Abstract
Most of the human genome is transcribed, yielding a complex network of transcripts that includes tens of thousands of long noncoding RNAs. Many of these transcripts have a 5′ cap and a poly(A) tail, yet some of the most abundant long noncoding RNAs are processed in unexpected ways and lack these canonical structures. Here, I highlight the mechanisms by which several of these well-characterized noncoding RNAs are generated, stabilized, and function. The MALAT1 and MEN β (NEAT1_2) long noncoding RNAs each accumulate to high levels in the nucleus, where they play critical roles in cancer progression and the formation of nuclear paraspeckles, respectively. Nevertheless, MALAT1 and MEN β are not polyadenylated as the tRNA biogenesis machinery generates their mature 3′ ends. In place of a poly(A) tail, these transcripts are stabilized by highly conserved triple helical structures. Sno-lncRNAs likewise lack poly(A) tails and instead have snoRNA structures at their 5′ and 3′ ends. Recent work has additionally identified a number of abundant circular RNAs generated by the pre-mRNA splicing machinery that are resistant to degradation by exonucleases. As these various transcripts use non-canonical strategies to ensure their stability, it is becoming increasingly clear that long noncoding RNAs may often be regulated by unique post-transcriptional control mechanisms. This article is part of a Special Issue entitled: Clues to long noncoding RNA taxonomy.
Keywords: MALAT1, MEN β, NEAT1, Triple helix, tRNA-like, Sno-lncRNA, Circular RNA, CircRNA, Pre-mRNA splicing, RNA stability, Polyadenylation
1. Introduction
Although proteins have long been thought to be the main structural and functional components of cells, many biological processes utilize and, in fact, require noncoding RNAs. For example, ribosomal RNAs (rRNAs) and transfer RNAs (tRNAs) are universally used in protein synthesis, small nuclear RNAs (snRNAs) catalyze pre-mRNA splicing, and telomerase RNA ensures that no genetic information is lost from the ends of eukaryotic chromosomes. With the recent realization that transcription is pervasive across eukaryotic genomes [1,2], it is now clear that these classic “housekeeping” noncoding RNAs represent only the tip of the iceberg. As most (~75%) of the human genome is transcribed, each of our cells likely generates tens of thousands of additional transcripts with little or no predicted protein-coding capacity [3–7]. Among these noncoding RNAs are well-studied small RNAs, such as microRNAs (reviewed in [8]), as well as many other classes of small (<200-nt) and long (>200-nt) transcripts whose functions and mechanisms of biogenesis are less clear. Interestingly, some of these long transcripts, including MALAT1 [9], MEN ε/β (NEAT1) [10], and certain circular RNAs [11,12], are expressed at levels higher than most protein-coding transcripts. Nevertheless, most long noncoding RNAs are expressed at low levels and are poorly conserved at the sequence level across species. This has led to some debate whether these transcripts are truly functional or simply represent transcriptional noise [13,14].
Because of the efforts of many laboratories, several dozen long noncoding RNAs now have proposed functions and multiple lines of evidence support the biological relevance of others (reviewed in [15–20]). For example, long noncoding RNAs are often stable [21], expressed in a tissue-specific manner [22,23], or mis-regulated in human diseases, including cancer and neurological disorders [24–26]. In addition, evolutionary selection within the promoter and/or transcribed regions is found for many long noncoding RNAs [27,28], with functional segments sometimes being 30-nucleotides or shorter [29]. In some cases, these segments likely correspond to binding sites for proteins, including histone modification complexes or RNA binding proteins, thereby allowing the formation of ribonucleoprotein (RNP) complexes [30–33]. In other cases, these motifs allow the long noncoding RNAs to base pair with DNA [34] or other RNAs, including microRNAs [35–37] and mRNAs [38–40], to regulate expression in trans. With the recent development of new strategies to probe how noncoding RNAs fold [41–44], we are also gaining an increasingly detailed view of how these transcripts look and function at the molecular level.
One major surprise that has come from studying the biogenesis of various long noncoding RNAs is that they can be processed in non-canonical ways (reviewed in [45–47]). Although most appear to be capped, spliced, and polyadenylated, some of the most abundant long noncoding RNAs transcribed by RNA polymerase II defy these dogmas. Here, several of these well-studied noncoding RNAs, which lack a 5′ cap, a poly(A) tail, or both terminal structures, are highlighted. Although such transcripts are normally thought to be rapidly degraded (reviewed in [48–50]), these particular long noncoding RNAs accumulate as stable transcripts and have key cellular roles. First, the MALAT1 and MEN ε/β (NEAT1) loci are discussed as they each generate abundant non-polyadenylated RNAs whose 3′ ends are protected by triple helical structures. I then discuss how the pre-mRNA splicing machinery is able to generate several classes of noncoding RNAs, including sno-lncRNAs that terminate in snoRNA structures at both ends. Splicing additionally allows numerous introns and exons to be processed into mature circular RNAs that have covalently linked ends and are resistant to exonucleolytic degradation. In total, these well-characterized noncoding RNAs serve as useful examples for future studies, especially since an increasing number of transcripts have been suggested to lack a poly(A) tail [51–54]. It is, therefore, likely that we are only beginning to understand the full variety of non-canonical mechanisms that control the biogenesis and functions of noncoding RNAs.
2. The long noncoding RNA MALAT1 is commonly mis-regulated in cancer
MALAT1 (metastasis-associated lung adenocarcinoma transcript 1), also known as NEAT2 [55], was originally identified as a prognostic marker in non-small cell lung cancer [56]. High MALAT1 expression in early-stage tumors was associated with increased risk for metastasis as well as decreased patient survival. Subsequent work has shown that MALAT1 is over-expressed in many human carcinomas, including those of the breast, pancreas, lung, colon, prostate, and liver [57–61]. Consistent with a role for the MALAT1 transcript in driving tumorigenesis, several studies (but not all [62]) have found that MALAT1 promotes cell proliferation [61,63–67]. Furthermore, over-expression of MALAT1 results in increased cell migration in vitro [61,64]. Depletion of MALAT1 (using antisense oligonucleotides, siRNAs, or gene knockout strategies), on the other hand, inhibits cell motility in vitro and significantly limits metastasis formation in mouse cancer models [61–63, 68–70]. For example, when human A549 lung cancer cells are injected into the tail vein of nude mice, cells lacking MALAT1 form 80–90% fewer lung metastases than wildtype A549 cells [62]. The MALAT1 locus thus represents both a promising cancer biomarker as well as a potential therapeutic target for limiting metastatic growth. As antisense oligonucleotides are able to efficiently knock-down the MALAT1 transcript in vivo [9,71], there exists the interesting potential to translate these findings into the clinic.
In addition to being commonly over-expressed in various cancers, the MALAT1 locus is often mutated [72,73]. Mutations map throughout MALAT1 and are most notably observed in bladder urothelial carcinomas (15.3% of cases, making MALAT1 the 8th most commonly mutated gene in these particular tumors), lung adenocarcinomas (9.7% of cases, 12th most commonly mutated gene), head and neck squamous cell carcinomas (6.3% of cases, 10th most commonly mutated gene), and lung squamous cell carcinomas (5.8% of cases, 20th most commonly mutated gene) [72]. The functional significance of these mutations is, however, still unknown. In some renal cell carcinomas, chromosomal translocations fuse the MALAT1 promoter upstream of the TFEB (transcription factor EB) open reading frame [74–77]. Besides presumably changing MALAT1 expression levels, these translocations result in a promoter swap and >30-fold over-expression of the TFEB transcription factor, which likely leads to the mis-regulation of key developmental pathways [75]. In total, these studies have revealed that the MALAT1 locus can be mis-regulated via multiple mechanisms in tumors, but the underlying molecular functions of MALAT1 that are altered in cancers remains a key unanswered question.
3. MALAT1 is processed to generate a long non-polyadenylated RNA and a tRNA-like small RNA
Even among non-cancerous cells, MALAT1 is broadly expressed and is among the most abundant long noncoding RNAs in mouse and human tissues [55,78]. This rarely spliced transcript (the dominant isoforms are unspliced and ~6.7-kb in mouse, ~7-kb in humans) is expressed at a level comparable with or higher than many protein-coding genes, including β-actin or GAPDH [9]. As the MALAT1 transcript generally has a long half-life [78,79], it was quite a surprise when it was determined that the mature MALAT1 transcript is not polyadenylated [79]. Although replication-dependent histone mRNAs are stable despite lacking poly(A) tails (reviewed in [80]), the vast majority of non-polyadenylated, long RNA polymerase II transcripts are generally thought to be rapidly degraded (reviewed in [48]).
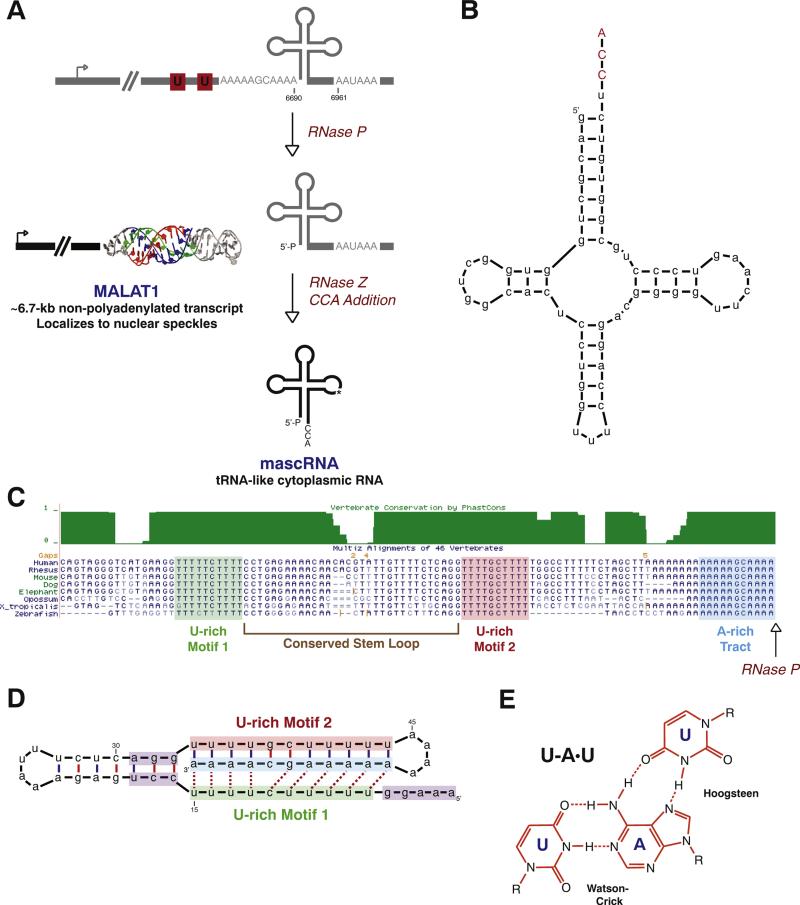
Rather than using the canonical cleavage/polyadenylation machinery, MALAT1 3′ end formation almost always occurs several hundred nucleotides upstream of the poly(A) signal (AAUAAA) (Fig. 1A). This upstream region is the most evolutionarily conserved part of the MALAT1 locus (with conservation extending from humans to fish) and, similar to canonical tRNAs, folds into a cloverleaf secondary structure [79]. The endonuclease RNase P recognizes this tRNA-like structure and cleaves to simultaneously generate the mature 3′ end of the long MALAT1 transcript and the 5′ end of a tRNA-like small RNA. Additional enzymes involved in tRNA biogenesis, including RNase Z and the CCA-adding enzyme, then further process the small RNA to generate a mature 61-nt transcript known as mascRNA (MALAT1-associated small cytoplasmic RNA) (Fig. 1B). mascRNA is exported to the cytoplasm, but likely does not read the genetic code as it has a small, poorly conserved anticodon loop. The exact biological function of mascRNA, however, remains undefined. Nevertheless, by taking advantage of the tRNA biogenesis machinery, the cell is able to process the primary MALAT1 transcript into two mature noncoding RNAs that have very different fates.
