size of the egl-4 mutant is larger than that of the gcy-12 mutant, suggesting that GCY-12 is not the only guanylyl cyclase involved in EGL-4 activation for body-size regulation.
Figure 3.
GCY-12 is likely to act through the EGL-4 kinase for body-size regulation. The body volumes at the adult stage (48 hr after the L4 stage) are shown for the indicated strains. Top: gcy-12(ks100);egl-4(ky185) double mutant was not bigger than the egl-4(ky185) single mutant. Bottom: gcy-12 overexpression (o.e.) reduced the size of the wild-type animal, but did not affect the body size of the egl-4(ky185) or egl-4(n478) mutants. Error bars indicate SEMs (n.s., not significant; *, P < 0.001; t-test).
To confirm that GCY-12 acts through EGL-4 activation, the effect of GCY-12 overexpression was examined. We introduced a genomic fragment containing the gcy-12 gene and its promoter region into wild-type and egl-4 animals. The overexpression of GCY-12 in wild-type animals led to a small body size (Figure 3). This appears to result from inadequate activation of the EGL-4 kinase in wild-type animals, because the GCY-12 overexpression did not cause body-size reduction in the egl-4(ky185) mutant (Figure 3). We confirmed this result with another egl-4 mutant, n478 (Figure 3). Together, these results suggest that GCY-12 acts upstream of EGL-4, possibly by providing cGMP to the kinase.
Characterization of a gcy-12 mutant
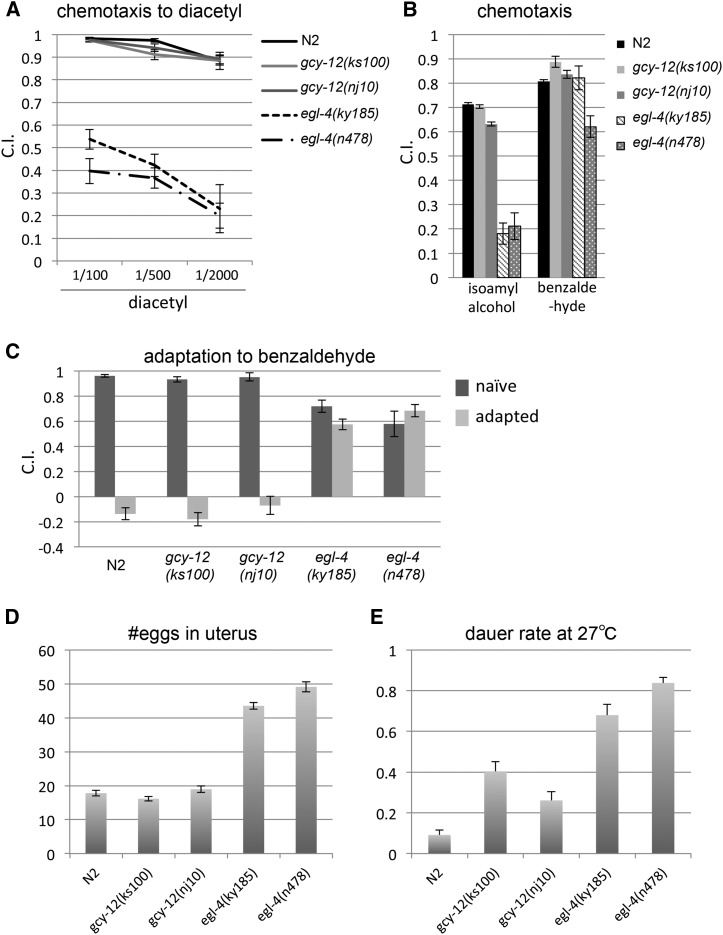
EGL-4 cGMP-dependent kinase is involved in the regulation of several aspects of behavior and development, as well as the regulation of body size and locomotory pattern. As shown above, gcy-12 mutants and egl-4 mutants share the suppressor phenotype of the small body and confined tracking pattern of the che-2 mutant, and GCY-12 seems to act upstream of EGL-4, at least for body-size regulation. Therefore, we next examined if gcy-12 mutants share the other phenotypes of egl-4 mutants, including egg-laying, chemotaxis, and odorant adaptation defects and inadequate dauer formation (Daniels et al. 2000). egl-4 mutants showed chemotaxis defects in responding to the volatile attractants, diacetyl and isoamylalcohol, but retained relatively high responsiveness to another volatile attractant, benzaldehyde (Figure 4, A and B), as previously reported (Daniels et al. 2000). On the other hand, gcy-12(ks100) and gcy-12(nj10) mutants showed almost normal responses to these three odorants, exhibiting a different chemotaxis trait from egl-4 mutants (Figure 4, A and B). egl-4 mutants also showed an adaptation defect to benzaldehyde (L’Etoile et al. 2002). gcy-12(ks100) and gcy-12(nj10) mutants showed normal adaptation, while egl-4 mutants retained high responsiveness to benzaldehyde after preexposure (Figure 4C). Next, we examined the egg-laying phenotype. While egl-4 mutants accumulated 40–50 eggs in the uterus, the egg retention of gcy-12(ks100) and of gcy-12(nj10) was comparable to wild-type animals (∼20) (Figure 4D). Another phenotype of egl-4 is the formation of dauer larvae, an alternative developmental stage of diapause, under noninducing conditions (Daniels et al. 2000). gcy-12(ks100) and gcy-12(nj10) mutants showed higher dauer rates than N2 under those conditions (Figure 4D). egl-4 mutants, however, showed much higher dauer rates than N2 and gcy-12 mutants (Figure 4D). These results indicated that not all phenotypes of egl-4 mutants are shared by gcy-12 mutants. Therefore, guanylyl cyclases other than GCY-12 may activate EGL-4, at least for the regulation of chemotaxis, odorant adaptation, and egg laying. In terms of the control of dauer formation, GCY-12 might have a role in EGL-4 activation.
Figure 4.
