Abstract
Background and Objective:
Antibiotic resistance is increasing, especially in healthcare-associated infections causing significant public health concerns worldwide. National information is required to make appropriate policies, update list of essential drugs for treatment, and evaluate the effects of intervention strategies. A nationwide surveillance of antimicrobial resistant bacteria in nosocomial infections was established in Iran in 2008, so that the data obtained through the surveillance would enable us to construct a database.
Materials and Methods:
Seven major teaching hospitals in Shiraz, Tabriz, Sari, Mashhad, Sanandaj, Ahwaz and Isfahan participated in this study. A total of 858 strains isolated from blood and other sterile body fluids were tested. Identification at the species level was performed with conventional biochemical methods and the API system. Susceptibility tests were done using disk diffusion method. The methicillin-resistance in S. aureus (MRSA) was determined by the oxacillin agar screen plate and respective MIC values were assessed using the E-test strips. The confirmatory disk diffusion methods were applied for phenotypic identification of extended-spectrum β- lactamase (ESBL) production for E. coli and K. pneumoniae, according to CLSI guidelines.
Results:
Cultivation and re-identification of the strains yielded 858 isolates, consisting of 224 S. aureus, 148 Klebsiella spp., 105 Serratia spp., 146 E. coli, 67 Acinetobacter spp., 38 Enterobacter spp., 95 Pseudomonas spp., 71 P.aeruginosa. 35 Stenotrophomonas sp., and 8 other organisms. MRSA was detected in 37.5% of the isolates. No vancomycin-resistant or vancomycin-intermediate resistant S. aureus was detected. With the exception of Acinetobacter and Stenotrophomonas, 85% of the Gram-negative isolates were found to be susceptible in vitro to imipenem. Overall, about 61% of K. pneumoniae and 35% of E. coli isolates were ESBL producing.
Conclusion:
Multidrug resistant isolates of Gram-negative organisms and methicillin-resistant strains of S. aureus have been detected in many hospitals in this study.
Keywords: Blood, Sterile body fluids, Gram negative bacteria, Staphylococcus aureus, Antimicrobial resistance, Iran
INTRODUCTION
Nosocomial infections occur worldwide and affect both developed and developing countries. Bacterial infections acquired in health care settings are among the major causes of death and increased morbidity and mortality among hospitalized patients. Nosocomial infections can be defined as those occurring within 72 hours of hospital admission, 3 days of discharge or 30 days of an operation. Many different microorganisms including bacteria, viruses, fungi and parasites may cause nosocomial infections (1, 2). Emerging patterns of antibiotic resistance of bacteria have altered outcome for critically ill patients. Physicians increasingly are faced with challenges to provide their patients with effective regimens while using antibiotics so that it does not result in further drug resistance. Antibiotic resistance is increasing and as a result significant public health problems are emerging (1–3). Moreover, it is now important to define local and national resistance rates for a range of pathogens in the blood and other strile body fluids in order to provide baseline data capable of serving as important reference for monitoring changes in resistance and empirical therapy. Data on antimicrobial resistance amongst pathogens recovered from blood and sterile body fluid infections in a nation wide study are limited.
Antibiotic resistance in Gram-positive cocci is a persistent issue. Methicillin resistant S. aureus (MRSA) is currently recognized as a major problem in hospitals throughout the world (3–5). Non-fermenter bacteria with high multidrug resistance, pose a particular challenge for healthcare management (6). Resistance due to the production of extended-spectrum B-lactamases (ESBLs) is a difficulty in the handling of Enterobacteriaceae infections, but other mechanisms of resistance are also emerging, leading to multidrug resistance and threatening to create panresistant species (7–9).
The term multidrug-resistant (MDR) applies to a bacterium that is resistant to: (1) several antibiotics to which they would normally be susceptible, or (2) all but one or two antibiotic classes, regardless of the mechanism of resistance (and often susceptible to only one or two commercially available antibiotics) (8, 9).
National information is required to develop appropriate policy, update lists of essential drugs and national guidelines for treatment, and evaluate the effects of intervention strategies. The present study aims to investigate antibiotic resistance among S. aureus and Gram-negative rods isolated from bloodstream and sterile body fluids, and evalute reduced methicillin and vancomycin susceptibility in S. aureus and extended-spectrum beta lactams in K. pneumoniae and E. coli.
MATERIALS AND METHODS
Seven major teaching hospitals located in different geographic areas of Iran (Shiraz, Tabriz, Sari, Mashhad, Sababdah, Ahwaz, Isfahan) in collaboration with the professor Alborzi clinical Microbiology Center (PACMRC), participated in this multicentre collaborative study over the period 2008–2009. The study focused on the most important pathogens responsible for nosocomial infections. These centers sent all the bacteria isolated from blood and sterile body fluids every two weeks to PACMRC. Isolated bacteria were stored at −80°C until reidentification and the antimicrobial susceptibility testing was conducted in PACMRC. Cultivation and re-identification of the strains yielded 858 isolates, consisting of S. aureus, Klebsiella spp., E.coli., Serratia spp., Acinetobacter spp., Enterobacter spp., Pseudomonas spp., Stenotrophomonas sp. Data regarding the antimicrobial susceptibility results were analysed by SPSS version 13.
