Abstract
The incidence and severity of Clostridium difficile infection (CDI) in North America and Europe has increased significantly since the 2000s. However, CDI is not widely recognized in China and other developing countries due to limited laboratory diagnostic capacity and low awareness. Most published studies on laboratory workflows for CDI diagnosis are from developed countries, and thus may not be suitable for most developing countries. Therefore, an alternative strategy for developing countries is needed. In this study, we evaluated the performance of the Glutamate Dehydrogenase (GDH) test and its associated workflow on 416 fecal specimens from suspected CDI cases. The assay exhibited excellent sensitivity (100.0%) and specificity (92.8%), compared to culture based method, and thus could be a good screening marker for C. difficile but not for indication of toxin production. The VIDAS CDAB assay, which can detect toxin A/B directly from fecal specimens, showed good specificity (99.7%) and positive predictive value (97.2%), but low sensitivity (45.0%) and negative predictive value (88.3%), compared with PCR-based toxin gene detection. Therefore, we propose a practical and efficient GDH test based workflow strategy for the laboratory diagnosis of CDI in developing countries like China. By applying this new workflow, the CDI laboratory diagnosis rate was notably improved in our center, yet the increasing cost was kept at a minimum level. Furthermore, to gain some insights into the genetic population structure of C. difficile isolates from our hospital, we performed MLST and PCR toxin gene typing.
Introduction
Clostridium difficile is a Gram-positive, spore-forming, anaerobic rod, which is an important cause of antibiotic associated diarrhea. C. difficile infection (CDI) is a major nosocomial disease, with clinical symptoms ranging from asymptomatic colonization, diarrhea to toxic megacolon, bowel perforation, sepsis, shock and death [1]. Since the 2000s, there has been a significant increase in the incidence and severity of CDI in North America and Europe, placing a huge financial burden on health care systems worldwide [2]. Consequently, in 2013, CDI was considered an urgent public health threat by the U. S. Centers for Disease Control and Prevention [3]. However, due to low awareness, possibly emanating from limited laboratory diagnostic capacity, low sample submission rate, and lack of high-quality surveillance systems [4, 5], CDI is not widely recognized in Asia, including China. Most publications on workflow for the laboratory diagnosis of CDI are from Europe, and thus may not be suitable for developing countries with limited resources. As such, alternative workflow strategies are urgently needed to address this issue in developing countries.
CDI diagnosis is based on a combination of clinical history and laboratory detection of C. difficile toxin in the feces or cultured isolates [6, 7]. PCR-based commercial kits, which offer high sensitivity and specificity [8, 9], are widely used for CDI diagnosis in developed countries, but their costs exceed the affordability of many laboratories in developing countries. In China, PCR-based tests are not approved for use, and thus culture remains the main diagnostic method.
Glutamate dehydrogenase (GDH) is a constitutive enzyme produced in large amounts by all strains of C. difficile independent of toxigenicity. GDH is, therefore, easily detected in feces, which makes it a good screening marker for C. difficile. In order to improve the laboratory diagnostic capacity for CDI investigation, previous studies have recommended the use of C. difficile GDH as a preliminary screening test, followed by further confirmatory tests for toxin production and possession of toxin genes [10–13]. Due to low awareness of CDI, the GDH assay has not been approved for use in China, though indications are that this will happen before the end of 2015.
Moreover, there is limited data on the effectiveness of the assay in the diagnosis of CDI in developing countries. To address these concerns, we evaluated the performance of GDH assay in detecting C. difficile from fecal samples in a Chinese hospital clinical laboratory. And, based on a comprehensive consideration of the sensitivity, specificity, turnaround time and costs of different C. difficile testing assays, we proposed a practical workflow for future laboratory diagnosis of CDI in developing countries. Finally, to gain some insight into the genetic population structure of C. difficile isolates, we studied the MLST and PCR toxin gene types of our isolates.
