Abstract
Cutaneous and mucosal leishmaniasis, caused in South America by Leishmania braziliensis, is difficult to cure by chemotherapy (primarily pentavalent antimonials [SbV]). Treatment failure does not correlate well with resistance in vitro, and the factors responsible for treatment failure in patients are not well understood. Many isolates of L. braziliensis (>25%) contain a double-stranded RNA virus named Leishmaniavirus 1 (LRV1), which has also been reported in Leishmania guyanensis, for which an association with increased pathology, metastasis, and parasite replication was found in murine models. Here we probed the relationship of LRV1 to drug treatment success and disease in 97 L. braziliensis–infected patients from Peru and Bolivia. In vitro cultures were established, parasites were typed as L. braziliensis, and the presence of LRV1 was determined by reverse transcription–polymerase chain reaction, followed by sequence analysis. LRV1 was associated significantly with an increased risk of treatment failure (odds ratio, 3.99; P = .04). There was no significant association with intrinsic SbV resistance among parasites, suggesting that treatment failure arises from LRV1-mediated effects on host metabolism and/or parasite survival. The association of LRV1 with clinical drug treatment failure could serve to guide more-effective treatment of tegumentary disease caused by L. braziliensis.
Keywords: leishmaniasis, L. braziliensis, antimony drug treatment, RNA viruses, Viannia, drug treatment failure, drug resistance, Totivirus
(See the major article by Bourreau et al on pages 105–11.)
Leishmania are widespread protozoan parasites transmitted by phlebotomine sand flies. Leishmania infections afflict >12 million people worldwide, with 1.2 million new cases/year [1]. The true incidence is likely far higher, as most infections are persistent and asymptomatic, only emerging as disease following immune compromise [2, 3]. Leishmaniasis can be viewed as a spectral disease, with a range of manifestations, including tegumentary disease (cutaneous leishmaniasis [CL] or mucosal leishmaniasis [ML]) and visceral disease (visceral leishmaniasis [VL]); these manifestations are typically associated with different parasite species [4, 5]. Among the different species, Leishmania braziliensis is considered one of the most important in North and South America because of its prevalence, the difficulty of curing the disease it causes, and its public health importance. Notably, L. braziliensis is the most frequent cause of ML, which typically manifests first as CL and progresses to ML in up to 10% of the cases [6]. The factors responsible for the progression from CL to ML are not well understood and likely involve both host and parasite factors [5].
As yet there is no effective vaccination against L. braziliensis, and treatment depends on diagnosis and chemotherapy. Pentavalent antimonials (SbV), typically sodium stibogluconate (Pentostam) or meglumine antimoniate (Glucantime), are presently the primary treatment. However, in Latin America, SbV treatment is characterized by a variable outcome, with treatment failure rates reaching 39% [7, 8]. While in some Leishmania species, SbV resistance has been linked to intrinsic changes in parasite susceptibility, this does not appear to be the case in L. braziliensis in Peru [9, 10]. Risk factors identified thus far include the presence of concomitant distant lesions and factors associated with the immunological response [11, 12]. For example, the persistence of high levels of interleukin 10 in the lesions is associated with a poor response to treatment, and it is well known that the efficacy of antimonials is strongly influenced by immune responses [5, 10, 13, 14]. Other factors contributing to the relative insensitivity of L. braziliensis to SbV chemotherapy are likely.
Several species of Leishmania show the presence of a persistent, monosegmented, double-stranded RNA (dsRNA) virus named Leishmaniavirus (LRV), a member of the family Totiviridae [15–17]. LRV1 is most often seen in the Leishmania (Viannia) species L. braziliensis and Leishmania guyanensis, in which the overall occurrence is about 20%–30%, with some populations showing a prevalence of >50% (see below) [18–22]. While the biological relevance of LRV1 had been elusive since its discovery >20 years ago, in murine models LRV1 is now known to be associated with increased parasite replication, pathology, and metastasis following infection with either L. guyanensis [23] or L. braziliensis (unpublished data). There, LRV1 can act as an immunomodulator through the interactions of its dsRNA genome with the host Toll-like receptor 3 (TLR3), leading to a hyperinflammatory response [24]. Similar responses in humans likewise would be expected to result in increased disease severity, as well, but currently the clinical impact of LRV1 is uncertain. Two studies reported little association of LRV1 with cutaneous versus mucocutaneous presentation [20, 25].
The studies above prompted us to consider the potential for a link between LRV1 and treatment success. Here, we performed a cross-sectional analysis of collections of L. braziliensis isolates from patients in Peru and Bolivia exhibiting various forms of tegumentary leishmaniasis (CL, ML, or both [MCL]). Importantly, a significant association was seen between the presence of LRV1 and therapeutic failure with SbV or amphotericin B.
MATERIALS AND METHODS
Ethics Statement
Research in this study was subject to ethical review by the European Commission and was approved as part of contract negotiation for LeishBolPe (an epidemiological study in Bolivia and Peru) and LeishNatDrug-R (a multicenter study on SbV treatment failure); the work conformed to all relevant European regulations. The research was also reviewed and approved by the ethics committees of the Universidad Peruana Cayetano Heredia (Lima, Peru), the Hospital Nacional Cayetano Heredia (Lima), and the Universidad Mayor de San Simón (Cochabamba, Bolivia). All human strains of Leishmania had been isolated from patients as part of normal diagnosis and treatment, with no unnecessary invasive procedures, and with written and/or verbal informed consent recorded at the time of clinical examination. Data on human isolates were coded and anonymized.
Patients
Patients were recruited at the Instituto de Medicina Tropical Alexander von Humboldt (Lima) and the Universidad Mayor de San Simón (Cochabamba, Bolivia) as part of 2 prospective studies: LeishBolPe (Bolivia and Peru, 1994–1998), an epidemiological study aiming to discriminate factors underlying clinical variability in infection and disease; and LeishNatDrug-R (Peru, 2001–2004), a case-control study of incident cases to understand risk factors of treatment failure. Here we focused on L. braziliensis, owing to its prevalence and association with a higher risk of treatment failure [11]. From both prior studies, 290 isolates were typed, and all 97 L. braziliensis isolates with adequate clinical and epidemiological documentation were considered (Table 1). Of these, 54 had been monitored for treatment outcomes for up to 1 year and were included in our analysis (Table 1 and Supplementary Figures 1 and 2). Patients were classified clinically as manifesting CL lesions, ML lesions, or both CL and ML (MCL) lesions. Patients underwent standard supervised treatment with intravenous or intramuscular meglumine antimoniate (Glucantime; Sanofi Aventis) or generic sodium stibogluconate (Viteco, Colombia; or Albert-David, India), depending on availability; both drugs are considered equally effective [35]. We used dosages of 20 mg/kg/day for 20 days (for CL) or 30 days (for ML or MCL) or conventional amphotericin B (Fungizone, Bristol-Myers Squibb) at dosages of 0.6 mg/kg/day for 30–41 days [9]. Follow-up visits were scheduled at 1, 2, 3, 6, and 12 months after treatment ended. The clinical outcomes were as follows: cure, defined as complete reepithelialization with a characteristic scar and no inflammation at the time point of follow-up assessment (3–12 months after treatment, which depended on patients returning for their medical evaluation); primary unresponsive, defined as the absence or incomplete scarring of lesion(s) and/or the persistence of inflammatory signs at 3 months after treatment or the worsening of existing lesion(s) or the appearance of new lesion(s) ≤3 months after treatment; and relapse, defined as the reappearance of an ulcer or nodule and/or local signs of inflammation after initial cure [9]. Cured patients were still observed until 12 months after treatment, to detect possible relapses. Patients with treatment failure received either a repeat course of antimonials with or without topical imiquimod (Aldara; 3 M Pharmaceuticals) or intravenous amphotericin B deoxycholate (Bristol-Myers Squibb) [9, 11]. Some patients had previously been treated for leishmaniasis; these were classified retrospectively as primary unresponsive if the second treatment led to cure or as secondary unresponsive if it did not. For statistical analysis, treatment failure was defined as either unresponsiveness or relapse (Table 1 and Supplementary Figures 1 and 2).
