Abstract
Here we describe a protocol for using force-clamp spectroscopy to precisely quantify the effect of force on biochemical reactions. A calibrated force is used to control the exposure of reactive sites in a single polyprotein substrate composed of repeated domains. The use of polyproteins allows the identification of successful single-molecule recordings from unambiguous mechanical unfolding fingerprints. Biochemical reactions are then measured directly by detecting the length changes of the substrate held at a constant force. We present the layout of a force-clamp spectrometer along with protocols to design and conduct experiments. These experiments measure reaction kinetics as a function of applied force. We show sample data of the force dependency of two different reactions, protein unfolding and disulfide reduction. These data, which can be acquired in just a few days, reveal mechanistic details of the reactions that currently cannot be resolved by any other technique.
Introduction
Single-molecule force-clamp spectroscopy is an expanding field of research in which molecules are studied under mechanical force. Force-spectroscopy techniques were initially developed to measure the effect of a mechanical force on individual biomolecules stretched under constant velocity conditions. From these early experiments, it was clear that mechanical forces trigger conformational changes in a diverse set of molecules, including proteins1, polysaccharides2, 3 and DNA4. Atomic force microscopy (AFM) has been especially successful for studying the mechanical properties of recombinant polyproteins, which are typically <50 nm in length. By using single-molecule AFM, it became possible to stretch single polyprotein molecules tethered between a gold-coated surface and the tip of an AFM cantilever probe1, 5. These initial measurements were acquired in the force-extension AFM mode, in which the separation between the cantilever and the surface is changed linearly with time. These experiments measured the force applied to the protein as a function of its extension, giving rise to the now-familiar sawtooth pattern traces of unfolding5. These observations revealed fundamental properties of proteins exposed to mechanical perturbations, which could not be captured using classical bulk biochemistry techniques. For example, the unfolding patterns of fibronectin and titin revealed that modular proteins unfold under force according to a mechanical hierarchy6, 7 and that the mechanical stability of a protein changes depending on where the force is being applied8, 9, 10.
Those discoveries notwithstanding, force-extension experiments are of very limited use in the study of chemical reactions. For example, studies of reactions involving buried (cryptic) disulfide bonds using force-extension experiments proved to be problematic for the following reasons. In the absence of reducing agents, disulfide bonds are expected to rupture under mechanical stress alone at forces of ~3,500 pN (ref. 11). Hence, in the absence of DTT, sawtooth patterns showed unfolding of an I27 mutant polyprotein at ~180 pN, with each domain extending right up to the disulfide bond, whereas in the presence of DTT a second regime was observed at higher forces (~300 pN), which corresponded to the rupture of the disulfide bonds12. From such data, one may reach the incorrect conclusion that the rupture forces measured the mechanical stability of a disulfide bond. In reality, mechanical unfolding of a domain exposed its cryptic disulfide and started the clock for the SN2 chemical reaction between the disulfide and the DTT13. At the same time, the protein continued to be extended, reaching increasingly higher forces until the disulfide bond was reduced. Hence, the observed rupture force, far from measuring mechanical stability of a disulfide bond, represents a complex convolution of chemical kinetics, force, length and time, which can be impossible to unravel. Owing to these limitations, the force-sensitivity of a chemical reaction cannot be studied effectively using force-extension techniques. In contrast, in force-clamp AFM, an active feedback maintains the force at a constant value. A force protocol is programmed to simultaneously expose the reactive sites, readily separating force, length and time, and also allowing for the accurate measurement of reaction rates and their sensitivity to force.
Although it was evident to us that studies on mechanical biochemistry had to be done under force-clamp conditions, developing suitable instrumentation and methods of analysis has been a great challenge over the years. For example, our first effort to measure the force dependency of protein unfolding using force-clamp techniques failed, owing to the poor performance of the piezoelectric actuators and AFM cantilevers available at the time14. The introduction of the picocube piezoelectric actuator by Physik Instrumente in 2003 greatly increased the accuracy of the instrument, making it possible to measure the force dependency of a chemical reaction13, 15, 16. More recently, the manufacturing of AFM cantilevers by companies such as Olympus and Bruker has further increased the reliability of these measurements17, 18, 19, 20, 21, 22, 23. Although our initial efforts have been concentrated in studying the redox chemistry of disulfide bonds13, 16, 24, 25, 26, 27, 28, we anticipate that our techniques will be extended to studying reactions that involve many other types of chemical bonds in polysaccharides, nucleic acids and other molecules.
The force-clamp technique now makes it possible to tackle key scientific questions that lie at the interface between physics, biology and chemistry. The technique is now available to scientists interested in new fields of inquiry from a single-molecule perspective. There are a number of key considerations in sample preparation, data collection, selection and analysis, which need to be taken into account in order to obtain reliable and reproducible results when one is using these techniques. By distilling a decade of experience, here we provide a practical guide of the details required to complete single-molecule force-clamp spectroscopy experiments with AFM.
An overview of force-clamp AFM
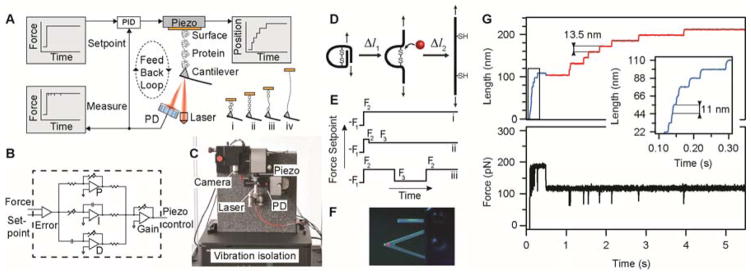
The technological improvements and our accumulated expertise in force-clamp spectroscopy have crystallized in a new commercial setup designed in collaboration with Luigs & Neumann (Germany) (Fig. 1a–c). This instrument is specifically built to enable fully automated operation in force-clamp mode. Part of the data shown in this protocol was acquired using a prototype of this instrument. There are other commercial instruments that are capable of operating in force-clamp mode; these are manufactured by companies such as Asylum Research and JPK Instruments.
Figure 1.
In a typical measurement, the proportional-integral-derivative (PID) feedback loop follows a given setpoint force protocol that is appropriate for the studied reaction. Depending on the reaction under investigation, this protocol may have one or several constant pulling forces (Fig. 1d,e). The cantilever (Fig. 1f) is initially pushed onto the surface in order to attach to a polyprotein chain. Once a polyprotein is tethered between the cantilever and the surface, the length of the polyprotein is monitored over time. Each time an unfolding or disulfide reduction event takes place, the unraveling of the trapped amino acids leads to a sudden increase in extension, which in turn causes a relaxation of the cantilever (Fig. 1g). The forces used are high enough to ensure an immediate extension after the breaking of the chemical bond. A possible limitation might arise at low forces that are less efficient in unraveling collapsed polypeptides.
When bond rupture has occurred, causing cantilever relaxation, the PID feedback loop moves the surface away from the cantilever until the measured force again matches the setpoint. Polyproteins show in length-versus-time plots staircase-like changes, with each step corresponding to the extension of one domain in the polyprotein (Fig. 1g).
The main components of our force-clamp AFM setup are described in brief below.
Piezoelectric actuator
The piezoelectric actuator (denoted as ‘piezo’ in Fig. 1a) is the moving stage that controls the extension of the molecule. The basis for this positioning stage is the piezoelectric effect, in which an applied voltage induces displacements with Ångström resolution. Typical piezoelectric actuators have travel ranges from hundreds of nanometers to few micrometers. Piezo actuators show hysteresis in the form of a history-dependent displacement with the driving voltage, which can cause errors in length. Thus, piezos showing hysteresis require an independent measurement of displacement, using, for instance, a capacitive or resistive sensor. Recently, Physik Instrumente developed a new type of piezo that addresses the above-mentioned problem: the P-313 PicoCube. This piezo incorporates a mechanical mechanism to correct for hysteresis, thereby removing the need for an independent measurement of the position and thus eliminating associated noise.
Force detection
The force experienced by the cantilever is measured through the position of a laser beam reflection, which is monitored by a quadrant photodiode (PD, First Sensor). Our setup uses a 670-nm fiber-coupled laser diode (Schäfter + Kirchhoff), with a 12-mm fiber collimator, which is capable of giving a sharply focused beam on the back of the cantilever. Sharp focusing leads to less light leakage, limiting the effect of interference when you are using reflective surfaces such as gold (see TROUBLESHOOTING section). The laser diode has good beam stability, which is important in order to minimize the time-dependent noise in the force measurement. The output of the sensor is proportional to the force on the cantilever, assuming that all responses are in a linear regime. The setup includes a video camera that is used to align and focus the laser on the cantilever (Fig. 1c,f).
Fluid cell
The cantilever is held in place by a spring-based mechanism implemented inside the fluid cell. The laser beam passes through the quartz surface of the fluid cell and bounces back from the cantilever through the same surface. A silicon O-ring seals the fluid cell, thus slowing the evaporation of the solution.
Control of the AFM
Instrument control and data acquisition are done on a PC using an USB-connected data acquisition card (National Instruments). Our AFM control software is custom-written in Igor Pro (Wavemetrics). Force-clamp operation is implemented using a PID feedback system that drives the AFM to follow a given force protocol (Fig. 1e). The analog PID features two inputs—the force setpoint (from the PC) and the measured force (from the cantilever)—and one output that controls the piezo displacement (Fig. 1a). Inside the PID controller (Fig. 1b), the two inputs are first fed into a differential amplifier, generating an error signal. This error signal is then processed in three parallel gain stages (proportional, integral and derivative). The outputs of these three stages are summed, amplified and sent to the piezo, thus driving the AFM to attain the required setpoint force. The individual gains of the feedback system are set to minimize the response time while still ensuring stability (meaning that the system converges at the desired setpoint). As shown in Figure 1b, the individual gain parameters can be controlled through appropriately placed variable resistors. To avoid artifacts resulting from rapid transients, the output signal can be passed through a low-pass filter (5 kHz) before being sent to the piezo.