Materials and Methods
Ethics
The study was approved by the Human Research Ethics Committee of Peking Union Medical College Hospital (No. S-263). Written informed consent was obtained from patients for the use of the samples in research.
Sample collection and study design
Consecutive, non-repetitive fecal specimens (n = 416) were collected from patients presenting with diarrheal symptoms at the Peking Union Medical College Hospital, from August 2012 to July 2014. All the fecal specimens were stored at 4°C before testing, and were simultaneously tested by VIDAS C. difficile GDH assay (bioMérieux, Marcy l'Etoile, France), VIDAS C. difficile Toxin A&B (bioMérieux, Marcy l'Etoile, France), and culture-based method followed by molecular detection of toxin genes (see below for details).
VIDAS C. difficile GDH and Toxin A&B (CDAB) detection
All the specimens were tested simultaneously for GDH and CDAB by commercial VIDAS kits (bioMérieux, Marcy l'Etoile, France), according to manufacturer’s instructions. Negative results for GDH assay were defined as optical density (OD) 450/630 nm < 0.10, and positive results were defined as OD450/630 nm ≥ 0.10. The fluorescence intensity for CDAB of < 0.13, ≥ 0.13 to < 0.37 and ≥ 0.37 were reported as negative, equivocal and positive, respectively.
Sample culture and MALDI-TOF MS identification
All the specimens were cultured in appropriate media and identification of C. difficile was done according to the laboratory’s current routine laboratory workflows. Generally, the specimens were cultured on cycloserine-cefoxitin fructose agar (CCFA) in anaerobe condition at 35°C for 48 h. Suspect C. difficile colonies were tested by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) with MALDI Biotyper version 3.1 (Bruker Daltonics GmbH, Bremen, Germany) according to the manufacturer’s instructions. Isolates with identification scores of ≥2.0 were considered to be accurately identified to species level.
Toxin gene detection and multilocus sequence typing (MLST)
The genomic DNA of the C. difficile isolates was extracted by QIAamp DNA Mini Kit (QIAGEN, Hilden, Germany) following the manufacturer’s instructions. A 5-plex PCR was performed to detect 4 toxin genes, namely tcdA, tcdB, cdtA and cdtB, and the 16S rDNA of C. difficile, as previously described by Persson et al. [14]. MLST was performed by sequencing seven gene loci (adk, atpA, dxr, glyA, recA, sodA and tpi) as previously developed by Griffiths et al. [15]. The seven gene loci sequences were submitted to the PubMLST sequence query page (http://pubmlst.org/cdifficile/) to obtain the allele, clade and sequence types (ST).
Statistical analysis
Statistical analyses were performed using SPSS software (version 17.0, IBM, New York, USA). The sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV) were calculated.
Results
General clinical information
About 64% (264/416) of the patients were from Medical Department, 18.5% (77/416) from Outpatient and Emergency Departments, 7.2% (30/416) from Surgery Department, 5.0% (21/416) from ICU, and 5.8% (24/416) were from other Departments (Gynecology, Obstetrics and Pediatrics Departments). The average age of the 416 patients, which included 220 males and 196 female patients, was 43.8±21.3 years (ranged from 3–95).
Performance of the VIDAS GDH assay for detection of C. difficile compared with routine culture
Among the 416 fecal specimens tested, 32.2% (134/416) were positive for GDH. In comparison to the GDH assay, 26.9% (112/416) of the specimens were C. difficile culture positive. About 5% (22 of 416) specimens were positive for GDH but negative for culture. Compared with routine C. difficile culture, the sensitivity, specificity, PPV and NPV of VIDAS GDH assay were 100.0%, 92.8%, 83.6% and 100.0%, respectively (Table 1).