Fig. 1.
The MALAT1 locus generates a nuclear-retained long noncoding RNA and a tRNA-like small RNA. (A) Rather than using the canonical cleavage/polyadenylation machinery, the 3′ end of MALAT1 is almost always generated by tRNA biogenesis factors. First, RNase P cleavage simultaneously generates the mature 3′ end of MALAT1 and the 5′ end of mascRNA. The tRNA-like small RNA is subsequently cleaved by RNase Z, subjected to CCA addition, and exported to the cytoplasm. In contrast, the mature MALAT1 transcript localizes to nuclear speckles and its 3′ terminus is protected by a triple helical structure (PDB code 4PLX [84]). The mouse coordinates (GenBank accession number FJ209304) are given. (B) Mouse mascRNA adopts a tRNA-like cloverleaf secondary structure. CCA (denoted in red) is post-transcriptionally added by the CCA-adding enzyme. (C) As shown by the Multiz Alignment track of the UCSC Genome Browser, highly conserved A- and U-rich tracts are present immediately upstream of the MALAT1 RNase P cleavage site (denoted by arrow). (D) These conserved sequence motifs form base triplets (denoted by dashed lines) that protect the 3′ end of MALAT1 from degradation. A minimal triple helix that supports both RNA stability and translation [82] is shown. Nucleotides that function in promoting translation are denoted in purple. (E) U–A·U base triplets form via Hoogsteen hydrogen bonds to the major groove of a Watson–Crick base-paired helix.
4. A triple helix stabilizes the mature 3′ end of MALAT1
Once the 3′ end of the long MALAT1 transcript has been generated by RNase P, no additional nucleotides are post-transcriptionally added to its end [79]. How then is the MALAT1 3′ end stabilized to allow this long noncoding RNA to accumulate to such high levels in cells? Mature MALAT1 ends in an evolutionarily conserved A-rich tract that is preceded by two U-rich tracts and a stem–loop structure [79] (Fig. 1C). Although the genomically encoded A-rich tract may look similar to a miniature poly(A) tail, it is disrupted by GC nucleotides and likely is too short to bind poly(A) binding protein [81]. Furthermore, the A-rich tract is necessary, but not sufficient for MALAT1 stability as both upstream U-rich motifs are also required in vitro [79] and in cells [82,83]. Surprisingly, the reason why all three motifs are required is that they interact through base pairing: U-rich Motif 1 forms Hoogsteen hydrogen bonds to a Watson–Crick base-paired helix that is formed between U-rich Motif 2 and the A-rich tract [82–84] (Fig. 1D). The 3′ end of MALAT1 thus folds into a bipartite triple helix containing runs of five and four U–A·U triplets (Fig. 1E) that are separated by a C–G·C triplet and a C–G doublet [82,83] (Fig. 1D). Interestingly, this C–G doublet is critical for allowing all 10 triplets to form as it prevents steric clashes between the Hoogsteen and Watson strands that otherwise occur when 6 or more triplets are present in a continuous row [84].
By forming the equivalent of a molecular “knot,” the MALAT1 triple helix elegantly functions to block exonucleolytic degradation from the 3′ end of the RNA. Even if the triple helix is somewhat weakened by mutations in either of the U-rich motifs, the cell still has some difficulty degrading through this structure [82,83]. 3′–5′ exonucleases appear to often stall within the triple helix, forcing the cell to post-transcriptionally add U-rich tails to these decay intermediates [82]. The short U-rich tails then likely serve as single-stranded sequences that additional exonucleases bind to re-start the decay process (reviewed in [85]).
A highly similar triple helix stabilizes the 3′ end of the MEN β long noncoding RNA (see below; Fig. 2A) [82,83]. In addition, PAN (polyadenylated nuclear) RNA, a long noncoding RNA generated by Kaposi's sarcoma-associated herpesvirus (KSHV) and related γ-herpesviruses, is protected from 3′–5′ degradation by a triple helical structure [86,87]. Unlike MALAT1 and MEN β, which encode their A tracts in the genomic DNA (and are thus part of the primary transcripts), PAN RNA ends in a canonical poly(A) tail [88]. After this poly(A) tail has been added post-transcriptionally, part of it reaches back to base pair with two upstream U-rich motifs to form five U–A·U triplets [86]. As U-rich motifs have recently been found to promote the stability of many yeast mRNAs [89], poly(A) tails may interact with upstream sequence motifs (possibly in a triple helical conformation) much more often than is currently appreciated.
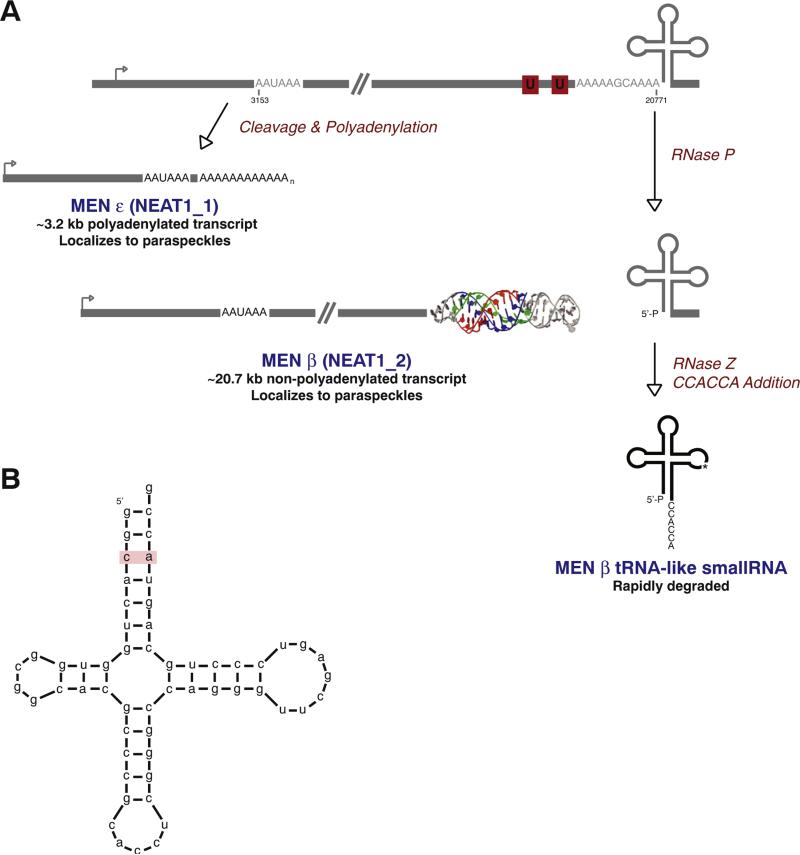
Fig. 2.
The MEN ε/β (NEAT1) locus is regulated by alternative 3′ end processing. (A) The MEN ε/β primary transcript can be cleaved by the canonical cleavage/polyadenylation machinery (to generate the polyadenylated MEN ε RNA) or by the tRNA biogenesis machinery (to generate the non-polyadenylated MEN β RNA). Analogous to the MALAT1 locus, an evolutionarily conserved triple helix is present immediately upstream of the RNase P cleavage site to protect the mature 3′ end of MEN β. The mouse coordinates (GenBank accession number GQ859163) are given. (B) The acceptor stem of the mouse MEN β tRNA-like small RNA is destabilized due to the presence of a C–A mismatch (denoted in red). This causes the CCA-adding enzyme to add CCACCA rather than CCA to the small RNA, triggering its efficient degradation.
5. The triple helix is a multi-functional structure with roles beyond RNA stability
Although the triple helix is structurally quite distinct from a poly(A) tail, it is able to carry out nearly all the functions that a poly(A) tail normally serves. When placed at the 3′ end of a reporter transcript, the MALAT1 and MEN β triple helices are each able to stabilize the RNA almost as efficiently as a poly(A) tail [82,83,90]. Remarkably, when an open reading frame is placed upstream of the triple helix, the reporter transcript is translated in mammalian cells at an efficiency similar to that obtained when the RNA is polyadenylated [82]. This indicates that the MALAT1 and MEN β 3′ ends can somehow function in RNA stability, nuclear export, and translation.
Whereas the 10 base triplets are critical for RNA stability, nucleotides flanking each side of the triple helical region are critical for supporting translation (Fig. 1D) [82]. Mutating these flanking nucleotides causes a reporter transcript to be poorly translated despite the mRNA being stable and localized in the cytoplasm. The key factor(s) that recognize the ends of MALAT1 and MEN β to support translation are still unknown, however. Neither poly(A) binding protein (PABP) [91] nor stem–loop binding protein (SLBP) [80], which recognize poly(A) tails and the histone stem–loop, respectively, are likely to directly bind these triple helices. This suggests that MALAT1, MEN β, and other RNAs ending in triple helices may be uniquely and specifically regulated by novel modes of post-transcriptional and translational control.
Whether endogenous MALAT1 or MEN β are ever translated is unclear as these transcripts are almost exclusively retained in the nucleus (see below). Curiously, ribosome profiling revealed that the 5′ end of MALAT1 appears to be bound by ribosomes in mouse embryonic stem cells [82,92,93]. As the MALAT1 putative ORFs are not evolutionarily conserved, additional work will be required to determine what this ribosome binding may mean. For example, it is possible that MALAT1 may generate short species-specific peptides, e.g. during mitosis when the nuclear membrane has broken down. Alternatively, MALAT1 may interact with ribosomes for reasons other than serving as a template for protein synthesis.
6. MALAT1 localizes to nuclear speckles, but its functions are still ambiguous
Once the primary MALAT1 transcript has been processed, mascRNA is exported to the cytoplasm [79], whereas the long MALAT1 transcript is retained in the nucleus in nuclear speckles [55,78]. These domains are enriched in pre-mRNA splicing factors and are thought to serve as sites for the assembly, modification, and/or storage of the pre-mRNA processing machinery [94]. Two distinct regions within human MALAT1 (nucleotides 1170–2249 and 5217–6220 of GenBank accession number FJ209305) are responsible for specifically targeting the RNA to nuclear speckles [95,96]. However, the exact function of MALAT1 in nuclear speckles is still rather unclear, especially since depletion of MALAT1 does not noticeably disrupt the architecture of this subnuclear domain [9,97,98].