Automation
Fully automated operation of the AFM is enabled by the addition of translation stages that correct for mechanical drift between experimental trials. The positions of the piezo actuator and the quadrant PD are adjusted using such auxiliary piezo stages (Agilis, Newport). In the absence of sample degradation, experiments can be run continuously for several days without user intervention.
Vibration isolation
The AFM setup is placed on top of an active vibration isolation platform (Accurion), which in turn is placed on a rigid table.
Experimental design
Polyprotein engineering for mechanical fingerprinting
The outcome of single-molecule measurements can be severely biased by the presence of contaminants such as other proteins, impurities from the surface or aggregates, as well as by the tethering of more than one molecule. Force-clamp AFM uses polyproteins to avoid these drawbacks. These polyproteins are typically homologous repeats of the protein of interest5, 29. Traces from these polyproteins show unique repetitive features that set them apart from recording of other present molecules. The mechanical unfolding of a polyprotein leads to identical step increases in the measured end-to-end length, resulting in a characteristic ‘staircase’. Such a staircase is virtually impossible to obtain from the rupture of multiple nonspecific interactions1. If more than one polyprotein is tethered at the same time, several populations of step sizes appear. The staircase profile thus represents the mechanical fingerprint of the proteins being measured.
In the most popular configuration (gold surface on the piezo, silicon nitride cantilever), the AFM tip can attach to the polyprotein at any point along its length, thus rendering traces that probe a varying number of domains5. Only recordings that have a minimum number of identical steps and that last long enough (see ‘Minimum duration of recordings’ below) should be included in the analysis. Unfolding traces may have additional steps apart from the expected fingerprint, which is indicative of attachment to more than one molecule, attachment to a dimer molecule (from the oxidation of the terminal thiol) or aggregation. However, as it is usually not possible to determine the source of such steps, these recordings are excluded from the analysis. One possible exception to this rule may be the steps detected at the beginning of a trace. In this case, nonspecific interactions between the cantilever and small surface-adsorbed contaminants are broken and, immediately after, the protein construct experiences the setpoint force5.
The polyprotein constructs used in force-clamp measurements are engineered through molecular biology techniques5. Natural polyproteins such as ubiquitin8, titin1 and filamin30 can also be used in these experiments with minimal modification, such as the addition of purification tags and terminal cysteines.
The polyprotein construction starts with amplification of the DNA encoding the protein of interest using PCR with specific restriction sites added. The DNA of this single monomeric protein is then sequentially digested and ligated using the restriction sites of a cloning vector, resulting in dimers, tetramers, octamers and so on5. Upon digestion with the appropriate enzymes, the DNA insert of the multimeric protein is cloned into a specific expression vector having a His6 tag. This expression vector endows transformed bacteria with antibiotic resistance and offers the possibility of triggering the expression of the protein of interest by the addition of IPTG. After protein expression, the cells are collected and lysed using enzymatic and mechanical treatments. The soluble fraction is passed through two separation columns: a His6 tag affinity column and a size-exclusion column. Depending on the expressed protein, the concentrated fractions are then stored at 4 or −80 °C until further use.
Cantilever calibration
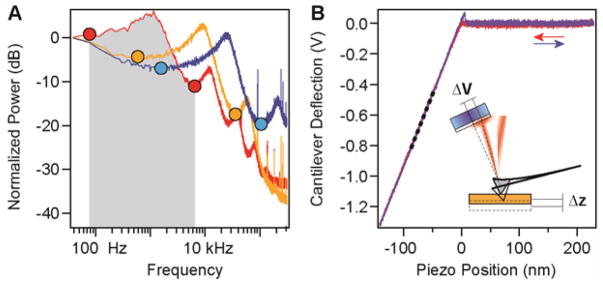
Cantilevers are typically manufactured from silicon, borosilicate glass or silicon nitride using microelectromechanical systems, and they may have a gold coating in order to improve their reflectivity. On the sample-facing side, the cantilevers have a tip with a sharpness of 10–30 nm, which increases the probability of attachment to only one molecule. For small bending angles, the cantilever behaves like a Hookean spring, with a spring constant that is determined by its material and dimensions. We calibrate each cantilever using the thermal fluctuations method, which is based on the equipartition theorem31.
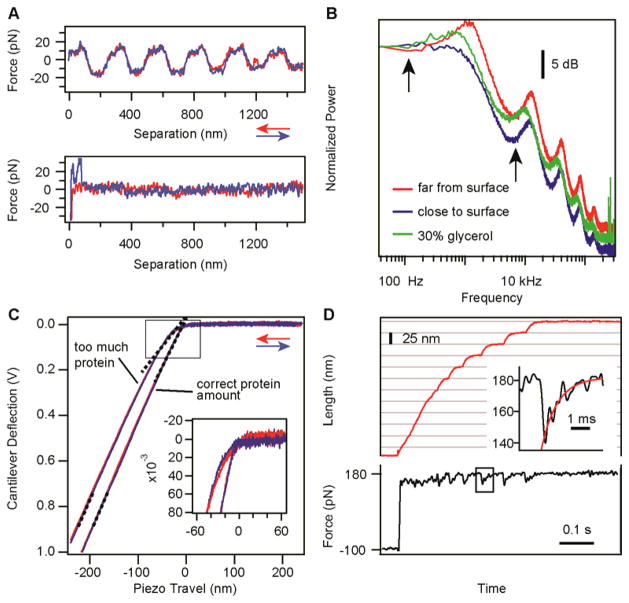
The equipartition theorem states that the kinetic energy of each degree of freedom (such as a vibrational mode) equals half the thermal energy kBT (= 4.11 pN·nm). The calibration of cantilevers comprises two steps (Fig. 2). In the first step, the thermal fluctuations of the cantilever far from the surface are measured and Fourier-transformed in order to separate the main vibration mode. By integrating the area below the first resonance peak, the mean squared displacement 〈x2〉 is measured in units of V2 (Fig. 2a). In the second step of the calibration procedure, the cantilever is brought close to the surface and a deflection-extension curve is measured (Fig. 2b). When in contact, the piezo and the cantilever travel identical distances. The slope s = Δz/ΔV of this constant compliance region of the curve yields a correlation between the change in voltage (ΔV) measured by the PD and the bending distance (Δz) measured from the movement of the piezo, in units of nm V−1. Finally, the spring constant of the cantilever in pN nm−1 units is obtained according to the following formula:
Figure 2.
Cantilever selection
The choice of cantilevers is critical for the experimental outcome, as cantilevers influence the signal-to-noise ratio, drift and feedback response times. The lateral dimensions of the cantilever have to be large enough to reflect the laser beam. Cantilevers with small spring constants give a better signal-to-noise ratio, as they deflect more for the same change in force. Cantilevers with small lateral dimensions typically have not only higher resonance frequencies and lower viscous drag, but also higher spring constants. The resonance frequency of the cantilever limits the response time of the feedback loop. Figure 2a shows the power spectra of three typically used cantilevers: MLCT-C (Bruker), BL-RC150VB-A (Olympus) and BL-AC40TS (Olympus). The MLCT cantilevers yield a stable signal, are linear over a wide range of forces, have a good price/performance ratio and give a response time of down to 1 ms. BL-RC150VB cantilevers have a higher spring constant, but they can yield feedback response times down to 150 μs and are therefore ideal for measuring fast processes21.
Sample surface preparation and covalent attachment
The surface of choice for adsorbing polyprotein constructs has traditionally been gold, which can bind to the thiol group of a terminal cysteine in a protein. The silicon-nitride cantilever attaches randomly to the polyprotein through unknown interactions, probably involving physisorption. The gold surfaces can be prepared by evaporating 40 nm of gold on top of a 20-nm ‘gluing’ layer of a 1:1 chromium/nickel (Cr/Ni) mixture deposited on a clean glass surface.
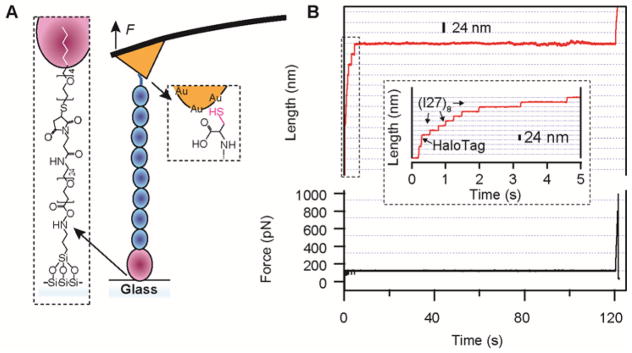
A recently developed method for dual specific covalent attachment holds the promise of vastly increasing the probability of having proteins tethered precisely from their termini and of holding on to a single protein for long durations, even over several minutes (Fig. 3; Box 1). This attachment chemistry was first described by Taniguchi and Kawakami32, and it has been successfully implemented in our laboratory (I.P. et al., unpublished data). In this approach, polyprotein constructs are engineered to have a HaloTag protein (a mutant haloalkane dehalogenase) at one terminus and a cysteine at the opposite end. The HaloTag forms an ester bond with a chloroalkane ligand linked covalently to the glass surface. Cantilevers can be similarly functionalized or coated with a gold layer. As opposed to the gold surface–silicon nitride cantilever method presented in the current protocol, the chloroalkane surface–gold cantilever approach (from Box 1) is more sensitive to variations in sample preparation because of the complexity of the functionalization procedure.
Figure 3.
BOX 1. Preparation of the surfaces and cantilevers for covalent attachment of proteins via HaloTag chemistry.
The functionalization of glass coverslips with a chloroalkane ligand specific for HaloTag involves three stages: 1. amino-functionalization of the surface (glass coverslips are reacted with an aminosilane)39; 2. reaction of the amino-functionalized glass coverslips with a bifunctional crosslinker40 (SM(PEG)24); 3. final reaction with a thiolated Halotag ligand. In the final step the bifunctional thiol-chloroalkane ligand is covalently attached. Gold coated cantilevers are obtained using standard evaporation techniques in low pressure, of ~ 5·10−6 torr or can be bought already coated with gold. The amount of gold and intermediate chromium/nickel layer affects the radius and spring constant of the cantilever.
