Abstract
Inorganic nanowires are among the most attractive functional materials emerged in the past two decades and have demonstrated applications to information technology and energy conversion, but the utility in biological or biomedical research remains relatively under-explored. Although nanowire-based sensors have been frequently reported for biomolecular detection, interfacing nanowire arrays and living mammalian cells for direct analysis of cellular functions was a very recent endeavor. Cell-penetrating nanowires enabled effective delivery of biomolecules, electrical and optical stimulation and recording of intra-cellular signals over a long period of time. Non-penetrating, high-density nanowire arrays display rich interactions between the nanostructured substrate and the micro/nano-scale features of cell surface. Such interactions enabled efficient capture of rare cells including circulating tumor cells and trafficking leukocytes from complex biospecimens. It also served as a platform for probing cell traction force and neuronal guidance. This article reviews the most recent advance in the field that exploits nanowire arrays (both penetrating and non-penetrating) to perform rapid analysis of cellular functions potentially for disease diagnosis and monitoring.
Keywords: nanowire array, intracellular delivery, rare cell analysis, cell-substrate interaction
1. Introduction
In the past two decades, inorganic nanowires have been a class of new material with intense research interest owing to the potential applications in nanoelectronics, nanophotonics, photovoltaics, electrochemical energy conversion, and catalytic water splitting.[1,2] The application to biological and biomedical research remains relatively limited. The majority of the studies were biomolecular detection using nanowire-based field effect or electrochemical sensors.[3,4] The utility of nanowires in functional interrogation of living cells emerged very recently. In 2007, Kim et al. reported the first demonstration of cell-penetrating nanowire arrays for physical delivery of genes into living mammalian stem cells.[5] Since then, a range of nanowires either synthetically or lithographically fabricated, and their variants including nanoneedles, nanopillars and nanostraws, were explored for intracellular delivery, electrical or optical stimulation and probing (Figure 1 red). All these works took advantage of the phenomenon that nanometer-sized wires permit full penetration into living cells but cause minimal disruption of cell membrane integrity and thereby negligible cytotoxic effect. In 2009, Wang et al. found that the non-penetrating high-density nanowire array functionalized with antibodies against cell surface antigen allowed for high efficiency capture of target cells, e.g., rare circulating tumor cells, presumably due to the enhanced interaction between nanotopographic structures and the micro/nanoscale structures on cell surface such as microvilli.[6] Lee et al. reported the bulk-scale separation of primary CD4+ T lymphocytes from a mixture of splenocytes.[7] These two studies evoked a new direction of research that utilizes the interfacing of live cell surface with non-penetrating nanowire arrays to conduct efficient capture, separation, and subsequent molecular and biomechanical characterization of rare cells including a range of pathophysiologically important cell types that were difficult to study due to their paucity (Figure 1 blue). Although it has been known for over fifteen years that the nanometer-scale physical or chemical cues dictate cell adhesion and fate decision that was covered by other review articles,[8–11] and the use of nanostructured surface for basic cell behavior analysis,[12,13] the utilization of nanowires or nanotopography for rapid analysis of cells and cellular functions potentially for disease diagnosis and monitoring represents a new and differentiated direction, which is the main topic of this paper. In addition, we would like to provide a retrospective view of the history of this field and our opinion on the future outlooks.
Figure 1.
Summary - interfacing inorganic nanowire arrays and living cells for a wide range of biological and biomedical applications. In general, this can be classified into two major categories: (1) cell-penetrating nanowire array (red) for biomolecular delivery, intracellular stimulation and probing; (2) non-penetrating nanowire array (blue) for high efficiency capture, separation and molecular phenotyping of rare cells and the biomechanical characterization.
2. Cell Penetrating Nanowires and Nanostraws for Gene and Biomolecular Delivery
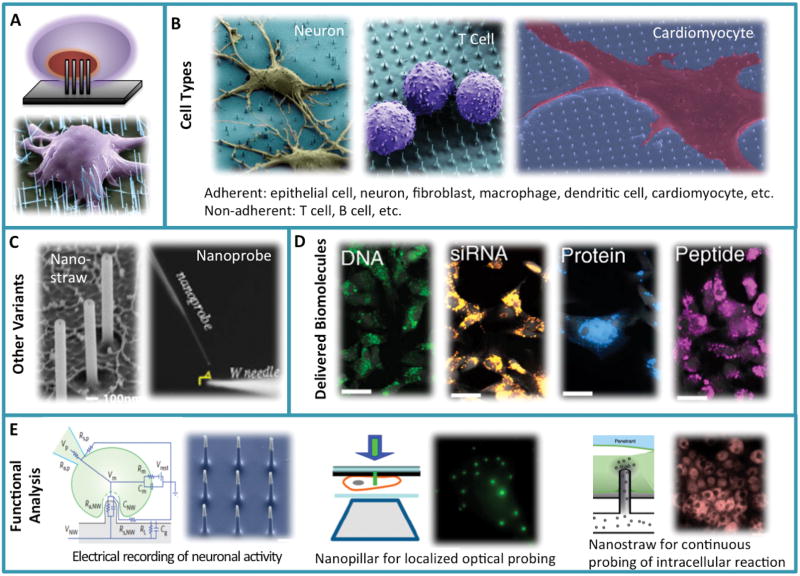
It was not so intuitive to believe nanometer-sized wires can penetrate living mammalian cells without “killing” or damaging them until the report by Kim and colleagues in 2007 that demonstrated, for the first time, the placement of mammalian cells on a bed of diluted vertical silicon nanowires resulted in minimally “invasive” penetration and successful delivery of gene constructs from the nanowire surface directly to the nucleus (Figure 2A).[5] Mouse embryonic stem (mES) cell–derived cardiomyocytes interfaced with an array of silicon nanowires showed the differentiation timeline comparable to the same cells grown on gelatin coated tissue culture flask. Nanowires functionalized with a polymer sheath and then loaded with the bare plasmid DNA encoding green fluorescence protein (GFP) can penetrate and successful transfect HEK 293T cells without the use of any viral delivery vesicles. In 2010, Shalek et al. further developed this technology and reported the efficient and universal delivery of a range of biomolecules into immortalized and primary mammalian cells including neurons and immune cells using surface-modified vertical silicon nanowires (Figure 2B left two panels).[14] This generalized platform was used to guide neuronal progenitor growth with small molecules, knock down the transcript levels by delivering siRNAs, inhibit apoptosis by delivering anti-apoptotic peptides, and introduce targeted proteins to specific organelles (Figure 2D). Using a microarray printer to dispense siRNA in the specific microscale regions further enabled spatially controlled delivery into select cells. Thus vertical nanowire arrays provide a powerful delivery modality for administering biomolecular perturbants directly into the cytoplasm or even nucleus. The negligible toxicity of nanowire penetration is advantageous for transfection of immune cells for a circuit level understanding of immune response, systematic discovery of signaling components, i.e., involved in the Toll-like receptor (TLR) activation in dendritic cells (DCs) and investigating the role of intracellular signaling pathway in chronic lymphocytic leukemia (CLL),[15,16] and systematical identification of key genes and pathways in leukemogenesis.[17]
Figure 2.