Mutant phenotypes of gcy-12. The gcy-12 mutants do not share all phenotypes exhibited by the egl-4 mutants. (A and B) Chemotaxis indexes to diacetyl (1/100, 1/500, 1/2000 dilutions), isoamyl alcohol (1/1000 dilution), and benzaldehyde (1/500 dilution) are shown. (C) Chemotaxis indexes to benzaldehyde (1/300 dilution) prior to and after conditioning (exposure to 1/50000 diluted benzaldehyde for 90 min) are shown. (D) The average number of eggs in an adult (27 hr after the L4 stage). (E) Dauer formation rates in culture conditions at 27°. Error bars indicate SEMs.
In addition to the phenotypes shown by egl-4 mutants, we examined the gcy-12 mutant for other phenotypes. Based on the visualization of several sensory neurons by fluorescent proteins, gcy-12(nj10) did not exhibit any apparent defects in neuronal morphology, including sensory cilia (Figure S3). Differentiation markers of sensory neurons, including sra-6p::gfp, srb-6p::mRFP, odr-1p::gfp, gcy-5p::gfp, gcy-7p::gfp, and str-2p::gfp, were expressed correctly in gcy-12(nj10), indicating no apparent defect in the differentiation of the following neurons: ASH, ASI, PHA, PHB, AWB, AWC, ASE, and AWCon (Troemel et al. 1995; Troemel et al. 1999) (data not shown).
gcy-12 is expressed in sensory neurons
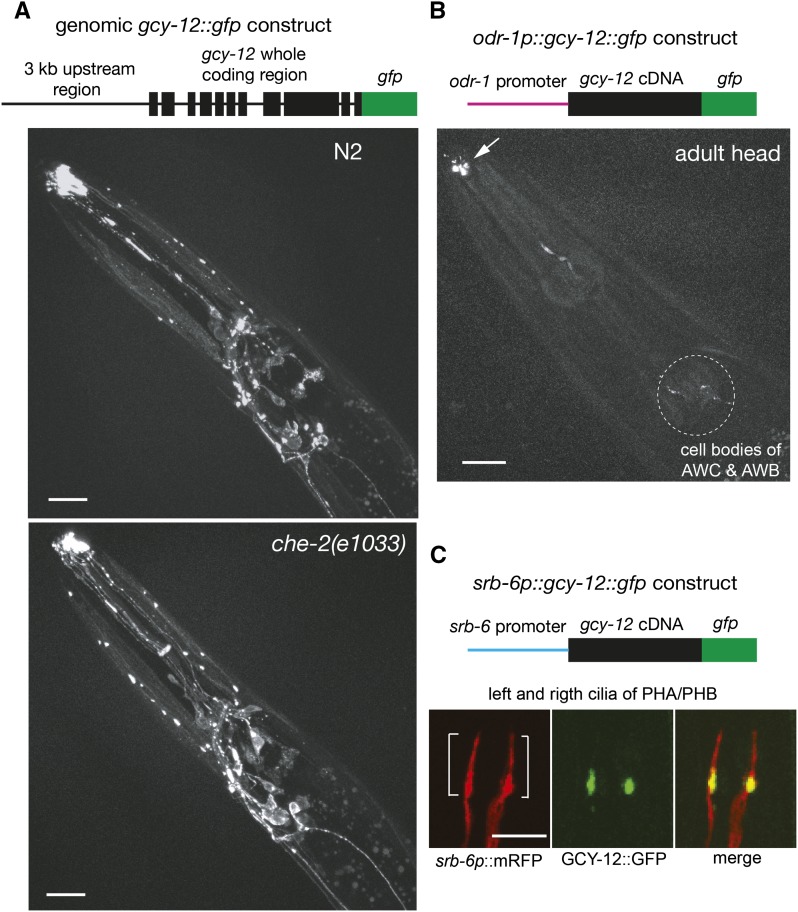
The GCY-12 expression pattern was determined by fusing GFP to the C terminus of GCY-12 in a genomic fusion (genomic gcy-12::gfp). This construct was made by inserting gfp into a genomic construct that included the whole coding region of gcy-12 and a 3-kb upstream region. We confirmed that the genomic gcy-12::gfp construct retained rescue activity (Figure S4). This clone directs GFP expression in chemosensory neurons (AWC, ASE, ASJ, AUA, PHA, PHB), an interneuron (PVQ), several other unidentified head neurons, an excretory gland cell, and adult head muscles (Figure 5A). In the head muscles, GFP shows a punctate pattern. The strongest GFP expression was observed at the tip of the nose, which appeared to be sensory cilia. To observe clear subcellular localization in sensory neurons, we drove the expression of GFP-tagged GCY-12 only in subsets of chemosensory neurons. gfp was ligated to the 3′ terminus of a gcy-12 cDNA and expressed under the control of an odr-1 promoter (for expression in AWB and AWC chemosensory neurons) or an srb-6 promoter (for expression in PHA and PHB chemosensory neurons) (Troemel et al. 1995; L’Etoile and Bargmann 2000) (Figure 5, B and C). With odr-1p::gcy-12::gfp, we observed a strong localization of GFP around the distal dendrite tip of AWC and AWB neurons. We also observed weak GFP expression in the cell bodies of these neurons as small dots around the nuclei, apparently reflecting transport through the Golgi apparatus (Figure 5B). With srb-6p::gcy-12::gfp, the relatively simple morphology of the cilia of PHA and PHB neurons allowed us to observe GCY-12 enriched mostly at the proximal segment of cilia or at the distal dendrite tip that is just close to cilia (Figure 5C).
Figure 5.
Expression analyses of gcy-12. Schematic of gfp expression constructs and the projections of confocal images are shown. An LSM510 confocal microscope (Carl Zeiss) was used to scan GFP and mRFP fluorescence. The nose tip is to the left (A and B). (A) The expression patterns of genomic gcy-12::gfp in the head regions of wild-type and che-2(e1033) animals at the L4 stage. Scale bar, 20 µm. (B) The expression patterns of odr-1p::gcy-12::gfp in the head region of a wild-type animal at the adult stage. The arrow indicates the dendrite tip and the dashed circle indicates cell bodies of AWC and AWB neurons. Scale bar, 20 µm. (C) The localization patterns of GCY-12::GFP in the sensory cilia of PHA/PHB neurons in the tail. The cilia (brackets) were visualized with srb-6p::mRFP. Scale bar, 5 µm.
