Abstract
Mortalin, an essential mitochondrial chaperone protein, has previously been implicated in the pathogenesis of a wide array of diseases, including neurodegenerative conditions such as Parkinson's disease (PD) and Alzheimer's disease. Previous reports have consistently described mortalin protein levels to be lower in the brain tissue of patients with neurodegenerative disease, with expression demonstrated to be lower in neurons of post‐mortem PD brain specimens. However, to date, mortalin expression has not yet been evaluated in astrocytes of post‐mortem brain tissue from either normal or PD subjects. Mortalin expression was demonstrated in mouse primary astrocyte cultures by Western blot and quantitative polymerase chain reaction (PCR). Furthermore, confocal microscopy studies in human post‐mortem tissue indicated co‐localization of mortalin within astrocytes. Utilizing a quantitative immunofluorescence staining approach, the protein was found to be moderately reduced (∼35%) in this cell type in the substantia nigra pars compacta, but not structures of the corpus striatum, in PD subjects as compared to age‐/gender‐matched controls. These findings highlight the potential contribution of disrupted astroglial function in the pathogenesis of PD.
Keywords: astrocytes, fluorescence microscopy, human brain tissue, mortalin, Parkinson's disease
Introduction
The pathologic and clinical features defining Parkinson's disease (PD) have been established for several decades; however, the underlying mechanisms that lead to disease initiation and progression remain poorly understood. Recent advances in genetic screening technology have led to the identification of at least 18 loci for which mutation and/or aberrant expression are associated with disease incidence 12. While the study of these genes and their protein products has led to considerable insight into dysfunctional processes and pathways that may contribute to disease development, the mechanistic etiology of the vast majority of idiopathic PD incidence is incompletely understood. Additionally, to date, mechanistic investigations regarding PD pathogenesis have largely focused on neurons, whose dysfunction and eventual death are responsible for the cardinal clinical signs of the disease. Yet, a growing body of evidence implicates critical roles for astrocytes, which play vital roles in supporting the health of neurons under normal conditions 2 and in disease pathogenesis 8.
In the current investigation, we focused on mortalin, a multi‐functional chaperone protein previously implicated to play a role in PD pathology 3, 4, 6, 10, 11, 17. A member of the HSP70 family of proteins 22, mortalin, has been described as being functionally important in the import of nuclear proteins into the mitochondria, maintenance of protein integrity and mitochondrial biogenesis 7. Its functionality is not exclusive to the mitochondrion, as mortalin may translocate to the cytosol where it participates in a number of cellular processes, including signal transduction 14, 15, stress response 19, cell proliferation 23 and cell survival 20. The protein has been demonstrated to be decreased in the mitochondrial fraction of whole tissue lysates in the substantia nigra pars compacta (SNpc) of PD patients 10 and was found to be associated with disease severity 17. In vitro mechanistic studies have demonstrated the direct interaction of mortalin with α‐synuclein 11, a protein intricately linked to PD pathogenesis. Further, mutated forms of mortalin 3 and its reduced expression 10 enhance neuronal toxicity in cell model systems.
While it has been shown that mortalin is decreased in neuronal cell bodies of PD patients 10, mortalin expression within astrocytes has not been quantitatively evaluated to date. In this study, we first demonstrate that mortalin is expressed in brain astrocytes, and subsequently measure the relative levels of mortalin protein in the SNpc and structures of the corpus striatum in the post‐mortem tissue of PD patients and healthy age‐ and gender‐matched controls.