Identification and confirmation of the isoltated bacteria.
Gram positive cocci isolates were identified as S. aureus by traditional biochemical tests, including catalase, coagulase, and acid production from D-mannitol (10). Identification of Gram negative bacteria at the species level was performed using conventional biochemical methods and tests incorporated in the API system (bio Merieux SA, Marcy-1, Etoile, France) (10).
Antimicrobial susceptibility testing.
Susceptibility tests were performed by the disk diffusion method, according to CLSI recommendations (11). Results were evaluated based on the respective standards for antimicrobial susceptibility testing.
Susceptibility of S. aureus isolates were tested for: clindamycin (CD, 2 μg), erythromycin (E, 15μg), linezolid (LZD, 30 μg), penicillin G (PG, 10μg), co-trimoxazole (SXT, 1.25/23.75μg), rifampin (RP, 5μg), oxacillin (OX, 1μg), ciprofloxacin (CIP, 5μg), chloramphenicol (C, 30μg), cephalothin (KF, 30μg), amikacin (AK, 30μg), tetracycline (T, 30μg), vancomycin (VA, 30μg), quinupristin-dalfopristin (SYN, 15μg), gentamicin (GM, 10μg) and fusidic acid (FC, 10μg).
Susceptibility of Gram negative rods were tested for the following 16 antimicrobial agents: Imipenem (IMP, 10μg), meropenem (MEM, 10μg), piperacillintazobactam (PTZ, 100/10μg), ciprofloxacin (CIP, 5μg), levofloxacin (LEV, 5μg), co-trimoxazole (SXT, 1.25/23.75μg), amoxicillin (A, 25 μg), nitrofurantion (NI, 200μg), cephalotin (KF, 30μg), amikacin (AK, 30μg), gentamicin (GM, 10μg), tobramycin (TB, 10μg), ceftriaxone (CRO, 30μg), cefixime (CFM, 5 μg), cefotaxime (CTX, 30μg), cefepime (CPM, 30μg), ceftazidime (CAZ, 30μg), aztreonam (ATM, 30μg) and ticarcillin (TC, 75μg).
Detection of methicillin resistant S. aureus (MRSA): Oxacillin agar screen plate: The oxacillin agar screen plate, prepared in-house, performed well in the detection of methicillin resistance in S. aureus. Ten microliters of the 106 CFU/ml bacterial inocula (final concentration=104 CFU/ml) was dropped onto MHA plates containing 4% NaCl and 6 μg/ml of oxacillin (11). If any growth occurred within 48h incubation at 33–35°C, the isolate was considered to be oxacillin resistant.
E-test Method: Methicillin (oxacillin) MICs were determined using the E-test strips (AB Biodisk, Solna, Sweden), according to the manufacturer's instructions on 150-mm-diameter MHA plates inoculated with 0.5 MacFarland density by swabbing in three directions. In case of heterogeneous growth, the highest MIC (inner limit of the inhibition zone) was read. S. aureus ATCC 25923 was tested with each batch of medium, as the standard strain.
Detection of vancomycin resistant S. aureus (VRSA)
BHI Agar Screen Plate: All S. aureus isolates were examined for reduced vancomycin susceptibility by an agar incorporation. Ten μL of a 0.5 Macfarland bacterial suspension (final concentration=106 CFU/ml) was spotted on the brain heart infusion (BHI) agar (Merck, Germany) containing 6 μg/ml vancomycin, allowed to air dry for approximately 5 min, and incubated at 35°C (11). Plates were examined at 24 and 48 h for any growth.
E-test Method: Standard E-test procedure was performed using a 2.0 McFarland inoculum on Mueller-Hinton agar (MHA) plates (Merck, Germany), according to the manufacturer's manual, using vancomycin E-test strips. Plates were incubated at 35°C for full 24h period. MIC endpoints were read, according to the manufacturer's recommendations. If heterogeneous growth occurred, the highest MIC (inner limit of the inhibition zone) was read. For quality control, Entercococcus faecalis ATCC 29212 as the susceptible control and E. faecalis ATCC 51299 as the resistant control, were used.
Detection of extended-spectrum β- lactamase (ESBL) in E.coli and K. pneumoniae
Combination Disc Diffusion Method: All E. coli and K. pneumoniae isolates were screened for extended-spectrum β- lactamase (ESBL) production, according to CLSI guidelines using confirmatory disk diffusion methods (11). A cefotaxime (30μg) and a cefotaxime + clavulanic acid (30μg+10μg), ceftazidime (30 μg) and ceftazidime + clavulanic acid (30μg+10μg) discs (Mast, UK) were placed at a distance of 25 mm on a Mueller-Hinton Agar plate, inoculated with a bacterial suspension of 0.5 McFarland turbidity standards and incubated overnight at 37°C. A ≥ 5mm increase in the diameter of inhibition zone for the combination disc versus ceftazidime disc, confirmed ESBL production. ESBL producing strain K. pneumoneae ATCC 700603 and non-ESBL producing strain E. coli ATCC 25922 were used as positive and negative controls.