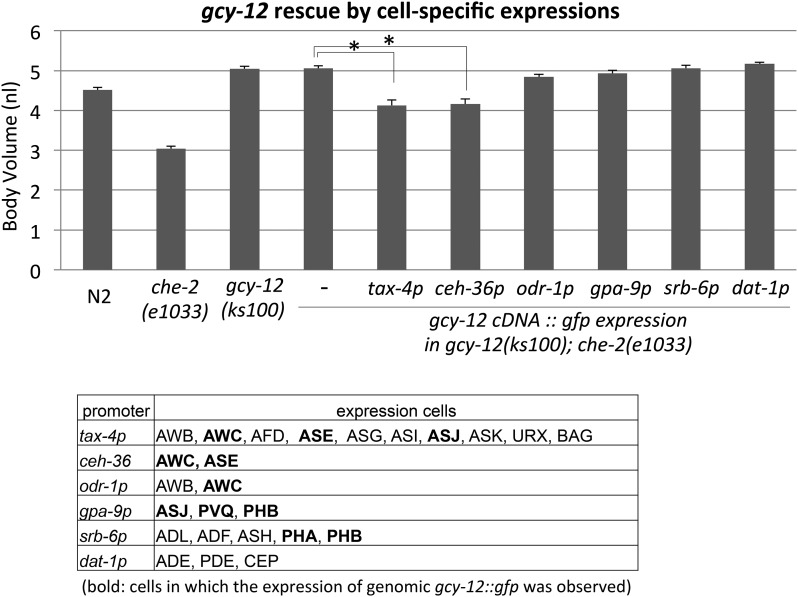
We observed GFP expression in several different tissues; therefore, we next sought to determine the identity of the cells in which GCY-12 acts to control body size. We hypothesized that expression in sensory neurons is important, because it was shown that the Chb phenotype of egl-4 animals can be rescued by sensory expression of egl-4 under a tax-4 promoter (Fujiwara et al. 2002). Therefore, we examined if the expression of gcy-12 under a tax-4 promoter can rescue the Chb phenotype of gcy-12 animals. As shown in Figure 6, we observed that the expression of gcy-12 cDNA, which was fused to gfp, under the tax-4 promoter can reduce the body size of the gcy-12;che-2 double mutant (Figure 6). This finding suggests that GCY-12 and EGL-4 act in the same sensory neurons to control body size and is consistent with the predicted role of GCY-12 in the activation of EGL-4. Furthermore, we tried further rescue experiments using an additional five promoters that promote expression in reduced subsets of the sensory neurons. Among them, we found that gcy-12 expression in AWC and ASE neurons under a ceh-36 promoter reduced the body size to a similar level as that produced by expression under the tax-4 promoter (Figure 6). Because gcy-12 expression in AWC (and AWB) under an odr-1 promoter did not have a significant effect, the expression of gcy-12 in ASE or in both neurons, ASE and AWC, seems to be important for the body size regulation. The expression of gcy-12 in other sensory neurons under gpa-9, srb-6, and dat-1 promoters did not show any significant effects on body size.
Figure 6.
GCY-12 acts in a set of sensory neurons. gcy-12 cDNA::gfp was expressed under the control of various sensory promoters in gcy-12(ks100);che-2(e1033) animals, and the body volumes were measured at the adult stage (48 hr after the L4 stage). Error bars indicate SEMs. (*, P < 0.01; t-test with Bonferroni correction). The table shows the cellular expression patterns induced by each promoter.
Genetic analyses of the relationship between GCY-12 and other GCs
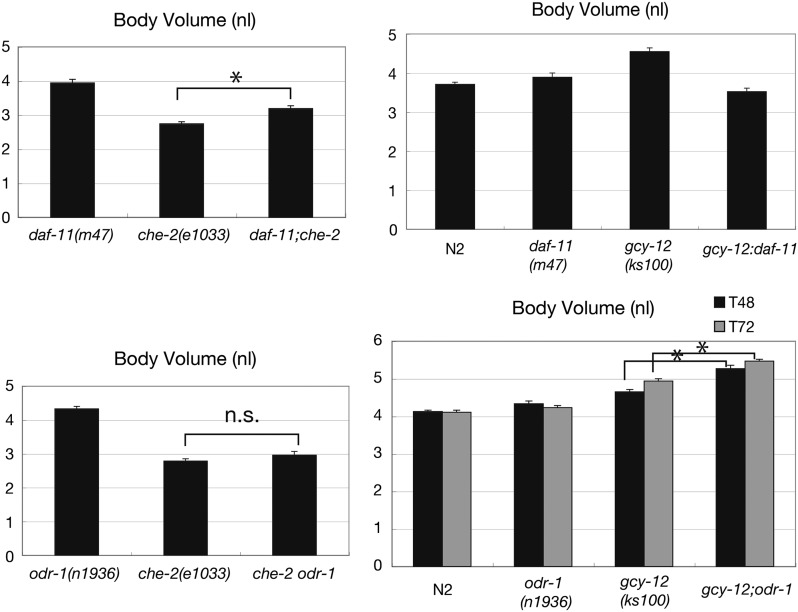
In C. elegans, 27 receptor-type guanylyl cyclase genes have been identified in the complete genome sequence (Ortiz et al. 2006). Some are known to function in sensory neurons by activating cyclic nucleotide-gated channels encoded by tax-2 and tax-4 (Coburn and Bargmann 1996; Komatsu et al. 1996; Inada et al. 2006; Ortiz et al. 2009; Hallem et al. 2011; Murayama et al. 2013). For example, two guanylyl cyclase genes, daf-11 and odr-1, play a key role in regulating chemotaxis in AWC and ASE chemosensory neurons (Birnby et al. 2000; L’etoile and Bargmann 2000). Our analyses suggest that GCY-12 acts in tax-4 expressing neurons, particularly in ASE and possibly in AWC, to activate EGL-4 and regulate body size. Therefore, we asked if the other GCs in tax-4-expressing neurons, such as DAF-11 and ODR-1, are also involved in sensory-dependent body size regulation. First, we examined if a daf-11 or odr-1 mutation exhibits the Chb phenotype in a che-2(e1033) background. daf-11(m47) caused a significant but small increase in the body size of che-2(e1033) animals, while odr-1(n1936) did not (Figure 7). Next, we examined if loss of DAF-11 or ODR-1 enhances the gcy-12 body size phenotype. While a gcy-12(ks100) mutant exhibits a large body size, daf-11(m47) and odr-1(n1936) mutants exhibit a normal body size (Figure 7). We predicted that double mutants would be larger than the gcy-12 single mutant, if ODR-1 and DAF-11 can complement the supply of cGMP in the absence of GCY-12. We found that gcy-12(ks100);daf-11(m47) becomes even smaller than each single mutant (Figure 7). This might reflect detrimental effects due to the specific combination of these two mutations. On the other hand, gcy-12(ks100);odr-1(n1936) was larger than each single mutant (Figure 7). Taken together, ODR-1 may partially complement GCY-12 for body-size regulation, possibly through supplying cGMP to the EGL-4 kinase in the absence of GCY-12, while DAF-11 seems to have little effect on the regulation of body size.