Cell-penetrating nanowire arrays. (A) Schematic illustration of nanowire penetration into living cells and the first demonstration. Reproduced with permission.[5] 2007, American Chemical Society. (B) A range of adherent and non-adherent cells can be interfaced with and penetrated by nanowire arrays. Reproduced with permission.[14, 34] 2010, National Academy of Sciences, USA. (C) Variants of cell penetrating nanowires including nanostraws (Reproduced with permission.[25] 2011, American Chemical Society.) and nanoprobe integrated on a scanning optical tip (Reproduced with permission.[33] 2012, Macmillan Publishers Limited.). (D) A range of biomolecules including DNA, siRNA, protein and peptide can be delivered via the generic nanowire array delivery platform Reproduced with permission.[14] 2010, National Academy of Sciences, USA. (E) Functional study of intracellular signals. Left: cell-penetrating nanoneedle array for electrical stimulation and recording of intracellular action potential. Reproduced with permission.[40] 2012, Macmillan Publishers Limited. Middle: cell-penetrating nanopillars act as subwavelength waveguides and enabled highly localized excitation and fluorescence imaging of single molecule events inside the living cells. Reproduced with permission.[32] 2011, National Academy of Sciences, USA. Right: nanostraws integrated with a microfluidic system permits direct fluid access and molecular delivery into cytosol in a continuous or repeated manner. Reproduced with permission.[46] 2014, Macmillan Publishers Limited.
Nanowire arrays opened an entirely new avenue for intracellular functional analysis and interrogation due to the ability for direct cell membrane penetration[5,18–21] and effective biomolecular delivery into cell interior,[14,22–24] but these methods are restricted to molecules that can be conjugated or otherwise immobilized on the nanowire surface, and offer little precise temporal or concentration control. A variant of cell-penetrating nanowires, nanostraws, were developed to improve the delivery efficiency and the control of delivery kinetics (Figure 2C). VanDersarl et al. fabricated vertical nanostraw array with the hollow fluid conduit inside all the way through both the nanostraw and the handling substrate, giving the cells cultured on top a direct and continuous fluid access to the outside environment.[25] His group then demonstrated long-term, sequential delivery of different biomolecular species and continuous monitoring of cell signaling response without the need for repeated rupture of cell membrane. Further integration of nanostraws and a microfluidic system permits more precise control and measurement of signaling dynamics in individual cells. This integrated nanostraw array microfluidic system was also utilized for electroporation with enhanced delivery efficiency, transfection uniformity, and cell viability.[26] Transfection by electroporation relies on applied electrical fields to create transient holes in cell membrane and drive biomolecules into the cytosol.[27–31] Due to the tight interface between the cell membrane and each nanostraw, the electric field is localized and induces only transient and focused membrane permeability which increased transfection efficiency and reduced cell damage. This platform allows for spatial and temporal control of electroporation of individual cells as opposed to the entire cell population.
3. Functional Nanowires Penetrating Cells for Optical or Electrical Stimulation and Recording
In addition to passive penetration of cells with inert nanowires, optically or electrically active materials can be used to fabricate nanowire arrays and perform functional interrogation of biomolecular signals inside cells (Figure 2E). Xie et al. fabricated a transparent vertical silicon dioxide nanopillar array that act as sub-wavelength waveguides to channel light into or out of the cell to perform stimulation and probing of localized intracellular fluorescence signals.[32] When illuminating the nanopillar with an external light source, the nanopillars restrict the propagation of light and generate an evanescent wave in the cell interior tens of nanometers within the vicinity of nanopillars, enabling highly localized single-molecule detection at high fluorophore concentrations. Upon functionalization with antibodies or ligands, the nanopillars can locally recruit proteins of interest, excite through nanopillar waveguide, and probe the protein dynamics in living cells with high specificity. Yan et al. combined the advantages of nanowire waveguide and a fibre-optic fluorescence imaging technique to develop a tin oxide nanowire-based single-cell endoscopy system for optical probing inside live cells and the spatiotemporal delivery of payloads into intracellular sites with minimal perturbation to the normal cellular functions.[33]
Semiconducting or metallic nanowires penetrating into cells allow for intracellular electrical stimulation and recoding of action potential in living cells with submicrometer resolution. Xie et al. used platinum nanopillar electrodes to record intracellular action potentials from HL-1 cardiomyocytes.[34] Interestingly, they found that cell membrane lipid layer could wrap around the short nanopillars and reduce electrical conductivity, they further designed a hollow nanotube geometry that gave rise to enhanced cell-nanopillar electrical coupling and resulted in substantially increased electrical signals compared to solid nanopillars.[35] Cell membrane not only curves inward to wrap around nano-objects but also protrudes outward into the holes as small as 1nm in diameter.[36,37] Such strong interaction between the plasma membrane and the nanotube electrodes leads to an increase in the recorded signal amplitude and one-to-two orders of magnitude increase of the intracellular access duration for nanotube electrodes compared to nanopillar electrodes. In addition, electrically assisted nanowire/cell interfacing makes nanowire arrays a promising tool to decipher the neuronal codes within a neural network.[38,39] Silicon nanofabrication techniques enable the scalability of such a platform to provide simultaneous, high-fidelity interfacing with a large array (hundreds of) of interconnected neurons. Robinson et al. fabricated vertical nanowire electrode arrays for intracellular interfacing to neuronal circuits.[40] Silicon nanowires were functionalized by sputter deposition of metal on the tips to realize excellent electrical conduction for neural stimulation and recording. The ability to perform multiplex stimulation and nanometer-resolution recording and the compatibility with conventional patch-clamp and fluorescence microscopy techniques enabled their nanowire electrode array for measuring functional connectivity in neuronal circuits.[41] Surface patterned horizontal electrode arrays were also used for recording neuronal activities, but several challenges exist with that approach including the difficulty for the electrodes to consistently locate the same neuron and measure the activity over a long period due in part to the mobility of neurons on the substrate and the lack of physical neuron-to-electrode registry.[42] Xie et al. used nanopillar arrays to pin individual neurons, prevent the migration, and enabled extracellular measurement of neural activity over a prolonged period of time using the patterned multielectrode arrays.[43]
4. Mechanistic Insights: How do Inorganic Nanowires Penetrate Living Cells?
Despite the success of biomolecular delivery and intracellular probing using vertically aligned nanowires, the exact structure of the nanowire-cell membrane interface is yet to be fully elucidated. There has been a debate on whether nanowires can truly penetrate through the cell membrane to gain full access to the cell interior or just deform the cell membrane. Hanson et al. examined neurons grown on nanopillars with diameter 50–500nm and heights 5–2μm.[44] A cross-sectional view of the cell/nanopillar interface with transmission electron microscopy (TEM) showed that cell membrane wrapped around the entirety of the nanopillar in most cases without full penetration into the interior of the cell when the nanopillars are less than 300 nm in diameter. It was possible that the cell-nanopillar interaction is time-dependent; there is a transient full penetration shortly after seeding cells on the nanowire arrays but it was later re-sealed with the lipid bilayer drift driven by surface tension. Aalipour et al. further account for the interaction of plasma membrane and actin cytoskeleton as a synergistic barriers to nanowire cell penetration.[45] Nanostraws were studied in this case and several modes of interface interaction were observed: (a) non-penetrant but in contact with an intact lipid bilayer, (b) passing through the cell membrane but not into the cytoskeleton, and (c) passing through the cell membrane and into the cytoskeleton. Their study confirmed that only ~7% of nanostraws successfully penetrated cells to provide cytosolic access, which was measured on a nanostraw-microfluidic platform through the delivery of Co2+.[46] They presented a mechanical continuum model of elastic cell membrane penetration through two mechanisms, namely through “impaling” as cells land onto a bed of nanowires, and through “adhesion-mediated” penetration, which occurs as cells spread on the substrate ant generate adhesion force.[47] In “impaling mechanism”, the penetration is driven by cellular gravitational force during the initial cell plating. Impaling penetration is inefficient without the assistance of externally force. In “adhesion mechanism”, cells adhere to the substrate and the adhesion force induces penetration.[48] Cell-substrate adhesion can generate stronger forces, making penetration more likely, yet this is highly dependent on the geometry of nanowire arrays and cell rigidity. The surface charge density of nanowires plays an important role in cell-substrate interaction. It could completely change the impaling mechanism and/or the adhesion mechanism. The surface charge property of a cell varies substantially with the cell type and state. Matching the electric charge characteristics of nanowires with the cell surface property can enhance the interaction between cell membrane and nanowire arrays.[49] Further studies using new tools with nanometer resolution (e.g., super-resolution imaging) are highly desired to better understand the mechanism of nanowire penetration into living cells.