Table 1.
Properties of Leishmania braziliensis Isolates From Peru and Bolivia, Including LRV1 Status
| International Code | Origin (Department, Province)a | Lesion Type | Treatment Outcome | Classification | LRV1 Present | Reference |
|---|---|---|---|---|---|---|
| Pentavalent antimonial treatment | ||||||
| MHOM/PE/03/PER260 | Madre de Dios, Tahuamanu | ML | Cure (12 mo) | Cure | − | [9] |
| MHOM/PE/02/PER094 | Huanuco, Puerto Inca | CL | Cure (12 mo) | Cure | − | [9] |
| MHOM/PE/02/PER122 | Madre de Dios, Tambopata | CL | Cure (12 mo) | Cure | − | [9] |
| MHOM/PE/03/PER163 | Huanuco, Leoncio Prado | CL | Cure (12 mo) | Cure | − | [9] |
| MHOM/PE/03/PER157 | Madre de Dios, Tambopata | CL | Cure (6 mo) | Cure | − | [9] |
| MHOM/PE/03/PER182 | Ayacucho, La Mar | CL | Cure (6 mo) | Cure | − | [9] |
| MHOM/PE/03/PER164 | Ucayali, Coronel Portillo | CL | Cure (3 mo) | Cure | − | [9] |
| MHOM/PE/03/ PER215 | Ucayali, Coronel Portillo | ML | Cure (6 mo) | Cure | − | [9] |
| MHOM/PE/84/LC03 cl6 | Madre de Dios, Tambopata | CL | NA | … | − | [26] |
| MHOM/PE/91/LC1409 | Huanuco, Huanuco | CL | NA | … | − | [27] |
| MHOM/PE/91/LC1412 | Huanuco, Huanuco | CL | NA | … | − | [27] |
| MHOM/PE/91/LC1565 | Cusco, Paucartambo | CL | NA | … | − | [28] |
| MHOM/PE/91/LC1580 | Cusco, Paucartambo | MCL | NA | … | − | [29] |
| MHOM/PE/91/LC2123 | Cusco, Paucartambo | CL | NA | … | − | [30] |
| MHOM/PE/91/LC2125 | Cusco, Paucartambo | MCL | NA | … | − | [29] |
| MHOM/PE/91/LC2141 | Cusco, Paucartambo | MCL | NA | … | − | [29] |
| MHOM/PE/93/LC2143 | Cusco, Paucartambo | CL | NA | … | − | [30] |
| MHOM/PE/91/LC2147 | Cusco, Paucartambo | CL | NA | … | − | [28] |
| MHOM/PE/91/LC2177 | Cusco, Paucartambo | CL | NA | … | − | [30] |
| MHOM/PE/91/LC2289 | Cusco, Paucartambo | CL | NA | … | − | [29] |
| MHOM/PE/91/LC2320 | Cusco, Paucartambo | MCL | NA | … | − | [30] |
| MHOM/PE/94/LC2353 | Cusco, Paucartambo | CL | NA | … | − | [29] |
| MHOM/PE/94/LC2355 | Cusco, Paucartambo | CL | NA | … | − | [30] |
| MHOM/PE/94/LC2367 | Cusco, Paucartambo | CL | NA | … | − | [30] |
| MHOM/PE/94/LC2368 | Cusco, Paucartambo | MCL | NA | … | − | [30] |
| MHOM/PE/91/LC2398 | Cusco, Paucartambo | CL | NA | … | − | [29] |
| MHOM/PE/00/LH699 | Madre de Dios, Manu | CL | NA | … | − | [28] |
| MHOM/PE/00/LH800 | Madre de Dios, Tambopata | CL | NA | … | − | [28] |
| MHOM/PE/01/PER005 | Loreto, Ucayali | CL | Primary unresponsiveness (p) | Failure | − | [9] |
| MHOM/PE/01/PER006 | Junin, Satipo | CL | Primary unresponsiveness (p) | Failure | − | [9] |
| MHOM/PE/02/PER015 | Ucayali, Coronel Portillo | CL | Primary unresponsiveness (p) | Failure | − | [9] |
| MHOM/PE/02/PER086b | Pasco, Oxapampa | CL | Relapse (re) + secondary unresponsiveness | Failure | − | [9] |
| MHOM/PE/02/PER104c | Madre de Dios, Tambopata | CL | Secondary unresponsiveness (re + p) | Failure | − | [9] |
| MHOM/PE/03/PER201 | Loreto, Requena | ML | Cure (12 mo) | Cure | + | [9] |
| MHOM/PE/02/PER016 | Huanuco, Puerto Inca | CL | Cure (12 mo) | Cure | + | [9] |
| MHOM/PE/02/PER096 | Madre de Dios, Manu | CL | Cure (12 mo) | Cure | + | [31] |
| MHOM/PE/03/PER207 | Madre de Dios, Tambopata | CL | Cure (12 mo) | Cure | + | [31] |
| MHOM/PE/03/PER231 | Junin, Satipo | ML | Cure (12 mo) | Cure | + | [9] |
| MHOM/PE/02/PER069c | Madre de Dios, Manu | ML | Incomplete treatment, lost | … | + | [9] |
| MHOM/PE/02/PER010 | Cajamarca, Jaen | CL | Cure (3 mo) | Cure | + | [9] |
| MHOM/PE/91/LC2041 | Cusco, Paucartambo | CL | NA | … | + | [28] |
| MHOM/PE/91/LC1568 | Cusco, Paucartambo | CL | NA | … | + | [32] |
| MHOM/PE/91/LC1569 | Cusco, Paucartambo | CL | NA | … | + | [28] |
| MHOM/PE/91/LC1578 | Cusco, Paucartambo | CL | NA | … | + | [28] |
| MHOM/PE/91/LC1586 | Cusco, Paucartambo | CL | NA | … | + | [29] |
| MHOM/PE/91/LC2043 | Cusco, Paucartambo | MCL | NA | … | + | [29] |
| MHOM/PE/91/LC2176 | Cusco, Paucartambo | CL | NA | … | + | [30] |
| MHOM/PE/94/LC2284 | Cusco, Paucartambo | CL | NA | … | + | [30] |
| MHOM/PE/91/LC2318 | Cusco, Paucartambo | CL | NA | … | + | [32] |
| MHOM/PE/91/LC2319 | Cusco, Paucartambo | CL | NA | … | + | [32] |
| MHOM/PE/91/LC2321 | Cusco, Paucartambo | CL | NA | … | + | [32] |
| MHOM/PE/90/LH825 | Ucayali, Padre Abad | CL | NA | … | + | [28] |
| MHOM/PE/01/PER002 | Madre de Dios, Tambopata | CL | Primary unresponsiveness (p) | Failure | + | [9] |
| MHOM/PE/01/PER012 | Cusco, Calca | CL | Primary unresponsiveness (p) | Failure | + | [9] |
| MHOM/PE/01/PER014c | Junin, Satipo | CL | Primary unresponsiveness (re) | Failure | + | [9] |
| MHOM/PE/03/PER130c | Cusco, Echarate | CL | Primary unresponsiveness (re) | Failure | + | [9] |
| MHOM/PE/03/PER186c | Junin, Satipo | CL | Primary unresponsiveness (re) | Failure | + | [9] |
| MHOM/PE/02/PER065 | Cusco, La Convencion | CL | Relapse (p) | Failure | + | This work |
| MHOM/PE/03/PER212c | Madre de Dios, Tambopata | CL | Secondary unresponsiveness (p) | Failure | + | This work |
