Abstract
Electrohydrodynamics (EHD) deals with the fluid motion induced by an electric field. This phenomenon originally developed in physical science, and engineering is currently experiencing a renaissance in microfluidics. Investigations by Taylor on Gilbert's theory proposed in 1600 have evolved to include multiple contributions including the promising effects arising from electric field interactions with cells and particles to influence their behaviour on electrode surfaces. Theoretical modelling of electric fields in microsystems and the ability to determine shear forces have certainly reached an advanced state. The ability to deftly manipulate microscopic fluid flow in bulk fluid and at solid/liquid interfaces has enabled the controlled assembly, coagulation, or removal of microstructures, nanostructures, cells, and molecules on surfaces. Furthermore, the ability of electrohydrodynamics to generate fluid flow using surface shear forces generated within nanometers from the surface and their application in bioassays has led to recent advancements in biomolecule, vesicle and cellular detection across different length scales. With the integration of Alternating Current Electrohydrodynamics (AC-EHD) in cellular and molecular assays proving to be highly fruitful, challenges still remain with respect to understanding the discrepancies between each of the associated ac-induced fluid flow phenomena, extending their utility towards clinical diagnostic development, and utilising them in tandem as a standard tool for disease monitoring. In this regard, this article will review the history of electrohydrodynamics, followed by some of the recent developments in the field including a new dimension of electrohydrodynamics that deals with the utilization of surface shear forces for the manipulation of biological cells or molecules on electrode surfaces. Recent advances and challenges in the use of electrohydrodynamic forces such as dielectrophoresis and ac electrosmosis for the detection of biological analytes are also reviewed. Additionally, the fundamental mechanisms of fluid flow using electrohydrodynamics forces, which are still evolving, are reviewed. Challenges and future directions are discussed from the perspective of both fundamental understanding and potential applications of these nanoscaled shear forces in diagnostics.
I. INTRODUCTION
A. Brief history of electrohydrodynamics (EHD)
EHD, generally referred to as the motion of fluid induced by an electric field, is an extremely useful technique to transport and manipulate small liquid volumes in microdevices. EHD is a very old physical phenomenon with the first experimental investigations being carried out back in 1600 by Gilbert.1 He observed that a spherical drop of water is drawn up into a cone when a charged rod (e.g., one made of rubbed amber) is held at a particular distance above it. However, it was not until 1964 that these observations were subjected to theoretical treatment by Taylor,2,3 where he performed similar experiments as Gilbert to understand the behaviour of droplets within an electric field. Taylor was the first to coin the term electrohydrodynamics as a generic label for this fluid flow behaviour arising from applied electric fields. In his landmark paper which included current flow in the bulk liquid, he accurately modelled the observations made by Gilbert 400 years ago. This model was later utilised by Melcher4 to further develop the field of electrohydrodynamics. Since then, several investigations have been carried out by Saville,5,6 Castellanos,7,8 Tadros,9 and others10,11 to extensively explore this phenomenon for a wide range of applications including spray atomization, fluid motion of bubble drop, electrostatic spinning, and printing. Further, in the mid 1990s, Saville and Trau utilised this phenomenon for the manipulation of nanoscaled colloidal objects in bulk fluid and on the surface of flat electrodes for the fabrication of nanostructured materials.12–15 In these studies, microscopic fluid flow arising from electrical body forces acting on free charges in solution was utilized for the assembly, coagulation, or removal of controlled micro- and nano-structures. These experiments were controlled by the same forces as observed by Gilbert. However, they now did not involve the effects of surface tension. These developments have been extensively reviewed (e.g., Saville)16 in literature and have deepened the understanding of electric field-induced fluid behaviour.
As reviewed by Saville16 in 1997, Electrohydrodynamics is a general term which also covers a combination of electrokinetic forces that drive the manipulation of colloidal particles and fluid transport phenomena including electrophoresis, electrokinesis, electro-osmosis (EO), dielectrophoresis (DEP), induced-charge electro-osmosis (ICEO), and electrorotation. Electrokinetics is a branch of electrohydrodynamics and a generic term applied to effects associated with the movement of ionic solutions near charged interfaces and particle movement under the influence of electric fields (e.g., electrophoresis).17,18 Although the discovery of electrokinetics is believed to have been in the early nineteenth century,19 our detailed understanding of electrokinetic motion begins only with Smoluchowski's studies in the early part of the twentieth century. Since then, investigations on the various electrokinetic-based fluid flow phenomena (listed above) have proceeded more or less independently. Nevertheless, the underlying processes share many similar characteristics.
Over the past two decades, the development and utility of EHD forces such as DEP, EO, and ICEO have deeply influenced the field of microfluidics and its integration into microfluidic-based platforms has driven the development of a myriad of technological platforms for a broad range of applications including manipulation of particles, colloids, droplets, and biological molecules across different length scales. These advances are well-documented in the literature and have been reviewed extensively.20–25 Fig. 1 represents the advances in electrically driven fluid flow systems right from Taylor-Melcher's model to recent applications in diagnostics. The use of alternating current (AC) induced fluid flow is widely preferred for fluid handling in microsystems over its direct current (DC) counterpart due to: (i) avoiding electrolysis or electrode damage, and (ii) the ability to operate at relatively lower voltage regimes, thereby making it highly suitable for the manipulation of biomolecules (e.g., electrorotation, manipulation based on dielectric properties of cells) such as cells or proteins.25,26 Moreover, the integration of electric field-induced fluid flow systems scale favourably with miniaturization and offer (i) the ability to modulate analyte transport (e.g., drive fluid across microchannels) without the need for external fluid manipulation systems,27–29 and (ii) the ability to tune the electrically induced forces via the applied ac field, which can potentially influence the transport of particles or target molecules on the electrode surface.25 Although several existing biological assays have been developed using traditional fluid control units (i.e., syringe pumps, fluidic valves), the ability to transport fluid across microchannels using applied electric field holds tremendous potential for numerous biological applications. This is of particular interest for the development of assays in resource-limited settings that can integrate fluid transport units within the micro or nano device (see Section V for specific examples) to generate similar flow rates to that under hydrodynamic flow with the use of a simple AC signal generator. However, the integration of AC-induced fluid systems for specific applications (e.g., biomarker binding, DNA hybridization, etc.) is dependent on solution conditions (i.e., conductivity), except possibly those utilizing negative DEP at high frequencies or those using DC electrophoresis. Thus, such systems require several optimization studies before being utilised for such applications. Further, numerous other applications include cell isolation systems based on the dielectric properties of cells or electrorotation units where particles rotate as a result of the interaction of the induced dipole and a rotating field. In addition to this, the ability to tune these forces has led to recent studies in addressing major problems associated with typically any diagnostic assay such as sensitivity and nonspecific adsorption of non-target species present in the sample.30–32 This has been achieved by externally tuning the fluid flow that can indirectly alter the magnitude of fluid shear forces responsible for sensor-target affinity interactions and has provided a new capability to physically displace weakly (nonspecifically) bound biomolecules from the electrode surface.30,31 In this article, we review the origin of EHD forces such as EO, DEP, and ICEO and briefly summarise the advances in the field with regards to fundamental mechanisms, applications in fluid flow systems and biomolecular detection. We also discuss the recent progress and technological advancements in the development of EHD based detection systems and highlight several new high-performance systems for sensitive detection of biomolecules.
FIG. 1.
Schematic representation of the progressive development in the field of electrohydrodynamics—from discovery and theoretical modelling to recent applications in clinical diagnostic development.
II. ORIGIN OF EO
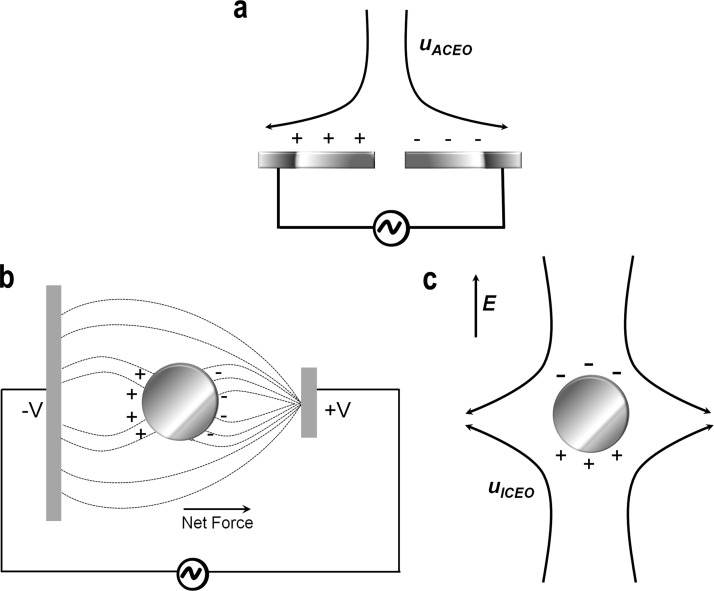
EO is a type of surface force. When a solid is in contact with a fluid solution, surface charge builds up at the interface.33 In order for the interface to remain neutral, the surface charge is balanced by the redistribution of ions close to the solid surface, which leads to the accumulation of counter-ions at the electrode surface.34 This can be viewed as the migration of ions with opposite charges from the bulk to surface area by electrostatic force. The ion aggregation process screens the surface and keeps the fluid bulk electro-neutral while at the same time it forms a capacitive like relaxed double layer due to the affinity of charge-determining ions to a surface (Fig. 2(a)). This phenomenon, known as electro-osmosis, was discovered in the eighteenth century when only limited resources or methods were available for the generation of an electric current.
FIG. 2.
(a) AC Electro-osmosis over a pair of microelectrodes, (b) Schematic of dielectrophoretic movement of a polarisable particle suspended within a point-plane electrode system under an applied electric field, and (c) Induced-charge electro-osmosis (ICEO) around a particle. (a) and (c) Reprinted with permission from Bazant et al., Adv. Colloid Interface Sci. 152, 48–88 (2009). Copyright 2009 Elsevier.
In 1798, the discovery of an electrical battery well-known as Volta pile by Volta was considered a more reliable and stable source of electric current.35 The device comprised of zinc and silver plates connected with paper soaked in salt (e.g., sodium chloride). Following this, in 1808, Reuss carried out experiments using a U-tube with two electrodes connected to an electrical battery containing 92 silver coins, with the same number of zinc plates.19 He put a plug of clay in the U-tube and discovered that an applied voltage resulted in a rise in the water level inside the tube. This marked the discovery of electro-osmosis, an important fluid flow phenomenon utilised to induce fluid motion across any porous material, capillary tube, membrane, or microchannel. Similarly, another set of experiments performed by Reuss marked the discovery of electrophoresis where he placed quartz sand above the clay plug and observed the migration of clay particles through the sand layer under an applied voltage. Later, in 1852, the first quantitative experiments were performed by Wiedemann36 when he investigated fluid movement through a charged tube. He observed that ratio of mass of the fluid to the electric current was independent of the applied voltage and inner diameter of the tube. These experiments were important and found that practically every surface became charged when in contact with an aqueous solution. However, the basic mechanism governing this fluid flow remained unknown.
These explanations and experimental observations gave rise to the concept of the electrical double layer which was proposed by Georg Quincke37 in 1859 and later derived by Hermann Helmholtz38 in 1879. Helmholtz's theory described the relation between fluid flow in a tube and the electric surface potential of the inner surface of the tube. However, at the end of the nineteenth century, Smoluchowski39 improved Helmholtz's model by incorporating dielectric constants to better describe the relation between fluid velocity in tube and electric potential. This relation was henceforth named the Helmholtz-Smoluchowski equation and formed the basis for the understanding of interactions between charged particles or surfaces. However, the Helmholtz-Smoluchowski theory of electro-osmosis was independent of the structure of the diffusive layer. Following this, in 1910, Gouy40 proposed a theory for electrical double layer by considering the precise structure of the diffuse layer. An equivalent theory from Chapman41 in 1913 gave rise to the Gouy-Chapman model that described an important parameter for characterising the thickness of the diffusive layer. This parameter has fundamental importance in almost all theories related to electrically driven fluid flow and also for generic colloidal phenomenon such as colloidal stability. These advances have been reviewed in detail and well-documented in the literature.42,43
Since then, EO has been widely used for investigations on colloidal stability and associated fluid flow behavior using various microelectrode geometries.33,44 Further, its potential for low-voltage microfluidic fluid flow handling was extensively explored by Brown, Ramos, Green, and Castellanos, respectively.20,23,27–29,45–52 These advances are described and reviewed in Section V. The use of alternating current electro-osmosis (ACEO) is a preferred technique over DCEO for microfluidic fluid handling with important applications in biomedical devices and portable electronics. The use of the ACEO technique to generate fluid flow has two major advantages: (i) The use of AC voltages avoids any unwanted electrochemical reactions and can generate faster fluid flows in comparison to DCEO.33 Eq. (1) in Table I represents the time-averaged ACEO velocity on two parallel electrodes assuming a linear relationship between the surface charge and potential, and (ii) ACEO has the capability of depositing particles on specific locations of electrodes, resulting from the formation of counter rotating vortices.53
TABLE I.
Key equations governing fluid flow in non-linear electrohydrodynamic phenomena. = initial potential; Ω = non-dimensional frequency; η = dynamic viscosity; = distance between the electrodes; = reciprocal of Debye length; = permittivity of the medium; σ = conductivity of the medium; ω = angular frequency; Re = Real part; p = dipole moment; E = electric field; * = complex conjugation; = volume of the particle; α = effective polarizability of the particle; a = radius of the particle; = permittivity of the particle; ζeq = steady-state (equilibrium) zeta potential; ζi = induced zeta potential; μ = electrolyte viscosity; E0 = zero-order field; = Coulomb force; = dielectric force.
| ACEO | AC Electroosmotic slip velocity on two parallel electrodes | |
| ||
| ||
| DEP | Time-averaged DEP force | |
| ||
| ||
| Dielectrophoresis under a frequency dependent AC field | ||
| ||
| ICEO | Time averaged ICEO flow velocity | |
| ||
| AC-EHD | Average electrical volume force | |
|
III. ORIGIN OF DIELECTROPHORESIS
DEP is the translational movement of neutral matter as a result of polarisation effects in non‐uniform electric fields.22,54 For instance, under a uniform DC field, a charged particle will experience a net force toward the electrode of opposite polarity. In contrast, a neutral particle is polarised as a result of the electric field, but will experience no net movement. Eq. (3) in Table I represents the time-averaged force on a particle under a spatially varying field magnitude. Under an applied field, the charge redistribution around the particle results in equal amounts of oppositely charged ions being induced on either side of the particle. However, as the field is non-uniform, the forces experienced by opposite ends of the neutral particle have different magnitudes. Under AC electric fields, the neutral particle will always move towards the high or low field regions as described above, irrespective of the polarity of the electrodes. The time-averaged DEP force on a particle and the variation in magnitude of the force with applied frequency is given by the Clausius-Mossotti factor (Eq. (5), Table I). At low frequencies, the charged particle will move toward the electrode of opposite polarity under the influence of electrophoresis, its direction oscillating with the field direction. At higher frequencies, the DEP force will move the charged particle towards the region of high or low field strength depending upon the relative polarisabilities of the particle and the surrounding medium. Thus, DEP force is a function of particle volume, polarizability difference between particle and medium, electric field gradient, and signal frequency.
