Abstract
The risks of contracting staphylococci food poisoning by the consumption of improperly manufactured salami and the possibility of this food being reservoirs for antibiotic resistance were evaluated. Nineteen coagulase-negative staphylococci (CNS) strains were found in commercial and artisanal salami. The species in commercial salami were S. saprophyticus, S. sciuri, S. xylosus, and S. carnosus. Artisanal salami showed S. succinus, S. epidermidis, and S. hominis but no S. carnosus. Phylogenetic analyses grouped the strains into three major staphylococcal species groups, comprised of 4 refined clusters with similarities superior to 90%. Fifteen strains harbored multiple enterotoxin genes, with high incidence of seb/sec and sea, 57% and 50%, respectively, intermediate incidence of sed/seh/selm and sei/seln/tst-H, 33% and 27%, correspondingly, and low incidence of see/selj/selo and seg, of respectively 13% and 1%. Real time RT-PCR and enzyme-linked-immunosorbent assays confirmed the enterotoxigenicity of the strains, which expressed and produced enterotoxins in vitro. The CNS strains showed multiresistance to several antimicrobials of therapeutic importance in both human and veterinarian medicine, such as β-lactams, vancomycin, and linezolid. The effective control of undue staphylococci in fermented meat products should be adopted to prevent or limit the risk of food poisoning and the spread of antimicrobial-resistant strains.
1. Introduction
Staphylococcal food poisoning is an illness caused by the ingestion of contaminated food containing enterotoxins produced by bacteria belonging to this genus. Enterotoxins that exhibit superantigenic activities are heat stable proteins and may not be destroyed even during cooking conditions.
In Brazil, according to data from the Ministério da Saúde (Ministry of Health), staphylococcal poisoning is the second most common foodborne disease, ranking only after outbreaks involving Salmonella spp. [1]. Staphylococcus classified as coagulase-positive are considered potential food enterotoxin-producing strains [1], although, recently, the enterotoxigenic potential of coagulase-negative staphylococci (CNS) species in food poisoning has also been recognized [2].
Initially, enterotoxin SEs family members were divided into five serological types (sea through see) based on their antigenicity [3, 4]. In recent years, however, newly described types of SEs—SEG, SEH, SEI, SElJ, SElK, SElL, SElM, SElN, SElO, SElP, SElQ, SElR, and SElU—with amino acid sequences similar to the classical SEs, were discovered. These newly described enterotoxins are designated as SE or SE-like (SEl), according to their emetic properties displayed in a primate model following oral administration [5].
The toxic shock syndrome toxin-1 (TSST-1) is also a member of the SE-related toxin family and has the ability to stimulate large populations of T cells containing a particular Vβ element in their T-cell receptors (TCR). Like other superantigenic toxins, it bypasses normal antigen presentation by binding to class II major histocompatibility complex molecules on antigen-presenting cells and to specific variable regions on the beta-chain of the T-cell antigen receptor. Through this interaction, a massive proliferation of T cells at orders of magnitude above antigen-specific activation occurs, resulting in a massive cytokine release that is believed to be responsible for the most severe features of TSST [6].
Enterotoxin (SE) genes are encoded in mobile genetic elements, such as plasmids, prophages, and Staphylococcus pathogenic islands (SaPIs) [7].
Salami is a kind of dry sausage obtained by the microbial fermentation of raw pork meat, using Staphylococcus starter cultures as technological accessories to ferment the product and give it its organoleptic characteristics. In Brazil, Italian type salami, similar to salami produced in Southern Europe, is avidly consumed, with a production trade of 13.093 tons between 2000 and 2014 [8].
S. xylosus, S. equorum, and S. carnosus are part of the starter culture microbiota that participate in the reactions required for creating the flavor and aroma during the maturing period of fermented meat production [9]. In addition, other species, such as S. epidermidis, S. pasteuri, S. sciuri, and S. succinus, may also occasionally be present in meaningful amounts [10].
However, even the combination of physical and chemical barriers cannot always guarantee the stability and microbial safety of starter cultures. Contamination of salami fermentation starter culture microbiota by pathogenic coagulase-negative staphylococci (CNS) strains is perhaps the most harmful factor in the production of cured meat products, since these pathogens are able to produce heat-stable enterotoxins with superantigenic activities in food matrices [11, 12].
Staphylococci species are not usually identified at the species level by routine laboratory testing and commercial kits, since phenotypic discrimination cannot reliably identify these species due to the variable expression of some phenotypic traits [13]. For this purpose, molecular techniques, including nucleotide sequencing within the 16S rDNA, hsp60, tuf, sodA, and rpoB genes, have been successfully used to identify Staphylococcus species [14].
Depending on the conditions, some species of coagulase-negative staphylococci can present health risks, since they have shown resistance to several antibiotics of therapeutic importance, such as β-lactams [15].
The aim of the present study was to identify the members of the CNS microbiota from salami. We sampled the salami marketed in Brazil, comparing the CNS microbiota in salami produced by industrial companies and in artisanal salami manufactured by small producers. The CNS strains were identified by sequencing of a 16S rDNA region and the phylogenic relationships between the observed species were established. The presence of multiple genes encoding the classical and newly described se/sel and tstH1 toxins in the CNS genomes was investigated. The risk of food poisoning was assessed by evaluating the ability of the CNS strains in transcribing and expressing the classical and newly described enterotoxins in vitro by using real time RT-PCR and enzyme-linked immunosorbent assay (ELISA). The resistance of the isolated strains to antimicrobial agents of therapeutic importance in staphylococci infections was also evaluated.
2. Materials and Methods
2.1. Isolation of Bacterial Strains
Six samples of distinct brands of salami, 03 from the meat industry and 03 from small artisanal producers, were collected in the municipality of Rio de Janeiro, Brazil. Twenty-five grams of salami was added to 225 mL of 0.1% peptone water. The suspensions were transferred to homogenizer bags (Interscience, Saint Nom, France) and coupled to a Stomacher@ 400 circulator (Seward, Worthing, West Sussex, UK) at 260 rpm for 1 min. The suspensions were serial-diluted from 10−6 to 100 and 100 μL of each dilution was transferred onto 20 μL of Baird-Parker agar containing egg yolk tellurite emulsion (BPAþ RPF, bioMerieux, France). Eighty presumptive coagulase-negative staphylococci colonies were tested by Gram-staining, catalase, coagulase, and thermostable DNAse activities according to Bergey's Manual of Systematic Bacteriology. Sixty presumptive CNS strains were stored at −80°C in tryptone soy agar (TSA, Franklin Lakes, New Jersey, USA) plus 45% v/v glycerol.
2.2. DNA Preparation
The strains were cultured aerobically overnight in 10 mL Brain Heart Infusion broth (BD BBL, Le Pont de Claix, France) at 37°C for 24 h. The suggestive CNS colonies were adjusted to 106 UFC/mL in a spectrophotometer and harvested by centrifugation at 5,700 × g for 1 min. The cell pellet was used for DNA extraction using the DNeasy blood and tissue kit (Qiagen, Dusseldorf, Germany), following the manufacturer's instructions. Genomic DNA was quantified using the Qubit fluorometer (Invitrogen, Grand Island, New York, USA) and Qubit assay kits.
2.3. PCR Tests
2.3.1. Primer Sequences and Target Genes
Primer sets flanking the sea, seb, sec, sed, see, seg, seh, sei, selk, selm, seln, selo, selq, selr, selu, and tstH1 sequences are listed in Table 1.
Table 1.
Primer set for V3 16S rDNA sequencing and PCR/real time RT-PCR tests targeting the classical and newly described staphylococcal enterotoxin genes.
| Primers set and sequences (5′-3′) | Gene | Amplicon (bp) | References | |
|---|---|---|---|---|
| SEAf | TTGGAAACGGTTAAAACGAA | sea | 120 | [17] |
| SEAr | GAACCTTCCCATCAAAAACA | |||
|
| ||||
| SEBf | TCGCATCAAACTGACAAACG | seb | 478 | [17] |
| SEBr | GCAGGTACTCTATAAGTGCC | |||
|
| ||||
| SECf | GACATAAAAGCTAGGAATTT | sec | 257 | [17] |
| SECr | AAATCGGATTAACATTATCC | |||
|
| ||||
| SEDf | CTAGTTTGGTAATATCTCCT | sed | 317 | [17] |
| SEDr | TAATGCTATATCTTATAGGG | |||
|
| ||||
| SEEf | TAGATAAAGTTAAAACAAGC | see | 170 | [17] |
| SEEr | TAACTTACCGTGGACCCTTC | |||
|
| ||||
| SEGf | TGCTATCGACACACTACAACC | seg | 704 | [18] |
| SEGr | CCAGATTCAAATGCAGAACC | |||
|
| ||||
| SEHf | CGAAAGCAGAAGATTTACACG | seh | 495 | [18] |
| SEHr | GACCTTTACTTATTTCGCTGTC | |||
|
| ||||
| SEIf | GACAACAAAACTGTCGAAACTG | sei | 630 | [18] |
| SEIr | CCATATTCTTTGCCTTTACCAG | |||
|
| ||||
| SElJf | CAGCGATAGCAAAAATGAAACA | selj | 426 | [19] |
| SElJr | TCTAGCGGAACAACAGTTCTGA | |||
|
| ||||
| SElMf | CCAATTGAAGACCACCAAAG | selm | 517 | [20] |
| SElMr | CTTGTCCTGTTCCAGTATCA | |||
|
| ||||
| SElNf | ATTGTTCTACATAGCTGCAA | seln | 682 | [20] |
| SElNr | TTGAAAAAACTCTGCTCCCA | |||
|
| ||||
| SElOf | AGTCAAGTGTAGACCCTATT | selo | 534 | [20] |
| SElOr | TATGCTCCGAATGAGAATGA | |||
|
| ||||
| SElKf | ATGAATCTTATGATTTAATTTCAGAATCAA | selk | 545 | [21] |
| SElKr | ATTTATATCGTTTCTTTATAAGAAATATCG | |||
|
| ||||
| SElQf | GGAAAATACACTTTATATTCACAGTTTCA | selq | 539 | [21] |
| SElQr | ATTTATTCAGTTTTCTCATATGAAATCTC | |||
|
| ||||
| SElRf | AATGGCTCTAAAATTGATGG | selr | 363 | [22] |
| SElRr | TCTTGTACCGTAACCGTTTT | |||
|
| ||||
| SElUf | AATGGCTCTAAAATTGATGG | selu | 215 | [22] |
| SElUr | ATTTGATTTCCATCATGCTC | |||
|
| ||||
| TSST-1f | ATGGCAGCATCAGCTTGATA | tstH1 | 350 | [17] |
| TSST-1r | TTTCCAATAACCACCCGTTT | |||
|
| ||||
| 16S rDNAf | ATA AGA CTG GGA TAA CTT CGG G | 16SrDNA | 500 | [23] |
| 16S rDNAr | CTT TGA GTT TCA ACC TTG CGG TCG | |||
f: forward; r: reverse.