| MHOM/PE/02/PER067c | Cusco, La Convencion | CL | Secondary unresponsiveness (re + p) | Failure | + | [9] |
| MHOM/BO/94/CUM153 | Parque Isiboro, Limoncitos | CL | Cure/scar | Cure | − | [30] |
| MHOM/BO/94/CUM25 | Parque Isiboro, Moleto | CL | Cure/scar | Cure | − | [32] |
| MHOM/BO/94/CUM27 | Parque Isiboro, Moleto | MCL | Cure/scar | Cure | − | [32] |
| MHOM/BO/94/CUM29 | Parque Isiboro, Moleto | MCL | Cure/scar | Cure | − | [30] |
| MHOM/BO/94/CUM31 | Parque Isiboro, Moleto | CL | Cure/scar | Cure | − | [32] |
| MHOM/BO/94/CUM34 | Parque Isiboro, Moleto | CL | Cure/scar | Cure | − | [32] |
| MHOM/BO/94/CUM43 | Parque Isiboro, Moleto | MCL | Cure/scar | Cure | − | [30] |
| MHOM/BO/94/CUM45 | Parque Isiboro, NA | MCL | Cure/scar | Cure | − | [30] |
| MHOM/BO/94/CUM49 | Parque Isiboro, Isinuta | MCL | Cure/scar | Cure | − | [30] |
| MHOM/BO/94/CUM50 | Parque Isiboro, Primavera | MCL | Cure/scar | Cure | − | [32] |
| MHOM/BO/94/CUM55 | Parque Isiboro, Isinuta | MCL | Cure/scar | Cure | − | [32] |
| MHOM/BO/94/CUM57 | Parque Isiboro, NA | CL | Cure/scar | Cure | − | [33] |
| MHOM/BO/94/CUM59 | Parque Isiboro, Moleto | CL | Cure/scar | Cure | − | [32] |
| MHOM/BO/94/CUM67 | Shinahota | MCL | Cure/scar | Cure | − | [32] |
| MHOM/BO/94/CUM96 | Parque Isiboro, Moleto | CL | Cure/scar | Cure | − | [32] |
| MHOM/BO/94/CUM152 | Parque Isiboro, Limoncitos | MCL | Cure/scar | Cure | − | [30] |
| MHOM/BO/96/CUM180 | Parque Isiboro, Primavera | MCL | Cure/scar | Cure | − | [34] |
| MHOM/BO/94/CUM42b | Parque Isiboro, Primavera | CL | Secondary unresponsiveness | Failure | − | [30] |
| MHOM/BO/2002/CUM623 | Parque Isiboro, Llallagua | CL | Primary unresponsiveness | Failure | − | [33] |
| MHOM/BO/2002/CUM700 | Parque Isiboro, Llallagua | CL | Primary unresponsiveness | Failure | − | [33] |
| MHOM/BO/2002/CUM704 | Parque Isiboro, Moleto | CL | Primary unresponsiveness | Failure | − | This work |
| MHOM/BO/84/CEN002 | NA | CL | NA | … | − | [26] |
| MHOM/BO/85/CEN007 | NA | CL | NA | … | − | [26] |
| MHOM/BO/94/CUM97 | Parque Isiboro, Primavera | CL | NA | … | − | [30] |
| MHOM/BO/94/CUM138 | Parque Isiboro, Isinuta | MCL | NA | … | − | [29] |
| MHOM/BO/96/CUM181 | Parque Isiboro, NA | MCL | NA | … | − | This work |
| MHOM/BO/--/CUM363 | Parque Isiboro, NA | CL | NA | … | − | This work |
| MHOM/BO/--/CUM505 | NA | MCL | NA | … | − | [33] |
| MHOM/BO/94/CUM52 | Parque Isiboro, Isinuta | MCL | NA | … | − | [32] |
| MHOM/BO/94/CUM65 | Parque Isiboro, Moleto | MCL | Cure/scar | Cure | + | [32] |
| MHOM/BO/94/CUM68 | Shinahota | MCL | Cure/scar | Cure | + | [30] |
| MHOM/BO/94/CUM24 | Parque Isiboro, Isinuta | MCL | Primary unresponsiveness | Failure | + | [32] |
| MHOM/BO/94/CUM41 | Parque Isiboro, Moleto | CL | NA | … | + | [30] |
| Amphotericin B treatment | ||||||
| MHOM/BO/2002/CUM637 | Parque Isiboro, Primavera | MCL | Cure/scar | Cure | − | This work |
| MHOM/BO/2002/CUM639 | Parque Isiboro, Primavera | CL | Cure/scar | Cure | − | This work |
| MHOM/PE/02/PER011 | Huanuco, Huanuco | MCL | NA | … | + | [9] |
| MHOM/PE/03/PER136 | Ucayali, Coronel Portillo | ML | Cure (12 mo) | Cure | − | [9] |
Unless otherwise indicated, ‘Cure’ signifies that the patient was monitored for 12 months. In some cases, patients could only be monitored for 3 or 6 months. For analysis, these were classified as “cured” because previous studies showed that the cure rate assessed at 3 months was very nearly that seen at 12 months [11].
Abbreviations: CL, cutaneous leishmaniasis; MCL, mucocutaneous leishmaniasis; ML, mucosal leishmaniasis; NA, not available; p, prospective (within the LeishNatDrug-R study); re, retrospective (previous leishmaniasis episode).
a Region, Town in Bolivia.
b Patients with a history of previous treatment, but for whom the drug used was not known.
c Patients with a history of previous treatment with antimonials.
Parasite Isolates
The LRV1-positive L. guyanensis strain Lg5313 (World Health Organization code WHI/BR/78/M5313) and the LRV1-deficient line Lg17 [23] were obtained from Nicolas Fasel (University of Lausanne, Switzerland). Ninety-seven isolates of L. braziliensis (62 from Peru, 35 from Bolivia) were available for analysis; the designation, geographical origins, and clinical features of the isolates used in this study are summarized in Table 1, Figure 1, and Supplementary Figures 1 and 2. The Peruvian strains constitute an allopatric sample spanning the geographical range of L. braziliensis, mainly in the jungle. Conversely, the Bolivian strains comprise a sympatric sample, originating from the Indigenous Territory and National Park Isiboro Sécure at Cochabamba (Figure 1). In this study, parasites were recovered from patients before treatment, cryopreserved, and later revived for culture for RNA and DNA isolation. The isolates studied were typed as L. braziliensis by polymerase chain reaction–restriction fragment-length polymorphism analysis targeting gp63, Hsp70, cpb, and/or H2b genes as described elsewhere [29, 30].
Figure 1.