DEP originated from investigations on the electrophoretic mobility of charged particles under an applied field. According to Mottelay,55 this effect was described by Thales of Miletus in approximately 600 B.C. in observations that suggested vigorously rubbed pieces of amber can attract straws, dried leaves, and other light bodies in the same way that a magnet attracts iron. However, actual experimental investigations and theoretical treatments were not performed until the early twentieth century. In 1923, Hatschek and Thorne studied nickel sols in anhydrous toluene in which rubber acted as a protective colloid.56 With the use of parallel plate electrodes, they observed equal quantities of precipitate forming at each electrode, with considerable rubber present in the precipitate. Further, alternating potential did not cause coagulation as did static voltage in their large apparatus, an observation which led them to interpret the phenomenon in this instance to be electrophoresis. Following this, in 1931, Soyenoff 57 observed the coalescence of coal dust in toluene occurring to be equally effective under AC and DC voltages. However, he attributed this behaviour as dielectric polarization and suggested that a particle or body of higher conductivity or dielectric constant than the medium tends to move toward the region of highest field intensity. Similarly, in 1937, Reising58 investigated the movement of pigment particles in paint vehicles and observed equal quantities of particles being precipitated on the electrodes. However, all these investigations related to particle movement under an applied field failed to provide clear descriptions or evidence related to the observed particle behaviour. Significant further progress in the field was made with investigations of Pohl in 1950 related to the motion of particle relative to that of the solvent resulting from polarization forces produced by an inhomogeneous electric field.59 Pohl coined the term dielectrophoresis based on his study involving the removal of carbon-black filler from polymer samples (e.g., polyvinyl chloride) with the use of applied electric fields. His later investigations60 suggested DEP to be significantly different from electrophoresis considering that the particle does not require a net electric charge for motion to be induced, and either an AC or DC electrical signal could be employed to impose an electric field on a particle.
Since then, DEP is widely recognised as a nondestructive electrokinetic phenomenon with great potential to manipulate different nano/bio-particles including cells, viruses, proteins, and DNA molecules.61–64 Recently, Pethig65 reviewed the history and advances of DEP phenomenon with respect to fundamentals and applications in numerous biomedical applications. In addition to this, numerous electrode designs for DEP manipulation of biomolecules have been reported in the literature, including polynomial electrodes,61 castellated electrodes,61 interdigitated electrodes,63,64 orthogonal electrodes (T electrodes),66 and three-dimensional (3D) electrode structures.62 The behaviour of particles under an applied field is well documented in the literature and involves the movement of particles relative to the field intensity and polarizability.65
IV. ORIGIN OF ICEO
Induced-charge electro-osmotic flow (EOF) occurs around polarizable surfaces in which an applied electric field induces a double layer and drives the induced double layer into motion.21,67 In a typical ICEO flow, ions in the double layer are forced to move by the tangential component of the field which results in an electro-osmotic flow directed from the edges of the electrode to the centre (Fig. 2(c)). The essential difference between traditional and ICEO flows concerns the origin of the diffuse-layer charge.68 In traditional electrokinetics, charges on the solid/electrolyte interface arise in equilibrium, due to the adsorption or dissolution of specific ions or groups. On the contrary in ICEO, the double layer is induced by the applied field.
ICEO is a recently discovered fluid flow phenomenon, and its discovery was stimulated by earlier investigations on the effect of particle separation using asymmetric AC fields,60 later followed by some pioneering work done by Murtsovkin69,70 who studied fluid flow around tin, quartz, and ionite particles and liquid mercury drops in AC fields. His observations were compared with the theoretical treatments and suggested steady quadrupolar (e.g., alternating positive and negative charges on either side of the particle) flow around a polarizable particle.
Earlier, electro-osmotic fluid flow investigations have assumed linear response in the applied voltage, based on the hypothesis of fixed surface charge (or fixed “zeta potential” relative to the bulk solution).42 Later, in 2004, Bazant and Squires focused on the nonlinear phenomenon of electro-osmosis at a polarisable (metal or dielectric) surface. They described an electro-osmotic fluid flow phenomenon in which an applied field was found to induce an ionic charge cloud around a surface and, subsequently, forces that induced charge cloud into fluid motion. They coined the term “induced-charge electro-osmosis”21 to describe it and suggested that ICEO flow can be used to generate steady electro-osmotic flows using AC or DC fields. The nonlinearity also allows larger fluid velocities and a richer, geometry-dependent flow structure. This phenomenon thus unified ACEO with other seemingly unrelated phenomena, such as AC electrohydrodynamic (AC-EHD) interactions and self-assembly of dielectric colloids on electrodes, and hydrodynamic interactions among polarisable particles. These advances are extensively reviewed and documented in the literature.68,71,72
The use of ICEO offers several advantages over EOF. First, as with traditional electrokinetic flows, the full ICEO velocity is established just outside the double-layer.73 Stronger flows are, in principle, more possible with ICEO than with traditional EOF in that stronger applied fields give rise to higher potential. The time-averaged ICEO slip velocity under a low-frequency AC field is derived under the condition where the induced-double layer varies in phase with the field (Eq. (6), Table I). Second, electrochemical reactions can play a less significant role in ICEO phenomena, because AC potentials “reverse” the reactions and because applied potentials are generally much smaller than in DC electrokinetic systems.74 As a result, electrodes may be brought closer to the region of interest (and to each other) with less concern about fouling or contaminating the fluid of interest. Subsequently, strong electric fields can be established even with small applied voltages. Finally, unlike traditional EOF, for which the EOF velocity field is proportional to the electric field, ICEO flows can be deliberately sculpted and designed.75 For instance, one can design an inducing surface that is anisotropic in some way to give a directed induced charge electrophoretic (ICEP) velocity/rotation or induced charge electro-osmotic flow under an AC field. Furthermore, modulating electrode height and geometry in ICEO fluid flow can enhance flow rates dramatically.46
Despite these advantages, their integration into lab-on-chip systems is limited by their theoretical understanding and electrode contamination layers (e.g., corrosion, oxidation or adsorption of solute molecules) that can impede ICEO flow.68 Further, ICEO flows are the strongest immediately over inducing surfaces (generally metal), whose length scales typically range from 10–100 mm, whereas lab-on-a-chip devices require flow manipulation over millimetres to centimetres. Thus, in order to truly exploit ICEO phenomena for practical lab-on-a-chip applications, the mechanisms behind these discrepancies must be understood and addressed.
V. APPLICATIONS OF AC ELECTROHYDRODYNAMIC-BASED FLUID FLOW PHENOMENON
Initial investigations on fluid flow were carried out using capillary electro-osmosis involving the application of an electric field across the chip to drive plug flows through microchannels by acting on the equilibrium double-layer charge. However, since the effect is linear in the applied field, a direct current must be maintained with Faradaic reactions, which can produce gas bubbles, unwanted reactions, electrode dissolution, and/or hydrodynamic instability. Moreover, a rather large voltage is needed to obtain a relatively small velocity which exacerbates these problems and limits portability.
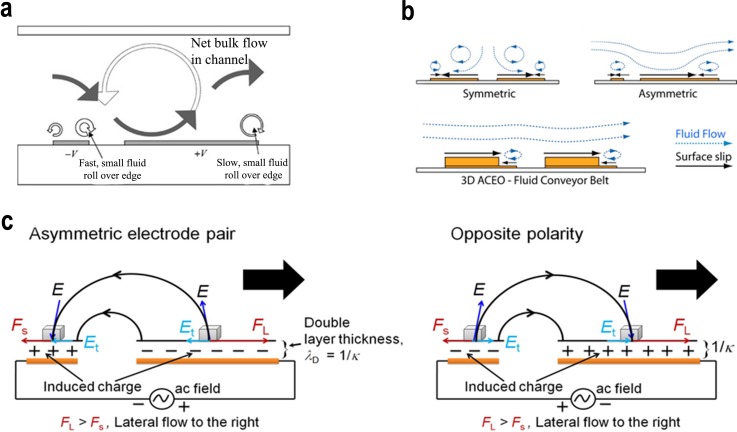
Since the late 1990s, several groups have begun to address these drawbacks by developing microfluidic devices based on nonlinear electro-osmotic flow. Experimental observations of nonlinear electro-osmotic flow, varying as the square of the applied voltage, led to the application of AC electro-osmosis.33,44,47 Further, it was predicted that the same effect could be used to drive fluids over a microelectrode array by taking advantage of broken symmetry within each period, either by modifying the surface capacitance or by modulating the surface height.22,23 With these predictions not being put into practice, alternatively breaking symmetry in the widths and spacings of each electrode pair (Fig. 3(a)) in the array was proposed.27 This planar design became the focus of experimental and theoretical studies of ACEO based fluid flow devices. The mechanism responsible for the flow is the interaction of the tangential component of the electric field and the induced charge in the diffuse double layer on the electrode surface.27 Consider two coplanar electrodes separated by a thin gap, subjected to an AC potential difference and covered in an electrolyte. At a certain time, induced charge accumulates in the diffuse double layer with a sign opposite to the electrode charge. This induced charge is subjected to the action of the tangential component of the electric field, giving rise to a force directed from the centre of the gap onto the electrode surface. This force drives the fluid at the level of the electrodes and has a direction that is independent of the sign of the electrode potential, so that the fluid flow has a non-zero time average. It should be noted that the mechanism requires a non-uniform electric field, thus ensuring that a tangential field component exists in the diffuse double layer on the electrodes. Alternatively, numerous other observations have suggested that spatial variations in the normal current distribution on the electrodes cause lateral fluid motion either in AC or DC fields (Fig. 3(b)). Concentration gradients arising from electrode reactions produce a distribution of free charge adjacent to the electrodes, and the interaction of these charges with any lateral electric field generates electrohydrodynamic flow. Eq. (7) and Table I represent the non-zero time-average electrical force required for fluid motion under AC-EHD field. Faster fluid flows, with a wider frequency range, were achieved with 3D electrode arrays (Fig. 3(b)), consisting of asymmetrically placed steps electroplated on a symmetric planar array.46 These investigations were motivated by studies of induced-charge electro-osmosis around 3D metal structures21,67 and suggested the possibility of achieving significant improvements in AC electrokinetic fluid flow by regulating the height of the electrodes. More recently, the asymmetry in electrode geometry has been utilised to manipulate biological entities on electrode surfaces to enable their capture and subsequent enumeration. In such systems, the application of an ac field E across an asymmetric electrode pair induces charges within the electrical double layer. The asymmetric geometry gives rise to a lateral variation in the total amount of free (double layer) charges and spatial distribution of charges on the electrode surface (Fig. 3(c)). Consequently, the free charges on the larger electrode create stronger lateral forces than those on the smaller electrode, resulting in a lateral flow towards the large electrode. This fluid flow is predicted to be electrohydrodynamic in nature,30,31 and, since all free charges in solution occur only within the double layer of the electrode, all of the ac-EHD body forces on the fluid also occur strictly within this region. Thus, this ability to engender fluid flow within molecular distances of the electrode surface was termed nanoshearing.30,31
FIG. 3.
Schematic representation of (a) physical mechanism of ac electro-osmosis in an asymmetric electrode array inside a microfluidic channel. Reprinted with permission from Ramos et al., Phys. Rev. E 67, 056302 (2003). Copyright 2003 American Physical Society. (b) Fluid flow generated by an AC field applied across two planar symmetric, asymmetric, and partially raised electrodes. Reprinted with permission from Appl. Phys. Lett. 89(14), 143508 (2006). Copyright 2006 AIP Publishing LLC. (c) Mechanism of AC-EHD induced fluid flow using asymmetric electrode pair. Reversing the polarity of the AC field also reverses the sign of the charges in the induced double layer, and since electrical body forces are the product of the charges and the applied field, a steady flow can be maintained towards the large electrode. Reprinted with permission from Shiddiky et al., Sci. Rep. 4, 3716 (2014). Copyright 2014 Nature Publishing Group.
Over the years, several key technological developments have leveraged this potential of AC-EHD and electrokinetic forces for biomolecule manipulation and sensitive detection of clinically relevant biomarkers from heterogeneous biological samples. Sections V A–V E will initially emphasise the key challenges associated with development of diagnostic tools for routine clinical diagnosis and also highlight some of the key technological advances in biomolecule manipulation utilising AC-EHD and electrokinetic forces to achieve clinical detection limits for cell, protein, DNA, and vesicular populations.
A. Technological challenges in biomolecular detection
Recent technological advances in nanotechnology, microfabriaction, and microfluidics have markedly improved the standards of diagnostic tools and point towards new opportunities for diagnosing disease. Similarly, advances in the level of automation for diagnostic based commercial platforms have enabled molecular testing and analysis even in moderately sophisticated laboratories. However, many of the latest innovations are yet to be utilised for routine diagnostic testing, owing to factors such as sensitivity, specificity, robustness, and portability. Critically, early stage disease diagnosis requires highly specific measurement of biomarkers from biological samples (e.g., blood or serum) at concentrations of 10−16 to 10−12 M for proteins or as low as 1–100 cancer cells in 106 blood cells per ml of blood, to provide clinically useful information.76 Of particular interest is the emphasis on stringent specificity to achieve accurate isolation of rare target molecules among numerous proteins, cells, and other small molecules, many of which have a tendency to adsorb onto a solid support without any specific receptor-recognition interaction. Thus, the “Holy Grail” clinical testing of molecular diagnostic tools requires simple, accurate, automated, and rapid analysis to aid clinicians during diagnosis and monitoring disease recurrence.
Enzyme-linked immunosorbent assay (ELISA) is recognised as the generic method for immuno-affinity based biomolecule isolation.77 This method utilising antibodies as capture agents is limited by its low sensitivity and fails to detect clinically relevant biomarkers often found in very low concentrations in most disease conditions such as cancer, cardiovascular disease, and neurodegenerative disorders.78 Alternatively, new materials and assays have been developed using signal-based amplification and detection. Most of these assays rely on the use of nanoscale materials, which have unique and controllable size-dependent properties, tunable chemical compositions, and robust structures.79 Similarly, over the last decade, the inception of microfluidic-based approaches for diagnostic technologies has emerged as one of the most promising solutions to address these issues. A microfluidic ensemble derives its potential application in such settings from its ability to handle complex fluids and their precise movement or relocation within the fluidic channel. Laminar flow based approaches for cell and protein biomarker detection have not advanced significantly to address clinical detection limits owing to the slow diffusion of biomolecules that limit the number of sensor-target interactions. Thus, the inception of nano- and micro-scale materials or electrocatalytic agents into microfluidic set-ups for signal amplification has been viewed as an effective alternative to achieve higher sensitivity.
B. Applications in cell analysis
1. Manipulation of cells and microorganisms
Common scientific challenges, such as answering a biological question related to a particular cell type (e.g., a key cell developmental pathway) or the development of precise biotechnological or biomedical cell-based applications (e.g., disease diagnosis on the basis of identification of a particular cell-type or a stem cell-based therapy), require the isolation of pure cell populations from biological samples. As a result, efficient capture and separation of cells from other populations or components within a biological sample (i.e., cell sorting) is not only the first step but a major goal for many applications.80 In some cases, this can also represent a true challenge. For example, bacteria cell detection is necessary for safety control of pathogens growing in food products and for disease diagnosis.81,82 Despite conventional microbiological methods being reliable and able to differentiate between live and dead cells, they are limited by their ability to distinguish different strains and levels of cell functionality or development. Moreover, their detection limits typically vary from 102–107 cfu/ml, therefore they are limited in detecting a few number of these cells.