2.3.2. Uniplex-, Duplex-, and Multiplex-PCR Tests
Uniplex-PCR tests targeting the tstH1 sequence and duplex-PCR targeting the sea/seb and sec/sed sequences were performed. PCR mixtures contained 25 μL of 20 mM MgCl2, 10x PCR buffer (Invitrogen, Grand Island, New York, USA), 100 mM dNTP mix (Fermentas Thermo Scientific, Vilnius, Lithuania), 0.2 mM of each primer (Table 1), 0.5 U Taq DNA polymerase (Invitrogen, Grand Island, New York, USA), and 100 ng of DNA templates. Uniplex- and duplex-PCR assays were performed under the following conditions: 94°C for 5 min followed by 35 cycles of 94°C for 2 min, 53°C for 2 min, and 72°C for 1 min for extension, ending with a final extension at 72°C after 7 min [16], with modifications in the annealing temperature, using a thermal cycler (MyCycler, Bio-Rad, Hercules, CA, USA). The amplified fragments were visualized on 1.0% agarose gels (Sigma) stained with GelRed (dilution 1 : 1000) (BioAmerica, Tel Aviv, Israel) and documented on a transilluminator (MiniLumi Imaging Bio-Systems, BioAmerica, Tel Aviv, Israel).
2.3.3. Multiplex-PCR Tests
Multiplex-PCR assays were performed by the simultaneous amplification of the see, seg, seh, sei, selj, selm, seln, selo, selk, selq, selr, and selu sequences using the primer sets listed in Table 1. Each reaction contained 50 μL of a mix containing 0.5 U Taq DNA polymerase, 10x PCR buffer, 100 mM dNTP, 0.2 μM of each primer, and 100 ng of DNA template. DNA amplification of see, seg, seh, and sei was carried out as follows: 95°C for 5 min, 35 cycles of 95°C for 30 s, 53°C for 90s and 72°C for 90 s, and a final extension at 72°C for 10 min. The DNA amplifications of the selj, selm, seln, and selo group and the selk, selq, selr, and selu group were carried out in the same conditions [3]. PCR products were visualized by electrophoresis on 1.2% agarose gels (Uniscience do Brasil, São Paulo, Brazil) in 1x TAE (Tris-boric acid-EDTA) buffer stained by 0.5 μg mL−1 of GelRed (BioAmerica, Tel Aviv, Israel) and documented on a transilluminator (MiniLumi Imaging Bio-Systems, BioAmerica, Tel Aviv, Israel).
DNA templates from the following reference strains were used: S. aureus ATCC 29231 (sea), S. aureus ATCC 14458 (seb, tstH, selk, selq, selr, and selu); S. aureus ATCC 19095 (sec, seg, seh, and sei), S. aureus ATCC 13563 (sed), S. aureus ATCC 27664 (see), and S. aureus ATCC 27154 (selj, selm, seln, and selo) and S. xylosus ATCC 29971.
2.4. Enterotoxin Expression Assays
The observed strains were cultured aerobically overnight in 10 mL Brain Heart Infusion Broth (BD BBL, Le Pont de Claix, France) at 37°C for 72 h. Bacteria supernatants were collected by centrifugation at 4,000 ×g for 10 min and used for the detection of sea, seb, sec, sed, and see by an ELISA assay using a commercial detection kit (RIDASCREEN SET A, B, C, D, E Art. number R4101, R-Biopharm AG, Germany). The assay was performed according to the manufacturer's recommendation and as described elsewhere [16]. The mean lower limit of detection of the assay was 0.25 ng mL−1. The threshold is defined as the average OD of two negative controls plus 0.15, a constant established by the kit. Samples containing SEs showed absorbance values equal to or greater than the threshold value. All experiments were performed in duplicate.
2.5. Real Time RT-PCR Assays
Total RNA was extracted by using the QIAGEN RiboPure Bacteria kit (Life Technologies, Carlsbad, California, USA) following the manufacturer's instructions and quantified using the Qubit fluorometer (Invitrogen, Grand Island, New York, USA) and Qubit assay kits. The cDNA synthesis was performed by using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, California, USA) and the ABI PRISM 7500 Fast RT-PCR system (Applied Biosystems, California, USA). Samples were plated in triplicate in 96-well plates as follows: 12 μL of the SYBR Green PCR Master Mix; 1 μL of primer mix (sea, seb, sec, sed, and see); and 4.5 μL of the cDNA ultrapure water in each well. Amplification was performed under the following conditions: 95°C for 15 min, 40 cycles at 95°C for 15 s, 54°C for 30s, and 72°C for 30 s. The dissociation curve was performed at 95°C for 15 sec, 54°C for 30 sec, and 95°C for 15 sec. CT means, the standard deviations, and the cDNA semiquantification were calculated using the GraphPad Prism 5 software package. Calibration curves based on five points were constructed in triplicate corresponding to serial dilutions (1, 1 : 10, 1 : 100, 1 : 1000, and 1 : 10000) from 100 ng of a DNA template stock solution.
2.6. 16S rDNA Sequencing
Amplification of the V5 region of 16S rDNA fragment was performed using 50 ng of DNA templates from the 65 strains found in the salami samples. PCR was performed under the following conditions: 95°C for 10 min, followed by 30 cycles at 95°C for 30 s, 60°C for 30 s and 72°C for 45 s, and a final extension at 72°C for 10 min. PCR products were purified using the PCR DNA Purification Kit (Applied Biosystems, California, USA) and sequenced using 20 ng purified DNA and 13 μL of primer sets in a final volume of 20 μL. After amplification, products were purified according to the protocol of the BigDye Terminator Purification X Kit (Applied Biosystems, California, USA) and sequenced on a 3130 sequencer Genetic Analyzer (Applied Biosystems, California, USA). The sequences were compared to the 16S rDNA gene sequences of Staphylococcus species available at the GenBank database (http://www.ncbi.nlm.nih.gov/Genbank/index.html). Multiple sequence alignments were performed using Clustal W (Kyoto University, Bioinformatics Center; http://www.genome.jp/tools/clustalw/).
2.7. Sequencing of Enterotoxin PCR Products
PCR products were purified using the PCR DNA Purification Kit (Applied Biosystems, California, USA) and sequenced using 10 ng of purified DNA and 3.2 pmoles of each primer set in a final volume of 20 μL. After amplification in the same conditions as the PCR step (Section 2.3.3), products were purified according to the BigDye Terminator Purification X Kit protocol (Applied Biosystems, California, USA) and sequenced on a 3130 sequencer Genetic Analyzer (Applied Biosystems, California, USA). The sequences were compared to Staphylococcus aureus and Staphylococcus pasteuri gene sequences available at the GenBank database (http://www.ncbi.nlm.nih.gov/Genbank/index.html). Multiple sequence alignments were performed using Clustal W (Kyoto University, Bioinformatics Center; http://www.genome.jp/tools/clustalw/).
2.8. Phylogenetic Analyses
Phylogenetic relationships between the CNS strains were performed by sequence alignments using the Clustal X 2.0 software package [24]. The phylogenetic trees were constructed using the software Mega 6.0 and UPGMA methods [25].
2.9. Antibiotic Susceptibility Tests
An inoculum of each strain equivalent to a 0.5 McFarland scale was swabbed onto a Mueller-Hinton agar plate (BD BBL Franklin Lakes, New Jersey, USA) and the antibiotic disc was then placed on the plate followed by overnight incubation at 37°C. The inhibition zone was interpreted according to the Clinical Laboratory Standard (CLSI) Guidelines, formerly known as the National Committee for Clinical Laboratory Standards. The tested antibiotics were penicillin G (10 U), oxacillin (1 μg), neomycin (30 μg), sulfamethoprim (5 μg), clindamycin (2 μg), gentamicin (10 μg), cefoxitin (30 μg), rifampicin (5 μg), erythromycin (15 μg), tetracycline (30 μg), vancomycin (30 μg), ciprofloxacin (5 μg), sulfazothrim (23 μg), cefepime (30 μg), linezolid (30 μg), and chloramphenicol (30 μg).
