Abstract
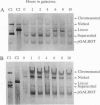
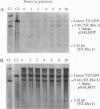
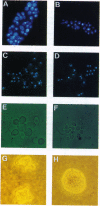
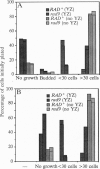
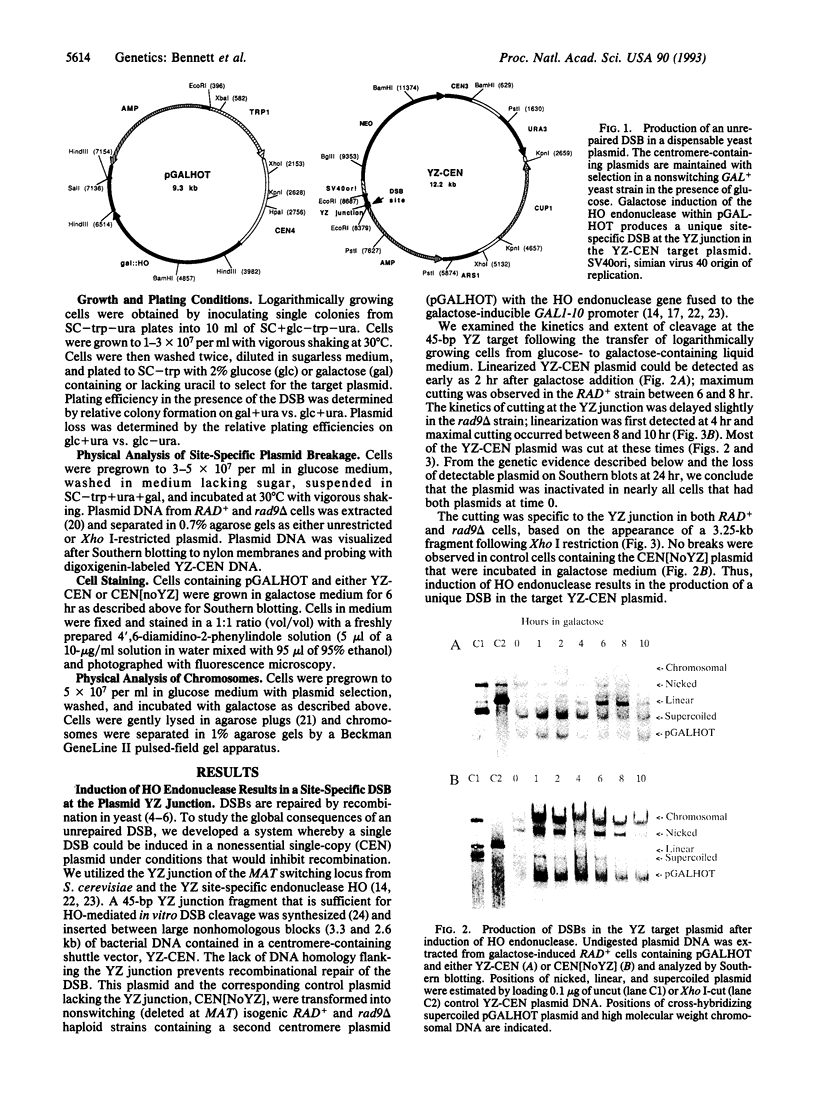
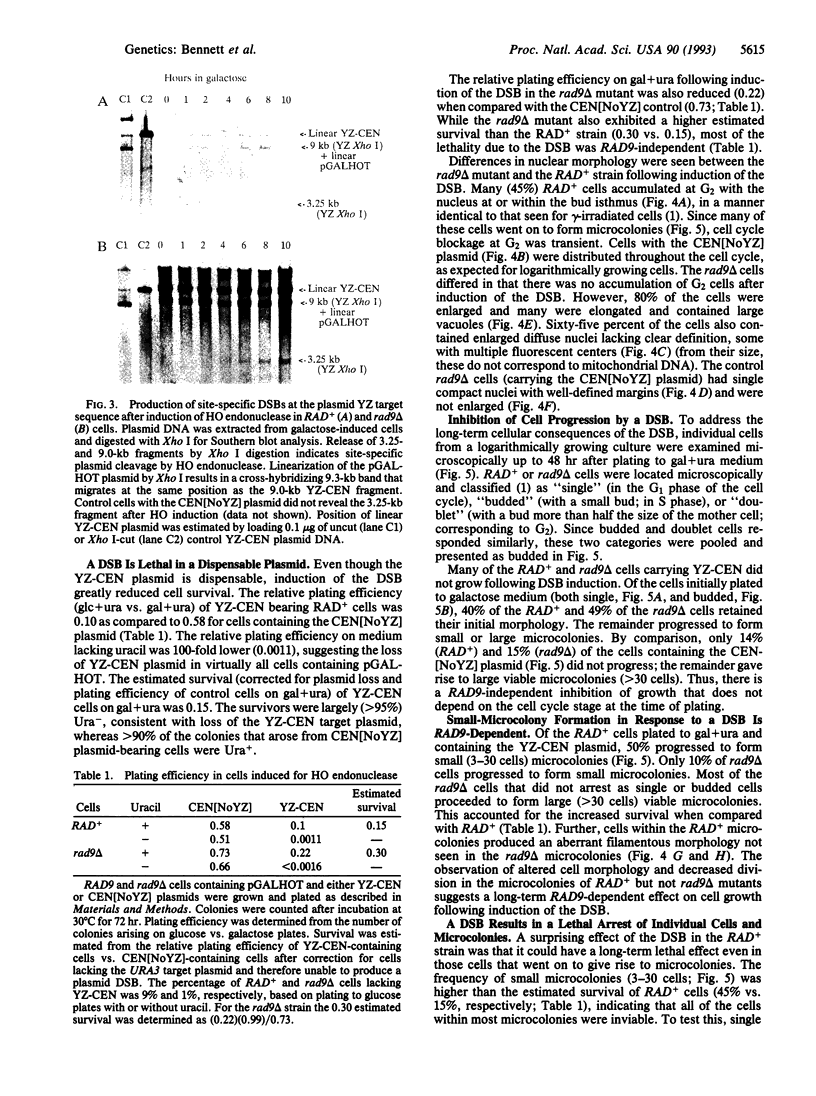
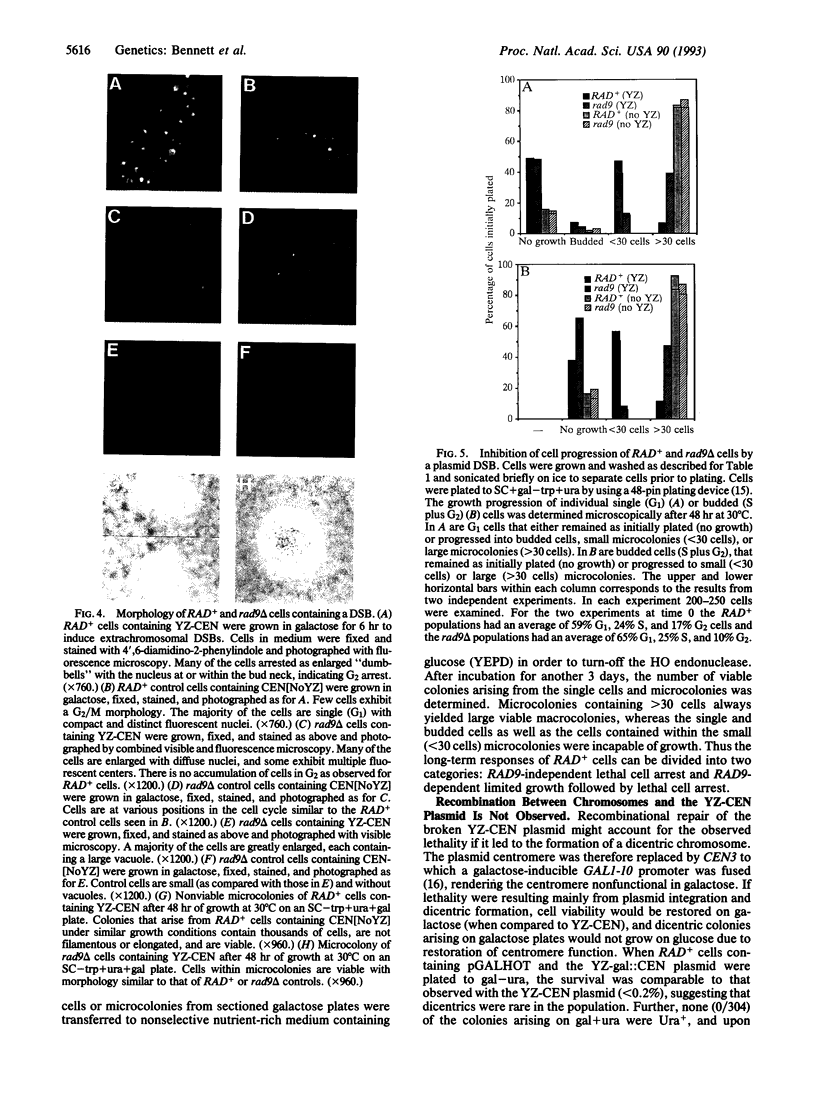
Cells of the yeast Saccharomyces cerevisiae are delayed in the G2 phase of the cell cycle following chromosomal DNA damage. This arrest is RAD9-dependent and suggests a signaling mechanism(s) between chromosomal lesions and cell cycling. We examined the global nature of growth inhibition caused by an HO endonuclease-induced double-strand break (DSB) at a 45-bp YZ sequence (from MAT YZ) in a non-yeast region of a dispensable single-copy plasmid. The presence of an unrepaired DSB results in cellular death even though the plasmid is dispensable. Loss of cell viability is partially dependent on the RAD9 gene product. Following induction of the DSB, 40% of RAD+ and 49% of rad9 delta cells [including both unbudded (G1) and budded (S plus G2) cells] did not progress further in the cell cycle. The remaining RAD+ cells progressed to form microcolonies (< 30 cells) containing aberrantly shaped inviable cells. For the rad9 delta mutant, the majority of the remaining cells produced viable colonies accounting for the greater survival of the rad9 delta strain. Based on the profound effects of a single nonchromosomal DNA lesion, this system provides a convenient means for studying the signaling effects of a DNA lesion, as well as for designing strategies for modulating cell proliferation.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barry M. A., Behnke C. A., Eastman A. Activation of programmed cell death (apoptosis) by cisplatin, other anticancer drugs, toxins and hyperthermia. Biochem Pharmacol. 1990 Nov 15;40(10):2353–2362. doi: 10.1016/0006-2952(90)90733-2. [DOI] [PubMed] [Google Scholar]