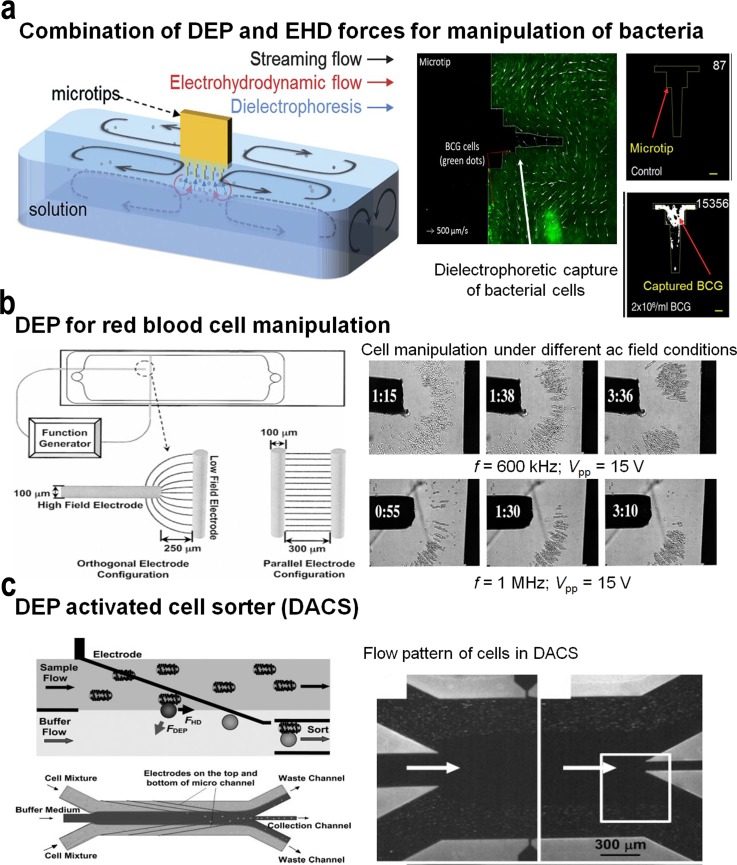
Electrokinetic methods such as dielectrophoresis can exploit the differences in conductivity between alive and dead bacteria cells (the conductivity of a live cell membrane is around 10−7 S/m, whereas death causes a cell's membrane to become permeable increasing its conductivity by a factor of 104) for inducing their effective separation under electric fields.82 By exploiting distinctive differences in dielectric properties between the target cells and non-target cells, electrokinetics acts directly on the targeted cell population. As a result, target cells are pulled down towards the electrode surface breaking the cell's inherent slow diffusion and increasing the cell's capture efficiency. In particular, differences in cell size are easy to exploit by these methods, and, whenever that size difference is small, strategies such as agglutination can be employed to artificially increase the size of one cell population with regard to the other, leading to high sensitivity at detection. These features coupled with antibody recognition can increase cell capture efficiency by several folds, as demonstrated in a number of applications such as bacteria or rare cancer cell detection. In spite of the complex nature of physical, chemical, and biological mechanisms, the integration of streaming flow, EHD flow, and dielectrophoresis has enabled the rapid concentration of target bacteria (Fig. 4(a)) by “dipping and withdrawal” of microfabricated tips.81 The technology has rapidly advanced away from simple wire configurations, and DEP is now steadily developing into a powerful cellular characterization, separation, manipulation, and cellular patterning tool utilizing complex microfluidics and automated microfabricated electrode arrays.
FIG. 4.
(a) Concentration mechanism and immunofluorescence detection of target bacteria in sputum samples using streaming flow, EHD flow, and dielectrophoresis. Reprinted with permission from Kim et al., Lab Chip 12(8), 1437–1440 (2012). Copyright 2012 The Royal Society of Chemistry. (b) DEP microdevice for the manipulation and characterization of red blood cells under different ac field conditions. Reprinted with permission from Minerick et al., Electrophoresis 24(21), 3703–3717 (2003). Copyright 2003 Wiley-VCH. (c) DEP activated cell sorter. Cells entering in the sample stream are only deflected into the collection stream if they are labelled with a dielectrophoretically responsive label (i.e., beads). The DEP particles entering the collection channel after being focused into the center of the stream. The arrows indicate the direction of fluid flow. The electrode region of the microchannels comprise of sample and buffer inlets, as well as waste and collection outlets. Reprinted with permission from Hu et al., Proc. Natl. Acad. Sci. U. S. A. 102(44), 15757–15761 (2005). Copyright 2005 National Academy of Sciences, USA.
Castellated electrode designs (e.g., built with turret block shaped electrodes) can generate distinctive field maxima and minima, and can be used to trap and characterize latex particles61 as well as biological cells. With a change in frequency, latex particles experienced a transition from negative DEP to positive DEP. Different particles and cells have different frequency responses in specific solutions, so at a certain condition (frequency and fluid conductivity), a mixture of particles can be separated and trapped to different regions of electrode patterns. Similarly, an orthogonal electrode pattern consists of one high field electrode and one low field electrode that are arranged perpendicular to each other. The high field electrode tip is the p-DEP (positive) region, and the low field electrode gap is the n-DEP (negative) region. Using this device, erythrocytes (red blood cells, RBCs) were characterized to have the optimal mobilities at approximately 1 MHz, in a 0.1 S/m isotonic phosphate buffer saline (PBS) medium (Fig. 4(b)).66 Alternatively, in the case of single wall nanotubes (SWNT), self-assembly was confined to a thin boundary near the electrode and highly branched, while at 1 MHz frequency, SWNT self-assembled rapidly from both electrodes to form thin uniform wires that bridged the gap.83 Bulk carbon nanotube (CNT)-pathogen docking was achieved using this method. Bacteria were trapped 10 times more entangled within the SWNT in the solution than without the SWNT present. Alternatively, strip electrodes on the top and bottom glass were aligned perpendicular to each other forming a grid pattern. By controlling the signal frequency and phase on electrodes, a biological cell can be trapped on the grid junction by p-DEP or released by n-DEP.62
Continuous-flow DEP devices have been used to sort bacterial cells in an integrated chip that utilises angled electrodes to deflect cells into individual channels based on their negative DEP mobilities.84 In these individual channels, the sorted cells were trapped and concentrated using a negative DEP electrode gate for further detection by surface enhanced Raman spectroscopy (SERS). The optimal ac field parameters were determined for each bacterium, and a specific positive and negative DEP frequency was determined for bacterium to sort individual cells.
Similarly, a DEP-well separation system comprising a scalable structure based on 3D wells with approximately unity height-to-width ratios (based on tubes with electrodes on the sides) was utilised to enrich yeast cell populations.85 Based on the device developed by Fatoyinboet et al.,86 it contains chambers with electrodes “striped” around the perimeter, allowing a much higher DEP throughput and also providing the ability to efficiently recover cells processed through the device. DEP forces have also been utilised for the separation of live and dead cells that could be of particular importance during early stage diagnosis and drug testing.87 The device continuously traps dead cells at a reservoir-microchannel junction based on the inherent electric field gradient to separate them from live ones right inside the reservoir. This approach is therefore termed reservoir-based dielectrophoresis (rDEP). This approach eliminates the need for any external mechanical parts and occupies zero channel space. All the above mentioned methods are applicable when target cells possess a significantly different dielectrophoretic response from that of other cells. However, in certain applications, target and non-target cells exhibit similar dielectrophoretic ability, thereby precluding sorting based on the intrinsic dielectrophoretic phenotypes. To address this, Hu et al.88 labelled cells with polymeric beads to achieve significant differences in dielectrophoretic amplitude response between the target (e.g., cells bound to beads) and unlabeled cells (Fig. 4(c)). This approach, referred to as dielectrophoresis activated cell sorter (DACS), demonstrated the enrichment of rare Escherichia coli that displays a specific surface marker from an excess of non-target bacteria of the same species.
2. Cancer cell isolation and analysis
The isolation and purification of cancer cells also present similar challenges in terms of recovering a rare population of cells from complex sample mixtures. In particular, the capture of specialized subpopulations of tumor-initiating cancer stem cells (CSC) or even single tumor cells within tissues and body-fluids (i.e., rare cells or circulating tumor cells (CTCs)) is extremely important for early detection of cancer and for monitoring the effectiveness of cancer therapies as they associate to treatment failure and risk of metastasis.76,89 Further, analysis of CTCs is considered a “liquid biospy” of the tumor that can directly correlate to the tumor's phenotype and metastatic potential. However, isolation of CTCs has been technically challenging due to the extremely low abundance (a few to 100 cells per ml) of CTCs among a billion-fold higher concentration of hematologic cells in blood.76,89 Despite the limitations on specificity and sensitivity associated with commercial immunomagnetic platforms, their integration has potentially identified CTCs as mainstream cancer biomarkers with excellent diagnostic potential.90 Further, the heterogeneity of CTCs, in terms of morphology and expression levels, limits the accuracy of both marker based and marker-free cell capture methods.91–93 Thus, there is a need for more versatile (e.g., more specific markers for capture) and advanced platforms to implement routine clinical analysis and monitoring based on CTCs.
a. Isolation based on biophysical characteristics
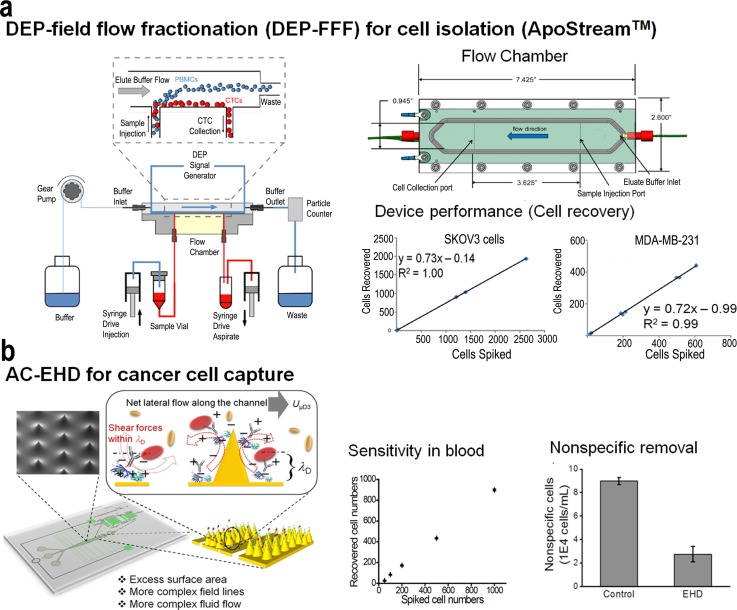
Current technologies for CTC isolation exploit differences in cell density, immunologic targets, or receptor-ligand interactions. The dielectric properties of cells and their behaviour under an applied field represent a simple label-free approach to isolate cells. When a particle or a cell is suspended in a medium with different dielectric properties, it becomes electrically polarized under an applied field and its interaction with the field gives rise to interesting electrokinetic effects. In this regard, Gupta et al.94 recently developed a label-free microfluidic approach that utilises dielectrophoretic forces to isolate circulating tumor cells based on differences in dielectric properties between blood cells (lymphocytes, monocytes, and granulocytes) and cancer cells (Fig. 5(a)). This approach, referred to as ApoStream, operates under a modified form of DEP-field flow fractionation (DEP-FFF) in which CTCs are drawn towards the electrode plane due to positive DEP force whilst healthy blood cells are levitated by negative DEP into the hydrodynamic flow velocity profile. The separation DEP forces are dependent on the conductivity and permittivity of the medium, and cell separation was most efficient at the crossover frequency where DEP force transitions from negative to a positive force. This approach operating in continuous separation enabled the efficient isolation and enrichment of viable CTCs from blood. Similar cell isolation approaches have been developed95–98 based on a continuous flow separation process in which the peripheral blood mononuclear cell fractions are slowly injected, deionized by diffusion, and then subjected to a balance of DEP, sedimentation, and hydrodynamic lift forces. These approaches work based on a principle similar to that of ApoStream in which DEP forces cause tumor cells to be transported close to the surface, while blood cells are carried out of the chamber as waste.
FIG. 5.
(a) Schematic diagram of the ApoStream device. The entire set-up is computer controlled allowing dynamic feedback control and monitoring. The sample injection and collection flow rates are controlled by high precision syringe pumps. When cells encounter the DEP field, the DEP forces pull cancer cells towards the chamber floor and repel other cells as they traverse the electrode. Cancer cells travelling close to the chamber floor are withdrawn through the collection port, while other blood cells travelling at greater heights are carried beyond this port and exit the chamber to the waste container via a second outlet port. Reprinted with permission from Biomicrofluidics 6(2), 024133 (2012). Copyright 2012 AIP Publishing LLC. AC-EHD induced tunable surface shear forces for highly sensitive and specific isolation of rare cancer cells from blood samples. Reprinted with permission from Vaidyanathan et al., Anal. Chem. 86(4), 2042–2049 (2014). Copyright 2014 American Chemical Society.
b. Immunoaffinity based isolation of cells
Microchip technologies that exploit immunoaffinity based isolation also combine electrokinetic effects to mediate fluid flow and control fluid shear forces on surfaces to enhance capture. In this regard, Dharmasiri et al.,99 developed a microfluidic device containing an electrokinetic manipulation unit interfaced to a fluidic chip designed for the high-throughput processing of whole blood and referred to as a high-throughput microsampling unit (HTMSU). This electrokinetic manipulation unit utilised a combination of hydrodynamic and electrokinetic forces to direct CTCs released from the selection surface (i.e., antibody based isolation) into a reservoir for further molecular analysis. The device deemed to be suitable for mass-limited sample analysis also demonstrated the analysis of KRAS mutations in the isolated CTCs. Similarly, the use of tunable nanoshear surface forces (Fig. 5(b)) has enabled the selective capture of target cancer cells whilst physically displacing nonspecific cells from the electrode surface.31,100 This tunable control of surface shear forces (Fig. 4(c)) and concomitant fluid micromixing facilitates two critical improvements to the traditional immunocapture of cellular targets: (i) enhanced capture efficiency due to increased number of sensor-target collisions, which is a result of improved transport, and (ii) enhanced specificity resulting from the ability to tune nanoscopic fluid shear forces at the electrode interface, which serves to shear away loosely bound, nonspecific species present in biological samples.31,100 Further, the incorporation of three-dimensional electrodes provides a very sensitive methodology to control AC-EHD flow vortices and to specifically capture target cells. Thus, a change in aspect ratio of the planar electrodes can result in additional asymmetry in the electrode structure, which can also produce more complex fluid vortices. Recently, Smith et al.101 demonstrated the combination of differential DEP response in a device containing obstacle arrays to capture target cells on these obstacle surfaces using antibody-antigen binding. In this device, a pair of electrodes offset from an array of dielectric obstacles generate a spatially variable electric field around these obstacles to enable cell capture. Such a configuration attracts cells with positive DEP (pDEP) response to the leading edge, where the shear stress is low and residence time is long, resulting in a high capture probability. Thus, cells eliciting a negative DEP (nDEP) are repelled from these regions and removed from the devices.