2.10. Minimal Inhibitory Concentration (MIC) Determinations
The MICs of vancomycin, linezolid, methicillin, and ampicillin were determined by the macrodilution broth method based on CLSI recommendations, using in-house-prepared panels [26]. Antibiotic concentrations of 0.03, 0.06, 0.125, 0.25, 0.5, 1.0, and 2.0 mg mL−1 were tested. One mL of broth was transferred to the tubes and 100 μL of the bacteria suspension was adjusted to 106 CFU/mL in saline 0.85% according to a 0.5 McFarland scale and transferred to tubes containing 1 mL of each antimicrobial. Strains were grown in Mueller-Hinton broth (BD BBL Franklin Lakes, New Jersey, USA) and the MIC was estimated as the lowest antibiotic concentration that inhibits visible growth after 24 h [26].
3. Results and Discussion
3.1. Isolation and Identification of Coagulase-Negative Strains from Salami
Sixty-five presumable coagulase-negative staphylococci microorganisms from salami were isolated by colony morphology, coagulase slide test, subsequent tube test, and biochemical tests. The sequencing of the V5 region of the 16S rDNA fragment of the strains was discriminative enough to differentiate the Staphylococcus isolated from salami at the subspecies level, with the exception of 1 strain, identified up to the Staphylococcus spp. genus.
Nineteen distinct strains were identified as CNS in salami with 08 of 19 (42%) identified as S. saprophyticus, the predominant species, followed by 05 strains of S. xylosus (26%), 02 strains of S. carnosus (11%), and 01 strain of each of the following species: S. succinus, S. sciuri, S. epidermidis, and S. hominis (5% each) (Table 2).
Table 2.
Genotypic and phenotypic characterization of CNS strains from salami.
| Strains identification and characterization | ||||
|---|---|---|---|---|
| Salami origin | Staphylococcus species | Genotypic | Phenotypic | |
| GenBank accession number and similarity (%) | Presence of enterotoxin genes | mRNA detection | Enterotoxin production (ng mL−1) | |
| Commercial | Staphylococcus spp. KF135445.1 (96) | sea, seb | — | — |
| S. carnosus KJ862002.1 (96) | sea, seb, sed, seh, sei,and selj | — | 1.2 ± 0.1 | |
| S. carnosus NR116434.1 (98) | seb, sec, sed, and see | see | 0.3 | |
| S. saprophyticus AB697717.1 (98) | seh, sei, and selm | sei, selm | 0.3 | |
| S. saprophyticus EU430992.1 (99) | sea, seb, selm, seln, selo,and tstH1 | seb, selm, and selo | 1.3 ± 0.1 | |
| S. saprophyticus HQ699510.1 (97) | — | — | — | |
| S. saprophyticus JX490122.1 (99) | sea, seh | seh | 0.5 ± 0.1 | |
| S. saprophyticus KJ004623.1 (96) | sec | — | 0.4 | |
| S. saprophyticus KJ949606.1 (98) | sea, seb, sec,and seh | sea, seb, and seh | — | |
| S. sciuri JX966436.1 (98) | sed, sei | sei | 1.0 ± 0.1 | |
| S. xylosus AM882700.1 (97) | — | — | — | |
| S. xylosus CP007208.1 (99) | — | — | — | |
| S. xylosus CP008724.1 (98) | sea, tstH1 | sea | 1.4 | |
| S. xylosus KC456590.1 (98) | — | — | — | |
|
| ||||
| Artisanal | S. epidermidis KF600589.1 (97) | sec, sed, see, seg, seh,and sei | see, sei | 0.7 ± 0.1 |
| S. hominis JX519988.1 (97) | sea, seb, sec, sed, selj, selm, seln, and selo | seln, selo | 0.9 ± 0.1 | |
| S. saprophyticus HF937252.1 (97) | seb | seb | 0.5 | |
| S. saprophyticus subsp. bovis KJ699151.1 (98) | sec, selm, seln,and tstH1 | seln | 0.9 ± 0.1 | |
| S. succinus KC329824.1 (99) | sea, seb, sec, selj,and seln | seb, sec | 1.3 | |
| S. xylosus KF198080.1 (97) | sec, selm,and tstH1 | — | 0.5 ± 0.1 | |
The presence of enterotoxin genes sea, seb, sec, sed, see, seg, seh, sei, selj, selk, selm, seln, selo, selq, selr, selu, and tstH1 was tested by PCR using the specific set of primers.
sea-see enterotoxin production was evaluated by immune-sorbent assays (ELISA) using the detection kit RIDASCREEN SET A, B, C, D, E. Values are displayed as the means ± SD of assays performed in duplicate.
mRNA transcripts for all enterotoxin genes were evaluated by real time RT-PCR tests.
CT values are displayed as means ± SD of RT-PCR tests performed in duplicate: sea—S. xylosus CP008724.1 30 ± 0.4; seb—S. xylosus CP008724.1 33 ± 2.0; S. saprophyticus JX 490122.1 30.2 ± 0.4; S. saprophyticus subsp. bovis KJ699151.1 32 ± 0.1; S. xylosus KF198080.1 36 ± 1.0; and S. hominis JX519988.1 34 ± 1.0; sec—S. saprophyticus subsp. bovis KJ699151.1 31 ± 1.1; see—S. saprophyticus subsp. bovis KJ699151.1 30.6 ± 0.5; S. succinus KC329824.1 28.5 ± 0.1; and S. saprophyticus HF937252.1 32 ± 0.4; seh—S. xylosus CP008724.1 31 ± 1.0; S. saprophyticus JX966436.1 33.2 ± 0.6; sei—S. saprophyticus AB697717.1 30 ± 1.0; S. saprophyticus JX966436.1 30.6 ± 1.0; seln—S. hominis JX519988.1 30.3 ± 1.0; S. saprophyticus EU430992.1 31 ± 0.3; and selo—S. hominis JX519988.1 32 ± 1.0.
The 08 S. saprophyticus strains were identified as KJ699151.1, AB697717.1, JX490122.1, KJ004623.1, and HQ699510.1, EU430992.1, HF937252.1, and KJ949606.1, with the latter two and S. sciuri JX966436.1 being homologous (96–98%) to strains from the environment. Five S. xylosus strains, CP007208.1, KF198080.1, CP008724.1, AM882700.1, and KC456590.1, and a single S. succinus strain, KC329824.1, were identified. The last three S. xylosus strains and the S. succinus strain are homologous (97-98%) to strains found in fermented meat or meat starter cultures. S. xylosus CP007208.1 showed homology (98%) to potential opportunistic pathogenic strains from mammal species.
The S. carnosus strains identified were KJ862002.1 and NR116434.1, and the latter, as well as the single S. hominis JX519988.1 (96-97%) identified, were homologous (97%) to species from human microbiota.
S. carnosus and S. xylosus are commonly used as commercial starter cultures for sausage manufacturing [26], but S. succinus has also been observed in dry fermented sausages and its use as a starter culture has been already proposed [27].
The diversity of CNS microbiota found in the samples analyzed in the present study could be related to the origin of the salami. The microbiota from the artisanal salami showed greater biodiversity when compared to the commercial salami. In commercial salami, S. saprophyticus was the predominant species, but S. xylosus and S. carnosus, also observed in commercial salami, are commonly isolated from starter cultures [26]. The predominance of S. saprophyticus followed by S. xylosus has also been reported in salami from South Italy [28], similar to Belgian sausages, where S. saprophyticus was the most frequently detected species [29].
S. carnosus was not observed in artisanal salami, but S. succinus, S. epidermidis, and S. hominis were detected.
The biodiversity of the CNS staphylococci species found in microbiota depends on the kind of meat-fermented product, but CNS strains such as S. saprophyticus, S. auricularis, S. xylosus, S. capitis, S. hominis, S. carnosus, S. haemolyticus, S. warneri, S. equorum, S. cohnii, S. capitis, and S. intermedius have been described in Napoli-type salami, Sremska sausages, dry sausages, raw meat, and naturally fermented meat [30–33].
During the last decades S. epidermidis and S. saprophyticus have been described as emerging pathogens [34]. S. saprophyticus is considered a frequent contaminant of fermented sausages and raw meats and has been isolated from rectal swabs of cattle carcasses and pigs. In humans, the main reservoir of S. saprophyticus is the gastrointestinal tract [35]. S. saprophyticus and S. epidermidis can be opportunistic pathogens, isolated from the human urinary tract, and the presence of these species in food should be taken into account concerning possible contamination of the starter inoculum and/or improvements in the salami manufacturing process [13].
Other CNS species observed in the present study are mainly associated with ordinary food contaminants, with S. epidermidis and S. hominis being the dominant species in human skin and occasionally isolated from the skin of domestic animals [36].
