Abstract
In the nearly five decades since its accidental discovery, adeno-associated virus (AAV) has emerged as a highly versatile vector system for both research and clinical applications. A broad range of natural serotypes, as well as an increasing number of capsid variants, has combined to produce a repertoire of vectors with different tissue tropisms, immunogenic profiles and transduction efficiencies. The story of AAV is one of continued progress and surprising discoveries in a viral system that, at first glance, is deceptively simple. This apparent simplicity has enabled the advancement of AAV into the clinic, where despite some challenges it has provided hope for patients and a promising new tool for physicians. Although a great deal of work remains to be done, both in studying the basic biology of AAV and in optimizing its clinical application, AAV vectors are currently the safest and most efficient platform for gene transfer in mammalian cells.
WILD-TYPE ADENO-ASSOCIATED VIRUS DISCOVERY TO VECTORIZATION
Adeno-associated virus discovery
In 1965, Atchinson and colleagues observed a small (25 nm) contaminant particle within electron micrographs of their adenovirus preparations (1). These contaminants were purified from adenovirus and applied to cells. They were shown to be nonautonomous, as particle production also required adenovirus coinfection (1). These defective, small particles were therefore named adeno-associated virus (AAV) which, even to this day (over 50 years later), remains one of the smallest viruses known to man. Consequently, AAV is a very simple virus with a protein capsid composed of 60 capsid subunits, and a ~4.7-kb single-stranded linear DNA genome that is framed by inverted terminal repeat sequences (ITRs) (2). Both polarities of the single-stranded genome are individually packaged at similar efficiencies (3). There are three AAV genes identified to date, which collectively mediate genome replication, site-specific integration, capsid production, and genome packaging (4–8). Twelve natural serotypes of AAV have been reported with many additional variants; however, the last 30 years have seen the most work done with AAV serotype 2 because of its amenability to cell culture.
In the case of wild-type AAV, the coordinated expression of viral genes involves complex self-regulatory feedback loops and appears to be mediated primarily by AAV gene products (9). The employment of these regulatory schemes controls the lysogenic or replicative phase of the viral life cycle which is induced by the presence of a helper virus. Although, AAV was originally described as an adenovirus contaminant, it was later determined that additional viruses, and cellular stress in general, also provide helper functions to induce the AAV replication phase decision (10). AAV is unique because it represents the only known case of site-specific integration in the human chromosome (5, 6); a process that is initiated when AAV infection occurs without a helper virus. In this process the AAV Rep protein interacts with the ITRs on the AAV genome and a very similar sequence (termed AAVS1) uniquely found on human chromosome 19 (reviewed in reference 10). It is thought that the endonuclease activity of the Rep protein mediates strand scission at AAVS1, facilitating host DNA polymerase strand switching, viral genome replication, and subsequent imprecise integration as both a monomer and concatamers in an approximate 1-kb region of AAVS1 (10, 11). The latent AAV genome remains primarily quiescent until favorable helper conditions shift viral gene expression in a manner that favors genome excision and entry into the replication phase (10). AAV particle escape relies on the disruption of cellular membranes by helper-virus-induced cell lysis and initiates a new round of the AAV life cycle.
ADENO-ASSOCIATED VIRUS COMPONENTS/VECTOROLOGY
Adeno-associated virus capsid
AAV virions have icosahedral symmetry with a triangulation number of 1 (T = 1), representing the simplest structure of all viruses. The capsid surface contains three axes of symmetry; two-fold, three-fold and five-fold. The two-fold axis is characterized by a valley-like depression and represents the thinnest capsid cross-section with the lowest number of contacts between capsid protein monomers in the AAV capsid. The three-fold axis is characterized by prominent spike-like protrusions and contains the known receptor binding sites (12, 13) as well as many of the identified antibody recognition sites (14, 15). Finally the five-fold axis is comprised of a low, flat pentamer with a prominent central pore that is surrounded by an elevated rim. This pore is the site of genome packaging (16) and is thought to be necessary for the externalization of N-terminal minor capsid protein motifs necessary for AAV infection (16, 17). Three AAV capsid proteins (VPs) have been identified that are transcribed from the P40 promoter (Figure 1). The P40 transcripts undergo slicing, with one major and one minor splice variant produced. The VPs of all AAVs share a common C-terminus, with VP1 (87 kDa), and VP2 (73 kDa) each having additional N-terminal sequences when compared with VP3 (62 kDa) (18, 19). VP3 represents nearly ~90% of the total protein of intact virions, whereas VP1 and VP2 together represent the remaining ~10% (18, 20, 21). Only the minor splice variant from the P40 transcript contains the VP1 translational start codon (AUG), so explaining its lower production levels compared with VP3. Likewise, whereas the major splice variant contains translational initiation sites for VP2 and VP3, only the latter uses a traditional AUG whereas the former uses an ACG, which is frequently skipped; this accounts for VP2's lower production levels. Although VP3 alone is capable of assembling into a capsid, the presence of VP1 is critical for successful infection because of the enzymatic and targeting motifs in the 135-amino-acid N-terminal region that is unique to VP1. In contrast, VP2 has a much lower effect on infection, to the point that it is considered nonessential, and can tolerate the addition of large peptides to its N-terminus (22) (a fact further discussed in the AAV receptors section). To date, 12 serotypes (AAV1 to AAV12) and more than 100 variants have been isolated from human and nonhuman primate tissue samples (23). Among the 12 serotypes, AAV serotypes 1–9 are currently being developed as gene therapy vectors because of their broad tissue tropisms. A review of these serotypes and their receptor preferences is given in Table 1.