5. Dense Nanowire Arrays for Rare Cell Capture, Separation, and Analysis
Dense nanowire arrays can be fabricated not only by chemical process but also precisely controlled nanolithographic patterning.[50] Placing live cells on such nanowire arrays often do not cause penetration through cell membrane (Figure 3A), but the biomechanical cues from cell-nanostructure interaction can mediate a range of cellular response including mechanotransduction, altered gene expression and release of tissue factors or cytokines, and alter the fate decision of a cell in the long term such as proliferation, differentiation, and cell death.[51,52] However, the early stage interaction between nanowire array and cell surface has not been found particularly useful until recently it was shown to give rise to enhanced efficiency for surface-bound immunocapture of specific cell types as compared to the conventional panning methods, opening new opportunities to isolate and characterize rare cell populations.
Figure 3.
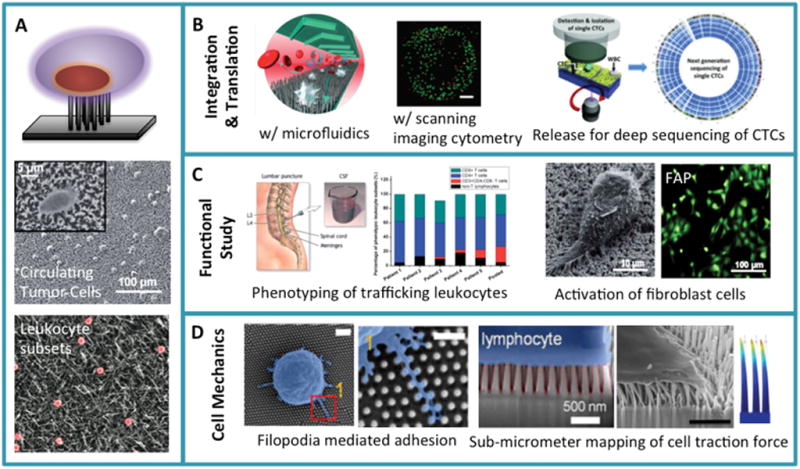
Non-penetrating nanowire arrays. (A) Schematic illustration of interfacing living cells with a dense array of nanowires that do not penetrate into a cell’s interior. It also shows the two pioneer works demonstrating the use of such nanowire arrays for high efficiency capture of circulating tumor cells (Reproduced with permission.[6] 2009, Wiley-VCH.) and the separation of immune cell subsets (Reproduced with permission.[7] 2010, American Chemical Society.). (B) Integration of nanowire rare cell capture with microfluidics (Reproduced with permission.[67] 2011, Wiley-VCH.), laser scanning cytometry (Reproduced with permission.[73] 2012, American Chemical Society.), or informative molecular characterization (Reproduced with permission.[70] 2013, Wiley-VCH.) represents important endeavors towards the translation to clinical applications. (C) Phenotypic or functional characterization of rare cells using nanowire arrays. Examples include the molecular typing of trafficking leukocytes from cerebrospinal fluid of patients with Alzheimer’s Disease (Reproduced with permission.[82] 2014, The Royal Society of Chemistry.) and the study of fibroblast activation (Reproduced with permission.[88] 2014, The Royal Society of Chemistry.). (D) Study of cell surface microstructure (microvilli, filopodia, etc) interacting with the nanopillar surface (left two panels. Reproduced with permission.[93] 2014, American Chemical Society.) and the measurement of nanopillar deflection to map cell traction force generated by T lymphocytes (third panel. Reproduced with permission.[90] 2013, Springer.) or fibroblasts. The right most panel show the finite element simulation of nanorod deflection upon interfacial traction force.
Efficient separation of specific cell populations are a critical process in numerous clinical and biological applications as quantity and functions of specific cell types have become important ‘biomarkers’ for disease progression or therapeutic outcome.[53] The existing methods for cell separation include size-based filtration, nanomagnetic bead enrichment, and fluorescence activated cell sorting[54] and a number of microfluidic-based sorting platforms,[55–57] but none of these methods are effective for separating very rare cell populations (below 1 per 1000) from highly heterogeneous samples. Surface-bound immuno-capture is dependent on the antibody loading density and the effective surface contact area between the substrate and the cells. It was not intuitive to propose using nanostructured surface for immune-capture because the size of mammalian cells is several to tens of micrometers rather than nanometers. Interestingly, the surface of many types of cells is not smooth but comprised of nanometer-sized features such as microvilli. The nanostructured surface, which shares nanoscale feature dimensions comparable to microvilli, may induce enhanced local topographic interactions and thereby promote the antibody-surface antigen binding, enhance adhesion, reduce motility, and increase cell capture efficiency.[48] Based upon this hypothesis, Wang et al. reported on the use of a nanostructured substrate functionalized with anti-EpCAM to capture circulating tumor cells with significantly improved capture efficiency and specificity (Figure 3A).[6] Kim et al. used a streptavidin-functionalized silicon nanowire array to separate at the bulk scale the CD4+ helper T lymphocytes labeled with biotinylated anti-mouse CD4 antibody (Figure 3A).[7]
5.1. Circulating Tumor Cells
It is the metastatic not the primary tumors that cause incurable disease and account for more than 90% of cancer-related mortality.[58] Cancer cells that are shed from primary and travel to distant parts of the body via bloodstreams are known as circulating tumor cells (CTCs), which are thought to mediate the hematological spread of cancer and contribute to metastatic progression.[59] CTCs are becoming a potential alternative to invasive biopsies to detect and manage metastatic epithelial (non-hematological) cancers[60] but the major challenge arises due to the extreme scarcity of CTCs in whole blood.[61,62] Despite significant efforts to develop reliable technologies for detection and enumeration of CTCs, it remain difficult to efficiently isolate and analyze CTCs that are present at very low abundance (a few to hundred cells/mL) among a large number (109/mL) of hematologic cells.[63] The CellSearch System is the first and currently only test approved by FDA for the detection and enumeration of circulating epithelial cells, but the efficiency and accuracy are far less than ideal.[61,64] While recent studies have employed many different CTC enrichment mechanisms based on immunomagnetic separation,[65] microfluidic devices,[55] and size-based filtration,[66] these methods are still limited in clinical applications due to low capture efficiency and purity as well as difficulties in conducting downstream molecular analysis.
