Abstract
The transcription factor, nuclear factor erythroid 2 p45-related factor 2 (Nrf2), acts as a sensor of oxidative or electrophilic stresses and plays a pivotal role in redox homeostasis. Oxidative or electrophilic agents cause a conformational change in the Nrf2 inhibitory protein Keap1 inducing the nuclear translocation of the transcription factor which, through its binding to the antioxidant/electrophilic response element (ARE/EpRE), regulates the expression of antioxidant and detoxifying genes such as heme oxygenase 1 (HO-1). Nrf2 and HO-1 are frequently upregulated in different types of tumours and correlate with tumour progression, aggressiveness, resistance to therapy, and poor prognosis. This review focuses on the Nrf2/HO-1 stress response mechanism as a promising target for anticancer treatment which is able to overcome resistance to therapies.
1. Introduction
The availability of intracellular antioxidants is essential in maintaining redox homeostasis in living cells. In aerobic conditions, cells are constantly exposed to the generation of reactive oxygen species (ROS) that can impact proteins, lipids, and DNA, playing a pathological role in the development of various human diseases such as cancer [1]. Therefore, cells have evolved endogenous defence mechanisms so as to counteract oxidative stress and to maintain ROS at low physiological levels, and the redox sensitive transcription factor, nuclear factor erythroid 2 p45-related factor 2 (Nrf2), acts as a key regulator of antioxidant response, crucially involved in the preservation of the structure and the functioning of normal healthy cells [2–4]. However, cancer cells, differently from normal cells, show an increased rate of ROS generation as by-products of their metabolism [5] and, as “masters” of adaptation, they take advantage of the overactivation of antioxidant defences, in particular Nrf2-dependent genes [6–8]. This ability to adapt and survive under conditions of electrophilic, oxidative, and inflammatory stress is strongly dependent on the expression of a complex network comprising nearly 500 genes, induced by Nrf2, encoding proteins with different antioxidant and cytoprotective functions [9]. In particular, heme oxygenase 1 (HO-1) exerts a strong antioxidant and antiapoptotic effect favouring cancer cell growth and resistance to therapy. In this review, we focus our attention on the deleterious properties of Nrf2, and of its target gene HO-1, in relation to cancer cell growth and chemoresistance.
2. Nrf2: Structure and Regulation
The nuclear factor erythroid 2 p45-related factor 2 (Nrf2) is a transcription factor that plays a key role in the regulation of the cellular redox status. Indeed, Nrf2 controls not only the expression of antioxidants as well as phase I and phase II drug-metabolizing enzymes [10, 11], but also multidrug-resistance-associated protein transporters [12] (Table 1).
Table 1.
Genes regulated by Nrf2 in mice and humans.
| Symbol | Name | Species | References |
|---|---|---|---|
| Antioxidant genes | |||
| GCLC | Glutamate-cysteine ligase, catalytic subunit | m, h | [188, 189] |
| GCLM | Glutamate-cysteine ligase, modifier subunit | m, h | [188, 189] |
| GLRX | Glutaredoxin 1 | h | [190] |
| GPX2 | Glutathione peroxidase 2 | m, h | [190, 191] |
| GPX4 | Glutathione peroxidase 4 | m | [16] |
| GSR1 | Glutathione reductase | m, h | [188, 190] |
| SLC6A9 | Glycine transporter | m | [16] |
| SLC7A11 | Cysteine/glutamate transporter | m, h | [16, 188] |
| PRDX1-6 | Peroxiredoxins 1 and 6 | m, h | [10, 16] |
| SRXN1 | Sulfiredoxin-1 | m, h | [11, 188] |
| TXN1 | Thioredoxin | m, h | [10, 16] |
| TXNRD1 | Thioredoxin reductase 1 | m, h | [188, 190] |
|
| |||
| HO-1-related genes | |||
| HMOX1 | Heme oxygenase 1 | m, h | [10, 188] |
| BLVRA | Biliverdin reductase A | h | [190] |
| BLVRB | Biliverdin reductase B | m, h | [16, 190] |
| FECH | Ferrochelatase | h | [188] |
| FTH1 | Ferritin, heavy polypeptide 1 | m, h | [10] |
| FTHL12-17 | Ferritin, heavy polypeptides 12 and 17 | h | [188, 190] |
| FTL1 | Ferritin, light polypeptide | m, h | [188, 190] |
|
| |||
| Detoxifying enzymes | |||
| NQO1 | NAD(P)H:quinone oxidoreductase 1 | m, h | [188, 190] |
| GSTA1 | Glutathione S-transferase class Alpha 1 | m | [192] |
| GSTM1 | Glutathione S-transferase class Mu 1 | m | [192] |
| GSTP1 | Glutathione S-transferase class Pi 1 | m | [12] |
| UGT1A1 | UDP glucuronosyl- transferase 1 family |
h | [190] |
| UGT2B7 | UDP glucuronosyl- transferase 2 family |
m, h | [16, 190] |
|
| |||
| Drug transporters | |||
| ABCB6 | ATP-binding cassette, subfamily B(MDR/TAP) | m, h | [10, 11] |
| ABCC1 | ATP-binding cassette, subfamily C(CFTR/MRP) | m | [193] |
| ABCC2 | ATP-binding cassette, subfamily C(CFTR/MRP) | m, h | [193] |
| ABCC3 | ATP-binding cassette, subfamily C(CFTR/MRP) | m, h | [193] |
| ABCC4 | ATP-binding cassette, subfamily C(CFTR/MRP) | m | [193] |
| ABCC5 | ATP-binding cassette, subfamily C(CFTR/MRP) | m | [194] |
The human Nrf2 was first described, cloned, and characterised by Moi and coworkers in 1994 [13] and its cloned gene is encoded within a 2.2 kB transcript, predicting a protein of 589 amino acids with a molecular mass of 66.1 kDa [13].
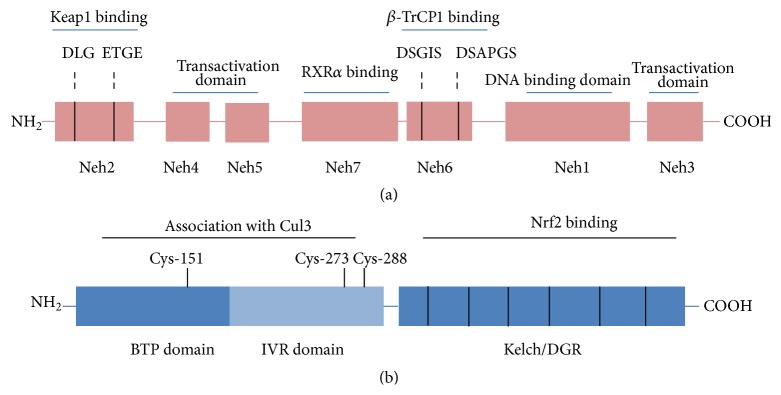
Nrf2 has seven functional domains named Nrf2-ECH homology (Neh) 1–7 (Figure 1(a)) [12, 14, 15]. The Neh1 domain has a basic region leucine zipper structure needed for the dimerisation with small Maf and binds to antioxidant/electrophile responsive elements (ARE/EpRE) [16]. Neh2 is the main negative regulatory domain which binds to the Kelch-like ECH-associated protein 1 (Keap1) via the DLG and ETGE motifs [17]. Neh3 is localised in the C-terminal region of Nrf2 and acts as a transactivation domain recruiting the chromo-ATPase/helicase DNA binding protein family (CHD6) [6], whereas both Neh4 and Neh5 are transactivation domains that recruit cAMP response element binding protein- (CREB-) binding protein (CBP) and/or the receptor-associated coactivator (RAC) [18]. The Neh6 domain mediates the Keap1-independent degradation of Nrf2 through recruitment by the DSGIS and DSAPGS motifs of the dimeric β-transducin repeat-containing protein (β-TrCP) [19]. Lastly, the Neh7 domain, recently identified by Wang and coworkers, interacts with the retinoid X receptor alpha (RXRα), a repressor of Nrf2 [20].
Figure 1.
Schematic representation of Nrf2 and Keap1 structures. (a) Nrf2 contains seven domains, Neh1–Neh7. The Neh2 domain contains two binding motifs, DLG and ETGE, responsible for the interaction with Keap1. The Neh4, Neh5, and Neh3 domains are important for the transactivation activity of Nrf2. The Neh7 domain is critical for RXRα binding. The Neh6 domain regulates Nrf2 degradation by β-TrCP1. The Neh1 domain has a basic region leucine zipper motif for DNA binding. (b) Keap1 contains three major domains. The BTB domain mediates Keap1 homodimerisation and the IVR domain contains critical cysteine residues and together they associate with Cul3. The Kelch/DGR domain mediates the binding with the Neh2 domain of Nrf2.
2.1. Keap1-Dependent Posttranscriptional Regulation
2.1.1. Ubiquitination and Proteasomal Degradation of Nrf2
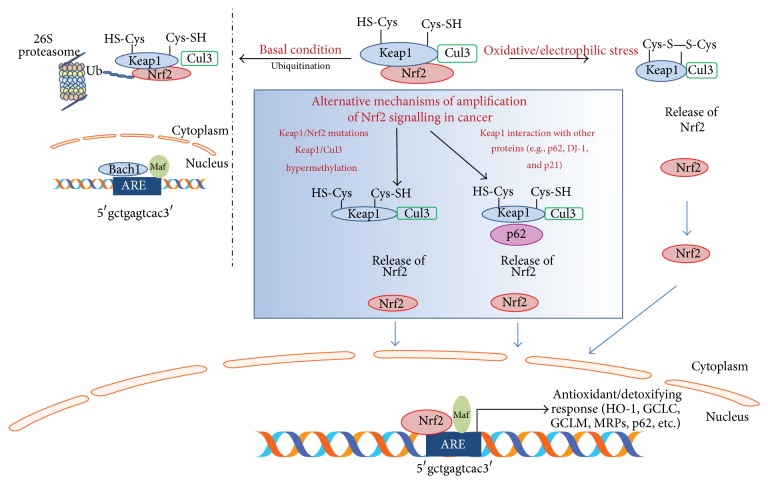
Under basal conditions, Nrf2 is localised in the cytosol associated with its negative regulator Keap1, an adaptor component of Cullin 3-based ubiquitin E3 ligase complex (Cul3) that promotes Nrf2 constant ubiquitination and proteasomal degradation, maintaining low basal levels [21, 22]. Nrf2 turnover is rapid, less than 20 minutes, and prevents the expression of Nrf2 target genes under normal conditions [23]. On exposure to oxidative or electrophilic stress, Keap1 is modified whereas the enzymatic activity of the E3 ubiquitin ligase is inhibited; Nrf2 is liberated from Keap1, accumulates in the nucleus, dimerises with small Maf protein, and then activates, after ARE-sequence binding, the transcription of its target genes [23, 24]. Thus, Keap1 is the main repressor of Nrf2, having three functional domains, namely, the broad complex, Tramtrack, and Bric-a-Brac (BTB) domain, the intervening region (IVR) domain, and the double glycine repeat (DGR)/Kelch domain [25] (Figure 1(b)).
Keap1 acts as a sensor for oxidative and electrophilic stress through the modification of 27 cysteine residues [26] (Figure 2). The main three cysteine residues for the regulation of Nrf2 activity are Cys151 in the BTB domain and Cys273 and Cys288 in the IVR domain, all of which are targets of oxidative and electrophilic modifications [27–29]. It has been shown that cells expressing Keap1 Cys151 point mutant protein show reduced activation of Nrf2 in response to a number of inducers (e.g., sulforaphane, tert-butylhydroquinone, and diethyl maleate) in comparison to the wild-type cells [30] and that Cys273 and Cys288 are critically required for the basal repression of Nrf2 [31]. In addition, the modification of a subset of cysteine residues in Keap1, by Nrf2 inducers, supports the hypothesis of a “cysteine code” which is critical in the activation of Nrf2 [32].
Figure 2.