Additional Materials
REAGENTS
Glass coverslips (Ted Pella Inc, cat. no. 26024)
(3-Aminopropyl)trimethoxysilane (Sigma-Aldrich, cat. no. 281778)
Succinimidyl-[(N-maleimidopropionamido) tetracosaethyleneglycol] ester,
SM(PEG)24 (Thermo Scientific, cat. no. 22114)
HaloTag Thiol (O4) Ligand, chloroalkane ligand (Promega, cat. no. P6761)
Dimethyl sulfoxide, DMSO (Sigma-Aldrich, cat. no. 276855)
Sulfuric acid, H2SO4 (EMD Chemicals, cat. no. SX1244-6)
Hydrogen Peroxide, 30% H2O2 (Fisher Scientific, cat. no. H325)
Borax (Sigma-Aldrich, cat. no. 71997)
2-Mercaptoethanol (Sigma-Aldrich, cat. no. M3148)
Chromium/Nickel powder, Cr/Ni (GoodFellow, cat. no. CR026010/2)
Gold wire, Au (GoodFellow, cat. no. AU005171/94)
Ethanol (Pharmco-Aaper)
Heptane (Sigma-Aldrich, cat. no. 246654)
Chloroform (Sigma-Aldrich, cat. no. 288306)
Double distilled water, DD water
EQUIPMENT
Desiccator (Wheaton, W365887)
Oven (Boekel Industries Inc, Model 107800)
Plasma Cleaner (Harrick Plasma, PDC-32G)
Evaporator (Edwards Auto 306)
REAGENT SETUP
SM(PEG)24 Dissolve SM(PEG)24 in DMSO to a concentration of 250 mM, aliquot in small volumes, freeze in liquid nitrogen and exchange the air with argon inside the recipients. Keep the aliquots at −80 °C until use.
HaloTag O4 chloroalkane ligand Dissolve HaloTag O4 chloroalkane ligand in DMSO to a concentration of 100 mM, aliquot in small volumes, freeze in liquid nitrogen and exchange the air with argon inside the recipients. Keep the aliquots at −80 °C until use.
PROCEDURE
Preparation of functionalized glass coverslips for HaloTag chemistry
-
1
Clean the glass coverslips using Piranha cleaning procedure (a 3:1 mixture of conc. H2SO4 and 30% H2O2 for 30 min at 80 °C).
CAUTION: Piranha solution is corrosive and can lead to violent reactions with organic solvents.
-
2
Wash the surfaces with water, dry them with nitrogen, expose them to oxygen plasma for 1 min on each side and heat them for 5 min in a preheated oven at 150 °C.
-
3
Add the surfaces one by one in a glass beaker containing 0.1% (3-aminopropyl)trimethoxysilane in heptane and leave them overnight in a desiccator.
-
4
Wash the surface by subsequently sonicating them for 5 min in heptane, water and chloroform.
-
5
Dry the surfaces in an oven for 1 h at 150 °C.
-
6
Sandwich silanized coverslips with freshly prepared SM(PEG)24 10 mM, dissolved in borax buffer pH 8.5 and incubate in the dark for 1 h.
-
7
Separate the sandwiches, wash the coverslips with DD water and dry them with nitrogen.
-
8
Sandwich the coverslips freshly prepared chloroalkane ligand 7.5 mM, dissolved in borax buffer pH 8.5 and incubate in the dark overnight.
-
9
Separate the sandwiches, wash the coverslips with DD water and dry them with nitrogen.
-
10
Quench the reaction with 2-mercaptoethanol 50 mM for 5 min.
-
11
Wash the coverslips with DD water, dry them with nitrogen and store them at −20 °C until use.
Preparation of functionalized gold cantilevers for HaloTag chemistry
-
12
Expose cantilevers to oxygen-plasma for 1 min.
-
13
Mount the cantilevers in the evaporator and apply vacuum.
-
14
Evaporate 2 nm of Ni/Cr mixture.
-
15
Evaporate 10–15 nm of Au.
-
16
Store the Au cantilevers until usage. Gold cantilevers can be washed with 100 % ethanol before experiment.
Other covalent attachment approaches for AFM experiments have also been explored in the past. Ikai and colleagues33 managed to attach a protein to an AFM cantilever by using thiol chemistry to tether carbonic anhydrase to AFM cantilevers and silicon substrates. Gaub and collaborators34 engineered a polyprotein containing a SNAP-tag enzyme at its C terminus, which reacts with benzylguanine, thereby forming a covalent bond. Zakeri et al.35 developed an attachment method based on the isopeptide bond by using a peptide chain to form an amide bond with its protein partner.
Minimum duration of recordings
A minimum duration for each recording should be defined to avoid biasing the measured reaction rates. Weak polyprotein anchoring leads to a high probability of detachment, which can take place before all the events have occurred. Without a strong selection criterion for minimum duration, slow events will be missed and the rate will be overestimated, creating an artificial plateau in the force dependency (see TROUBLESHOOTING section)22. The analysis must include only traces that are long enough for the rate being measured. A simple way to ensure that random detachment times are not introducing artifacts is to show that the measured rate is not affected by increasing the minimum trace duration22. With covalently tethered polyproteins and more stable cantilevers, it is now becoming possible to make unambiguous long-lasting recordings that easily meet these criteria. When you are analyzing these recordings, take extra care to detect and include fast early events that may be poorly resolved. By overlooking fast events, the measured rate can be artificially underestimated.
Data analysis
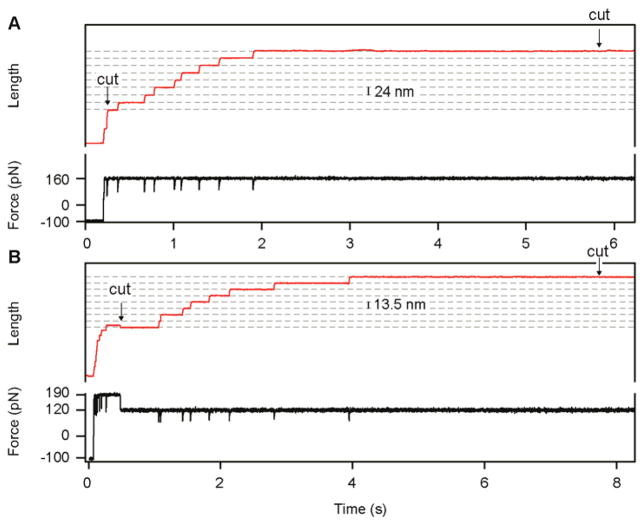
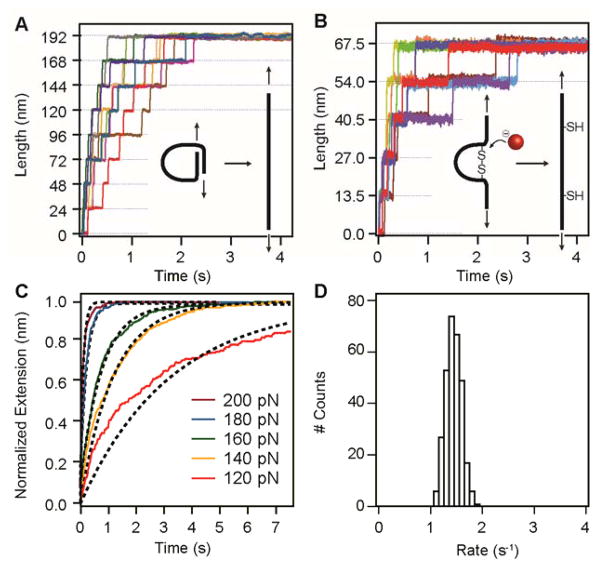
Recordings that pass all these selection criteria are used to calculate reaction rates. First, each trace is cropped so that it starts with the onset of the reaction and ends at the previously determined minimum duration requirement (Fig. 4). The cropped trace should consist of flat regions interrupted only by a series of steps corresponding to the studied reaction. For unfolding experiments, the first cut is made at the beginning of the plateau that precedes the first unfolding event (Fig. 4a). This cropping protocol leaves out the initial elastic extension of the polyprotein and any changes in length caused by nonspecific interactions with the surface, which are unrelated to the unfolding of the polyprotein. In disulfide reduction experiments, the first cut is made at the start of the reduction pulse (Fig. 4b). From this point onward, the cropped traces from unfolding and reduction experiments are analyzed in an identical way. For each force, all of the cropped traces are summed and normalized (Fig. 5a–c). The time course of the resulting summed trace reflects the progression of the studied process, as every length increment reports on a single reaction.
Figure 4.
Figure 5.
Dwell time analysis is an alternative to summed traces17, 36, 37, 38, 39. In this analysis, zero time is once again set at the beginning of the plateau preceding the first unfolding or reduction event. A dwell time is the time at which a single event occurs, measured from the zero time reference. The result of this method of analysis is a distribution of all dwell times for a given process.
Modeling
If the process follows simple exponential kinetics, the reaction rate r at a given set force can be extracted by fitting the normalized extension l to l = 1 – exp(−r · t) (Fig. 5c; ref. 15). For some reactions, the force dependency of the rate appears as a straight line when it is displayed in a semilogarithmic plot (see ANTICIPATED RESULTS). In those cases, data can be analyzed by assuming the simple Arrhenius kinetics, where the reaction rate changes according to
(where r0 is the rate in the absence of force, F is force, Δx is the distance to the transition state, kB is the Boltzmann constant and T is the absolute temperature)41, 42. Thus, in this model the distance to the transition state is obtained from the slope in the semilogarithmic plot. The height of the energy barrier of the reaction can be obtained from the extrapolated value in the absence of force combined with the pre-exponential factor that we measured to be A ~ 108 M−1 s−1 for chemical reactions26 and A ~ 109 s−1 for mechanical unfolding19. In some other cases, force dependencies may not be single exponential and other methods of analysis are needed to interpret the results16, 43.