Materials and Methods
Identification of mortalin in astrocytes
Primary cortical glial cell cultures were generated from postnatal mouse pup brains harvested at 1–3 days following birth. Following decapitation, brains were carefully removed and transferred into ice‐cold Dulbecco's Modified Eagle Medium: Nutrient Mixture F‐12 (DMEM/F12) culture media (Gibco #11330‐032, Life Technologies, Grand Island, NY, USA). Using a Leica L2 microscope (Leica Microsystems Inc., Buffalo Grove, IL, USA), both hemispheres of the cortex were isolated and meninges were carefully removed mechanically. Isolated cortical tissue was washed with plain DMEM/F12 media three times, transferred to digestion media [0.5 mM ethylenediaminetetraacetic acid (EDTA), 0.2 mg/mL L‐cysteine, 150 U/mL papain, 10 mg/mL DNase in DMEM/F12] and incubated at 37°C for 30 minutes. Following digestion, the tissue was washed three times with culture medium (10% fetal bovine serum and 1% penicillin/streptomycin in DMEM/F1) pre‐equilibrated to 37°C. After the final wash, the tissue was triturated using a flame‐polished glass pipette in 10 mL of culture medium, and the supernatant passed through a 100‐μm nylon cell strainer into a conical tube. This process was repeated in 5 mL of culture medium, and the resulting supernatant was transferred into 75 cm2 vented tissue culture flasks (BD Falcon #35136, BD Biosciences, San Jose, CA, USA) coated with poly‐D‐lysine hydrobromide (BD Biosciences #354210). Following overnight incubation at 37°C with a 5% CO2 atmosphere, the culture medium was replaced and the cultures were returned to the incubator for an additional 9–14 days before experimental use.
Following the incubation period, microglia were removed forcibly by striking the flask several times, aspirating the media, striking the flask again and washing with phosphate‐buffered saline (PBS) (pH 7.4). Trypsin (with 0.25% EDTA, Gibco #25200‐056) was then added to the cellular monolayer, the flask was returned to the incubator for 5 minutes and cells were collected by washing with 10 mL of culture medium. The supernatant was transferred to a conical tube, centrifuged at 1900 × g and the pellet was resuspended to a plating density of 5 × 105 cells/mL. This preparation resulted in a culture of primary mouse astrocytes of >95% purity (data not shown).
Transfection
Cells to be transfected were plated as described above in culture medium without antibiotics. Transfections were carried out using Lipofectamine 2000 reagent (Life Technologies #11668019) according to the manufacturer's protocol, with slight modifications. Briefly, Lipofectamine and 2× siRNA [either mortalin (Mm_Hspa9a_6 #SI02712710) or AllStars Negative Control (#1027201), Qiagen (Venlo, Limburg, The Netherlands)] were incubated in Opti MEM media (Life Technologies #31985070) for 5 minutes at room temperature, combined and incubated at room temperature for an additional 20 minutes. Following the incubation period, the Lipofectamine/siRNA complex was added to the antibiotic‐free media, stored in the incubator overnight and the media changed to that containing antibiotics.
Western blot
After allowing primary astrocytes to grow to confluency in a poly‐D‐lysine‐coated 6‐well plate, the cells were washed once with ice‐cold PBS, collected in 100 μL lysis buffer [150 μM NaCl, 500 μM Tris–HCl, 1% (v/v) NP‐40, 10% (v/v) 1× protease inhibitor cocktail, 1 μM PMSF, 10 μM NaF, 0.2 μM Na3VO4 in deionized water], immediately flash‐frozen on dry ice and stored at −80°C. After thawing on ice, cellular lysates were sonicated, centrifuged at 21 130 × g and the supernatants were transferred to a clean 1.5‐mL Eppendorf tube. Following protein quantification by biochronic acid assay according to the manufacturer's protocol (Pierce #23225, Thermo Scientific, Waltham, MA, USA), equal amounts of protein were diluted into 2× Laemmli sample buffer (#61‐0737, Bio‐Rad Laboratories, Inc., Hercules, CA, USA) containing 5% (v/v) 2‐mercaptoethanol, loaded onto a 10–20% Tris–HCl polyacrylamide gel (Bio‐Rad #345‐0042) and separated by electrophoresis (100 V for 10 minutes; 120 V for 110 minutes). Proteins were then transferred to a polyvinyl difluoride (PVDF) membrane (Bio‐Rad #162–0177) by applying a constant current of 0.36A for ∼18 h.