Nrf2 activity regulation. In a resting state, Nrf2 is sequestered in the cytoplasm through the binding with Keap1, responsible for Nrf2 ubiquitination and proteasomal degradation via Cul3. Oxidative/electrophilic stress causes a conformational change in Keap1-Cul3, by acting on specific cysteine residues in Keap1, leading to Nrf2 dissociation. Thus, free Nrf2 translocates to the nucleus, which dimerises with small Maf protein and binds to ARE/EpRE sequence within regulatory regions of a wide variety of target genes (e.g., HO-1, GCLC, GCLM, MRPs, and p62). In cancer cells (blue box), Keap1/Nrf2 mutations and Keap1/Cul3 aberrant hypermethylations as well as Keap1 interactions with ETGE motif-containing proteins lead to an increased Nrf2 activation and induction of target genes.
2.1.2. Autophagic Degradation of Keap1
Furthermore, other interacting protein partners, such as the sequestosome-1 protein (p62/SQSTM1), can modulate Nrf2 activity [33, 34]. p62/SQSTM1 is a scaffold protein that binds to polyubiquitinated proteins and targets protein aggregates for autophagic degradation. p62 contains an STGE-binding motif similar to the Nrf2 ETGE motif [33] which is needed for the direct interaction with Keap1 [35]. As a consequence, Keap1 is sequestered in autophagosomes and Nrf2 ubiquitination decreases, leading to a prolonged activation of Nrf2 in response to oxidative stress [34, 35] (Figure 2).
2.2. Keap1-Independent Posttranscriptional Regulation
Recently, alternative mechanisms for the degradation of Nrf2 have been identified. For example, a number of studies have demonstrated that glycogen synthase kinase 3β (GSK-3β) directs the ubiquitination and proteasomal degradation of various transcription factors, including Nrf2, through the activation of E3 ubiquitin ligase complex (β-TrCP-Skp1-Cul1-Rbx1) [19, 36]. Indeed, GSK-3β is able to phosphorylate Ser residues located in the Neh6 domain of Nrf2 which are then recognised by β-TrCP and, through the binding to Cullin 1 (Cul1) scaffold protein, lead to Nrf2 ubiquitination and degradation in a redox-independent manner [19, 37].
Moreover, a novel E3 ubiquitin ligase, namely, Hrd1, has been described by Wu and coworkers [36]. They showed that Hrd1 controls the Nrf2 stability by way of an interaction between the C-terminal domain of Hrd1 and the Neh4-5 domains of Nrf2. This Hrd1-mediated ubiquitination of Nrf2 is independent of both Keap1 and β-TrCP and compromises the Nrf2-mediated protection during liver cirrhosis [36].
2.3. Nrf2 Regulation at the Transcriptional Level
Although strong evidence shows that Nrf2 is primarily regulated at the protein level, it has also been demonstrated that the oncogenic KRAS transcriptionally upregulates the mRNA levels of Nrf2 through a TPA response element (TRE) located within the Nrf2 promoter. The oncogenic mutation of KRAS, or KRAS overexpression, indeed, is able to activate the Nrf2-dependent pathway [38]. In addition, an increase in Nrf2 mRNA levels has been shown to occur in response to the oncogenic activation of BRAF and C-MYC [39].
3. Nrf2 and Chemoprevention
It is well known that Nrf2 plays a key role in the cellular adaptation and protection against oxidative stress. The ability of Nrf2 to activate cytoprotective genes, which code for detoxifying enzymes, drug transporters, antioxidants, and anti-inflammatory proteins, plays a crucial role in reducing electrophiles and ROS, thus decreasing DNA damage and mutations and preventing genomic instability in normal cells. Several studies have shown that, in Nrf2 null mice, there is an enhanced susceptibility to chemical carcinogens, such as benzo(a)pyrene, compared to wild-type mice, due to a decreased expression of phase II detoxification and antioxidant enzymes [40]. In a similar way, after exposure to N-nitrosobutyl (4-hydroxybutyl) amine (BBN), Nrf2 knockout mice develop urinary bladder carcinoma [41] and show an increased incidence of skin, colorectal, and mammary tumours [42–44]. Moreover, the protective role of Nrf2 against carcinogenesis is highlighted from studies in humans on single-nucleotide polymorphism (SNP) in the promoter region of the Nrf2 gene [45]. As described by Suzuki and coworkers, A/A homozygotes of Nrf2 rSNP-617 showed decreased expression of Nrf2 and consequently an increased risk of developing lung cancers, especially in the case of individuals with smoking habits [46].
4. The Dark Side of Nrf2 in Cancer Biology
Several studies have shown a deleterious aspect of Nrf2 defined as the “dark side of Nrf2” [47]. Its high and prolonged activation in cancer cells has been long associated with progression, metastatic invasion, angiogenesis, and chemo- and radio-resistance in tumours and is considered a poor prognostic factor [15, 48]. Indeed, the stable overexpression of Nrf2 was found in various types of tumours such as lung [49, 50], breast [51], head and neck [52], ovarian [53], and endometrial cancer [54].
4.1. Molecular Mechanisms of Nrf2 Activation in Cancer Cells
Several mechanisms have been shown to be involved in the constitutive activation of Nrf2 in cancer cells (Figure 2), mainly gain-of-function mutations in Nrf2 and loss-of-function mutations in Keap1, leading to an impairment of the binding to Keap1. This results in the stabilisation of Nrf2 and in the activation of its target genes, as identified in patients with lung or head and neck cancer [55, 56], whereas the loss-of-function mutations of Keap1 are mainly located in the Kelch/DRG domain and in the IVR domain [14] as observed in gastric, hepatocellular, colorectal, lung, breast, and prostate carcinomas, as reviewed by Shelton and Jaiswal [57].
In addition, epigenetic modifications in Keap1 are responsible for the accumulation and activation of Nrf2. Aberrant hypermethylation, inhibiting Keap1 gene expression, results in the accumulation of Nrf2, as shown in lung and malignant glioma [58], prostate [59], and colorectal cancer [60]. Moreover, somatic mutations and hypermethylation in CUL3 have been identified as being responsible for Nrf2 activation [53, 61, 62], as shown in thyroid, head and neck, and ovarian cancers [63–65].
4.2. Nongenomic Alteration of Nrf2 in Cancer Cells
Increased levels of Nrf2 in cancer cells can also occur in the absence of genomic alterations. In fact, much evidence shows that different proteins can alter the Nrf2-Keap1 binding [14]. Nrf2 activity is subject to a positive regulation by p21 [66, 67], which interferes with Keap1-mediated ubiquitination, interacting with the DLG motif in Nrf2, leading to its stabilisation. Therefore, Nrf2 expression is significantly lower in the absence of p21, and conversely it is increased upon p21 overexpression [67].
It is a fact that DJ-1, a protein belonging to the Thi/PfpI superfamily, is able to stabilise Nrf2 by preventing its association with Keap1, thus reducing ubiquitination and subsequent proteasomal degradation [68, 69]. DJ-1 in cancer is often overexpressed and leads to increased detoxification enzymes, such as NQO1, providing a survival advantage [68, 70, 71].
As previously described, p62 sequesters Keap1 in autophagosomes, leading to Nrf2 activation [72, 73]. Moreover, it has been shown that p62 contains the ARE sequence in its promoter region which is responsible for its Nrf2-dependent induction in response to oxidative stress, thus generating a positive feedback loop [73].
The list of proteins which can interact with Nrf2 and Keap1 and, therefore, modulate their regulation is continuously expanding (e.g., WTX, PALB2, and DPP3) [74–76]. All of these proteins contain an ETGF motif, suggesting that they are capable of upregulating Nrf2 by competing for Keap1 binding and suppressing Keap1-mediated ubiquitination of Nrf2 [14].
4.3. Nrf2 Activation and Hallmarks of Cancer Malignancy
Different Nrf2 target genes are associated with cancer cell proliferation and death. Among these, those genes involved in the pentose phosphate pathway such as glucose-6-phosphate dehydrogenase, phosphogluconate dehydrogenase, transketolase, and transaldolase 1 are responsible for NADPH and purine regeneration and, therefore, accelerate cancer cell proliferation [14]. Nrf2 may also bind to the ARE sequence in the promoter region of Notch1, contributing directly to its expression and leading to a more malignant phenotype and a more aggressive growth [77]. Moreover, Nrf2 is directly involved in the basal expression of the p53 inhibitor Mdm2, through the binding to the ARE sequence located in the first intron of this gene. It has been demonstrated that, in Nrf2-deleted murine embryonic fibroblasts (MEFs), Mdm2 expression is repressed and, compared to wild-type cells, a high level of p53 is accumulated, favouring cell death [78].
In addition, elevated levels of Nrf2 have been observed in various tumours with high metastatic potential [79], characterised by epithelial-mesenchymal transition (EMT) and the degradation of extracellular matrix (ECM), exerted by metalloproteases (MMPs). A major step of EMT is the loss of E-cadherin and the gain of N-cadherin [80]. It has been shown that E-cadherin is able to bind to the C terminus of Nrf2, preventing its nuclear accumulation, and that, during EMT, the overexpression of N-cadherin reduces the Nrf2 inhibition, thus favouring its activity [81]. Moreover, the knockdown of the Nrf2 by short hairpin RNA (shRNA) in esophageal squamous cell carcinoma (ESCC) suppressed the expression of MMP-2 and enhanced E-cadherin mRNA levels, resulting in a decreased invasion and migration of cancer cells [82].
Furthermore, the upregulation of Nrf2 is also related to angiogenesis which is promoted by HIF-1α, a transcription factor that senses oxygen homeostasis and is deregulated in tumours in hypoxic environments [83]. Under hypoxic conditions, indeed, the O2-dependent regulator prolyl-hydrolase domain (PHD) is catalytically inactive and increases the stability of HIF-1α. Consequently, the expression of its target proteins, including the vascular endothelial growth factor (VEGF), is enhanced [84]. It has been shown that Nrf2 silencing blocks HIF-1α-dependent VEGF expression in HT29 colon cancer and suppresses tumour growth with a concomitant reduction in VEGF-induced angiogenesis in mouse xenograft models [84].
4.4. Nrf2 and Cancer Resistance to Therapies
Several studies have shown that cancer cells with high levels of Nrf2 are less sensitive to etoposide, cisplatin, and doxorubicin [14]. Doxorubicin-resistant human cancer cells, such as ovarian SKOV3 and OV90 and mammary MCF-7/DOX, have shown high levels of Nrf2-ARE binding and ARE-driven luciferase activity, as well as the upregulation of Nrf2 target genes compared to the respective sensitive cell lines A2780 and MCF-7 [85, 86]. Non-small cell lung carcinoma A549 cells, in which Nrf2 is strongly activated, have shown a higher resistance to cisplatin compared to NCI-H292 and LC-AI cells [87]. Furthermore, Jayakumar and coworkers have demonstrated the role of Nrf2 and its dependent genes in the radioresistance of prostate tumour cells. Specifically, the radioresistant DU145 cells show enhanced levels of Nrf2 and a high GSH/GSSG ratio in comparison to the radiosensitive PC3 cells which show a faster depletion of GSH after radiation exposure [88]. It has also been shown that radiotherapy significantly reduces cancer cell survival when applied in combination with the Nrf2 inhibitor 4-(2-cyclohexylethoxy)-aniline, IM3829 [89]. Moreover, our own studies demonstrate that Nrf2 activation plays a key role in the resistance of neuroblastoma cells to GSH depletion or proteasome inhibition [90, 91].
In addition, Nrf2 activity is related to the upregulation of several multidrug-resistant efflux pumps, such as the ATP-binding cassette, subfamily G, member A2 (ABCG2), which favours drug resistance. It has been shown, indeed, that Nrf2 silencing attenuates the expression of the ABCG2 transcript and protein and sensitises lung cancer cells to mitoxantrone and topotecan, two representative chemotherapeutic drugs effluxed mainly by the presence of ABCG2 [92]. Multidrug-resistant protein-3 (MRP3), MRP4, and MRP5 are all upregulated by Nrf2 [93, 94]. Upregulation of MRP3 and GSTs leads to the increased hydrophilicity and excretion of cytotoxic agents, such as cisplatin, etoposide, and doxorubicin [95].