The ability of AC-EHD and other electrokinetic methods for driving and controlling the movement of the operating fluid and the charged suspensions by electric fields has been critical for promoting manipulation of biomolecules in many applications involving microfluidic and bioanalytical systems. The integration of AC-EHD has enabled fine control of flow velocity and direction under purely electrical command by simply adjusting the applied voltage and frequency, and the electrode configuration.102,103 Because the magnitude of this fluid shear force can be tuned externally (e.g., via the application of an ac electric field), it provides the ability to achieve better detection performance due to the manipulation of the surface shear forces and greater complexity in the fluid flow vortices (fluid mixing). AC-EHD is also unique in terms of the extremely high pressures,104 it can generate even at low voltages,105,106 and it can be designed to reduce the formation of bubbles or gas blocking upon liquid flow, which is another drawback in many sensing read-outs.107,108 Importantly, since electrodes are easier to microfabricate and incorporate into microfluidic devices than pumps and valves, fabrication complexity is significantly reduced. This provides plasticity to current designs and greater miniaturization and parallelization degree to the analysis leading to versatile, cheap, and highly efficient microfluidic devices.109–111
c. Alternative technologies for CTC isolation
Simultaneously, over the years, a plethora of technologies based on hydrodynamic flow systems with integrated capture features have emerged for rare cell capture and concentration enrichment.112–119 CellSearch® system (Veridex) is the only FDA (Food and Drug Administration) approved method for CTC detection.90,120 It involves EpCAM conjugated with magnetic beads to enrich CTCs from the blood of patients with cancers of the breast, prostate, and colon. Although this platform is currently being tested for clinical applications, it is limited by a high level of “biological noise” associated with low sensitivity, purity, and specificity.90,120 In this context, microfluidic devices have many advantages to offer for enhancing cell capture efficiency or even for providing on-chip cell-detection (e.g., direct imaging on optically transparent channels121). A microfluidic ensemble derives its potential application in such settings from its ability to handle complex fluids, their precise movement, relocate within the fluidic channel, and also manipulate shear forces to preserve cell dormancy.122 Channel materials can in turn be derivatized with proper recognition elements such as antibodies, lectins, aptamers, or dendrimers for the multiplexed and/or selective enrichment of channels on the desired cell population. Moreover, microfluidics platforms are also highly amenable for having their channels structured with interesting patterns such as micro/nanopillars,117,123,124 microdams,125 microposts,92,114,116,121 microfilters,126–128 microcavities,123 nanotextures,129 chevron,117 or herringbones (HBs)116 using micro/nanofabrication techniques. These structures can be used for (i) increasing surface binding area and non-laminar fluid motion, rendering larger sensor-target collisions and cell-capture efficiencies or for (ii) cell isolation without prior knowledge of cell-surface biomarkers by patterning these structures on size-based wise manner. On top of these features, microfluidic devices have an intrinsic main advantage for cell capture over conventional methods, i.e., the hydrodynamic force resulting from fluid motion is itself a separation tool, and microfluidic devices are expert fluid motion players. This is because the flow rate can be optimized to generate a certain shear force strong enough to avoid attachment of non-specific cells, but mild enough to permit specific cell-capture.
In recent times, notably, Nagrath et al. developed a microfluidic platform (CTC chip) for single-step isolation of CTCs from unprocessed blood specimens.114 The CTC-chip consists of a silicon chamber etched with 78 000 microposts coated with an anti-EpCAM antibody. Flow kinetics was optimized for minimal shear stress on cells while enhancing contacts with the antibody-coated microposts. Subsequently, the same research group developed a HB chip for enhanced CTC isolation by inducing fluid microvortices for better fluid mixing.116 In addition to the increased target cell capture efficiency, the less complex design of the HB-chip is more amenable to high throughput manufacture and reliable coating of the inner surface with antibodies and allows for the chambers to be made out of transparent materials, which greatly enhance high resolution imaging, including the use of transmitted light microscopy. Similar studies including the integration of a chaotic mixing channel with a patterned nanostructured substrate also facilitated highly efficient CTC capture resulting in the synergistic effects of enhanced cell–substrate contact frequency as well as affinity.
Cell separation using inertial focusing offers rapid size-based separation, and trapping of particles by size in laminar vortices has been utilised extensively for the isolation of CTCS. Recently, Ozkumur et al.130 demonstrated the isolation and enrichment of CTCs using an inertial focusing-enhanced microfluidic CTC capture platform, termed “CTC-iChip,” that is capable of sorting rare CTCs from whole blood. This integrated microfluidic platform incorporates three individual components including initial size based separation of cells from blood followed by inertial focussing for alignment of cells and finally defection of magnetically tagged cells into a collection channel. This platform has demonstrated the isolation of CTCs in suspension using both tumor antigen-dependent and tumor-antigen-independent modes. The use of iChip has been demonstrated in an expanded set of both epithelial and non-epithelial cancers including lung, prostate, pancreas, breast, and melanoma. Similarly, the irreversible migration of particles into microscale vortices was demonstrated for the isolation of CTCs using parallel expansion-contraction trapping reservoirs.131 The approach utilised microscale laminar vortices in combination with inertial focusing to selectively isolate and trap larger cells of interest while non-target cells are removed from the device. This technology demonstrates size-based separation without clogging mechanical filters, employing only a simple single-layered microfluidic device for rapid analysis. Recently, Lv et al.,132 demonstrated the utility of a microfluidic system to segregate, enumerate, and recover CTCs based on the physical differences of CTCs with the main constituents of the human peripheral blood. This size based separation technique involved the initial lysis of RBCs to improve flow rates and reduce blood clogging. Also, Dean drag forces derived from centrifugal effects on spiral microchannels can be exploited for size-dependent focusing of cells at distinct equilibrium positions within the microchannel cross-section.133 Furthermore, a number of technologies91,99,112,115,134–149 for CTC capture have been developed focusing on cell-substrate interactions with the development of integrated microfluidic platforms and extensively reviewed in the literature.150–152
C. Applications in protein biomarker analysis
Protein biomarkers represent simple non-invasive screening tools, with development in microfluidics and nanomaterials enabling detection of clinically relevant limits in combination with high sensitivity, specificity, and speed. Despite this rapid progress, most biomarker detection systems operate with very small sample volumes, and it is highly likely that certain important biomarkers present in ultralow concentrations could remain undetected in light of the achieved sensitivity limits. Thus, enhancing analyte transport towards the transducer surface is of critical importance particularly in the case of protein biomarkers that diffuse slowly, thereby increasing the assay time. Microfluidic-based sensors, characterized by low Reynolds number flow, eliminate any associated turbulent mixing. Thus, the integration of electrokinetically controlled devices can facilitate diffusive mass transport whilst inducing fluid mixing to enhance the scale of sensitivity achieved for protein detection.
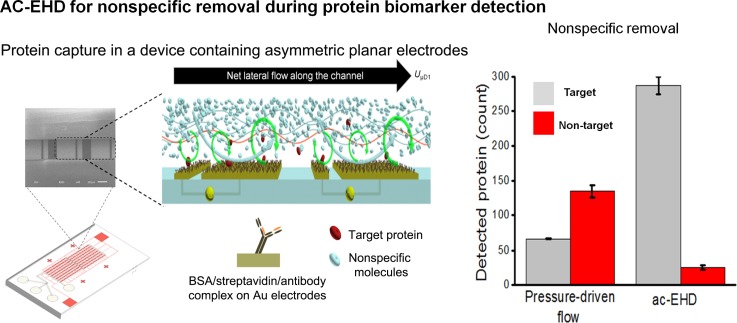
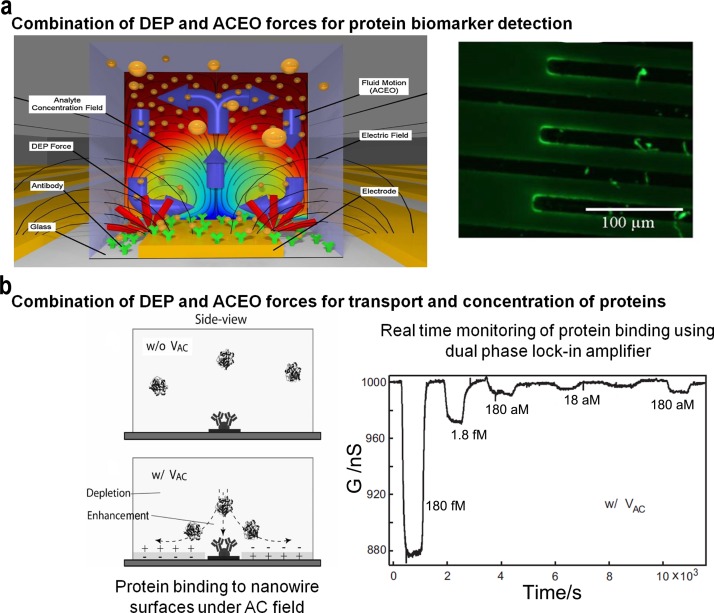
Recently, the use of nanoshear surface shear forces combined with integrated microfluidic units has produced significant advances in sensitivity that can be achieved for detection of clinically relevant protein biomarkers (Fig. 6). This capability can possibly enable clinical monitoring and potentially recognize biomarkers present at very low levels. This methodology offers a unique ability to shear-off loosely bound molecules from the solid/liquid interface and significantly improved sensitivity and specificity in comparison to hydrodynamic flow based approaches.30,153 This detection performance is also attributed to the presence of 3D geometries that accentuate the surface shear forces and concomitant micromixing effect via increased surface area and more complex and asymmetric electric field lines resulting in a more complex fluid flow. Similarly, enhancement of heterogeneous immunoassays using AC electro-osmosis was demonstrated using interdigitated planar electrodes built directly onto, or near, the binding region of a transducer.154 The binding of antibodies on a functionalized surface was significantly enhanced, especially at the ends of each electrode without noticeable nonspecific binding. Finite element modelling and fluorescent immunoassays were established to understand the effect of applied voltage, electrode geometry, and fluidic channel on assay performance. Further improvements in sensitivity were achieved using utilizing ACEO (Fig. 7(a)) on specially modified quartz crystal microbalances (QCM), known as electrokinetic QCMs (EKQCMs).155 Immunoassays were performed on electrodes fabricated on glass surfaces to ensure antibody function was not significantly degraded by the enhancement technique.
FIG. 6.
Molecular nanoshearing for highly specific capture of cancer biomarkers. This AC-EHD induced fluid flow phenomenon generates micro- and nano-scopic shear forces within the double layer, which can shear away nonspecifically adsorbed molecules thereby enhancing target protein capture. Reprinted with permission from Shiddiky et al., Sci. Rep. 4, 3716 (2014). Copyright 2014 Nature Publishing Group.
FIG. 7.
(a) Fluorescent immunoassays performed using AC electrokinetic mixing on interdigitated electrodes printed on a QCM. Immunoglobulin G (IgG), a ubiquitous was used in a direct adsorption assay on the surface of an EKQCM. Stirring antibody-antigen reactions with ACEO enhanced the detection sensitivity. Reprinted with permission from Hart et al., Sens. Actuators, B 147(1), 366–375 (2010). Copyright 2010 Elsevier. (b) Schematic image of the microfluidic channel cross section showing the protein binding at the original injected concentration near the NW surface with and without AC excitation. Reprinted with permission from J.-R. Gong, Small 6(8), 967–973 (2010). Copyright 2010 Wiley-VCH.
Protein pre-concentration on integrated microchips has also been achieved to concentrate proteins prior to enzymatic modifications or separations. Advancements in nanofabrication have enabled the possibility of manipulating protein molecules in constricted nanoscale channels. Under an applied field, an ion exclusion-enrichment effect caused by electrical double layer overlapping induces surface charge-based selectivity of proteins within a nanochannel.156 Subsequent separation of two different concentrated proteins was achieved by switching the direction of the electric field in the direction parallel to the thin-walled section of the nanochannel. Similar electrokinetic protein trapping methods have been demonstrated under uniform DC electric fields, where migration behaviours are the result of a balance between the electrophoretic force and a drag force induced by a recirculating electro-osmotic flow generated across the channel.157 These approaches based on the electrophoretic mobility of proteins could potentially be applicable as a preconditioning step to concentrate or separate other analytes for downstream processes.
Dielectrophoretic manipulation of molecules has been utilised for the development of cell-based protein detection in a microsystem wherein genetically engineered bacterial cells are engineered to express the desired capture proteins on the membrane surface and are spatially arrayed as sensing elements in a microfluidic device.158 This represents a typical microarray technology where each sensor element is electrically addressable to assemble cells and the co-expression of peptide-based capture ligands on the cell surface enabled protein capture. Protein capture was verified using fluorescence intensity analysis and presents an attractive method for simple proteomic analysis. Further, the use of integrated nanoelectronic detection systems combined with electrokinetic devices has produced significant advances in sensitivity and enabled label-free attomolar detection (Fig. 7(b)) of proteins.159 Electrically addressable silicon nanowire field-effect transistors and electrodes for electrokinetic transport are integrated onto a common sensor chip platform. Enhancement in assay performance is achieved through streaming dielectrophoresis and corresponding electrostatic contribution to the binding affinity of protein onto nanowires upon the application of an AC electric field.
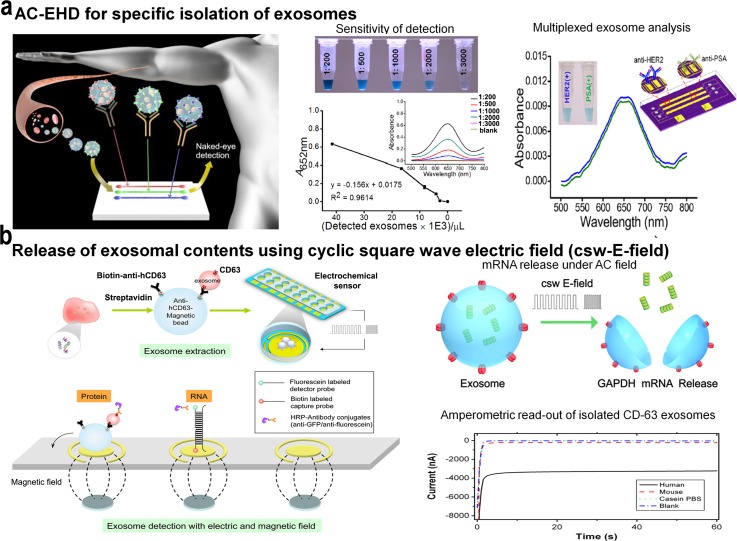
Simultaneous measurement of multiple biomarkers is desirable for understanding more complex diseases such as cancer. A simple methodology for the routine assessment of multiple protein biomarkers promises to transform disease diagnosis, particularly in resource-constrained settings. To this end, the use of tunable nanoshearing forces has also been extended towards achieving sensitive detection of multiple protein targets from human serum.160 Under AC-EHD fluid flow, this method provides the capability to simultaneously detect multiple exosome targets using a simple and rapid on-chip naked eye detection readout based on the catalytic oxidation of peroxidase substrate 3,3′,5,5′-Tetramethylbenzidine (TMB) and potentially represent a simple diagnostic tool that can be integrated into resource-limited settings. Alternatively, integrated electrokinetically controlled microfluidics has been manifested for the development of a heterogeneous immunoassay for multiplexed analysis.161 These platforms have been tested for the detection of multiple pathogen targets using an indirect approach of capturing antibody molecules to detect the immobilized antigen on the sensor surface.
The capture performance demonstrated by the aforementioned methods is comparable to that of the traditional bio-barcode,162 immuno-polymerase chain reaction (PCR),163 liposome-PCR,164 and redox-cycling165 based bioassays. Similar detection limits for serum protein detection were also reported using non-electrokinetic microfluidics based platforms.166–168 However, their practical application is restricted due to their complex detection procedures, complicated coupling chemistries, pre-concentration/modification steps, and operational control systems. In contrast, engendering fluid flow and also shearing off non-target proteins via alternating voltage on an electrokinetic-based microfluidic system could be a simple and powerful tool to reduce nonspecific adsorption and also enhance capture performance of proteomic assays. Thus, integration of electrokinetics represents an ideal approach for routine clinical disease management.