By using dwell time analysis, we have shown that the time course of mechanical unfolding of the ubiquitin and I27 proteins does not follow simple exponential kinetics17, 37, 40, 44. For both proteins, we have recently proposed that the origin of the nonexponential behavior can be explained in terms of a static disorder scenario17, 40. In this framework, static disorder originates from the existence of an ensemble of conformations that populate the transition state of the unfolding process and that cannot interconvert during the time scale of the experiment. Dwell time analysis of the unfolding and reduction processes taking place within the same protein show the presence of static disorder for the unfolding process and the lack of it for the chemical reaction40. This finding also excludes the possibility of static disorder being a consequence of instrument errors. Despite these findings, single exponential fits of the measured extensions are good approximations with which to evaluate reaction rates (Fig. 5c).
The force dependency of a reaction is determined by the underlying free-energy landscape of the molecule45. We anticipate that more sophisticated models for analyzing force dependencies will become available in the next few years, which will allow for a better description of free-energy landscapes.
Error analysis
The error of a measured rate is determined through a bootstrapping procedure16, 46. The bootstrap analysis uses the number of iterations NBS as an input parameter, and it carries out the following procedure for each experimental condition (e.g., for each force). In the pool of experimental traces selected for analysis, each trace is considered an independent observation. On the basis of this original data set, a chosen number (NBS) of artificial data sets are then created, each containing the same number of traces as the original data set. Individual traces from the original data set are randomly selected to populate the artificial data sets (a trace can be selected more than once). If one assumes simple exponential kinetics, the traces in each artificial data set can be summed and fitted to a single exponential to yield a rate. These NBS rates obtained from the chosen number of data sets give the spread of the data and hence the s.e.m. (Fig. 5d). The s.e.m. measured by this method converges to a single value as the number of bootstrap iterations (NBS) increases, and a sufficiently large number of iterations should therefore be used. We have found empirically that 300 iterations (NBS = 300) are usually sufficient to converge into a reliable result.
Sources of error
The main sources of experimental errors arise from laser interference, errors in the thermal fluctuations spectrum, nonlinearity in the contact slope measured during cantilever calibration, and errors due to feedback response time (Figs. 6 and 7). These errors and some other possible issues, such as those regarding data selection and analysis, are discussed in the TROUBLESHOOTING section.
Figure 6.
Figure 7.
Materials
Reagents
HEPES (Sigma-Aldrich, cat. no. H3375)
Sodium phosphate, (Na2HPO4; Sigma-Aldrich, cat. no. S0751)
Sodium chloride (NaCl; Sigma-Aldrich, cat. no. S7653)
EDTA (Sigma-Aldrich, cat. no. EDS)
BLR(DE3)-competent cells (Novagen, cat. no. 69053-3)
ERL-competent cells (not commercially available)
LB medium (BD, cat. no. 244620)
Protease inhibitors (Novagen, cat. no. 539134-1SET)
Lysozyme (Calbiochem, cat. no. 4403)
DNase (Roche, cat. no. 104 159)
RNase (Roche, cat. no. 109 169)
Protease inhibitors (Calbiochem, cat. no. 539137)
Magnesium chloride (MgCl2; Sigma-Aldrich, cat. no. M8266)
IPTG (Calbiochem, cat. no. 420322)
Imidazole (Sigma-Aldrich, cat. no. I2399)
Tris-(2-carboxyethyl)phosphine, hydrochloride (TCEP; Molecular Probes, cat. no. T-2556)
Glycerol, molecular biology grade (Promega, cat. no. H5433)
Apiezon N cryogenic high-vacuum grease (Apiezon, 25 g tube)
Equipment
Force-clamp AFM setup (Luigs & Neumann)
Spectrophotometer (DU 730, Beckman Coulter)
French press (SLM Aminco, Spectronic Instruments)
AktaFPLC with a Superdex 200 column (Amersham Pharmacia Biotech)
Centrifuge (5424 R, Eppendorf and RC-5B, Sorvall)
Ni-NTA agarose (Qiagen)
Filters (F2504-10, Thermo Scientific)
Reagent setup
HEPES buffer Combine 10 mM HEPES, 150 mM NaCl and 1 mM EDTA (pH 7.2). Filter the buffer, degas it and cool it at 4 °C. Store it in the dark at 4 °C for up to 1 month.
Phosphate buffer Combine 50 mM Na2HPO4 and 150 mM NaCl (pH 7.2). Filter the buffer, degas it and cool it at 4 °C. Store it at 4 °C for up to 1 month.
Elution/wash (E/W) buffer Prepare the buffer by mixing 50 mM Na2HPO4 and 300 mM NaCl (pH 7.0). Store at 4 °C for up to 1 month.
Protein expression and purification Grow ERL- or BLR(DE3)-competent cells containing the expression vector in LB medium with appropriate antibiotics at 37 °C. Induce the expression of the vectors with 1mM IPTG when the culture reaches an optical density at 600 nm (OD600) of 0.6, and allow protein expression to take place at 25 °C overnight. Collect the cells by centrifugation and resuspend them in E/W buffer. Lyse the cells using a French press after treatment with 1mg ml−1 lysozyme, 5 μg ml−1 DNase I and 5 μg ml−1 RNase A in the presence of 10 mM MgCl2 and protease inhibitors. Centrifuge the lysate and transfer the soluble fraction onto the Ni-NTA Agarose column. Wash the column with E/W buffer including a low concentration of imidazole according to the manufacturer’s guidelines (e.g., 10 mM imidazole). Elute the protein with 250 mM imidazole-E/W buffer. Inject the fractions of protein having the highest concentration in the Superdex 200 size-exclusion FPLC column. Store the protein at 4 or −80 °C. Use 0.5 mM TCEP (except in the final FPLC buffer) and 10% (vol/vol) glycerol in the purification buffers for the proteins containing the HaloTag domain to prevent aggregation and a decrease in HaloTag activity.
Procedure
Experiment preparation
-
1
Mount the cantilever in the fluid cell. Turn on the laser and move the camera such that the cantilever is in focus. Align the laser beam at the tip of the cantilever and focus it using the laser-positioning screws until the laser spot looks small compared with the size of the cantilever (Fig. 1f). Optionally, align the laser focus on the cantilever and compare the total intensity of the reflected beam in different focusing positions until the maximum is obtained.
Critical step To minimize light leakage, the diameter of the laser should not exceed the surface dimensions of the cantilever. When you are using a small cantilever, this requirement might not be possible to fulfill. In this case, align and focus the laser as much as possible onto the free end of the cantilever.
-
2
Centrifuge the polyprotein sample at 20,000g for 10 min at 4 °C to get rid of possible aggregates.
-
3
Position the gold or glass surface on top of the piezo actuator and firmly attach it by using vacuum grease. Add a small volume of polyprotein solution (2–10 μl) to the surface and spread it with the pipette tip without scratching the surface. Wait for 10–20 min to allow the protein to adsorb, but do not let the solution dry out completely. (When you are using silicon or silicon nitride cantilevers, 10–20 μl of protein solution can instead be added directly to the cantilever, skipping Steps 4 and 5. This approach increases the probability of having proteins adsorbed on the surface area close to the cantilever).
-
4
Gently wash the nonspecifically adsorbed proteins with buffer. Skip this step if the protein is very dilute or not very sticky (for example, if it lacks terminal cysteines, or if a reductive environment is used).
-
5
Add a drop of buffer solution (typically 20–100 μl of HEPES or phosphate buffer) onto the cantilever tip and check for air bubbles.
Critical step Air bubbles can attach to the cantilever in this step. Focus the camera on the cantilever and look for air bubbles. If air bubbles are present, clean and dry the fluid cell and repeat Step 5.
-
6
Close the fluid cell by approaching the piezo toward the cantilever until the Oring seals it.
-
7
Align the PD so that the reflected laser beam is aimed at the center of the detector.
Critical step Verify that the experimental solution is completely free of particulate matter by examining the force signal, as even low concentrations of micrometer-sized particles can have a strong adverse effect on the stability of the measurements by scattering the laser beam used to monitor the position of the cantilever.
Critical step After closing the fluid cell, wait for 10–60 min for the system to thermally equilibrate and for the drift of the cantilever (measured from the PD output) to settle.
Check troubleshooting section
Experiment preparation
-
8
Measure the thermal fluctuations (power) spectrum of the oscillations of the cantilever by using a fast Fourier transform analyzer. Select the main vibrational mode and integrate the resonance peak.
Critical step The thermal fluctuations spectrum contains information about the quality of the experiment. Mechanical drift affects the low-frequency region of the power spectrum, whereas electronic noise will yield sharp peaks in the high- frequency range. A change in the position of the resonance peak within the same class of cantilevers can be indicative of the presence of a bubble or of structural damage to the cantilever. Record the power spectrum far from the surface in order to avoid dampening from the surface.
Check troubleshooting section
-
9
Bring the cantilever close to the surface by monitoring the PD signal. An automatic procedure is implemented in our setup, by using the motorized stage to control the distance between the cantilever and the surface.
-
10
Move the piezo actuator such that the cantilever touches the surface and then retracts to break the contact. Perform a deflection-extension curve. Fit a line in the region where the cantilever is in contact with the piezo-moving surface and has a linear behavior (this region is often referred to as ‘the constant compliance region’; Figs. 2b and 6c). The slope in this region relates the PD signal in V with cantilever tip displacement in nm (see the calibration section above).
Critical step A good practice for checking whether the calibration is correct is to run the spectrometer in force- extension mode for a few cycles. The force-extension recordings using a correctly calibrated cantilever will show the expected unfolding force (which depends also on the approach-retract velocity) and expected change of the contour length of the protein (when fitted with a phenomenological model for polymer elasticity, such as the worm-like chain).
Critical step Analyze the part of the force curve before the cantilever comes in contact with the surface in order to detect laser interference; this is normally evident as a sinusoidal curve (Fig. 6a). Interference affects the force applied to the protein in force-clamp mode.
Check troubleshooting section
-
11
Follow options A or B for assigning the force protocol.