RESULTS
Cultivation and re-identification of the strains yielded 858 isolates, consisting of 224 S. aureus, 148 Klebsiella spp., 146 E. coli., 105 Serratia spp., 67 Acinetobacter spp., 38 Enterobacter spp., 95 Pseudomonas spp., 71 Pseudomonas aeruginosa, 35 Stenotrophomonas sp. and 8 other organisms (Table 1, 2).
Table 1.
Frequencies of isolates obtained from positive sterile body fluid cultures in different cities (N=858).
| City | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Shiraz | Sari | Tabriz | Mashhad | Sanandaj | Ahwaz | Esfahan | Total | ||
| Gram positive cocci (N=224) | |||||||||
| S. aureus | 54(24%) | 11(5%) | 35(15.5%) | 1(0.5%) | 36(16%) | 85(38%) | 2(1%) | 224 | |
| Gram negative bacilli (N=634) | |||||||||
| Klebsiella spp. | 22(15%) | 47(32%) | 12(8%) | 31(21%) | 16(11%) | 8(5%) | 12(8%) | 148 | |
| E. coli | 36(24.5%) | 39(27%) | 13(9%) | 25(17%) | 21(14.5%) | 2(1%) | 10(7%) | 146 | |
| Serratia spp. | 11(10.5%) | 17(16%) | 5(5%) | 16(15%) | 33(31.5%) | 22(21%) | 1(1%) | 105 | |
| Enterobacter spp. | 8(21%) | 14(37%) | 2(5%) | 2(5%) | 5(13.5%) | 4(10.5%) | 3(8%) | 38 | |
| Pseudomonas spp. | 19(20%) | 45(47.5%) | 5(5.3%) | 16(16.8%) | 4(4.1%) | 3(3.2%) | 3(3.2%) | 95 | |
| Acinetobacter spp. | 20(30%) | 4(6%) | 1(1.5%) | 24(35.5%) | 5(7.5%) | 3(4.5%) | 10(15%) | 67 | |
| Stenotrophomonas spp. | 5(14%) | 10(28.5%) | 1(3%) | 14(40%) | 1(3%) | 3(8.5%) | 1(3%) | 35 | |
| Total | 175 | 187 | 74 | 129 | 121 | 130 | 42 | ||
Table 2.
Distribution of isolates obtained from positive sterile body fluid cultures (N=858).
| Hospital Units | |||||||
|---|---|---|---|---|---|---|---|
| Blood | CSF | Pleural Fluid | Ascitic Fluid | Joint Fluid | Peritunium | Total | |
| Gram positive cocci (N=224) | |||||||
| S. aureus | 210(94%) | 5(2%) | 5(2%) | 3(1.5%) | 1(0.5%) | - | 224 |
| Gram negative bacilli (N=634) | |||||||
| Klebsiella spp. | 121(82%) | 4(2.5%) | 11(7.5%) | 6(4%) | 1(0.5%) | 5(3.5%) | 148 |
| E. coli | 112(77%) | 11(7.5%) | 6(4%) | 13(9%) | 1(0.5%) | 3(2%) | 146 |
| Serratia spp. | 97(92%) | 3(3%) | 2(2%) | 3(3%) | - | - | 105 |
| Enterobacter spp. | 24(63%) | 2(5%) | 6(16%) | 4(11%) | - | 2(5%) | 38 |
| Pseudomonas spp. | 65(68.5%) | 6(6%) | 4(4.5%) | 7(7.5%) | 7(7.5%) | 6(6%) | 95 |
| Acinetobacter spp. | 60(89.5%) | 2(3%) | 4(6%) | 1(1.5%) | - | - | 67 |
| Stenotrophomonas spp. | 26(74%) | - | 1(3%) | 1(3%) | 7(20%) | - | 35 |
| Total | 715 | 33 | 39 | 38 | 17 | 16 | |
Findings for Staphylococcus isolates.