The nanowire array CTC capture method reported by Wang et al. was one of the seminal studies.[6] It demonstrated that the capture efficiency (45–65%) was almost 10 times higher than that with a flat silicon substrate (4–14%) in the stationary immuno-capture mode. It was further integrated with a microfluidic chaotic mixing system to increase the capture yield to >90% through synergistic effect of enhanced cell-substrate interaction strength and contact frequency (Figure 3B left).[67] The integrated platform was applied to capture and quantitation of CTCs from the blood samples of patients with prostate cancer and the results were compared to the CellSearch assay (FDA-approved). In general, it was found the nanowire array method captured a greater number of CTCs compared to CellSearch. In the meantime, the separation-less method was explored for CTC countin by using the large-area imaging instrument to scan all cells from a blood sample. Kuhn et al. reported a CTC detection method that simply employs immunofluorescence staining and imaging with a high-content digital microscopy.[68, 69] Although this is a good move toward practical application of CTC counting in the clinic, the complexity of blood composition and the extreme rarity of CTCs make it a lengthy and costly approach. In addition, all cells detected by this method were already fixed and stained precluding further in-depth molecular characterization such as whole exome sequencing (Figure 3B right) and functional characterization such as the test for drug response.[70] Our group utilized a laser scanning cytometry (LSC) technique[71, 72] and the nanowire-substrate for rare tumor cell capture and quantitation.[73] The uniqueness of this study is the integration of the nanowire substrate to enrich tumor cells and LSC for rapid, automated, high-content characterization of immobilized tumor cells. It can also perform rapid quantification of morphometric parameters such as cell size and aspect ratio (Figure 3B middle). Nanowire substrates with micro-patterns were shown to capture tumor cells only on the regions with nanowires, clearly manifesting the substantial enhancement of rare cell capture efficiency via the nanostructured surface as opposed to a flat substrate.[74] Further efforts have been made towards the development of multifunctional platforms and performance improvement. Park et al. employed rapid thermal chemical vapor deposition to achieve high-density and uniform deposition of gold nanoclusters (Au NCs) on nanowire surface.[75] The AuNC-coated silicon nanowires exhibited a superb capture yield for breast cancer cells (~88%) and the plasmonic excitation of AuNCs can be exploited to kill captured cancer cells using the photothermal effect. Electrospun nanofiber-deposited substrates grafted with capture agents were also employed for efficient CTC capture. Zhang et al. showed horizontally aligned, densely packed nanofibers can enhance substrate-cell topographic interactions, resulting in increased yield for capturing CTCs from patient blood.[76]
5.2. Trafficking Leukocytes and Immune Cell Subsets
Detection and enumeration of specific phenotypes of immune cells can provide valuable information for diagnosis and monitoring of a variety of human diseases. For example, the absolute count of CD4+ T lymphocytes and the ratio of CD4+/CD8+ T cells are used as indicators for the onset of AIDS.[77] While flow cytometry is the gold standard for leukocyte immunophenotyping, it has several shortcomings: 1) it requires a relative large sample input (~1×106 cells) making flow cytometry inadequate for analysis of rare cell populations, 2) the degree of multiplexing is often limited by spectral overlap.[78] There is still an urgent need for new tools to analyze rare or small subsets of immune cells from low quantities of clinical specimens. One of the pioneer works demonstrating nanowire arrays for separation of rare cells was the study by Kim et al. that reported on specific and efficient separation of CD4+ T lymphocytes from a highly heterogeneous mixture of immune cells harvested from spleen (spleenocytes).[7] Another study achieved excellent capture efficiency for CD4+ T cells using nanohole arrays.[79] To perform downstream analysis of leukocytes for cell-based disease study, both isolation and release with high efficiency and specificity should be realized. Chen et al. developed a new nanostructured platform for T lymphocyte capture and release mediated by DNA-aptamer on silicon nanowire arrays.[80] DNA-aptamers can be designed by incorporating a sequence that can be quickly cleaved by exonuclease to specifically release the target cells and this mild release process causes little damage to vulnerable primary cells.[81]
The simultaneous detection and analysis of multiple phenotypic subsets of immune cells is essential to understanding the complex immune responses. However, this is challenging for the analysis of extremely low abundance cells such as trafficking leukocytes in cerebrospinal fluid (CSF). Trafficking leukocytes in CSF can provide critical information for clinical diagnosis of the inflammatory conditions in deep brain without surgically removing and examining brain tissues. Kwak et al. developed a novel immune cell separation platform which enabled multiplexed and quantitative analysis of ultra-rare leukocyte samples.[82] In the study, the six distinct silicon nanowire surfaces were created, each of which was functionalized with different capture agent directed against specific leukocyte phenotypic markers such as CD4, CD8, CD45, and CD11 (Figure 3C left). Successfully employed for capture and analyses of trafficking leukocytes in CSF from the patients with Alzheimer’s disease, the platform demonstrated the detection of the changes in total leukocyte count and distribution of multiple major T cell subsets associated with neurodegenerative pathology, paving a new route toward non-invasive measurement or diagnosis of neurodegenerative pathology.
6. Nanowire Arrays for Tuning and Characterizing Cell Adhesion, Spreading, and Mechanics
Mechanical properties, especially substrate rigidity, can have dramatic effects on cellular functions such as adhesion, proliferation, and differentiation.[83,84] Interestingly, the dense silicone rubber micropillar array with the identical surface contact area but different micropost height serves as a new type of “material” with tunable nominal substrate rigidity.[85] Fu et al. showed that the normal rigidity influences cell spreading, focal adhesion, and cytoskeletal contractility.[86] Moreover, varying micropost rigidity could impact stem cell differentiation; the “rigid” micropost arrays favored osteogenic lineage while “soft” assays promoted adipogenic differentiation. More recently, this was applied to the study of motor neuron differentiation in order to accelerate neural induction and caudalization from human pluripotent stem cells (hPSCs).[87] Further reduction of the size of the posts led to the idea that nanowire arrays can function in a similar manner to modulate cell adhesion, spreading and response but with a nanometer scale precision. For example, fibroblasts cultured on SiNW arrays showed altered adhesion, spreading and substantial elevation of fibroblast activation protein (FAP) expression (Figure 3C right).[88]
During nanoscale topographic interactions between cells and nanopatterned substrates, cellular traction force (CTF) plays a crucial role in directing cell adhesion and function. Therefore, technologies for measurement and regulation of CTF on nanostructured substrates are critical in understanding cell-nanosubstrate interactions. Li et al. first demonstrated the quantification of subcellular CTFs by seeding adherent cells on a bed of vertical silicon nanowires via analyzing the observed nanowire deflection compared to the force-displacement curve calculated from finite element simulation.[89] Kim et al. further extended this approach to quantify the lateral force distribution at the interface of non-adherent CD4+ T cells captured on a quartz nanopillar array via measuring the deflection of the nanopillars using focused ion beam milling and imaging. The results showed a strong correlation between the lateral extension of microvilli on the surface of substrate and the magnitude of traction force (Figure 3D left & middle).[90] Nanowires or nanorods can be made of different materials such as bulk metallic glass (BMG) and employed as the substrate to investigate mechanosensing and the response of various cell types to nanotopographic features.[91] The crossectional scanning electron microscopy images of fibroblasts in contact with nanopatterned BMG displayed obvious nanorod deflections at the interface. Quantitative analysis of cellular traction forces that cells exert on the substrates could be calculated from these nanorod deflections using a simple force-displacement relationship and previously described modeling algorithm (Figure 3D right).[85,92] These studies demonstrate that micro- and nano-engineering of soft materials can serve as unique platforms to direct/modulate cellular functions for a range of biomedical applications.