Recently, it has been proven that Nrf2 also plays a critical role in the drug resistance of cancer stem cells [96–98]. The resistance of glioblastoma is due to the presence of glioblastoma stem cells (GSCs) which confer tumourigenic potential and a survival advantage against chemotherapy [99]. Moreover, it has been shown that while knocking down Nrf2 decreases the self-renewing activity of GSCs [100, 101], the enhancement of Nrf2 levels and of its downstream genes, that is, HO-1, GCLM, and NQO1, is related to increased tumourigenic activity of the human mammospheres in comparison to their adherent counterparts, MCF-7 and MDA-MB-231 cells [102].
5. HO-1 as a Key Effector of Nrf2 Upregulation in Tumour Progression
Heme oxygenase 1 (HO-1) is considered one of the main effectors of Nrf2-dependent cell responses [48]. HO-1 is the inducible form of heme oxygenase, the first rate-limiting enzyme in the degradation of heme into biliverdin/bilirubin, carbon monoxide (CO), and ferritin induced by free iron release [103–105] (Figure 3). It is a 32 kDa stress protein present at low levels in most mammalian tissues [106] and its expression is induced by a wide variety of stress stimuli, including its substrate, as well as heavy metals [107], UV irradiation, ROS [108], nitric oxide [109], and inflammatory cytokines [110]. HO-1 and its metabolic products are involved in the maintenance of cellular homeostasis and they play a key role in the adaptive response to cellular stress as well as in the protection of healthy cells, preventing them from being transformed into neoplastic cells by counteracting ROS-mediated carcinogenesis [111–115].
Figure 3.
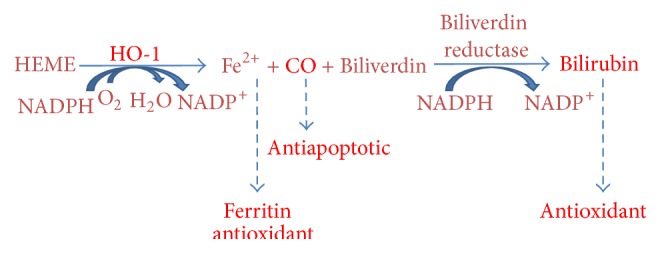
Heme catabolic pathway. HO-1 catalyses the degradation of heme into biliverdin/bilirubin (antioxidant), carbon monoxide (antiapoptotic), and ferritin (antioxidant) induced by free iron release.
However, HO-1 has been widely recognised as playing an important role in the malignant transformation of cancer cells. High levels of HO-1 have been found in various human tumours, inducing survival advantage, aggressiveness, and poor outcome [116–122]. HO-1 overexpression has been considered to be involved in invasive and metastatic mechanisms [123–125], and “in vitro” and “in vivo” studies, including clinical data, have shown that the inhibition or silencing of HO-1 inhibits this behaviour [125–128]. Moreover, a proangiogenic role of HO-1 in cancer has been reported “in vitro” and “in vivo” [123, 124, 127–130]. Finally, HO-1 has been shown to be correlated with resistance to chemo-, radio-, and photodynamic therapies [116, 119, 131–136] and its inhibition is able to sensitize cancer cells to death [90, 91, 137–139].
However, the role of HO-1 in cancer biology is not completely understood and some disputes in literature remain about its role in tumour progression, especially with regard to different types of tumours. It should be taken into consideration that several studies have reported that HO-1 activation prevents breast cancer proliferation [140] and prostate cancer angiogenesis [141] and mediates the anticancer activity of some drugs such as andrographolide by reducing the MMP-9 expression in breast cancer cells [142]. Moreover, considering the complex cross talk between HO-1 activity and cellular metabolic pathways, reviewed in depth by Wegiel and coauthors [143], it would seem conceivable that HO-1 can be subject to different modulations in different tumours, since the various metabolic statuses of cancer cells may influence how HO-1 activity modulates tumour growth. Therefore, it is important to note that HO-1 expression is controlled by other transcription factors other than Nrf2. Indeed, specific consensus sequences for both NF-kB and AP-1 are present in the promoter region of HO-1 [144–148] which, then, may be activated in response to different stimuli through the activation of different intracellular signaling pathways, as widely reviewed in [136, 149–151], suggesting a highly complex regulation which, up to now, is far from being fully understood.
However, what seems most interesting is the association between the upregulation of Nrf2 and the activation of HO-1 in tumour progression which correlates with cancer aggressiveness and malignancy. Interestingly, for instance, in human samples from gallbladder cancers, the upregulation of both Nrf2 and HO-1 correlates with tumour aggressiveness and a poor clinical outcome [79]. Moreover, Nrf2/HO-1 association has been widely reported, for instance, in non-small lung cancer, cervical cancer, hepatoma [152], esophageal squamous carcinoma [82], and multiple myeloma [153] (Table 2). In particular, malignant transformations have been associated with Nrf2-dependent HO-1 activation in B lymphocytes exposed to prostaglandin J2 [154]. Therefore, the gain of metastatic phenotypes is correlated with the overexpression of Nrf2 in association with the activation of HO-1, as shown in osteopontin-induced glioma cell invasiveness [155]. Furthermore, resistance to therapies has been related to the activation of Nrf2 together with HO-1, as shown by our group in neuroblastoma cells after GSH depletion or bortezomib exposure [90, 91] and by others in cisplatin-treated ovarian carcinoma cells [156] and in doxorubicin-resistant breast cancer cells [86].
Table 2.
Nrf2 and HO-1 upregulation in tumours.
| Type of tumor | Nrf2 | HO-1 | Reference |
|---|---|---|---|
| Glioblastoma stem cells (GSCs) | ↑ | — | [101] |
| Lung cancer (NSCLC) | ↑ | — | [92, 94] |
| Ovarian carcinoma | ↑ | — | [85] |
|
| |||
| Bladder cancer | — | ↑ | [121] |
| Chronic myeloid leukemia | — | ↑ | [139] |
| Colon adenocarcinoma | — | ↑ | [135] |
| Colorectal cancer | — | ↑ | [118] |
| Fibrosarcoma | — | ↑ | [131] |
| Gastric cancer | — | ↑ | [138] |
| Hepatocellular carcinoma (HCC) | — | ↑ | [127] |
| Kaposi sarcoma | — | ↑ | [128] |
| Lung cancer (NSCLC) | — | ↑ | [120, 122, 134, 137] |
| Melanoma | — | ↑ | [123, 131] |
| Oral squamous cell carcinoma | — | ↑ | [125] |
| Pancreatic cancer | — | ↑ | [119, 124, 126, 133] |
|
| |||
| Breast cancer | ↑ | ↑ | [86] |
| Cervical cancer | ↑ | ↑ | [152] |
| Chronic myeloid leukemia | ↑ | ↑ | [136] |
| Esophageal squamous carcinoma | ↑ | ↑ | [82] |
| Gallbladder cancer | ↑ | ↑ | [79] |
| Glioblastoma | ↑ | ↑ | [100, 155] |
| Glioblastoma stem cells (GSCs) | ↑ | ↑ | [100] |
| Hepatoma | ↑ | ↑ | [152] |
| Lung cancer (NSCLC) | ↑ | ↑ | [87, 89, 152] |
| Malignant B lymphocytes | ↑ | ↑ | [154] |
| Mammosphere stem cells (MSC) | ↑ | ↑ | [102] |
| Multiple myeloma | ↑ | ↑ | [153] |
| Neuroblastoma | ↑ | ↑ | [90, 91] |
| Ovarian carcinoma cells | ↑ | ↑ | [156] |
| Prostate cancer | ↑ | ↑ | [88] |
| Renal cancer | ↑ | ↑ | [117] |
Therefore, HO-1 activation provides tumour cells with strong survival advantage exerted by the antioxidant and antiapoptotic properties of its metabolic products. Moreover, when HO-1 activation is dependent on Nrf2 activity, generally this leads to highly aggressive cancer phenotypes. This can also account for the parallel activation of other Nrf2-dependent genes which can contribute to the prosurviving effect, for instance, by inducing HIFα [82], increasing multidrug-resistance-related proteins [79], or increasing the synthesis of GSH [8]. Unpublished data from our laboratory show that HO-1-dependent bilirubin generation and increasing amounts of GSH are key factors in inducing resistance to bortezomib in high-risk neuroblastoma.
6. Pharmacological Modulation of Nrf2/HO-1 Axis as a Strategy in Anticancer Treatment
The role of Nrf2/HO-1 axis in protecting cells by overcoming environmental stresses has already been demonstrated and compounds that are able to modulate this activity are well worth being considered. The activation of this pathway in normal cells can prevent tumour formation while its inhibition can be useful in improving cancer therapies.
As far as cancer prevention is concerned, different activators of Nrf2 have been proposed and have been intensively reviewed over the past 10 years [42, 157–166]. Many of these compounds are plant-derived phytochemicals, such as sulforaphane, curcumin, epigallocatechin-3-gallate, resveratrol, cinnamoyl-based compounds, garlic organosulfur compounds, and lycopene. These molecules have been considered to be chemopreventive due to their ability to induce antioxidant/detoxification enzymes, including HO-1, and xenobiotic transporters, through the activation of Nrf2 [18, 41, 50, 167–170]. On the other hand, the pharmacological inhibition of Nrf2/HO-1 axis has also recently emerged as a promising approach for cancer therapy.
Starting from the modulation of HO-1, the major effector of the pathway considered, several “in vivo” studies have confirmed the usefulness of the HO-1 competitive inhibitor zinc (II) protoporphyrin IX, ZnPPIX, in the reduction of hepatoma, sarcoma, lung cancer, and B-cell lymphoma growth in mice [129, 171]. Moreover, the PEG conjugation of ZnPPIX, which increases water solubility of the inhibitor, is able to improve its clinical application [172]. Another highly water-soluble micellar form of ZnPPIX, the amphiphilic styrene maleic acid copolymer (SMA-ZnPPIX) with a potent antitumour activity both “in vitro” and “in vivo,” has been additionally proposed [173].
However, it is important to note that pharmacological HO-1 inhibitors, as well as HO-1 activators, are responsible for strong HO-1 independent activities due to some nonspecific properties of these compounds [174, 175] and therefore the employment of siRNA could be more specific. Indeed, this approach is able to induce apoptosis of colon carcinoma cells [176, 177] and to diminish proliferation, growth, and angiogenesis in orthotopic hepatocellular tumours in mice [127].
As already discussed in the previous paragraph, there is also some evidence that HO-1 activation, induced by cobalt protoporphyrin IX (Co PPIX) or heme [140], as well as the overexpression of HO-1 can block tumour growth and invasion in “in vitro” studies, and this seems to be dependent on the cancer cell type and experimental model used.
Furthermore, the inhibition of Nrf2, acting upstream from HO-1 activation and involving also other downstream targets in addition to HO-1, could be a successful therapeutic approach.
Unfortunately, only a few inhibitors of Nrf2 have been developed so far. Among them, brusatol, extracts from Brucea javanica growing in Southeast Asia and Northern Australia, is able to decrease Nrf2 protein levels as well as decreasing the expression of its target genes, thus enhancing the cytotoxic effect of several chemotherapeutic agents, both “in vitro” and “in vivo” [178, 179].
In addition, flavonoid luteolin (3′,4′,5,7-tetrahydroxyflavone) found at high concentrations in celery, green pepper, parsley, perilla leaf, and chamomile tea has been shown to be another strong and selective Nrf2 inhibitor, which is able to reduce the constitutive expression of NQO1 in HepG2, Hepa1c1c7, and RL-34 cells in a time- and dose-dependent manner [180]. At physiological concentrations, luteolin inhibits Nrf2 activity by enhancing Nrf2 mRNA turnover, and it has been shown to sensitise NSCLC A549 cells to therapeutic drugs [181]. Similar results have been observed in the sensitisation of colorectal cancer cell lines to oxaliplatin “in vitro” [182] and “in vivo” in the chemotherapy of non-small cell lung cancer, NSCLC. Moreover, its oral administration, either alone or combined with an intraperitoneal injection of cisplatin, is seen to greatly inhibit the growth of xenograft tumours from the NSCLC cell line A549 in athymic nude mice [183].