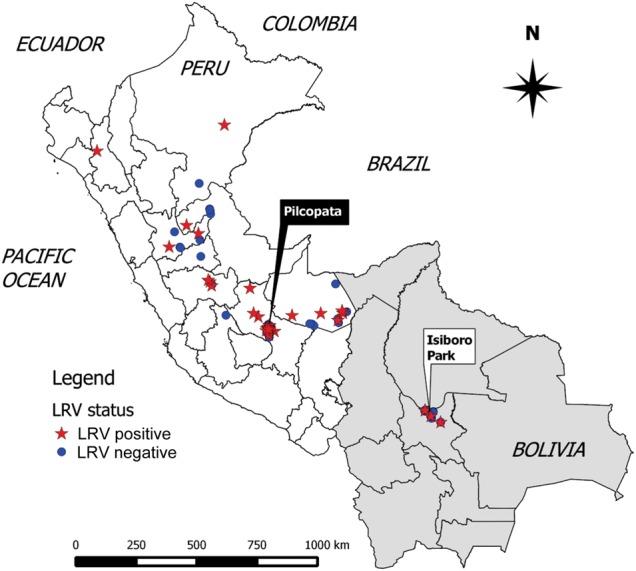
Geographical distribution of LRV1-positive Leishmania braziliensis isolates from Peru and Bolivia. The origins of L. braziliensis lines summarized in Table 1 are displayed on a map of Peru and Bolivia, created using the software package Quantum GIS, version 2.0.1 (available at: http://www.qgis.org/en/site/forusers/download.html), and the latitude and longitude coordinates of each locality. Both LRV-positive (star) and LRV-negative (circle) isolates occurred in the same geographic areas; in Peru, mostly in the jungle. Most Bolivian L. braziliensis isolates (33 of 35) originated from the Indigenous Territory and National Park Isiboro Sécure (municipality of Villa Tunari), and 2 isolates (CUM67 and CUM68) originated from the town of Shinahota (municipality of Tiraque), all located in the department of Cochabamba.
RNA Purification
Parasites were thawed and grown in Schneider's medium containing fetal bovine serum until reaching late log/early stationary phase. A total of 3 × 108 promastigotes were washed with ice-cold phosphate-buffered saline, collected by centrifugation, and resuspended and lysed in 1 mL of Trizol (Invitrogen). Cell lysates were stored at −80°C prior to shipment or processing. Total RNA was isolated according to the manufacturer's instructions (Life Technologies, California). Residual DNA was removed by treatment with DNase I (Life Technologies, California) at 37°C for 45 minutes, and RNA was further purified with the Zymo RCC-25 kit, using the manufacturer's instructions (Zymo Research, Irvine, California). The integrity of the purified RNA was verified by electrophoresis in 0.8% agarose gels in TAE buffer (40 mM Tris, 20 mM acetic acid, and 1 mM ethylenediaminetetraacetic acid, pH 8) at 4°C.
LRV1 Detection and Sequencing
These methods have been reported previously [17, 36, 37]. Complementary DNA (cDNA) was prepared from total RNA by priming with random hexamers and was then subjected to 30 cycles of PCR, using universal LRV degenerate primers that amplified a 488-nucleotide segment within the LRV1 capsid gene. Controls included buffer only, mock cDNA lacking reverse transcriptase, and both LRV1-positive and LRV1-negative strains of L. guyanensis. Reverse transcription–PCR (RT-PCR) products were analyzed on a 1.5% native agarose gel in TAE buffer, and LRV1 amplicons were purified and subjected to automated sequencing. The sequence was obtained from both strands, assembled and trimmed to remove low quality bases and primer sequences, and edited and aligned using DNAStar Lasergene software. Molecular phylogenies were constructed on a 299-nucleotide segment, using MEGA 6 analysis software [38]. The final LRV1 data set has been deposited in GenBank (KP682453-KP682484).
The presence of LRV1 was confirmed independently by the presence of an appropriately sized dsRNA following digestion with single-stranded nucleases (data not shown) [37, 39]. Following a recent proposal to the International Committee on the Taxonomy of Viruses, LRVs are referred to as “LRV1” or “LRV2,” followed by a species and then strain designation [40]. Thus, LRV1-Lguy-M4147 is the preferred name for M4147 LRV1–4, and LRV1-Lbr-CUM24 is the preferred name for the LRV occurring within strain MHOM/BO/94/CUM24.
Statistical Analysis
The type of lesion was treated as a nonordered 3-level categorical variable. Simple exact logistic regression was used to independently model the total (unadjusted) effect of the presence of LRV1 and lesion type on the probability of treatment failure in scenarios involving a small sample size. Multiple exact logistic regression was used to evaluate the direct effect on the probability of treatment failure of the presence of LRV1, after adjustment for the type of lesion. Statistical tests were performed under a 5% significance level, using the statistical software Stata 13.
RESULTS
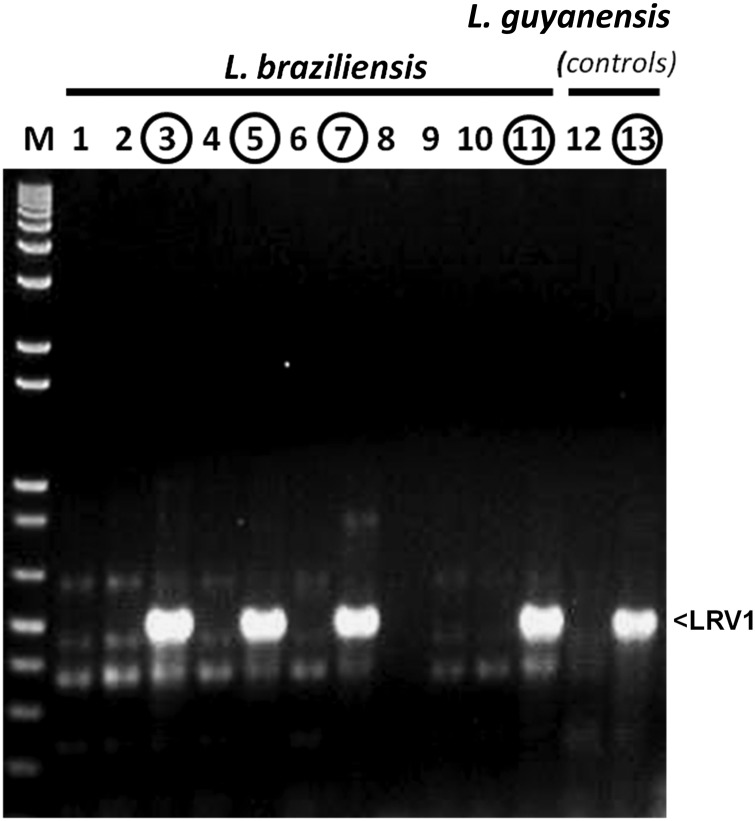
We analyzed a collection of 97 isolates of L. braziliensis obtained from patients exhibiting CL, ML, or MCL in Peru and Bolivia (Table 1 and Supplementary Figures 1 and 2). Axenic promastigote cultures were established in vitro following biopsy. Subsequently, patients were treated with SbV (93) or amphotericin B (4), and we were able to monitor the treatment outcome of 54 patients for up to 1 year. The patient response was classified as described previously [9] and, for statistical analysis, was further classified as “cure” or “failure,” with the latter including both instances of unresponsiveness and relapses (Table 1 and Supplementary Figures 1 and 2). Parasites were confirmed as L. braziliensis by molecular typing, and RT-PCR with universal LRV primers was used to detect LRV1 (Figure 2 and Table 1). When present, the levels of individual LRV1 amplicons were similar (Figure 2), and the sequence of each was determined.
Figure 2.
Reverse transcription–polymerase chain reaction (PCR) detection of LRV1 in Leishmania braziliensis. Agarose gel electrophoresis of PCR products obtained using the LRV universal primers SMB4647 and SMB4648 with randomly primed complementary DNA derived from RNA from the species/strains is shown, as described in “Materials and Methods” section. M, double-stranded DNA (dsDNA) molecular weight marker (1 kb plus; Life Technologies, CA). Lanes 1–11: L. braziliensis isolates LC2143 (lane 1), LC2147 (lane 2), LC2176 (lane 3), LC2177 (lane 4), LC2284 (lane 5), LC2289 (lane 6), LC2321 (lane 7), LC2353 (lane 8), LC2367 (lane 9), LC2398 (lane 10), and LC2318 (lane 11). Lanes 12 and 13, L. guyanensis isolates: Lg17 (LRV1 negative; lane 12) and Lg5313 (LRV1 positive; lane 13).