- Chow T. Y., Perkins E. L., Resnick M. A. Yeast RNC1 encodes a chimeric protein, RhoNUC, with a human rho motif and deoxyribonuclease activity. Nucleic Acids Res. 1992 Oct 11;20(19):5215–5221. doi: 10.1093/nar/20.19.5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L. H., Weinert T. A. Checkpoints: controls that ensure the order of cell cycle events. Science. 1989 Nov 3;246(4930):629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- Hill A., Bloom K. Acquisition and processing of a conditional dicentric chromosome in Saccharomyces cerevisiae. Mol Cell Biol. 1989 Mar;9(3):1368–1370. doi: 10.1128/mcb.9.3.1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho K. S., Mortimer R. K. Induction of dominant lethality by x-rays in radiosensitive strain of yeast. Mutat Res. 1973 Oct;20(1):45–51. doi: 10.1016/0027-5107(73)90096-1. [DOI] [PubMed] [Google Scholar]
- Hoffman C. S., Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57(2-3):267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- Kastan M. B., Onyekwere O., Sidransky D., Vogelstein B., Craig R. W. Participation of p53 protein in the cellular response to DNA damage. Cancer Res. 1991 Dec 1;51(23 Pt 1):6304–6311. [PubMed] [Google Scholar]
- Kostriken R., Heffron F. The product of the HO gene is a nuclease: purification and characterization of the enzyme. Cold Spring Harb Symp Quant Biol. 1984;49:89–96. doi: 10.1101/sqb.1984.049.01.012. [DOI] [PubMed] [Google Scholar]
- Lutkenhaus J. Regulation of cell division in E. coli. Trends Genet. 1990 Jan;6(1):22–25. doi: 10.1016/0168-9525(90)90045-8. [DOI] [PubMed] [Google Scholar]
- Nickoloff J. A., Chen E. Y., Heffron F. A 24-base-pair DNA sequence from the MAT locus stimulates intergenic recombination in yeast. Proc Natl Acad Sci U S A. 1986 Oct;83(20):7831–7835. doi: 10.1073/pnas.83.20.7831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickoloff J. A., Singer J. D., Hoekstra M. F., Heffron F. Double-strand breaks stimulate alternative mechanisms of recombination repair. J Mol Biol. 1989 Jun 5;207(3):527–541. doi: 10.1016/0022-2836(89)90462-2. [DOI] [PubMed] [Google Scholar]
- Orr-Weaver T. L., Szostak J. W. Fungal recombination. Microbiol Rev. 1985 Mar;49(1):33–58. doi: 10.1128/mr.49.1.33-58.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed S. I. G1-specific cyclins: in search of an S-phase-promoting factor. Trends Genet. 1991 Mar;7(3):95–99. doi: 10.1016/0168-9525(91)90279-Y. [DOI] [PubMed] [Google Scholar]