Furthermore, all-trans-retinoic acid (ATRA) is able to suppress the Nrf2 pathway [184]. ATRA like other agonists of RA receptor α (RARα) and retinoid X receptor α (RXRα) was shown to inhibit the basal and the inducible activity of Nrf2 both “in vitro” and “in vivo” [12, 185]. After ATRA treatment, Nrf2 forms a complex with RARα. This complex is unable to bind to the ARE sequences, thus decreasing the ability of Nrf2 to activate its target genes [15]. In acute promyelocytic leukaemia cells, the cytotoxic drug arsenic trioxide (ATO) induces an antioxidant response characterised by Nrf2 nuclear translocation and enhances the transcription of its downstream target genes such as HO-1, NQO1, GCLM, and ferritin. It has been shown that, after cotreatment of ATO plus ATRA, the Nrf2 nuclear translocation is prevented and the cytotoxic effects of ATO treatment are enhanced [186].
Lastly, a recent paper shows that metformin, which was previously associated with a better survival of diabetic patients with pancreatic cancer [187], exerts its antitumour activity by suppressing HO-1 expression in cancer cells. In this paper, metformin is reported to inhibit Nrf2 through a Keap1-independent mechanism by inactivating Raf and ERK signaling [152]. The excellent therapeutic index of metformin, with few side-effects associated even with long-term treatment, could increase the chances of its application in cancer therapy.
7. Conclusions
The activation of Nrf2/HO-1 axis plays a central role in cellular adaptive responses to oxidative stress and cytotoxic insults representing a crucial point in the prevention of carcinogenesis. On the contrary, in tumour tissues, a prolonged activation of Nrf2 and HO-1 contributes to the gain of malignant phenotypes. Consequently, the Nrf2/HO-1 axis can be used by cancer cells to promote their growth advantage, metastatic potential, and resistance to therapy. Therefore, the therapeutic usefulness of inhibitors of Nrf2, and of its target gene, HO-1, especially in combination with conventional antineoplastic therapies, may well represent a potential and promising approach in the fight against cancer.
Acknowledgments
The authors are thankful for grants from MIUR-PRIN (no. 20125S38FA) (M. Nitti) and Genoa University.
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Gupta S. C., Hevia D., Patchva S., Park B., Koh W., Aggarwal B. B. Upsides and downsides of reactive oxygen species for Cancer: the roles of reactive oxygen species in tumorigenesis, prevention, and therapy. Antioxidants and Redox Signaling. 2012;16(11):1295–1322. doi: 10.1089/ars.2011.4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ishii T., Itoh K., Takahashi S., et al. Transcription factor Nrf2 coordinately regulates a group of oxidative stress-inducible genes in macrophages. Journal of Biological Chemistry. 2000;275(21):16023–16029. doi: 10.1074/jbc.275.21.16023. [DOI] [PubMed] [Google Scholar]
- 3.Ishii T., Itoh K., Ruiz E., et al. Role of Nrf2 in the regulation of CD36 and stress protein expression in murine macrophages: activation by oxidatively modified LDL and 4-hydroxynonenal. Circulation Research. 2004;94(5):609–616. doi: 10.1161/01.res.0000119171.44657.45. [DOI] [PubMed] [Google Scholar]
- 4.Itoh K., Mimura J., Yamamoto M. Discovery of the negative regulator of Nrf2, keap1: a historical overview. Antioxidants and Redox Signaling. 2010;13(11):1665–1678. doi: 10.1089/ars.2010.3222. [DOI] [PubMed] [Google Scholar]
- 5.Weinberg F., Chandel N. S. Reactive oxygen species-dependent signaling regulates cancer. Cellular and Molecular Life Sciences. 2009;66(23):3663–3673. doi: 10.1007/s00018-009-0099-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hayes J. D., McMahon M. NRF2 and KEAP1 mutations: permanent activation of an adaptive response in cancer. Trends in Biochemical Sciences. 2009;34(4):176–188. doi: 10.1016/j.tibs.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 7.Taguchi K., Motohashi H., Yamamoto M. Molecular mechanisms of the Keap1-Nrf2 pathway in stress response and cancer evolution. Genes to Cells. 2011;16(2):123–140. doi: 10.1111/j.1365-2443.2010.01473.x. [DOI] [PubMed] [Google Scholar]
- 8.Traverso N., Ricciarelli R., Nitti M., et al. Role of glutathione in cancer progression and chemoresistance. Oxidative Medicine and Cellular Longevity. 2013;2013:10. doi: 10.1155/2013/972913.972913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kumar H., Kim I.-S., More S. V., Kim B.-W., Choi D.-K. Natural product-derived pharmacological modulators of Nrf2/ARE pathway for chronic diseases. Natural Product Reports. 2014;31(1):109–139. doi: 10.1039/c3np70065h. [DOI] [PubMed] [Google Scholar]
- 10.Malhotra D., Portales-Casamar E., Singh A., et al. Global mapping of binding sites for Nrf2 identifies novel targets in cell survival response through ChIP-Seq profiling and network analysis. Nucleic Acids Research. 2010;38(17):5718–5734. doi: 10.1093/nar/gkq212.gkq212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chorley B. N., Campbell M. R., Wang X., et al. Identification of novel NRF2-regulated genes by ChiP-Seq: influence on retinoid X receptor alpha. Nucleic Acids Research. 2012;40(15):7416–7429. doi: 10.1093/nar/gks409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hayes J. D., Dinkova-Kostova A. T. The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends in Biochemical Sciences. 2014;39(4):199–218. doi: 10.1016/j.tibs.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 13.Moi P., Chan K., Asunis I., Cao A., Kan Y. W. Isolation of NF-E2-related factor 2 (Nrf2), a NF-E2-like basic leucine zipper transcriptional activator that binds to the tandem NF-E2/AP1 repeat of the beta-globin locus control region. Proceedings of the National Academy of Sciences of the United States of America. 1994;91(21):9926–9930. doi: 10.1073/pnas.91.21.9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jaramillo M. C., Zhang D. D. The emerging role of the Nrf2-Keap1 signaling pathway in cancer. Genes and Development. 2013;27(20):2179–2191. doi: 10.1101/gad.225680.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moon E. J., Giaccia A. Dual roles of NRF2 in tumor prevention and progression: possible implications in cancer treatment. Free Radical Biology and Medicine. 2015;79:292–299. doi: 10.1016/j.freeradbiomed.2014.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hirotsu Y., Katsuoka F., Funayama R., et al. Nrf2-MafG heterodimers contribute globally to antioxidant and metabolic networks. Nucleic Acids Research. 2012;40(20):10228–10239. doi: 10.1093/nar/gks827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tong K. I., Katoh Y., Kusunoki H., Itoh K., Tanaka T., Yamamoto M. Keap1 recruits Neh2 through binding to ETGE and DLG motifs: characterization of the two-site molecular recognition model. Molecular and Cellular Biology. 2006;26(8):2887–2900. doi: 10.1128/mcb.26.8.2887-2900.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim J.-H., Yu S., Chen J. D., Kong A. N. The nuclear cofactor RAC3/AIB1/SRC-3 enhances Nrf2 signaling by interacting with transactivation domains. Oncogene. 2013;32(4):514–527. doi: 10.1038/onc.2012.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rada P., Rojo A. I., Chowdhry S., McMahon M., Hayes J. D., Cuadrado A. SCF/β-TrCP promotes glycogen synthase kinase 3-dependent degradation of the Nrf2 transcription factor in a Keap1-independent manner. Molecular and Cellular Biology. 2011;31(6):1121–1133. doi: 10.1128/mcb.01204-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang H., Liu K., Geng M., et al. RXRα inhibits the NRF2-ARE signaling pathway through a direct interaction with the Neh7 domain of NRF2. Cancer Research. 2013;73(10):3097–3108. doi: 10.1158/0008-5472.can-12-3386. [DOI] [PubMed] [Google Scholar]
- 21.Cullinan S. B., Gordan J. D., Jin J., Harper J. W., Diehl J. A. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: oxidative stress sensing by a Cul3-Keap1 ligase. Molecular and Cellular Biology. 2004;24(19):8477–8486. doi: 10.1128/mcb.24.19.8477-8486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kobayashi A., Kang M.-I., Okawa H., et al. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Molecular and Cellular Biology. 2004;24(16):7130–7139. doi: 10.1128/mcb.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kobayashi A., Kang M.-I., Watai Y., et al. Oxidative and electrophilic stresses activate Nrf2 through inhibition of ubiquitination activity of Keap1. Molecular and Cellular Biology. 2006;26(1):221–229. doi: 10.1128/MCB.26.1.221-229.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Osburn W. O., Kensler T. W. Nrf2 signaling: an adaptive response pathway for protection against environmental toxic insults. Mutation Research—Reviews in Mutation Research. 2008;659(1-2):31–39. doi: 10.1016/j.mrrev.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Itoh K., Wakabayashi N., Katoh Y., et al. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes & Development. 1999;13(1):76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holland R., Fishbein J. C. Chemistry of the cysteine sensors in kelch-like ECH-associated protein 1. Antioxidants and Redox Signaling. 2010;13(11):1749–1761. doi: 10.1089/ars.2010.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamamoto T., Suzuki T., Kobayashi A., et al. Physiological significance of reactive cysteine residues of Keap1 in determining Nrf2 activity. Molecular and Cellular Biology. 2008;28(8):2758–2770. doi: 10.1128/MCB.01704-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dinkova-Kostova A. T., Holtzclaw W. D., Cole R. N., et al. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(18):11908–11913. doi: 10.1073/pnas.172398899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wakabayashi N., Dinkova-Kostova A. T., Holtzclaw W. D., et al. Protection against electrophile and oxidant stress by induction of the phase 2 response: fate of cysteines of the Keap1 sensor modified by inducers. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(7):2040–2045. doi: 10.1073/pnas.0307301101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takaya K., Suzuki T., Motohashi H., et al. Validation of the multiple sensor mechanism of the Keap1-Nrf2 system. Free Radical Biology and Medicine. 2012;53(4):817–827. doi: 10.1016/j.freeradbiomed.2012.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bryan H. K., Olayanju A., Goldring C. E., Park B. K. The Nrf2 cell defence pathway: Keap1-dependent and -independent mechanisms of regulation. Biochemical Pharmacology. 2013;85(6):705–717. doi: 10.1016/j.bcp.2012.11.016. [DOI] [PubMed] [Google Scholar]