Some reports have indicated that MALAT1 interacts with splicing factors, such as SR proteins, to regulate alternative pre-mRNA splicing patterns in human HeLa and fibroblast cells [65,96,99]. Nevertheless, significant splicing changes were not observed when MALAT1 was knocked-down in lung cancer cells [62], endothelial cells [67], or in MALAT1 knockout mice [9]. MALAT1 has also been reported to bind unmethylated Polycomb 2 (Pc2) protein to control the expression of growth-signal responsive genes, in part by helping these genes relocate from Polycomb bodies to nuclear speckles [66]. Somewhat consistent with this idea, MALAT1 appears to bind many nascent pre-mRNAs derived from actively transcribed gene loci [100,101]. In neurons, MALAT1 has additionally been reported to regulate synapse formation [78] as well as bind TDP-43 [102,103], a DNA/RNA-binding protein that has been implicated in the pathogenesis of amyotrophic lateral sclerosis (ALS) and other neurodegenerative disorders [104]. It thus appears that MALAT1 may regulate gene expression via multiple mechanisms.
Despite all these reported phenotypes using various cell lines, MALAT1 loss-of-function mice surprisingly have almost no phenotype [9,97,105]. These mice develop normally without any gross abnormalities and are fertile. The only notable change that one study could identify is that the expression of 12 genes, most of which are located adjacent to MALAT1 in the genome, was weakly altered upon MALAT1 loss [9]. This suggests that the mouse genome may encode other genes (protein-coding or noncoding) that may compensate for MALAT1 loss or are functionally redundant with MALAT1. Alternatively, it may be that MALAT1 has an essential function only when cells are stressed or are changing their state, e.g. during cellular transformation events in cancer. It is nevertheless important to emphasize that depletion of MALAT1 clearly affects tumor growth in mice [62] and thus MALAT1 has important roles in vivo. Future studies will hopefully clarify why even normal cells bother to generate so much of the MALAT1 transcript.
7. The MEN ε/β (NEAT1) locus is subjected to alternative 3′ end processing to generate multiple noncoding RNAs
Like MALAT1, the MEN ε/β long noncoding RNAs accumulate to high levels in the nucleus of many mammalian cells [55]. MEN ε (also known as NEAT1_1) and MEN β (also known as NEAT1_2) are both predominately unspliced and are transcribed from the same promoter, but differ in the location of their 3′ ends (Fig. 2A). Whereas MEN ε (~3.2-kb in mouse and ~3.7-kb in humans) is subjected to canonical cleavage/polyadenylation, the mature 3′ end of MEN β (~20.7-kb in mouse and ~22.7-kb in humans) is generated via a mechanism very similar to that used at the MALAT1 locus [10]. RNase P recognizes and cleaves immediately upstream of a tRNA-like structure (Fig. 2B) that is encoded at the end of the MEN β locus, releasing a tRNA-like small RNA as well as the mature MEN β transcript.
The decision to generate MEN ε vs. MEN β is at least partially controlled by the opposing actions of CFIm (cleavage factor Im, which is composed of a heterodimer of NUDT21 and CPSF6) and hnRNP K [106]. Binding of CFIm upstream of the MEN ε poly(A) signal recruits the cleavage/polyadenylation machinery (including CPSF and poly(A) polymerase), thereby promoting the biogenesis of MEN ε. hnRNP K is also able to bind near the MEN ε poly(A) signal, where it inhibits CFIm binding and acts to disassemble the CFIm heterodimer [106]. hnRNP K thus promotes the production of the long MEN β transcript, and the competition between CFIm and hnRNP K helps ensure that cells produce the appropriate amounts of each noncoding RNA. The expression of MEN ε is further controlled post-transcriptionally by an RNA decay pathway that is mediated by PABN1 (poly(A)-binding protein nuclear 1) [107,108].
Whereas the 3′ end of MEN ε is protected by a poly(A) tail, MEN β ends in a triple helical structure that is very similar to that at the 3′ end of MALAT1 (Fig. 2A) [82,83]. However, unlike mascRNA, the MEN β tRNA-like small RNA is structurally unstable (due to presence of a mismatch or multiple G–U wobbles in its acceptor stem) (Fig. 2B), causing it to be marked with CCACCA and rapidly degraded in most mouse and human cells [109,110]. Interestingly, the MEN β tRNA-like small RNA is structurally stable in many other species, including monkeys, allowing it to be marked with CCA and accumulate in cells [109]. As its function is unknown, it is still unclear why the MEN β tRNA-like small RNA is stable in only some species.
8. The MEN ε/β (NEAT1) long noncoding RNAs function as scaffolds for paraspeckle formation
Within the nucleus, the MEN ε and MEN β transcripts specifically localize to paraspeckles [10,98,111,112], which are commonly found in close proximity to nuclear speckles (where MALAT1 localizes) (reviewed in [113]). Paraspeckles contain many RNA-binding proteins, including paraspeckle protein 1 (PSPC1), NONO (also known as p54/nrb), and SFPQ (also known as PSF), SWI/SNF chromatin-remodeling complexes, as well as a number of RNAs that have been subjected to adenosine-to-inosine (A-to-I) hyper-editing (generally in inverted repeat sequences) [106,114–118]. Beyond retaining RNAs in the nucleus [116,117], paraspeckles have been proposed to regulate transcription (for example, via the sequestration of key transcription factors) [101,119,120], to control alternative splicing patterns [121], and to modulate the DNA damage response [122].
Interestingly, RNase A treatment disrupts the structural integrity of paraspeckles [117,123], and it is now clear that MEN ε/β are the critical RNAs responsible for the biogenesis and maintenance of these nuclear structures [10,98,111,112,124,125]. In fact, paraspeckles generally co-localize with the MEN ε/β gene loci, and inducing MEN ε/β expression is sufficient for the co-transcriptional assembly of paraspeckles [124]. Both MEN ε and MEN β are able to directly bind to paraspeckle proteins [10,98,111,112], thereby functioning as platforms for the formation of the nuclear body. Consistent with this model, electron microscopy has revealed that the MEN β transcript localizes to the paraspeckle core [126].
9. The MEN ε/β (NEAT1) long noncoding RNAs are tightly regulated in vivo
MEN ε/β levels are dynamically regulated, resulting in altered numbers of paraspeckles in different cell types. In addition, these noncoding RNAs are induced upon hypoxia [127], viral infection [128,129], treatment with the innate immune activator poly(I:C) [130], as well as during differentiation of human embryonic stem cells [112] and mouse myoblasts [10]. As expected, increased MEN ε/β levels results in increased numbers of paraspeckles as well as enhanced nuclear retention of RNAs that contain edited inverted repeats [112,130]. Interestingly, the arginine methyltransferase CARM1 appears to be a critical regulator of paraspeckles as it regulates MEN ε/β transcription as well as the ability of RNAs to bind to the paraspeckle protein NONO [130]. Notably, at least one of the RNAs retained in paraspeckles (CTN-RNA, which is generated from the mouse CAT2 gene) can be specifically released from this subnuclear body in response to stress via a poorly characterized post-transcriptional cleavage mechanism [117]. Paraspeckles thus appear to be highly dynamic structures that act to refine and reprogram gene expression patterns, thereby helping cells to properly differentiate and respond to various stresses. When paraspeckles and MEN ε/β are misregulated, they likely contribute to numerous pathological conditions, including breast and prostate cancers [127,131].
The MEN ε/β locus was originally suggested to be dispensable in vivo [132], although recent work has now revealed that these particular long noncoding RNAs may play key roles in fertility [133] and mammary gland development [134]. Despite normal ovulation, nearly half of all mated MEN ε/β knockout mice failed to become pregnant, likely due to corpus luteum dysfunction and low progesterone levels [133]. Likewise, MEN ε/β knockout female mice have aberrant mammary glands and reduced lactation [134]. This resulted in only 24% of pups born to MEN ε/β knockout females surviving beyond 5 days after birth (compared to >85% from wildtype mothers). Future work will hopefully reveal the underlying molecular processes (e.g. a failure to retain some RNAs in the nucleus) that are misregulated when the MEN ε/β long noncoding RNAs and paraspeckles are absent in vivo.
10. Long noncoding RNAs with snoRNA ends regulate pre-mRNA splicing patterns
Besides MALAT1 and MEN β, an increasing number of stable non-polyadenylated noncoding RNAs have been identified [7,45,46,52–54,135]. Although these mature transcripts often look and function very differently from one another, the pre-mRNA splicing machinery generally plays a critical role in their biogenesis. For example, the Carmichael and Chen groups identified a class of long noncoding RNAs known as sno-lncRNAs (snoRNA-related long noncoding RNAs) that are derived from introns and have either box C/D or box H/ACA small nucleolar RNA (snoRNA) structures at their 5′ and 3′ ends [136]. These terminal structures are used in place of a canonical 5′ cap and poly(A) tail, and allow these noncoding RNAs to accumulate to high levels (expression similar to that of some histone mRNAs). Interestingly, sno-lncRNAs are processed by the same machinery that processes snoRNAs, but differences in the sequence content among introns (namely, the number of encoded snoRNA genes) allows some introns to generate long noncoding RNAs, whereas others generate snoRNAs that are only ~70–200-nt.
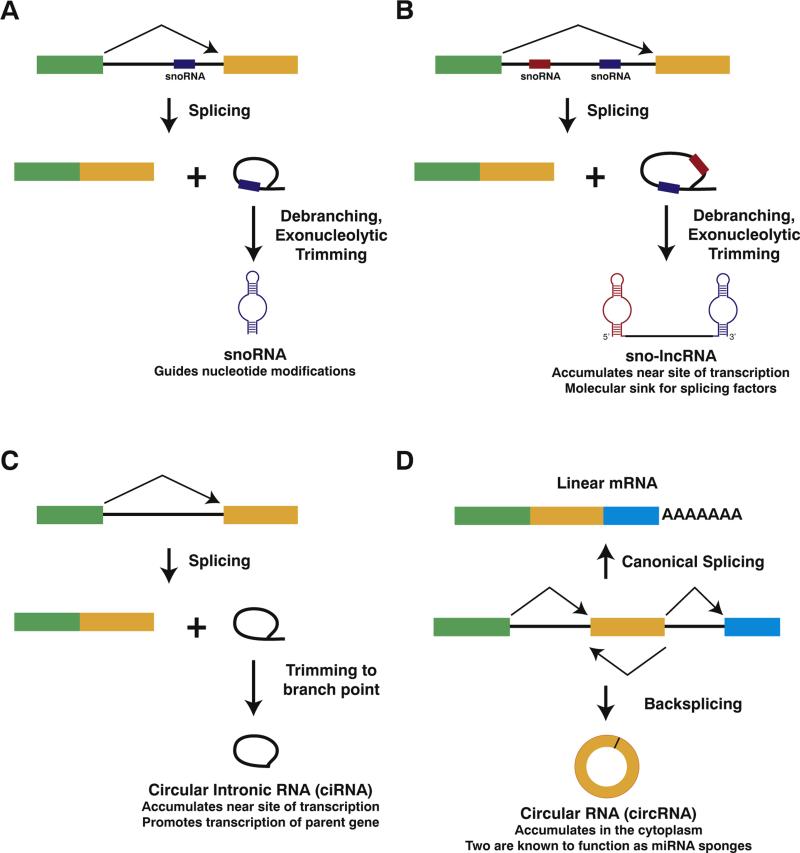
In vertebrates, snoRNA sequences are generally encoded in the introns of protein-coding genes and are processed out of the excised introns following pre-mRNA splicing (Fig. 3A) (reviewed in [137,138]). Exonucleases degrade from each end of the debranched intron until they reach the 5′ and 3′ ends of the snoRNA structure, which abruptly inhibit further degradation. The mature snoRNA is subsequently released and functions to modify other noncoding RNAs, including ribosomal RNAs and small nuclear RNAs (snRNAs). However, if two snoRNA sequences are present within the same intron, the exonucleases are unable to degrade the internal region between the snoRNAs, and a long noncoding RNA with snoRNA ends is produced (Fig. 3B) [136]. Consistent with this biogenesis mechanism, the terminal snoRNAs, but not the internal sequence between the snoRNAs, are highly conserved across evolution [139]. In fact, the internal region can be replaced with unrelated sequences to generate “designer” sno-lncRNAs [140].