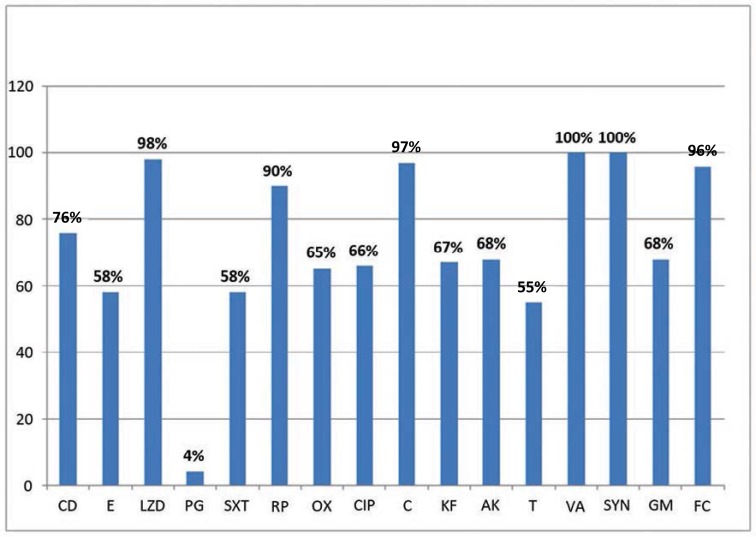
Of 224 S. aureus isolates, 210 were from blood, 5 from pleural fluid, 5 from CSF, 3 from ascitic fluid and 1 isolate from joint fluid. S.aureus was the species with the highest frequency (224/858; 26%) among the isolates in this study. The in vitro antimicrobial susceptibilities for S. aureus isolates to 16 antibacterial agents are shown in Table 2. Of the 224 S. aureus isolates, 84 (37.5%) were methicillin resistant (MRSA). No vancomycin resistance (VRSA), defined as MIC VA>8μg/ml, and vancomycin-intermediate resistance (VISA), defined as MIC VA 4–8μg/ml, were detected among S. aureus isolates. Also, no resistance to quinupristin-dalfopristin was observed. Susceptibility rates for linezolid, chloramphenicol and fusidic acid were 98%, 97% and 96%, respectively, and susceptibility rate for both erythromycin and co-trimoxazole was 58%. Among aminoglycosides the susceptibility rate for amikacin and gentamicin was found to be 68% (Fig. 1).
Fig. 1.
Antimicrobial susceptibilities profile of S.aureus isolates.
CD, clindamycin; E, erythromycin; LZD, linezolid; PG, penicillin G; SXT, co-trimoxazole; RP, rifampin; OX, oxacillin; CIP, ciprofloxacin; C, chloramphenicol; KF, cephalothin; AK, amikacin; T, tetracycline; VA, vancomycin; SYN, quinupristin-dalfopristin; GM, gentamicin; FC, fusidic acid.
Findings for Gram-negative isolates.
Of the 634 Gram-negative isolates, 511 were from the blood, 48 from acitic fluid, 32 from pleural fluid, 27 from CSF and 16 isolates from synovial fluid. The distribution of bacterial species and the in vitro antimicrobial susceptibilities for Gram-negative isolates against different antimicrobial agents are shown in Table 3. The most frequent Gram-negative isolates were Klebsiella spp. (17.2%), E.coli (17%), Serratia spp. (12.2%) and Pseudomonas spp. (11%).
Table 3.
Antimicrobial susceptibilities profile of Gram negative isolates
| Klebsiella spp. N (%) | E.coli N (%) | Serratia spp. N (%) | Pseudomonas spp. N (%) | Acinetobacter spp. N (%) | Enterobacter spp. N (%) | Stenotrophomonas spp. N (%) | |
|---|---|---|---|---|---|---|---|
| Imipenem | 145(98) | 145(99.3) | 105(100) | 81(85) | 43(64) | 38(100) | 10(29) |
| Ciprofloxacin | 109(74) | 84(57) | 98(93) | 72(76) | 18(27) | 33(87) | 23(66) |
| Levofloxacin | 118(80) | 92(63) | 101(96) | 73(77) | 16(24) | 35(92) | 22(63) |
| Co-trimoxazole | 48(32) | 41(28) | 70(67) | 17(18) | 20(30) | 22(58) | 29(83) |
| Amoxicillin | 2(1) | 16(11) | 2(2) | - | 2(3) | 4(11) | - |
| Ampicillin | 5(3) | 15(10) | 2(2) | - | 4(6) | 4(11) | - |
| Nitrofurantion | 103(70) | 141(96) | 23(22) | - | 1(2) | 22(58) | - |
| Cephalotin | 39 (26) | 50(34) | 3(3) | - | 4(6) | 5(14) | - |
| Amikacin | 94(64) | 133(91) | 68(65) | 76(80) | 24(36) | 26(69) | 21(60) |
| Gentamicin | 69(46) | 105(72) | 55(52) | 64(67) | 13(21) | 24(63) | 19(54) |
| Tobramycin | - | - | - | 67(71) | - | - | 17(49) |
| Ceftriaxone | 48(32) | 73(50) | 50(48) | - | 1(2) | 18(47) | - |
| Cefixime | 50(34) | 73(49) | 50(48) | - | 1(2) | 13(34) | - |
| Cefotaxime | 44(30) | 73(50) | 45(43) | - | 2(3) | 19(50) | - |
| Cefepime | 69(47) | 101(69) | 67(64) | 38(40) | 17(25) | 24(63) | 22(63) |
| Ceftazidime | 67(45) | 91(62) | 69(66) | 70(74) | 20(30) | 20 (52) | 28(80) |
| Piperacillin-Tazobactam | 104(70) | 116(80) | 88(84) | 90(95) | 27(40) | 30(79) | 29(83) |
With the exception of Acinetobacter and Stenotrophomonas, imipenem retained acceptable in vitro activites against the other Gram-negative isolates (>85%, susceptibilities). The susceptibility rate for Serratia and Enterobacter to imipenem was 100%. About 71% of Stenotrophomonas and 36% of Acinetobacter strains were resistant or intermediate- resistant to imipenem. The sensitivity of Acientobacter to imipenem was 64%.