D. Applications in DNA trapping and analysis
DNA being a highly charged polyelectrolyte,169,170 studies on its physical and mechanical properties provide fundamental insights and scope for numerous biological applications. The central role of DNA in modern molecular biology necessitates the need for new methods to trap, size, and separate DNA molecules. Recent advances in genetic approaches have attracted widespread attention in a wide variety of applications in molecular diagnosis. Since conventional lab based techniques such as microarrays, Southern blot and real time PCR involve sophisticated protocols and instrumentation, rendering them inappropriate for applications in resource-limited settings,171,172 the use of electric field induced forces to manipulate DNA molecules could be an effective alternative considering the charged nature of DNA that can readily be polarized under an electric field. A dipole can be induced upon the application of an electric field and under a spatially nonuniform oscillating electric field; DNA molecules may experience positive or negative dielectrophoretic forces depending on the field strength, field gradient, and frequency as well as the dielectric properties of DNA and surrounding medium.173
Washizu and Kurosawa were the first to demonstrate the use of dielectrophoretic forces to manipulate DNA molecules within a miniaturized platform.174 Later, Asbury and van den Engh utilised thin gold film stripes to trap and manipulate DNA molecules across confined structures under a combination of static and oscillating fields.173,175 Further, they extended this capability to measure the strength and capacity of dielectrophoretic traps using a CCD camera and microfluidic channel placed over the trapping electrodes. The trapping efficiency was measured as a characteristic of strand length, the strength and frequency of the applied field, and the ionic concentration of the solution. Kreft et al. demonstrated the use of uniform attractive potential to trap DNA molecules within a stagnation region created by counter-rotating vortices.176 Numerical simulation studies based on the lattice-Boltzmann method suggested the elongation of DNA along the stagnation point. This was attributed to the conformation of DNA, and the trapping rate was found to be independent of the applied potential.
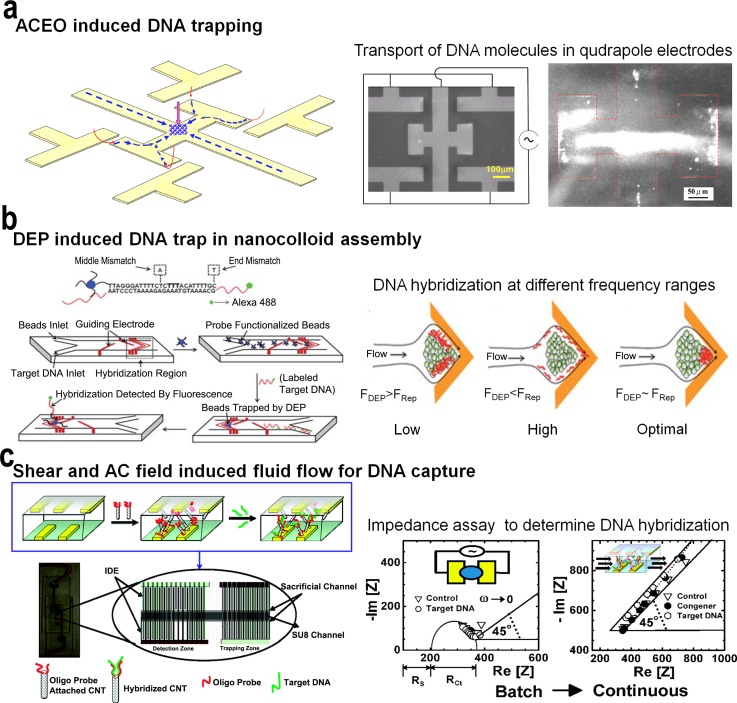
Du et al. demonstrated the trapping and concentration of trace amounts of DNA molecules using a microfluidic platform operating under nonlinear electro-osmotic flow (Fig. 8(a)).177 Under an applied field, the asymmetric quadrupole electrode design facilitates focussing effects that can transform into a robust funnel to collect DNA molecules distantly from the bulk and tightly condense them into a compact cone. This trapping phenomenon was identified to be the combined result of the formation of two pre-focused DNA jets flowing toward each other, dipole-induced attraction between focused DNA molecules, and dielectrophoretic trap on the spot. The method enabled rapid concentration of pM concentrations of DNA demonstrating the long-range trapping capability of this funnel. Similarly, single-stranded DNA (ssDNA) molecules were trapped using DEP under a cusp-shaped nanocolloid assembly on a chip with a locally amplified AC electric field gradient (Fig. 8(b)).178 The tunable nature of AC frequency and electrophoretic mobility of DNA resulted in a converging flow that enabled mismatch-specific binding of DNA at the cusp. The optimum flow rate provided enhanced discrimination between target DNA sequences and a single mismatch sequence and high shear force associated with the fluid flow also removed any nonspecifically bound molecules. The approach was also able to locate the region of mismatch within the target DNA hybridization region (i.e., in this case 26 base docking sequence). The enhanced DEP DNA mobility enabled rapid and sensitive detection (e.g., pM levels) of DNA from a large sample volume.
FIG. 8.
(a) Schematic representation of focussing and trapping of DNA using a device containing quadrapole electrodes. Initially DNA molecules (blue) are prefocused by converging streams generated by ACEO vortices and then undergo head-on collision to trap DNA at the center of the system, with the assistance of dipole-induced association between focused DNA and the holding of the trapped spot by the downward DEP force (pink). Reprinted with permission from Du et al., Biomicrofluidics 2(4), 044103 (2008). Copyright 2008 AIP Publishing LLC. (b) Rapid dielectrophoresis induced assay to trap ssDNA in a cusp-shaped nanocolloidal assembly. The microfluidic platform traps ssDNA in a nanocolloidal assembly functionalized with a capture probe under a locally amplified gradient field. The DNA molecule rapidly concentrates under optimal frequency, and the hybridized DNA is detected using a fluorescence detector. Reprinted with permission from Cheng et al., Lab Chip 10(7), 828–831 (2010). Copyright 2010 The Royal Society of Chemistry. (c) AC field induced DNA transport to capture DNA molecules on oligo functionalized CNTs. CNTs are trapped by dielectrophoresis by an AC field supplied by interdigitated electrodes (IDE). Sample containing target DNA is passed through the trapped DNA and detected using the observed change in impedance. Reprinted with permission from Basuray et al., ACS Nano 3(7), 1823–1830 (2009). Copyright 2009 American Chemical Society.
With biomolecular assays requiring the detection of picomolar or lower target DNA concentration ranges, detection platforms require methods to direct the transport of target biomolecules towards the sensor surface. In this regard, constriction-based DEP methods179 were developed for the directed delivery of DNA within a micro-constricted fluidic channel onto a functionalized microelectrode sensor with nanostructured edges. Numerical simulation analysis followed by electric field optimization enabled the measurement of the effect of DNA pre-concentration on hybridization kinetics and minimized any degradation of the capture probe due to high field strengths. Thus, this approach enabled rapid pre-concentration of DNA and resulted in a 10-fold enhancement of the DNA hybridization kinetics with a sensitivity limit of 10 pM for the sensor platform. Similarly, Basuray et al.180 demonstrated the pre-concentration and hybridization of DNA on carbon nanotube surfaces (CNTs) under high-frequency AC electric field (Fig. 8(c)). In a microchannel containing interdigitated electrodes, ssDNA passed through CNTs was trapped in DEP field and detected using a change in impedance caused by the hybridization of DNA to the oligo probe immobilised on the CNT surface. This is achieved due to the enhanced charge-transfer rate across the CNT and shear-enhanced transport of DNA that decouples the AC impedance signal from the charge-transfer signal due to hybridization. The DEP force on the CNTs was strong enough to hold them against the fluid flow, thereby eliminating the need for their immobilisation onto electrodes or a microfilter trap. Using this approach, the authors demonstrate label-free and sensitive detection of picomolar target DNA hybridization events in less than 20 min. In addition to this, the manipulation of nucleic acids (e.g., DNA and miRNA) under an applied electric field have also been demonstrated using combinations of nanoscale structures. The applied field attracts nucleic acids and other bioparticles towards the vicinity of the nanotip or cone arrays to enable ultrasensitive detection of nucleic acids. Yeo et al. demonstrated DEP-induced concentration and size-specific separation of extracellular DNA using a nanostructured tip.181 This technique enabled rapid concentration (<1 min) of 6.7 pg/ml (210 aM) of DNA from a sample mixture containing λ-DNA and Drosophila cells. Similarly, Wang et al. utilised a conical fiber array platform containing metal coated nanocone tips to concentrate small nucleic acids (miRNA of 20 bases) with singular scattering effect.182 The scattering effect produced by the tip with a fluorescent signal enabled preferential and sensitive capture (∼100 molecules per cone tip) of nucleic acids. Unlike plasmonic structures, the use of nanostructured tips avoided any conduction loss or quenching of fluorescent reporters, making it suitable for ultrasensitive detection of target molecules.
Thus, the emerging field of molecular nanotechnology requires techniques with the ability to manipulate on the molecular scale in order to understand fundamental properties of molecular systems. Some of the prominent tools for molecular manipulation include optical and magnetic tweezers,183 atomic force microscopy,184,185 and the use of hydrodynamic forces186 to shear molecules off the surface. While the use of tweezers relies on trapping labelled molecules using a laser beam or magnetic field, scanning probe techniques such as AFM can directly trap molecules without the need for any labelling steps. However, the use of these techniques is limited for on-field or portable DNA detection due to their sophisticated instrumentation and reagents. Further, field assays also require rapid and specific detection without the use of repeated wash steps. AC electrokinetics provides a powerful mechanism for both positioning and inducing conformational changes in molecules with only minimal instrumentation.178,187 Thus, the above discussed approaches along with numerous other AC field induced DNA manipulation techniques have certainly enabled the development of simple point-of-care platforms for rapid DNA detection.
E. Applications in exosome isolation and detection
Cells and proteins represent important blood-based biomarkers for non-invasive screening and, in most cases, require the detection of low cell numbers or ultralow concentrations of proteins from blood. Recent evidence suggests that certain nanovesicles released by most, if not all, cells into body fluids (e.g., blood, saliva, urine, etc.) carry vital molecular information (e.g., mRNA, microRNA, and proteins) representative of the parent cell or tumor.188,189 Generally recognised as exosomes, these vesicles derived from tumor cells contribute to cancer progression by mediating local and systemic cell communication between the primary tumor and bone marrow cells. Unlike other blood-based markers such as CTCs (1–100 cells/ml of blood), exosomes are generally present in large numbers (8.0 × 103 to 5.0 × 105 exosomes/μl in biological fluids such as serum, blood, plasma, etc.)190,191 and represent a simple and non-invasive source of profiling the primary tumor. For over three decades, these vesicles were considered “cellular waste” with no specific function.192 The discovery of their role in mediating immune responses gathered attention and rekindled widespread interest in understanding their biological significance. Since then, exosomes have been recognised as membrane nanovesicles (40–100 nm) of endocytic origin, released by cells into body fluids.189 Exosomes shed from tumor cells have been predicted to have a role in cross talk between the primary tumor and bone marrow cells by mediating local and systemic cell communication through the horizontal transfer of molecular information. Thus, exosomes can be regarded as novel disease markers for non-invasive detection, and unravelling the process of exosome release could possibly yield new targets in anti-metastatic therapy.
Over the years, a plethora of technologies and methods have been developed for the isolation of exosomes from biological fluids, including ultracentrifugation, electron microscopy, fluorescence activated cell sorter (FACS), and conventional isolation kits based on buoyant density or polymer based sedimentation.193 While these bulk analysis methods successfully isolate nano-sized vesicular populations from biological fluids, they tend to induce precipitation of cellular and protein debris within the isolates, rendering them less suited for diagnostic or therapeutic applications. To the other end, integration of microfluidics and optical sensors has significantly enhanced the accuracy and specificity of exosome isolation from heterogeneous, biological samples.194–199 The foremost among these approaches include the use of a microfluidic device for isolating exosomes using an immuno-affinity approach,194,197,198 use of a porous silicon nanowire-on-micropillar structure,199 or isolating exosomes from whole blood using in situ prepared nanoporous membranes. These new techniques provide faster separation than the standard approaches; however, optimization of these microfluidic platforms is needed for applications in clinical settings. Similarly, the use of periodic plasmonic nanoholes that readily match vesicle size has been demonstrated for improved sensitivity.196 The array of nanoholes coated with different exosomal protein markers displays spectral shifts or intensity changes proportional to target marker protein levels. Despite these advances in achieving enhanced sensitivity, no current technique has been integrated with standard bio-analytical systems for simultaneous profiling and quantification of the exosomes.
Taking into account the need for a significantly improved approach for exosome isolation, the use of ac electrohydrodynamic-induced surface shear forces can be used to accentuate the capture performance and also profile exosomes in a more efficient manner. Recently, the use of nanoshear surface forces was demonstrated in a multiplexed microfluidic device (Fig. 9(a)) for highly specific capture and detection of multiple exosome targets.200 This approach demonstrates the analysis of exosomes derived from cells expressing HER2 and prostate specific antigen (PSA), and was also capable of specifically isolating exosomes from breast cancer patient samples. This method was capable of simultaneously detecting multiple exosome targets using a simple and rapid on-chip naked eye detection readout (i.e., avoids use of any sophisticated instrumental readouts) based on the catalytic oxidation of peroxidase (e.g., from horseradish peroxidise (HRP) conjugated detection antibody) substrate TMB. The device also exhibited a significant enhancement in detection sensitivity in comparison to hydrodynamic flow (i.e., syringe pump) based assays. This approach clearly demonstrates the versatility of using ac electrohydrodynamics with the use of asymmetric microelectrode pairs as fluid flow units (avoids the use of additional pumps, valves, etc.) and capture/detection domain during simultaneous capture of multiple target exosomes under AC-EHD induced fluid flow. Further, this multiplexed approach can potentially be applied for essentially any biochemical assay based on immunocapture (via modifying the device with any antibody specific to any disease biomarker).
FIG. 9.
(a) A multiplexed device based on ac-EHD-induced nanoshearing for the isolation of multiple exosome targets. When samples containing target exosomes are driven through antibody-functionalized devices under ac-EHD flow, it provides the capability to specifically capture these exosomes by increasing the number of exosome−antibody (surface bound) collisions, which is a result of improved analyte transport. Reprinted with permission from Vaidyanathan et al., Anal. Chem. 86(22), 11125–11132 (2014). Copyright 2014 American Chemical Society. (b) Electric field-induced release and measurement (EFIRM) of exosome biomarkers. Exosomes isolated using a immunomagnetic enrichment method are subjected to an electric field to release its contents and subsequently monitored using a amperometric readout method. Reprinted with permission from Wei et al., Biosens. Bioelectron. 44, 115–121 (2013). Copyright 2013 Elsevier.
A means of combining nanoparticles or traditional magnetic bead based exosome isolation with the potential of electric field induced detection of exosomal contents is viewed as a simple method to profile exosomes. The approach (Fig. 9(b)) termed electric field-induced release and measurement (EFIRM) can simultaneously disrupt exosomes to release the contents and monitor the harboured exosomal RNA/proteins biomarkers.201 The underlying principle involves the application of a cyclic square wave electrical field (csw E-field) that can cause redistribution or polarization of lipid vesicular structures causing membrane rupture to release exosomal contents. This approach utilizes a magnetic bead based isolation of exosomes from saliva using vesicular membrane markers and, subsequently, releases exosomal contents onto a gold surface. Exosomal markers were detected using oligonucleotide probes and monitored using an amperometric readout method.