Thirty-two isolates clearly evidenced LRV1 (33%), with the proportion significantly higher in Peru (28 of 62 [45%]), compared with Bolivia (4 of 35 [11%]). LRV1-positive parasites were found widely across Peru, with some regions showing a higher prevalence than others (Figure 1). Similar variation among localities was reported previously [19–22].
LRV1 is Associated With a Significant Increase in the Risk of Treatment Failure
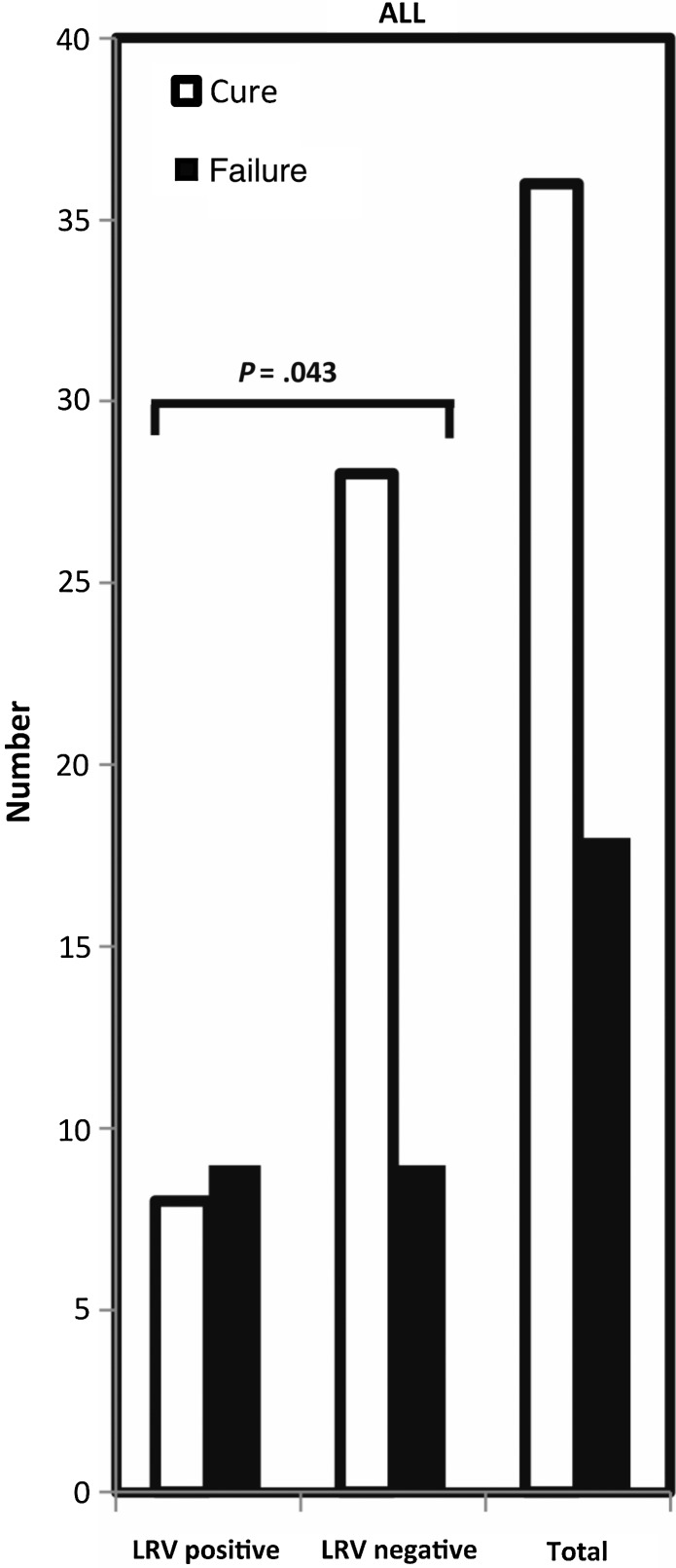
We next examined the association of LRV1 with treatment outcome for all patients. Extensive clinical data were available for 54 patients, including treatment history and outcome; all but 4 patients had been treated with SbV. The association of LRV1 and treatment outcome for the entire data set is shown in Figure 3; findings subdivided by country are shown in Supplementary Figure 3. Overall, 33% (18 of 54) were classified as failures; importantly, the percentage of failure was less in the LRV-negative isolates than in the LRV-positive isolates (24% [9 of 37] vs 53% [9 of 17]). Exact logistic regression showed this difference to be significant (P = .043) and having a notably high odds ratio (OR) of 3.5, associating the risk of failure with LRV1. This finding was seen within both the Peruvian and Bolivian isolates, although the number of treatment failures among the latter group was too few for statistical significance. Exclusion of the 3 patients treated with amphotericin B (all with LRV1-negative isolates) caused the overall significance to decrease (OR, 3.12; P = .067).
Figure 3.
Treatment failure versus LRV1 prevalence among Leishmania isolates. The number of cures (open bars) and treatment failures (closed bars) following chemotherapy is shown for the complete data set (n = 54). Within each grouping, the number of isolates positive or negative for LRV1 are shown. Data are taken from Table 1.
We further assessed the impact of LRV1 after adjustment for lesion type (both variables showing no significant interaction), using multiple exact logistic regression analysis. Again the presence of LRV1 was associated with an increased risk of treatment failure (OR, 3.99; P = .05). Interestingly, patients with CL showed a higher risk of treatment failure than those showing mucosal involvement (ie, patients with ML or MCL; OR, 18.5; P = .009). This was unexpected, as prior studies had not revealed a consistent difference [41–46]. In our studies, patients with ML or MCL received a longer course of SbV treatment than patients with CL (30 vs 20 days [9]), perhaps accounting for this outcome. Given the implications for the success of SbV treatments, this warrants further controlled studies in the future.
LRV1 Does Not Confer Intrinsic Parasite Antimony Resistance in Infected Macrophages
We considered the hypothesis that, in some manner, the presence of LRV1 conferred intrinsic drug resistance to the parasites. In a previous study, 26 of the Peruvian isolates had been examined for in vitro resistance to SbV as intracellular amastigotes in macrophage infections [9]. Of the SbV-resistant lines, 10 were LRV1 positive, while 12 were LRV1 negative; of the SbV -sensitive lines, 2 were LRV1 positive, while 2 were LRV1 negative. Thus, LRV1 was not significantly associated with SbV resistance directly (P = .43).
LRV1 Subtypes Are Not Associated With Treatment Outcome
We considered the possibility that the association between LRV1 and treatment failure arose not from the presence of LRV1, but from other parasite genetic factors. LRV1, like most other Totiviridae, are not shed or infectious and are transmitted only during cell division; thus by coinheritance, isolates that bear closely related LRV1s are closely related at the nuclear DNA level [47, 48]. If observed, clustering of treatment failures by LRV1 and, presumably, nuclear DNA relationship could signify that shared ancestry of other genetic factors was responsible, rather than LRV1. Differences in LRV1 sequence are unlikely to play a role, as it is the viral dsRNA itself (rather than any specific sequence motif) that serves to mediate virulence through interactions with TLR3 [23].