Figure 7.
Genetic interaction analyses of daf-11(m47) and odr-1(n1936) with the body-size mutants, che-2(e1033) and gcy-12(ks100). daf-11(m47) and the compared strains were cultured at 15° to prevent dauer entry and the body volumes were measured at the adult stage (99 hr from the L4 stage). odr-1(n1936) and the compared strains were cultured at 20° as usual and the body volumes were measured at the adult stage (48 and 72 hr after the L4 stage). Error bars indicate SEMs (n.s., not significant; *, P < 0.001; t-test).
Role of the extracellular domain of GCY-12
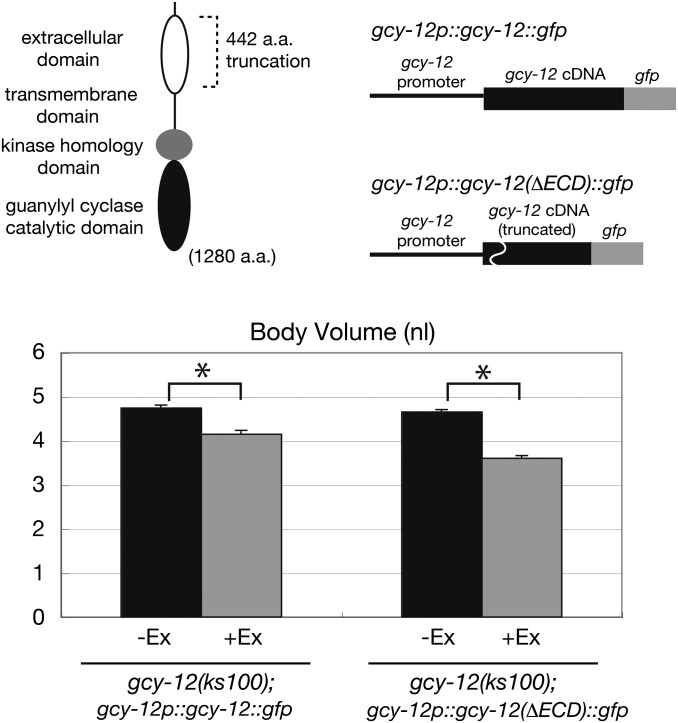
GCY-12 has an extracellular domain that is conserved from bacteria to mammals (Ortiz et al. 2006). The domain is termed RFLBR (receptor family ligand-binding region) and is speculated to be required for ligand binding. We next sought to determine if the RFLBR is required for the control of GCY-12 activity. A large part of the GCY-12 extracellular domain (442 aa) was deleted and this truncated construct was introduced into the gcy-12(ks100) mutant. As shown in Figure 8, the increased body size of gcy-12 animals was rescued by the truncated form of GCY-12. The effect is comparable to that of full-length GCY-12. This result indicates that the extracellular domain is not required for the activation of GCY-12.
Figure 8.
Extracellular domain of GCY-12 is dispensable for body size regulation. Top left: domain structure of the GCY-12 receptor-type guanylyl cyclase. Top right: schematic images of the control and deletion constructs. Each construct was introduced into gcy-12(ks100) animals and the body volumes of animal without (−Ex) or with (+Ex) the transgene were measured at the adult stage (48 hr after the L4 stage). Error bars indicate SEMs (*, P < 0.001; t-test).