Fig. 3.
The pre-mRNA splicing machinery generates a number of non-polyadenylated noncoding RNAs. (A) When a snoRNA sequence is encoded in an intron, pre-mRNA splicing releases the excised intron, which is subsequently debranched and trimmed to produce the mature ~70–200 nt snoRNA. In this case, a box C/D snoRNA is shown. (B) In contrast, when two snoRNA sequences are present in a single intron, debranching and trimming of the excised intron produce a long noncoding RNA with snoRNA ends. (C) After splicing, some introns fail to be debranched and accumulate as stable circular intronic RNAs in the nucleus. These transcripts are covalent circles due to the 2′,5′-phosphodiester bond between the 5′ end of the intron and the branch point adenosine. (D) Pre-mRNA splicing can generate linear or circular RNAs comprised of exons. If the splice sites are joined in the canonical order, a mature linear mRNA is generated that is subsequently polyadenylated (top). Alternatively, the splicing machinery can backsplice and join a splice donor to an upstream splice acceptor, generating a circular RNA whose ends are covalently linked by a 3′,5′-phosphodiester bond (bottom).
At least 19 endogenous sno-lncRNAs have now been identified in human, rhesus monkey, or mouse, with most showing tissue- and species-specific expression patterns [139]. Sno-lncRNAs are most notably generated from the 15q11–q13 region of human chromosome 15 [136], which is subjected to genomic imprinting and implicated in Prader–Willi Syndrome (PWS) [141,142]. These PWS region sno-lncRNAs accumulate in the nucleus at their sites of transcription and have multiple predicted binding sites (ranging from 4 to 20) for RbFox2, a protein known to regulate alternative splicing and many other post-transcriptional events [136]. Indeed, RbFox2 strongly binds these sno-lncRNAs, and modulating the levels of the noncoding RNAs induced changes in RbFox2-regulated splicing events. Therefore, these sno-lncRNAs likely act, at least in part, as molecular sinks that prevent RbFox2 from binding its mRNA targets [136]. As the genomic region encoding the sno-lncRNAs is deleted in patients with Prader–Willi Syndrome, future work will hopefully reveal novel insights into why these long noncoding RNAs are required for proper development.
11. Circular intronic long noncoding RNAs can regulate the transcription of their parent genes
The vast majority of introns are rapidly debranched and degraded, although some are processed to produce stable noncoding RNAs, such as sno-lncRNAs (discussed above), snoRNAs, or microRNAs [143]. Surprisingly, recent work has indicated that many full-length introns accumulate to high levels in the nuclei of Xenopus oocytes [144] and in human cells [145]. The situation is Xenopus is particularly remarkable as the germinal vesicle (nucleus) stores stable intronic sequences from most (~92%) of its transcribed genes [144]. These intron-derived sequences are stable (and, hence, called stable intronic sequence RNA [sisRNA]) and appear to be transmitted to the developing embryo, although their exact functions are still unknown.
In humans, at least 100 different introns have been shown to have their 3′ tails (the region between the branch point sequence and the 3′ splice site) removed, but the 2′,5′-phosphodiester bond at the branch point was not hydrolyzed (Fig. 3C) [145,146]. This failure to completely process these introns allows these sequences to accumulate as circular intronic long noncoding RNAs (so-called ciRNAs) that are often more stable than their parent linear mRNAs [145]. The exact reason why these introns escape debranching is still unclear, although a 7-nt G/U-rich motif near the 5′ splice site and an 11-nt C-rich motif near the branch point appear to play critical roles. Once generated, ciRNAs accumulate near their site of synthesis and appear to promote the transcription of their parental genes in cis by interacting with elongating RNA polymerase II complexes [145]. In addition, a portion of the ciRNA transcripts localize to other sites in the nucleus, suggesting they may have additional trans effects. Whether the stable introns produced in Xenopus function similarly to regulate transcription is an open question.
12. Backsplicing generates cytoplasmic circular RNAs that can function as microRNA sponges
Although it was long assumed that eukaryotic pre-mRNAs are almost always spliced to generate a linear mRNA, it is now clear that thousands of protein-coding genes can be non-canonically spliced to generate circular RNAs (so-called circRNAs) [11,12,37,147–150]. Unlike ciRNAs, circRNAs are almost exclusively derived from exons and are products of alternative splicing events known as “backsplicing” (Fig. 3D). Rather than joining the exons in the canonical order (joining exon 1 to exon 2 to exon 3, etc.), backsplicing joins a splice donor to an upstream splice acceptor, e.g. joining the end of exon 2 to the beginning of exon 2. CircRNAs are observed across eukaryotes [151] and occur at a surprisingly high frequency. For example, deep sequencing the transcriptome of a single human cell type revealed that ~15% of actively transcribed genes may produce circRNAs, generating over 25,000 distinct circular transcripts [11].
Recent work has revealed that intronic repetitive elements, including sequences derived from transposons, are critical determinants of whether a pre-mRNA is subjected to canonical splicing or backsplicing. In most cases (but not all [152,153]), circular RNA biogenesis is initiated when complementary sequences from two different introns base pair to one another [11,154–158]. This brings the splice sites from the intervening exon(s) into close proximity, facilitating backsplicing. As pre-mRNAs generally contain multiple intronic repeats, distinct circRNAs can be produced depending on which repeats base pair to one another [155]. This allows the functional output of a protein-coding gene to be finely tuned [148,159]. Surprisingly, some genes produce circRNAs at levels that exceed the associated linear mRNAs by a factor of 10 [11,12]. This is at least partly due to the fact that circRNAs are highly stable, as their covalently joined ends make the transcripts naturally resistant to exonucleases. Exactly how circRNAs are generally degraded is unknown, although the ciRS-7/CDR1as circular RNA can be endonucleolytically cleaved by Argonaute-2 to trigger its decay [160]. It should nevertheless be noted that this mechanism does not appear to be generally used to post-transcriptionally regulate the levels of other RNA circles.
Once produced, most circRNAs accumulate in the cytoplasm, but do not associate with ribosomes [11,147]. The two most highly studied circular RNAs, ciRS-7/CDR1as and Sry, each function to modulate the activity of microRNAs [36,37]. These particular circles contain many binding sites for specific microRNAs (miR-7 and miR-138, respectively), allowing the transcripts to act as sponges that titrate the microRNAs from their other RNA targets. However, most other circular RNAs contain few microRNA binding sites and likely have a different function [147]. They may, for example, allow the formation of large RNA–protein complexes or perhaps work in the same pathway as the protein produced from its parental gene (analogous to how bacterial operons are organized and function).
13. Summary and perspectives
Although long RNA polymerase II transcripts are generally thought to have 5′ caps and poly(A) tails, MALAT1, MEN β, sno-lncRNAs, ciRNAs, and circRNAs all defy this dogma. Nevertheless, one of the main reasons why all of these noncoding transcripts are stable and accumulate to high levels is that they either have alternative structures that protect their ends or they, in fact, have no ends. MALAT1 and MEN β end in triple helical structures, sno-lncRNAs end in elaborately folded snoRNA structures, and ciRNAs as well as circRNAs have no ends that can be recognized by exonucleases.
In each of the cases described, well-studied RNA processing factors catalyzed the biogenesis reactions, yet the resulting mature RNAs have non-canonical features. This is not because the proteins were behaving in unusual ways, but because of the unique organization of these particular genomic loci. For example, RNase P recognizes and cleaves the tRNA-like structures present within the primary MALAT1 and MEN β transcripts, just as the enzyme recognizes and cleaves primary tRNA transcripts generated by RNA polymerase III. However, because these particular tRNA-like structures are at the ends of long RNA polymerase II transcripts, cleavage results in the generation of long noncoding RNAs. It will, therefore, be informative to determine if there are other non-canonical RNAs being produced in cells, especially as this may reveal other sequences/structures that perform key functions in unusual ways. Notably, such RNAs would be depleted from deep sequencing libraries that use an oligo(dT) selection step, and thus they may have been missed in many studies.
By employing distinct strategies to protect the ends of RNAs, cells are likely able to mark certain classes of RNAs as being different from others. In this way, cells ensure that these various RNAs are stable, but look different at their termini so that they can be regulated by unique mechanisms. Therefore, it will be of great interest to identify the unique factors that recognize each of these ends, as this will reveal a much clearer picture of how these transcripts are stabilized, degraded, trafficked around the cell, and function. Just as the histone stem–loop structure dictates that histone mRNAs are only stable and translated during S phase (reviewed in [80]), these other non-canonical sequences/structures likely impart unique regulation on the noncoding transcripts. In summary, it is clear that a number of abundant long noncoding RNAs are generated in non-canonical ways, and it seems certain that future research will provide many more unexpected insights into how non-coding RNAs are generated and regulated.
Acknowledgments
I thank Jeff Wilusz as well as Deirdre Tatomer and the other members of my laboratory for suggestions and discussions. Supported by NIH R00-GM104166.
Footnotes
This article is part of a Special Issue entitled: Clues to long noncoding RNA taxonomy. wilusz@mail.med.upenn.edu.
