Abstract
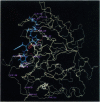
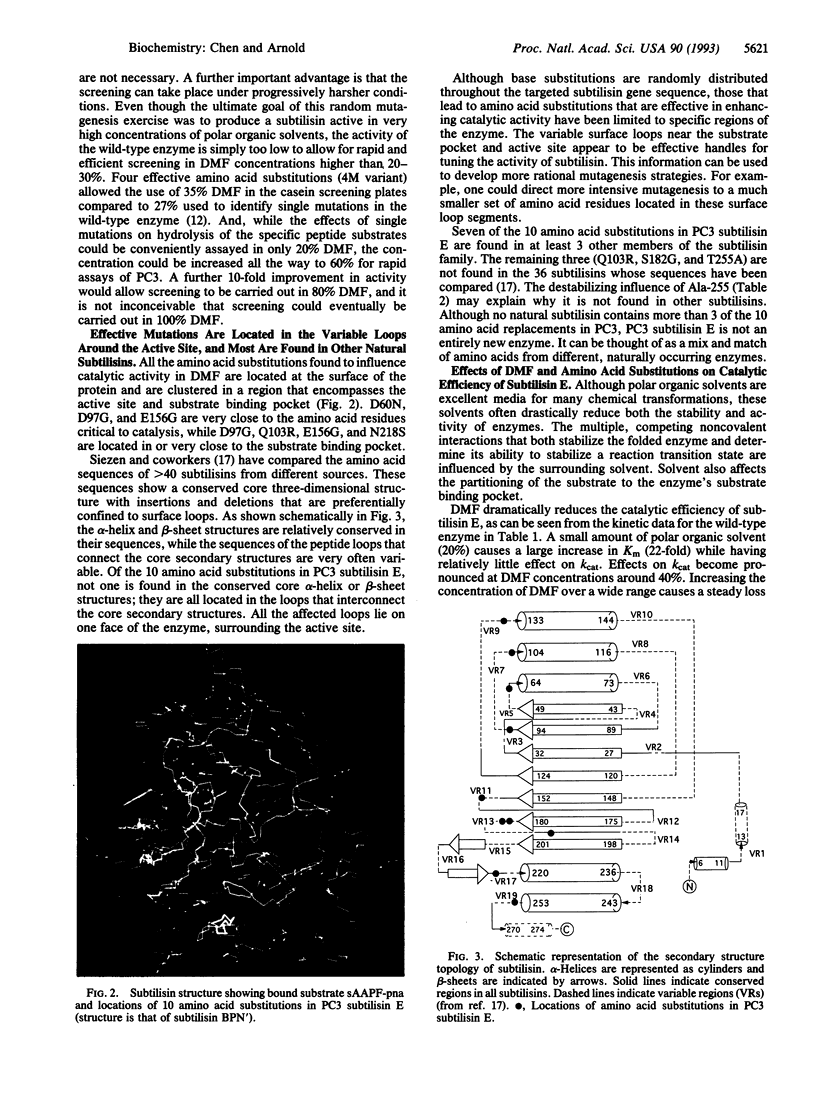
Random mutagenesis has been used to engineer the protease subtilisin E to function in a highly nonnatural environment--high concentrations of a polar organic solvent. Sequential rounds of mutagenesis and screening have yielded a variant (PC3) that hydrolyzes a peptide substrate 256 times more efficiently than wild-type subtilisin in 60% dimethylformamide. PC3 subtilisin E and other variants containing different combinations of amino acid substitutions are effective catalysts for transesterification and peptide synthesis in dimethylformamide and other organic media. Starting with a variant containing four effective amino acid substitutions (D60N, D97G, Q103R, and N218S; where, for example, D60N represents Asp-60-->Asn), six additional mutations (G131D, E156G, N181S, S182G, S188P, and T255A) were generated during three sequential rounds of mutagenesis and screening. The 10 substitutions are clustered on one face of the enzyme, near the active site and substrate binding pocket, and all are located in loops that connect core secondary structure elements and exhibit considerable sequence variability in subtilisins from different sources. These variable surface loops are effective handles for "tuning" the activity of subtilisin. Seven of the 10 amino acid substitutions in PC3 are found in other natural subtilisins. Great variability is exhibited among naturally occurring sequences that code for similar three-dimensional structures--it is possible to make use of this sequence flexibility to engineer enzymes to exhibit features not previously developed (or required) for function in vivo.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bryan P. N., Rollence M. L., Pantoliano M. W., Wood J., Finzel B. C., Gilliland G. L., Howard A. J., Poulos T. L. Proteases of enhanced stability: characterization of a thermostable variant of subtilisin. Proteins. 1986 Dec;1(4):326–334. doi: 10.1002/prot.340010406. [DOI] [PubMed] [Google Scholar]
- Chen K. Q., Arnold F. H. Enzyme engineering for nonaqueous solvents: random mutagenesis to enhance activity of subtilisin E in polar organic media. Biotechnology (N Y) 1991 Nov;9(11):1073–1077. doi: 10.1038/nbt1191-1073. [DOI] [PubMed] [Google Scholar]
- Cunningham B. C., Wells J. A. Improvement in the alkaline stability of subtilisin using an efficient random mutagenesis and screening procedure. Protein Eng. 1987 Aug-Sep;1(4):319–325. doi: 10.1093/protein/1.4.319. [DOI] [PubMed] [Google Scholar]