In addition to imipenem, only piperacillin-tazobactam, nitrofurantion and amikacin had acceptable in vitro activities (≥80%) against E. coli isolates. Imipenem and levofloxacin were the most effective agents against (≥80%), followed by ciprofloxacin, pieracillin-tazobactam, nitrofurantion and amikacin which exhibited (60–80%) efficacy against Klebsiella isolates. As revealed, third generation ceplalosporines had poor activity against Klebsiella.
Overall, about 61% of K. pneumoniae isolates (90 out of the 148 isolates tested) and 35% of E. coli isolates (51/146) were ESBL producing. Imipenem (100%), piperacillin-tazobactam (84%), levofloxacin (96%) and ciprofloxacin (93%) functioned actively against Serratia isolates. Most strains of Serratia were resistant to amoxicillin, ampicillin, cephalothin and nitrofurantoin and most third generation cephalosporins except ceftazidime showed poor activity against Serratia isolates.
Most strains of Enterobacter were found susceptible to imipenem, levofloxacin, ciprofloxacin and piperacillin-tazobactam (100%, 92%, 87%, and 79%, respectively). Third generation cephalosporins were not active enough against the Enterobacter isolates tested (60%). Piperacillin-tazobactam exhibitied excellent (95%) and meropenem, imipenem and ticarcillin good activity (>80%) against Pseudomonas isolates. Only piperacillin-tazobactam, ceftazidime and co-trimoxazole had acceptable in vitro activities against Stenotrophomonas isolates (>75% susceptibilities) that were highly resistant to carbapenems (imipenem an meropenem). Levofloxacin, ciprofloxacin, amikacin, cefepime, piperacillin, carbencillin and tricarcillin were fairly effective (60–80%) against Stenotrophomonas (Table 3).
DISCUSSION
To arrive at a more accurate view about the most common bacteria isolated from patients with nosocomial infections and their antimicrobial susceptibility, we established a multicenter surveillance program in 2008 in Iran. Despite a large number of sporadic reports on antimicrobial susceptibity of bacteria from single hospitals in Iran, there is no accessible and comprehensive database about the susceptibility of bacterial pathogens to the currently used antibacterial agents. This study was the first nationwide one to address the problem of drug resistance and susceptibility among nosocomial bacterial pathogens in Iran. The antimicrobial susceptibility rates of S. aureus and Gram-negative rods strains isolated from the blood and other sterile body fluids, were determined in the present study.
As observed, E. coli was the most common isolated Gram negative bacterium in some of the centers including Shiraz, and Tabriz, Klebsiella spp. was most frequent bacterium causing nosocomial infections in Esfahan and Mashhad, and finally, Serratia spp. was the most common in Sanandaj and Ahvaz. Ampicillin and co-trimoxazole were the least effective antibiotics against all the isolates, except Stenotrophomonas. More than 90% of E. coli isolates were sensitive to amikacin, and carbapenems (imipenem and meropenem). In aminoglycoside class of antibiotics, amikacin was the most effective agent against different isolated Gram negative bacteria. Among the isolates, E. coli was the most sensitive one to amikacin (91%). Imipenem was the most effective agent against Klebsiella spp. and only 20% of Klebsiella spp. strains showed susceptibility to all the tested antibiotics. Twenty-nine (83%) of Stenotrophomonas spp. isolates were susceptible to co-trimoxazole, which can act as the drug of choice against this bacterium.
The data analysis indicates that the antibiotics such as Beta-lactam agents, used extensively in the treatment of different infections, were active only against about 50% of total Enterobacter species tested. In some studies (12), cefepime showed a good activity against Enterobacter species, but we didn't find such a result. Approximately, 48% of Enterobacter species were resistant to ceftazidime, according to the studies of ICU isolates in the United States between 1987 and 1991 (13) and between 1994 and 1995 (14) and in 5 European countries (15), in agreement with the present study findings. Previous use of third generation cephalosporins is more likely to cause resistance to β-lactams in certain isolates of Enterobacter species.