For protein visualization, membranes were first incubated for 1 h at room temperature in blocking buffer (5% non‐fat milk in TBS‐T), followed by incubation with the primary antibody (mortalin #ADI‐SPS‐825, Enzo Life Sciences, Inc., Farmingdale, NY, USA; β‐actin #ab8226, Abcam plc, Cambridge, MA, USA) and diluted into blocking buffer overnight at 4°C. Membranes were then washed three times with TBS‐T, and the membrane was incubated with the secondary antibody (anti‐mouse IgG, HRP‐linked #50‐195‐914, Cell Signaling Technology, Beverly, MA, USA) for 1 h at room temperature. After washing three times with TBS‐T, enhanced chemiluminescent reagents (Amersham ECL #RPN2209, GE Healthcare Bio‐Sciences, Pittsburgh, PA, USA) were applied to the membrane for 2 minutes and the signal visualized on X‐ray film.
Quantitative polymerase chain reaction (PCR)
Primary astrocytes were plated into 6‐well plates as described earlier. After reaching confluency, the cells were washed once with room temperature PBS, collected in Trizol reagent (Invitrogen #15596‐026, Life Technologies) and total RNA were isolated via a phenol–chloroform extraction method with slight modifications 5. Genomic DNA was removed using a TURBO DNA‐free Kit (Life Technologies #AM1907), isolated RNA was quantified using a NanoDrop ND 1000 spectrophotometer (Thermo Scientific), and cDNA was generated using equal inputs of RNA template using a high‐capacity cDNA reverse transcription kit (Life Technologies #4368814). Quantitative real‐time PCR was performed using TaqMan reagents (Life Technologies) with off‐the‐shelf primer sets [Life Technologies #4331185; Hsap9 Mm00477716_g1 (mortalin); Gfap Mm01253033_m1 (GFAP); Rbfox3 Mm01248771_m1 (NeuN); Cd68 Mm03047340_m1 (CD68)] according to the manufacture's protocol on a ViiA7 PCR system (Life Technologies).
Human brain specimen preparation
This study was approved by the University of Washington Institutional Review Board. Formalin‐fixed paraffin embedded human brain specimens were provided from six PD patients and six age‐/gender‐matched controls for this study courtesy of the University of Washington Alzheimer's Disease Research Center neuropathology core. Specimens from the midbrain and the corpus striatum were cut to a thickness of 4–5 μm and tissue sections were mounted onto glass microscope slides.
Immunofluorescent staining
Human tissue sections were prepared as described previously 9, with slight modifications. Briefly, the tissues were deparaffinized in 100% xylenes (4 × 10 minutes), washed in a 1:1 mixture of xylenes and ethanol (2 × 5 minutes) and rehydrated by washing in 100% (2 × 5 minutes), 95% (1 × 3 minutes), 70% (1 × 3 minutes) and 50% (1 × 3 minutes) ethanol. Tissues were then washed in deionized water (briefly) and PBS (2 × 5 minutes) and antigen retrieval was carried out by heating specimens submerged in 10 mM citric acid (pH 6.0) to a boil and maintaining them at a high temperature for 15 minutes. After cooling for 30 minutes at room temperature, tissues were washed with 50 mM Tris‐buffered saline containing 0.05% Tween‐20 (TBS‐T; 3 × 10 min) and incubated with blocking buffer (5% normal goat serum, 2% bovine serum albumin, 0.1% Triton 100‐X diluted in TBS‐T) overnight at 4°C. The following day, the primary antibodies [rabbit anti‐mortalin (Sigma SQ‐15 #G4045) and chicken anti‐GFAP (Millipore, Billerica, MA, USA, #AB5541)] were diluted (anti‐mortalin 1:100; anti‐GFAP 1:500) into blocking buffer and again incubated with tissues overnight at 4°C. Following the primary antibody incubation period, tissues were washed with TBS‐T containing 5% normal goat serum and 2% bovine serum albumin (3 × 10 min), and secondary antibodies conjugated with Alexa fluorophores (568 goat anti‐chicken and 488 goat anti‐rabbit, Life Sciences, Inc.) diluted 1:500 in washing buffer and were incubated at 4°C overnight. The following day, the tissues were washed in TBS‐T (3 × 10 minutes) and gently rocked in a solution of 70% ethanol containing 0.3% Sudan Black B to reduce autofluorescence. Finally, tissues were briefly rinsed with 70% ethanol, washed in TBS‐T (3 × 10 minutes), covered with Vectashield mounting medium with DAPI (Vector Laboratories, Inc., Burlingame, CA, USA), affixed with a cover glass and sealed with nail polish.