- Heinz D. W., Baase W. A., Matthews B. W. Folding and function of a T4 lysozyme containing 10 consecutive alanines illustrate the redundancy of information in an amino acid sequence. Proc Natl Acad Sci U S A. 1992 May 1;89(9):3751–3755. doi: 10.1073/pnas.89.9.3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermes J. D., Blacklow S. C., Knowles J. R. Searching sequence space by definably random mutagenesis: improving the catalytic potency of an enzyme. Proc Natl Acad Sci U S A. 1990 Jan;87(2):696–700. doi: 10.1073/pnas.87.2.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyet P., Declerck N., Gaillardin C. Hyperthermostable variants of a highly thermostable alpha-amylase. Biotechnology (N Y) 1992 Dec;10(12):1579–1583. doi: 10.1038/nbt1292-1579. [DOI] [PubMed] [Google Scholar]
- Liao H., McKenzie T., Hageman R. Isolation of a thermostable enzyme variant by cloning and selection in a thermophile. Proc Natl Acad Sci U S A. 1986 Feb;83(3):576–580. doi: 10.1073/pnas.83.3.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliphant A. R., Struhl K. An efficient method for generating proteins with altered enzymatic properties: application to beta-lactamase. Proc Natl Acad Sci U S A. 1989 Dec;86(23):9094–9098. doi: 10.1073/pnas.86.23.9094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantoliano M. W., Whitlow M., Wood J. F., Dodd S. W., Hardman K. D., Rollence M. L., Bryan P. N. Large increases in general stability for subtilisin BPN' through incremental changes in the free energy of unfolding. Biochemistry. 1989 Sep 5;28(18):7205–7213. doi: 10.1021/bi00444a012. [DOI] [PubMed] [Google Scholar]
- Poteete A. R., Rennell D., Bouvier S. E. Functional significance of conserved amino acid residues. Proteins. 1992 May;13(1):38–40. doi: 10.1002/prot.340130104. [DOI] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Siezen R. J., de Vos W. M., Leunissen J. A., Dijkstra B. W. Homology modelling and protein engineering strategy of subtilases, the family of subtilisin-like serine proteinases. Protein Eng. 1991 Oct;4(7):719–737. doi: 10.1093/protein/4.7.719. [DOI] [PubMed] [Google Scholar]
- Wong C. H., Wang K. T. New developments in enzymatic peptide synthesis. Experientia. 1991 Dec 1;47(11-12):1123–1129. doi: 10.1007/BF01918376. [DOI] [PubMed] [Google Scholar]
- Zhong Z. Y., Wong C. H. Development of new enzymatic catalysts for peptide synthesis in aqueous and organic solvents. Biomed Biochim Acta. 1991;50(10-11):S9–14. [PubMed] [Google Scholar]