Possible role of PDE-2 for the regulation of EGL-4 in controlling body size
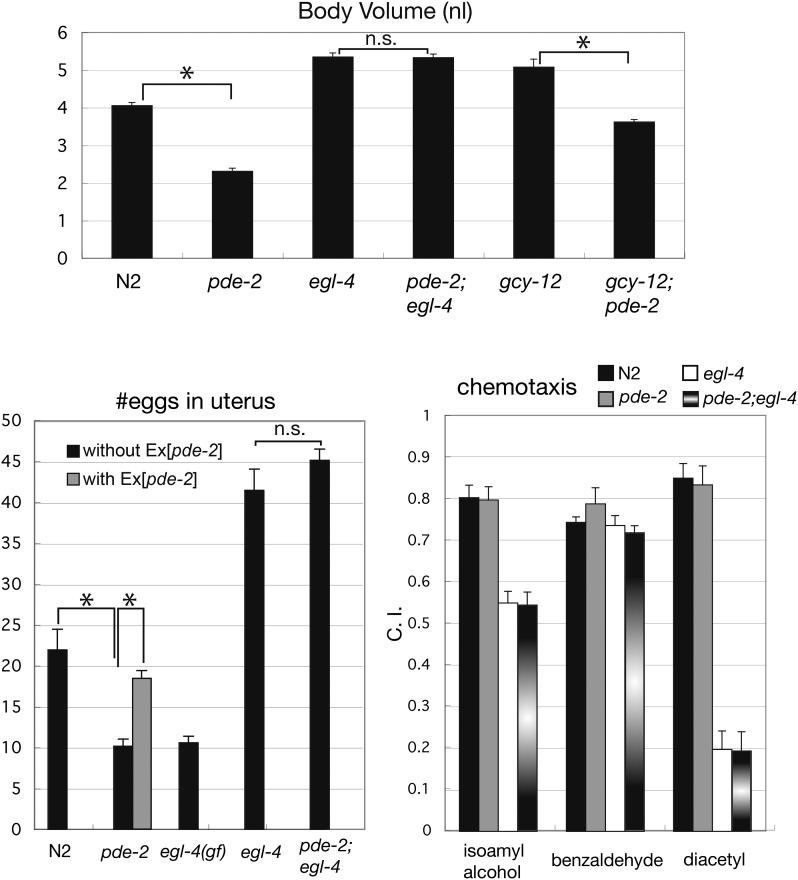
The intracellular cGMP level is known to be tightly controlled both by its rate of synthesis by GCs and by its rate of hydrolysis by cyclic nucleotide phosphodiesterases (PDEs). By RNAi and/or mutant analyses of the six PDE homologs in C. elegans (Charlie et al. 2006), we found that the pde-2 gene is required for normal body size regulation (Figure 9 and data not shown). pde-2 encodes the closest C. elegans homolog of human cGMP-dependent 3′,5′-cyclic phosphodiesterase 2A (PDE2A). Human PDE2A is stimulated by cGMP binding to its GAF domain and hydrolyzes both cGMP and cAMP (Omori and Kotera 2007). C. elegans PDE-2 retains the conserved phosphodiesterase catalytic domain and the GAF domain. The pde-2(qj6) mutation, a deletion allele that is expected to produce a truncated protein lacking the whole catalytic domain (see Materials and Methods), caused a small-body-size phenotype (Figure 9), and the phenotype was rescued by introduction of the wild-type pde-2 gene (Figure S5). Importantly, the pde-2(qj6) mutation did not affect the body size of the egl-4(ky185) mutant, indicating PDE-2 action upstream of EGL-4 (Figure 9). The gcy-12(ks100);pde-2(qj6) double mutant exhibited an intermediate body size relative to each single mutant (Figure 9). These results indicate that PDE-2 antagonizes GCY-12 and negatively regulates the EGL-4 kinase in controlling body size.
Figure 9.
Phenotypes of the pde-2(qj6) mutant. Body volumes of wild-type, pde-2(qj6), egl-4(ky185), pde-2(qj6);egl-4(ky185), gcy-12(ks100), and gcy-12(ks100);pde-2(qj6) animals were measured at the adult stage (48 hr after the L4 stage). The egg-laying phenotype and chemotaxis behavior of wild-type, pde-2(qj6), egl-4(ad450gf), egl-4(ky185), and pde-2(qj6);egl-4(ky185) animals were examined. For the chemotaxis assay, isoamyl alcohol (1/100 dilution), benzaldehyde (1/500 dilution), diacetyl (1/1000 dilution) were used. The Egl-c phenotype of pde-2(qj6) was rescued by the transgene containing the wild-type pde-2 gene (Ex[pde-2]). Error bars indicate SEMs (n.s., not significant; *, P < 0.001; t-test).
The pde-2(qj6) animals retain fewer eggs in the uterus than wild-type animals, and the phenotype can be rescued by introduction of the wild-type pde-2 gene (Figure 9). This phenotype, termed Egl-c (egg-laying constitutive), is opposite to the egg-accumulation phenotype (Egl-d; egg-laying defective) exhibited by the egl-4(ky185) loss-of-function mutant. Since an egl-4 (ad450) gain-of-function mutant also shows the Egl-c phenotype (Raizen et al. 2006), PDE-2 is likely to negatively regulate the EGL-4 kinase in egg-laying control. The pde-2(qj6) mutation did not decrease the number of eggs retained in the uterus in the egl-4(ky185) background (pde-2;egl-4 in Figure 9), supporting the role of PDE-2 upstream of EGL-4. In the regulation of chemotaxis behavior, PDE-2 appears to play a dispensable role; the pde-2(qj6) mutant showed normal responsiveness to all the attractive odorants examined, and the pde-2(qj6);egl-4 (ky185) double mutant showed the same chemotactic defects as were observed for the egl-4(ky185) single mutant (Figure 9).
Discussion
To investigate the mechanism of sensory-dependent regulation of body size in C. elegans, we performed a mutant screen for suppressors of che-2 small body size and identified the gene, gcy-12, which encodes a receptor-type guanylyl cyclase. The gcy-12 gene was previously predicted from the C. elegans genome sequence database (Yu et al. 1997; Ortiz et al. 2006), and biochemical analysis of gcy-12 by expression in COS-M6 cells also demonstrated its guanylyl cyclase activity (Yu et al. 1997). In the current study, we report extensive analyses of the function of this gene in vivo.
GCY-12 is the guanylyl cyclase that controls EGL-4 kinase activity in body size regulation
C. elegans body size is controlled by the EGL-4 cGMP-dependent protein kinase, whose activity is negatively regulated by sensory inputs (Fujiwara et al. 2002). Here, we have identified gcy-12, as another gene involved in sensory-dependent regulation of body size. Similar to egl-4 mutations, mutations in gcy-12 increased body size and suppressed the small-body phenotype of the che-2 cilium-defective mutant. The gcy-12 mutations, as well as the egl-4 mutations, were classified as class I suppressors, indicating that the che-2 mutation causes the small-body phenotype through altered GCY-12 function. Biochemical analyses showed that EGL-4 indeed requires cGMP for its kinase activity (Stansberry et al. 2001; Hirose et al. 2003); therefore, the simplest interpretation is that GCY-12 is the guanylyl cyclase that provides cGMP to EGL-4 in the pathway regulating body size. The studies of this article provide several lines of evidence to support this hypothesis. First, the double-mutant analysis suggested that gcy-12 and egl-4 act in the same pathway. Second, the overexpression analysis of gcy-12 indicated that GCY-12 affects body size through EGL-4. Finally, rescue of the che-2 phenotype by gcy-12 expression under the control of the tax-4 promoter suggested that GCY-12 and EGL-4 act in the same set of sensory neurons.