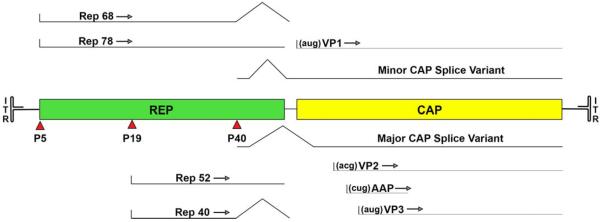
FIGURE 1.
The adeno-associated virus (AAV) genome is a linear ~4.7-kb single-stranded DNA which is flanked by inverted terminal repeats. The genome contains three promoters that drive transcription of the viruses replication (REP), capsid (CAP) and assembly (AAP) genes. The first two promoters, p5 and p19, drive transcription of the large (78/68 kDa) and small (52/40 kDa) Rep proteins, respectively. In each case an alternate splice at the end of each transcript results in the smaller variant of each protein (Rep68 and Rep40). Transcription from the p40 promoter produces one large mRNA with an intron. A minor splice variant of this message contains the translational initiation codon (AUG) for the largest capsid protein (VP1), while the major splice variant truncates this sequence. The major splice variant contains a nontraditional translation initiation codon (ACG) at the start of VP2, which is often skipped for the downstream AUG of VP3, the smallest and most abundant capsid protein. The p40 transcripts also contain an alternate reading frame that encodes the assembly activating protein (AAP), which is translated from a nontraditional CUG. doi:10.1128/microbiolspec.MDNA3-0052-2014.f1
TABLE 1.
Known receptor usage for adeno-associated virus (AAV) serotypes 1–12
| Serotype | Receptor/Co-Receptor |
|---|---|
| AAV1 | α2–3/α2–6 N-linked SA (69, 178, 226) |
| AAV2 | HSPG (70), FGFR1 (227), HGFR (228), Integrin αvβ5/α5β1 (229, 230), 37/67 kDa LamR (231) |
| AAV3 | HSPG (70), 37/67 kDa LamR (231), HGFR (232) |
| AAV4 | α2–3 O-linked SA (71, 72) |
| AAV5 | α2–3 N-linked SA (71, 72), PDGFR (233) |
| AAV6 | HSPG, α2–3/ α2–6 N-linked SA (69, 234), EGFR (235) |
| AAV7 | Undetermined |
| AAV8 | 37/67 kDa LamR (231) |
| AAV9 | Galactose (73, 236), 37/67 kDa LamR (231) |
| AAVrh10 | Undetermined |
| AAV11 | Undetermined |
| AAV12 | Undetermined |
Note: Does not include mutated variants with alternate receptor usage.
FGFR1, fibroblast growth factor receptor; HGFR, hepatocyte growth factor receptor; HSPG, heparan sulfate proteoglycan; PDGFR, platelet-derived growth factor receptor; SA, sialic acid.
To date, the crystal structure of AAVs 1–9 have been determined using X-ray crystallography (24–36). Co-crystal structures of AAV serotypes 2, 5, and 6 with respective capsid receptors (e.g., heparin sulfate and sialic acid) have furthered the molecular understanding of AAV virus/receptor interactions (37–40). In solving the structures of intact AAV capsids, only the common C-terminal-most ~520 amino acids are typically observed (VP3 common region), with the remainder of the sequence (VP1 and VP2 unique regions) believed to either be disordered or in a nonicosahedral symmetry, which is assumed when solving these structures. When the AAV capsid structure is compared to other parvoviruses (adenovirus, parvovirus B19, canine parvovirus, feline parvovirus, minute virus of mice, and porcine parvovirus), the core of the protein is comprised of a remarkably conserved eight-stranded anti-parallel β-barrel motif among all parvoviruses. Structural differences between AAVs and other parvoviruses can primarily be mapped to their surface topology, which accounts for their binding to different receptors and the resulting tissue tropisms (Table 1). The surface loops equivalent to those making up the three-fold protrusions were found to be the most variable among serotypes. This variable region spans the center of the primary capsid sequence (residues ~440 to 600, AAV2 VP1 numbering); whereas residues located at the N- and C-termini are conserved (27). Variations within these loop regions are also thought to eliminate antibody recognition of serotypes AAV1 and AAV3–9 (41, 42). These observations highlight the conservation in capsid structure and yet illustrate the significant diversity of AAV serotype capsids and their ability to exploit multiple cell surface receptors, mediate successful intracellular trafficking with altered efficiencies/kinetics, and elicit specific humoral immunity.