Table 1. Comparison of the diagnostic methods for C. difficile infections in this study.
| Testing methods | Percentage of (95% confidential interval) | TAT (h) | Costs (US$) | |||
|---|---|---|---|---|---|---|
| Sensitivity | Specificity | PPV | NPV | |||
| GDH | 100 (96.8–100.0) | 92.8 (89.3–95.4) | 83.6 (76.2–89.4) | 100 (98.7–100.0) | 2 | 8 |
| Culture | Reference method for GDH | 48 | 15 | |||
| CDAB | 45 (33.9–56.5) | 99.7 (98.6–99.9) | 97.2 (85.8–99.9) | 88.3 (84.7–91.4) | 2 | 8 |
| Toxin gene detection | Reference method for CDAB | 50 | 20 | |||
| Proposed workflow | Not applicable | 15.1 a | 15.6 a | |||
Abbreviations: GDH, glutamate dehydrogenase; PPV, positive predictive value; NPV, negative predictive value; TAT, turnaround time.
a The TAT and costs of the proposed workflow were calculated by average value of the 416 fecal specimens.
Performance of VIDAS CDAB assay compared with molecular assay for toxin detection
Of the 416 fecal samples tested, 19.2% (80/416) were positive for toxin genes by PCR-based detection, and 36 (45.0%) of these samples were CDAB positive or equivocal. Furthermore, among the 336 PCR negative samples, one (0.3%) sample was equivocal for CDAB, and the rest negative. Compared with PCR-based toxin gene detection, the sensitivity, specificity, PPV and NPV of VIDAS CDAB were 45.0%, 99.7%, 97.2% and 88.3%, respectively (Table 1).
Amongst the 134 GDH positive fecal samples, 22.4% (30/134) and 9.0% (12/134) were positive and equivocal for VIDAS CDAB testing, respectively. All the GDH negative fecal samples (n = 282), were also negative for VIDAS CDAB assay.
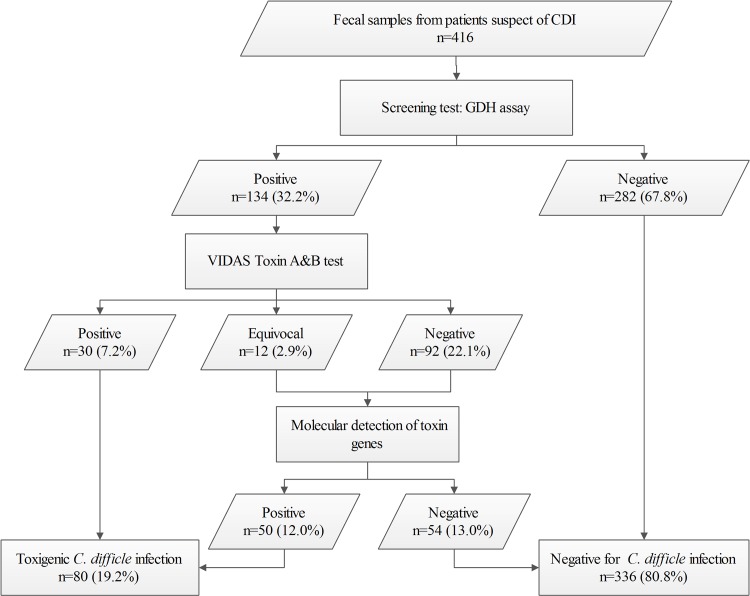
A proposed GDH-based diagnostic workflow for improved laboratory diagnosis of CDI in developing countries
To improve the laboratory diagnostic capacity for CDI in developing countries, we propose a practical and efficient workflow, based on a combination of testing methods in laboratory diagnosis of CDI (Fig 1). The first step is to screen the suspect CDI fecal specimens by GDH assay, with negative results directly reported to physicians. For GDH positive specimens, CDAB testing should be performed subsequently to detect toxin production. If the CDAB results are positive, laboratory diagnosis of CDI can be made. Samples with equivocal or negative CDAB results should be referred for further testing, such as molecular detection of toxin genes, toxigenic culture (TC) or cell cytotoxicity neutralization assay (CCNA). By applying this new workflow in our lab, the CDI diagnosis rate was notably improved from 8.2% (30/416) to 19.2% (80/416), whilst keeping the rising costs at a reasonable minimum (from US$8 to US$15.6) (Table 1).