Confocal microscopy
Images of human astrocytes labeled for GFAP and mortalin as described earlier were acquired using an Olympus (Waltham, MA, USA) Fluoview‐1000 (version 3.1b) scanning confocal microscope [488 nm argon ion laser (green) and 561 nm DPSS laser (red)] at the Center on Human Development and Disability at the University of Washington. 60× (numerical aperture 1.35) and 100× (numerical aperture 1.40) PlanSApo oil immersion objectives were used to acquire 1024 × 1024 pixel images with a 2× zoom. Channel images were obtained sequentially by frame, using Kalman averaging. A step size of 0.2 μm was used to obtain a Z‐series of 26–38 image planes per channel. Single‐plane TIFF images were deconvolved using NIS‐Elements Software (Nikon Elements Inc., Melville, NY, USA).
Quantitative fluorescence microscopy
Relative quantification of astroglial mortalin signal intensity within the SNpc was performed as described previously 9, with slight modifications. Imaging was carried out using a Nikon Eclipse Ti (Nikon Instruments Inc., Melville, NY, USA) instrument under 60× magnification, and exposure times for each channel were determined by randomly using the auto‐exposure feature over several random fields across several unique subjects. The chosen exposure times were 250 ms for the 568‐nm signal (GFAP) and 450 ms for the 488‐nm signal (mortalin). Z‐series images were acquired from 15 randomly selected astroglial‐containing fields based on GFAP‐positive signals. Following deconvolution, regions of interest were defined by their GFAP signal and mortalin fluorescence intensity was quantified within the astrocytes using NIS‐Elements software (Nikon).
Relative quantification of astroglial mortalin signal intensity within the structures of the corpus striatum (caudate, putamen, globus pallidus internal and external segments) was performed as above, but using a DeltaVision Elite imaging system (GE Healthcare, Issaquah, WA, USA), which offers the advantage of extremely short exposure times and very fast data acquisition. Using the auto‐exposure feature in eight randomly selected fields for three unique subjects, the exposure times were chosen to be 18.0 ms for the 56‐nm signal and 18.2 ms for the 488‐nm signal. Data were processed and analyzed as described previously, but using softWoRx software (GE Healthcare).
Results
Identification of mortalin in astrocytes
As mentioned earlier, mortalin has not previously been identified in astrocytes of healthy individuals and its involvement in neurodegenerative diseases has only been described in neurons. Given that mortalin antibody could, at least in theory, cross‐interact with other proteins, we first sought to show that mortalin is definitively expressed in astrocytes. We first utilized mouse primary astrocytes to accomplish this. The presence of mortalin within purified mouse primary astrocytes was identified by both Western blot and quantitative PCR (Figure 1). The positive identification of the protein as mortalin was further confirmed by knocking down the HSPA9 gene with siRNA (Figure 1A). Furthermore, the purity of the mouse primary astrocyte cultures was confirmed by high gene expression of GFAP (astroglial marker) and very low expression (Ct > 35) of NeuN (neuronal marker) and CD68 (microglial marker; Figure 1B).
Figure 1.
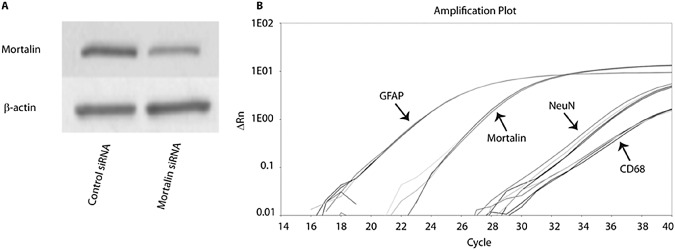
Identification of mortalin in mouse primary astrocytes. Astroglial expression of native mortalin was demonstrated by Western blot and quantitative polymerase chain reaction (qPCR). A. Detection of mortalin by Western blot along with confirmation following HSPA9 siRNA transfection. B. Mortalin gene expression was detected by qPCR along with high expression of GFAP and low expression of NeuN and CD68, demonstrating purity of the culture and therefore high probability of the presence of mortalin in astrocytes.