- Resnick M. A., Skaanild M., Nilsson-Tillgren T. Lack of DNA homology in a pair of divergent chromosomes greatly sensitizes them to loss by DNA damage. Proc Natl Acad Sci U S A. 1989 Apr;86(7):2276–2280. doi: 10.1073/pnas.86.7.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnick M. A. The repair of double-strand breaks in DNA; a model involving recombination. J Theor Biol. 1976 Jun;59(1):97–106. doi: 10.1016/s0022-5193(76)80025-2. [DOI] [PubMed] [Google Scholar]
- Resnick M. A., Westmoreland J., Bloom K. Heterogeneity and maintenance of centromere plasmid copy number in Saccharomyces cerevisiae. Chromosoma. 1990 Aug;99(4):281–288. doi: 10.1007/BF01731704. [DOI] [PubMed] [Google Scholar]
- Rudin N., Haber J. E. Efficient repair of HO-induced chromosomal breaks in Saccharomyces cerevisiae by recombination between flanking homologous sequences. Mol Cell Biol. 1988 Sep;8(9):3918–3928. doi: 10.1128/mcb.8.9.3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassanfar M., Roberts J. W. Nature of the SOS-inducing signal in Escherichia coli. The involvement of DNA replication. J Mol Biol. 1990 Mar 5;212(1):79–96. doi: 10.1016/0022-2836(90)90306-7. [DOI] [PubMed] [Google Scholar]
- Schiestl R. H., Gietz R. D. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet. 1989 Dec;16(5-6):339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- Schiestl R. H., Reynolds P., Prakash S., Prakash L. Cloning and sequence analysis of the Saccharomyces cerevisiae RAD9 gene and further evidence that its product is required for cell cycle arrest induced by DNA damage. Mol Cell Biol. 1989 May;9(5):1882–1896. doi: 10.1128/mcb.9.5.1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz D. C., Cantor C. R. Separation of yeast chromosome-sized DNAs by pulsed field gradient gel electrophoresis. Cell. 1984 May;37(1):67–75. doi: 10.1016/0092-8674(84)90301-5. [DOI] [PubMed] [Google Scholar]
- Strathern J. N., Klar A. J., Hicks J. B., Abraham J. A., Ivy J. M., Nasmyth K. A., McGill C. Homothallic switching of yeast mating type cassettes is initiated by a double-stranded cut in the MAT locus. Cell. 1982 Nov;31(1):183–192. doi: 10.1016/0092-8674(82)90418-4. [DOI] [PubMed] [Google Scholar]