Fig 1. A proposed GDH-based workflow for laboratory diagnosis of C. difficile infections (CDI).
Abbreviations in the figure: GDH, glutamate dehydrogenase. Cell cytotoxicity neutralization assay or toxigenic culture could be used as alternative testing methods for determining toxigenicity of C. difficile where molecular methods were not available.
Toxin genotypes and MLST of the 112 C. difficile culture positive isolates
Of the C. difficile culture positive 112 isolates, 54 (48.2%) were tcdA-positive, tcdB-positive and cdtA/cdtB-negative (A+B+CDT-); 32 (28.6%) were tcdA-negative, tcdB-negative and cdtA/cdtB-negative (A-B-CDT-); and 20 (17.9%) were tcdA-negative, tcdB-positive and cdtA/cdtB-negative (A−B+CDT−). The remaining six (5.3%) isolates were tcdA-positive, tcdB-positive and cdtA/cdtB-positive (A+B+CDT+).
Using MLST, the 112 culture positive C. difficile isolates were classified into 23 STs, with the majority (68.8%) belonging to clade 1. ST-3 was the most common ST (18.8%, 21/112), followed by ST-54 (13.4%, 15/112) and ST-37 (9.8%, 11/112). The six A+B+CDT+ isolates were grouped into ST-1 and ST-5. In addation, all the 20 A−B+CDT− isolates were ST-37 and ST-81 (Table 2). Generally, there were no obvious correlations between clonal clusters generated and the isolate hospital source departments.
Table 2. MLST sequence types (STs), allelic profiles and toxin gene profiles of the 112 C. difficile culture positive clinical isolates.
| ST (no. of isolates) | Clades | Allelic profile | Toxin genotype | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| adk | atpA | dxr | glyA | recA | sodA | tpi | tcdA | tcdB | cdtA/cdtB | ||
| ST-1 (3) | 2 | 1 | 1 | 1 | 10 | 1 | 3 | 5 | + | + | +/+ |
| ST-2 (8) | 1 | 1 | 1 | 2 | 1 | 5 | 3 | 1 | + | + | -/- |
| ST-3 (12) | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | + | + | -/- |
| ST-3 (9) | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | - | - | -/- |
| ST-5 (3) | 3 | 1 | 6 | 4 | 7 | 2 | 8 | 7 | + | + | +/+ |
| ST-8 (2) | 1 | 1 | 1 | 2 | 6 | 1 | 5 | 1 | + | + | -/- |
| ST-15 (5) | 1 | 1 | 1 | 6 | 1 | 8 | 5 | 1 | - | - | -/- |
| ST-26 (4) | 1 | 1 | 1 | 6 | 1 | 4 | 3 | 4 | - | - | -/- |
| ST-35 (6) | 1 | 2 | 5 | 8 | 1 | 1 | 3 | 6 | + | + | -/- |
| ST-37 (11) | 4 | 3 | 7 | 3 | 8 | 6 | 9 | 11 | - | + | -/- |
| ST-39 (8) | 4 | 3 | 7 | 10 | 8 | 7 | 2 | 10 | - | - | -/- |
| ST-42 (3) | 1 | 1 | 1 | 2 | 1 | 1 | 7 | 1 | + | + | -/- |
| ST-54 (15) | 1 | 1 | 4 | 7 | 1 | 1 | 3 | 3 | + | + | -/- |
| ST-55 (2) | 1 | 1 | 1 | 6 | 6 | 1 | 12 | 12 | + | + | -/- |
| ST-81 (9) | 4 | 3 | 1 | 3 | 8 | 6 | 9 | 11 | - | + | -/- |
| ST-91 (1) | 1 | 1 | 1 | 6 | 6 | 1 | 6 | 1 | + | + | -/- |
| ST-98 (1) | 1 | 1 | 1 | 2 | 6 | 1 | 1 | 3 | + | + | -/- |
| ST-100 (2) | 1 | 1 | 1 | 6 | 19 | 2 | 24 | 1 | - | - | -/- |
| ST-101 (1) | 1 | 1 | 2 | 2 | 1 | 1 | 23 | 1 | - | - | -/- |
| ST-117 (2) | 1 | 1 | 1 | 7 | 1 | 2 | 5 | 1 | - | - | -/- |
| ST-129 (2) | 1 | 1 | 3 | 6 | 1 | 1 | 1 | 3 | + | + | -/- |
| ST-286 (1) | 1 | 1 | 1 | 2 | 3 | 1 | 5 | 3 | + | + | -/- |
| ST-289 (1) | 1 | 1 | 1 | 7 | 1 | 5 | 3 | 1 | + | + | -/- |
| ST-320 (1) | 4 | 11 | 7 | 10 | 43 | 6 | 20 | 15 | - | - | -/- |