3.2. Phylogenetic Relationships of the CNS Identified in Salami
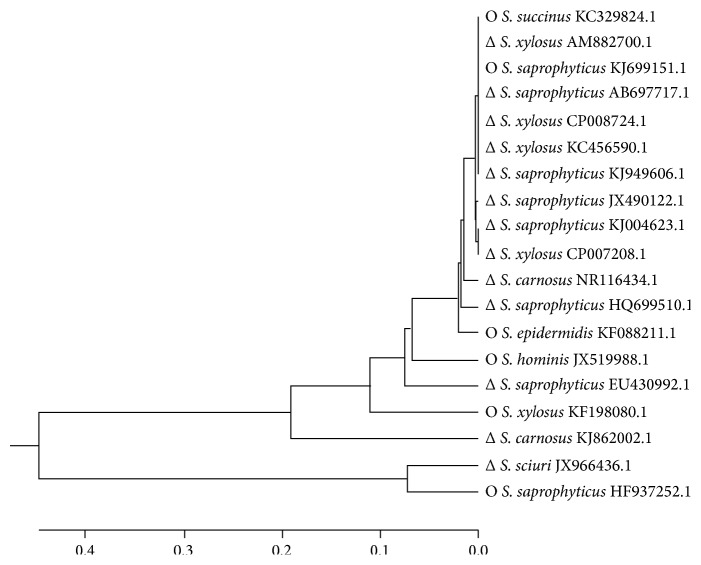
CNS strains can be grouped into four species groups: saprophyticus, simulans, epidermidis, and haemolyticus. Frequently, the saprophyticus species group includes S. xylosus and S. saprophyticus, while the simulans species group is comprised of S. carnosus and S. piscifermentans; the epidermidis species group is composed of S. epidermidis, S. capitis, S. caprae, and S. saccharolyticus and the haemolyticus species group encompasses S. haemolyticus, S. hominis, and S. devriesei [37, 38]. In the present study, the main cluster grouped several S. saprophyticus strains, namely, KJ699151.1, AB697517.1, KJ949606.1, JX490122.1, and KJ004623.1, four S. xylosus strains CP008724.1, CP007208.1. AM882700.1, and KC456590.1, and the S. succinus KC329824.1 strain (Figure 1). These strains are homologous to those from fermented meat microbiota.
Figure 1.
Phylogenetic tree generated from the multiple alignments of the 16S rDNA sequences of CNS strains found in salami using the ClustalX 2.0 software. The phylogenetic tree was constructed by using the Mega 6.0 software and the unweighted pair group method (UPGMA). Bootstrap values ranged from 0.0 to 0.4. Strains found in commercial (Δ) or artisanal salami (O).
The subclusters of those clusters showed mismatches of species belonging to the four species groups. The first subcluster grouped S. epidermidis HF088211.1, S. saprophyticus HQ699510.1, and S. carnosus NR116434.1 and the second subcluster grouped S. hominis JX519988.1 and the S. saprophyticus EU430992.1, all of them originally isolated from animals and human beings. The third subcluster includes species previously isolated from the marine environment, such as S. xylosus KF198080.1, while S. carnosus KJ862002.1 is utilized as a probiotic organism in foods. The fourth cluster grouped the S. sciuri JX966436.1 and S. saprophyticus HF937252.1 strains, which are homologous to species found in soil.
The CNS strains clustered into groups near the bottom of the phylogenetic tree are mostly strains found in artisanal salami, whereas the species at the top of the tree are mainly CNS strains found in commercial salami (Figure 1).
The close similarities between the S. saprophyticus AB697517.1 and KJ949606.1 strains, the S. succinus KC329824.1, S. xylosus AM882700.1, KC456590.1, and CP008724.1 strains, the S. saprophyticus KJ004623.1 and KJ699151.1 strains, and S. xylosus CP007208.1 are supported by a bootstrap value of 100%. The interspecies similarities were over 90%, which demonstrates close phylogenies between these CNS strains. Suzuki et al., 2012, demonstrated a bootstrap value higher than 90% for interspecies similarities between S. saprophyticus, S. epidermidis, S. hominis, and S. carnosus.
3.3. Genotypic and Phenotypic Characterization of CNS Strains
Twelve distinct combinations of staphylococcal enterotoxins genes were found in the 15 CNS strains, comprising SEs A–E, G–J, and also the enterotoxin-like toxins (SElL) K–R and U (Table 2).
Fifteen strains (79%) carried at least one gene encoding enterotoxins in their genomes (Figure 2). The seb and sec genes were the most predominant, harbored by 57% of the strains, followed by sea, carried by 50%, whereas the sed/seh/selm genes showed intermediate incidence harbored by 33% of the strains, while sei/seln and tsH1 were found in 27% of strains. Finally, see, selj, and selo showed low incidence (13%) and seg was carried by only 1% of strains (Table 2).
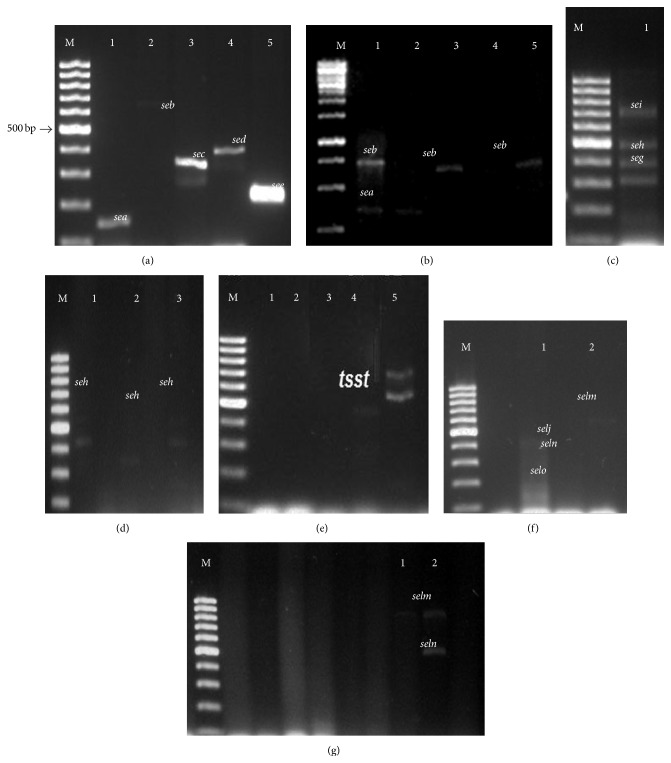
Figure 2.
Uniplex, duplex, and multiplex PCR screening for the detection of enterotoxin genes in CNS strains from salami. (a) Lane M, 100 bp DNA ladder plus (Fermentas, Foster City, CA, USA); lane 1, S. aureus ATCC 29231 harboring sea gene; lane 2, S. aureus NCTC 10654 harboring seb gene; lane 3, S. aureus ATCC19095 harboring the sec gene; lane 4, S. aureus ATCC 13563 harboring the sed gene; and lane 5, S. aureus ATCC 27664 harboring the see gene. (b) Lane M 100 bp DNA ladder plus; lane 1, Staphylococcus spp.; lane 2, S. carnosus NR116434; lane 3, S. carnosus KJ862002; lane 4, S. carnosus KJ862002; and lane 5, S. carnosus KJ862002.1. (c) Lane M, 100 bp DNA ladder plus; lane 1, S. aureus ATCC 19095 harboring seg, seh, and sei genes. (d) Lane M, 100 bp DNA ladder plus; lane 1, S. saprophyticus AB697717.1; lane 2, S. epidermidis KF 600589.1; and lane 3, S. sciuri JX966436.1. (e) Lane M, 100 bp DNA ladder plus; lane 1, S. xylosus KF198080.1; lane 2, S. saprophyticus KJ699151.1. (f) Lane M, 100 bp DNA ladder plus; lane 1, S. aureus ATCC 27154 harboring selj, slem, seln, and selo genes. (g) Lane M, 100 bp DNA ladder plus; lane 1, S. xylosus KF198080.1; and lane 2, S. saprophyticus subsp. bovis KJ699151.1.
The relationships between superantigenic toxin genotypes and toxin gene-encoding mobile genetic elements in CNS strains were evaluated. Distinct combinations of SaPI and plasmids or plasmids and genes of egc operon enterotoxins were found in the CNS strains obtained from salami.
Four strains presented only the classical enterotoxin genes, namely, S. saprophyticus KJ004623.1 and HF937252.1, S. xylosus CP008724.1, and S. carnosus NR116434.1. Another strain, S. saprophyticus AB697717.1, presented only the newly described enterotoxins seh, sei, and selm and, finally, 10 strains presented a combination of classical and newly described enterotoxin genes in their genomes, with at least one of each enterotoxin type (Table 2).
Previous studies have shown that sea is the most common toxin associated with Staphylococcus food poisoning, followed by sed and see, with SEH and SEI being considered as playing only a minor role [39]. Among the 15 enterotoxigenic strains found in salami, 73% of them harbored at least the sea gene or combinations of these 05 genes. Strains S. carnosus KJ862002.1, found in commercial salami, and S. epidermidis KF600589.1, found in artisanal salami, carry 04 of these genes. The sea and seb enterotoxin genes are known to occupy the same locus on the chromosome, which may explain why these enterotoxins are commonly found together in food poisoning outbreaks [36]. The combination of sea and seb genes was found in 05 strains: S. saprophyticus EU430992.1, S. carnosus KJ862002.1, S. hominis JX519988.1, S. saprophyticus KJ949606.1, and S. succinus KC329824.1. A single strain, S. carnosus KJ862002.1, showed the combination of sea and sei genes.