Control of GCY-12 activity in cilia
GCY-12, together with most of the other receptor-type guanylyl cyclases in C. elegans, contains a conserved extracellular domain, the receptor family ligand-binding region [RFLBR (PF01094)]. The presence of this conserved domain indicates that ligand binding may control GCY-12 activity and consequently regulate body size. The distinct localization of GFP-tagged GCY-12 at sensory cilia supports this idea. However, the deletion analysis of the extracellular domain (ECD) of GCY-12 indicated that the domain is dispensable for body-size regulation. It is possible that GCY-12 heterodimerizes with other guanylyl cyclases, whose extracellular domains bind to ligands. In fact, analysis of the amino acid sequence suggested that GCY-12 may form an obligate heterodimer (Morton 2004). Another possibility is that GCY-12 does not require the ECD for its activation, because GC activity is regulated independently of signals through the ECD. We prefer this possibility, because it may explain why mutants with malformed cilia, such as che-2, are small. Suppression of the small body size of the che-2 mutant by the mutations in gcy-12 and egl-4 indicates that the GCY-12-EGL-4 signaling cascade is inadequately activated when the cilia are malformed. We observed that GCY-12::GFP is still localized to dendrite tips in the che-2 mutant (Figure 5A). An electron microscopy analysis showed that the distal and middle segments but not the proximal segments of cilia are missing in the che-2(e1033) mutant (Lewis and Hodgkin 1977). Because GCY-12::GFP is mostly enriched around the base of the cilia (Figure 5C), GCY-12 in the che-2 animals may still be capable of activating EGL-4. One possibility is that EGL-4 kinase activity in cilia may be regulated positively by GCY-12 and negatively by other signaling molecules. If the positive regulator, GCY-12, does not require extracellular signals for activation, the ciliary defect in the che-2 mutant might inhibit only the negative regulation signaling, leading to hyperactivation of the EGL-4 kinase and consequently resulting in the small body size phenotype.
A candidate component of the negative regulation pathway is pde-2, mutation of which causes the small body phenotype in an EGL-4 dependent manner, as observed for the che-2 mutation. Although a detailed expression analysis has not been performed, pde-2 appears to be widely expressed, including in many head neurons (Hunt-Newbury et al. 2007; Wang et al. 2013). Similar to the mechanism of phototransduction signaling, sensory inputs may activate PDE-2 and negatively regulate EGL-4 in the control of body size. It would be interesting to examine cGMP levels in the cilia of pde-2 and gcy-12 mutants, as well as in the malformed cilia of a che-2 mutant. Recent developments in imaging technology have made it possible to visualize cGMP levels in neurons in vivo, although it is still difficult to observe at subcellular levels, such as in cilia (Couto et al. 2013). We showed that GCY-12 expression in ASE sensory neurons is important for body-size regulation. Because ASE sensory neurons act as a main sensor for various salt ions (Bargmann and Horvitz 1991), it is possible that the salt sensation is involved in body-size regulation, for example, by affecting PDE-2 activity.
Biochemical analysis by Yu et al. (1997) showed that the guanylyl cyclase activity of GCY-12 is dependent on temperature, with activity higher at 25° than at 15°. Given the action of GCY-12 in body-size regulation, this suggests that animals cultivated at 25° may be smaller than those cultivated at 15°. However, we did not observe such temperature-dependent body size changes in either wild-type or egl-4 animals (data not shown).
Function-specific regulation of EGL-4 cGMP-dependent kinase activity
EGL-4 is involved in various biological processes, including chemotaxis, odorant adaptation, dauer formation, and egg laying (Daniels et al. 2000). Our analyses indicated that GCY-12 is required for EGL-4 activation only in a limited number of EGL-4 functions, including body size regulation, but not in other functions including chemotaxis, odorant adaptation, and egg laying. Our analysis with the mutants of ODR-1 and DAF-11, other GCs expressed in the same neurons as GCY-12 is expressed, also suggests that these GCs do not play major roles in the body size regulation pathway. Given the existence of an unusually large number of guanylyl cyclases in C. elegans (∼34), EGL-4 may have different GC partners for each function. This may occur through the expression of different GCs in different cells or through the localization of different GC partners in distinct functional units in the same cell. Indeed, the localization of GCY-12 to cilia is tightly controlled and its mislocalization results in the body-size defect (Fujiwara et al. 2010). Other GCs are also known to localize to specific subcellular compartments for compartmentalized signaling (Gross et al. 2014).
Using different GCs for different EGL-4 functions may enable elaborate control of the kinase. For example, in the process of odorant adaptation, activation of EGL-4 by ODR-1 is thought to induce the downregulation of ODR-1 (Juang et al. 2013). Without independent GC partners, such a feedback regulation would affect other processes in which EGL-4 is involved. Our study suggests that there are multiple function-specific modes of regulation of kinase activity during signal transduction for complex behavior and development.
Supplementary Material
Acknowledgments
We thank the C. elegans Genetics Center for worm strains, the Sanger Center for cosmids clones, Andy Fire for the various vectors, and Yuji Kohara for the cDNA clones. We also thank Isao Katsura for comments on the manuscript. This work was supported by Grants-in-Aid for Scientific Research (C) (23570007 to M.F.) and for Young Scientists (B) (15770002 to M.F.) and for Scientific Research on Innovative Areas “Memory dynamism” (25115009 to T.I.), “Molecular Ethology”(20115003 to T.I.), and “Comprehensive Brain Science Network” and PRESTO, Japan Science and Technology Corporation (to M.K.).
Footnotes
Communicating editor: P. Sengupta
Supporting information is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.177543/-/DC1.