Discussion
CDI poses a great public health threat to health care facilities worldwide, with a significant rise in incidence, severity and mortality worldwide [16, 17]. Accurate and timely diagnosis of CDI is necessary for appropriate clinical management of the patients and infection control interventions [18, 19]. However, CDI is not widely recognized in Asia, including China. Although several C. difficile testing assays are now available worldwide, especially in North America and Europe, the VIDAS CDAB kit was the only commercial assay approved by China Food and Drug Administration as of December 2014. Furthermore, few laboratories routinely culture or carry out molecular-based detection of C. difficile. Therefore, the current laboratory diagnostic capacity for CDI detection in China, is inadequate to meet clinical demands, and thus there is an urgent need to introduce more practical C. difficile testing assays, including updating the current workflow, which would help in raising CDI awareness amongst clinicians [20, 21].
Several studies have demonstrated that GDH is a good screening test for C. difficile in fecal samples [22–25]. Our study is in agreement with these studies as GDH exhibited high sensitivity (100%) and favorable NPV (100%), compared with the culture based method. GDH testing can efficiently detect samples that are negative for CDI, with minimal hands-on time and cost. However, the detection of GDH simply indicates the presence of the organism and is not indicative of toxin production and thus cannot be used to diagnose CDI alone [11, 26].
The pathogenesis of CDI is mainly attributed to toxin A/B, which can be detected by VIDAS CDAB assay directly from fecal specimens. In the present study, the VIDAS CDAB assay showed good specificity (99.7%) and PPV (97.2%), but with low sensitivity (45%) and NPV (88.3%), compared with PCR-based toxin gene detection, which is in agreement to other studies [27]. Thus some CDI positive patients would be missed by using this test alone as a confirmatory test. PCR-based assays have high sensitivity and specificity [7, 8], but their costs exceed the affordability of many laboratories in developing countries.
To improve the sensitivity and cost-effectiveness of CDI diagnosis, we propose a GDH-based practical and efficient workflow, with combined testing methods in laboratory diagnosis of CDI. In this workflow, the VIDAS GDH assay is used as an initial screening assay, followed by detection of toxin production by VIDAS CDAB assay, and any discordant results between GDH and CDAB assays are confirmed by a third method such as molecular detection of toxin genes, toxigenic culture or cell culture cytotoxicity neutralization assay. By applying this proposed GDH-based workflow in this study, the CDI laboratory diagnosis rate was notably improved from 8.2% to 19.2%, yet the increasing cost was kept at the minimum level. This provides a good balance between increasing the CDI detection capacity for most China diagnostic clinical labs, and not using PCR, which as of October 2015 was not a legally authorized test.