Concerning the alarming types of resistance (i.e., resistance to third generation cephalosporins in Klebsiella pneumoniae, to quinolones in E. coli, and to methicillin in S.aureus) (16–18), our data showed that resistance to third generation cepholosporins tested was common (55% on average) among Klebsiella spp. As revealed, significant resistance to ciprofloxacin in E. coli (43%) and the percentage of MRSA strains (37.5%) were alarming. In international studies on antimicrobial resistance, the susceptibility of Klebsiella pneumoniae has been quite variable. In contrast to some reports that indicated low incidence of ESBL producing K. pneumoniae (19), about 61% of K. pneumoniae isolates in this study were ESBL producing, consistent with some other studies (20), which could be an alarming situation, too.
Our results for ceftazidime resistance among E. coli (38%), which is thought to be the result of ESBL production, are not in agreement with some reports from other countries like USA, which showed resistance to ceftazidime to be 4% (21). In the present study, 35% of E. coli isolates were ESBL positive.
In some studies, Acinetobacter and Stenotrophomonas maltophilia were the most resistant pathogens to many antibiotics (22,23). It was also the case with the present study. Acinetobacter is an increasingly infectious threat, especially in patients receiving broad spectrum antimicrobial therapy and requiring life support (24, 25). A Spanish study has shown that Acinetobacter isolates, usually acquired in the ICU, are multiresistant and may cause severe infections associated with a high mortality rate (26). Riely et al. recently described the failure to stop the spread of gentamicin resistant A. baumannii in an Australian ICU despite infection control measures (27).
P. aeruginosa species are naturally resistant to a number of antimicrobials and their resistance to the commonly used therapeutic agents has increased in recent years. Strains resistant to all available antimicrobial agents (pan-resistant strains) have emerged in hospitalized patients (28).
In our study, more than 95% of Pseudomonas isolates were sensitive to piperacillin-tazbactam and more than 80% of the isolates were sensitive to meropenem, imipenem and ticarcillin. As such a resistance can readily spread within a hospital setting and cause protracted outbreaks with high mortality rates, strict infection control procedures are highly recommended. Compared with other studies, imipenem exhibited excellent activity (100%) against Serratia isolates, in this study. piperacillin-tazobactam, levofloxacin and ciprofloxacin were highly active (84%, 96%, 93%, respectively) against Serratia isolates.
About 37.5% of S. aureus isolates in our study were MRSA, which is much higher, compared to the report from other countries (12.8%) between 2000 and 2001 as well as from studies performed in Tehran (Iran) (29, 30). The present study shows antibiotic resistance pattern within Iran, and has been conducted specifically on sterile body fluids, whereas others were limited to certain regions (30) and on a wide variety of sterile and non sterile clinical specimens (30, 31). Therefore, the present findings can serve as an index of actual antibiotic resistance across the nation. The prevalence of MRSA continues to increase worldwide, sometimes accounting for approximately 40–60% of all hospital acquired strains (32). No vancomycin resistant (VRSA) or vancomycin-intermdiate resistant S.aureus (VISA) isolates were detected. There could be many explanations for such differences, including: infection control measures, antibiotic prophylaxis and treatments used in each ward/hospital and last but not least, the clonal and epidemic nature of these microorganisms. While there are reports aroud the world indicating a tendency toward decreasing susceptibility to vancomycin in S.aureus (33), we had no VRSA of VISA isolates. This may be due to judicious and controlled use of vancomycin in our hospitals.
Multidrug resistant strains of Gram-negative organisms have emerged in many hospitals and has caused restrictions in the choice of antibiotics for empirical therapy and control of the increasing incidence of such organisms. There is a progressive increase in MRSA prevalence in Iran and an extremely low prevalence of VRSA or VISA was confirmed among S.aureus clinical isolates. Vancomycin can serve as the drug of choice for treating multidrug resistant MRSA infections. It should be also noted that in many cases antibiotic resistance is transmited to humans and hospital environment through other sourses including food animals, plants, poultries, fish, and other industries, in which antibiotics are used for different purposes and may lead to emerging resistant strains (34–36). Isolation, identification and antimicrobial susceptibility of pathogens can be helpful in optimizing the antimicrobial use. It is, therefore, crucial to implement the rational use of available antimicrobials in everyday clinical practice to prevent selective pressure and the further development of resistance in these pathogens. Also, a regular surveillance of hospital associated infections, monitoring of antibiotic susceptibility pattern and formulation of practical antibiotic policy are suggested.
Acknowledgments
This work was supported by research grant from the Professor Alborzi Clinical Microbiology Research Center (PACMRC). Our thanks go to Hassan Khajehei, PhD, for copy editing of the manuscript. Our deep gratitude goes to Fariba Miri, MD, for collecting patients data.