References
- 1.Kapranov P, Willingham AT, Gingeras TR. Genome-wide transcription and the implications for genomic organization. Nat. Rev. Genet. 2007;8:413–423. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- 2.Clark MB, Amaral PP, Schlesinger FJ, Dinger ME, Taft RJ, Rinn JL, Ponting CP, Stadler PF, Morris KV, Morillon A, Rozowsky JS, Gerstein MB, Wahlestedt C, Hayashizaki Y, Carninci P, Gingeras TR, Mattick JS. The reality of pervasive transcription. PLoS Biol. 2011;9:e1000625. doi: 10.1371/journal.pbio.1000625. (discussion e1001102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, Kodzius R, Shimokawa K, Bajic VB, Brenner SE, Batalov S, Forrest AR, Zavolan M, Davis MJ, Wilming LG, Aidinis V, Allen JE, Ambesi-Impiombato A, Apweiler R, Aturaliya RN, Bailey TL, Bansal M, Baxter L, Beisel KW, Bersano T, Bono H, Chalk AM, Chiu KP, Choudhary V, Christoffels A, Clutterbuck DR, Crowe ML, Dalla E, Dalrymple BP, de Bono B, Della Gatta G, di Bernardo D, Down T, Engstrom P, Fagiolini M, Faulkner G, Fletcher CF, Fukushima T, Furuno M, Futaki S, Gariboldi M, Georgii-Hemming P, Gingeras TR, Gojobori T, Green RE, Gustincich S, Harbers M, Hayashi Y, Hensch TK, Hirokawa N, Hill D, Huminiecki L, Iacono M, Ikeo K, Iwama A, Ishikawa T, Jakt M, Kanapin A, Katoh M, Kawasawa Y, Kelso J, Kitamura H, Kitano H, Kollias G, Krishnan SP, Kruger A, Kummerfeld SK, Kurochkin IV, Lareau LF, Lazarevic D, Lipovich L, Liu J, Liuni S, McWilliam S, Madan Babu M, Madera M, Marchionni L, Matsuda H, Matsuzawa S, Miki H, Mignone F, Miyake S, Morris K, Mottagui-Tabar S, Mulder N, Nakano N, Nakauchi H, Ng P, Nilsson R, Nishiguchi S, Nishikawa S, Nori F, Ohara O, Okazaki Y, Orlando V, Pang KC, Pavan WJ, Pavesi G, Pesole G, Petrovsky N, Piazza S, Reed J, Reid JF, Ring BZ, Ringwald M, Rost B, Ruan Y, Salzberg SL, Sandelin A, Schneider C, Schonbach C, Sekiguchi K, Semple CA, Seno S, Sessa L, Sheng Y, Shibata Y, Shimada H, Shimada K, Silva D, Sinclair B, Sperling S, Stupka E, Sugiura K, Sultana R, Takenaka Y, Taki K, Tammoja K, Tan SL, Tang S, Taylor MS, Tegner J, Teichmann SA, Ueda HR, van Nimwegen E, Verardo R, Wei CL, Yagi K, Yamanishi H, Zabarovsky E, Zhu S, Zimmer A, Hide W, Bult C, Grimmond SM, Teasdale RD, Liu ET, Brusic V, Quackenbush J, Wahlestedt C, Mattick JS, Hume DA, Kai C, Sasaki D, Tomaru Y, Fukuda S, Kanamori-Katayama M, Suzuki M, Aoki J, Arakawa T, Iida J, Imamura K, Itoh M, Kato T, Kawaji H, Kawagashira N, Kawashima T, Kojima M, Kondo S, Konno H, Nakano K, Ninomiya N, Nishio T, Okada M, Plessy C, Shibata K, Shiraki T, Suzuki S, Tagami M, Waki K, Watahiki A, Okamura-Oho Y, Suzuki H, Kawai J, Hayashizaki Y. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 4.Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, Kuehn MS, Taylor CM, Neph S, Koch CM, Asthana S, Malhotra A, Adzhubei I, Greenbaum JA, Andrews RM, Flicek P, Boyle PJ, Cao H, Carter NP, Clelland GK, Davis S, Day N, Dhami P, Dillon SC, Dorschner MO, Fiegler H, Giresi PG, Goldy J, Hawrylycz M, Haydock A, Humbert R, James KD, Johnson BE, Johnson EM, Frum TT, Rosenzweig ER, Karnani N, Lee K, Lefebvre GC, Navas PA, Neri F, Parker SC, Sabo PJ, Sandstrom R, Shafer A, Vetrie D, Weaver M, Wilcox S, Yu M, Collins FS, Dekker J, Lieb JD, Tullius TD, Crawford GE, Sunyaev S, Noble WS, Dunham I, Denoeud F, Reymond A, Kapranov P, Rozowsky J, Zheng D, Castelo R, Frankish A, Harrow J, Ghosh S, Sandelin A, Hofacker IL, Baertsch R, Keefe D, Dike S, Cheng J, Hirsch HA, Sekinger EA, Lagarde J, Abril JF, Shahab A, Flamm C, Fried C, Hackermuller J, Hertel J, Lindemeyer M, Missal K, Tanzer A, Washietl S, Korbel J, Emanuelsson O, Pedersen JS, Holroyd N, Taylor R, Swarbreck D, Matthews N, Dickson MC, Thomas DJ, Weirauch MT, Gilbert J, Drenkow J, Bell I, Zhao X, Srinivasan KG, Sung WK, Ooi HS, Chiu KP, Foissac S, Alioto T, Brent M, Pachter L, Tress ML, Valencia A, Choo SW, Choo CY, Ucla C, Manzano C, Wyss C, Cheung E, Clark TG, Brown JB, Ganesh M, Patel S, Tammana H, Chrast J, Henrichsen CN, Kai C, Kawai J, Nagalakshmi U, Wu J, Lian Z, Lian J, Newburger P, Zhang X, Bickel P, Mattick JS, Carninci P, Hayashizaki Y, Weissman S, Hubbard T, Myers RM, Rogers J, Stadler PF, Lowe TM, Wei CL, Ruan Y, Struhl K, Gerstein M, Antonarakis SE, Fu Y, Green ED, Karaoz U, Siepel A, Taylor J, Liefer LA, Wetterstrand KA, Good PJ, Feingold EA, Guyer MS, Cooper GM, Asimenos G, Dewey CN, Hou M, Nikolaev S, Montoya-Burgos JI, Loytynoja A, Whelan S, Pardi F, Massingham T, Huang H, Zhang NR, Holmes I, Mullikin JC, Ureta-Vidal A, Paten B, Seringhaus M, Church D, Rosenbloom K, Kent WJ, Stone EA, Batzoglou S, Goldman N, Hardison RC, Haussler D, Miller W, Sidow A, Trinklein ND, Zhang ZD, Barrera L, Stuart R, King DC, Ameur A, Enroth S, Bieda MC, Kim J, Bhinge AA, Jiang N, Liu J, Yao F, Vega VB, Lee CW, Ng P, Yang A, Moqtaderi Z, Zhu Z, Xu X, Squazzo S, Oberley MJ, Inman D, Singer MA, Richmond TA, Munn KJ, Rada-Iglesias A, Wallerman O, Komorowski J, Fowler JC, Couttet P, Bruce AW, Dovey OM, Ellis PD, Langford CF, Nix DA, Euskirchen G, Hartman S, Urban AE, Kraus P, Van Calcar S, Heintzman N, Kim TH, Wang K, Qu C, Hon G, Luna R, Glass CK, Rosenfeld MG, Aldred SF, Cooper SJ, Halees A, Lin JM, Shulha HP, Xu M, Haidar JN, Yu Y, Iyer VR, Green RD, Wadelius C, Farnham PJ, Ren B, Harte RA, Hinrichs AS, Trumbower H, Clawson H, Hillman-Jackson J, Zweig AS, Smith K, Thakkapallayil A, Barber G, Kuhn RM, Karolchik D, Armengol L, Bird CP, de Bakker PI, Kern AD, Lopez-Bigas N, Martin JD, Stranger BE, Woodroffe A, Davydov E, Dimas A, Eyras E, Hallgrimsdottir IB, Huppert J, Zody MC, Abecasis GR, Estivill X, Bouffard GG, Guan X, Hansen NF, Idol JR, Maduro VV, Maskeri B, McDowell JC, Park M, Thomas PJ, Young AC, Blakesley RW, Muzny DM, Sodergren E, Wheeler DA, Worley KC, Jiang H, Weinstock GM, Gibbs RA, Graves T, Fulton R, Mardis ER, Wilson RK, Clamp M, Cuff J, Gnerre S, Jaffe DB, Chang JL, Lindblad-Toh K, Lander ES, Koriabine M, Nefedov M, Osoegawa K, Yoshinaga Y, Zhu B, de Jong PJ. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, Bell I, Cheung E, Drenkow J, Dumais E, Patel S, Helt G, Ganesh M, Ghosh S, Piccolboni A, Sementchenko V, Tammana H, Gingeras TR. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 6.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, Cabili MN, Jaenisch R, Mikkelsen TS, Jacks T, Hacohen N, Bernstein BE, Kellis M, Regev A, Rinn JL, Lander ES. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, Xue C, Marinov GK, Khatun J, Williams BA, Zaleski C, Rozowsky J, Roder M, Kokocinski F, Abdelhamid RF, Alioto T, Antoshechkin I, Baer MT, Bar NS, Batut P, Bell K, Bell I, Chakrabortty S, Chen X, Chrast J, Curado J, Derrien T, Drenkow J, Dumais E, Dumais J, Duttagupta R, Falconnet E, Fastuca M, Fejes-Toth K, Ferreira P, Foissac S, Fullwood MJ, Gao H, Gonzalez D, Gordon A, Gunawardena H, Howald C, Jha S, Johnson R, Kapranov P, King B, Kingswood C, Luo OJ, Park E, Persaud K, Preall JB, Ribeca P, Risk B, Robyr D, Sammeth M, Schaffer L, See LH, Shahab A, Skancke J, Suzuki AM, Takahashi H, Tilgner H, Trout D, Walters N, Wang H, Wrobel J, Yu Y, Ruan X, Hayashizaki Y, Harrow J, Gerstein M, Hubbard T, Reymond A, Antonarakis SE, Hannon G, Giddings MC, Ruan Y, Wold B, Carninci P, Guigo R, Gingeras TR. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang B, Arun G, Mao YS, Lazar Z, Hung G, Bhattacharjee G, Xiao X, Booth CJ, Wu J, Zhang C, Spector DL. The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep. 2012;2:111–123. doi: 10.1016/j.celrep.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sunwoo H, Dinger ME, Wilusz JE, Amaral PP, Mattick JS, Spector DL. MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 2009;19:347–359. doi: 10.1101/gr.087775.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7:e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kowalczyk MS, Higgs DR, Gingeras TR. Molecular biology: RNA discrimination. Nature. 2012;482:310–311. doi: 10.1038/482310a. [DOI] [PubMed] [Google Scholar]
- 14.Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157:77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 17.Yang L, Froberg JE, Lee JT. Long noncoding RNAs: fresh perspectives into the RNA world. Trends Biochem. Sci. 2014;39:35–43. doi: 10.1016/j.tibs.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bergmann JH, Spector DL. Long non-coding RNAs: modulators of nuclear structure and function. Curr. Opin. Cell Biol. 2014;26:10–18. doi: 10.1016/j.ceb.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat. Struct. Mol. Biol. 2013;20:300–307. doi: 10.1038/nsmb.2480. [DOI] [PubMed] [Google Scholar]
- 21.Clark MB, Johnston RL, Inostroza-Ponta M, Fox AH, Fortini E, Moscato P, Dinger ME, Mattick JS. Genome-wide analysis of long noncoding RNA stability. Genome Res. 2012;22:885–898. doi: 10.1101/gr.131037.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ravasi T, Suzuki H, Pang KC, Katayama S, Furuno M, Okunishi R, Fukuda S, Ru K, Frith MC, Gongora MM, Grimmond SM, Hume DA, Hayashizaki Y, Mattick JS. Experimental validation of the regulated expression of large numbers of non-coding RNAs from the mouse genome. Genome Res. 2006;16:11–19. doi: 10.1101/gr.4200206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 25.Qureshi IA, Mehler MF. Long non-coding RNAs: novel targets for nervous system disease diagnosis and therapy. Neurotherapeutics. 2013;10:632–646. doi: 10.1007/s13311-013-0199-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gutschner T, Hammerle M, Diederichs S. MALAT1 — a paradigm for long noncoding RNA function in cancer. J. Mol. Med. 2013;91:791–801. doi: 10.1007/s00109-013-1028-y. [DOI] [PubMed] [Google Scholar]
- 27.Ponjavic J, Ponting CP, Lunter G. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res. 2007;17:556–565. doi: 10.1101/gr.6036807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, Lagarde J, Veeravalli L, Ruan X, Ruan Y, Lassmann T, Carninci P, Brown JB, Lipovich L, Gonzalez JM, Thomas M, Davis CA, Shiekhattar R, Gingeras TR, Hubbard TJ, Notredame C, Harrow J, Guigo R. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell. 2011;147:1537–1550. doi: 10.1016/j.cell.2011.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ilik IA, Quinn JJ, Georgiev P, Tavares-Cadete F, Maticzka D, Toscano S, Wan Y, Spitale RC, Luscombe N, Backofen R, Chang HY, Akhtar A. Tandem stem–loops in roX RNAs act together to mediate X chromosome dosage compensation in Drosophila. Mol. Cell. 2013;51:156–173. doi: 10.1016/j.molcel.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maenner S, Muller M, Frohlich J, Langer D, Becker PB. ATP-dependent roX RNA remodeling by the helicase maleless enables specific association of MSL proteins. Mol. Cell. 2013;51:174–184. doi: 10.1016/j.molcel.2013.06.011. [DOI] [PubMed] [Google Scholar]