In order to have some insight into the molecular epidemiology of C. difficile isolates in a part of China, MLST and toxin genotyping were performed. Our study revealed that the majority (71.4%) of the 112 isolates was toxin gene positive, and six isolates possessed the binary toxin gene, which is associated with higher mortality and recurrence rates [28]. MLST allows an easier inter-laboratory comparison of data and appears reasonably discriminatory for studying the transmission of C. difficile [15]. To date, this is the second study on MLST of C. difficile strains in China. Our findings showed that ST-3, ST-54 and ST-37 were the most common types at our hospital, which is in agreement to the first MLST study on 104 isolates in China [29], but different from other countries. For instance, in a tertiary care hospital in Spain, 34% of the isolates were ST-2, whilst in a teaching hospital in Japan, ST-17 (21.8%) was the most predominant ST [30, 31]. There was no obvious sign of outbreak as all the STs were distributed sporadically among different departments in our hospital.
This study has some limitations. First, all the specimens were collected from one single center and thus the efficiency and utility of the proposed workflow should be validated by more hospitals. Second, due to unavailability of commercial PCR kits in China for legal use, toxin gene detection was performed on C. difficile isolates after culture, instead of directly from fecal samples, which prolonged the turnaround time in the workflow.
The high positive rate of toxigenic strains in our study indicates that CDI may be an under-estimated problem in China. By introducing GDH as the screening test and properly applying the workflow, the diagnostic capacity and control of potential outbreaks of CDI may be improved significantly. Although GDH assay is not currently legalized for use in China, the licensing process is in the final stages, with indications of approval before end of 2015. Thus this proposed workflow is part of the preparatory work for best utilization of this test when it becomes available in China. The C. difficile strains in our study have been well preserved, and we are doing molecular epidemiology study of CDI. We hope our further study can provide more data and increase awareness and surveillance of CDI in China.
Acknowledgments
This study was financially supported by a National Natural Science Foundation of China (grant number 81501807) and National Research Special Fund for Public Welfare Industry of Health of China (grant number 201402001).
Data Availability
All relevant data are within the paper.
Funding Statement
This study was financially supported by a National Natural Science Foundation of China (grant number 81501807) and National Research Special Fund for Public Welfare Industry of Health of China (grant number 201402001). Website: http://www.nhfpc.gov.cn/zwgkzt/pkjjy1/201302/9cb220c8a5704c0a9ebf5ef65a337544.shtml. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Rupnik M, Wilcox MH, Gerding DN. Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nat Rev Microbiol. 2009; 7: 526–536. 10.1038/nrmicro2164 [DOI] [PubMed] [Google Scholar]
- 2. Jones AM, Kuijper EJ, Wilcox MH. Clostridium difficile: a European perspective. J Infect. 2013; 66: 115–128. 10.1016/j.jinf.2012.10.019 [DOI] [PubMed] [Google Scholar]
- 3.U.S. Center for Disease Control and Prevention. Antibiotic resistance threats in the United States, 2013