7. Mechanistic Insights: How do cell surface microstructures interact with inorganic nanowire arrays?
While a variety of nanostructured surface platforms demonstrated high efficiency immunocapture and separation of rare cells, the mechanism underlying such performance improvement remained elusive. Several mechanistic studies were reported very recently and the results revealed several threads of new possible mechanisms that were not expected although still far from conclusive. The microvilli-nanowire interaction was proposed to account for increased efficiency of immunocapture of tumor cells,but several studies using scanning electron or ion beam imaging of the cell-substrate contact interface showed very few microvilli that successfully penetrated into interstitial or inter-nanowire space. Live cell imaging of cell adhesion showed that when cells were captured on a nanowire substrate the cells quickly develop diverse subcellular surface features including filopodia and lamelliapodia. The dimensions of these nanofilaments strongly depend on the feature size of the nanostructured substrate. Quantitative analysis showed that the capture efficiency is correlated with the number of filopodial fibers, suggesting a possible role for mechanosensing and cell spreading to determine the cell capture efficiency.[79,93] Such interaction between neurons and isolated nanopillars led to successful cell pinning for improved electrical recording.[48,94] Bonde et al. investigated how the cells adhere strongly to nanowires by labeling actin cytoskeleton and measuring focal adhesion. While cytoskeleton remains diffused across the cytoplasm in controls, actin-rich cell edges mainly co-localize around nanowires, suggesting the stabilization of cytoskeleton around nanowires might contribute to the physical entrapment of cells on the substrate, leading to upregulated focal adhesion formation and subsequent capture of cells on the nanowire arrays.[95]
8. Beyond Nanowires – new structures, materials, chemistries, applications and future outlook
The demonstration of nanowire arrays for rare cell capture and analysis has inspired a range of research endeavors to further improve the capture performance or to introduce new functionality by employing other nanopatterned materials that confer unique advantages over current inorganic nanowires. Different patterns of nanostructure such as nanoholes, nanopores, and nanofibers and polymeric nanomaterials have been studied. Many also exhibited enhanced efficiency for immunocapture of rare cells.[76,79,96–98] A new CTC-separation platform was developed with a transparent polymer nanofiber substrate that gave rise to the same capture efficiency but allows for the recovery of captured tumor cells using laser microdissection.[98] Hou et al. reported the use of thermoresponsive polymers coated on nanowires for efficient separation followed by highly specific release of CTCs. The polymer brush, which allows cell adhesion at 37°C, induce detachment of adhered cells via a surface hydrophobic-to-hydrophilic switch that occurred when it was cooled down to the critical temperature of 4°C.[96]
While EpCAM-antibody has been most commonly used for nanowire-based CTC capture, the surface antigens of CTCs are very heterogeneous and EpCAM may not be universally expressed in CTCs of all epithelial cancer types, in particular, due to the epithelial-to-mesenchymal transition that occurs in cancer metastatic progression. Therefore, it is still a challenge to accurately detect and capture the metastasis-initiating CTCs that may express different or more diverse repertoire of phenotypic surface markers. Chen et al. reported a simple and effective CTC purification strategy that does not require capture antibodies.[99] Because of the differential adhesion preference of cancer cells to nanotolologic surfaces compared to normal cells, a nanorough glass substrate was fabricated for efficient cancer cell capture without the use of capture antibodies. While the aforementioned platforms are mostly based on inorganic, rigid nanowire substrates, the integration with soft materials with tissue-like properties would create a microenvironment more resembling the native environment of tumors. Liu et al. developed a soft polymer-nanotube substrate that demonstrated ~80% capture efficiency with improved cell viability.[100,101] These platforms would not only serve as the substrate for immunocapture and separation of rare cells, but also provide new insights into the biology of the cell-material interface.
Although nanowire gene delivery has been reported by numerous groups for in vitro gene delivery, yet extending this platform to in vivo delivery has been difficult. Recently, it was shown that a tunable array of biodegradable silicon nanoneedles can access the cytosol to co-deliver DNA and siRNA with an efficiency greater than 90%, and that in vivo the nanoneedles transfect the VEGF-165, inducing sustained neovascularization in a target region of the muscle (Figure 4).[102] This work opens a whole new field of applications of nanowire arrays and successfully pushed to vivo applications.
Figure 4.
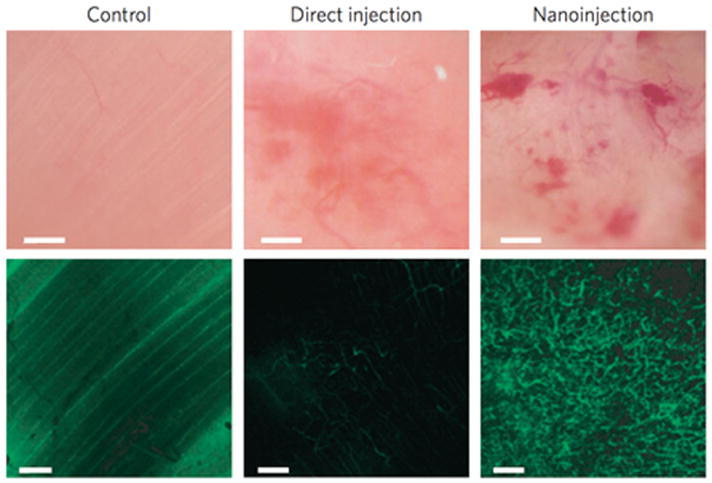
First demonstration of nanoneedle arrays to deliver proangeiogentic factor and promote in vivo neovascularization. Intravital bright-field (top panels) and confocal (bottom panels) microscopy images of the vasculature of untreated (left) and hVEGF-165-treated muscles with either direct injection (centre) or nanoneedle array-mediated delivery (called nanoinjection, right). The fluorescence signal originates from systemically injected FITC–dextran. Scale bars, bright-field 100 μm; confocal 50 μm. (Reproduced with permission.[102] 2015, Macmillan Publishers Limited.)
In brief, despite the wide-spread appreciation of the important role for nanoscale cures (mechanical, structural, biophysical and biochemical) to guide the cell fate and the demonstration of nanostructured surfaces for engineering cell behavior, differentiation and proliferation in the long term, the application of an important class of nanostructures – vertical nanowire arrays – for rapid analysis and interrogation of cellular functions emerged very recently. This includes both i) cell-penetrating nanowires for biomolecular delivery, stimulation and probing of intracellular signaling dynamics and ii) non-penetrating nanowire arrays as a tool for highly efficient capture of rare cell populations and the study of biomechanical properties of cell-nanotopologic surface interaction. Combined with new functional materials for nanowires, responsible materials for surface chemistry, and microfluidic integration for more accurate control spatially and temporally, this platform is gaining a wide range of applications to and adapted to addressing several major challenges in biology and medicine.
Acknowledgments
We acknowledge the support from the Dana Farber Physical Sciences Oncology Center - Single Cell Profiling Core (NIH U54 CA143798 subaward to R.F.), the Alzheimer Association Early Investigator Award, and the U.S. National Cancer Institute Howard Temin Pathway to Independence Award (NIH R00 CA136759 to R.F.).
Biography
Rong Fan is Associate Professor of Biomedical Engineering at Yale Univeristy. He received his Ph.D. in Chemistry from the University of California at Berkeley in 2006 and conducted postdoctoral training at California Institute of Technology. His current research interest is centered on single-cell analysis micro-/nano-technologies to measure functional cellular heterogeneity in human cancers and the immune system. He is the recipient of the Howard Temin Pathway to Independence award from the U.S. National Cancer Institute, the U.S. National Science Foundation’s Early Stage Faculty Career Development Award and the Packard Fellowship for Science and Engineering.
Contributor Information
Minsuk Kwak, Department of Biomedical Engineering, Yale University, New Haven, CT 06520, USA.