- 32.Kobayashi M., Li L., Iwamoto N., et al. The antioxidant defense system Keap1-Nrf2 comprises a multiple sensing mechanism for responding to a wide range of chemical compounds. Molecular and Cellular Biology. 2009;29(2):493–502. doi: 10.1128/mcb.01080-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Komatsu M., Kurokawa H., Waguri S., et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nature Cell Biology. 2010;12(3):213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- 34.Ichimura Y., Waguri S., Sou Y.-S., et al. Phosphorylation of p62 activates the Keap1-Nrf2 pathway during selective autophagy. Molecular Cell. 2013;51(5):618–631. doi: 10.1016/j.molcel.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 35.Taguchi K., Fujikawa N., Komatsu M., et al. Keap1 degradation by autophagy for the maintenance of redox homeostasis. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(34):13561–13566. doi: 10.1073/pnas.1121572109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu T., Zhao F., Gao B., et al. Hrd1 suppresses Nrf2-mediated cellular protection during liver cirrhosis. Genes and Development. 2014;28(7):708–722. doi: 10.1101/gad.238246.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chowdhry S., Zhang Y., McMahon M., Sutherland C., Cuadrado A., Hayes J. D. Nrf2 is controlled by two distinct beta-TrCP recognition motifs in its Neh6 domain, one of which can be modulated by GSK-3 activity. Oncogene. 2013;32(32):3765–3781. doi: 10.1038/onc.2012.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tao S., Wang S., Moghaddam S. J., et al. Oncogenic KRAS confers chemoresistance by upregulating NRF2. Cancer Research. 2014;74(24):7430–7441. doi: 10.1158/0008-5472.CAN-14-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Denicola G. M., Karreth F. A., Humpton T. J., et al. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature. 2011;475(7354):106–109. doi: 10.1038/nature10189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramos-Gomez M., Kwak M.-K., Dolan P. M., et al. Sensitivity to carcinogenesis is increased and chemoprotective efficacy of enzyme inducers is lost in nrf2 transcription factor-deficient mice. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(6):3410–3415. doi: 10.1073/pnas.051618798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iida K., Itoh K., Kumagai Y., et al. Nrf2 is essential for the chemopreventive efficacy of oltipraz against urinary bladder carcinogenesis. Cancer Research. 2004;64(18):6424–6431. doi: 10.1158/0008-5472.CAN-04-1906. [DOI] [PubMed] [Google Scholar]
- 42.Xu C., Huang M.-T., Shen G., et al. Inhibition of 7,12-dimethylbenz(a)anthracene-induced skin tumorigenesis in C57BL/6 mice by sulforaphane is mediated by nuclear factor E2-related factor 2. Cancer Research. 2006;66(16):8293–8296. doi: 10.1158/0008-5472.CAN-06-0300. [DOI] [PubMed] [Google Scholar]
- 43.Khor T. O., Huang M.-T., Prawan A., et al. Increased susceptibility of Nrf2 knockout mice to colitis-associated colorectal cancer. Cancer Prevention Research. 2008;1(3):187–191. doi: 10.1158/1940-6207.capr-08-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Becks L., Prince M., Burson H., et al. Aggressive mammary carcinoma progression in Nrf2 knockout mice treated with 7,12-dimethylbenz[a]anthracene. BMC Cancer. 2010;10, article 540 doi: 10.1186/1471-2407-10-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamamoto T., Yoh K., Kobayashi A., et al. Identification of polymorphisms in the promoter region of the human NRF2 gene. Biochemical and Biophysical Research Communications. 2004;321(1):72–79. doi: 10.1016/j.bbrc.2004.06.112. [DOI] [PubMed] [Google Scholar]
- 46.Suzuki T., Shibata T., Takaya K., et al. Regulatory nexus of synthesis and degradation deciphers cellular Nrf2 expression levels. Molecular and Cellular Biology. 2013;33(12):2402–2412. doi: 10.1128/mcb.00065-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang X.-J., Sun Z., Villeneuve N. F., et al. Nrf2 enhances resistance of cancer cells to chemotherapeutic drugs, the dark side of Nrf2. Carcinogenesis. 2008;29(6):1235–1243. doi: 10.1093/carcin/bgn095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Na H.-K., Surh Y.-J. Oncogenic potential of Nrf2 and its principal target protein heme oxygenase-1. Free Radical Biology and Medicine. 2014;67:353–365. doi: 10.1016/j.freeradbiomed.2013.10.819. [DOI] [PubMed] [Google Scholar]
- 49.Shibata T., Ohta T., Tong K. I., et al. Cancer related mutations in NRF2 impair its recognition by Keap1-Cul3 E3 ligase and promote malignancy. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(36):13568–13573. doi: 10.1073/pnas.0806268105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang D. D. Mechanistic studies of the Nrf2-Keap1 signaling pathway. Drug Metabolism Reviews. 2006;38(4):769–789. doi: 10.1080/03602530600971974. [DOI] [PubMed] [Google Scholar]
- 51.Nioi P., Nguyen T. A mutation of Keap1 found in breast cancer impairs its ability to repress Nrf2 activity. Biochemical and Biophysical Research Communications. 2007;362(4):816–821. doi: 10.1016/j.bbrc.2007.08.051. [DOI] [PubMed] [Google Scholar]
- 52.Stacy D. R., Ely K., Massion P. P., et al. Increased expression of nuclear factor E2 p45-related factor 2 (NRF2) in head and neck squamous cell carcinomas. Head and Neck. 2006;28(9):813–818. doi: 10.1002/hed.20430. [DOI] [PubMed] [Google Scholar]
- 53.van der Wijst M. G. P., Brown R., Rots M. G. Nrf2, the master redox switch: the Achilles' heel of ovarian cancer? Biochimica et Biophysica Acta—Reviews on Cancer. 2014;1846(2):494–509. doi: 10.1016/j.bbcan.2014.09.004. [DOI] [PubMed] [Google Scholar]
- 54.Jiang T., Chen N., Zhao F., et al. High levels of Nrf2 determine chemoresistance in type II endometrial cancer. Cancer Research. 2010;70(13):5486–5496. doi: 10.1158/0008-5472.CAN-10-0713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mitsuishi Y., Motohashi H., Yamamoto M. The Keap1–Nrf2 system in cancers: stress response and anabolic metabolism. Frontiers in Oncology. 2012;2, article 200 doi: 10.3389/fonc.2012.00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shibata T., Saito S., Kokubu A., Suzuki T., Yamamoto M., Hirohashi S. Global downstream pathway analysis reveals a dependence of oncogenic NF—E2-related factor 2 mutation on the mTOR growth signaling pathway. Cancer Research. 2010;70(22):9095–9105. doi: 10.1158/0008-5472.can-10-0384. [DOI] [PubMed] [Google Scholar]
- 57.Shelton P., Jaiswal A. K. The transcription factor NF-E2-related factor 2 (nrf2): a protooncogene? The FASEB Journal. 2013;27(2):414–423. doi: 10.1096/fj.12-217257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Muscarella L. A., Parrella P., D'Alessandro V., et al. Frequent epigenetics inactivation of KEAP1 gene in non-small cell lung cancer. Epigenetics. 2011;6(6):710–719. doi: 10.4161/epi.6.6.15773. [DOI] [PubMed] [Google Scholar]
- 59.Zhang P., Singh A., Yegnasubramanian S., et al. Loss of kelch-like ECH-associated protein 1 function in prostate cancer cells causes chemoresistance and radioresistance and promotes tumor growth. Molecular Cancer Therapeutics. 2010;9(2):336–346. doi: 10.1158/1535-7163.MCT-09-0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hanada N., Takahata T., Zhou Q., et al. Methylation of the KEAP1 gene promoter region in human colorectal cancer. BMC Cancer. 2012;12, article 66 doi: 10.1186/1471-2407-12-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ooi A., Dykema K., Ansari A., et al. CUL3 and NRF2 mutations confer an NRF2 activation phenotype in a sporadic form of papillary renal cell carcinoma. Cancer Research. 2013;73(7):2044–2051. doi: 10.1158/0008-5472.can-12-3227. [DOI] [PubMed] [Google Scholar]
- 62.Loignon M., Miao W., Hu L., et al. Cul3 overexpression depletes Nrf2 in breast cancer and is associated with sensitivity to carcinogens, to oxidative stress, and to chemotherapy. Molecular Cancer Therapeutics. 2009;8(8):2432–2440. doi: 10.1158/1535-7163.MCT-08-1186. [DOI] [PubMed] [Google Scholar]
- 63.Martinez V. D., Vucic E. A., Thu K. L., Pikor L. A., Lam S., Lam W. L. Disruption of KEAP1/CUL3/RBX1 E3-ubiquitin ligase complex components by multiple genetic mechanisms: association with poor prognosis in head and neck cancer. Head and Neck. 2015;37(5):727–734. doi: 10.1002/hed.23663. [DOI] [PubMed] [Google Scholar]
- 64.Martinez V. D., Vucic E. A., Thu K. L., Pikor L. A., Hubaux R., Lam W. L. Unique pattern of component gene disruption in the NRF2 inhibitor KEAP1/CUL3/RBX1 E3-ubiquitin ligase complex in serous ovarian cancer. BioMed Research International. 2014;2014:10. doi: 10.1155/2014/159459.159459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Martinez V. D., Vucic E. A., Pikor L. A., Thu K. L., Hubaux R., Lam W. L. Frequent concerted genetic mechanisms disrupt multiple components of the NRF2 inhibitor KEAP1/CUL3/RBX1 E3-ubiquitin ligase complex in thyroid cancer. Molecular Cancer. 2013;12(1, article 124) doi: 10.1186/1476-4598-12-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Villeneuve N. F., Sun Z., Chen W., Zhang D. D. Nrf2 and p21 regulate the fine balance between life and death by controlling ROS levels. Cell Cycle. 2009;8(20):3255–3256. doi: 10.4161/cc.8.20.9565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen W., Sun Z., Wang X.-J., et al. Direct interaction between Nrf2 and p21Cip1/WAF1 upregulates the Nrf2-mediated antioxidant response. Molecular Cell. 2009;34(6):663–673. doi: 10.1016/j.molcel.2009.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Clements C. M., McNally R. S., Conti B. J., Mak T. W., Ting J. P.-Y. DJ-1, a cancer- and Parkinson's disease-associated protein, stabilizes the antioxidant transcriptional master regulator Nrf2. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(41):15091–15096. doi: 10.1073/pnas.0607260103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gan L., Johnson D. A., Johnson J. A. Keap1-Nrf2 activation in the presence and absence of DJ-1. European Journal of Neuroscience. 2010;31(6):967–977. doi: 10.1111/j.1460-9568.2010.07138.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.MacKeigan J. P., Clements C. M., Lich J. D., Pope R. M., Hod Y., Ting J. P.-Y. Proteomic profiling drug-induced apoptosis in non-small cell lung carcinoma: identification of RS/DJ-1 and RhoGDIalpha. Cancer Research. 2003;63(20):6928–6934. [PubMed] [Google Scholar]
- 71.Taira T., Saito Y., Niki T., Iguchi-Ariga S. M. M., Takahashi K., Ariga H. DJ-1 has a role in antioxidative stress to prevent cell death. The EMBO Reports. 2004;5(2):213–218. doi: 10.1038/sj.embor.7400074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Copple I. M., Lister A., Obeng A. D., et al. Physical and functional interaction of sequestosome 1 with Keap1 regulates the Keap1-Nrf2 cell defense pathway. The Journal of Biological Chemistry. 2010;285(22):16782–16788. doi: 10.1074/jbc.m109.096545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jain A., Lamark T., Sjøttem E., et al. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. Journal of Biological Chemistry. 2010;285(29):22576–22591. doi: 10.1074/jbc.m110.118976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Camp N. D., James R. G., Dawson D. W., et al. Wilms tumor gene on X chromosome (WTX) inhibits degradation of NRF2 protein through competitive binding to KEAP1 protein. Journal of Biological Chemistry. 2012;287(9):6539–6550. doi: 10.1074/jbc.m111.316471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ma J., Cai H., Wu T., et al. PALB2 interacts with KEAP1 to promote NRF2 nuclear accumulation and function. Molecular and Cellular Biology. 2012;32(8):1506–1517. doi: 10.1128/mcb.06271-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hast B. E., Goldfarb D., Mulvaney K. M., et al. Proteomic analysis of ubiquitin ligase KEAP1 reveals associated proteins that inhibit NRF2 ubiquitination. Cancer Research. 2013;73(7):2199–2210. doi: 10.1158/0008-5472.can-12-4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wakabayashi N., Shin S., Slocum S. L., et al. Regulation of Notch1 signaling by Nrf2: implications for tissue regeneration. Science Signaling. 2010;3(130, article ra52) doi: 10.1126/scisignal.2000762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.You A., Nam C.-W., Wakabayashi N., Yamamoto M., Kensler T. W., Kwak M.-K. Transcription factor Nrf2 maintains the basal expression of Mdm2: an implication of the regulation of p53 signaling by Nrf2. Archives of Biochemistry and Biophysics. 2011;507(2):356–364. doi: 10.1016/j.abb.2010.12.034. [DOI] [PubMed] [Google Scholar]