-
unfolding experiments at constant force
Set the AFM to push the cantilever into the protein layer adsorbed on the surface with −1000 pN for 1 s. (ii) Apply a short 0.2-s pulse with a pushing force of −100 pN to record the contact location. (iii) Choose a pulling force that is sufficiently high to unfold the protein under investigation. (iv) Increase the pulling force to a high value (~1,000 pN) to break the tether.
-
chemical reactions under force
Set the AFM to push the cantilever into the protein layer adsorbed on the surface with −1,000 pN for 1 s.
Apply a short 0.2-s pulse with a pushing force of −100 pN to record the contact location.
Apply a short pulling pulse (typically ~0.3 s) at a pulling force that is high enough to unfold all the protein domains and expose the cryptic bonds.
Adjust the force in the second pull pulse to measure the force dependency of the chemical reaction.
Increase the pulling force to a high value (~1,000 pN) to break the tether.
-
-
12
After setting the desired pulse protocol, optimize the feedback gain constants to obtain optimal response time. Typically, the integral gain is the only parameter that needs adjustment from the standard values. Start with a small gain value and increase in small increments until the system goes close to resonance (high-frequency oscillations in force and length), and then reduce the gain with one step.
Check troubleshooting section
-
13
Once it is started, the experiment can automatically run for extended amounts of time, up to several days. During each attempt to attach a protein, the experimental pulse protocol is automatically canceled if the attachment is unsuccessful. Each complete successful protocol ends with a high pulling force (~1,000 pN) that causes detachment, allowing for a new trial. Data are saved after each trace.
Critical step Traces can be selectively saved only if they comply with the requirements set by the user, such as the number of measured steps and/or the duration of the trace.
Check troubleshooting section
-
14
Change the value of the force in order to obtain the force dependency of the studied reaction.
Data analysis
-
15
Select traces having clear, equal unfolding steps and crop them between the beginning of the plateau that precedes the first unfolding event and a point close to their detachment point (Fig. 4a); for disulfide reduction experiments, the first cut is made at the start of the reduction pulse (Fig. 4b). Discard traces with mixed unfolding and reduction steps in the second pulse.
-
16
Superimpose all selected and cropped traces obtained at a given force and choose an adequate duration limit. Exclude all traces where detachment occurred before this duration.
-
17
Sum the final set of cropped traces and normalize by dividing by the final summed length (Fig. 5c).
-
18
Calculate the measured rate and error using a bootstrapping analysis procedure that assumes a specific model for the process (such as an exponential constrained through the origin, specific for a two-state process (Figure 5c,d) or a more complex model, taking into account factors such as static disorder17).
-
19
Plot the measured rates in logarithmic scale as a function of force. Use the Arrhenius or other kinetic models to extract the activation energy and the distance to transition state (Fig. 8).
Check troubleshooting section
Figure 8.
Timing
Steps 1–7, Experiment preparation: 1–1.5 h
Steps 8–10, Cantilever calibration: 5 min
Steps 11–14, Data acquisition: 5 h – 2 d
Steps 15–19, Data analysis: 1–2 h.
TROUBLESHOOTING
Problem: ‘shaky’ signal (step 7)
There is unbiased random noise in the recordings, on a subsecond-to-second time scale, which is usually indicative of particulate diffusion in the solution. Particles present in the measuring buffer scatter the laser beam and add noise to the force measurement (in force extension) or length measurement (in force clamp). Dismantle the experiment, clean the fluid cell and replace the surface, and then restart from Step 2.
Problem: thermal fluctuations spectrum has an unusual shape (step 8)
The shape of the thermal fluctuations spectrum can indicate possible problems. A broken arm of a V-shaped cantilever or an air bubble will change the position of the resonance peak and can be solved by either replacing the cantilever or the buffer. We recommend using boxes of individual cantilevers rather than wafers to avoid damaging the cantilever in the process of carving. A thermal spectrum that is acquired too close to the surface affects the calibration, resulting in incorrect estimates of <x2> and the spring constant of the cantilever (Fig. 6b, blue trace). The separation between the surface and the cantilever should be increased in such a case. The presence of viscous compounds such as glycerol in the measuring solvent leads to a shift in all the fluctuation peaks that must be taken into account in order to integrate the full range of the resonant mode (Fig. 6b, green trace).
Problem: interference pattern is detected in the noncontact part of the force-extension traces (step 10)
Interference effects can appear when you are using reflective surfaces such as gold (Fig. 6a). The presence of interference induces a change in the real applied force as a function of surface-tip separation during the force-clamp experiment. Measurements obtained in the presence of interference have an error in the force proportional to the magnitude of the measured interference in force-extension curves. The use of nonreflective surfaces such as glass and mica may eliminate interference, but attachment of proteins to these surfaces is generally poorer than that on gold (with the possible exception of attachment to glass using covalent chemistry methods, such as the HaloTag method presented in Box 1). If you are working with reflective surfaces (such as gold), reduce or eliminate the interference either by slightly changing the position of the laser on the cantilever without severely affecting the total reflected voltage, or by refocusing the laser so that there is no light leakage. Alternatively, slightly tilt the surface so that the surface-reflected light does not hit the PD, or replace the cantilever.
Problem: constant compliance region is not linear (step 10)
As shown in Figure 6c, the constant compliance region may not have a linear domain when too much protein is adsorbed on the surface or when small cantilevers are used. If too much protein is adsorbed, restart the experiment using less protein. For small cantilevers, a clean uncoated surface can be used to obtain the slope of the PD signal with cantilever bending distance. Owing to the lack of a protein layer, measurements with this surface will have a well-defined inflection point when the cantilever touches the surface. In this case, the deflection-extension curve might also show a high-force detachment peak, characteristic for the cantilever-glass attraction. This peak does not interfere with estimation of the slope.
Problem: piezo hysteresis (step 10)
Piezo hysteresis can have a profound effect on cantilever calibration and measured sample extension. The deflection-extension curve used to obtain the constant compliance region might, in this case, show a different slope for the approach and retract part. The measured distance might also have errors arising from the linear extrapolation of the applied voltage versus piezo moving distance. Use linear piezo actuators or piezo actuators with position feedback. The presence of different slopes on the approach and redraw part of the constant compliance region can be indicative of a loosely attached surface (solution: glue the surface to the piezo again), a thick protein layer (solution: set up a new experiment with less protein), a bubble (solution: replace the buffer) or even a broken piezo (solution: send the piezo to repair or replace the piezo with a better one).
Problem: piezo calibration (step 10)
The calibration of the capacitive sensor of the piezo as a function of applied voltage is reported by the manufacturer, and this may cause substantial error due to the limitations of the calibration method used. The linear piezo actuator used in our setup is less prone to this kind of error. Whenever you are using a new piezo, verify and adjust the calibration of the positioning sensor from the unfolding steps of a known protein (for example, unfolding of ubiquitin at 100 pN shows step increases of 20 nm).
Large variations in the measured spring constant
Nowadays, most cantilevers are reproducibly manufactured to have the same spring constant, and little variation is expected from cantilevers within the same box. table 1 shows measured values for the spring constant of Bruker MLCT-C cantilevers on the same instrument.
Table 1.
Calibration parameters obtained on the same setup using different cantilevers from different boxes
| Box Number | s (nm/V) | <x2> (V2) | kc (pN/nm) |
|---|---|---|---|
| 1 | 146.7 | 1.30E-05 | 15.2 |
| 128.8 | 1.80E-05 | 14.1 | |
| 134.9 | 1.50E-05 | 15.4 | |
| 129.3 | 1.50E-05 | 15.8 | |
| 135.7 | 1.60E-05 | 14.0 | |
| 127.1 | 1.70E-05 | 14.9 | |
| 2 | 133.7 | 1.60E-05 | 14.7 |
| 140.0 | 1.60E-05 | 13.9 | |
| 120.4 | 2.00E-05 | 14.0 | |
| 117.2 | 1.90E-05 | 15.4 | |
| 3 | 140.8 | 1.40E-05 | 14.9 |
| 157.9 | 1.20E-05 | 13.3 | |
| 163.8 | 1.10E-05 | 13.3 | |
| 149.0 | 1.30E-05 | 13.9 | |
| 153.9 | 1.00E-05 | 16.9 | |
| 141.5 | 1.50E-05 | 13.9 | |
| 132.2 | 1.60E-05 | 14.6 | |
| 142.1 | 1.50E-05 | 13.9 | |
| 137.3 | 1.60E-05 | 14.6 | |
| 146.3 | 1.40E-05 | 13.5 | |
| 4 | 146.8 | 1.30E-05 | 14.5 |
| 136.7 | 1.60E-05 | 13.6 | |
| 115.8 | 1.80E-05 | 16.8 | |
| 113.4 | 2.20E-05 | 14.6 | |
| 130.5 | 1.80E-05 | 13.7 | |
| 116.4 | 2.30E-05 | 13.4 | |
| 116.2 | 2.30E-05 | 13.4 | |
| 5 | 117.0 | 2.00E-05 | 15.0 |
| 120.3 | 2.00E-05 | 14.1 | |
| 128.0 | 1.70E-05 | 14.5 | |
| 121.3 | 1.90E-05 | 14.4 | |
| 115.6 | 2.20E-05 | 14.2 | |
| 103.8 | 2.70E-05 | 14.3 | |
| 6 | 148.9 | 1.40E-05 | 13.3 |
| 138.3 | 1.50E-05 | 14.0 | |
| 126.8 | 2.00E-05 | 12.6 |
A significant deviation from the average spring constant of the cantilever usually indicates a broken cantilever arm or the presence of a bubble close to the laser reflection point. Try changing the buffer. If the anomalous value remains, change the cantilever. Other possible reasons for errors in the calibration of cantilevers include an incorrectly calibrated piezo (as explained above). Calibration data obtained for the same cantilever on three instruments available in our laboratory are shown in table 2, which shows that very similar values are obtained for the same cantilever in different setups.
Table 2.