Dr. Lin Han, Department of Biomedical Engineering, Yale University, New Haven, CT 06520, USA
Jonathan J. Chen, Department of Biomedical Engineering, Yale University, New Haven, CT 06520, USA
Prof. Dr. Rong Fan, Email: rong.fan@yale.edu, Department of Biomedical Engineering, Yale University, New Haven, CT 06520, USA. Yale Cancer Center, New Haven, CT 06520, USA
References
- 1.Hochbaum AI, Yang P. Chemical Reviews. 2010;110(1):527. doi: 10.1021/cr900075v. [DOI] [PubMed] [Google Scholar]
- 2.Li Y, Qian F, Xiang J, Lieber CM. Materials Today. 2006;9(10):18. doi: 10.1016/S1369-7021(06)71650-9. [DOI] [Google Scholar]
- 3.Cui Y, Wei Q, Park H, Lieber CM. Science. 2001;293(5533):1289. doi: 10.1126/science.1062711. [DOI] [PubMed] [Google Scholar]
- 4.Yogeswaran U, Chen S-M. Sensors. 2008;8(1):290. doi: 10.3390/s8010290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim W, Ng JK, Kunitake ME, Conklin BR, Yang PD. Journal of the American Chemical Society. 2007;129(23):7228. doi: 10.1021/Ja071456k. [DOI] [PubMed] [Google Scholar]
- 6.Wang S, Wang H, Jiao J, Chen KJ, Owens GE, Kamei K, Sun J, Sherman DJ, Behrenbruch CP, Wu H, Tseng HR. Angewandte Chemie (International ed in English) 2009;48(47):8970. doi: 10.1002/anie.200901668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim ST, Kim DJ, Kim TJ, Seo DW, Kim TH, Lee SY, Kim K, Lee KM, Lee SK. Nano Letters. 2010;10(8):2877. doi: 10.1021/nl100942p. [DOI] [PubMed] [Google Scholar]
- 8.Lange K. Journal of Cellular Physiology. 2011;226(4):896. doi: 10.1002/jcp.22302. [DOI] [PubMed] [Google Scholar]
- 9.Bettinger CJ, Langer R, Borenstein JT. Angewandte Chemie International Edition. 2009;48(30):5406. doi: 10.1002/anie.200805179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Biggs MJP, Richards RG, Dalby MJ. Nanomedicine: Nanotechnology, Biology and Medicine. 2010;6(5):619. doi: 10.1016/j.nano.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu XL, Wang ST. Chemical Society Reviews. 2014;43(8):2385. doi: 10.1039/C3cs60419e. [DOI] [PubMed] [Google Scholar]
- 12.Bonde S, Buch-Manson N, Rostgaard KR, Andersen TK, Berthing T, Martinez KL. Nanotechnology. 2014;25:36. doi: 10.1088/0957-4484/25/36/362001. [DOI] [PubMed] [Google Scholar]
- 13.Mumm F, Beckwith KM, Bonde S, Martinez KL, Sikorski P. Small. 2013;9(2):263. doi: 10.1002/smll.201201314. [DOI] [PubMed] [Google Scholar]
- 14.Shalek AK, Robinson JT, Karp ES, Lee JS, Ahn DR, Yoon MH, Sutton A, Jorgolli M, Gertner RS, Gujral TS, MacBeath G, Yang EG, Park H. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(5):1870. doi: 10.1073/pnas.0909350107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chevrier N, Mertins P, Artyomov MN, Shalek AK, Iannacone M, Ciaccio MF, Gat-Viks I, Tonti E, DeGrace MM, Clauser KR, Garber M, Eisenhaure TM, Yosef N, Robinson J, Sutton A, Andersen MS, Root DE, von Andrian U, Jones RB, Park H, Carr SA, Regev A, Amit I, Hacohen N. Cell. 2011;147(4):853. doi: 10.1016/j.cell.2011.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shalek AK, Gaublomme JT, Wang LL, Yosef N, Chevrier N, Andersen MS, Robinson JT, Pochet N, Neuberg D, Gertner RS, Amit I, Brown JR, Hacohen N, Regev A, Wu CJ, Park H. Nano Letters. 2012;12(12):6498. doi: 10.1021/Nl3042917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang LL, Shalek AK, Lawrence M, Ding RH, Gaublomme JT, Pochet N, Stojanov P, Sougnez C, Shukla SA, Stevenson KE, Zhang WD, Wong J, Sievers QL, MacDonald BT, Vartanov AR, Goldstein NR, Neuberg D, He X, Lander E, Hacohen N, Regev A, Getz G, Brown JR, Park H, Wu CJ. Blood. 2014;124(7):1089. doi: 10.1182/blood-2014-01-552067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tian BZ, Cohen-Karni T, Qing Q, Duan XJ, Xie P, Lieber CM. Science. 2010;329(5993):830. doi: 10.1126/science.1192033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang PD, Yan RX, Fardy M. Nano Letters. 2010;10(5):1529. doi: 10.1021/Nl100665r. [DOI] [PubMed] [Google Scholar]
- 20.Pearton SJ, Lele T, Tseng Y, Ren F. Trends Biotechnol. 2007;25(11):481. doi: 10.1016/j.tibtech.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 21.Yoo SM, Kang M, Kang T, Kim DM, Lee SY, Kim B. Nano Letters. 2013;13(6):2431. doi: 10.1021/Nl4003393. [DOI] [PubMed] [Google Scholar]
- 22.McKnight TE, Melechko AV, Hensley DK, Mann DGJ, Griffin GD, Simpson ML. Nano Letters. 2004;4(7):1213. doi: 10.1021/Nl049504b. [DOI] [Google Scholar]
- 23.Lampert L, Timonen B, Smith S, Davidge B, Li HY, Conley JF, Singer JD, Jiao J. Chem Commun. 2014;50(10):1234. doi: 10.1039/C3cc48088g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pan JJ, Lyu ZL, Jiang WW, Wang HW, Liu Q, Tan M, Yuan L, Chen H. Acs Appl Mater Inter. 2014;6(16):14391. doi: 10.1021/Am5036626. [DOI] [PubMed] [Google Scholar]
- 25.VanDersarl JJ, Xu AM, Melosh NA. Nano Letters. 2012;12(8):3881. doi: 10.1021/Nl204051v. [DOI] [PubMed] [Google Scholar]
- 26.Xie X, Xu AM, Leal-Ortiz S, Cao YH, Garner CC, Melosh NA. ACS Nano. 2013;7(5):4351. doi: 10.1021/Nn400874a. [DOI] [PubMed] [Google Scholar]
- 27.Gehl J. Acta Physiol Scand. 2003;177(4):437. doi: 10.1046/j.1365-201X.2003.01093.x. [DOI] [PubMed] [Google Scholar]
- 28.Golzio M, Teissie J, Rols MP. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(3):1292. doi: 10.1073/pnas.022646499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boukany PE, Morss A, Liao WC, Henslee B, Jung HC, Zhang XL, Yu B, Wang XM, Wu Y, Li L, Gao KL, Hu X, Zhao X, Hemminger O, Lu W, Lafyatis GP, Lee LJ. Nat Nanotechnol. 2011;6(11):747. doi: 10.1038/Nnano.2011.164. [DOI] [PubMed] [Google Scholar]
- 30.Khine M, Lau A, Ionescu-Zanetti C, Seo J, Lee LP. Lab on a chip. 2005;5(1):38. doi: 10.1039/B408352k. [DOI] [PubMed] [Google Scholar]
- 31.Guignet EG, Meyer T. Nature Methods. 2008;5(5):393. doi: 10.1038/Nmeth.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xie C, Hanson L, Cui Y, Cui BX. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(10):3894. doi: 10.1073/pnas.1015589108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yan RX, Park JH, Choi Y, Heo CJ, Yang SM, Lee LP, Yang PD. Nat Nanotechnol. 2012;7(3):191. doi: 10.1038/Nnano.2011.226. [DOI] [PubMed] [Google Scholar]