REFERENCES
- 1. Karam GH, Heffner JE. Emerging issues in Antibiotic Resistance in Blood-borne Infections. Am J Respir Crit Care Med 2000; 162: 1610– 1616. [DOI] [PubMed] [Google Scholar]
- 2. Laxminarayan R, Duse A, Wattal C, Zaidi AK, Wertheim HF, Sumpradit N, et al. Antibiotic resistance-the need for global solutions. Lancet Infect Dis 2013; 13: 1057–1098. [DOI] [PubMed] [Google Scholar]
- 3. Rice LB. Antimicrobial Resistance in Gram-Positive Bacteria. Am J Med 2006; 119 (6 Suppl 1): S11– 9. [DOI] [PubMed] [Google Scholar]
- 4. Boucher HW, Ralph Corey G. Epidemiology of Methicillin-Resistant Staphylococcus aureus. Clin Infect Dis 2008; 46: S344– 349. [DOI] [PubMed] [Google Scholar]
- 5. Udobi CE, Obajuluwa AF, Onaolapo JA. Prevalence and Antibiotic Resistance Pattern of Methicillin-Resistant Staphylococcus aureus from an Orthopaedic Hospital in Nigeria. BioMed Research International 2013; 2013: 1– 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Latif S, Saeed Anwar M, Ahmad I. Bacterial Pathogens Responsible For Blood Stream Infection (Bsi) And Pattern Of Drug Resistance In A Tertiary Care Hospital Of Lahore. Biomedica 2009; 25: 101– 105. [Google Scholar]
- 7. Poole K. Resistance to β-lactam antibiotics. CMLS 2004; 61: 2200– 2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 2012; 18: 268– 281. [DOI] [PubMed] [Google Scholar]
- 9. Pasricha J, Koessler T, Harbarth S, Schrenzel J, Camus V, Cohen J, et al. Carriage of extended-spectrum beta-lactamase-producing enterobacteriacae among internal medicine patients in Switzerland. Antimicrob Resist Infect Control 2013; 2: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Grundmann H, Hahn A, Ehrenstein B, Geiger K, Just H, Daschner FD. Detection of cross-transmission of multiresistant Gram-negative bacilli and Staphylococcus aureus in adult intensive care units by routine typing of clinical isolates. Clin Microbiol Infect 1999; 5: 355– 363. [PubMed] [Google Scholar]
- 11. Clinical and Laboratory Standards Institute (CLSI) . 2011, M100–S21. Vol. 31 No. 1. [Google Scholar]
- 12. Tamma PD, Girdwood SCT, Gopaul R, Tekle T, Roberts AA, Harris AD, et al. The Use of Cefepime for Treating AmpC β-Lactamase–Producing Enterobacteriaceae. Clin Infect Dis 2013. [DOI] [PubMed] [Google Scholar]
- 13. Burwen DR, Banerjee SN, Gaynes RP. Ceftazidime resistance among selected nosocomial Gram-negative bacilli in the United-States. J Infect Dis 1994;170: 1622– 1625. [DOI] [PubMed] [Google Scholar]
- 14. Archibald L, Phillips L, Monnet D, McGowan JE, Tenover F, Gaynes R. Antimicrobial resistance in isolates from inpatients and outpatients in the United States: increasing importance of the intensive care unit. Clin Infect Dis 1997; 24: 211– 215. [DOI] [PubMed] [Google Scholar]
- 15. Hanberger H, Garcia-Rodriguez JA, Gobernado M, Gossens H, ilsson LE, Struelens MJ, et al. Enterobacter cloacae were resistant to ceftazidime, according to studies of ICU isolates in the United States between 1987 and 1991. JAMA 1999; 281: 67– 71. [DOI] [PubMed] [Google Scholar]
- 16. Braykov NP, Eber MR, Klein EY, Morgan DJ, Laxminarayan R. Trends in Resistance to Carbapenems and Third-Generation Cephalosporins among Clinical Isolates of Klebsiella pneumoniae in the United States, 1999–2010. Infect Control Hosp Epidemiol 2013; 34. [DOI] [PubMed] [Google Scholar]
- 17. Ponsa MJ, Mosquitob S, Gomesa C, del Vallec LJ, Ochoab TJ, Ruiz J. Analysis of quinolone-resistance in commensal and diarrheagenic Escherichia coli isolates from infants in Lima, Peru. Trans R Soc Trop Med Hyg 2014; 108: 22– 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu C, Bayer A, Cosgrove SE, Daum RS, Fridkin SK, Gorwitz RJ, et al. Clinical Practice Guidelines by the Infectious Diseases Society of America for the Treatment of Methicillin-Resistant Staphylococcus aureus Infections in Adults and Children: Executive Summary. Clin Infect Dis 2011; 52: 285– 292. [DOI] [PubMed] [Google Scholar]