Literature Cited
- Alcedo J., Kenyon C., 2004. Regulation of C. elegans longevity by specific gustatory and olfactory neurons. Neuron 41: 45–55. [DOI] [PubMed] [Google Scholar]
- Apfeld J., Kenyon C., 1999. Regulation of lifespan by sensory perception in Caenorhabditis elegans. Nature 402: 804–809. [DOI] [PubMed] [Google Scholar]
- Bargmann C. I., Horvitz H. R., 1991. Control of larval development by chemosensory neurons in Caenorhabditis elegans. Science 251: 1243–1246. [DOI] [PubMed] [Google Scholar]
- Bargmann C. I., Hartwieg E., Horvitz H. R., 1993. Odorant-selective genes and neurons mediate olfaction in C. elegans. Cell 74: 515–527. [DOI] [PubMed] [Google Scholar]
- Birnby D. A., Link E. M., Vowels J. J., Tian H., Colacurcio P. L., et al. , 2000. A transmembrane guanylyl cyclase (DAF-11) and Hsp90 (DAF-21) regulate a common set of chemosensory behaviors in Caenorhabditis elegans. Genetics 155: 85–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlie N. K., Thomure A. M., Schade M. A., Miller K. G., 2006. The Dunce cAMP phosphodiesterase PDE-4 negatively regulates G alpha(s)-dependent and G alpha(s)-independent cAMP pools in the Caenorhabditis elegans synaptic signaling network. Genetics 173: 111–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coburn C. M., Bargmann C. I., 1996. A putative cyclic nucleotide-gated channel is required for sensory development and function in C. elegans. Neuron 17: 695–706. [DOI] [PubMed] [Google Scholar]
- Couto A., Oda S., Nikolaev V. O., Soltesz Z., de Bono M., 2013. In vivo genetic dissection of O2-evoked cGMP dynamics in a Caenorhabditis elegans gas sensor. Proc. Natl. Acad. Sci. USA 110: E3301–E3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels S. A., Ailion M., Thomas J. H., Sengupta P., 2000. egl-4 acts through a transforming growth factor-beta/SMAD pathway in Caenorhabditis elegans to regulate multiple neuronal circuits in response to sensory cues. Genetics 156: 123–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara M., Ishihara T., Katsura I., 1999. A novel WD40 protein, CHE-2, acts cell-autonomously in the formation of C. elegans sensory cilia. Development 126: 4839–4848. [DOI] [PubMed] [Google Scholar]
- Fujiwara M., Sengupta P., McIntire S. L., 2002. Regulation of body size and behavioral state of C. elegans by sensory perception and the EGL-4 cGMP-dependent protein kinase. Neuron 36: 1091–1102. [DOI] [PubMed] [Google Scholar]
- Fujiwara M., Teramoto T., Ishihara T., Ohshima Y., McIntire S. L., 2010. A novel zf-MYND protein, CHB-3, mediates guanylyl cyclase localization to sensory cilia and controls body size of Caenorhabditis elegans. PLoS Genet. 6: e1001211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gengyo-Ando K., Mitani S., 2000. Characterization of mutations induced by ethyl methanesulfonate, UV, and trimethylpsoralen in the nematode Caenorhabditis elegans. Biochem. Biophys. Res. Commun. 269: 64–69. [DOI] [PubMed] [Google Scholar]
- Gross E., Soltesz Z., Oda S., Zelmanovich V., Abergel Z., et al. , 2014. GLOBIN-5-dependent O2 responses are regulated by PDL-1/PrBP that targets prenylated soluble guanylate cyclases to dendritic endings. J. Neurosci. 34: 16726–16738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallem E. A., Spencer W. C., McWhirter R. D., Zeller G., Henz S. R., et al. , 2011. Receptor-type guanylate cyclase is required for carbon dioxide sensation by Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 108: 254–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y., Xu N., Box A. C., Schaefer L., Kannan K., et al. , 2011. Nuclear cGMP-dependent kinase regulates gene expression via activity-dependent recruitment of a conserved histone deacetylase complex. PLoS Genet. 7: e1002065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose T., Nakano Y., Nagamatsu Y., Misumi T., Ohta H., et al. , 2003. Cyclic GMP-dependent protein kinase EGL-4 controls body size and lifespan in C. elegans. Development 130: 1089–1099. [DOI] [PubMed] [Google Scholar]
- Hunt-Newbury R., Viveiros R., Johnsen R., Mah A., Anastas D., et al. , 2007. High-throughput in vivo analysis of gene expression in Caenorhabditis elegans. PLoS Biol. 5: e237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inada H., Ito H., Satterlee J., Sengupta P., Matsumoto K., et al. , 2006. Identification of guanylyl cyclases that function in thermosensory neurons of Caenorhabditis elegans. Genetics 172: 2239–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juang B. T., Gu C., Starnes L., Palladino F., Goga A., et al. , 2013. Endogenous nuclear RNAi mediates behavioral adaptation to odor. Cell 154: 1010–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu H., Mori I., Rhee J. S., Akaike N., Ohshima Y., 1996. Mutations in a cyclic nucleotide-gated channel lead to abnormal thermosensation and chemosensation in C. elegans. Neuron 17: 707–718. [DOI] [PubMed] [Google Scholar]
- Kuhara A., Inada H., Katsura I., Mori I., 2002. Negative regulation and gain control of sensory neurons by the C. elegans calcineurin TAX-6. Neuron 33: 751–763. [DOI] [PubMed] [Google Scholar]
- Lanjuin A., Sengupta P., 2002. Regulation of chemosensory receptor expression and sensory signaling by the KIN-29 Ser/Thr kinase. Neuron 33: 369–381. [DOI] [PubMed] [Google Scholar]
- L’Etoile N. D., Bargmann C. I., 2000. Olfaction and odor discrimination are mediated by the C. elegans guanylyl cyclase ODR-1. Neuron 25: 575–586. [DOI] [PubMed] [Google Scholar]
- L’Etoile N. D., Coburn C. M., Eastham J., Kistler A., Gallegos G., et al. , 2002. The cyclic GMP-dependent protein kinase EGL-4 regulates olfactory adaptation in C. elegans. Neuron 36: 1079–1089. [DOI] [PubMed] [Google Scholar]