- 34.Xie C, Lin ZL, Hanson L, Cui Y, Cui BX. Nat Nanotechnol. 2012;7(3):185. doi: 10.1038/Nnano.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin ZLC, Xie C, Osakada Y, Cui Y, Cui BX. Nat Commun. 2014;5:Artn 3206. doi: 10.1038/Ncomms4206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moran-Mirabal JM, Torres AJ, Samiee KT, Baird BA, Craighead HG. Nanotechnology. 2007;18(19) doi: 10.1088/0957-4484/18/19/195101. [DOI] [Google Scholar]
- 37.Richards CI, Luong K, Srinivasan R, Turner SW, Dougherty DA, Korlach J, Lester HA. Nano Letters. 2012;12(7):3690. doi: 10.1021/Nl301480h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yuste R. Frontiers in Neuroscience. 2008;2(1):6. doi: 10.3389/neuro.01.017.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bock DD, Lee WC, Kerlin AM, Andermann ML, Hood G, Wetzel AW, Yurgenson S, Soucy ER, Kim HS, Reid RC. Nature. 2011;471(7337):177. doi: 10.1038/nature09802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robinson JT, Jorgolli M, Shalek AK, Yoon MH, Gertner RS, Park H. Nat Nanotechnol. 2012;7(3):180. doi: 10.1038/Nnano.2011.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dan Y, Poo MM. Physiol Rev. 2006;86(3):1033. doi: 10.1152/physrev.00030.2005. [DOI] [PubMed] [Google Scholar]
- 42.Erickson J, Tooker A, Tai YC, Pine J. J Neurosci Meth. 2008;175(1):1. doi: 10.1016/j.jneumeth.2008.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xie C, Hanson L, Xie WJ, Lin ZL, Cui BX, Cui Y. Nano Letters. 2010;10(10):4020. doi: 10.1021/Nl101950x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hanson L, Lin ZC, Xie C, Cui Y, Cui BX. Nano Letters. 2012;12(11):5815. doi: 10.1021/Nl303163y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aalipour A, Xu AM, Leal-Ortiz S, Garner CC, Melosh NA. Langmuir. 2014;30(41):12362. doi: 10.1021/La502273f. [DOI] [PubMed] [Google Scholar]
- 46.Xu AM, Aalipour A, Leal-Ortiz S, Mekhdjian AH, Xie X, Dunn AR, Garner CC, Melosh NA. Nat Commun. 2014;5 doi: 10.1038/Ncomms4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xie X, Xu AM, Angle MR, Tayebi N, Verma P, Melosh NA. Nano Letters. 2013;13(12):6002. doi: 10.1021/nl403201a. [DOI] [PubMed] [Google Scholar]
- 48.Qi S, Yi C, Ji S, Fong CC, Yang M. ACS applied materials & interfaces. 2009;1(1):30. doi: 10.1021/am800027d. [DOI] [PubMed] [Google Scholar]
- 49.Ning RZ, Wang SQ, Wu J, Wang F, Lin JM. Small. 2014;10(20):4113. doi: 10.1002/smll.201400734. [DOI] [PubMed] [Google Scholar]
- 50.Choi CH, Hagvall SH, Wu BM, Dunn JC, Beygui RE, CJKCJ Biomaterials. 2007;28(9):1672. doi: 10.1016/j.biomaterials.2006.11.031. [DOI] [PubMed] [Google Scholar]
- 51.Park J, Bauer S, von der Mark K, Schmuki P. Nano Letters. 2007;7(6):1686. doi: 10.1021/nl070678d. [DOI] [PubMed] [Google Scholar]
- 52.Teo BK, Ankam S, Chan LY, Yim EK. Methods in Cell Biology. 2010;98:241. doi: 10.1016/s0091-679x(10)98011-4. [DOI] [PubMed] [Google Scholar]
- 53.Pantel K, Brakenhoff RH, Brandt B. Nature Reviews Cancer. 2008;8(5):329. doi: 10.1038/nrc2375. [DOI] [PubMed] [Google Scholar]
- 54.Kim JS, Yoon TJ, Yu KN, Kim BG, Park SJ, Kim HW, Lee KH, Park SB, Lee JK, Cho MH. Toxicological Sciences: An Official Journal of the Society of Toxicology. 2006;89(1):338. doi: 10.1093/toxsci/kfj027. [DOI] [PubMed] [Google Scholar]
- 55.Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky A, Ryan P, Balis UJ, Tompkins RG, Haber DA, Toner M. Nature. 2007;450(7173):1235. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang Z, Chin SY, Chin CD, Sarik J, Harper M, Justman J, Sia SK. Analytical Chemistry. 2010;82(1):36. doi: 10.1021/ac902144w. [DOI] [PubMed] [Google Scholar]
- 57.Xu Y, Phillips JA, Yan J, Li Q, Fan ZH, Tan W. Analytical Chemistry. 2009;81(17):7436. doi: 10.1021/ac9012072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weigelt B, Peterse JL, van’t Veer LJ. Nature Reviews Cancer. 2005;5(8):591. doi: 10.1038/nrc1670. [DOI] [PubMed] [Google Scholar]
- 59.Steeg PS. Nature Medicine. 2006;12(8):895. doi: 10.1038/nm1469. [DOI] [PubMed] [Google Scholar]
- 60.Mocellin S, Hoon D, Ambrosi A, Nitti D, Rossi CR. Clinical Cancer Research : An Official Journal of the American Association for Cancer Research. 2006;12(15):4605. doi: 10.1158/1078-0432.ccr-06-0823. [DOI] [PubMed] [Google Scholar]
- 61.Cristofanilli M. Seminars in Oncology. 2006;33(3 Suppl 9):S9. doi: 10.1053/j.seminoncol.2006.03.016. [DOI] [PubMed] [Google Scholar]
- 62.Cristofanilli M, Hayes DF, Budd GT, Ellis MJ, Stopeck A, Reuben JM, Doyle GV, Matera J, Allard WJ, Miller MC, Fritsche HA, Hortobagyi GN, Terstappen LW. Journal of Clinical Oncology : official journal of the American Society of Clinical Oncology. 2005;23(7):1420. doi: 10.1200/jco.2005.08.140. [DOI] [PubMed] [Google Scholar]
- 63.Racila E, Euhus D, Weiss AJ, Rao C, McConnell J, Terstappen LW, Uhr JW. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(8):4589. doi: 10.1073/pnas.95.8.4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Olmos D, Arkenau HT, Ang JE, Ledaki I, Attard G, Carden CP, Reid AH, A’Hern R, Fong PC, Oomen NB, Molife R, Dearnaley D, Parker C, Terstappen LW, de Bono JS. Annals of oncology : official journal of the European Society for Medical Oncology / ESMO. 2009;20(1):27. doi: 10.1093/annonc/mdn544. [DOI] [PubMed] [Google Scholar]
- 65.Talasaz AH, Powell AA, Huber DE, Berbee JG, Roh KH, Yu W, Xiao W, Davis MM, Pease RF, Mindrinos MN, Jeffrey SS, Davis RW. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(10):3970. doi: 10.1073/pnas.0813188106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vona G, Sabile A, Louha M, Sitruk V, Romana S, Schutze K, Capron F, Franco D, Pazzagli M, Vekemans M, Lacour B, Brechot C, Paterlini-Brechot P. The American Journal of Pathology. 2000;156(1):57. doi: 10.1016/s0002-9440(10)64706-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang S, Liu K, Liu J, Yu ZT, Xu X, Zhao L, Lee T, Lee EK, Reiss J, Lee YK, Chung LW, Huang J, Rettig M, Seligson D, Duraiswamy KN, Shen CK, Tseng HR. Angewandte Chemie (International ed in English) 2011;50(13):3084. doi: 10.1002/anie.201005853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Marrinucci D, Bethel K, Lazar D, Fisher J, Huynh E, Clark P, Bruce R, Nieva J, Kuhn P. Journal of Oncology. 2010;2010:861341. doi: 10.1155/2010/861341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nieva J, Wendel M, Luttgen MS, Marrinucci D, Bazhenova L, Kolatkar A, Santala R, Whittenberger B, Burke J, Torrey M, Bethel K, Kuhn P. Physical Biology. 2012;9(1):016004. doi: 10.1088/1478-3975/9/1/016004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhao L, Lu Y-T, Li F, Wu K, Hou S, Yu J, Shen Q, Wu D, Song M, OuYang W-H, Luo Z, Lee T, Fang X, Shao C, Xu X, Garcia MA, Chung LWK, Rettig M, Tseng H-R, Posadas EM. Advanced Materials. 2013;25(21):2897. doi: 10.1002/adma.201205237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhou J, Wu Y, Lee SK, Fan R. Lab on a Chip. 2012;12(23):5025. doi: 10.1039/c2lc40309a. [DOI] [PubMed] [Google Scholar]