Additionally, some CNS are able to produce TSST-1 alone or in combination with other enterotoxins. Herein, 04 of the 19 strains (21%), S. saprophyticus EU430992.1, S. saprophyticus subsp. bovis KJ699151.1, and S. xylosus CP008724.1 and KF198080.1, were shown to harbor the tstH1 combined with se and/or sel enterotoxin genes (Table 2).
The staphylococci enterotoxins genes seg and sei [40] are part of a chromosomal operon gene cluster (egc), comprising five genes designated as selo, selm, sei, seln, and seg. Two of the CNS strains determined in the present study, S. saprophyticus EU430992.1 and S. hominis JX519988.1, were shown to carry the selm, seln, and selo genes and the selm and seln sequences were detected in the genome of a single strain, S. saprophyticus subsp. bovis KJ699151.1. S. epidermidis KF600589.1 showed a combination of seg and sei genes and 03 strains harbored a single gene, sei or selm or seln, perhaps due to the high degree of genetic polymorphism in the chromosomal assembly [41]. Indrawattana et al., 2013, detected the seg-sei-selm-seln-selo of the highly prevalent egc locus was 26.1% in contrast to this study where 13% CNS presents the combination of selm, seln, and selo genes [42].
The CNS strains clustered into groups near the bottom of the phylogenetic tree carried the classical enterotoxin genes, while the species at the top of the tree showed high diversity among the enterotoxin genes, combining the classical and the newly described genes in their genomes (Figure 1).
To assess the risk of staphylococcal food poisoning, the ability of the identified strains in harboring the se and sel genes and in expressing and producing enterotoxins in vitro was evaluated. The mRNA for each enterotoxin gene was evaluated by real time RT-PCR assays and enterotoxin content was estimated by a sandwich enzyme immunoassay for the combined detection of Staphylococcus enterotoxins (SET) A, B, C, D, and E.
The fifteen enterotoxigenic CNS strains were able to express the classical enterotoxins SEA, seb, sec, sed, and see, in concentrations ranging from 0.3 ng mL−1 to 1.4 ng mL−1, as assessed by the in vitro assays (Table 2). The S. saprophyticus AB697717.1, KJ699151.1, JX490122.1, KJ004623.1, and HF937252.1 strains, S. xylosus KF198080.1, and S. carnosus NR116434.1 produced low amount of enterotoxins, lower than 0.5 ng mL−1. S. epidermidis KF600589.1, S. saprophyticus bovis KJ699151.1, and S. hominis JX519988.1 produced intermediate amount of enterotoxins, ranging from 0.7 to 0.90 ng mL−1, while S. sciuri JX966436.1, S. saprophyticus EU430992.1, S. carnosus KJ862002.1, S. xylosus CP008724.1, and S. succinus KC329824.1 produced enterotoxins in concentrations ≥1.0 ng mL−1.
Although the sandwich enzyme immunoassay is considered the most sensitive method to detect sea–see enterotoxins, able to detect 0.125 ng mL−1, differences in the specificity and sensitivity of the assays for the detection of staphylococcal enterotoxins from foods are expected [43]. A single strain, Staphylococcus spp. KF135445.1, which harbors both the sea and seb genes, was unable to produce sea–see enterotoxins.
The mRNA levels evaluated by the real time RT-PCR assays for enterotoxins were detected in 12 of 15 strains (86%) (Table 2). Transcripts for the classical enterotoxins genes were detected when S. saprophyticus HF937252.1, S. carnosus NR116434.1, S. xylosus CP008724.1, and S. succinus KC329824.1 were assayed. Transcripts for newly described enterotoxins were observed in S. saprophyticus JX490122.1 and AB697717.1, S. sciuri JX966436.1, S. saprophyticus subsp. bovis KJ699151.1, and S. hominis JX519988.1. Transcripts from both the classical and newly described enterotoxins were detected in S. saprophyticus EU430992.1 and KJ949606.1 and S. epidermidis KF600589.1.
No mRNA transcripts were obtained for S. saprophyticus KJ0046232.1 and S. carnosus KJ862002.1, although enterotoxin production was detected by the enzyme-linked immunosorbent tests (Table 2).
The 04 strains that carry tstH1 in their genome, S. saprophyticus EU430992.1 and KJ699151.1, and S. xylosus CP008724.1 and KF98080.1 were not able to produce mRNA for TSST-1 in the assay conditions.
The production of classical enterotoxins in vitro (immunologic test) matched the results shown by real time RT-PCR assays for the following strains: S. saprophyticus JX490122.1, S. sciuri JX966436.1, S. saprophyticus EU430992.1, S. saprophyticus HF937252.1, S. saprophyticus subsp. bovis KJ699151.1, S. hominis JX519988.1, S. epidermidis KF600589.1, and S. succinus KC329824.1.
Ten strains, namely, S. saprophyticus AB697717.1, S. saprophyticus EU430992.1, S. saprophyticus HF937252.1, S. saprophyticus KJ699151.1, S. saprophyticus KJ949606.1, S. succinus KC329824.1, S. hominis JX519988.1, S. xylosus KF198080.1 and S. xylosus CP008724.1, and S. epidermidis KF600589.1 expressed mRNA for multiple se and/or sel genes. There is a differential transcription among these genes, where the most frequent among the classical ones were seb and see/sea, transcribed by 04 and 02 strains, respectively, and the most frequent among the newly described genes were sei and seh/seln/selo, transcribed by 03 and 02 strains, respectively.
No mRNA for the sec gene was detected, although S. saprophyticus AB697717.1 was able to produce the enterotoxin in vitro, as detected by the ELISA assays.
3.4. Enterotoxin Gene Homologies between Salami CNS and CPS Strains
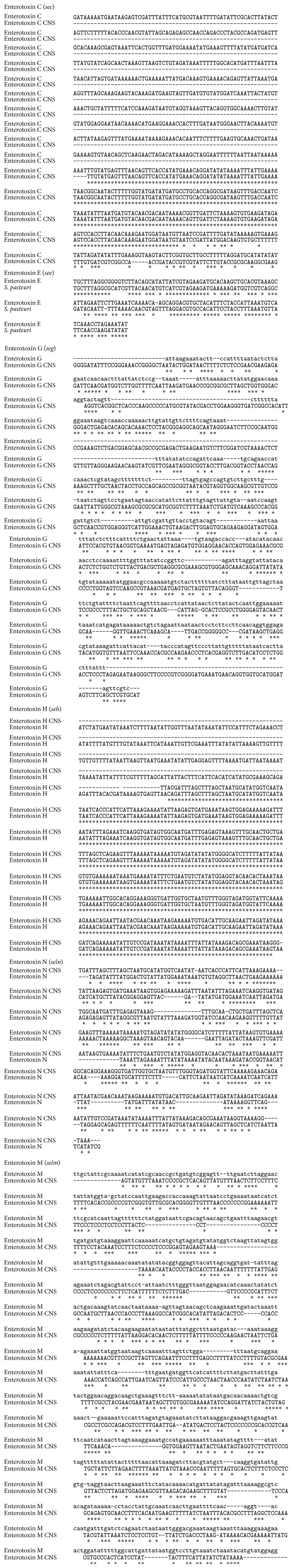
The nucleotide sequencing of six enterotoxin genes—two of them, sec and see, encoding classical enterotoxins—and four of them, encoding the newly described genes, seg, seh, selm, and seln from CNS strains, was compared to the enterotoxin genes from S. aureus or S. pasteuri, two coagulase-positive staphylococci strains (Figure 3). The homology between the CNS and CPS enterotoxin genes varied from 65% to 98%. The homology of sec, seg, seh, selm, and seln between CNS from salami and S. aureus was 98%, 60%, 98%, 65%, and 70%, respectively. The homology between see from CNS found in salami and in S. pasteuri was 98%.
Figure 3.
Alignment of the enterotoxin gene sequences found in coagulase-negative staphylococci (CNS) and in coagulase-positive staphylococci (CPS).
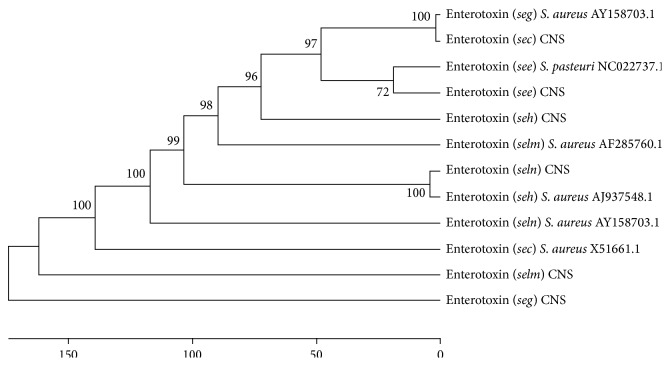
Although further studies should be performed, it seems that the sequences encoding enterotoxins can be conserved among coagulase-negative and coagulase-positive staphylococci, as shown in the phylogenetic analysis of the 06 enterotoxin genes (Figure 4).
Figure 4.
Phylogenetic tree generated from the multiple alignments of the enterotoxin sequences of CNS and CPS strains using the ClustalX 2.0 software package. The phylogenetic tree was constructed using the Mega 6.0 software and the unweighted pair group method (UPGMA).