Over the last decade, these nanovesicles have garnered attention as potential disease biomarkers and the field of exosome isolation is certainly in its budding stages. Inherent characteristics of exosomes may make them ideal next-generation biomarkers for 21st century research and therapy. While advances in micro-nanofabrication and microfluidics have improved the accuracy of analysis, key challenges that remain include the demonstration of the robustness of direct exosome isolation strategies in a clinical setting. With only a handful of technologies developed, the field of exosome research certainly does offer plenty of scope for exploring the utility of AC-EHD and electrokinetic forces for their isolation and to develop novel applications for exosome analysis. The field is certainly progressing towards understanding the clinical potential of exosomes and their role in disease progression. In particular, the involvement of exosomes in intracellular communication and the dynamic nature of their composition, for example, have allowed investigators to explore their tumor-modulating potential. Because of the multifunctional nature of exosomes, it is important to understand the balance between healthy and oncogenic exosome signaling. One way to use exosomes for therapeutic purposes is to remove these vesicles to prevent metastasis and tumorigenesis. However, the technical and financial challenges involved in removing exosomes have prevented the clinical implementation of this technique to date.
VI. CONCLUSIONS
Advances in colloidal science and the increasing need for new approaches to handle fluids or particles in microsystems have driven a revival in electrohydrodynamics, a technique originally described 415 years ago. We have reviewed the origins of electrohydrodynamics and other fluid phenomenon covered by EHD such as electro-osmosis, dielectrophoresis, and induced-charge electro-osmosis. It is evident that several key discoveries that lead up to the origin of these phenomena and their underlying characteristics overlap with each other. However, advances in each of these fields have occurred more or less independently. These advances in ac electrohydrodynamics and electrokinetic phenomena with regards to theoretical and experimental, fundamental, and practical understanding have been key to their utilization in practical systems. Thus, their integration into microfluidics and the ability to handle fluids with precise control garnered widespread attention in biomolecular analysis. The technologies outlined in this review have demonstrated the utility of electrohydrodynamics for the detection and quantitation of clinical biomarkers such as cells, proteins, DNA, and exosomes with the desired level of accuracy and sensitivity. Further, this new dimension of electrohydrodynamics reviewed here with regards to biomolecule detection presents numerous opportunities for the development of more robust platforms for biomarker analysis. Despite being in its early stages, the above discussed approaches represent significant advances in micro-nanotechnology-biology interfacing, and will serve as the foundation for new fundamental studies and novel directions in biomedical research and applications.
ACKNOWLEDGMENTS
The authors gratefully thank all the colleagues who have contributed over the years to the cited joint research work. The authors acknowledge financial support from the ARC DECRA (Grant No. DE120102503) and ARC DP (Grant No. DP140104006). We gratefully acknowledge funding received from the National Breast Cancer Foundation of Australia (Grant Nos. CG-08-07 and CG-12-07). These grants have significantly contributed to the environment to stimulate the research described here.
References
- 1. Gilbert W., de Magnete, edited by Mottelay T. B. P. F. ( Peter Short, London, 1600), Vol. 2. [Google Scholar]
- 2. Taylor G. I., Proc. R. Soc. A 280, 383–397 (1964). 10.1098/rspa.1964.0151 [DOI] [Google Scholar]
- 3. Taylor G., Proc. R. Soc. Lond. A 291, 159–166 (1966). 10.1098/rspa.1966.0086 [DOI] [Google Scholar]
- 4. Melcher J. R. and Taylor G. I., Annu. Rev. Fluid Mech. 1(1), 111–146 (1969). 10.1146/annurev.fl.01.010169.000551 [DOI] [Google Scholar]
- 5. Saville D. A., Phys. Fluids (1958–1988) 17(1), 54–60 (1974). 10.1063/1.1694613 [DOI] [Google Scholar]
- 6. Saville D. A., Phys. Rev. Lett. 71(18), 2907–2910 (1993). 10.1103/PhysRevLett.71.2907 [DOI] [PubMed] [Google Scholar]
- 7. Ramos A. and Castellanos A., Phys. Lett. A 184(3), 268–272 (1994). 10.1016/0375-9601(94)90387-5 [DOI] [Google Scholar]
- 8. Ramos A., González H., and Castellanos A., Phys. Fluids 6(9), 3206–3208 (1994). 10.1063/1.868103 [DOI] [Google Scholar]
- 9. Hayati I., Bailey A. I., and Tadros T. F., Nature 319(6048), 41–43 (1986). 10.1038/319041a0 [DOI] [Google Scholar]
- 10. Torza S., Cox R. G., and Mason S. G., Phil. Trans. R. Soc. Lond. A 269, 295–319 (1971) 10.1098/rsta.1971.0032. [DOI] [Google Scholar]
- 11. Sherwood J. D., J. Fluid Mech. 188, 133–146 (1988). 10.1017/S0022112088000667 [DOI] [Google Scholar]
- 12. Trau M., Saville D. A., and Aksay I. A., Science 272(5262), 706–709 (1996). 10.1126/science.272.5262.706 [DOI] [PubMed] [Google Scholar]
- 13. Trau M., Sankaran S., Saville D., and Aksay I., Nature 374(6521), 437–439 (1995). 10.1038/374437a0 [DOI] [Google Scholar]
- 14. Trau M., Sankaran S., Saville D. A., and Aksay I. A., Langmuir 11(12), 4665–4672 (1995). 10.1021/la00012a015 [DOI] [Google Scholar]
- 15. Trau M., Saville D., and Aksay I., Langmuir 13(24), 6375–6381 (1997). 10.1021/la970568u [DOI] [Google Scholar]
- 16. Saville D., Annu. Rev. Fluid Mech. 29(1), 27–64 (1997). 10.1146/annurev.fluid.29.1.27 [DOI] [Google Scholar]
- 17. Russel W. B., Saville D. A., and Schowalter W. R., Colloidal Dispersions ( Cambridge University Press, 1989). [Google Scholar]
- 18. Saville D. A., Annual Rev. Fluid Mech. 9(1), 321–337 (1977). 10.1146/annurev.fl.09.010177.001541 [DOI] [Google Scholar]
- 19. Reuss F. F., Sur un Novel Effet de l'Électricité Galvanique ( Mémoires de la Société Impériale des Naturalistes de Moskou, 1809). [Google Scholar]
- 20. Castellanos A., Ramos A., Gonzalez A., Green N. G., and Morgan H., J. Phys. D: Appl. Phys. 36(20), 2584–2597 (2003). 10.1088/0022-3727/36/20/023 [DOI] [Google Scholar]
- 21. Bazant M. Z. and Squires T. M., Phys. Rev. Lett. 92(6), 066101 (2004). 10.1103/PhysRevLett.92.066101 [DOI] [PubMed] [Google Scholar]
- 22. Ramos A., Morgan H., Green N. G., and Castellanos A., J. Phys. D: Appl. Phys. 31(18), 2338 (1998). 10.1088/0022-3727/31/18/021 [DOI] [Google Scholar]
- 23. Ramos A., Morgan H., Green N. G., and Castellanos A., J. Colloid Interface Sci. 217(2), 420–422 (1999). 10.1006/jcis.1999.6346 [DOI] [PubMed] [Google Scholar]
- 24. Saville D. A., Phys. Fluids (1958–1988) 13(12), 2987–2994 (1970). 10.1063/1.1692890 [DOI] [Google Scholar]
- 25. Velev O. D. and Bhatt K. H., Soft Matter 2(9), 738 (2006). 10.1039/b605052b [DOI] [PubMed] [Google Scholar]
- 26. Pycraft H. M., Nanotechnology 11(2), 124 (2000). 10.1088/0957-4484/11/2/314 [DOI] [Google Scholar]
- 27. Brown A. B. D., Smith C. G., and Rennie A. R., Phys. Rev. E 63(1), 016305 (2000). 10.1103/PhysRevE.63.016305 [DOI] [PubMed] [Google Scholar]
- 28. Gonzalez A., Ramos A., Green N. G., Castellanos A., and Morgan H., Phys. Rev. E 61(4), 4019–4028 (2000). 10.1103/PhysRevE.61.4019 [DOI] [PubMed] [Google Scholar]
- 29. Green N. G., Ramos A., González A., Morgan H., and Castellanos A., Phys. Rev. E 66(2), 026305 (2002). 10.1103/PhysRevE.66.026305 [DOI] [PubMed] [Google Scholar]
- 30. Shiddiky M. J. A., Vaidyanathan R., Rauf S., Tay Z., and Trau M., Sci. Rep. 4, 3716 (2014). 10.1038/srep03716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vaidyanathan R., Shiddiky M. J. A., Rauf S., Dray E., Tay Z., and Trau M., Anal. Chem. 86(4), 2042–2049 (2014). 10.1021/ac4032516 [DOI] [PubMed] [Google Scholar]
- 32. Rauf S., Shiddiky M. J. A., and Trau M., Chem. Commun. 50, 4813–4815 (2014). 10.1039/c4cc01357c [DOI] [PubMed] [Google Scholar]
- 33. Ajdari A., Phys. Rev. Lett. 75(4), 755–758 (1995). 10.1103/PhysRevLett.75.755 [DOI] [PubMed] [Google Scholar]
- 34. Scott M., Kaler K. V. I. S., and Paul R., J. Colloid Interface Sci. 238(2), 449–451 (2001). 10.1006/jcis.2001.7651 [DOI] [PubMed] [Google Scholar]
- 35. Volta A., Philos. Trans. R. Soc. London 90, 403–431 (1800). 10.1098/rstl.1800.0018 [DOI] [Google Scholar]
- 36. Wiedemann G., Ann. Phys. 163, 321–352 (1852). 10.1002/andp.18521631102 [DOI] [Google Scholar]
- 37. Quincke G., Ann. Phys. 189, 513–598 (1861). 10.1002/andp.18611890802 [DOI] [Google Scholar]
- 38. Helmholtz H., Ann. Phys. 243, 337–382 (1879). 10.1002/andp.18792430702 [DOI] [Google Scholar]
- 39. Smoluchowski M., “ Contribution à la théorie de l'endosmose électrique et de quelques phénomènes corrélatifs (ut supra),” Bull. Int. Acad. Sci. Cracovie, Cl. Sc. Math. Nat. 8, 182–199 (1903). [Google Scholar]
- 40. Gouy L. G., J. Phys. Theor. Appl. 9, 457–468 (1910) 10.1051/jphystap:019100090045700. [DOI] [Google Scholar]
- 41. Chapman D. L., Philos. Mag. 25, 475–481 (1913). 10.1080/14786440408634187 [DOI] [Google Scholar]
- 42. Wall S., Curr. Opin. Colloid Interface Sci. 15(3), 119–124 (2010). 10.1016/j.cocis.2009.12.005 [DOI] [Google Scholar]
- 43. Chang H.-C. and Yeo L. Y., Electrokinetically Driven Microfluidics and Nanofluidics ( Cambridge University Press, New York, 2010). [Google Scholar]
- 44. Ajdari A., Phys. Rev. E 53(5), 4996–5005 (1996). 10.1103/PhysRevE.53.4996 [DOI] [PubMed] [Google Scholar]
- 45. Olesen L. H., Bruus H., and Ajdari A., Phys. Rev. E 73(5), 056313 (2006). 10.1103/PhysRevE.73.056313 [DOI] [PubMed] [Google Scholar]
- 46. Urbanski J. P., Thorsen T., Levitan J. A., and Bazant M. Z., Appl. Phys. Lett. 89(14), 143508 (2006). 10.1063/1.2358823 [DOI] [Google Scholar]
- 47. Ajdari A., Phys. Rev. E 61(1), R45–R48 (2000). 10.1103/PhysRevE.61.R45 [DOI] [PubMed] [Google Scholar]
- 48. Castellanos A., Gonzalez A., Ramos A., Green N. G., and Morgan H., Electrostatics 2003, 175–180 (2003). [Google Scholar]
- 49. Green N. G., Nanotech 2003: Technical Proceedings of the 2003 Nanotechnology Conference and Trade Show, San Francisco, CA ( Nano Science and Technology Institute, 2003), Vol. 1, pp. 63–66. [Google Scholar]
- 50. Prost J., Chauwin J.-F., Peliti L., and Ajdari A., Phys. Rev. Lett. 72(16), 2652–2655 (1994). 10.1103/PhysRevLett.72.2652 [DOI] [PubMed] [Google Scholar]
- 51. Studer V., Pepin A., Chen. Y., and Ajdari A., Analyst 129(10), 944–949 (2004). 10.1039/b408382m [DOI] [PubMed] [Google Scholar]
- 52. Ramos A., González A., Castellanos A., Green N. G., and Morgan H., Phys. Rev. E 67, 056302 (2003). 10.1103/PhysRevE.67.056302 [DOI] [PubMed] [Google Scholar]
- 53. Lian M. and Wu J., Appl. Phys. Lett. 94(6), 064101 (2009). 10.1063/1.3080681 [DOI] [Google Scholar]
- 54. Pethig R., Crit. Rev. Biotechnol. 16(4), 331–348 (1996). 10.3109/07388559609147425 [DOI] [Google Scholar]
- 55. Mottelay P. F., Bibliographical History of Electricity and Magnetism (Charles Griffin, London, 1922), p. 720. [Google Scholar]
- 56. Hatschek E. and Thorne P. C. L., Proc. R. Soc. Lond. A 103, 276–284 (1923) 10.1098/rspa.1923.0056. [DOI] [Google Scholar]
- 57. Soyenkoff B. C., J. Phys. Chem. 35(10), 2993–3010 (1930). 10.1021/j150328a010 [DOI] [Google Scholar]
- 58. Reising J. A., Ind. Eng. Chem. 29(5), 565–571 (1937). 10.1021/ie50329a018 [DOI] [Google Scholar]
- 59. Pohl H. A., J. Appl. Phys. 22(7), 869–871 (1951). 10.1063/1.1700065 [DOI] [Google Scholar]
- 60. Pohl H. A., Dielectrophoresis ( Cambridge University Press, Cambridge, 1978). [Google Scholar]
- 61. Green N. G., Ramos A., and Morgan H., J. Phys. D: Appl. Phys. 33(6), 632 (2000). 10.1088/0022-3727/33/6/308 [DOI] [Google Scholar]
- 62. Junya S. and Ronald P., J. Phys. D: Appl. Phys. 31(22), 3298 (1998). 10.1088/0022-3727/31/22/019 [DOI] [Google Scholar]
- 63. Li H. and Bashir R., Sens. Actuators, B 86(2–3), 215–221 (2002). 10.1016/S0925-4005(02)00172-7 [DOI] [Google Scholar]
- 64. Markarian N., Yeksel M., Khusid B., Farmer K., and Acrivos A., Appl. Phys. Lett. 82(26), 4839–4841 (2003). 10.1063/1.1587882 [DOI] [Google Scholar]