- 4. Collins DA, Hawkey PM, Riley TV. Epidemiology of Clostridium difficile infection in Asia. Antimicrob Resist Infect Control. 2013; 2: 21 10.1186/2047-2994-2-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hawkey PM, Marriott C, Liu WE, Jian ZJ, Gao Q, Ling TK, et al. Molecular epidemiology of Clostridium difficile infection in a major Chinese hospital: an underrecognized problem in Asia? J Clin Microbiol. 2013; 51: 3308–3313. 10.1128/JCM.00587-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Burnham CA, Carroll KC. Diagnosis of Clostridium difficile infection: an ongoing conundrum for clinicians and for clinical laboratories. Clin Microbiol Rev. 2013; 26: 604–630. 10.1128/CMR.00016-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bassetti M, Villa G, Pecori D, Arzese A, Wilcox M. Epidemiology, diagnosis and treatment of Clostridium difficile infection. Expert Rev Anti Infect Ther. 2012; 10: 1405–1423. 10.1586/eri.12.135 [DOI] [PubMed] [Google Scholar]
- 8. Novak-Weekley SM, Marlowe EM, Miller JM, Cumpio J, Nomura JH, Vance PH, et al. Clostridium difficile testing in the clinical laboratory by use of multiple testing algorithms. J Clin Microbiol. 2010; 48: 889–893. 10.1128/JCM.01801-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lalande V, Barrault L, Wadel S, Eckert C, Petit JC, Barbut F. Evaluation of a loop-mediated isothermal amplification assay for diagnosis of Clostridium difficile infections. J Clin Microbiol. 2011; 49: 2714–2716. 10.1128/JCM.01835-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ticehurst JR, Aird DZ, Dam LM, Borek AP, Hargrove JT, Carroll KC. Effective detection of toxigenic Clostridium difficile by a two-step algorithm including tests for antigen and cytotoxin. J Clin Microbiol. 2006. 44: 1145–1149. 10.1128/JCM.44.3.1145-1149.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Crobach MJ, Dekkers OM, Wilcox MH, Kuijper EJ. European Society of Clinical Microbiology and Infectious Diseases (ESCMID): data review and recommendations for diagnosing Clostridium difficile-infection (CDI). Clin Microbiol Infect. 2009; 15: 1053–1066. 10.1111/j.1469-0691.2009.03098.x [DOI] [PubMed] [Google Scholar]
- 12. Culbreath K, Ager E, Nemeyer RJ, Kerr A, Gilligan PH. Evolution of testing algorithms at a university hospital for detection of Clostridium difficile infections. J Clin Microbiol. 2012; 50: 3073–3076. 10.1128/JCM.00992-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Surawicz CM, Brandt LJ, Binion DG, Ananthakrishnan AN, Curry SR, Gilligan PH, et al. Guidelines for diagnosis, treatment, and prevention of Clostridium difficile infections. Am J Gastroenterol. 2013; 108: 478–498. 10.1038/ajg.2013.4 [DOI] [PubMed] [Google Scholar]
- 14. Persson S, Torpdahl M, Olsen KE. New multiplex PCR method for the detection of Clostridium difficile toxin A (tcdA) and toxin B (tcdB) and the binary toxin (cdtA/cdtB) genes applied to a Danish strain collection. Clin Microbiol Infect. 2008; 14: 1057–1064. 10.1111/j.1469-0691.2008.02092.x [DOI] [PubMed] [Google Scholar]
- 15. Griffiths D, Fawley W, Kachrimanidou M, Bowden R, Crook DW, Fung R, et al. Multilocus sequence typing of Clostridium difficile . J Clin Microbiol. 2010; 48: 770–778. 10.1128/JCM.01796-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Murabata M, Kato H, Yano H, Ogura M, Shibayama J, Wakimoto Y, et al. Intestinal colonization and nosocomial spread of Clostridium difficile in pediatric cancer patients under long-term hospitalization. Kansenshogaku Zasshi. 2008; 82: 419–426. [DOI] [PubMed] [Google Scholar]
- 17. Bartlett JG, Perl TM. The new Clostridium difficile—what does it mean? N Engl J Med. 2005; 353: 2503–2505. 10.1056/NEJMe058221 [DOI] [PubMed] [Google Scholar]
- 18. Muto CA, Blank MK, Marsh JW, Vergis EN, O'Leary MM, Shutt KA, et al. Control of an outbreak of infection with the hypervirulent Clostridium difficile BI strain in a university hospital using a comprehensive "bundle" approach. Clin Infect Dis. 2007; 45: 1266–1273. 10.1086/522654 [DOI] [PubMed] [Google Scholar]