3.5. Antimicrobial Multiresistance of CNS Strains (MRCNS)
Another safety hazard associated with CNS strains besides the ability of producing enterotoxins in food matrices is the antimicrobial resistance to antimicrobial agents commonly used to treat staphylococci infections. The antimicrobial resistance carried by CNS strains from food matrices can be spread to the population by the consumption of an apparently safe food.
Among the 19 CNS strains identified in salami, 14 showed multiresistance to antimicrobial agents. Three strains showed the highest MAR indices, 0.93 and 0.80; 07 strains presented MAR indices varying from 0.66 to 0.46, and the remaining 04 strains showed MAR indices ≤0.26 (Table 3).
Table 3.
Multiple resistance to antimicrobial as found in CNS strains from salami.
| Salami origin | CNS strains | Antimicrobial agent resistance | Multiple antimicrobial resistance (MAR) index∗ |
|---|---|---|---|
| Commercial | Staphylococcus spp. KF135445.1 | CIP, CLO, CPM, GEN, NEO, OXA, PEN, SXT, TET, and VAN | 0.66 |
| Commercial | S. carnosus KJ862002.1 | CIP, GEN, LZD, NEO, OXA, SXT, and TET | 0.46 |
| Commercial | S. saprophyticus AB697717.1 | CFO, CLO, ERI, PEN, OXA, TET, and VAN | 0.46 |
| Commercial | S. saprophyticus EU430992.1 | CFO, CLO, ERI, GEN, LZD, NEO, OXA, PEN, SXT, TET, and VAN | 0.80 |
| Commercial | S. saprophyticus JX490122.1 | CFO, CLO, ERI, GEN, NEO, PEN, OXA, SXT, TET, and VAN | 0.66 |
| Commercial | S. saprophyticus KJ004623.1 | CFO, OXA, and PEN | 0.20 |
| Commercial | S. sciuri JX966436.1 | CFO, CLO, GEN, NEO, OXA, PEN, and TET | 0.46 |
| Commercial | S. xylosus CP007208.1 | OXA, LZD, PEN, and VAN | 0.26 |
| Commercial | S. xylosus CP008724.1 | CFO, CLO, ERI, GEN, NEO, OXA, PEN, TET, and VAN | 0.60 |
| Artisanal | S. epidermidis KF600589.1 | CFO, CLI, CLO, CPM, ERI, GEN, LZD, NEO, OXA, PEN, RIF, SXT, and TET | 0.93 |
| Artisanal | S. hominis JX519988.1 | CIP, CPM, CPO, ERI, GEN, LZD, NEO, OXA, PEN, RIF, SXT, and VAN | 0.80 |
| Artisanal | S. saprophyticus HF937252.1 | CFO, OXA, PEN, and SXT | 0.26 |
| Artisanal | S. saprophyticus KJ699151.1 | OXA, PEN, and VAN | 0.20 |
| Artisanal | S. succinus KC329824.1 | CFO, CLO, ERI, GEN, OXA, PEN, RIF, SXT, TET, and VAN | 0.66 |
| Artisanal | S. xylosus KF198080.1 | CFO, CLO, ERI, GEN, NEO, OXA, PEN, TET, and VAN | 0.60 |
∗The MAR index of an isolate is defined as a/b, where a represents the number of antimicrobials to which the isolate was resistant and b represents the number of antimicrobials to which the isolate was subjected.
S. aureus strains ATCC WB81 (sea), ATCC 13563 (sed), and ATCC 27664 (see) showing a MAR index of 0.5 and S. aureus strains ATCC14458 (seb) and ATCCWB72 (sec) and S. xylosus ATCC 29971 showing a MAR index of 0.3 were used as reference strains.
CPM: cefepime, CFO: cefoxitin, CLO: chloramphenicol, CIP: ciprofloxacin, CLI: clindamycin, ERI: erythromycin, GEN: gentamycin, NEO: neomycin, LZD: linezolid, RIF; rifampicin, TET: tetracycline, OXA; oxacillin, PEN: penicillin, SXT: sulfamethoprim, and VAN: vancomycin.
The 14 MRCNS strains were resistant to β-lactams (oxacillin, penicillin, and/or cefoxitin) and to vancomycin, corresponding to 73% of the total CNS strains identified, while 09 strains (64%) showed resistance to tetracycline and gentamicin, 08 strains (57%) were resistant to neomycin, erythromycin, and chloramphenicol, 07 strains (50%) were resistant to sulfamethoprim, 05 strains (36%) were resistant to linezolid, 03 strains (21%) were resistant to rifampicin, 02 strains (14%) were resistant to ciprofloxacin and cefepime, and 01 strain (7%) was resistant to clindamycin (Table 3).
The multiresistance of CNS strains demonstrated herein is consistent with previous studies on coagulase-negative and coagulase-positive staphylococci that have found several resistant and multiresistant Staphylococcus aureus strains in raw milk, meat, and fermented meat products [43].
Surprisingly, the resistance to chloramphenicol is very similar to that estimated for MRCNS strains isolated from human clinical samples. Indeed, there is no direct correlation between the researched features and the origin of the staphylococci strains, since different virulence factors are widespread, such as antibiotic resistance [44], reinforcing the fact that the ability of food matrices strains to produce SE and SEl and their multiresistance character must be considered when evaluating the safety hazards of food poisoning.
S. epidermidis KF600589.1 and S. hominis JX519988.1 showed the highest MAR indexes (0.93 and 0.80, resp.). These strains found in artisanal salami are homologous to strains isolated from human skin microbiota and should be carefully considered among the potential pathogenic staphylococci found in food from animal origins, which can be caused by contamination by poor hygienic conditions during salami manufacturing.
Depending on the conditions, some species of coagulase-negative staphylococci can present health risks, since they have shown resistance to antibiotics of therapeutic importance, such as beta lactams [15]. However, multiresistant strains like S. carnosus KJ1862002.1 and S. xylosus CP007208.1 and CP008724.1 were intentionally introduced in the food matrix, since they are part of the culture starter used in Italian-salami manufacturing. As previously discussed, food can be a reservoir of multiresistant microorganisms that can spread by the consumption of an apparently safe food. Due to the intensive and indiscriminate use of antibiotics for human and veterinarian therapeutic purposes, multiresistant staphylococci strains are being selected and reproduced in food matrices [45].
The cefoxitin and oxacillin disk diffusion tests are recommended for determining the resistance and susceptibility breakpoint for MRSA surveillance cultures [46]. In this study, there was a good correlation between the disc test zone diameters for oxacillin and cefoxitin (Table 3). The MICs for ampicillin and methicillin/oxacillin/cefoxitin for all susceptible CNS strains were determined by the diffusion disc test, using 0.03 to 2 mg mL−1 of each antimicrobial agent.
S. saprophyticus HF937252.1 showed an MIC of 0.03 mg mL−1 for methicillin, strain S. carnosus KJ862002.1, S. xylosus strains CP008724.1 and CP007208.1, S. saprophyticus JX490122.1, S. hominis JX519988.1, and S. succinus KC329824.1 were resistant to 0.06 mg mL−1 and two strains, S. saprophyticus KJ699151.1 and S. saprophyticus KJ004623.1, showed an MIC of 0.5 mg mL−1 (Table 4). Two of the methicillin-resistant strains, S. saprophyticus KJ004623.1 and S. xylosus CP007208.1, harbor no enterotoxin gene.
Table 4.
Minimal inhibitory concentration (MIC) of compounds used in antimicrobial therapy against staphylococci infections.
| Salami origin | GenBank accession number and similarity (%) | MIC mg mL−1 | |||
|---|---|---|---|---|---|
| Methicillin | Ampicillin | Vancomycin | Linezolid | ||
| Commercial | S. carnosus KJ862002.1 | 0.06 | 0.03 | — | — |
| S. xylosus CP008724.1 | 0.06 | 0.03 | 0.06 | — | |
| S. saprophyticus JX490122.1 | 0.06 | 0.03 | 0.5 | — | |
| S. saprophyticus AB697717.1 | — | — | 0.5 | — | |
| S. saprophyticus EU430992.1 | — | — | 0.25 | 0.125 | |
| S. succinus KC329824.1 | 0.06 | 0.25 | 0.03 | — | |
|
| |||||
| Artisanal | S. hominis. JX519988.1 | 0.06 | 0.5 | — | — |
| S. saprophyticus HF937252.1 | 0.03 | — | — | — | |
| S. xylosus KF198080.1 | 0.03 | 0.5 | — | ||
| S. saprophyticus KJ699151.1 | 0.5 | 0.25 | 0.03 | — | |
| S. saprophyticus KJ004623.1 | 0.5 | 0.03 | 0.03 | — | |
| S. xylosus CP007208.1 | 0.06 | — | — | 0.25 | |
Strains S. epidermidis KF600589.1 and Staphylococcus spp. KF135445.1 were not susceptible to the antimicrobial concentrations tested in the present study.
Five strains, S. saprophyticus JX490122.1, S. carnosus KJ862002.1, S. xylosus CP008724.1, S. xylosus KF198080.1, and S. saprophyticus KJ004623.1, showed an MIC for ampicillin of 0.03 mg mL−1, two strains, S. succinus KC329824.1 and S. saprophyticus KJ699151.1, showed an MIC of 0.25 mg mL−1, and S. hominis JX519988.1 showed an MIC of 0.5 mg mL−1 (Table 4). S. saprophyticus KJ004623.1 also showed an MIC of 0.03 mg mL−1 but does not harbor enterotoxin genes. It is important to note that most of the CNS strains that showed resistance to ampicillin were also resistant to methicillin, as highlighted by the MIC tests.