- 34.Martianov I, Ramadass A, Serra Barros A, Chow N, Akoulitchev A. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature. 2007;445:666–670. doi: 10.1038/nature05519. [DOI] [PubMed] [Google Scholar]
- 35.Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 37.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, le Noble F, Rajewsky N. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 38.Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature. 2011;470:284–288. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beltran M, Puig I, Pena C, Garcia JM, Alvarez AB, Pena R, Bonilla F, de Herreros AG. A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial–mesenchymal transition. Genes Dev. 2008;22:756–769. doi: 10.1101/gad.455708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee N, Moss WN, Yario TA, Steitz JA. EBV noncoding RNA binds nascent RNA to drive host PAX5 to viral DNA. Cell. 2015;160:607–618. doi: 10.1016/j.cell.2015.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spitale RC, Flynn RA, Zhang QC, Crisalli P, Lee B, Jung JW, Kuchelmeister HY, Batista PJ, Torre EA, Kool ET, Chang HY. Structural imprints in vivo decode RNA regulatory mechanisms. Nature. 2015;519:486–490. doi: 10.1038/nature14263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chu C, Spitale RC, Chang HY. Technologies to probe functions and mechanisms of long noncoding RNAs. Nat. Struct. Mol. Biol. 2015;22:29–35. doi: 10.1038/nsmb.2921. [DOI] [PubMed] [Google Scholar]
- 43.Novikova IV, Hennelly SP, Sanbonmatsu KY. Structural architecture of the human long non-coding RNA, steroid receptor RNA activator. Nucleic Acids Res. 2012;40:5034–5051. doi: 10.1093/nar/gks071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rouskin S, Zubradt M, Washietl S, Kellis M, Weissman JS. Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature. 2014;505:701–705. doi: 10.1038/nature12894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilusz JE, Spector DL. An unexpected ending: noncanonical 3′ end processing mechanisms. RNA. 2010;16:259–266. doi: 10.1261/rna.1907510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Y, Yang L, Chen LL. Life without A tail: new formats of long noncoding RNAs. Int. J. Biochem. Cell Biol. 2014;54:338–349. doi: 10.1016/j.biocel.2013.10.009. [DOI] [PubMed] [Google Scholar]
- 47.Peart N, Sataluri A, Baillat D, Wagner EJ. Non-mRNA 3′ end formation: how the other half lives. Wiley Interdiscip. Rev. RNA. 2013;4:491–506. doi: 10.1002/wrna.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat. Rev. Mol. Cell Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- 49.Chen CY, Shyu AB. Mechanisms of deadenylation-dependent decay. Wiley Interdiscip. Rev. RNA. 2011;2:167–183. doi: 10.1002/wrna.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li Y, Kiledjian M. Regulation of mRNA decapping. Wiley Interdiscip. Rev. RNA. 2010;1:253–265. doi: 10.1002/wrna.15. [DOI] [PubMed] [Google Scholar]
- 51.Cheng J, Kapranov P, Drenkow J, Dike S, Brubaker S, Patel S, Long J, Stern D, Tammana H, Helt G, Sementchenko V, Piccolboni A, Bekiranov S, Bailey DK, Ganesh M, Ghosh S, Bell I, Gerhard DS, Gingeras TR. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308:1149–1154. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- 52.Wu Q, Kim YC, Lu J, Xuan Z, Chen J, Zheng Y, Zhou T, Zhang MQ, Wu CI, Wang SM. Poly A-transcripts expressed in HeLa cells. PLoS One. 2008;3:e2803. doi: 10.1371/journal.pone.0002803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang L, Duff MO, Graveley BR, Carmichael GG, Chen LL. Genomewide characterization of non-polyadenylated RNAs. Genome Biol. 2011;12:R16. doi: 10.1186/gb-2011-12-2-r16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, Markenscoff-Papadimitriou E, Kuhl D, Bito H, Worley PF, Kreiman G, Greenberg ME. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hutchinson JN, Ensminger AW, Clemson CM, Lynch CR, Lawrence JB, Chess A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 2007;8:39. doi: 10.1186/1471-2164-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ji P, Diederichs S, Wang W, Boing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, Thomas M, Berdel WE, Serve H, Muller-Tidow C. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 57.Luo JH, Ren B, Keryanov S, Tseng GC, Rao UN, Monga SP, Strom S, Demetris AJ, Nalesnik M, Yu YP, Ranganathan S, Michalopoulos GK. Transcriptomic and genomic analysis of human hepatocellular carcinomas and hepatoblastomas. Hepatology. 2006;44:1012–1024. doi: 10.1002/hep.21328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lin R, Maeda S, Liu C, Karin M, Edgington TS. A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. Oncogene. 2007;26:851–858. doi: 10.1038/sj.onc.1209846. [DOI] [PubMed] [Google Scholar]
- 59.Guffanti A, Iacono M, Pelucchi P, Kim N, Solda G, Croft LJ, Taft RJ, Rizzi E, Askarian-Amiri M, Bonnal RJ, Callari M, Mignone F, Pesole G, Bertalot G, Bernardi LR, Albertini A, Lee C, Mattick JS, Zucchi I, De Bellis G. A transcriptional sketch of a primary human breast cancer by 454 deep sequencing. BMC Genomics. 2009;10:163. doi: 10.1186/1471-2164-10-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lai MC, Yang Z, Zhou L, Zhu QQ, Xie HY, Zhang F, Wu LM, Chen LM, Zheng SS. Long non-coding RNA MALAT-1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation. Med. Oncol. 2011;29:1810–1816. doi: 10.1007/s12032-011-0004-z. [DOI] [PubMed] [Google Scholar]
- 61.Schmidt LH, Spieker T, Koschmieder S, Schaffers S, Humberg J, Jungen D, Bulk E, Hascher A, Wittmer D, Marra A, Hillejan L, Wiebe K, Berdel WE, Wiewrodt R, Muller-Tidow C. The long noncoding MALAT-1 RNA indicates a poor prognosis in non-small cell lung cancer and induces migration and tumor growth. J. Thorac. Oncol. 2011;6:1984–1992. doi: 10.1097/JTO.0b013e3182307eac. [DOI] [PubMed] [Google Scholar]
- 62.Gutschner T, Hammerle M, Eissmann M, Hsu J, Kim Y, Hung G, Revenko A, Arun G, Stentrup M, Gross M, Zornig M, MacLeod AR, Spector DL, Diederichs S. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73:1180–1189. doi: 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guo F, Li Y, Liu Y, Wang J, Li G. Inhibition of metastasis-associated lung adenocarcinoma transcript 1 in CaSki human cervical cancer cells suppresses cell proliferation and invasion. Acta Biochim. Biophys. Sin. (Shanghai) 2010;42:224–229. doi: 10.1093/abbs/gmq008. [DOI] [PubMed] [Google Scholar]
- 64.Xu C, Yang M, Tian J, Wang X, Li Z. MALAT-1: a long non-coding RNA and its important 3′ end functional motif in colorectal cancer metastasis. Int. J. Oncol. 2011;39:169–175. doi: 10.3892/ijo.2011.1007. [DOI] [PubMed] [Google Scholar]
- 65.Tripathi V, Shen Z, Chakraborty A, Giri S, Freier SM, Wu X, Zhang Y, Gorospe M, Prasanth SG, Lal A, Prasanth KV. Long noncoding RNA MALAT1 controls cell cycle progression by regulating the expression of oncogenic transcription factor B-MYB. PLoS Genet. 2013;9:e1003368. doi: 10.1371/journal.pgen.1003368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang L, Lin C, Liu W, Zhang J, Ohgi KA, Grinstein JD, Dorrestein PC, Rosenfeld MG. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell. 2011;147:773–788. doi: 10.1016/j.cell.2011.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Michalik KM, You X, Manavski Y, Doddaballapur A, Zornig M, Braun T, John D, Ponomareva Y, Chen W, Uchida S, Boon RA, Dimmeler S. Long noncoding RNA MALAT1 regulates endothelial cell function and vessel growth. Circ. Res. 2014;114:1389–1397. doi: 10.1161/CIRCRESAHA.114.303265. [DOI] [PubMed] [Google Scholar]
- 68.Tano K, Mizuno R, Okada T, Rakwal R, Shibato J, Masuo Y, Ijiri K, Akimitsu N. MALAT-1 enhances cell motility of lung adenocarcinoma cells by influencing the expression of motility-related genes. FEBS Lett. 2010;584:4575–4580. doi: 10.1016/j.febslet.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 69.Tseng JJ, Hsieh YT, Hsu SL, Chou MM. Metastasis associated lung adenocarcinoma transcript 1 is up-regulated in placenta previa increta/percreta and strongly associated with trophoblast-like cell invasion in vitro. Mol. Hum. Reprod. 2009;15:725–731. doi: 10.1093/molehr/gap071. [DOI] [PubMed] [Google Scholar]
- 70.Gutschner T, Baas M, Diederichs S. Noncoding RNA gene silencing through genomic integration of RNA destabilizing elements using zinc finger nucleases. Genome Res. 2011;21:1944–1954. doi: 10.1101/gr.122358.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wheeler TM, Leger AJ, Pandey SK, MacLeod AR, Nakamori M, Cheng SH, Wentworth BM, Bennett CF, Thornton CA. Targeting nuclear RNA for in vivo correction of myotonic dystrophy. Nature. 2012;488:111–115. doi: 10.1038/nature11362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, Xie M, Zhang Q, McMichael JF, Wyczalkowski MA, Leiserson MD, Miller CA, Welch JS, Walter MJ, Wendl MC, Ley TJ, Wilson RK, Raphael BJ, Ding L. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ellis MJ, Ding L, Shen D, Luo J, Suman VJ, Wallis JW, Van Tine BA, Hoog J, Goiffon RJ, Goldstein TC, Ng S, Lin L, Crowder R, Snider J, Ballman K, Weber J, Chen K, Koboldt DC, Kandoth C, Schierding WS, McMichael JF, Miller CA, Lu C, Harris CC, McLellan MD, Wendl MC, DeSchryver K, Allred DC, Esserman L, Unzeitig G, Margenthaler J, Babiera GV, Marcom PK, Guenther JM, Leitch M, Hunt K, Olson J, Tao Y, Maher CA, Fulton LL, Fulton RS, Harrison M, Oberkfell B, Du F, Demeter R, Vickery TL, Elhammali A, Piwnica-Worms H, McDonald S, Watson M, Dooling DJ, Ota D, Chang LW, Bose R, Ley TJ, Piwnica-Worms D, Stuart JM, Wilson RK, Mardis ER. Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature. 2012;486:353–360. doi: 10.1038/nature11143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Davis IJ, Hsi BL, Arroyo JD, Vargas SO, Yeh YA, Motyckova G, Valencia P, Perez-Atayde AR, Argani P, Ladanyi M, Fletcher JA, Fisher DE. Cloning of an Alpha-TFEB fusion in renal tumors harboring the t(6;11)(p21;q13) chromosome translocation. Proc. Natl. Acad. Sci. U. S. A. 2003;100:6051–6056. doi: 10.1073/pnas.0931430100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kuiper RP, Schepens M, Thijssen J, van Asseldonk M, van den Berg E, Bridge J, Schuuring E, Schoenmakers EF, van Kessel AG. Upregulation of the transcription factor TFEB in t(6;11)(p21;q13)-positive renal cell carcinomas due to promoter substitution. Hum. Mol. Genet. 2003;12:1661–1669. doi: 10.1093/hmg/ddg178. [DOI] [PubMed] [Google Scholar]