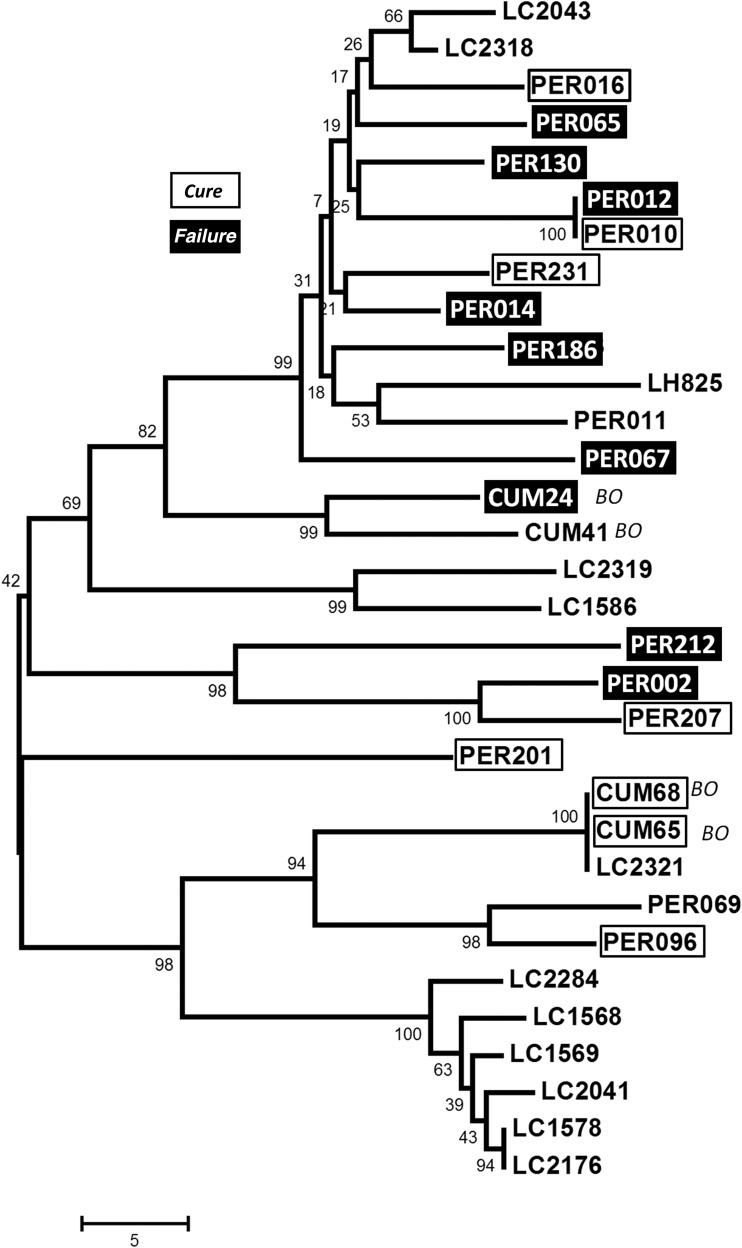
We constructed a dendrogram depicting LRV1 sequence relationships, onto which we displayed drug treatment outcomes where available (Figure 4). It was clear that treatment failures (Figure 4) did not cluster preferentially by the degree of LRV1 relationship. Instead, failures were interspersed among cures in most LRV1 lineages, including 2 bearing identical LRV1s (PER012 and PER010). Where known from microsatellite typing [31], the relationships of LRV1s were consistent with those of the underlying parasite genomes, including the close relationship of PER010 and PER012. While these data cannot rule out a direct contribution of other genetic factors, they suggest that the LRV1 effect seen here is independent of these, if present. Future studies using high resolution methods to probe the relationships of the isolates studied here may further test and extend these findings.
Figure 4.
LRV1 molecular phylogeny and drug treatment outcomes. The figure shows a molecular tree based on comparisons of a 299-nucleotide region of LRV1 (described in “Materials and Methods” section). When known for a given isolate, the clinical outcome of pentavalent antimonial therapy is shown (no patients treated with amphotericin B yielded strains bearing LRV1). The tree was constructed using the neighbor-joining algorithm, based on the uncorrected number of nucleotide differences and uniform rate assumptions. The scale shows a branch length of 5-nucleotide differences. Bootstrap values calculated from 5000 replicas are shown at each node. Cure, open boxes; treatment failure, dark boxes (see Table 1 for data and classification).
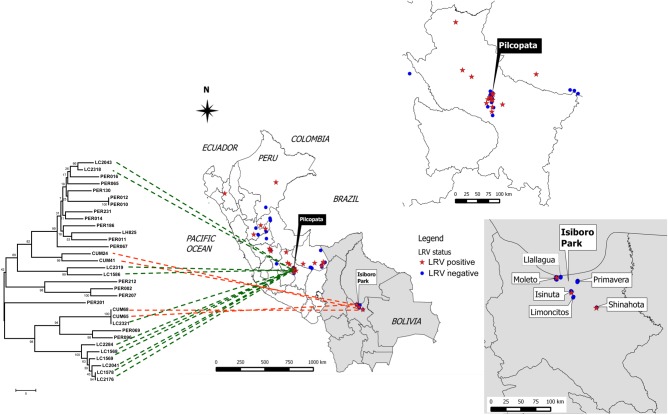
We examined the LRV1 phylogeny for geographic associations; however, there was no clear cline of LRV1 across Peru or Bolivia. This is perhaps best illustrated by closer examination of 2 sympatric populations, one occurring in Pilcopata (the Amazonian foothills, Cusco) in Peru and the other in Parque Isiboro (the Amazonian lowlands) in Bolivia. Both populations displayed considerable LRV1 diversity, spanning (or nearly so) the limits of the evolutionary tree (Figure 5). These data further suggest considerable diversity of L. braziliensis parasite populations in these localities.
Figure 5.
LRV1 relationships for 2 sympatric populations of Leishmania braziliensis in Peru and Bolivia. The figure associates the LRV1 sequence relationships depicted in Figure 4 with the geographical relationships shown in Figure 1. The fine distribution of LRV1 genotypes is shown in the insets for the district of Pilcopata at Paucartambo, Cusco, Peru; Isiboro Sécure National Park; and the municipality of Shinahota at Cochabamba, Bolivia. From this and the data in Figure 1, it can be seen that both LRV1-positive and LRV1-negative lines occur within both populations (insets), including LRV1s whose sequences differ considerably.
LRV1 is Not Preferentially Associated With MCL or ML
For CL presentations, 28 of 67 (42%) were LRV1 positive; for MCL and ML presentations, 1 of 13 (8%) and 7 of 17 (41%), respectively, were positive; and for ML/MCL combined, 8 of 30 (27%) were positive. These values did not differ significantly when analyzed separately or after combining the ML and MCL groups (P = .29 and P = .57, respectively). These findings are consistent with studies of other Leishmania populations [19, 20]; there was no significant association between disease status at the time of biopsy and the presence of LRV1 in axenic cultured parasites.
DISCUSSION
Here we examined a large panel of isolates of L. braziliensis and probed for associations between the presence of LRV1 and response to treatment or disease manifestations. Our data show a significant association between the presence of LRV1 and treatment failure (Figure 3). We ruled out the possibility that this arose by intrinsic LRV1-mediated SbV resistance, as there was no correlation between the presence of LRV1 and parasite SbV resistance that manifested during infections of macrophages in vitro. Similarly, we ruled out a significant contribution from other parasite genetic factors, using LRV1 sequence relationships as a surrogate measure of parasite genetic relatedness (due to LRV1-parasite coevolution [48]), rather than the presence of LRV1 itself, to assess whether the treatment failures were clustered preferentially into one or a few lineages. This analysis provided no evidence for preferential genetic clustering of treatment failures, further pointing to the presence of LRV1 itself as key risk factor.