- 19. Gerding DN, Muto CA, Owens RC Jr. Measures to control and prevent Clostridium difficile infection. Clin Infect Dis. 2008; 46 Suppl 1:S43–49. 10.1086/521861 [DOI] [PubMed] [Google Scholar]
- 20. Chen X, Lamont JT. Overview of Clostridium difficile infection: implications for China. Gastroenterol Rep (Oxf). 2013; 1: 153–158. 10.1093/gastro/got029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jin K, Wang S, Huang Z, Lu S. Clostridium difficile infections in China. J Biomed Res. 2010; 24: 411–416. 10.1016/S1674-8301(10)60055-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Eastwood K, Else P, Charlett A, Wilcox M. Comparison of nine commercially available Clostridium difficile toxin detection assays, a real-time PCR assay for C. difficile tcdB, and a glutamate dehydrogenase detection assay to cytotoxin testing and cytotoxigenic culture methods. J Clin Microbiol. 2009; 47: 3211–3217. 10.1128/JCM.01082-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sharp SE, Ruden LO, Pohl JC, Hatcher PA, Jayne LM, Ivie WM. Evaluation of the C.Diff Quik Chek Complete Assay, a new glutamate dehydrogenase and A/B toxin combination lateral flow assay for use in rapid, simple diagnosis of Clostridium difficile disease. J Clin Microbiol. 2010; 48: 2082–2086. 10.1128/JCM.00129-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kawada M, Annaka M, Kato H, Shibasaki S, Hikosaka K, Mizuno H, et al. Evaluation of a simultaneous detection kit for the glutamate dehydrogenase antigen and toxin A/B in feces for diagnosis of Clostridium difficile infection. J Infect Chemother. 2011; 17: 807–811. 10.1007/s10156-011-0267-5 [DOI] [PubMed] [Google Scholar]
- 25. Kim H, Kim WH, Kim M, Jeong SH, Lee K. Evaluation of a rapid membrane enzyme immunoassay for the simultaneous detection of glutamate dehydrogenase and toxin for the diagnosis of Clostridium difficile infection. Ann Lab Med. 2014; 34: 235–239. 10.3343/alm.2014.34.3.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cheng AC, Ferguson JK, Richards MJ, Robson JM, Gilbert GL, McGregor A, et al. Australasian Society for Infectious Diseases guidelines for the diagnosis and treatment of Clostridium difficile infection. Med J Aust. 2011; 194: 353–358 [DOI] [PubMed] [Google Scholar]
- 27. Planche T, Aghaizu A, Holliman R, Riley P, Poloniecki J, Breathnach A, et al. Diagnosis of Clostridium difficile infection by toxin detection kits: a systematic review. Lancet Infect Dis. 2008; 8: 777–784. 10.1016/S1473-3099(08)70233-0 [DOI] [PubMed] [Google Scholar]
- 28. Stewart DB, Berg A, Hegarty J. Predicting recurrence of C. difficile colitis using bacterial virulence factors: binary toxin is the key. J Gastrointest Surg. 2013; 17: 118–124. 10.1007/s11605-012-2056-6 [DOI] [PubMed] [Google Scholar]
- 29. Yan Q, Zhang J, Chen C, Zhou H, Du P, Cui Z, et al. Multilocus sequence typing (MLST) analysis of 104 Clostridium difficile strains isolated from China. Epidemiol Infect. 2013; 141: 195–199. 10.1017/S0950268812000453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kuwata Y, Tanimoto S, Sawabe E, Shima M, Takahashi Y, Ushizawa H, et al. Molecular epidemiology and antimicrobial susceptibility of Clostridium difficile isolated from a university teaching hospital in Japan. Eur J Clin Microbiol Infect Dis. 2015; 34: 763–772. 10.1007/s10096-014-2290-9 [DOI] [PubMed] [Google Scholar]
- 31. Weber I, Riera E, Deniz C, Perez JL, Oliver A, Mena A. Molecular epidemiology and resistance profiles of Clostridium difficile in a tertiary care hospital in Spain. Int J Med Microbiol. 2013; 303: 128–133. 10.1016/j.ijmm.2013.02.016 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.