- 76.Rajaram V, Knezevich S, Bove KE, Perry A, Pfeifer JD. DNA sequence of the translocation breakpoints in undifferentiated embryonal sarcoma arising in mesenchymal hamartoma of the liver harboring the t(11;19)(q11;q13.4) translocation. Genes Chromosome. Cancer. 2007;46:508–513. doi: 10.1002/gcc.20437. [DOI] [PubMed] [Google Scholar]
- 77.Argani P, Yonescu R, Morsberger L, Morris K, Netto GJ, Smith N, Gonzalez N, Illei PB, Ladanyi M, Griffin CA. Molecular confirmation of t(6;11)(p21;q12) renal cell carcinoma in archival paraffin-embedded material using a break-apart TFEB FISH assay expands its clinicopathologic spectrum. Am. J. Surg. Pathol. 2012;36:1516–1526. doi: 10.1097/PAS.0b013e3182613d8f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bernard D, Prasanth KV, Tripathi V, Colasse S, Nakamura T, Xuan Z, Zhang MQ, Sedel F, Jourdren L, Coulpier F, Triller A, Spector DL, Bessis A. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J. 2010;29:3082–3093. doi: 10.1038/emboj.2010.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wilusz JE, Freier SM, Spector DL. 3′ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell. 2008;135:919–932. doi: 10.1016/j.cell.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Marzluff WF, Wagner EJ, Duronio RJ. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat. Rev. Genet. 2008;9:843–854. doi: 10.1038/nrg2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sachs AB, Davis RW, Kornberg RD. A single domain of yeast poly(A)-binding protein is necessary and sufficient for RNA binding and cell viability. Mol. Cell. Biol. 1987;7:3268–3276. doi: 10.1128/mcb.7.9.3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wilusz JE, JnBaptiste CK, Lu LY, Kuhn CD, Joshua-Tor L, Sharp PA. A triple helix stabilizes the 3′ ends of long noncoding RNAs that lack poly(A) tails. Genes Dev. 2012;26:2392–2407. doi: 10.1101/gad.204438.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Brown JA, Valenstein ML, Yario TA, Tycowski KT, Steitz JA. Formation of triple-helical structures by the 3′-end sequences of MALAT1 and MENbeta noncoding RNAs. Proc. Natl. Acad. Sci. U. S. A. 2012;109:19202–19207. doi: 10.1073/pnas.1217338109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Brown JA, Bulkley D, Wang J, Valenstein ML, Yario TA, Steitz TA, Steitz JA. Structural insights into the stabilization of MALAT1 noncoding RNA by a bipartite triple helix. Nat. Struct. Mol. Biol. 2014;21:633–640. doi: 10.1038/nsmb.2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Houseley J, LaCava J, Tollervey D. RNA-quality control by the exosome. Nat. Rev. Mol. Cell Biol. 2006;7:529–539. doi: 10.1038/nrm1964. [DOI] [PubMed] [Google Scholar]
- 86.Mitton-Fry RM, DeGregorio SJ, Wang J, Steitz TA, Steitz JA. Poly(A) tail recognition by a viral RNA element through assembly of a triple helix. Science. 2010;330:1244–1247. doi: 10.1126/science.1195858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tycowski KT, Shu MD, Borah S, Shi M, Steitz JA. Conservation of a triple-helix-forming RNA stability element in noncoding and genomic RNAs of diverse viruses. Cell Rep. 2012;2:26–32. doi: 10.1016/j.celrep.2012.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Conrad NK, Shu MD, Uyhazi KE, Steitz JA. Mutational analysis of a viral RNA element that counteracts rapid RNA decay by interaction with the polyadenylate tail. Proc. Natl. Acad. Sci. U. S. A. 2007;104:10412–10417. doi: 10.1073/pnas.0704187104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Geisberg JV, Moqtaderi Z, Fan X, Ozsolak F, Struhl K. Global analysis of mRNA isoform half-lives reveals stabilizing and destabilizing elements in yeast. Cell. 2014;156:812–824. doi: 10.1016/j.cell.2013.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bhandari D, Raisch T, Weichenrieder O, Jonas S, Izaurralde E. Structural basis for the Nanos-mediated recruitment of the CCR4–NOT complex and translational repression. Genes Dev. 2014;28:888–901. doi: 10.1101/gad.237289.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kahvejian A, Roy G, Sonenberg N. The mRNA closed-loop model: the function of PABP and PABP-interacting proteins in mRNA translation. Cold Spring Harb. Symp. Quant. Biol. 2001;66:293–300. doi: 10.1101/sqb.2001.66.293. [DOI] [PubMed] [Google Scholar]
- 92.Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ingolia NT, Brar GA, Stern-Ginossar N, Harris MS, Talhouarne GJ, Jackson SE, Wills MR, Weissman JS. Ribosome profiling reveals pervasive translation outside of annotated protein-coding genes. Cell Rep. 2014;8:1365–1379. doi: 10.1016/j.celrep.2014.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Spector DL, Lamond AI. Nuclear speckles. Cold Spring Harb. Perspect. Biol. 2011;3 doi: 10.1101/cshperspect.a000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Miyagawa R, Tano K, Mizuno R, Nakamura Y, Ijiri K, Rakwal R, Shibato J, Masuo Y, Mayeda A, Hirose T, Akimitsu N. Identification of cis- and trans-acting factors involved in the localization of MALAT-1 noncoding RNA to nuclear speckles. RNA. 2012;18:738–751. doi: 10.1261/rna.028639.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, Blencowe BJ, Prasanth SG, Prasanth KV. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nakagawa S, Ip JY, Shioi G, Tripathi V, Zong X, Hirose T, Prasanth KV. Malat1 is not an essential component of nuclear speckles in mice. RNA. 2012;18:1487–1499. doi: 10.1261/rna.033217.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol. Cell. 2009;33:717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lin R, Roychowdhury-Saha M, Black C, Watt AT, Marcusson EG, Freier SM, Edgington TS. Control of RNA processing by a large non-coding RNA over-expressed in carcinomas. FEBS Lett. 2011;585:671–676. doi: 10.1016/j.febslet.2011.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Engreitz JM, Sirokman K, McDonel P, Shishkin AA, Surka C, Russell P, Grossman SR, Chow AY, Guttman M, Lander ES. RNA–RNA interactions enable specific targeting of noncoding RNAs to nascent Pre-mRNAs and chromatin sites. Cell. 2014;159:188–199. doi: 10.1016/j.cell.2014.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.West JA, Davis CP, Sunwoo H, Simon MD, Sadreyev RI, Wang PI, Tolstorukov MY, Kingston RE. The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol. Cell. 2014;55:791–802. doi: 10.1016/j.molcel.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tollervey JR, Curk T, Rogelj B, Briese M, Cereda M, Kayikci M, Konig J, Hortobagyi T, Nishimura AL, Zupunski V, Patani R, Chandran S, Rot G, Zupan B, Shaw CE, Ule J. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat. Neurosci. 2011;14:452–458. doi: 10.1038/nn.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Polymenidou M, Lagier-Tourenne C, Hutt KR, Huelga SC, Moran J, Liang TY, Ling SC, Sun E, Wancewicz E, Mazur C, Kordasiewicz H, Sedaghat Y, Donohue JP, Shiue L, Bennett CF, Yeo GW, Cleveland DW. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat. Neurosci. 2011;14:459–468. doi: 10.1038/nn.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lagier-Tourenne C, Polymenidou M, Cleveland DW. TDP-43 and FUS/TLS: emerging roles in RNA processing and neurodegeneration. Hum. Mol. Genet. 2010;19:R46–R64. doi: 10.1093/hmg/ddq137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Eissmann M, Gutschner T, Hammerle M, Gunther S, Caudron-Herger M, Gross M, Schirmacher P, Rippe K, Braun T, Zornig M, Diederichs S. Loss of the abundant nuclear non-coding RNA MALAT1 is compatible with life and development. RNA Biol. 2012;9:1076–1087. doi: 10.4161/rna.21089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Naganuma T, Nakagawa S, Tanigawa A, Sasaki YF, Goshima N, Hirose T. Alternative 3′-end processing of long noncoding RNA initiates construction of nuclear paraspeckles. EMBO J. 2012;31:4020–4034. doi: 10.1038/emboj.2012.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bresson SM, Conrad NK. The human nuclear poly(a)-binding protein promotes RNA hyperadenylation and decay. PLoS Genet. 2013;9:e1003893. doi: 10.1371/journal.pgen.1003893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Beaulieu YB, Kleinman CL, Landry-Voyer AM, Majewski J, Bachand F. Polyadenylation-dependent control of long noncoding RNA expression by the poly(A)-binding protein nuclear 1. PLoS Genet. 2012;8:e1003078. doi: 10.1371/journal.pgen.1003078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wilusz JE, Whipple JM, Phizicky EM, Sharp PA. tRNAs marked with CCACCA are targeted for degradation. Science. 2011;334:817–821. doi: 10.1126/science.1213671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kuhn CD, Wilusz JE, Zheng Y, Beal PA, Joshua-Tor L. On-enzyme refolding permits small RNA and tRNA surveillance by the CCA-adding enzyme. Cell. 2015;160:644–658. doi: 10.1016/j.cell.2015.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sasaki YT, Ideue T, Sano M, Mituyama T, Hirose T. MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc. Natl. Acad. Sci. U. S. A. 2009;106:2525–2530. doi: 10.1073/pnas.0807899106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chen LL, Carmichael GG. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol. Cell. 2009;35:467–478. doi: 10.1016/j.molcel.2009.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Fox AH, Lamond AI. Paraspeckles. Cold Spring Harb. Perspect. Biol. 2010;2:a000687. doi: 10.1101/cshperspect.a000687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Fox AH, Lam YW, Leung AK, Lyon CE, Andersen J, Mann M, Lamond AI. Paraspeckles: a novel nuclear domain. Curr. Biol. 2002;12:13–25. doi: 10.1016/s0960-9822(01)00632-7. [DOI] [PubMed] [Google Scholar]
- 115.Yamazaki T, Hirose T. The building process of the functional paraspeckle with long non-coding RNAs. Front. Biosci. 2015;7:1–41. doi: 10.2741/715. [DOI] [PubMed] [Google Scholar]
- 116.Chen LL, DeCerbo JN, Carmichael GG. Alu element-mediated gene silencing. EMBO J. 2008;27:1694–1705. doi: 10.1038/emboj.2008.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Prasanth KV, Prasanth SG, Xuan Z, Hearn S, Freier SM, Bennett CF, Zhang MQ, Spector DL. Regulating gene expression through RNA nuclear retention. Cell. 2005;123:249–263. doi: 10.1016/j.cell.2005.08.033. [DOI] [PubMed] [Google Scholar]