- 19. Søraas A, Sundsfjord A, Sandven I, Brunborg C, Jenum PA. Risk Factors for Community-Acquired Urinary Tract Infections Caused by ESBL-Producing Enterobacteriaceae-A Case-Control Study in a Low Prevalence Country. PLoS ONE 2013; 8: e69581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hawser SP, Bouchillon SK, Hoban DJ, Badal RE, Hsueh PR, Paterson DL. Emergence of High Levels of Extended-Spectrum-β-Lactamase-Producing Gram-Negative Bacilli in the Asia-Pacific Region: Data from the Study for Monitoring Antimicrobial Resistance Trends (SMART) Program, 2007. Antimicrob Agents Chemother 2009; 53: 3280– 3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Robicsek A, Strahilevitz J, Sahm DF, Jacoby GA, Hooper DC. qnr Prevalence in Ceftazidime-Resistant Enterobacteriaceae Isolates from the United States. Antimicrob Agents Chemothert 2006; 50: 2872– 2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Brooke JS. Stenotrophomonas maltophilia: an Emerging Global Opportunistic Pathogen. Clin Microbiol Rev January 2012; 25: 2– 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Decker K, Rather PN, Bonomo Federico Perez RA, Hujer AM, Hujer KM, et al. Global Challenge of Multidrug-Resistant Acinetobacter baumannii. Antimicrob Agents Chemother 2007; 51: 3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kempf M, Rolain JM. Emergence of resistance to carbapenems in Acinetobacter baumannii in Europe: clinical impact and therapeutic options. Int J Antimicrob Agents 2012; 39: 105– 114. [DOI] [PubMed] [Google Scholar]
- 25. Katragkou A, Roilides E. Successful Treatment of Multidrug-Resistant Acinetobacter baumannii Central Nervous System Infections with Colistin. J Clin Microbiol 2005; 43: 4916– 4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Acosta J, Merino M, Viedma E, Poza M, Sanz F, Otero JR, et al. Multidrug-resistant Acinetobacter baumannii harboring OXA-24 carbapenemase, Spain. Emerg Infect Dis 2011; 17: 1064– 1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Riley TV, Webb SAR, Cadwallader H, Briggs BD, Christiansen L, Bowman RA. Outbreak of gentamicin-resistant Acinetobacter baumanii in an intensive care unit: Clinical, epidemiological and microbiological features. Pathology 1996; 28: 359– 63. [DOI] [PubMed] [Google Scholar]
- 28. Tuon FF, Gortz LW, Rocha JL. Risk factors for pan-resistant Pseudomonas aeruginosa bacteremia and the adequacy of antibiotic therapy. Braz J Infect Dis 2012; 16: 351–356. [DOI] [PubMed] [Google Scholar]
- 29. Akpaka PE, Kissoon S, Rutherford C, Swanston WH, Jayaratne P. Molecular epidemiology of methicillin-resistant Staphylococcus aureus isolates from regional hospitals in Trinidad and Tobago. Int J Infec Dis 2007; 11: 544– 548. [DOI] [PubMed] [Google Scholar]
- 30. Pourakbari B, Sadr A, Ashtiani MT, Mamishi S, Dehghani M, Mahmoudi S, et al. Five-year evaluation of the antimicrobial susceptibility patterns of bacteria causing bloodstream infections in Iran. J Infect Dev Ctries 2012; 6: 120– 125. [DOI] [PubMed] [Google Scholar]
- 31. Anvarinejad M, Japoni A, Rafaatpour N, Mardaneh J, Abbasi P, Amin Shahidi M, et al. Burn patients infected with metallo-beta-lactamase-producing Pseudomonas aeruginosa: Multidrug-resistant strains. Arch Trauma Res 2014; 3: e18182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chen CJ, Huang YC. Community-acquired methicillin-resistant Staphylococcus aureus in Taiwan. Microbiol Immunol Infect 2005; 38: 376– 382. [PubMed] [Google Scholar]
- 33. Hu J, Ma XX, Tian Y, Pang L, Cui LZ, Shang H. Reduced vancomycin susceptibility found in methicillin-resistant and methicillin-sensitive Staphylococcus aureus clinical isolates in Northeast China. PLoS ONE 2013; 8: e73300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mardaneh J, Dallal MM. Isolation, identification and antimicrobial susceptibility of Pantoea (Enterobacter) agglomerans isolated from consumed powdered infant formula milk (PIF) in NICU ward: First report from Iran. Iran J Microbiol 2013; 5: 263– 267. [PMC free article] [PubMed] [Google Scholar]
- 35. Mardaneh J, Soltan-Dallal MM. Isolation and identification of E. cowanii from powdered infant formula in NICU and determination of antimicrobial susceptibility of isolates. Iran J Pediatr 2014; 24: 261– 266. [PMC free article] [PubMed] [Google Scholar]
- 36. Mardaneh J, Soltan Dallal MM, Taheripoor M, Rajabi Z. Isolation, identification and antimicrobial susceptibility pattern of Tatumella ptyseos strains isolated from powdered infant formula milk consumed in neonatal intensive care unit: First report from Iran. Jundishapur J Microbiol 2014; 7(6): e10608. [DOI] [PMC free article] [PubMed] [Google Scholar]