Importantly, a companion article by Bourreau et al reports a similar association of LRV1 with pentamidine treatment failures in cases of L. guyanensis infection, in the absence of intrinsic parasite resistance [18]. Thus, current data suggest that LRV1 may act across species and drug classes to thwart efforts to treat leishmaniasis. These remarkable findings prompted us to consider potential mechanisms by which this occurs.
In the L. guyanensis murine model, LRV1-bearing parasites induce the expression of a distinctive set of macrophage inflammatory markers constituting a hyperinflammatory response, resulting ultimately in a TLR3-dependent increase in parasite numbers and disease severity [23, 24]. Correspondingly, many studies have shown a critical role for the host immune system in mediating SbV activity [10, 13, 14]. Thus, LRV1-mediated changes in the human host response could potentially serve to dampen the efficacy of SbV action. A second and nonexclusive model suggests the elevated parasite burden associated with LRV1 would act to compromise the efficacy for any given drug treatment regimen, even in the absence of intrinsic parasite resistance or drug-specific host interactions. Indeed, this may be especially likely for most antileishmanial compounds, whose efficacy and selective index is far from optimal [49, 50]. One key prediction of the higher parasite load model is independence from the specific mode of drug action and/or drug-specific involvement of host metabolism, which differ considerably among SbV, amphotericin B, and pentamidine. It also provides a potential framework for viewing the preferential association of treatment failures in CL (if this finding is confirmed in the future), as parasite numbers are generally much higher in this form of the disease than in chronic forms of ML.
The evidence presented here and in the companion work by Bourreau et al [18] provides a strong rationale implicating LRV1 in important aspects of human–parasite biology. Current data do not permit a firm determination of the mechanism by which the presence of LRV1 leads to treatment failures, and further studies will be required to unravel this process. Regardless of the mechanism, these findings have important implications for antileishmanial therapy, as they suggest that knowledge of the LRV1 status in L. braziliensis and L. guyanensis could support prognostics and follow-up. Our findings should also guide further research on new options for combination therapy, including targeting LRV1.
Supplementary Data
Supplementary materials are available at http://jid.oxfordjournals.org. Consisting of data provided by the author to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the author, so questions or comments should be addressed to the author.
Notes
Acknowledgments. We thank Catherine Ronet and Nicolas Fasel (University of Lausanne, Switzerland), for discussions and communicating unpublished results; F. Matthew Kuhlmann, for advice on the presentation of clinical data; and Jonathan Berman, for discussions concerning SbV treatment failures.
Disclosure. The sponsors had no role in study design, data collection, data analysis, data interpretation, or report writing.
Financial support. This work was supported by the European Commission (to projects LeishBolPe [contract ERBIC18CT960123] and LeishNatDrug-R [contract ICA4-CT-2001-10076], for parasite collection), the Directorate-General for Development Cooperation of the Belgian Government (framework agreement 03 [project 95502], for parasite characterization and data analysis), and the National Institutes of Health (grant AID R56 AI099364, for LRV1 analysis).
Potential conflicts of interest. All authors: No reported conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Alvar J, Velez ID, Bern C et al. Leishmaniasis worldwide and global estimates of its incidence. PLoS One 2012; 7:e35671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gollob KJ, Viana AG, Dutra WO. Immunoregulation in human American leishmaniasis: balancing pathology and protection. Parasite Immunol 2014; 36:367–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singh OP, Hasker E, Sacks D, Boelaert M, Sundar S. Asymptomatic Leishmania infection: a new challenge for Leishmania control. Clin Infect Dis 2014; 58:1424–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schriefer A, Wilson ME, Carvalho EM. Recent developments leading toward a paradigm switch in the diagnostic and therapeutic approach to human leishmaniasis. Curr Opin Infect Dis 2008; 21:483–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Castellucci LC, Almeida LF, Jamieson SE, Fakiola M, Carvalho EM, Blackwell JM. Host genetic factors in American cutaneous leishmaniasis: a critical appraisal of studies conducted in an endemic area of Brazil. Mem Inst Oswaldo Cruz 2014; 109:279–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reithinger R, Dujardin JC, Louzir H, Pirmez C, Alexander B, Brooker S. Cutaneous leishmaniasis. Lancet Infect Dis 2007; 7:581–96. [DOI] [PubMed] [Google Scholar]
- 7.Palacios R, Osorio LE, Grajalew LF, Ochoa MT. Treatment failure in children in a randomized clinical trial with 10 and 20 days of meglumine antimonate for cutaneous leishmaniasis due to Leishmania viannia species. Am J Trop Med Hyg 2001; 64:187–93. [DOI] [PubMed] [Google Scholar]
- 8.Tuon FF, Gomes-Silva A, Da-Cruz AM, Duarte MI, Neto VA, Amato VS. Local immunological factors associated with recurrence of mucosal leishmaniasis. Clin Immunol 2008; 128:442–6. [DOI] [PubMed] [Google Scholar]
- 9.Yardley V, Ortuno N, Llanos-Cuentas A et al. American tegumentary leishmaniasis: Is antimonial treatment outcome related to parasite drug susceptibility? J Infect Dis 2006; 194:1168–75. [DOI] [PubMed] [Google Scholar]
- 10.Croft SL, Sundar S, Fairlamb AH. Drug resistance in leishmaniasis. Clin Microbiol Rev 2006; 19:111–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Llanos-Cuentas A, Tulliano G, Araujo-Castillo R et al. Clinical and parasite species risk factors for pentavalent antimonial treatment failure in cutaneous leishmaniasis in Peru. Clin Infect Dis 2008; 46:223–31. [DOI] [PubMed] [Google Scholar]
- 12.Valencia C, Arevalo J, Dujardin JC, Llanos-Cuentas A, Chappuis F, Zimic M. Prediction score for antimony treatment failure in patients with ulcerative leishmaniasis lesions. PLoS Negl Trop Dis 2012; 6:e1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maurer-Cecchini A, Decuypere S, Chappuis F et al. Immunological determinants of clinical outcome in Peruvian patients with tegumentary leishmaniasis treated with pentavalent antimonials. Infect Immun 2009; 77:2022–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amato VS, Tuon FF, Bacha HA, Neto VA, Nicodemo AC. Mucosal leishmaniasis. Current scenario and prospects for treatment. Acta Trop 2008; 105:1–9. [DOI] [PubMed] [Google Scholar]
- 15.Widmer G, Comeau AM, Furlong DB, Wirth DF, Patterson JL. Characterization of a RNA virus from the parasite Leishmania. Proc Natl Acad Sci U S A 1989; 86:5979–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tarr PI, Aline RF Jr, Smiley BL, Scholler J, Keithly J, Stuart K. LR1: a candidate RNA virus of Leishmania. Proc Natl Acad Sci U S A 1988; 85:9572–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zangger H, Hailu A, Desponds C et al. Leishmania aethiopica field isolates bearing an endosymbiontic dsRNA virus induce pro-inflammatory cytokine response. PLoS Negl Trop Dis 2014; 8:e2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bourreau E, Marine Ginouves M, Prévot G et al. Leishmania RNA virus presence in L. guyanensis parasites increases the risk of first-line treatment failure and symptomatic relapse. J Infect Dis 2016; 213:105–11. [DOI] [PubMed] [Google Scholar]