- 72.Harnett MM. Nature Reviews Immunology. 2007;7(11):897. doi: 10.1038/nri2188. [DOI] [PubMed] [Google Scholar]
- 73.Lee SK, Kim GS, Wu Y, Kim DJ, Lu Y, Kwak M, Han L, Hyung JH, Seol JK, Sander C, Gonzalez A, Li J, Fan R. Nano Letters. 2012;12(6):2697. doi: 10.1021/nl2041707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lee SK, Kim DJ, Lee G, Kim GS, Kwak M, Fan R. Biosensors & Bioelectronics. 2014;54:181. doi: 10.1016/j.bios.2013.10.048. [DOI] [PubMed] [Google Scholar]
- 75.Park GS, Kwon H, Kwak DW, Park SY, Kim M, Lee JH, Han H, Heo S, Li XS, Lee JH, Kim YH, Lee JG, Yang W, Cho HY, Kim SK, Kim K. Nano Letters. 2012;12(3):1638. doi: 10.1021/nl2045759. [DOI] [PubMed] [Google Scholar]
- 76.Zhang N, Deng Y, Tai Q, Cheng B, Zhao L, Shen Q, He R, Hong L, Liu W, Guo S, Liu K, Tseng HR, Xiong B, Zhao XZ. Advanced Materials (Deerfield Beach, Fla) 2012;24(20):2756. doi: 10.1002/adma.201200155. [DOI] [PubMed] [Google Scholar]
- 77.Guarner J, Montoya P, del Rio C, Hernandez-Tepichin G. Cytometry. 1997;30(4):178. doi: 10.1002/(sici)1097-0320(19970815)30:4<178::aid-cyto3>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 78.Zhu H, Macal M, Jones CN, George MD, Dandekar S, Revzin A. Analytica Chimica Acta. 2008;608(2):186. doi: 10.1016/j.aca.2007.12.021. [DOI] [PubMed] [Google Scholar]
- 79.Kim DJ, Kim GS, Seol JK, Hyung JH, Park NW, Lee MR, Lee MK, Fan R, Lee SK. Journal of Biomedical Nanotechnology. 2014;10(6):1030. doi: 10.1166/jbn.2014.1814. [DOI] [PubMed] [Google Scholar]
- 80.Chen L, Liu X, Su B, Li J, Jiang L, Han D, Wang S. Advanced Materials. 2011;23(38):4376. doi: 10.1002/adma.201102435. [DOI] [PubMed] [Google Scholar]
- 81.Kwong GA, Radu CG, Hwang K, Shu CJ, Ma C, Koya RC, Comin-Anduix B, Hadrup SR, Bailey RC, Witte ON, Schumacher TN, Ribas A, Heath JR. Journal of the American Chemical Society. 2009;131(28):9695. doi: 10.1021/ja9006707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kwak M, Kim DJ, Lee MR, Wu Y, Han L, Lee SK, Fan R. Nanoscale. 2014;6(12):6537. doi: 10.1039/c3nr06465d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Discher DE, Janmey P, Wang YL. Science. 2005;310(5751):1139. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 84.Vogel V, Sheetz M. Nature reviews Molecular Cell Biology. 2006;7(4):265. doi: 10.1038/nrm1890. [DOI] [PubMed] [Google Scholar]
- 85.Yang MT, Sniadecki NJ, Chen CS. Advanced Materials. 2007;19(20):3119. doi: 10.1002/adma.200701956. [DOI] [Google Scholar]
- 86.Fu J, Wang YK, Yang MT, Desai RA, Yu X, Liu Z, Chen CS. Nature Methods. 2010;7(9):733. doi: 10.1038/nmeth.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sun Y, Yong KM, Villa-Diaz LG, Zhang X, Chen W, Philson R, Weng S, Xu H, Krebsbach PH, Fu J. Nature Materials. 2014;13(6):599. doi: 10.1038/nmat3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ha Q, Yang G, Ao Z, Han D, Niu F, Wang S. Nanoscale. 2014;6(14):8318. doi: 10.1039/C4NR01415D. [DOI] [PubMed] [Google Scholar]
- 89.Li Z, Song J, Mantini G, Lu MY, Fang H, Falconi C, Chen LJ, Wang ZL. Nano Letters. 2009;9(10):3575. doi: 10.1021/nl901774m. [DOI] [PubMed] [Google Scholar]
- 90.Kim DJ, Kim GS, Hyung JH, Lee WY, Hong CH, Lee SK. Nanoscale Research Letters. 2013;8(1):332. doi: 10.1186/1556-276x-8-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Padmanabhan J, Kinser ER, Stalter MA, Duncan-Lewis C, Balestrini JL, Sawyer AJ, Schroers J, Kyriakides TR. ACS Nano. 2014;8(5):4366. doi: 10.1021/nn501874q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tan JL, Tien J, Pirone DM, Gray DS, Bhadriraju K, Chen CS. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(4):1484. doi: 10.1073/pnas.0235407100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kim GS, Kim DJ, Hyung JH, Lee MK, Lee SK. Analytical Chemistry. 2014;86(11):5330. doi: 10.1021/ac5001916. [DOI] [PubMed] [Google Scholar]
- 94.Xie C, Hanson L, Xie W, Lin Z, Cui B, Cui Y. Nano Letters. 2010;10(10):4020. doi: 10.1021/nl101950x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bonde S, Berthing T, Madsen MH, Andersen TK, Buch-Manson N, Guo L, Li X, Badique F, Anselme K, Nygard J, Martinez KL. ACS Applied Materials & Interfaces. 2013;5(21):10510. doi: 10.1021/am402070k. [DOI] [PubMed] [Google Scholar]
- 96.Hou S, Zhao H, Zhao L, Shen Q, Wei KS, Suh DY, Nakao A, Garcia MA, Song M, Lee T, Xiong B, Luo SC, Tseng HR, Yu HH. Advanced Materials. 2013;25(11):1547. doi: 10.1002/adma.201203185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sekine J, Luo SC, Wang S, Zhu B, Tseng HR, Yu HH. Advanced Materials. 2011;23(41):4788. doi: 10.1002/adma.201102151. [DOI] [PubMed] [Google Scholar]
- 98.Hou S, Zhao L, Shen Q, Yu J, Ng C, Kong X, Wu D, Song M, Shi X, Xu X, OuYang WH, He R, Zhao XZ, Lee T, Brunicardi FC, Garcia MA, Ribas A, Lo RS, Tseng HR. Angewandte Chemie (International ed in English) 2013;52(12):3379. doi: 10.1002/anie.201208452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chen W, Weng S, Zhang F, Allen S, Li X, Bao L, Lam RH, Macoska JA, Merajver SD, Fu J. ACS Nano. 2013;7(1):566. doi: 10.1021/nn304719q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Liu X, Chen L, Liu H, Yang G, Zhang P, Han D, Wang S, Jiang L. NPG Asia Mater. 2013;5:e63. doi: 10.1038/am.2013.43. [DOI] [Google Scholar]
- 101.Wan Y, Winter M, Delalat B, Hardingham JE, Grover PK, Wrin J, Voelcker NH, Price TJ, Thierry B. ACS Applied Materials & Interfaces. 2014;6(23):20828. doi: 10.1021/am505201s. [DOI] [PubMed] [Google Scholar]
- 102.Chiappini C, De Rosa E, Martinez JO, Liu X, Steele J, Stevens MM, Tasciotti E. Nat Mater. 2015;14(5):532. doi: 10.1038/nmat4249. [DOI] [PMC free article] [PubMed] [Google Scholar]