- 65. Pethig R., Biomicrofluidics 4(2), 022811 (2010). 10.1063/1.3456626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Minerick A. R., Zhou R., Takhistov P., and Chang H.-C., Electrophoresis 24(21), 3703–3717 (2003). 10.1002/elps.200305644 [DOI] [PubMed] [Google Scholar]
- 67. Squires T. M. and Bazant M. Z., J. Fluid Mech. 560, 65 (2006). 10.1017/S0022112006000371 [DOI] [Google Scholar]
- 68. Squires T. M., Lab Chip 9(17), 2477–2483 (2009). 10.1039/b906909g [DOI] [PubMed] [Google Scholar]
- 69. Murtsovkin V. A. and Mantrov G. I., Colloid J. USSR 53, 240–244 (1991). [Google Scholar]
- 70. Murtsovkin V. A., Colloid J. 58, 341–349 (1996). [Google Scholar]
- 71. Bazant M. Z., Kilic M. S., Storey B., and Ajdari A., Adv. Colloid Interface Sci. 152, 48–88 (2009). 10.1016/j.cis.2009.10.001 [DOI] [PubMed] [Google Scholar]
- 72. Bazant M. Z. and Squires T. M., Curr. Opin. Colloid Interface Sci. 15(3), 203–213 (2010). 10.1016/j.cocis.2010.01.003 [DOI] [Google Scholar]
- 73. Bazant M. Z. and Ben Y., Lab Chip 6(11), 1455–1461 (2006). 10.1039/b608092h [DOI] [PubMed] [Google Scholar]
- 74. Bazant M. Z., Kilic M. S., Storey B. D., and Ajdari A., Adv. Colloid Interface Sci. 152(1–2), 48–88 (2009). [DOI] [PubMed] [Google Scholar]
- 75. Levitan J. A., Devasenathipathy S., Studer V., Ben Y., Thorsen T., Squires T. M., and Bazant M. Z., Colloid Surf., A 267(1–3), 122–132 (2005) 10.1016/j.colsurfa.2005.06.050. [DOI] [Google Scholar]
- 76. Alix-Panabieres C. and Pantel K., Nat. Rev. Cancer 14(9), 623–631 (2014). 10.1038/nrc3820 [DOI] [PubMed] [Google Scholar]
- 77. Gosling J. P., Clin. Chem. 36(8), 1408–1427 (1990); available at http://www.clinchem.org/content/36/8/1408.2.abstract. [PubMed] [Google Scholar]
- 78. Giljohann D. A. and Mirkin C. A., Nature 462(7272), 461–464 (2009). 10.1038/nature08605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Rosi N. L. and Mirkin C. A., Chem. Rev. 105(4), 1547–1562 (2005). 10.1021/cr030067f [DOI] [PubMed] [Google Scholar]
- 80. Kumar A., Galaev I. Y., and Mattiasson B., Cell Separation: Fundamentals, Analytical and Preparative Methods, Advances in Biochemical Engineering, Biotechnology Series, edited by Scheper T. ( Springer, 2007), pp. 1–203. [Google Scholar]
- 81. Kim J.-H., Yeo W.-H., Shu Z., Soelberg S. D., Inoue S., Kalyanasundaram D., Ludwig J., Furlong C. E., Riley J. J., and Weigel K. M., Lab Chip 12(8), 1437–1440 (2012). 10.1039/c2lc21131a [DOI] [PubMed] [Google Scholar]
- 82. Li H. and Bashir R., Sens. Actuators, B 86(2), 215–221 (2002). 10.1016/S0925-4005(02)00172-7 [DOI] [Google Scholar]
- 83. Chang H.-C., Can. J. Chem. Eng. 84, 1–15 (2006). [Google Scholar]
- 84. Cheng I.-F., Chang H.-C., Hou D., and Chang H.-C., Biomicrofluidics 1(2), 021503 (2007). 10.1063/1.2723669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Abdul Razak M. A., Hoettges K. F., Fatoyinbo H. O., Labeed F. H., and Hughes M. P., Biomicrofluidics 7(6), 064110 (2013). 10.1063/1.4842395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Fatoyinbo H. O., Kamchis D., Whattingham R., Ogin S. L., and Hughes M. P., IEEE Trans. Biomed. Eng. 52(7), 1347–1349 (2005). 10.1109/TBME.2005.847553 [DOI] [PubMed] [Google Scholar]
- 87. Patel S., Showers D., Vedantam P., Tzeng T.-R., Qian S., and Xuan X., Biomicrofluidics 6(3), 034102 (2012). 10.1063/1.4732800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Hu X., Bessette P. H., Qian J., Meinhart C. D., Daugherty P. S., and Soh H. T., Proc. Natl. Acad. Sci. U. S. A. 102(44), 15757–15761 (2005). 10.1073/pnas.0507719102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Cristofanilli M., Budd G. T., Ellis M. J., Stopeck A., Matera J., Miller M. C., Reuben J. M., Doyle G. V., Allard W. J., Terstappen L. W. M. M., and Hayes D. F., New England J. Med. 351(8), 781–791 (2004). 10.1056/NEJMoa040766 [DOI] [PubMed] [Google Scholar]
- 90. Allard W. J., Matera J., Miller M. C., Repollet M., Connelly M. C., Rao C., Tibbe A. G. J., Uhr J. W., and Terstappen L. W. M. M., Clin. Cancer Res. 10(20), 6897–6904 (2004). 10.1158/1078-0432.CCR-04-0378 [DOI] [PubMed] [Google Scholar]
- 91. Song E.–Q., Hu J., Wen C.-Y., Tian Z.-Q., Yu X., Zhang Z.-L., Shi Y.-B., and Pang D.-W., ACS Nano 5(2), 761–770 (2011). 10.1021/nn1011336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Talasaz A. H., Powell A. A., Huber D. E., Berbee J. G., Roh K.-H., Yu W., Xiao W., Davis M. M., Pease R. F., and Mindrinos M. N., Proc. Natl. Acad. Sci. 106(10), 3970–3975 (2009). 10.1073/pnas.0813188106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Wang X., Qian X., Beitler J. J., Chen Z. G., Khuri F. R., Lewis M. M., Shin H. J. C., Nie S., and Shin D. M., Cancer Res. 71(5), 1526–1532 (2011). 10.1158/0008-5472.CAN-10-3069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Gupta V., Jafferji I., Garza M., Melnikova V. O., Hasegawa D. K., Pethig R., and Davis D. W., Biomicrofluidics 6(2), 024133 (2012). 10.1063/1.4731647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Shim S., Stemke-Hale K., Tsimberidou A. M., Noshari J., Anderson T. E., and Gascoyne P. R. C., Biomicrofluidics 7(1), 011807 (2013). 10.1063/1.4774304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Gascoyne P. R. C., Noshari J., Anderson T. J., and Becker F. F., Electrophoresis 30(8), 1388–1398 (2009). 10.1002/elps.200800373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Shim S., Gascoyne P., Noshari J., and Hale K. S., Integrative Biol. 3(8), 850–862 (2011). 10.1039/c1ib00032b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Yang F., Yang X., Jiang H., Bulkhaults P., Wood P., Hrushesky W., and Wang G., Biomicrofluidics 4(1), 013204 (2010). 10.1063/1.3279786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Dharmasiri U., Njoroge S. K., Witek M. A., Adebiyi M. G., Kamande J. W., Hupert M. L., Barany F., and Soper S. A., Anal. Chem. 83(6), 2301–2309 (2011). 10.1021/ac103172y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Vaidyanathan R., Rauf S., Dray E., Shiddiky M. J. A., and Trau M., Chem.- Eur. J. 20(13), 3724–3729 (2014). 10.1002/chem.201304590 [DOI] [PubMed] [Google Scholar]
- 101. Smith J. P., Huang C., and Kirby B. J., Biomicrofluidics 9(1), 014116 (2015). 10.1063/1.4908049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. García-Sánchez P., Ramos A., Green N., and Morgan H., IEEE Trans. Dielectr. Electr. Insul. 13(3), 670–677 (2006). 10.1109/TDEI.2006.1657983 [DOI] [Google Scholar]
- 103. Yang H., Jiang H., Ramos A., and García-Sánchez P., Microfluid. Nanofluid. 7(6), 767–772 (2009). 10.1007/s10404-009-0434-5 [DOI] [Google Scholar]
- 104. Paul P. H., Arnold D. W., and Rakestraw D. J., paper presented at the Micro Total Analysis Systems' 98, 1998. [Google Scholar]
- 105. Brask A., Goranović G., and Bruus H., Sens. Actuators, B 92(1), 127–132 (2003). 10.1016/S0925-4005(03)00130-8 [DOI] [Google Scholar]
- 106. Studer V., Pepin A., Chen Y., and Ajdari A., Microelectron. Eng. 61, 915–920 (2002). 10.1016/S0167-9317(02)00518-X [DOI] [Google Scholar]
- 107. Arnold D. W., Paul P. H., and Schoeniger J. S., “ Method for eliminating gas blocking in electrokinetic pumping systems,” U.S. patent 6,287,440 B1 (2001).
- 108. Selvaganapathy P., Ki Y.-S. L., Renaud P., and Mastrangelo C. H., J. Microelectromech. Syst. 11(5), 448–453 (2002). 10.1109/JMEMS.2002.803415 [DOI] [Google Scholar]
- 109. Krishnamoorthy G., Carlen E. T., Bomer J. G., Wijnperlé D., deBoer H. L., van den Berg A., and Schasfoort R. B. M., Lab Chip 10(8), 986–990 (2010) 10.1039/C000705F. [DOI] [PubMed] [Google Scholar]
- 110. Dodge A., Fluri K., Verpoorte E., and de Rooij N. F., Anal. Chem. 73(14), 3400–3409 (2001). 10.1021/ac0015366 [DOI] [PubMed] [Google Scholar]
- 111. Whitaker K. W. and Sepaniak M. J., Electrophoresis 15(1), 1341–1345 (1994). 10.1002/elps.11501501205 [DOI] [PubMed] [Google Scholar]
- 112. Adams A. A., Okagbare P. I., Feng J., Hupert M. L., Patterson D., Göttert J., McCarley R. L., Nikitopoulos D., Murphy M. C., and Soper S. A., J. Am. Chem. Soc. 130(27), 8633–8641 (2008). 10.1021/ja8015022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Balasubramanian P., Yang L., Lang J. C., Jatana K. R., Schuller D., Agrawal A., Zborowski M., and Chalmers J. J., Mol. Pharmaceutics 6(5), 1402–1408 (2009). 10.1021/mp9000519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Nagrath S., Sequist L. V., Maheswaran S., Bell D. W., Irimia D., Ulkus L., Smith M. R., Kwak E. L., Digumarthy S., Muzikansky A., Ryan P., Balis U. J., Tompkins R. G., Haber D. A., and Toner M., Nature 450(7173), 1235–1239 (2007). 10.1038/nature06385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Schiro P. G., Zhao M., Kuo J. S., Koehler K. M., Sabath D. E., and Chiu D. T., Angew. Chem., Int. Ed. 51(19), 4618–4622 (2012). 10.1002/anie.201108695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Stott S. L., Hsu C.-H., Tsukrov D. I., Yu M., Miyamoto D. T., Waltman B. A., Rothenberg S. M., Shah A. M., Smas M. E., Korir G. K., Floyd F. P., Gilman A. J., Lord J. B., Winokur D., Springer S., Irimia D., Nagrath S., Sequist L. V., Lee R. J., Isselbacher K. J., Maheswaran S., Haber D. A., and Toner M., Proc. Natl. Acad. Sci. USA 107(43), 18392–18397 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Wang S., Liu K., Liu J., Yu Z. T. F., Xu X., Zhao L., Lee T., Lee E. K., Reiss J., Lee Y.-K., Chung L. W. K., Huang J., Rettig M., Seligson D., Duraiswamy K. N., Shen C. K. F., and Tseng H.-R., Angew. Chem. Int. Ed. 50(13), 3084–3088 (2011). 10.1002/anie.201005853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Wang S., Wang H., Jiao J., Chen K.-J., Owens G. E., Kamei K.-i., Sun J., Sherman D. J., Behrenbruch C. P., Wu H., and Tseng H.-R., Angew. Chem. Int. Ed. 48(47), 8970–8973 (2009). 10.1002/anie.200901668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Xu T., Lu B., Tai Y.-C., and Goldkorn A., Cancer Res. 70(16), 6420–6426 (2010). 10.1158/0008-5472.CAN-10-0686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Riethdorf S., Müller V., Zhang L., Rau T., Loibl S., Komor M., Roller M., Huober J., Fehm T., Schrader I., Hilfrich J., Holms F., Tesch H., Eidtmann H., Untch M., von Minckwitz G., and Pantel K., Clin. Cancer Res. 16(9), 2634–2645 (2010). 10.1158/1078-0432.CCR-09-2042 [DOI] [PubMed] [Google Scholar]
- 121. Kurkuri M. D., Al-Ejeh F., Shi J. Y., Palms D., Prestidge C., Griesser H. J., Brown M. P., and Thierry B., J Mater. Chem. 21(24), 8841–8848 (2011). 10.1039/c1jm10317b [DOI] [Google Scholar]
- 122. Gagnon Z., Mazur J., and Chang H.-C., Lab Chip 10(6), 718–726 (2010). 10.1039/b917220c [DOI] [PubMed] [Google Scholar]
- 123. Kang J. H., Krause S., Tobin H., Mammoto A., Kanapathipillai M., and Ingber D. E., Lab Chip 12(12), 2175–2181 (2012). 10.1039/c2lc40072c [DOI] [PubMed] [Google Scholar]
- 124. Wang S., Wang H., Jiao J., Chen K. J., Owens G. E., Kamei K. i., Sun J., Sherman D. J., Behrenbruch C. P., and Wu H., Angew. Chem. 121(47), 9132–9135 (2009). 10.1002/ange.200901668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Li L., Liu W., Wang J., Tu Q., Liu R., and Wang J., Electrophoresis 31(18), 3159–3166 (2010). 10.1002/elps.201000139 [DOI] [PubMed] [Google Scholar]
- 126. Kim M. S., Sim T. S., Kim Y. J., Kim S. S., Jeong H., Park J.-M., Moon H.-S., Kim S. I., Gurel O., and Lee S. S., Lab Chip 12(16), 2874–2880 (2012). 10.1039/c2lc40065k [DOI] [PubMed] [Google Scholar]
- 127. Lin H. K., Zheng S., Williams A. J., Balic M., Groshen S., Scher H. I., Fleisher M., Stadler W., Datar R. H., and Tai Y.-C., Clin. Cancer Res. 16(20), 5011–5018 (2010). 10.1158/1078-0432.CCR-10-1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Zheng S., Lin H., Liu J.-Q., Balic M., Datar R., Cote R. J., and Tai Y.-C., J. Chromatogr. A 1162(2), 154–161 (2007). 10.1016/j.chroma.2007.05.064 [DOI] [PubMed] [Google Scholar]