Human subject overview
Human brain tissue specimens from the SNpc and the corpus striatum were obtained from six PD subjects and six age/gender‐matched controls (Table 1). On average, controls were slightly older (73.7 years of age) upon autopsy as compared to PD subjects (71.8 years of age), but differences were not statistically significant. Each group was composed of four males and two females, reflecting the relative predisposition of PD in males as compared to females. The post‐mortem interval was slightly higher in PD patients (11.6 h) compared with controls (6.2 h) but again was not statistically significant. PD disease duration ranged from 2 to 43 years.
Table 1.
Human subjects. Abbreviations: HC = healthy controls; PD = Parkinson's disease; PMI = post‐mortem interval
| HC (n = 6) | PD (n = 6) | |
|---|---|---|
| Age (years) | 73.67 ± 9.91 | 71.86 ± 11.55 |
| Sex (female/male) | 4/2 | 4/2 |
| PMI (h) | 6.22 ± 2.60 | 11.60 ± 7.80 |
| Age at onset (years) | 48.00 ± 14.34 | |
| Disease duration (years) | 22.40 ± 16.13 |
Data presented as mean ± standard deviation.
Subjects were matched on age and gender.
Differential mortalin protein expression human astrocytes
Building upon our findings in mouse astrocytes, we found that the mortalin protein resides within human astrocytes in both control and PD subjects utilizing confocal microscopy techniques (Figure 2). To the best of our knowledge, these data represent the first report of the presence of native mortalin protein within human astrocytes that are not tumorigenic.
Figure 2.
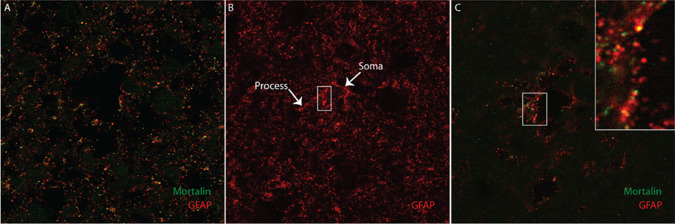
Identification of mortalin in human astrocytes. Human brain tissue was stained for GFAP (red) and mortalin (green). Co‐localization was observed by fluorescent confocal microscopy, represented by distinct yellow puncta. A. Representative full field image at 60× magnification. B. Representative stack projection of a 60× field stained for GFAP demonstrating the appearance of an astrocyte. White arrowheads indicate the astroglial soma and associated process. C. Representative single plane image at 100× magnification, with 3× zoom inset in the area indicated by the white box.
To characterize potential differences in astroglial mortalin protein levels between PD subjects and controls, a quantitative fluorescence microscopy approach was employed. A significant decrease of mortalin protein fluorescent signal was observed within astrocytes in the SNpc of PD subjects (n = 248 astrocytes) as compared to controls (n = 206 astrocytes). Conversely, no significant change in mortalin protein was found in the structures of the corpus striatum of PD subjects as compared to controls (Figure 3).
Figure 3.