- Lewis J. A., Hodgkin J. A., 1977. Specific neuroanatomical changes in chemosensory mutants of the nematode Caenorhabditis elegans. J. Comp. Neurol. 172: 489–510. [DOI] [PubMed] [Google Scholar]
- Libert S., Zwiener J., Chu X., Vanvoorhies W., Roman G., et al. , 2007. Regulation of Drosophila life span by olfaction and food-derived odors. Science 315: 1133–1137. [DOI] [PubMed] [Google Scholar]
- Mak H. Y., Nelson L. S., Basson M., Johnson C. D., Ruvkun G., 2006. Polygenic control of Caenorhabditis elegans fat storage. Nat. Genet. 38: 363–368. [DOI] [PubMed] [Google Scholar]
- Mello C. C., Kramer J. M., Stinchcomb D., Ambros V., 1991. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10: 3959–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton D. B., 2004. Invertebrates yield a plethora of atypical guanylyl cyclases. Mol. Neurobiol. 29: 97–116. [DOI] [PubMed] [Google Scholar]
- Murayama T., Takayama J., Fujiwara M., Maruyama I. N., 2013. Environmental alkalinity sensing mediated by the transmembrane guanylyl cyclase GCY-14 in C. elegans. Curr. Biol. 23: 1007–1012. [DOI] [PubMed] [Google Scholar]
- Nakano Y., Nagamatsu Y., Ohshima Y., 2004. cGMP and a germ-line signal control body size in C. elegans through cGMP-dependent protein kinase EGL-4. Genes Cells 9: 773–779. [DOI] [PubMed] [Google Scholar]
- O’Halloran D. M., Hamilton O. S., Lee J. I., Gallegos M., L’Etoile N. D., 2012. Changes in cGMP levels affect the localization of EGL-4 in AWC in Caenorhabditis elegans. PLoS One 7: e31614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omori K., Kotera J., 2007. Overview of PDEs and their regulation. Circ. Res. 100: 309–327. [DOI] [PubMed] [Google Scholar]
- Ortiz C. O., Etchberger J. F., Posy S. L., Frokjaer-Jensen C., Lockery S., et al. , 2006. Searching for neuronal left/right asymmetry: genomewide analysis of nematode receptor-type guanylyl cyclases. Genetics 173: 131–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz C. O., Faumont S., Takayama J., Ahmed H. K., Goldsmith A. D., et al. , 2009. Lateralized gustatory behavior of C. elegans is controlled by specific receptor-type guanylyl cyclases. Curr. Biol. 19: 996–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins L. A., Hedgecock E. M., Thomson J. N., Culotti J. G., 1986. Mutant sensory cilia in the nematode Caenorhabditis elegans. Dev. Biol. 117: 456–487. [DOI] [PubMed] [Google Scholar]
- Raizen D. M., Cullison K. M., Pack A. I., Sundaram M. V., 2006. A novel gain-of-function mutant of the cyclic GMP-dependent protein kinase egl-4 affects multiple physiological processes in Caenorhabditis elegans. Genetics 173: 177–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raizen D. M., Zimmerman J. E., Maycock M. H., Ta U. D., You Y. J., et al. , 2008. Lethargus is a Caenorhabditis elegans sleep-like state. Nature 451: 569–572. [DOI] [PubMed] [Google Scholar]
- Silverman M. A., Leroux M. R., 2009. Intraflagellar transport and the generation of dynamic, structurally and functionally diverse cilia. Trends Cell Biol. 19: 306–316. [DOI] [PubMed] [Google Scholar]
- Stansberry J., Baude E. J., Taylor M. K., Chen P. J., Jin S. W., et al. , 2001. A cGMP-dependent protein kinase is implicated in wild-type motility in C. elegans. J. Neurochem. 76: 1177–1187. [DOI] [PubMed] [Google Scholar]
- Starich T. A., Herman R. K., Kari C. K., Yeh W. H., Schackwitz W. S., et al. , 1995. Mutations affecting the chemosensory neurons of Caenorhabditis elegans. Genetics 139: 171–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tain L. S., Lozano E., Saez A. G., Leroi A. M., 2008. Dietary regulation of hypodermal polyploidization in C. elegans. BMC Dev. Biol. 8: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trent C., Tsuing N., Horvitz H. R., 1983. Egg-laying defective mutants of the nematode Caenorhabditis elegans. Genetics 104: 619–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troemel E. R., Chou J. H., Dwyer N. D., Colbert H. A., Bargmann C. I., 1995. Divergent seven transmembrane receptors are candidate chemosensory receptors in C. elegans. Cell 83: 207–218. [DOI] [PubMed] [Google Scholar]
- Troemel E. R., Sagasti A., Bargmann C. I., 1999. Lateral signaling mediated by axon contact and calcium entry regulates asymmetric odorant receptor expression in C. elegans. Cell 99: 387–398. [DOI] [PubMed] [Google Scholar]
- van der Linden A. M., Wiener S., You Y. J., Kim K., Avery L., et al. , 2008. The EGL-4 PKG acts with KIN-29 salt-inducible kinase and protein kinase A to regulate chemoreceptor gene expression and sensory behaviors in Caenorhabditis elegans. Genetics 180: 1475–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., O’Halloran D., Goodman M. B., 2013. GCY-8, PDE-2, and NCS-1 are critical elements of the cGMP-dependent thermotransduction cascade in the AFD neurons responsible for C. elegans thermotaxis. J. Gen. Physiol. 142: 437–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicks S. R., Yeh R. T., Gish W. R., Waterston R. H., Plasterk R. H., 2001. Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nat. Genet. 28: 160–164. [DOI] [PubMed] [Google Scholar]
- You Y. J., Kim J., Raizen D. M., Avery L., 2008. Insulin, cGMP, and TGF-beta signals regulate food intake and quiescence in C. elegans: a model for satiety. Cell Metab. 7: 249–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S., Avery L., Baude E., Garbers D. L., 1997. Guanylyl cyclase expression in specific sensory neurons: a new family of chemosensory receptors. Proc. Natl. Acad. Sci. USA 94: 3384–3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All strains and plasmids described here and in the Supporting Information are available upon request.