Five strains, S. saprophyticus HF937252.1, S. saprophyticus KJ699151.1, S. saprophyticus JX490122.1, S. saprophyticus KJ004623.1, and S. succinus KC329824.1, showed an MIC for vancomycin of 0.03 mg mL−1, one strain, S. xylosus CP008724.1, showed an MIC of 0.06 mg mL−1, one strain, S. saprophyticus EU430992.1, showed an MIC of 0.25 mg mL−1, and two strains, S. xylosus KF198080.1 and S. saprophyticus AB697717.1, showed an MIC of 0.5 mg mL−1 (Table 4).
Additionally, five CNS strains, S. xylosus CP008724.1, S. saprophyticus JX 490122.1, S. succinus KC329824.1, S. saprophyticus KJ699151.1, and S. saprophyticus KJ004623.1, showed resistance to methicillin, as demonstrated by the MIC determinations (Table 4).
Previous studies have demonstrated that linezolid is active against Gram-positive bacteria, including methicillin-resistant staphylococci [47]. In the present study, 05 strains showed linezolid resistance in the disc diffusion test, but it was impossible to establish MIC values for S. epidermidis KF600589.1, Staphylococcus hominis JX519988.1, and Staphylococcus carnosus KJ862002.1. The remaining strains S. saprophyticus EU430992.1 and S. xylosus CP007208.1 presented an MIC for linezolid of 0.125 mg mL−1 and 0.25 mg mL−1, respectively.
The MIC of 0.03 mg mL−1 for penicillin is in accordance with the previous values estimated for resistant S. aureus strains found in several food matrices, such as meat, dairy products, and ready-to-eat food [48].
There is still a lack of information on the antimicrobial resistance of staphylococci strains from food matrices, although they are the most worrisome vehicles of dissemination of antibiotic-resistant pathogens [49].
The high resistance found for salami staphylococci strains can be ascribed to their inappropriate use as growth promoters of antimicrobial agents like oxacillin, vancomycin, chloramphenicol, neomycin, and erythromycin, which are commonly used in veterinary medicine to treat infections [50]. Antimicrobial therapy of infections staphylococci is based on results of susceptibility tests in vitro [51]. Methicillin-resistant coagulase-negative Staphylococcus spp. found in ready-to-eat products such as meats, fish, and dairy products also offer risks and, although they are not classical food poisoning bacteria, their presence in food offers significant risks to public health due to the possible spread of antibiotic resistance [52].
The standardization of salami quality should include diagnostic methods to screen and quantify the presence of classical and newly described enterotoxins directly in the food matrices as a routine procedure to be conducted by the meat product industry in Brazil.
The safety of salami consumption could also be enhanced by the inclusion of a microbial barrier such as the inclusion of probiotic strains producing natural antibiotics or competitive flora or even the addition of natural bioagents against spoilage or pathogenic microorganisms.
Acknowledgments
The authors acknowledge the financial support from Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ, Rio de Janeiro, Brazil), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, Brasília, Brazil), and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES, Brasília, Brazil).
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.ICMSF: International Comission on Microbiological Specifications for Foods. Microorganisms in Foods. Their Significance and Methods of Enumerations. 3rd. Toronto, Canada: University of Toronto Press; 1983. [Google Scholar]
- 2.Veras J. F., Carmo L. S., Tong L. C., et al. A study of the enterotoxigenicity of coagulase-negative and coagulase-positive staphylococcal isolates from food poisoning outbreaks in Minas Gerais, Brazil. International Journal of Infectious Diseases. 2008;12(4):410–415. doi: 10.1016/j.ijid.2007.09.018. [DOI] [PubMed] [Google Scholar]
- 3.Omoe K., Hu D.-L., Takahashi-Omoe H., Nakane A., Shinagawa K. Comprehensive analysis of classical and newly described staphylococcal superantigenic toxin genes in Staphylococcus aureus isolates. FEMS Microbiology Letters. 2005;246(2):191–198. doi: 10.1016/j.femsle.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 4.Kadariya J., Smith T. C., Thapaliya D. Staphylococcus aureus and staphylococcal food-borne disease: an ongoing challenge in public health. BioMed Research International. 2014;2014:9. doi: 10.1155/2014/827965.827965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Omoe K., Hu D.-L., Ono H. K., et al. Emetic potentials of newly identified staphylococcal enterotoxin-like toxins. Infection and Immunity. 2013;81(10):3627–3631. doi: 10.1128/iai.00550-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCormick J. K., Yarwood J. M., Schlievert P. M. Toxic shock syndrome and bacterial superantigens: an update. Annual Review of Microbiology. 2001;55:77–104. doi: 10.1146/annurev.micro.55.1.77. [DOI] [PubMed] [Google Scholar]
- 7.Baba T., Takeuchi F., Kuroda M., et al. Genome and virulence determinants of high virulence community-acquired MRSA. The Lancet. 2002;359(9320):1819–1827. doi: 10.1016/s0140-6736(02)08713-5. [DOI] [PubMed] [Google Scholar]
- 8.Associação Brasileira de Indústria Produtora e Exportadora da Carne Suínas (ABIPECS) Statistics slaughter ranking 2000–2014. 2014, http://abpa-br.com.br/setores/suinocultura/publicacoes/relatorios-anuais.
- 9.Zdolec N., Hadžiosmanović M., Kozačinski L., et al. Microbial and physicochemical succession in fermented sausages produced with bacteriocinogenic culture of Lactobacillus sakei and semi-purified bacteriocin mesenterocin Y. Meat Science. 2008;80(2):480–487. doi: 10.1016/j.meatsci.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 10.Ravyts F., Vuyst L. D., Leroy F. Bacterial diversity and functionalities in food fermentations. Engineering in Life Sciences. 2012;12(4):356–367. doi: 10.1002/elsc.201100119. [DOI] [Google Scholar]
- 11.Ohlsson T., Bengtsson N. The Hurdle Concept: Minimal Processing Technologies in the Food Industry. Woodhead Publishing; 2002. [Google Scholar]
- 12.Bernardi S., Golineli B. B., Contreras-Castillo C. J. Revisão: aspectos da aplicação de culturas starter na produção de embutidos cárneos fermentados. Brazilian Journal of Food Technology. 2010;13(2):133–140. doi: 10.4260/bjft2010130200018. [DOI] [Google Scholar]
- 13.Irlinger F. Safety assessment of dairy microorganisms: coagulase-negative staphylococci . International Journal of Food Microbiology. 2008;126(3):302–310. doi: 10.1016/j.ijfoodmicro.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 14.Ghebremedhin B., Layer F., König W., König B. Genetic classification and distinguishing of Staphylococcus species based on different partial gap, 16S rRNA, hsp60, rpoB, sodA, and tuf gene sequences. Journal of Clinical Microbiology. 2008;46(3):1019–1025. doi: 10.1128/jcm.02058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Babić I., Markov K., Kovačević D., et al. Identification and characterization of potential autochthonous starter cultures from a Croatian ‘brand’ product ‘slavonski kulen’. Meat Science. 2011;88(3):517–524. doi: 10.1016/j.meatsci.2011.02.003. [DOI] [PubMed] [Google Scholar]
- 16.Mehrotra M., Wang G., Johnson W. M. Multiplex PCR for the detection of genes for Staphylococcus aureus enterotoxins, exfoliative toxins, toxic shock syndrome toxin, and methicillin resistance. Journal of Clinical Microbiology. 2000;38(3):1032–1041. doi: 10.1128/jcm.38.3.1032-1035.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson W. M., Tyler S. D., Ewan E. P., Ashton F. E., Pollard D. R., Rozee K. R. Detection of genes for enterotoxins, exfoliative toxins, and toxic shock syndrome toxin 1 in Staphylococcus aureus by the polymerase chain reaction. Journal of Clinical Microbiology. 1991;29(3):426–430. doi: 10.1128/jcm.29.3.426-430.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mclauchlin J., Narayanan G. L., Mithani V., O'Neill G. The detection of enterotoxins and toxic shock syndrome toxin genes in Staphylococcus aureus by polymerase chain reaction. Journal of Food Protection. 2000;63(4):479–488. doi: 10.4315/0362-028x-63.4.479. [DOI] [PubMed] [Google Scholar]
- 19.Rosec J. P., Gigaud O. Staphylococcal enterotoxin genes of classical and new types detected by PCR in France. International Journal of Food Microbiology. 2002;77(1-2):61–70. doi: 10.1016/S0168-1605(02)00044-2. [DOI] [PubMed] [Google Scholar]