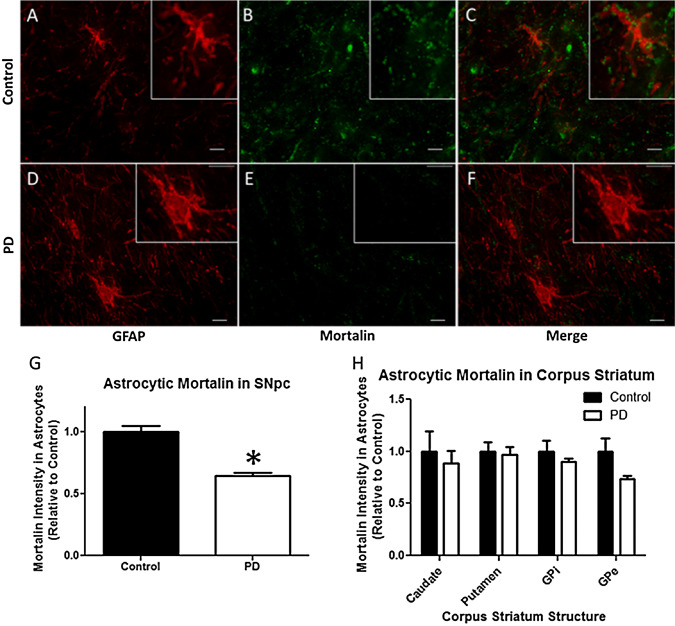
Astroglial mortalin is decreased in the substantia nigra pars compacta (SNpc) of Parkinson's disease (PD) patients. Human brain tissue from PD patients (n = 6) and age‐/gender‐matched controls (n = 6) was stained for GFAP (red) and mortalin (green), and the mortalin fluorescent signal within GFAP marked astrocytes quantified. A–C. Representative images from the SNpc of a control specimen and (D–F) for a PD specimen. G. Mortalin was found to be significantly lower in the astrocytes of the SNpc of PD subjects as compared to controls, whereas (H) no significant differences were observed in structures of the corpus striatum. Scale bar indicates 10 μm. *P < 0.05.
Discussion
In this study, we provide evidence of the presence of mortalin in both primary mouse astrocyte cultures and post‐mortem human brain tissue. To date, mortalin has been studied largely in the context of neurons, with only a few reports describing enhanced mortalin expression in gliomas 1, 13, 18 and not detected in normal astrocytes by immunohistochemistry 18. Here, we used immunofluorescence which may be significantly more sensitive than standard immunohistochemistry techniques. Further, we applied primary and secondary antibodies for an extended amount of time as compared to that reported previously 18. Taken together, these differences may explain why we were able to detect mortalin within astrocytes in this study. Increased mortalin expression in carcinogenic astrocytes is consistent with the general observation that the protein becomes overexpressed in tumorigenic cells 24. On the contrary, a role for mortalin in healthy astrocytes or its dysfunction in the astrocytes of neurodegenerative patients has not yet been described. Given that mortalin is known to be decreased in the brain tissue of not only PD patients but also AD patients 16, the demonstration of the presence of mortalin within astrocytes is significant in that it opens the possibility that the protein is modulated in neurodegenerative disease in cell types other than neurons.
To address the question of whether or not mortalin is altered in astrocytes under disease conditions, we utilized a quantitative immunofluorescent staining approach to investigate astroglial mortalin levels in PD patients as compared to age‐ and gender‐matched controls. Interestingly, we found a statistically significant 35% decrease in mortalin protein within astrocytes of the SNpc in PD patients, despite the fact that neurodegeneration is usually associated with increased astrogliosis. This result is consistent with a previous report on decreased mortalin in the mitochondrial fraction of whole tissue SNpc lysates in PD patients 10. Conversely, we found no significant difference within the structures of the corpus striatum. These results reflect the pathology typically observed in PD, with severe neuronal loss and the presence of Lewy bodies in the SNpc, while neurons of both the striatum and globus pallidus do not typically develop Lewy bodies and are far less affected in PD cases without accompanying dementia 21. It should be noted that the average age of the PD subjects studied is relatively young at 47 years of age, which is largely driven by the inclusion of one subject who was 28 years of age at diagnosis. The subject was negative for PARK2 (parkin) gene mutations that typically result in early onset of PD. More importantly, removing this subject from consideration had no significant impact on the overall results.