- 79.Wang J., Zhang M., Zhang L., et al. Correlation of Nrf2, HO-1, and MRP3 in gallbladder cancer and their relationships to clinicopathologic features and survival. Journal of Surgical Research. 2010;164(1):e99–e105. doi: 10.1016/j.jss.2010.05.058. [DOI] [PubMed] [Google Scholar]
- 80.Kim W. D., Kim Y. W., Cho I. J., Lee C. H., Kim S. G. E-cadherin inhibits nuclear accumulation of Nrf2: implications for chemoresistance of cancer cells. Journal of Cell Science. 2012;125(5):1284–1295. doi: 10.1242/jcs.095422. [DOI] [PubMed] [Google Scholar]
- 81.Jain A. K., Bloom D. A., Jaiswal A. K. Nuclear import and export signals in control of Nrf2. The Journal of Biological Chemistry. 2005;280(32):29158–29168. doi: 10.1074/jbc.m502083200. [DOI] [PubMed] [Google Scholar]
- 82.Shen H., Yang Y., Xia S., Rao B., Zhang J., Wang J. Blockage of Nrf2 suppresses the migration and invasion of esophageal squamous cell carcinoma cells in hypoxic microenvironment. Diseases of the Esophagus. 2014;27(7):685–692. doi: 10.1111/dote.12124. [DOI] [PubMed] [Google Scholar]
- 83.Semenza G. L. Intratumoral hypoxia, radiation resistance, and HIF-1. Cancer Cell. 2004;5(5):405–406. doi: 10.1016/S1535-6108(04)00118-7. [DOI] [PubMed] [Google Scholar]
- 84.Kim T.-H., Hur E.-G., Kang S.-J., et al. NRF2 blockade suppresses colon tumor angiogenesis by inhibiting hypoxia-induced activation of HIF-1α . Cancer Research. 2011;71(6):2260–2275. doi: 10.1158/0008-5472.can-10-3007. [DOI] [PubMed] [Google Scholar]
- 85.Shim G.-S., Manandhar S., Shin D.-H., Kim T.-H., Kwak M.-K. Acquisition of doxorubicin resistance in ovarian carcinoma cells accompanies activation of the NRF2 pathway. Free Radical Biology and Medicine. 2009;47(11):1619–1631. doi: 10.1016/j.freeradbiomed.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 86.Zhong Y., Zhang F., Sun Z., et al. Drug resistance associates with activation of Nrf2 in MCF-7/DOX cells, and wogonin reverses it by down-regulating Nrf2-mediated cellular defense response. Molecular Carcinogenesis. 2013;52(10):824–834. doi: 10.1002/mc.21921. [DOI] [PubMed] [Google Scholar]
- 87.Homma S., Ishii Y., Morishima Y., et al. Nrf2 enhances cell proliferation and resistance to anticancer drugs in human lung cancer. Clinical Cancer Research. 2009;15(10):3423–3432. doi: 10.1158/1078-0432.ccr-08-2822. [DOI] [PubMed] [Google Scholar]
- 88.Jayakumar S., Kunwar A., Sandur S. K., Pandey B. N., Chaubey R. C. Differential response of DU145 and PC3 prostate cancer cells to ionizing radiation: role of reactive oxygen species, GSH and Nrf2 in radiosensitivity. Biochimica et Biophysica Acta—General Subjects. 2014;1840(1):485–494. doi: 10.1016/j.bbagen.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 89.Lee S., Lim M.-J., Kim M.-H., et al. An effective strategy for increasing the radiosensitivity of Human lung Cancer cells by blocking Nrf2-dependent antioxidant responses. Free Radical Biology and Medicine. 2012;53(4):807–816. doi: 10.1016/j.freeradbiomed.2012.05.038. [DOI] [PubMed] [Google Scholar]
- 90.Furfaro A. L., MacAy J. R. Z., Marengo B., et al. Resistance of neuroblastoma GI-ME-N cell line to glutathione depletion involves Nrf2 and heme oxygenase-1. Free Radical Biology and Medicine. 2012;52(2):488–496. doi: 10.1016/j.freeradbiomed.2011.11.007. [DOI] [PubMed] [Google Scholar]
- 91.Furfaro A. L., Piras S., Passalacqua M., et al. HO-1 up-regulation: a key point in high-risk neuroblastoma resistance to bortezomib. Biochimica et Biophysica Acta. 2014;1842(4):613–622. doi: 10.1016/j.bbadis.2013.12.008. [DOI] [PubMed] [Google Scholar]
- 92.Singh A., Wu H., Zhang P., Happel C., Ma J., Biswal S. Expression of ABCG2 (BCRP) is regulated by Nrf2 in cancer cells that confers side population and chemoresistance phenotype. Molecular Cancer Therapeutics. 2010;9(8):2365–2376. doi: 10.1158/1535-7163.mct-10-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Geismann C., Arlt A., Sebens S., Schäfer H. Cytoprotection “gone astray”: Nrf2 and its role in cancer. OncoTargets and Therapy. 2014;7:1497–1518. doi: 10.2147/OTT.S36624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mahaffey C. M., Zhang H., Rinna A., Holland W., Mack P. C., Forman H. J. Multidrug-resistant protein-3 gene regulation by the transcription factor Nrf2 in human bronchial epithelial and non-small-cell lung carcinoma. Free Radical Biology and Medicine. 2009;46(12):1650–1657. doi: 10.1016/j.freeradbiomed.2009.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Slocum S. L., Kensler T. W. Nrf2: control of sensitivity to carcinogens. Archives of Toxicology. 2011;85(4):273–284. doi: 10.1007/s00204-011-0675-4. [DOI] [PubMed] [Google Scholar]
- 96.Tsai J. J., Dudakov J. A., Takahashi K., et al. Nrf2 regulates haematopoietic stem cell function. Nature Cell Biology. 2013;15(3):309–316. doi: 10.1038/ncb2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Santos D. M., Santos M. M. M., Moreira R., Solá S., Rodrigues C. M. P. Synthetic condensed 1,4-naphthoquinone derivative shifts neural stem cell differentiation by regulating redox state. Molecular Neurobiology. 2013;47(1):313–324. doi: 10.1007/s12035-012-8353-y. [DOI] [PubMed] [Google Scholar]
- 98.Murakami S., Motohashi H. Roles of NRF2 in cell proliferation and differentiation. Free Radical Biology and Medicine. 2015 doi: 10.1016/j.freeradbiomed.2015.06.030. [DOI] [PubMed] [Google Scholar]
- 99.Johannessen T.-C. A., Bjerkvig R., Tysnes B. B. DNA repair and cancer stem-like cells—potential partners in glioma drug resistance? Cancer Treatment Reviews. 2008;34(6):558–567. doi: 10.1016/j.ctrv.2008.03.125. [DOI] [PubMed] [Google Scholar]
- 100.Zhu J., Wang H., Sun Q., et al. Nrf2 is required to maintain the self-renewal of glioma stem cells. BMC Cancer. 2013;13, article 380 doi: 10.1186/1471-2407-13-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhu J., Wang H., Ji X., et al. Differential Nrf2 expression between glioma stem cells and non-stem-like cells in glioblastoma. Oncology Letters. 2014;7(3):693–698. doi: 10.3892/ol.2013.1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wu T., Harder B. G., Wong P. K., Lang J. E., Zhang D. D. Oxidative stress, mammospheres and Nrf2-new implication for breast cancer therapy? Molecular Carcinogenesis. 2014 doi: 10.1002/mc.22202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Trakshel G. M., Maines M. D. Multiplicity of heme oxygenase isozymes. HO-1 and HO-2 are different molecular species in rat and rabbit. Journal of Biological Chemistry. 1989;264(2):1323–1328. [PubMed] [Google Scholar]
- 104.Maines M. D. Heme oxygenase: function, multiplicity, regulatory mechanisms, and clinical applications. The FASEB Journal. 1988;2(10):2557–2568. [PubMed] [Google Scholar]
- 105.Ryter S. W., Choi A. M. K. Targeting heme oxygenase-1 and carbon monoxide for therapeutic modulation of inflammation. Translational Research. 2015 doi: 10.1016/j.trsl.2015.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Maines M. D., Trakshel G. M., Kutty R. K. Characterization of two constitutive forms of rat liver microsomal heme oxygenase. Only one molecular species of the enzyme is inducible. The Journal of Biological Chemistry. 1986;261(1):411–419. [PubMed] [Google Scholar]
- 107.Alam J., Shibahara S., Smith A. Transcriptional activation of the heme oxygenase gene by heme and cadmium in mouse hepatoma cells. Journal of Biological Chemistry. 1989;264(11):6371–6375. [PubMed] [Google Scholar]
- 108.Keyse S. M., Tyrrell R. M. Heme oxygenase is the major 32-kDa stress protein induced in human skin fibroblasts by UVA radiation, hydrogen peroxide, and sodium arsenite. Proceedings of the National Academy of Sciences of the United States of America. 1989;86(1):99–103. doi: 10.1073/pnas.86.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Foresti R., Clark J. E., Green C. J., Motterlini R. Thiol compounds interact with nitric oxide in regulating heme oxygenase-1 induction in endothelial cells: involvement of superoxide and peroxynitrite anions. The Journal of Biological Chemistry. 1997;272(29):18411–18417. doi: 10.1074/jbc.272.29.18411. [DOI] [PubMed] [Google Scholar]
- 110.Terry C. M., Clikeman J. A., Hoidal J. R., Callahan K. S. Effect of tumor necrosis factor-alpha and interleukin-1α on heme oxygenase-1 expression in human endothelial cells. American Journal of Physiology. 1998;274(3, part 2):H883–H891. doi: 10.1152/ajpheart.1998.274.3.H883. [DOI] [PubMed] [Google Scholar]
- 111.Stocker R., Yamamoto Y., McDonagh A. F., Glazer A. N., Ames B. N. Bilirubin is an antioxidant of possible physiological importance. Science. 1987;235(4792):1043–1046. doi: 10.1126/science.3029864. [DOI] [PubMed] [Google Scholar]
- 112.Brouard S., Otterbein L. E., Anrather J., et al. Carbon monoxide generated by heme oxygenase 1 suppresses endothelial cell apoptosis. Journal of Experimental Medicine. 2000;192(7):1015–1026. doi: 10.1084/jem.192.7.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ward R. J., Wilmet S., Legssyer R., Crichton R. R. The influence of iron homoeostasis on macrophage function. Biochemical Society Transactions. 2002;30(4):762–765. doi: 10.1042/BST0300762. [DOI] [PubMed] [Google Scholar]
- 114.Ryter S. W., Choi A. M. K. Heme oxygenase-1: redox regulation of a stress protein in lung and cell culture models. Antioxidants and Redox Signaling. 2005;7(1-2):80–91. doi: 10.1089/ars.2005.7.80. [DOI] [PubMed] [Google Scholar]
- 115.Prawan A., Kundu J. K., Surh Y.-J. Molecular basis of heme oxygenase-1 induction: implications for chemoprevention and chemoprotection. Antioxidants and Redox Signaling. 2005;7(11-12):1688–1703. doi: 10.1089/ars.2005.7.1688. [DOI] [PubMed] [Google Scholar]