- Sugawara N., Haber J. E. Characterization of double-strand break-induced recombination: homology requirements and single-stranded DNA formation. Mol Cell Biol. 1992 Feb;12(2):563–575. doi: 10.1128/mcb.12.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak J. W., Orr-Weaver T. L., Rothstein R. J., Stahl F. W. The double-strand-break repair model for recombination. Cell. 1983 May;33(1):25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- Thaler D. S., Stahl F. W. DNA double-chain breaks in recombination of phage lambda and of yeast. Annu Rev Genet. 1988;22:169–197. doi: 10.1146/annurev.ge.22.120188.001125. [DOI] [PubMed] [Google Scholar]
- Walker G. C. Mutagenesis and inducible responses to deoxyribonucleic acid damage in Escherichia coli. Microbiol Rev. 1984 Mar;48(1):60–93. doi: 10.1128/mr.48.1.60-93.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert T. A., Hartwell L. H. Characterization of RAD9 of Saccharomyces cerevisiae and evidence that its function acts posttranslationally in cell cycle arrest after DNA damage. Mol Cell Biol. 1990 Dec;10(12):6554–6564. doi: 10.1128/mcb.10.12.6554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert T. A., Hartwell L. H. The RAD9 gene controls the cell cycle response to DNA damage in Saccharomyces cerevisiae. Science. 1988 Jul 15;241(4863):317–322. doi: 10.1126/science.3291120. [DOI] [PubMed] [Google Scholar]
- White C. I., Haber J. E. Intermediates of recombination during mating type switching in Saccharomyces cerevisiae. EMBO J. 1990 Mar;9(3):663–673. doi: 10.1002/j.1460-2075.1990.tb08158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker S. G., Rockmill B. M., Blechl A. E., Maloney D. H., Resnick M. A., Fogel S. The detection of mitotic and meiotic aneuploidy in yeast using a gene dosage selection system. Mol Gen Genet. 1988 Dec;215(1):10–18. doi: 10.1007/BF00331296. [DOI] [PubMed] [Google Scholar]
- Williams G. T. Programmed cell death: apoptosis and oncogenesis. Cell. 1991 Jun 28;65(7):1097–1098. doi: 10.1016/0092-8674(91)90002-g. [DOI] [PubMed] [Google Scholar]