Spring constants measured for the same cantilever on three different instruments
| AFM Setup | ks (pN/nm) |
|---|---|
| 1 | 14.34 |
| 2 | 14.27 |
| 3 | 15.50 |
PID response slower than reaction rate
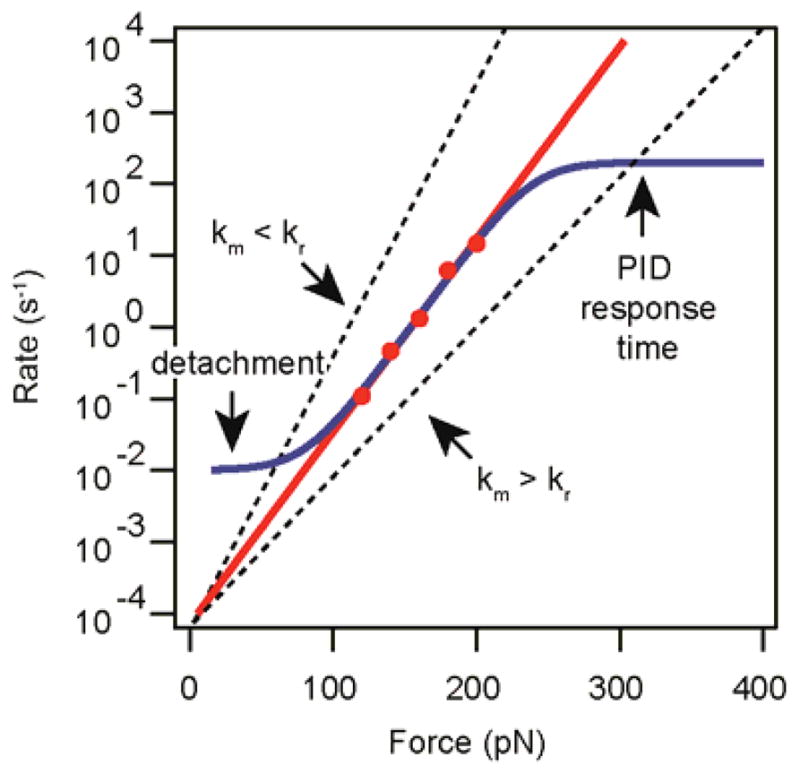
As shown in Figure 6d, the feedback response time might be too slow to capture fast events. Under these conditions, the setpoint force is reached in a time comparable to the rate of the measured process or is not reached at all for the fast-occurring events. The response time (i.e., the time it takes the feedback loop to recover the setpoint force after an extension event) depends on the properties of the cantilever and the settings of the PID loop. Increase the integral gain until it is close to resonance, or choose a faster cantilever. If the response time of a given cantilever model is longer than normal, replace the cantilever. A plateau in the high-rates region of the rate-force plot (Fig. 7) may be indicative of the fact that the PID response time is limiting the rates that can be measured.
Problem: the feedback creates oscillations (step 12)
Oscillations appear as high-frequency periodic noise in the recordings or as an audible high-pitch noise from the piezo. Decrease the gain factors in the feedback circuit. Try avoiding prolonged high-frequency oscillations, as they have a direct impact on the lifetime of the piezo actuator. This issue can also appear after prolonged use during the same experiment. Evaporation of the buffer can induce a change in the optimal feedback parameters.
Problem: too few pick-ups (step 13)
Try increasing the magnitude of the contact force (to −2 nN, for instance), or try increasing the contact time (to 2 s, for instance). If these changes do not work, open the fluid cell and add more protein sample to the surface.
Problem: many pick-ups but few clean fingerprints (step 13)
Traces lacking a clear unfolding fingerprint may originate from several proteins tethered in parallel, a degraded protein or contaminations. Try to reduce the magnitude and/or duration of the contact force pulse (−500 pN, 0.3 s for instance). If the problem persists, then set up the experiment again from Step 2 with less protein. If there are still no clean fingerprints, then the protein sample might be compromised. In our experience, protein preparations age and their performance in single- molecule AFM may decrease, even with no obvious changes in the behavior of the protein in SDS-PAGE or size-exclusion chromatography. In this case, purify a new batch of protein.
Problem: cantilever calibration changes over time (step 13)
Thermal drift and mechanical damage of the cantilever can occasionally lead to a change of the already calibrated parameters. We recommend obtaining a force-extension trace every few hours and checking that the contact part shows the same slope as for the original calibration. If a change in the slope of the constant compliance region is measured, the cantilever should be recalibrated by moving it away from the surface and repeating Steps 8–10. If a change in the total intensity of the laser takes place, reposition the laser to obtain maximum intensity and recalibrate the cantilever. If a substantial change in the value of the cantilever spring constant is measured, change the cantilever.
Problem: variation of the separation between the surface and the cantilever with time (step 13)
Mechanical drift can cause errors in the surface-tip distance measurement. This kind of drift can come from a poorly attached surface on the piezo, a poorly fixed cantilever or evaporation of the measuring solvent. Our instrument uses a piezo motor to adjust the separation between the cantilever and the surface before every attachment trial. If this kind of drift persists, check the attachment of the surface to the piezo and of the cantilever to the fluid cell. In addition, try adding more measuring buffer.
Problem: the force dependency shows a plateau at lower rates (step 19)
Measurement of slow rates can be biased by selecting traces whose duration is short compared with the kinetics of the reaction of interest22. Make sure that traces are long enough so that the majority of the reactions are completed before detachment (Fig. 7).
Problem: the force dependency of the studied process is not reproducible (step 19)
Errors in the calibration of the cantilever have a direct impact on the measured slope of the force dependency of the process (dotted lines in Fig. 7). A constant displacement of the cantilever can be expressed as Δz = Fr/kr = Fm/km, where Fr and kr are the real pulling force and spring constant, and Fm and km are the measured pulling force and spring constant. If we consider Arrhenius kinetics, the variation of the reaction rate with the measured force follows
Hence, an error in the calibration is directly reflected in the measured distance to the transition state through xm = (kr/km) · x. Therefore, overestimation of the spring constant (km > kr) leads to smaller experimental slopes (Fig. 7). In contrast, if the spring constant is underestimated (km < kr), x will be overestimated (Fig. 7). When you are using data from different miscalibrated experiments, the extrapolated rate at zero force may also be affected. To avoid calibration errors of this sort, cycle between forces within the same experiment. This approach minimizes any effect arising from time-dependent changes in calibration. Compare the force dependency measured for the same forces with different cantilevers. If the pick-up rate is too low to allow for measurement of the entire force range in a single experiment, choose at least one reference force.
If the calibration is correct, the rates of reaction at the reference force will be consistent for different experiments and/or cantilevers. If the results at the reference force deviate significantly from the average value, discard all data obtained for that particular experiment.
ANTICIPATED RESULTS
We have successfully used force-clamp AFM to study the force dependency of mechanical unfolding of proteins14,15, as well as chemical reactions including formation46, isomerization38 and cleavage of disulfide bonds13,16. Force-clamp AFM has also been used to study protein folding20,47, rupture of lipid bilayers48, mechanical strength of catch bonds49, avidin-biotin interaction50 and actin network growth51. For future studies, systems readily amenable for this technique include metal chelation52, receptor-ligand binding53, isopeptide bond formation54 and ring opening of cyclic compounds55.
Successful force-clamp recordings are fingerprinted by the appearance of staircases composed of characteristic step changes in the length of the protein, where each step marks a single unfolding event or a single chemical reaction. Owing to the probabilistic nature of processes taking place at the single-molecule level, no two traces are identical. Nevertheless, the aver- aged summed extensions of a sufficient number of individual traces (typically n > 20; Fig. 5c) render reproducible results.
In our experience, the proportion of successful recordings showing clean staircases varies greatly among proteins. Some proteins such as I27, protein L or ubiquitin, adsorbed on gold, can provide over 1,000 pickups per day, with an estimated 5–10% of traces showing clean fingerprints that can be used for further analysis. Although it is more difficult to implement, the covalent attachment chemistry presented in Box 1 yields higher pickup yield, and it has the advantage of an increased probability of tethering the polyprotein at its termini. Experiments using this chemistry show a 40–50% pickup efficiency, out of which ~25% are full-length unfolding traces. Therefore, a force dependency composed of four data points can be obtained in just one day or a few days of experiments. However, some proteins may aggregate or become inactive in a matter of days after their purification. The presence of reducing agents in the measuring buffer may also affect the pickup yield by disrupting the attachment to the gold surface. For proteins that are more challenging to measure, we recommend investing time in optimizing the experimental conditions (buffer, concentration of protein, incubation time and so on) so that at least 5–10 good traces are obtained per day.
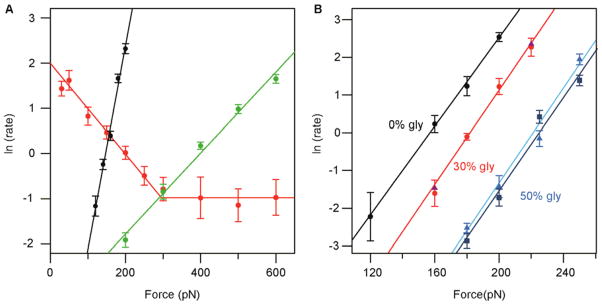
In Figure 8a, we show three different force dependencies for three different reactions: mechanical protein unfolding, chemical reduction of disulfides and enzyme-catalyzed reduction of disulfides. The different force dependencies report on the different nature of the energy landscape of the reactions. Protein unfolding shows a strong dependency with force, characterized by a large distance to the transition state, in the range of 1.6–2.5 Å (refs. 18,22,56,57; black points in Fig. 8a). The height of the unfolding energy barrier can be directly obtained by extrapolating the unfolding rate in the absence of force, by using the recently measured pre-exponential factor of 109 s−1 (ref. 19). The absolute value of the unfolding barrier changes from protein to protein, reminiscent of its different mechanical stability. Interestingly, the height of such an energy barrier is increased upon introduction of an osmolyte such as glycerol, which stabilizes the native conformation of the protein (Fig. 8b).