- 129. Wan Y., Mahmood M., Li N., Allen P. B., Kim Y. t., Bachoo R., Ellington A. D., and Iqbal S. M., Cancer 118(4), 1145–1154 (2012). 10.1002/cncr.26349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Ozkumur E., Shah A. M., Ciciliano J. C., Emmink B. L., Miyamoto D. T., Brachtel E., Yu M., Chen P.-I., Morgan B., Trautwein J., Kimura A., Sengupta S., Stott S. L., Karabacak N. M., Barber T. A., Walsh J. R., Smith K., Spuhler P. S., Sullivan J. P., Lee R. J., Ting D. T., Luo X., Shaw A. T., Bardia A., Sequist L. V., Louis D. N., Maheswaran S., Kapur R., Haber D. A., and Toner M., “ Inertial focusing for tumor antigen-dependent and -independent sorting of rare circulating tumor cells,” Sci. Trans. Med. 5, 179ra49 (2013). 10.1126/scitranslmed.3005616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Hur S. C., Mach A. J., and Di Carlo D., Biomicrofluidics 5(2), 022206 (2011). 10.1063/1.3576780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Lv P., Tang Z., Liang X., Guo M., and Han R. P. S., Biomicrofluidics 7(3), 034109 (2013). 10.1063/1.4808456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Kuntaegowdanahalli S. S., Bhagat A. A. S., Kumar G., and Papautsky I., Lab Chip 9(20), 2973–2980 (2009). 10.1039/b908271a [DOI] [PubMed] [Google Scholar]
- 134. Gleghorn J. P., Pratt E. D., Denning D., Liu H., Bander N. H., Tagawa S. T., Nanus D. M., Giannakakou P. A., and Kirby B. J., Lab Chip 10(1), 27–29 (2010). 10.1039/B917959C [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Lien K.-Y., Chuang Y.-H., Hung L.-Y., Hsu K.-F., Lai W.-W., Ho C.-L., Chou C.-Y., and Lee G.-B., Lab Chip 10(21), 2875–2886 (2010). 10.1039/c005178k [DOI] [PubMed] [Google Scholar]
- 136. Cheung L. S. L., Zheng X., Stopa A., Baygents J. C., Guzman R., Schroeder J. A., Heimark R. L., and Zohar Y., Lab Chip 9(12), 1721–1731 (2009). 10.1039/b822172c [DOI] [PubMed] [Google Scholar]
- 137. Wan Y., Tan J., Asghar W., Kim Y.-T., Liu Y., and Iqbal S. M., J. Phys. Chem. B 115(47), 13891–13896 (2011). 10.1021/jp205511m [DOI] [PubMed] [Google Scholar]
- 138. Balasubramanian S., Kagan D., Jack Hu C. M., Campuzano S., Lobo-Castañon M. J., Lim N., Kang D. Y., Zimmerman M., Zhang L., and Wang J., Angew. Chem. Int. Ed. 50(18), 4161–4164 (2011). 10.1002/anie.201100115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Benitez J. J., Topolancik J., Tian H. C., Wallin C. B., Latulippe D. R., Szeto K., Murphy P. J., Cipriany B. R., Levy S. L., Soloway P. D., and Craighead H. G., Lab Chip 12(22), 4848–4854 (2012). 10.1039/c2lc40955k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Castro C. M., Ghazani A. A., Chung J., Shao H., Issadore D., Yoon T. J., Weissleder R., and Lee H., Lab Chip 14(1), 14–23 (2014). 10.1039/C3LC50621E [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Cheung L. S.-L., Zheng X., Wang L., Baygents J. C., Guzman R., Schroeder J. A., Heimark R. L., and Zohar Y., J. Micromech. Microeng. 21(5), 054033 (2011). 10.1088/0960-1317/21/5/054033 [DOI] [Google Scholar]
- 142. Fan Z. H. and Beebe D. J., Lab Chip 14(1), 12–13 (2014). 10.1039/C3LC90125D [DOI] [PubMed] [Google Scholar]
- 143. Gleghorn J. P., Smith J. P., and Kirby B. J., Phys. Rev. E 88(3), 032136 (2013). 10.1103/PhysRevE.88.032136 [DOI] [PubMed] [Google Scholar]
- 144. Hou S., Zhao L., Shen Q., Yu J., Ng C., Kong X., Wu D., Song M., Shi X., Xu X., OuYang W. H., He R., Zhao X. Z., Lee T., Brunicardi F. C., Garcia M. A., Ribas A., Lo R. S., and Tseng H. R., Angew. Chem. Int. Ed. 52(12), 3379–3383 (2013). 10.1002/anie.201208452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Hur S. C., Mach A. J., and Di Carlo D., Biomicrofluidics 5(2), 22206 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. King J. D., Casavant B. P., and Lang J. M., Lab Chip 14(1), 24–31 (2014). 10.1039/C3LC50741F [DOI] [PubMed] [Google Scholar]
- 147. Li X., Chen Y., and Li P. C., Lab Chip 11(7), 1378–1384 (2011). 10.1039/c0lc00626b [DOI] [PubMed] [Google Scholar]
- 148. Sheng W., Chen T., Kamath R., Xiong X., Tan W., and Fan Z. H., Anal. Chem. 84(9), 4199–4206 (2012). 10.1021/ac3005633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Thierry B., Kurkuri M., Shi J. Y., Lwin L. E., and Palms D., Biomicrofluidics 4(3), 32205 (2010). 10.1063/1.3480573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Pratt E. D., Huang C., Hawkins B. G., Gleghorn J. P., and Kirby B. J., Chem. Eng. Sci. 66(7), 1508–1522 (2011). 10.1016/j.ces.2010.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Yoon H. J., Kozminsky M., and Nagrath S., ACS Nano 8(3), 1995–2017 (2014). 10.1021/nn5004277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Yu M., Stott S., Toner M., Maheswaran S., and Haber D. A., J. Cell Biol. 192(3), 373–382 (2011). 10.1083/jcb.201010021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Vaidyanathan R., Rauf S., Shiddiky M. J. A., and Trau M., Biosens. Bioelectron. 61(0), 184–191 (2014). 10.1016/j.bios.2014.03.061 [DOI] [PubMed] [Google Scholar]
- 154. Hart R., Lec R., and Noh H. M., Sens. Actuators, B 147(1), 366–375 (2010). 10.1016/j.snb.2010.02.027 [DOI] [Google Scholar]
- 155. Hart R., Ergezen E., Lec R., and Noh H. M., Biosens. Bioelectron. 26(8), 3391–3397 (2011). 10.1016/j.bios.2010.12.038 [DOI] [PubMed] [Google Scholar]
- 156. Kim S. M., Burns M. A., and Hasselbrink E. F., Anal. Chem. 78(14), 4779–4785 (2006). 10.1021/ac060031y [DOI] [PubMed] [Google Scholar]
- 157. Morales M. C., Lin H., and Zahn J. D., Lab Chip 12(1), 99–108 (2012). 10.1039/C1LC20605B [DOI] [PubMed] [Google Scholar]
- 158. Oh S.-H., Lee S.-H., Kenrick S. A., Daugherty P. S., and Soh H. T., J. Proteome Res. 5(12), 3433–3437 (2006). 10.1021/pr060193a [DOI] [PubMed] [Google Scholar]
- 159. Gong J.-R., Small 6(8), 967–973 (2010). 10.1002/smll.200902132 [DOI] [PubMed] [Google Scholar]
- 160. Vaidyanathan R., van Leeuwen L. M., Rauf S., Shiddiky M. J. A., and Trau M., Sci. Rep. 5, 9756 (2015). 10.1038/srep09756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Gao Y., Sherman P. M., Sun Y., and Li D., Anal. Chim. Acta 606(1), 98–107 (2008). 10.1016/j.aca.2007.10.052 [DOI] [PubMed] [Google Scholar]
- 162. Nam J.-M., Thaxton C. S., and Mirkin C. A., Science 301(5641), 1884–1886 (2003). 10.1126/science.1088755 [DOI] [PubMed] [Google Scholar]
- 163. Niemeyer C. M., Adler M., and Wacker R., Trends Biotechnol. 23(4), 208–216 (2005). 10.1016/j.tibtech.2005.02.006 [DOI] [PubMed] [Google Scholar]
- 164. Mason J. T., Xu L., Sheng Z.-M., and O'Leary T. J., Nat. Biotechnol. 24(5), 555–557 (2006). 10.1038/nbt1201 [DOI] [PubMed] [Google Scholar]
- 165. Das J., Aziz M. A., and Yang H., J. Am. Chem. Soc. 128(50), 16022–16023 (2006). 10.1021/ja0672167 [DOI] [PubMed] [Google Scholar]
- 166. Hu M., Yan J., He Y., Lu H., Weng L., Song S., Fan C., and Wang L., ACS Nano 4(1), 488–494 (2010). 10.1021/nn901404h [DOI] [PubMed] [Google Scholar]
- 167. Tekin H. C. and Gijs M. A. M., Lab Chip 13(24), 4711–4739 (2013). 10.1039/c3lc50477h [DOI] [PubMed] [Google Scholar]
- 168. Yan J., Hu M., Li D., He Y., Zhao R., Jiang X., Song S., Wang L., and Fan C., Nano Res. 1(6), 490–496 (2008). 10.1007/s12274-008-8052-1 [DOI] [Google Scholar]
- 169. Mandel M. and Odijk T., Annu. Rev. Phys. Chem. 35(1), 75–108 (1984). 10.1146/annurev.pc.35.100184.000451 [DOI] [Google Scholar]
- 170. Takashima S., Biopolymers 5(10), 899–913 (1967). 10.1002/bip.1967.360051004 [DOI] [PubMed] [Google Scholar]
- 171. Ng J. K.-K., Feng H., and Liu W.-T., Anal. Chim. Acta 582(2), 295–303 (2007). 10.1016/j.aca.2006.09.016 [DOI] [PubMed] [Google Scholar]
- 172. Tu I.-P., Schaner M., Diehn M., Sikic B., Brown P., Botstein D., and Fero M., BMC Genomics 5(1), 64 (2004). 10.1186/1471-2164-5-64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Asbury C. L. and van den Engh G., Biophys. J. 74(2 Pt 1), 1024–1030 (1998). 10.1016/S0006-3495(98)74027-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Washizu M. and Kurosawa O., paper presented at the Conference Record of the 1989 IEEE Industry Applications Society Annual Meeting, 1989. [Google Scholar]
- 175. Asbury C. L., Diercks A. H., and van den Engh G., Electrophoresis 23(16), 2658–2666 (2002). [DOI] [PubMed] [Google Scholar]
- 176. Kreft J., Chen Y.-L., and Chang H.-C., Phys. Rev. E 77(3), 030801 (2008). 10.1103/PhysRevE.77.030801 [DOI] [PubMed] [Google Scholar]
- 177. Du J.-R., Juang Y.-J., Wu J.-T., and Wei H.-H., Biomicrofluidics 2(4), 044103 (2008). 10.1063/1.3037326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Cheng I. F., Senapati S., Cheng X., Basuray S., Chang H.-C., and Chang H.-C., Lab Chip 10(7), 828–831 (2010). 10.1039/b925854j [DOI] [PubMed] [Google Scholar]
- 179. Swami N., Chou C.-F., Ramamurthy V., and Chaurey V., Lab Chip 9(22), 3212–3220 (2009). 10.1039/b910598k [DOI] [PubMed] [Google Scholar]
- 180. Basuray S., Senapati S., Aijian A., Mahon A. R., and Chang H.-C., ACS Nano 3(7), 1823–1830 (2009). 10.1021/nn9004632 [DOI] [PubMed] [Google Scholar]
- 181. Yeo W.-H., Chung J.-H., Liu Y., and Lee K.-H., J. Phys. Chem. B 113(31), 10849–10858 (2009). 10.1021/jp900618t [DOI] [PubMed] [Google Scholar]
- 182. Wang Y., Chang T.-C., Stoddart P. R., and Chang H.-C., Biomicrofluidics 8(2), 021101 (2014). 10.1063/1.4869694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Amblard F., Yurke B., Pargellis A., and Leibler S., Rev. Sci. Instrum. 67(3), 818–827 (1996). 10.1063/1.1146816 [DOI] [PubMed] [Google Scholar]
- 184. Binnig G., Quate C. F., and Gerber C., Phys. Rev. Lett. 56(9), 930–933 (1986). 10.1103/PhysRevLett.56.930 [DOI] [PubMed] [Google Scholar]
- 185. Rief M., Oesterhelt F., Heymann B., and Gaub H. E., Science 275(5304), 1295–1297 (1997). 10.1126/science.275.5304.1295 [DOI] [PubMed] [Google Scholar]
- 186. Perkins T., Smith D., Larson R., and Chu S., Science 268(5207), 83–87 (1995). 10.1126/science.7701345 [DOI] [PubMed] [Google Scholar]
- 187. Wälti C., Germishuizen W. A., Tosch P., Kaminski C. F., and Davies A. G., J. Phys. D: Appl. Phys. 40(1), 114 (2007). 10.1088/0022-3727/40/1/S16 [DOI] [Google Scholar]
- 188. Valadi H., Ekstrom K., Bossios A., Sjostrand M., Lee J. J., and Lotvall J. O., Nat. Cell Biol. 9(6), 654–659 (2007). 10.1038/ncb1596 [DOI] [PubMed] [Google Scholar]
- 189. Thery C., Ostrowski M., and Segura E., Nat. Rev Immunol. 9(8), 581–593 (2009). 10.1038/nri2567 [DOI] [PubMed] [Google Scholar]
- 190. de Vrij J., Maas S. L. N., van Nispen M., Sena-Esteves M., Limpens R. W. A., Koster A. J., Leenstra S., Lamfers M. L., and Broekman M. L. D., Nanomedicine 8(9), 1443–1458 (2013). 10.2217/nnm.12.173 [DOI] [PubMed] [Google Scholar]
- 191. Huang X., Yuan T., Tschannen M., Sun Z., Jacob H., Du M., Liang M., Dittmar R., Liu Y., Liang M., Kohli M., Thibodeau S., Boardman L., and Wang L., BMC Genomics 14(1), 319 (2013). 10.1186/1471-2164-14-319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Couzin J., Science 308(5730), 1862–1863 (2005). 10.1126/science.308.5730.1862 [DOI] [PubMed] [Google Scholar]
- 193. Lane R. E., Korbie D., Anderson W., Vaidyanathan R., and Trau M., Sci. Rep. 5, 7639 (2015). 10.1038/srep07639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Chen C., Skog J., Hsu C. H., Lessard R. T., Balaj L., Wurdinger T., Carter B. S., Breakefield X. O., Toner M., and Irimia D., Lab Chip 10(4), 505–511 (2010). 10.1039/B916199F [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Goda T., Masuno K., Nishida J., Kosaka N., Ochiya T., Matsumoto A., and Miyahara Y., Chem. Commun. 48(98), 11942–11944 (2012). 10.1039/c2cc36111f [DOI] [PubMed] [Google Scholar]
- 196. Im H., Shao H., Park Y. I., Peterson V. M., Castro C. M., Weissleder R., and Lee H., Nat. Biotechnol. 32(5), 490–495 (2014). 10.1038/nbt.2886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Kanwar S. S., Dunlay C. J., Simeone D. M., and Nagrath S., Lab Chip 14(11), 1891–1900 (2014). 10.1039/c4lc00136b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Shao H., Chung J., Balaj L., Charest A., Bigner D. D., Carter B. S., Hochberg F. H., Breakefield X. O., Weissleder R., and Lee H., Nat. Med. 18(12), 1835–1840 (2012). 10.1038/nm.2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Wang Z., Wu H. J., Fine D., Schmulen J., Hu Y., Godin B., Zhang J. X., and Liu X., Lab Chip 13(15), 2879–2882 (2013). 10.1039/c3lc41343h [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Vaidyanathan R., Naghibosadat M., Rauf S., Korbie D., Carrascosa L. G., Shiddiky M. J. A., and Trau M., Anal. Chem. 86(22), 11125–11132 (2014). 10.1021/ac502082b [DOI] [PubMed] [Google Scholar]
- 201. Wei F., Yang J., and Wong D. T., Biosens. Bioelectron. 44, 115–121 (2013). 10.1016/j.bios.2012.12.046 [DOI] [PMC free article] [PubMed] [Google Scholar]