- 19.Ogg MM, Carrion R Jr, Botelho AC, Mayrink W, Correa-Oliveira R, Patterson JL. Short report: quantification of leishmaniavirus RNA in clinical samples and its possible role in pathogenesis. Am J Trop Med Hyg 2003; 69:309–13. [PubMed] [Google Scholar]
- 20.Pereira OLD, Maretti-Mira AC, Rodrigues KM et al. Severity of tegumentary leishmaniasis is not exclusively associated with Leishmania RNA virus 1 infection in Brazil. Mem Inst Oswaldo Cruz 2013; 108:665–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salinas G, Zamora M, Stuart K, Saravia N. Leishmania RNA viruses in Leishmania of the Viannia subgenus. Am J Trop Med Hyg 1996; 54:425–9. [DOI] [PubMed] [Google Scholar]
- 22.Guilbride L, Myler PJ, Stuart K. Distribution and sequence divergence of LRV1 viruses among different Leishmania species. Mol Biochem Parasitol 1992; 54:101–4. [DOI] [PubMed] [Google Scholar]
- 23.Ives A, Ronet C, Prevel F et al. Leishmania RNA virus controls the severity of mucocutaneous leishmaniasis. Science 2011; 331:775–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hartley MA, Drexler S, Ronet C, Beverley SM, Fasel N. The immunological, environmental, and phylogenetic perpetrators of metastatic leishmaniasis. Trends Parasitol 2014; 30:412–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saiz M, Llanos-Cuentas A, Echevarria J et al. Short report: detection of Leishmaniavirus in human biopsy samples of leishmaniasis from Peru. Am J Trop Med Hyg 1998; 58:192–4. [DOI] [PubMed] [Google Scholar]
- 26.Dujardin JC, Gajendran N, Arevalo J et al. Karyotype polymorphism and conserved characters in the Leishmania (Viannia) braziliensis complex explored with chromosome-derived probes. Ann Soc Belg Med Trop 1993; 73:101–18. [PubMed] [Google Scholar]
- 27.Dujardin JC, Dujardin JP, Tibayrenc M et al. Karyotype plasticity in neotropical Leishmania: an index for measuring genomic distance among L. (V.) peruviana and L. (V.) braziliensis populations. Parasitology 1995; 110(Pt 1):21–30. [DOI] [PubMed] [Google Scholar]
- 28.Victoir K, Banuls AL, Arevalo J et al. The gp63 gene locus, a target for genetic characterization of Leishmania belonging to subgenus Viannia. Parasitology 1998; 117(Pt 1):1–13. [PubMed] [Google Scholar]
- 29.Victoir K, De Doncker S, Cabrera L et al. Direct identification of Leishmania species in biopsies from patients with American tegumentary leishmaniasis. Trans R Soc Trop Med Hyg 2003; 97:80–7. [DOI] [PubMed] [Google Scholar]
- 30.Garcia AL, Kindt A, Quispe-Tintaya KW et al. American tegumentary leishmaniasis: antigen-gene polymorphism, taxonomy and clinical pleomorphism. Infect Genet Evol 2005; 5:109–16. [DOI] [PubMed] [Google Scholar]
- 31.Adaui V, Maes I, Huyse T et al. Multilocus genotyping reveals a polyphyletic pattern among naturally antimony-resistant Leishmania braziliensis isolates from Peru. Infect Genet Evol 2011; 11:1873–80. [DOI] [PubMed] [Google Scholar]
- 32.Rougeron V, De Meeus T, Hide M et al. Extreme inbreeding in Leishmania braziliensis. Proc Natl Acad Sci U S A 2009; 106:10224–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Odiwuor S, Veland N, Maes I, Arevalo J, Dujardin JC, Van der Auwera G. Evolution of the Leishmania braziliensis species complex from amplified fragment length polymorphisms, and clinical implications. Infect Genet Evol 2012; 12:1994–2002. [DOI] [PubMed] [Google Scholar]
- 34.Fraga J, Montalvo AM, De Doncker S, Dujardin JC, Van der Auwera G. Phylogeny of Leishmania species based on the heat-shock protein 70 gene. Infect Genet Evol 2010; 10:238–45. [DOI] [PubMed] [Google Scholar]
- 35.Bermudez H, Rojas E, Garcia L et al. Generic sodium stibogluconate is as safe and effective as branded meglumine antimoniate, for the treatment of tegumentary leishmaniasis in Isiboro Secure Park, Bolivia. Ann Trop Med Parasitol 2006; 100:591–600. [DOI] [PubMed] [Google Scholar]
- 36.Lye LF, Owens KL, Shi H et al. Retention and loss of RNA interference pathways in Trypanosomatid protozoa. PLoS Pathog 2010; 6:e1001161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zangger H, Ronet C, Desponds C et al. Detection of Leishmania RNA virus in Leishmania parasites. PLoS Negl Trop Dis 2013; 7:e2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 2013; 30:2725–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beiting DP, Peixoto L, Akopyants NS et al. Differential induction of TLR3-dependent innate immune signaling by closely related parasite species. PLoS One 2014; 9:e88398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adams MJ, Lefkowitz EJ, King AM, Carstens EB. Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses (2014). Arch Virol 2014; 159:2831–41. [DOI] [PubMed] [Google Scholar]
- 41.Falqueto A, Sessa PA, Ferreira AL et al. Epidemiological and clinical features of Leishmania (Viannia) braziliensis American cutaneous and mucocutaneous leishmaniasis in the State of Espirito Santo, Brazil. Mem Inst Oswaldo Cruz 2003; 98:1003–10. [DOI] [PubMed] [Google Scholar]
- 42.Guimaraes LH, Machado PR, Lago EL et al. Atypical manifestations of tegumentary leishmaniasis in a transmission area of Leishmania braziliensis in the state of Bahia, Brazil. Trans R Soc Trop Med Hyg 2009; 103:712–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Machado PR, Ampuero J, Guimaraes LH et al. Miltefosine in the treatment of cutaneous leishmaniasis caused by Leishmania braziliensis in Brazil: a randomized and controlled trial. PLoS Negl Trop Dis 2010; 4:e912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Unger A, O'Neal S, Machado PR et al. Association of treatment of American cutaneous leishmaniasis prior to ulcer development with high rate of failure in northeastern Brazil. Am J Trop Med Hyg 2009; 80:574–9. [PMC free article] [PubMed] [Google Scholar]
- 45.Andersen EM, Cruz-Saldarriaga M, Llanos-Cuentas A et al. Comparison of meglumine antimoniate and pentamidine for peruvian cutaneous leishmaniasis. Am J Trop Med Hyg 2005; 72:133–7. [PubMed] [Google Scholar]
- 46.Velez I, Lopez L, Sanchez X, Mestra L, Rojas C, Rodriguez E. Efficacy of miltefosine for the treatment of American cutaneous leishmaniasis. Am J Trop Med Hyg 2010; 83:351–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wickner RB, Ghabrial SA, Nibert ML, Patterson JL, Wang CC. Totiviridae. In: King AMQ, Lefkowitz E, Adams MJ, Carstens EB, eds. Virus taxonomy: ninth report of the International Committee on Taxonomy of Viruses. San Diego: Elsevier, 2011:639–50. [Google Scholar]
- 48.Widmer G, Dooley S. Phylogenetic analysis of Leishmania RNA virus and Leishmania suggests ancient virus-parasite association. Nucleic Acids Res 1995; 23:2300–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pace D. Leishmaniasis. J Infect 2014; 69(suppl 1):S10–8. [DOI] [PubMed] [Google Scholar]
- 50.Amato VS, Tuon FF, Siqueira AM, Nicodemo AC, Neto VA. Treatment of mucosal leishmaniasis in Latin America: systematic review. Am J Trop Med Hyg 2007; 77:266–74. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.