Chemical reactions using small nucleophiles such as thiols, phosphines or hydroxyl ions have a markedly smaller distance to the transition state, spanning between 0.3 and 0.5 Å (refs. 13,24,25) depending on the chemistry of the attacking nucleophile. Notably, thiol-based reducing agents such as DTT induce a smaller distance to the transition state (Δx = 0.31 Å) compared with phosphine-based reducing agents (Δx = 0.44 Å; ref. 25). Interestingly, ab initio simulations showed that during the reaction with a thiol-based nucleophile, the disulfide bond extends at the transition state with approximately the same length as the measured distance to the transition state Δx (ref. 13). This trend was further confirmed by studying the atomistic mechanisms underlying the reduction of nucleophiles with different central atoms (P, O)25. Hence, these observations suggest a direct relationship between the slope of the force dependency of a chemical reaction experimentally measured with sub-Ångström resolution by force-clamp AFM and the conformation of the transition state of the process along the pulling coordinate.
In contrast to the simple exponential force dependency of disulfide reduction by small chemical nucleophiles (Fig. 8a, green data set), reduction by thioredoxin enzyme shows a complex force dependency composed of separate regimes (Fig. 8a, red data set). These regimes report on different chemical mechanisms and conformational rearrangements used by the enzyme during its catalytic activity28. In the low-force regime, the negative force dependency of thioredoxin-catalyzed reactions underlies a shortening of the substrate, whereas the transition state acquires the geometry required for the SN2 mechanism16. Alternative reaction mechanisms become prominent at forces over 200 pN, making thioredoxin enzymes active over the whole range of experimental forces28. The extrapolated rate at zero force also contains important information that can be used to estimate the free energy of the process using specific attempt frequencies19,26.
If the guidelines described in this paper are followed, results are free of potential pitfalls that were not unusual just 5–10 years ago. The production of cantilevers has been improved, such that commercial cantilevers are reliable and give consistent results from batch to batch. Even in the challenging conditions imposed by viscous solutions, the calibration of cantilevers is highly reproducible. Results obtained in different setups and/or by different experimenters are also consistent. For example, in Figure 8b we show the unfolding rates of wild-type I27 measured with different cantilevers and setups (denoted using different symbols) and in HEPES buffer containing 0, 30 and 50% (vol/vol) glycerol (different color codes). There is little variation of the force dependency of I27 with glycerol concentration, although we detect a clear stabilization of the native state induced by the osmolyte, as indicated by the slower rate of mechanical unfolding. We included in this protocol saved measurements from a typical experiment, alongside the number of traces selected and a procedure for loading these traces in Igor Pro (Box 2 and supplementary Data). In summary, single-molecule force-clamp spectroscopy is a robust technique that provides insights into the molecular mechanisms governing biological reactions at the single-molecule level, such as protein folding or chemical reactions under mechanical force. These experiments have prompted a new field of computational analysis11,16,58, which will complement single-molecule experiments to provide an integrated framework in order to understand force-induced processes occurring within the core of an individual molecule.
BOX 2. Sample Data Set.
We have made available on-line the raw data from a single typical experiment that shows the full spectrum of challenges encountered when analyzing force-clamp measurements (Supplementary Materials). This data set probes the mechanical unfolding of I27 in the presence of 30% glycerol, measured at four different forces. After reading this practical guide, any reader should be able to i) select and cut traces showing clean fingerprints; ii) sum the traces at each force; iii) reproduce the force dependency of the reaction shown in Figure 8B (red squares). The reader is also encouraged to deviate from the protocols and observe many of the possible artifacts that we describe, for example when including traces with short detachment times. We also include the traces that we selected following the guidelines described in this paper. We used those selected traces to obtain the rates that appear in Figure 8B.
Force-clamp AFM has reached maturity. The commercialization of state-of-the-art setups will make the technique available to biochemists, chemists and biophysicists who want to explore the energy landscape of biological and chemical reactions. We expect to see improvement in the component parts of the AFM setup in the upcoming years, leading to an increase in sensitivity and speed. Our AFM setup already uses a linear piezo without hysteresis. Although this piezo is currently limited to relatively small travel distances of 800 nm, future models will increase this range. Smaller and softer cantilevers are expected to emerge alongside with better laser-focusing optics. These cantilevers will have a fast response time and little thermal noise. A change of the laser detection system to fluorescence or electromagnetic methods may allow for further reductions of the cantilever size and for further increases in the bandwidth of the AFM. Cantilever technology may expand the currently available force range and improve the system stability, allowing for measurement of hour-long traces. Furthermore, design of new protein substrates may allow for mechanochemical studies of covalent bonds besides disulfides. This approach requires the development of experimental systems that link a chemical reaction with a measured change in length. For example, specific residues in polyproteins can be cross-linked with small molecules containing reactive bonds that become exposed upon mechanical unfolding.
Supplementary Material
Acknowledgments
We acknowledge all the past and present members of the Fernandez laboratory for their contribution in developing the AFM force-clamp technique. We acknowledge Luigs & Neumann for the pictures of the AFM setup and R.T. Sauer from Massachusetts Institute of Technology for the ERL-competent cells. This work was supported by grants from the US National Institutes of Health (HL066030 and HL061228 to J.M.F.). I.P. acknowledges the Swiss National Science Foundation for a postdoctoral research grant. J.A-.C. acknowledges a Fellowship from Fundación Ibercaja.
Footnotes
Author contributions All the authors performed the measurements and wrote the manuscript.
Competing financial interests The authors declare competing financial interests: details are available in the online version of the paper.
Supplementary information is available in the online version of the paper. Raw data set for the mechanical unfolding of I27 in the presence of 30% glycerol, measured at four different forces. This is a single typical experiment that shows the full spectrum of challenges encountered when analyzing force-clamp measurements.
References
- 1.Rief M, Gautel M, Oesterhelt F, Fernandez JM, Gaub HE. Reversible unfolding of individual titin immunoglobulin domains by AFM. Science. 1997;276:1109–1112. doi: 10.1126/science.276.5315.1109. [DOI] [PubMed] [Google Scholar]
- 2.Oberhauser AF, Marszalek PE, Erickson HP, Fernandez JM. The molecular elasticity of the extracellular matrix protein tenascin. Nature. 1998;393:181–185. doi: 10.1038/30270. [DOI] [PubMed] [Google Scholar]
- 3.Rief M, Oesterhelt F, Heymann B, Gaub HE. Single-molecule force spectroscopy on polysaccharides by atomic force microscopy. Science. 1997;275:1295–1297. doi: 10.1126/science.275.5304.1295. [DOI] [PubMed] [Google Scholar]
- 4.Smith SB, Cui YJ, Bustamante C. Overstretching B-DNA: the elastic response of individual double-stranded and single-stranded DNA molecules. Science. 1996;271:795–799. doi: 10.1126/science.271.5250.795. [DOI] [PubMed] [Google Scholar]
- 5.Carrion-Vazquez M, et al. Mechanical design of proteins studied by single-molecule force spectroscopy and protein engineering. Prog Biophys Mol Biol. 2000;74:63–91. doi: 10.1016/s0079-6107(00)00017-1. [DOI] [PubMed] [Google Scholar]
- 6.Oberhauser AF, Badilla-Fernandez C, Carrion-Vazquez M, Fernandez JM. The mechanical hierarchies of fibronectin observed with single-molecule AFM. J Mol Biol. 2002;319:433–447. doi: 10.1016/S0022-2836(02)00306-6. [DOI] [PubMed] [Google Scholar]
- 7.Li H, et al. Reverse engineering of the giant muscle protein titin. Nature. 2002;418:998–1002. doi: 10.1038/nature00938. [DOI] [PubMed] [Google Scholar]
- 8.Carrion-Vazquez M, et al. The mechanical stability of ubiquitin is linkage dependent. Nat Struct Biol. 2003;10:738–743. doi: 10.1038/nsb965. [DOI] [PubMed] [Google Scholar]
- 9.Brockwell DJ, et al. Pulling geometry defines the mechanical resistance of a β-sheet protein. Nat Struct Biol. 2003;10:731–737. doi: 10.1038/nsb968. [DOI] [PubMed] [Google Scholar]
- 10.Dietz H, Rief M. Exploring the energy landscape of GFP by single-molecule mechanical experiments. Proc Natl Acad Sci USA. 2004;101:16192–16197. doi: 10.1073/pnas.0404549101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iozzi MF, Helgaker T, Uggerud E. Influence of external force on properties and reactivity of disulfide bonds. J Phys Chem A. 2011;115:2308–2315. doi: 10.1021/jp109428g. [DOI] [PubMed] [Google Scholar]