- 116.Jozkowicz A., Was H., Dulak J. Heme oxygenase-1 in tumors: Is it a false friend? Antioxidants and Redox Signaling. 2007;9(12):2099–2117. doi: 10.1089/ars.2007.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Banerjee P., Basu A., Datta D., Gasser M., Waaga-Gasser A. M., Pal S. The heme oxygenase-1 protein is overexpressed in human renal cancer cells following activation of the Ras-Raf-ERK pathway and mediates anti-apoptotic signal. The Journal of Biological Chemistry. 2011;286(38):33580–33590. doi: 10.1074/jbc.m111.248401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Yin H., Fang J., Liao L., Maeda H., Su Q. Upregulation of heme oxygenase-1 in colorectal cancer patients with increased circulation carbon monoxide levels, potentially affects chemotherapeutic sensitivity. BMC Cancer. 2014;14(1, article 436) doi: 10.1186/1471-2407-14-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Berberat P. O., Dambrauskas Z., Gulbinas A., et al. Inhibition of heme oxygenase-1 increases responsiveness of pancreatic cancer cells to anticancer treatment. Clinical Cancer Research. 2005;11(10):3790–3798. doi: 10.1158/1078-0432.CCR-04-2159. [DOI] [PubMed] [Google Scholar]
- 120.Degese M. S., Mendizabal J. E., Gandini N. A., et al. Expression of heme oxygenase-1 in non-small cell lung cancer (NSCLC) and its correlation with clinical data. Lung Cancer. 2012;77(1):168–175. doi: 10.1016/j.lungcan.2012.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Miyake M., Fujimoto K., Anai S., et al. Clinical significance of heme oxygenase-1 expression in non-muscle-invasive bladder cancer. Urologia Internationalis. 2010;85(3):355–363. doi: 10.1159/000317785. [DOI] [PubMed] [Google Scholar]
- 122.Tsai J.-R., Wang H.-M., Liu P.-L., et al. High expression of heme oxygenase-1 is associated with tumor invasiveness and poor clinical outcome in non-small cell lung cancer patients. Cellular Oncology. 2012;35(6):461–471. doi: 10.1007/s13402-012-0105-5. [DOI] [PubMed] [Google Scholar]
- 123.Was H., Cichon T., Smolarczyk R., et al. Overexpression of heme oxygenase-1 in murine melanoma: increased proliferation and viability of tumor cells, decreased survival of mice. The American Journal of Pathology. 2006;169(6):2181–2198. doi: 10.2353/ajpath.2006.051365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sunamura M., Duda D. G., Ghattas M. H., et al. Heme oxygenase-1 accelerates tumor angiogenesis of human pancreatic cancer. Angiogenesis. 2003;6(1):15–24. doi: 10.1023/A:1025803600840. [DOI] [PubMed] [Google Scholar]
- 125.Lee S.-S., Yang S.-F., Tsai C.-H., Chou M.-C., Chou M.-Y., Chang Y.-C. Upregulation of heme oxygenase-1 expression in areca-quid-chewing-associated oral squamous cell carcinoma. Journal of the Formosan Medical Association. 2008;107(5):355–363. doi: 10.1016/s0929-6646(08)60100-x. [DOI] [PubMed] [Google Scholar]
- 126.Alaoui-Jamali M. A., Bismar T. A., Gupta A., et al. A novel experimental heme oxygenase-1-targeted therapy for hormone-refractory prostate cancer. Cancer Research. 2009;69(20):8017–8024. doi: 10.1158/0008-5472.CAN-09-0419. [DOI] [PubMed] [Google Scholar]
- 127.Sass G., Leukel P., Schmitz V., et al. Inhibition of heme oxygenase 1 expression by small interfering RNA decreases orthotopic tumor growth in livers of mice. International Journal of Cancer. 2008;123(6):1269–1277. doi: 10.1002/ijc.23695. [DOI] [PubMed] [Google Scholar]
- 128.Marinissen M. J., Tanos T., Bolós M., De Sagarra M. R., Coso O. A., Cuadrado A. Inhibition of heme oxygenase-1 interferes with the transforming activity of the Kaposi sarcoma herpesvirus-encoded G protein-coupled receptor. The Journal of Biological Chemistry. 2006;281(16):11332–11346. doi: 10.1074/jbc.m512199200. [DOI] [PubMed] [Google Scholar]
- 129.Was H., Dulak J., Jozkowicz A. Heme oxygenase-1 in tumor biology and therapy. Current Drug Targets. 2010;11(12):1551–1570. doi: 10.2174/1389450111009011551. [DOI] [PubMed] [Google Scholar]
- 130.Loboda A., Jazwa A., Grochot-Przeczek A., et al. Heme oxygenase-1 and the vascular bed: from molecular mechanisms to therapeutic opportunities. Antioxidants & Redox Signaling. 2008;10(10):1767–1812. doi: 10.1089/ars.2008.2043. [DOI] [PubMed] [Google Scholar]
- 131.Fang J., Greish K., Qin H., et al. HSP32 (HO-1) inhibitor, copoly(styrene-maleic acid)-zinc protoporphyrin IX, a water-soluble micelle as anticancer agent: in vitro and in vivo anticancer effect. European Journal of Pharmaceutics and Biopharmaceutics. 2012;81(3):540–547. doi: 10.1016/j.ejpb.2012.04.016. [DOI] [PubMed] [Google Scholar]
- 132.Nakamura H., Liao L., Hitaka Y., et al. Micelles of zinc protoporphyrin conjugated to N-(2-hydroxypropyl)methacrylamide (HPMA) copolymer for imaging and light-induced antitumor effects in vivo. Journal of Controlled Release. 2013;165(3):191–198. doi: 10.1016/j.jconrel.2012.11.017. [DOI] [PubMed] [Google Scholar]
- 133.Nuhn P., Künzli B. M., Hennig R., et al. Heme oxygenase-1 and its metabolites affect pancreatic tumor growth in vivo . Molecular Cancer. 2009;8, article 37 doi: 10.1186/1476-4598-8-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zhang W., Qiao T., Zha L. Inhibition of heme oxygenase-1 enhances the radiosensitivity in human nonsmall cell lung cancer A549 cells. Cancer Biotherapy and Radiopharmaceuticals. 2011;26(5):639–645. doi: 10.1089/cbr.2010.0939. [DOI] [PubMed] [Google Scholar]
- 135.Nowis D., Legat M., Grzela T., et al. Heme oxygenase-1 protects tumor cells against photodynamic therapy-mediated cytotoxicity. Oncogene. 2006;25(24):3365–3374. doi: 10.1038/sj.onc.1209378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ma D., Fang Q., Wang P., et al. Induction of heme oxygenase-1 by Na+-H+ exchanger 1 protein plays a crucial role in imatinib-resistant chronic myeloid leukemia cells. The Journal of Biological Chemistry. 2015;290(20):12558–12571. doi: 10.1074/jbc.M114.626960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Jeon W.-K., Hong H.-Y., Seo W.-C., et al. Smad7 sensitizes A549 lung cancer cells to cisplatin-induced apoptosis through heme oxygenase-1 inhibition. Biochemical and Biophysical Research Communications. 2012;420(2):288–292. doi: 10.1016/j.bbrc.2012.02.151. [DOI] [PubMed] [Google Scholar]
- 138.Yin Y., Liu Q., Wang B., Chen G., Xu L., Zhou H. Expression and function of heme oxygenase-1 in human gastric cancer. Experimental Biology and Medicine. 2012;237(4):362–371. doi: 10.1258/ebm.2011.011193. [DOI] [PubMed] [Google Scholar]
- 139.Mayerhofer M., Florian S., Krauth M.-T., et al. Identification of heme oxygenase-1 as a novel BCR/ABL-dependent survival factor in chronic myeloid leukemia. Cancer Research. 2004;64(9):3148–3154. doi: 10.1158/0008-5472.can-03-1200. [DOI] [PubMed] [Google Scholar]
- 140.Hill M., Pereira V., Chauveau C., et al. Heme oxygenase-1 inhibits rat and human breast cancer cell proliferation: mutual cross inhibition with indoleamine 2,3-dioxygenase. The FASEB Journal. 2005;19(14):1957–1968. doi: 10.1096/fj.05-3875com. [DOI] [PubMed] [Google Scholar]
- 141.Ferrando M., Gueron G., Elguero B., et al. Heme oxygenase 1 (HO-1) challenges the angiogenic switch in prostate cancer. Angiogenesis. 2011;14(4):467–479. doi: 10.1007/s10456-011-9230-4. [DOI] [PubMed] [Google Scholar]
- 142.Chao C.-Y., Lii C.-K., Hsu Y.-T., et al. Induction of heme oxygenase-1 and inhibition of TPA-induced matrix metalloproteinase-9 expression by andrographolide in MCF-7 human breast cancer cells. Carcinogenesis. 2013;34(8):1843–1851. doi: 10.1093/carcin/bgt131. [DOI] [PubMed] [Google Scholar]
- 143.Wegiel B., Nemeth Z., Correa-Costa M., Bulmer A. C., Otterbein L. E. Heme oxygenase-1: a metabolic nike. Antioxidants and Redox Signaling. 2014;20(11):1709–1722. doi: 10.1089/ars.2013.5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ryter S. W., Xi S., Hartsfield C. L., Choi A. M. K. Mitogen activated protein kinase (MAPK) pathway regulates heme oxygenase-1 gene expression by hypoxia in vascular cells. Antioxidants and Redox Signaling. 2002;4(4):587–592. doi: 10.1089/15230860260220085. [DOI] [PubMed] [Google Scholar]
- 145.Lin C.-C., Chiang L.-L., Lin C.-H., et al. Transforming growth factor-beta1 stimulates heme oxygenase-1 expression via the PI3K/Akt and NF-kappaB pathways in human lung epithelial cells. European Journal of Pharmacology. 2007;560(2-3):101–109. doi: 10.1016/j.ejphar.2007.01.025. [DOI] [PubMed] [Google Scholar]
- 146.Lavrovsky Y., Schwartzman M. L., Levere R. D., Kappas A., Abraham N. G. Identification of binding sites for transcription factors NF-kappa B and AP-2 in the promoter region of the human heme oxygenase 1 gene. Proceedings of the National Academy of Sciences of the United States of America. 1994;91(13):5987–5991. doi: 10.1073/pnas.91.13.5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Rushworth S. A., Bowles K. M., Raninga P., MacEwan D. J. NF-κB-Lnhibited acute myeloid leukemia cells are rescued from apoptosis by heme oxygenase-1 induction. Cancer Research. 2010;70(7):2973–2983. doi: 10.1158/0008-5472.can-09-3407. [DOI] [PubMed] [Google Scholar]
- 148.Farombi E. O., Surh Y.-J. Heme oxygenase-1 as a potential therapeutic target for hepatoprotection. Journal of Biochemistry and Molecular Biology. 2006;39(5):479–491. doi: 10.5483/bmbrep.2006.39.5.479. [DOI] [PubMed] [Google Scholar]
- 149.Wang L.-H., Li Y., Yang S.-N., et al. Gambogic acid synergistically potentiates cisplatin-induced apoptosis in non-small-cell lung cancer through suppressing NF-κB and MAPK/HO-1 signalling. British Journal of Cancer. 2014;110(2):341–352. doi: 10.1038/bjc.2013.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Nemmiche S., Chabane-Sari D., Kadri M., Guiraud P. Cadmium-induced apoptosis in the BJAB human B cell line: involvement of PKC/ERK1/2/JNK signaling pathways in HO-1 expression. Toxicology. 2012;300(3):103–111. doi: 10.1016/j.tox.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 151.Cheng P.-Y., Lee Y.-M., Shih N.-L., Chen Y.-C., Yen M.-H. Heme oxygenase-1 contributes to the cytoprotection of alpha-lipoic acid via activation of p44/42 mitogen-activated protein kinase in vascular smooth muscle cells. Free Radical Biology and Medicine. 2006;40(8):1313–1322. doi: 10.1016/j.freeradbiomed.2005.11.024. [DOI] [PubMed] [Google Scholar]
- 152.Do M. T., Kim H. G., Khanal T., et al. Metformin inhibits heme oxygenase-1 expression in cancer cells through inactivation of Raf-ERK-Nrf2 signaling and AMPK-independent pathways. Toxicology and Applied Pharmacology. 2013;271(2):229–238. doi: 10.1016/j.taap.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 153.Barrera L. N., Rushworth S. A., Bowles K. M., MacEwan D. J. Bortezomib induces heme oxygenase-1 expression in multiple myeloma. Cell Cycle. 2012;11(12):2248–2252. doi: 10.4161/cc.20343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Bancos S., Baglole C. J., Rahman I., Phipps R. P. Induction of heme oxygenase-1 in normal and malignant B lymphocytes by 15-deoxy-∆12.14-prostaglandin J2 requires Nrf2. Cellular Immunology. 2010;262(1):18–27. doi: 10.1016/j.cellimm.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Lu D.-Y., Yeh W.-L., Huang S.-M., Tang C.-H., Lin H.-Y., Chou S.-J. Osteopontin increases heme oxygenase-1 expression and subsequently induces cell migration and invasion in glioma cells. Neuro-Oncology. 2012;14(11):1367–1378. doi: 10.1093/neuonc/nos262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Bao L.-J., Jaramillo M. C., Zhang Z.-B., et al. Nrf2 induces cisplatin resistance through activation of autophagy in ovarian carcinoma. International Journal of Clinical and Experimental Pathology. 2014;7(4):1502–1513. [PMC free article] [PubMed] [Google Scholar]