Whether a reduction of mortalin has an impact on astroglial function, and what effect this may have on neuronal health, is currently unclear. Mortalin is typically described as an important mitochondrial protein, playing an active role in the mitochondrial import of nuclear encoded proteins, bioenergetics and protein integrity maintenance 7. It is widely understood that astrocytes play a role in nurturing neuronal health by providing energy via glycogenesis, metabolic support and antioxidants, as well as regulating ions, neurotransmitters and synaptic transmission. Thus, it is plausible that impairment of mortalin expression in astrocytes could lead to altered mitochondrial function, which, in turn, could impair astrocyte function and have an ultimate adverse effect on proximal neurons. Further mechanistic studies are needed to address this possibility.
In conclusion, we demonstrate here for the first time the presence of mortalin within the astrocytes of both healthy subjects and PD patients. Furthermore, we found astroglial mortalin to be significantly reduced in the SNpc of PD patients as compared to controls. Further studies are needed to elucidate the potential mechanistic implications of reduced astroglial mortalin in PD pathology.
Conflict of Interest
The authors report no conflicts of interest.
Acknowledgments
This work would not have been possible without the generous donation made by the study patients and the support of their families. This work was supported in part by the UW NIEHS‐sponsored Biostatistics, Epidemiologic and Bioinformatics Training in Environmental Health (BEBTEH) Training Grant (Grant No. NIEHS TSES015459) and additionally by grants from the NIEHS (T32ES007032, ES016873 and ES019277). Autopsy materials used in this study were obtained from the University of Washington Neuropathology Core, which is supported by the Alzheimer's Disease Research Center (AG05136), the Adult Changes in Thought Study (AG006781) and Morris K Udall Center of Excellence for Parkinson's Disease Research (NS062684).
References
- 1. Baudet C, Perret E, Delpech B, Kaghad M, Brachet P, Wion D, Caput D (1998) Differentially expressed genes in C6.9 glioma cells during vitamin D‐induced cell death program. Cell Death Differ 5:116–125. [DOI] [PubMed] [Google Scholar]
- 2. Bélanger M, Allaman I, Magistretti PJ (2011) Brain energy metabolism: focus on astrocyte‐neuron metabolic cooperation. Cell Metab 14:724–738. [DOI] [PubMed] [Google Scholar]
- 3. Burbulla LF, Schelling C, Kato H, Rapaport D, Woitalla D, Schiesling C et al (2010) Dissecting the role of the mitochondrial chaperone mortalin in Parkinson's disease: functional impact of disease‐related variants on mitochondrial homeostasis. Hum Mol Genet 19:4437–4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chiasserini D, Tozzi A, de Iure A, Tantucci M, Susta F, Orvietani PL et al (2011) Mortalin inhibition in experimental Parkinson's disease. Mov Disord 26:1639–1647. [DOI] [PubMed] [Google Scholar]
- 5. Chomczynski P, Sacchi N (1987) Single‐step method of RNA isolation by acid guanidinium thiocyanate‐phenol‐chloroform extraction. Anal Biochem 162:156–159. [DOI] [PubMed] [Google Scholar]
- 6. De Mena L, Coto E, Sánchez‐Ferrero E, Ribacoba R, Guisasola LM, Salvador C et al (2009) Mutational screening of the mortalin gene (HSPA9) in Parkinson's disease. J Neural Transm 116:1289–1293. [DOI] [PubMed] [Google Scholar]
- 7. Deocaris CC, Kaul SC, Wadhwa R (2008) From proliferative to neurological role of an hsp70 stress chaperone, mortalin. Biogerontology 9:391–403. [DOI] [PubMed] [Google Scholar]
- 8. Halliday GM, Stevens CH (2011) Glia: initiators and progressors of pathology in Parkinson's disease. Mov Disord 26:6–17. [DOI] [PubMed] [Google Scholar]