- 20.Omoe K., Hu D.-L., Takahashi-Omoe H., Nakane A., Shinagawa K. Identification and characterization of a new staphylococcal enterotoxin-related putative toxin encoded by two kinds of plasmids. Infection and Immunity. 2003;71(10):6088–6094. doi: 10.1128/iai.71.10.6088-6094.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sergeev N., Volokhov D., Chizhikov V., Rasooly A. Simultaneous analysis of multiple staphylococcal enterotoxin genes by an oligonucleotide microarray assay. Journal of Clinical Microbiology. 2004;42(5):2134–2143. doi: 10.1128/jcm.42.5.2134-2143.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holtfreter S., Grumann D., Schmudde M., et al. Clonal distribution of superantigen genes in clinical Staphylococcus aureus isolates. Journal of Clinical Microbiology. 2007;45(8):2669–2680. doi: 10.1128/jcm.00204-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mason W. J., Blevins J. S., Beenken K., Wibowo N., Ojha N., Smeltzer M. S. Multiplex PCR protocol for the diagnosis of staphylococcal infection. Journal of Clinical Microbiology. 2001;39(9):3332–3338. doi: 10.1128/JCM.39.9.3332-3338.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rahimi E., Safai H. G. Detection of classical enterotoxins of Staphylococcus aureus strains isolated from bovine subclinical mastitis in Isfahan, Iran. Veterinary Microbiology. 2010;141(3-4):393–394. doi: 10.1016/j.vetmic.2009.09.023. [DOI] [PubMed] [Google Scholar]
- 25.Larkin M. A., Blackshields G., Brown N. P., et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 26.Sneath P. H. A., Sokal R. R. Unweighted Pair Group Method with Arithmetic Mean. Numerical Taxonomy. San Francisco, Calif, USA: W. H. Freeman; 1973. [Google Scholar]
- 27.NCCLS. M 100-S14. NCCLS; 2004. Performance standards for antimicrobial susceptibility testing; 14th informational supplement. [Google Scholar]
- 28.Corbiere Morot-Bizot S., Leroy S., Talon R. Monitoring of staphylococcal starters in two French processing plants manufacturing dry fermented sausages. Journal of Applied Microbiology. 2007;102(1):238–244. doi: 10.1111/j.1365-2672.2006.03041.x. [DOI] [PubMed] [Google Scholar]
- 29.Talon R., Leroy S., Lebert I., et al. Safety improvement and preservation of typical sensory qualities of traditional dry fermented sausages using autochthonous starter cultures. International Journal of Food Microbiology. 2008;126(1-2):227–234. doi: 10.1016/j.ijfoodmicro.2008.05.031. [DOI] [PubMed] [Google Scholar]
- 30.Mauriello G., Casaburi A., Blaiotta G., Villani F. Isolation and technological properties of coagulase negative staphylococci from fermented sausages of Southern Italy. Meat Science. 2004;67(1):149–158. doi: 10.1016/j.meatsci.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 31.Janssens M., Myter N., De Vuyst L., Leroy F. Species diversity and metabolic impact of the microbiota are low in spontaneously acidified Belgian sausages with an added starter culture of Staphylococcus carnosus . Food Microbiology. 2012;29(2):167–177. doi: 10.1016/j.fm.2011.07.005. [DOI] [PubMed] [Google Scholar]
- 32.Blaiotta G., Ercolini D., Mauriello G., Salzano G., Villani F. Rapid and reliable identification of Staphylococcus equorum by a species-specific PCR assay targeting the sodA gene. Systematic and Applied Microbiology. 2004;27(6):696–702. doi: 10.1078/0723202042369901. [DOI] [PubMed] [Google Scholar]
- 33.Kozačinski L., Drosinos E., Čaklovica F., Cocolin L., Gasparik-Reichardt J., Vesković S. Investigation of microbial association of traditionally fermented sausages. Food Technology and Biotechnology. 2008;46(1):93–106. [Google Scholar]
- 34.Leroy S., Giammarinaro P., Chacornac J.-P., Lebert I., Talon R. Biodiversity of indigenous staphylococci of naturally fermented dry sausages and manufacturing environments of small-scale processing units. Food Microbiology. 2010;27(2):294–301. doi: 10.1016/j.fm.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 35.Coton E., Desmonts M.-H., Leroy S., et al. Biodiversity of Coagulase-Negative Staphylococci in French cheeses, dry fermented sausages, processing environments and clinical samples. International Journal of Food Microbiology. 2010;137(2-3):221–229. doi: 10.1016/j.ijfoodmicro.2009.11.023. [DOI] [PubMed] [Google Scholar]
- 36.Hosseinzadeh S., Saei H. D. Staphylococcal species associated with bovine mastitis in the North West of Iran: emerging of coagulase-negative staphylococci. International Journal of Veterinary Science and Medicine. 2014;2(1):27–34. doi: 10.1016/j.ijvsm.2014.02.001. [DOI] [Google Scholar]
- 37.Piette A., Verschraegen G. Role of coagulase-negative staphylococci in human disease. Veterinary Microbiology. 2009;134(1-2):45–54. doi: 10.1016/j.vetmic.2008.09.009. [DOI] [PubMed] [Google Scholar]
- 38.Moura T. M., Campos F. S., D'Azevedo P. A., et al. Prevalence of enterotoxin-encoding genes and antimicrobial resistance in coagulase-negative and coagulase-positive Staphylococcus isolates from black pudding in Southern Brazil. Revista da Sociedade Brasileira de Medicina Tropical. 2012;45(5):579–585. doi: 10.1590/s0037-86822012000500008. [DOI] [PubMed] [Google Scholar]
- 39.Suzuki H. I., Lefébure T., Bitar P. P., Stanhope M. J. Comparative genomic analysis of the genus Staphylococcus including Staphylococcus aureus and its newly described sister species Staphylococcus simiae . BMC Genomics. 2012;13, article 38 doi: 10.1186/1471-2164-13-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lamers R. P., Muthukrishnan G., Castoe T. A., Tafur S., Cole A. M., Parkinson C. L. Phylogenetic relationships among Staphylococcus species and refinement of cluster groups based on multilocus data. BMC Evolutionary Biology. 2012;12, article 171 doi: 10.1186/1471-2148-12-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pinchuk I. V., Beswick E. J., Reyes V. E. Staphylococcal enterotoxins. Toxins. 2010;2(8):2177–2197. doi: 10.3390/toxins2082177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Monday S. R., Bohach G. A. Genes encoding staphylococcal enterotoxins G and I are linked and separated by DNA related to other staphylococcal enterotoxins. Journal of Natural Toxins. 2001;10(1):1–8. [PubMed] [Google Scholar]
- 43.Thomas D. Y., Jarraud S., Lemercier B., et al. Staphylococcal enterotoxin-like toxins U2 and V, two new staphylococcal superantigens arising from recombination within the enterotoxin gene cluster. Infection and Immunity. 2006;74:4724–4734. doi: 10.1128/IAI.00132-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Indrawattana N., Sungkhachat O., Sookrung N., et al. Staphylococcus aureus clinical isolates: antibiotic susceptibility, molecular characteristics, and ability to form biofilm. BioMed Research International. 2013;2013:11. doi: 10.1155/2013/314654.314654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pereira V., Lopes C., Castro A., Silva J., Gibbs P., Teixeira P. Characterization for enterotoxin production, virulence factors, and antibiotic susceptibility of Staphylococcus aureus isolates from various foods in Portugal. Food Microbiology. 2009;26(3):278–282. doi: 10.1016/j.fm.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 46.Anacarso I., Condò C., Sabia C., et al Antimicrobial resistance and other related virulence factors in staphylococcus spp isolated from food, environmental and humans in Italy. Universal Journal Microbiology Research. 2013;1(1):1–9. doi: 10.13189/ujmr.2013.010101. [DOI] [Google Scholar]
- 47.Martín B., Garriga M., Hugas M., Bover-Cid S., Veciana-Nogués M. T., Aymerich T. Molecular, technological and safety characterization of Gram-positive catalase-positive cocci from slightly fermented sausages. International Journal of Food Microbiology. 2006;107(2):148–158. doi: 10.1016/j.ijfoodmicro.2005.08.024. [DOI] [PubMed] [Google Scholar]
- 48.Broekema N. M., Van Tam T., Monson T. A., Marshall S. A., Warshauer D. M. Comparison of cefoxitin and oxacillin disk diffusion methods for detection of mecA-mediated resistance in Staphylococcus aureus in a large-scale study. Journal of Clinical Microbiology. 2009;47(1):217–219. doi: 10.1128/jcm.01506-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cercenado E., García-Garrote F., Bouza E. In vitro activity of linezolid against multiply resistant Gram-positive clinical isolates. Journal of Antimicrobial Chemotherapy. 2001;47(1):77–81. doi: 10.1093/jac/47.1.77. [DOI] [PubMed] [Google Scholar]
- 50.Aydin A., Muratoglu K., Sudagidan M., Bostan K., Okuklu B., Harsa S. Prevalence and antibiotic resistance of foodborne Staphylococcus aureus isolates in Turkey. Foodborne Pathogens and Disease. 2011;8(1):63–69. doi: 10.1089/fpd.2010.0613. [DOI] [PubMed] [Google Scholar]
- 51.Sergelidis D., Abrahim A., Papadopoulos T., et al. Isolation of methicillin-resistant Staphylococcus spp. from ready-to-eat fish products. Letters in Applied Microbiology. 2014;59(5):500–506. doi: 10.1111/lam.12304. [DOI] [PubMed] [Google Scholar]
- 52.Phillips I., Casewell M., Cox T., et al. Does the use of antibiotics in food animals pose a risk to human health? A critical review of published data. Journal of Antimicrobial Chemotherapy. 2004;53(1):28–52. doi: 10.1093/jac/dkg483. [DOI] [PubMed] [Google Scholar]