- 157.Kensler T. W., Curphey T. J., Maxiutenko Y., Roebuck B. D. Chemoprotection by organosulfur inducers of phase 2 enzymes: dithiolethiones and dithiins. Drug Metabolism and Drug Interactions. 2000;17(1–4):3–22. doi: 10.1515/dmdi.2000.17.1-4.3. [DOI] [PubMed] [Google Scholar]
- 158.Conaway C. C., Wang C.-X., Pittman B., et al. Phenethyl isothiocyanate and sulforaphane and their N-acetylcysteine conjugates inhibit malignant progression of lung adenomas induced by tobacco carcinogens in A/J mice. Cancer Research. 2005;65(18):8548–8557. doi: 10.1158/0008-5472.can-05-0237. [DOI] [PubMed] [Google Scholar]
- 159.Garg R., Gupta S., Maru G. B. Dietary curcumin modulates transcriptional regulators of phase I and phase II enzymes in benzo[a]pyrene-treated mice: mechanism of its anti-initiating action. Carcinogenesis. 2008;29(5):1022–1032. doi: 10.1093/carcin/bgn064. [DOI] [PubMed] [Google Scholar]
- 160.Thimmulappa R. K., Rangasamy T., Alam J., Biswal S. Dibenzoylmethane activates Nrf2-dependent detoxification pathway and inhibits benzo(a)pyrene induced DNA adducts in lungs. Medicinal Chemistry. 2008;4(5):473–481. doi: 10.2174/157340608785700199. [DOI] [PubMed] [Google Scholar]
- 161.Singh B., Shoulson R., Chatterjee A., et al. Resveratrol inhibits estrogen-induced breast carcinogenesis through induction of NRF2-mediated protective pathways. Carcinogenesis. 2014;35(8):1872–1880. doi: 10.1093/carcin/bgu120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Chang K. M., Liang F. P., Chen I. L., et al. Discovery of oxime-bearing naphthalene derivatives as a novel structural type of Nrf2 activators. Bioorganic & Medicinal Chemistry. 2015;23(13):3852–3859. doi: 10.1016/j.bmc.2015.03.046. [DOI] [PubMed] [Google Scholar]
- 163.Gan F.-F., Ling H., Ang X., et al. A novel shogaol analog suppresses cancer cell invasion and inflammation, and displays cytoprotective effects through modulation of NF-kappaB and Nrf2-Keap1 signaling pathways. Toxicology and Applied Pharmacology. 2013;272(3):852–862. doi: 10.1016/j.taap.2013.07.011. [DOI] [PubMed] [Google Scholar]
- 164.Hu R., Saw C. L.-L., Yu R., Kong A.-N. T. Regulation of NF-E2-related factor 2 signaling for cancer chemoprevention: antioxidant coupled with antiinflammatory. Antioxidants and Redox Signaling. 2010;13(11):1679–1698. doi: 10.1089/ars.2010.3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Li C., Xu X., Wang X. J., Pan Y. Imine resveratrol analogues: molecular design, Nrf2 activation and SAR analysis. PLoS ONE. 2014;9(7) doi: 10.1371/journal.pone.0101455.e101455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Zhao C. R., Gao Z. H., Qu X. J. Nrf2-ARE signaling pathway and natural products for cancer chemoprevention. Cancer Epidemiology. 2010;34(5):523–533. doi: 10.1016/j.canep.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 167.Lee J.-S., Surh Y.-J. Nrf2 as a novel molecular target for chemoprevention. Cancer Letters. 2005;224(2):171–184. doi: 10.1016/j.canlet.2004.09.042. [DOI] [PubMed] [Google Scholar]
- 168.Jeong W.-S., Jun M., Kong A.-N. T. Nrf2: a potential molecular target for cancer chemoprevention by natural compounds. Antioxidants & Redox Signaling. 2006;8(1-2):99–106. doi: 10.1089/ars.2006.8.99. [DOI] [PubMed] [Google Scholar]
- 169.Lau A., Villeneuve N. F., Sun Z., Wong P. K., Zhang D. D. Dual roles of Nrf2 in cancer. Pharmacological Research. 2008;58(5-6):262–270. doi: 10.1016/j.phrs.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Kensler T. W., Chen J.-G., Egner P. A., et al. Effects of glucosinolate-rich broccoli sprouts on urinary levels of aflatoxin-DNA adducts and phenanthrene tetraols in a randomized clinical trial in He Zuo Township, Qidong, People's Republic of China. Cancer Epidemiology Biomarkers and Prevention. 2005;14(11):2605–2613. doi: 10.1158/1055-9965.epi-05-0368. [DOI] [PubMed] [Google Scholar]
- 171.Shang F., Hui L., An X., Zhang X., Guo S., Kui Z. ZnPPIX inhibits peritoneal metastasis of gastric cancer via its antiangiogenic activity. Biomedicine & Pharmacotherapy. 2015;71:240–246. doi: 10.1016/j.biopha.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 172.Sahoo S. K., Sawa T., Fang J., et al. Pegylated zinc protoporphyrin: a water-soluble heme oxygenase inhibitor with tumor-targeting capacity. Bioconjugate Chemistry. 2002;13(5):1031–1038. doi: 10.1021/bc020010k. [DOI] [PubMed] [Google Scholar]
- 173.Iyer A. K., Greish K., Seki T., et al. Polymeric micelles of zinc protoporphyrin for tumor targeted delivery based on EPR effect and singlet oxygen generation. Journal of Drug Targeting. 2007;15(7-8):496–506. doi: 10.1080/10611860701498252. [DOI] [PubMed] [Google Scholar]
- 174.Józkowicz A., Dulak J. Effects of protoporphyrins on production of nitric oxide and expression of vascular endothelial growth factor in vascular smooth muscle cells and macrophages. Acta Biochimica Polonica. 2003;50(1):69–79. [PubMed] [Google Scholar]
- 175.Loboda A., Jazwa A., Wegiel B., Jozkowicz A., Dulak J. Heme oxygenase-1-dependent and -independent regulation of angiogenic genes expression: effect of cobalt protoporphyrin and cobalt chloride on VEGF and IL-8 synthesis in human microvascular endothelial cells. Cellular and Molecular Biology. 2005;51(4):347–355. doi: 10.1170/t637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Busserolles J., Megías J., Terencio M. C., Alcaraz M. J. Heme oxygenase-1 inhibits apoptosis in Caco-2 cells via activation of Akt pathway. International Journal of Biochemistry and Cell Biology. 2006;38(9):1510–1517. doi: 10.1016/j.biocel.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 177.Fang J., Sawa T., Akaike T., et al. In vivo antitumor activity of pegylated zinc protoporphyrin: targeted inhibition of heme oxygenase in solid tumor. Cancer Research. 2003;63(13):3567–3574. [PubMed] [Google Scholar]
- 178.Ren D., Villeneuve N. F., Jiang T., et al. Brusatol enhances the efficacy of chemotherapy by inhibiting the Nrf2-mediated defense mechanism. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(4):1433–1438. doi: 10.1073/pnas.1014275108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Olayanju A., Copple I. M., Bryan H. K., et al. Brusatol provokes a rapid and transient inhibition of Nrf2 signaling and sensitizes mammalian cells to chemical toxicity-implications for therapeutic targeting of Nrf2. Free Radical Biology and Medicine. 2015;78:202–212. doi: 10.1016/j.freeradbiomed.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Zhang T., Kimura Y., Jiang S., Harada K., Yamashita Y., Ashida H. Luteolin modulates expression of drug-metabolizing enzymes through the AhR and Nrf2 pathways in hepatic cells. Archives of Biochemistry and Biophysics. 2014;557:36–46. doi: 10.1016/j.abb.2014.05.023. [DOI] [PubMed] [Google Scholar]
- 181.Tang X., Wang H., Fan L., et al. Luteolin inhibits Nrf2 leading to negative regulation of the Nrf2/ARE pathway and sensitization of human lung carcinoma A549 cells to therapeutic drugs. Free Radical Biology and Medicine. 2011;50(11):1599–1609. doi: 10.1016/j.freeradbiomed.2011.03.008. [DOI] [PubMed] [Google Scholar]
- 182.Chian S., Li Y.-Y., Wang X.-J., Tang X.-W. Luteolin sensitizes two oxaliplatin-resistant colorectal cancer cell lines to chemotherapeutic drugs via inhibition of the Nrf2 pathway. Asian Pacific Journal of Cancer Prevention. 2014;15(6):2911–2916. doi: 10.7314/apjcp.2014.15.6.2911. [DOI] [PubMed] [Google Scholar]
- 183.Chian S., Thapa R., Chi Z., Wang X. J., Tang X. Luteolin inhibits the Nrf2 signaling pathway and tumor growth in vivo. Biochemical and Biophysical Research Communications. 2014;447(4):602–608. doi: 10.1016/j.bbrc.2014.04.039. [DOI] [PubMed] [Google Scholar]
- 184.Magesh S., Chen Y., Hu L. Small molecule modulators of Keap1-Nrf2-ARE pathway as potential preventive and therapeutic agents. Medicinal Research Reviews. 2012;32(4):687–726. doi: 10.1002/med.21257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Wang X. J., Hayes J. D., Henderson C. J., Wolf C. R. Identification of retinoic acid as an inhibitor of transcription factor Nrf2 through activation of retinoic acid receptor alpha. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(49):19589–19594. doi: 10.1073/pnas.0709483104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Valenzuela M., Glorieux C., Stockis J., et al. Retinoic acid synergizes ATO-mediated cytotoxicity by precluding Nrf2 activity in AML cells. British Journal of Cancer. 2014;111(5):874–882. doi: 10.1038/bjc.2014.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Sadeghi N., Abbruzzese J. L., Yeung S.-C. J., Hassan M., Li D. Metformin use is associated with better survival of diabetic patients with pancreatic cancer. Clinical Cancer Research. 2012;18(10):2905–2912. doi: 10.1158/1078-0432.CCR-11-2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.MacLeod A. K., Mcmahon M., Plummer S. M., et al. Characterization of the cancer chemopreventive NRF2-dependent gene battery in human keratinocytes: demonstration that the KEAP1-NRF2 pathway, and not the BACH1-NRF2 pathway, controls cytoprotection against electrophiles as well as redox-cycling compounds. Carcinogenesis. 2009;30(9):1571–1580. doi: 10.1093/carcin/bgp176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Jung K.-A., Choi B.-H., Nam C.-W., et al. Identification of aldo-keto reductases as NRF2-target marker genes in human cells. Toxicology Letters. 2013;218(1):39–49. doi: 10.1016/j.toxlet.2012.12.026. [DOI] [PubMed] [Google Scholar]
- 190.Agyeman A. S., Chaerkady R., Shaw P. G., et al. Transcriptomic and proteomic profiling of KEAP1 disrupted and sulforaphane-treated human breast epithelial cells reveals common expression profiles. Breast Cancer Research and Treatment. 2012;132(1):175–187. doi: 10.1007/s10549-011-1536-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Paek J., Lo J. Y., Narasimhan S. D., et al. Mitochondrial SKN-1/Nrf mediates a conserved starvation response. Cell Metabolism. 2012;16(4):526–537. doi: 10.1016/j.cmet.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Chanas S. A., Jiang Q., McMahon M., et al. Loss of the Nrf2 transcription factor causes a marked reduction in constitutive and inducible expression of the glutathione S-transferase Gsta1, Gsta2, Gstm1, Gstm2, Gstm3 and Gstm4 genes in the livers of male and female mice. Biochemical Journal. 2002;365(2):405–416. doi: 10.1042/bj20020320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Maher J. M., Dieter M. Z., Aleksunes L. M., et al. Oxidative and electrophilic stress induces multidrug resistance-associated protein transporters via the nuclear factor-E2-related factor-2 transcriptional pathway. Hepatology. 2007;46(5):1597–1610. doi: 10.1002/hep.21831. [DOI] [PubMed] [Google Scholar]
- 194.Yates M. S., Tran Q. T., Dolan P. M., et al. Genetic versus chemoprotective activation of Nrf2 signaling: overlapping yet distinct gene expression profiles between Keap1 knockout and triterpenoid-treated mice. Carcinogenesis. 2009;30(6):1024–1031. doi: 10.1093/carcin/bgp100. [DOI] [PMC free article] [PubMed] [Google Scholar]