- 12.Ainavarapu SR, et al. Contour length and refolding rate of a small protein controlled by engineered disulfide bonds. Biophys J. 2007;92:225–233. doi: 10.1529/biophysj.106.091561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wiita AP, Ainavarapu SR, Huang HH, Fernandez JM. Force-dependent chemical kinetics of disulfide bond reduction observed with single-molecule techniques. Proc Natl Acad Sci USA. 2006;103:7222–7227. doi: 10.1073/pnas.0511035103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oberhauser AF, Hansma PK, Carrion-Vazquez M, Fernandez JM. Stepwise unfolding of titin under force-clamp atomic force microscopy. Proc Natl Acad Sci USA. 2001;98:468–472. doi: 10.1073/pnas.021321798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schlierf M, Li H, Fernandez JM. The unfolding kinetics of ubiquitin captured with single-molecule force-clamp techniques. Proc Natl Acad Sci USA. 2004;101:7299–7304. doi: 10.1073/pnas.0400033101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wiita AP, et al. Probing the chemistry of thioredoxin catalysis with force. Nature. 2007;450:124–127. doi: 10.1038/nature06231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuo TL, et al. Probing static disorder in Arrhenius kinetics by single-molecule force spectroscopy. Proc Natl Acad Sci USA. 2010;107:11336–11340. doi: 10.1073/pnas.1006517107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia-Manyes S, Dougan L, Fernandez JM. Osmolyte-induced separation of the mechanical folding phases of ubiquitin. Proc Natl Acad Sci USA. 2009;106:10540–10545. doi: 10.1073/pnas.0902090106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Popa I, Fernandez JM, Garcia-Manyes S. Direct quantification of the attempt frequency determining the mechanical unfolding of ubiquitin protein. J Biol Chem. 2011;286:31072–31079. doi: 10.1074/jbc.M111.264093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garcia-Manyes S, Dougan L, Badilla CL, Brujic J, Fernandez JM. Direct observation of an ensemble of stable collapsed states in the mechanical folding of ubiquitin. Proc Natl Acad Sci USA. 2009;106:10534–10539. doi: 10.1073/pnas.0901213106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berkovich R, et al. Rate limit of protein elastic response is tether dependent. Proc Natl Acad Sci USA. 2012;109:14416–14421. doi: 10.1073/pnas.1212167109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garcia-Manyes S, Brujic J, Badilla CL, Fernandez JM. Force-clamp spectroscopy of single-protein monomers reveals the individual unfolding and folding pathways of I27 and ubiquitin. Biophys J. 2007;93:2436–2446. doi: 10.1529/biophysj.107.104422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walther KA, et al. Signatures of hydrophobic collapse in extended proteins captured with force spectroscopy. Proc Natl Acad Sci USA. 2007;104:7916–7921. doi: 10.1073/pnas.0702179104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garcia-Manyes S, Liang J, Szoszkiewicz R, Kuo TL, Fernandez JM. Force-activated reactivity switch in a bimolecular chemical reaction. Nat Chem. 2009;1:236–242. doi: 10.1038/nchem.207. [DOI] [PubMed] [Google Scholar]
- 25.Ainavarapu SRK, Wiita AP, Dougan L, Uggerud E, Fernandez JM. Single-molecule force spectroscopy measurements of bond elongation during a bimolecular reaction. J Am Chem Soc. 2008;130:6479–6487. doi: 10.1021/ja800180u. [DOI] [PubMed] [Google Scholar]
- 26.Liang J, Fernandez JM. Kinetic measurements on single-molecule disulfide bond cleavage. J Am Chem Soc. 2011;133:3528–3534. doi: 10.1021/ja109684q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perez-Jimenez R, et al. Single-molecule paleoenzymology probes the chemistry of resurrected enzymes. Nat Struct Mol Biol. 2011;18:U592–U599. doi: 10.1038/nsmb.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perez-Jimenez R, et al. Diversity of chemical mechanisms in thioredoxin catalysis revealed by single-molecule force spectroscopy. Nat Struct Mol Biol. 2009;16:890–896. doi: 10.1038/nsmb.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carrion-Vazquez M, et al. Mechanical and chemical unfolding of a single protein: a comparison. Proc Natl Acad Sci USA. 1999;96:3694–3699. doi: 10.1073/pnas.96.7.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Furuike S, Ito T, Yamazaki M. Mechanical unfolding of single filamin A (ABP-280) molecules detected by atomic force microscopy. Febs Lett. 2001;498:72–75. doi: 10.1016/s0014-5793(01)02497-8. [DOI] [PubMed] [Google Scholar]
- 31.Hutter JL, Bechhoefer J. Calibration of atomic-force microscope tips. Rev Sci Instrum. 1993;64:1868–1873. [Google Scholar]
- 32.Taniguchi Y, Kawakami M. Application of HaloTag protein to covalent immobilization of recombinant proteins for single molecule force spectroscopy. Langmuir. 2010;26:10433–10436. doi: 10.1021/la101658a. [DOI] [PubMed] [Google Scholar]
- 33.Wang T, Arakawa H, Ikai A. Force measurement and inhibitor binding assay of monomer and engineered dimer of bovine carbonic anhydrase B. Biochem Biophys Res Commun. 2001;285:9–14. doi: 10.1006/bbrc.2001.5118. [DOI] [PubMed] [Google Scholar]
- 34.Kufer SK, et al. Covalent immobilization of recombinant fusion proteins with hAGT for single-molecule force spectroscopy. Eur Biophys J Biophys Lett. 2005;35:72–78. doi: 10.1007/s00249-005-0010-1. [DOI] [PubMed] [Google Scholar]
- 35.Zakeri B, et al. Peptide tag forming a rapid covalent bond to a protein, through engineering a bacterial adhesin. Proc Natl Acad Sci USA. 2012;109:E690–E697. doi: 10.1073/pnas.1115485109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brujic J, Hermans RIZ, Garcia-Manyes S, Walther KA, Fernandez JM. Dwell-time distribution analysis of polyprotein unfolding using force-clamp spectroscopy. Biophys J. 2007;92:2896–2903. doi: 10.1529/biophysj.106.099481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Szoszkiewicz R, et al. Dwell time analysis of a single-molecule mechanochemical reaction. Langmuir. 2008;24:1356–1364. doi: 10.1021/la702368b. [DOI] [PubMed] [Google Scholar]
- 38.Alegre-Cebollada J, Kosuri P, Rivas-Pardo JA, Fernandez JM. Direct observation of disulfide isomerization in a single protein. Nat Chem. 2011;3:882–887. doi: 10.1038/nchem.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garcia-Manyes S, Kuo TL, Fernandez JM. Contrasting the individual reactive pathways in protein unfolding and disulfide bond reduction observed within a single protein. J Am Chem Soc. 2011;133:3104–3113. doi: 10.1021/ja109865z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bell GI. Models for the specific adhesion of cells to cells. Science. 1978;200:618–627. doi: 10.1126/science.347575. [DOI] [PubMed] [Google Scholar]
- 41.Evans E. Probing the relation between force—lifetime—and chemistry in single molecular bonds. Annu Rev Biophys Biomol. 2001;30:105–128. doi: 10.1146/annurev.biophys.30.1.105. [DOI] [PubMed] [Google Scholar]
- 42.Dudko OK, Hummer G, Szabo A. Intrinsic rates and activation free energies from single-molecule pulling experiments. Phys Rev Lett. 2006;96:108101. doi: 10.1103/PhysRevLett.96.108101. [DOI] [PubMed] [Google Scholar]
- 43.Brujic J, Hermans RI, Walther KA, Fernandez JM. Single-molecule force spectroscopy reveals signatures of glassy dynamics in the energy landscape of ubiquitin. Nat Phys. 2006;2:282–286. [Google Scholar]
- 44.Berkovich R, Garcia-Manyes S, Urbakh M, Klafter J, Fernandez JM. Collapse dynamics of single proteins extended by force. Biophys J. 2010;98:2692–2701. doi: 10.1016/j.bpj.2010.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Efron B. The Jackknife, the Bootstrap, and Other Resampling Plans. Society for Industrial and Applied Mathematics; 1982. [Google Scholar]
- 46.Kosuri P, et al. Protein folding drives disulfide formation. Cell. 2012;151:794–806. doi: 10.1016/j.cell.2012.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fernandez JM, Li H. Force-clamp spectroscopy monitors the folding trajectory of a single protein. Science. 2004;303:1674–1678. doi: 10.1126/science.1092497. [DOI] [PubMed] [Google Scholar]
- 48.Redondo-Morata L, Giannotti MI, Sanz F. AFM-based force-clamp monitors lipid bilayer failure kinetics. Langmuir. 2012;28:6403–6410. doi: 10.1021/la3005147. [DOI] [PubMed] [Google Scholar]
- 49.Le Trong I, et al. Structural basis for mechanical force regulation of the adhesin FimH via finger trap-like β sheet twisting. Cell. 2010;141:645–655. doi: 10.1016/j.cell.2010.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Favre M, et al. Force-clamp spectroscopy with a small dithering of AFM tip, and its application to explore the energy landscape of single avidin-biotin complex. Ultramicroscopy. 2007;107:882–886. doi: 10.1016/j.ultramic.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 51.Choy JL, et al. Differential force microscope for long time-scale biophysical measurements. Rev Sci Instrum. 2007;78:043711. doi: 10.1063/1.2727478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zheng P, Li HB. Highly covalent ferric-thiolate bonds exhibit surprisingly low mechanical stability. J Am Chem Soc. 2011;133:6791–6798. doi: 10.1021/ja200715h. [DOI] [PubMed] [Google Scholar]
- 53.Kim J, Zhang CZ, Zhang XH, Springer TA. A mechanically stabilized receptor-ligand flex-bond important in the vasculature. Nature. 2010;466:U992–U123. doi: 10.1038/nature09295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alegre-Cebollada J, Badilla CL, Fernandez JM. Isopeptide bonds block the mechanical extension of pili in pathogenic Streptococcus pyogenes. J Biol Chem. 2010;285:11235–11242. doi: 10.1074/jbc.M110.102962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Klukovich HM, Kouznetsova TB, Kean ZS, Lenhardt JM, Craig SL. A backbone lever-arm effect enhances polymer mechanochemistry. Nat Chem. 2013;5:110–114. doi: 10.1038/nchem.1540. [DOI] [PubMed] [Google Scholar]
- 56.Liu RC, Garcia-Manyes S, Sarkar A, Badilla CL, Fernandez JM. Mechanical characterization of protein L in the low-force regime by electromagnetic tweezers/evanescent nanometry. Biophys J. 2009;96:3810–3821. doi: 10.1016/j.bpj.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cao Y, Kuske R, Li HB. Direct observation of Markovian behavior of the mechanical unfolding of individual proteins. Biophys J. 2008;95:782–788. doi: 10.1529/biophysj.107.128298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li WJ, Grater F. atomistic evidence of how force dynamically regulates thiol/disulfide exchange. J Am Chem Soc. 2010;132:16790–16795. doi: 10.1021/ja104763q. [DOI] [PubMed] [Google Scholar]
- 59.Zimmermann JL, Nicolaus T, Neuert G, Blank K. Thiol-based, site-specific and covalent immobilization of biomolecules for single-molecule experiments. Nat Protoc. 2010;5:975–985. doi: 10.1038/nprot.2010.49. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.