- 9. Hoekstra JG, Cook TJ, Stewart T, Mattison H, Dreisbach MT, Hoffer ZS, Zhang J (2015) Astrocytic dynamin‐like protein 1 regulates neuronal protection against excitotoxicity in Parkinson disease. Am J Pathol 185:536–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jin J, Hulette C, Wang Y, Zhang T, Pan C, Wadhwa R, Zhang J (2006) Proteomic identification of a stress protein, mortalin/mthsp70/GRP75: relevance to Parkinson disease. Mol Cell Proteomics 5:1193–1204. [DOI] [PubMed] [Google Scholar]
- 11. Jin J, Li GJ, Davis J, Zhu D, Wang Y, Pan C, Zhang J (2007) Identification of novel proteins associated with both alpha‐synuclein and DJ‐1. Mol Cell Proteomics 6:845–859. [DOI] [PubMed] [Google Scholar]
- 12. Klein C, Westenberger A (2012) Genetics of Parkinson's disease. Cold Spring Harb Perspect Med 2:a008888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mishra R, Kaur G (2013) Aqueous ethanolic extract of Tinospora cordifolia as a potential candidate for differentiation based therapy of glioblastomas. PLoS ONE 8:e78764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mizukoshi E, Suzuki M, Loupatov A, Uruno T, Hayashi H, Misono T et al (1999) Fibroblast growth factor‐1 interacts with the glucose‐regulated protein GRP75/mortalin. Biochem J 343 (Pt 2):461–466. [PMC free article] [PubMed] [Google Scholar]
- 15. Mizukoshi E, Suzuki M, Misono T, Loupatov A, Munekata E, Kaul SC et al (2001) Cell‐cycle dependent tyrosine phosphorylation on mortalin regulates its interaction with fibroblast growth factor‐1. Biochem Biophys Res Commun 280:1203–1209. [DOI] [PubMed] [Google Scholar]
- 16. Park SJ, Shin JH, Jeong JI, Song JH, Jo YK, Kim ES et al (2014) Down‐regulation of mortalin exacerbates Aβ‐mediated mitochondrial fragmentation and dysfunction. J Biol Chem 289:2195–2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shi M, Jin J, Wang Y, Beyer RP, Kitsou E, Albin RL et al (2008) Mortalin: a protein associated with progression of Parkinson disease? J Neuropathol Exp Neurol 67:117–124. [DOI] [PubMed] [Google Scholar]
- 18. Takano S, Wadhwa R, Yoshii Y, Nose T, Kaul SC, Mitsui Y (1997) Elevated levels of mortalin expression in human brain tumors. Exp Cell Res 237:38–45. [DOI] [PubMed] [Google Scholar]
- 19. Takano S, Wadhwa R, Mitsui Y, Kaul SC (2001) Identification and characterization of molecular interactions between glucose‐regulated proteins (GRPs) mortalin/GRP75/peptide‐binding protein 74 (PBP74) and GRP94. Biochem J 357 (Pt 2):393–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Taurin S, Seyrantepe V, Orlov SN, Tremblay TL, Thibault P, Bennett MR et al (2002) Proteome analysis and functional expression identify mortalin as an antiapoptotic gene induced by elevation of [Na+]i/[K+]i ratio in cultured vascular smooth muscle cells. Circ Res 91:915–922. [DOI] [PubMed] [Google Scholar]
- 21. Tsuboi Y, Uchikado H, Dickson DW (2007) Neuropathology of Parkinson's disease dementia and dementia with Lewy bodies with reference to striatal pathology. Parkinsonism Relat Disord 13 (Suppl. 3):S221–S224. [DOI] [PubMed] [Google Scholar]
- 22. Wadhwa R, Taira K, Kaul SC (2002) An Hsp70 family chaperone, mortalin/mthsp70/PBP74/Grp75: what, when, and where? Cell Stress Chaperones 7:309–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wadhwa R, Yaguchi T, Hasan MK, Taira K, Kaul SC (2003) Mortalin‐MPD (mevalonate pyrophosphate decarboxylase) interactions and their role in control of cellular proliferation. Biochem Biophys Res Commun 302:735–742. [DOI] [PubMed] [Google Scholar]
- 24. Wadhwa R, Takano S, Kaur K, Deocaris CC, Pereira‐Smith OM, Reddel RR, Kaul SC (2006) Upregulation of mortalin/mthsp70/Grp75 contributes to human carcinogenesis. Int J Cancer 118:2973–2980. [DOI] [PubMed] [Google Scholar]