- 118.Kawaguchi T, Tanigawa A, Naganuma T, Ohkawa Y, Souquere S, Pierron G, Hirose T. SWI/SNF chromatin-remodeling complexes function in noncoding RNA-dependent assembly of nuclear bodies. Proc. Natl. Acad. Sci. U. S. A. 2015;112:4304–4309. doi: 10.1073/pnas.1423819112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Imamura K, Imamachi N, Akizuki G, Kumakura M, Kawaguchi A, Nagata K, Kato A, Kawaguchi Y, Sato H, Yoneda M, Kai C, Yada T, Suzuki Y, Yamada T, Ozawa T, Kaneki K, Inoue T, Kobayashi M, Kodama T, Wada Y, Sekimizu K, Akimitsu N. Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol. Cell. 2014;53:393–406. doi: 10.1016/j.molcel.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 120.Hirose T, Virnicchi G, Tanigawa A, Naganuma T, Li R, Kimura H, Yokoi T, Nakagawa S, Benard M, Fox AH, Pierron G. NEAT1 long noncoding RNA regulates transcription via protein sequestration within subnuclear bodies. Mol. Biol. Cell. 2014;25:169–183. doi: 10.1091/mbc.E13-09-0558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Cooper DR, Carter G, Li P, Patel R, Watson JE, Patel NA. Long non-coding RNA NEAT1 associates with SRp40 to temporally regulate PPARgamma2 splicing during adipogenesis in 3T3-L1 cells. Genes. 2014;5:1050–1063. doi: 10.3390/genes5041050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Gao X, Kong L, Lu X, Zhang G, Chi L, Jiang Y, Wu Y, Yan C, Duerksen-Hughes P, Zhu X, Yang J. Paraspeckle protein 1 (PSPC1) is involved in the cisplatin induced DNA damage response—role in G1/S checkpoint. PLoS One. 2014;9:e97174. doi: 10.1371/journal.pone.0097174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Fox AH, Bond CS, Lamond AI. P54nrb forms a heterodimer with PSP1 that localizes to paraspeckles in an RNA-dependent manner. Mol. Biol. Cell. 2005;16:5304–5315. doi: 10.1091/mbc.E05-06-0587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mao YS, Sunwoo H, Zhang B, Spector DL. Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat. Cell Biol. 2011;13:95–101. doi: 10.1038/ncb2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Shevtsov SP, Dundr M. Nucleation of nuclear bodies by RNA. Nat. Cell Biol. 2011;13:167–173. doi: 10.1038/ncb2157. [DOI] [PubMed] [Google Scholar]
- 126.Souquere S, Beauclair G, Harper F, Fox A, Pierron G. Highly ordered spatial organization of the structural long noncoding NEAT1 RNAs within paraspeckle nuclear bodies. Mol. Biol. Cell. 2010;21:4020–4027. doi: 10.1091/mbc.E10-08-0690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Choudhry H, Albukhari A, Morotti M, Hider S, Moralli D, Smythies J, Schodel J, Green CM, Camps C, Buffa F, Ratcliffe P, Ragoussis J, Harris AL, Mole DR. Tumor hypoxia induces nuclear paraspeckle formation through HIF-2alpha dependent transcriptional activation of NEAT1 leading to cancer cell survival. Oncogene. 2014 http://dx.doi.org/10.1038/onc.2014.378.
- 128.Saha S, Murthy S, Rangarajan PN. Identification and characterization of a virus-inducible non-coding RNA in mouse brain. J. Gen. Virol. 2006;87:1991–1995. doi: 10.1099/vir.0.81768-0. [DOI] [PubMed] [Google Scholar]
- 129.Zhang Q, Chen CY, Yedavalli VS, Jeang KT. NEAT1 long noncoding RNA and paraspeckle bodies modulate HIV-1 posttranscriptional expression. mBio. 2013;4:e00512–e00596. doi: 10.1128/mBio.00596-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hu SB, Xiang JF, Li X, Xu Y, Xue W, Huang M, Wong CC, Sagum CA, Bedford MT, Yang L, Cheng D, Chen LL. Protein arginine methyltransferase CARM1 attenuates the paraspeckle-mediated nuclear retention of mRNAs containing IRAlus. Genes Dev. 2015;29:630–645. doi: 10.1101/gad.257048.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Chakravarty D, Sboner A, Nair SS, Giannopoulou E, Li R, Hennig S, Mosquera JM, Pauwels J, Park K, Kossai M, MacDonald TY, Fontugne J, Erho N, Vergara IA, Ghadessi M, Davicioni E, Jenkins RB, Palanisamy N, Chen Z, Nakagawa S, Hirose T, Bander NH, Beltran H, Fox AH, Elemento O, Rubin MA. The oestrogen receptor alpha-regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nat. Commun. 2014;5:5383. doi: 10.1038/ncomms6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Nakagawa S, Naganuma T, Shioi G, Hirose T. Paraspeckles are subpopulation-specific nuclear bodies that are not essential in mice. J. Cell Biol. 2011;193:31–39. doi: 10.1083/jcb.201011110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Nakagawa S, Shimada M, Yanaka K, Mito M, Arai T, Takahashi E, Fujita Y, Fujimori T, Standaert L, Marine JC, Hirose T. The lncRNA Neat1 is required for corpus luteum formation and the establishment of pregnancy in a subpopulation of mice. Development. 2014;141:4618–4627. doi: 10.1242/dev.110544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Standaert L, Adriaens C, Radaelli E, Van Keymeulen A, Blanpain C, Hirose T, Nakagawa S, Marine JC. The long noncoding RNA Neat1 is required for mammary gland development and lactation. RNA. 2014;20:1844–1849. doi: 10.1261/rna.047332.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Box JA, Bunch JT, Tang W, Baumann P. Spliceosomal cleavage generates the 3′ end of telomerase RNA. Nature. 2008;456:910–914. doi: 10.1038/nature07584. [DOI] [PubMed] [Google Scholar]
- 136.Yin QF, Yang L, Zhang Y, Xiang JF, Wu YW, Carmichael GG, Chen LL. Long non-coding RNAs with snoRNA ends. Mol. Cell. 2012;48:219–230. doi: 10.1016/j.molcel.2012.07.033. [DOI] [PubMed] [Google Scholar]
- 137.Kiss T, Fayet E, Jady BE, Richard P, Weber M. Biogenesis and intranuclear trafficking of human box C/D and H/ACA RNPs. Cold Spring Harb. Symp. Quant. Biol. 2006;71:407–417. doi: 10.1101/sqb.2006.71.025. [DOI] [PubMed] [Google Scholar]
- 138.Matera AG, Terns RM, Terns MP. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat. Rev. Mol. Cell Biol. 2007;8:209–220. doi: 10.1038/nrm2124. [DOI] [PubMed] [Google Scholar]
- 139.Zhang XO, Yin QF, Wang HB, Zhang Y, Chen T, Zheng P, Lu X, Chen LL, Yang L. Species-specific alternative splicing leads to unique expression of sno-lncRNAs. BMC Genomics. 2014;15:287. doi: 10.1186/1471-2164-15-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Yin QF, Hu SB, Xu YF, Yang L, Carmichael GG, Chen LL. SnoVectors for nuclear expression of RNA. Nucleic Acids Res. 2015;43:e5. doi: 10.1093/nar/gku1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Sahoo T, del Gaudio D, German JR, Shinawi M, Peters SU, Person RE, Garnica A, Cheung SW, Beaudet AL. Prader–Willi phenotype caused by paternal deficiency for the HBII-85 C/D box small nucleolar RNA cluster. Nat. Genet. 2008;40:719–721. doi: 10.1038/ng.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Duker AL, Ballif BC, Bawle EV, Person RE, Mahadevan S, Alliman S, Thompson R, Traylor R, Bejjani BA, Shaffer LG, Rosenfeld JA, Lamb AN, Sahoo T. Paternally inherited microdeletion at 15q11.2 confirms a significant role for the SNORD116 C/D box snoRNA cluster in Prader–Willi syndrome. Eur. J. Hum. Genet. 2010;18:1196–1201. doi: 10.1038/ejhg.2010.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 2007;130:89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Gardner EJ, Nizami ZF, Talbot CC, Jr., Gall JG. Stable intronic sequence RNA (sisRNA), a new class of noncoding RNA from the oocyte nucleus of Xenopus tropicalis. Genes Dev. 2012;26:2550–2559. doi: 10.1101/gad.202184.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, Zhu S, Yang L, Chen LL. Circular intronic long noncoding RNAs. Mol. Cell. 2013;51:792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 146.Qian L, Vu MN, Carter M, Wilkinson MF. A spliced intron accumulates as a lariat in the nucleus of T cells. Nucleic Acids Res. 1992;20:5345–5350. doi: 10.1093/nar/20.20.5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15:409. doi: 10.1186/s13059-014-0409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9:e1003777. doi: 10.1371/journal.pgen.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wilusz JE, Sharp PA. Molecular biology. A circuitous route to noncoding RNA. Science. 2013;340:440–441. doi: 10.1126/science.1238522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA, Goodall GJ. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160:1125–1134. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 151.Wang PL, Bao Y, Yee MC, Barrett SP, Hogan GJ, Olsen MN, Dinneny JR, Brown PO, Salzman J. Circular RNA is expressed across the eukaryotic tree of life. PLoS One. 2014;9:e90859. doi: 10.1371/journal.pone.0090859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Westholm JO, Miura P, Olson S, Shenker S, Joseph B, Sanfilippo P, Celniker SE, Graveley BR, Lai EC. Genome-wide analysis of Drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep. 2014;9:1966–1980. doi: 10.1016/j.celrep.2014.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 154.Liang D, Wilusz JE. Short intronic repeat sequences facilitate circular RNA production. Genes Dev. 2014;28:2233–2247. doi: 10.1101/gad.251926.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L. Complementary sequence-mediated exon circularization. Cell. 2014;159:134–147. doi: 10.1016/j.cell.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 156.Ivanov A, Memczak S, Wyler E, Torti F, Porath HT, Orejuela MR, Piechotta M, Levanon EY, Landthaler M, Dieterich C, Rajewsky N. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 2015;10:170–177. doi: 10.1016/j.celrep.2014.12.019. [DOI] [PubMed] [Google Scholar]
- 157.Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L, Zhu P, Chang Z, Wu Q, Zhao Y, Jia Y, Xu P, Liu H, Shan G. Exon–intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015;22:256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 158.Dubin RA, Kazmi MA, Ostrer H. Inverted repeats are necessary for circularization of the mouse testis Sry transcript. Gene. 1995;167:245–248. doi: 10.1016/0378-1119(95)00639-7. [DOI] [PubMed] [Google Scholar]
- 159.Burd CE, Jeck WR, Liu Y, Sanoff HK, Wang Z, Sharpless NE. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet. 2010;6:e1001233. doi: 10.1371/journal.pgen.1001233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Hansen TB, Wiklund ED, Bramsen JB, Villadsen SB, Statham AL, Clark SJ, Kjems J. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 2011;30:4414–4422. doi: 10.1038/emboj.2011.359. [DOI] [PMC free article] [PubMed] [Google Scholar]