Adeno-associated virus genome
The 4.679-kb AAV2 genome displays the rep, cap, and aap genes in the sense orientation, flanked on either end by 145 nucleotide ITRs. There are three promoter sites (Figure 1), P5, P19, and P40. The first two promoters drive transcription of the large (78/68 kDa) and small (52/40 kDa) replication (Rep) proteins, with the large Rep proteins containing a unique N-terminal domain in addition to the sequence shared with the small Reps. The smaller variants of both the large and small Reps are produced by splicing the P5 and P19 transcripts (Figure 1). As previously mentioned, the P40 promoter is responsible for producing transcripts that are spliced and translated into VP1–3. Recently, an additional open reading frame involved in particle assembly, termed the assembly activating protein (AAP), was found nested within the cap gene (8) (Figure 3). AAP is thought to directly interact with the capsid protein's conserved C-terminus, serving as an assembly scaffold (8, 60).
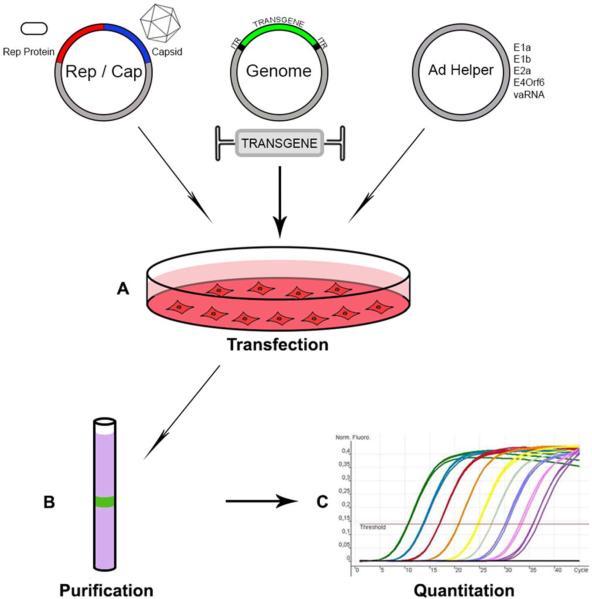
FIGURE 3.
Adeno-associated virus (AAV) vector production. (A) Tradition AAV vector production begins with transfection of mammalian cells (commonly HEK 293) with three plasmids. The first provides the Cap proteins from the chosen AAV serotype in conjunction with Rep from AAV2. This plasmid lacks inverted terminal repeats (ITRs), ensuring that the Rep/Cap sequences are not packaged into AAV capsids and no replication-competent virus is made. The Genome plasmid contains the chosen transgene sequence flanked by ITRs, which are necessary for packaging and genome replication. In the case of ssDNA vectors, transgene cassettes up to 5 kbp can be packaged with high efficiency, whereas scDNA vectors can accommodate cassettes half that size. The third plasmid provides in trans the adenovirus genes that are necessary for AAV replication. It is common to see this plasmid and the Rep/Cap plasmid combined in a single large construct for simplified production. (B) After 48–72 hr cells are harvested and lysed. Vectors can be purified by either column chromatography or density gradient centrifugation, which can separate AAV from contaminating cellular proteins as well as separating empty capsids from genome-containing particles. It is not uncommon to see at least two purification steps employed for high purity vectors and column-based methods can vary depending on the serotype being purified. (C) Vector quantitation is most often performed by measuring DNase-resistant genomes by real-time PCR. In cases where verification is needed for the separation of full and empty capsids, this genome titering is combined with an ELISA-based assay for capsid protein. doi:10.1128/microbiolspec.MDNA3-0052-2014.f3
The AAV ITRs can assume a T-shaped structure (43) and exist in two conformations termed “flip” or “flop” and generated during the rolling hairpin mechanism of replication (44). There are three salient features of the AAV ITR that are required for vector production: (i) a 16-nucleotide tetrameric repeat (GAGC) sequence that is specifically bound by the Rep68/78 proteins termed the Rep-binding element (RBE), (ii) a sequence within the ITR internal hairpin that serves to orient Rep68/78 towards its resolution site termed RBE' (45–49), and (iii) the terminal resolution sequence (trs), which is the site of the strand-specific Rep68/78 endonuclease activity (44, 50–52). The N-termini of the larger Rep proteins contain a DNA-binding domain, as well as strand-specific endonuclease activity, necessary for initiating replication during virion production and site-specific integration (46, 53–56). The C-terminal portion of Rep68/78 is required for multimerization and confers helicase activity which, in part, extrudes a putative nicking stem within the ITR sequence (54, 57, 58). This conformational change presents the trs loop structure to the endonuclease domain of RBE-bound Rep68/78. Rep then induces a strand-specific nick at the trs forming a 5′-phosphotyrosyl tether between Rep and the trs (52, 54, 59). In AAV genome replication, this allows for the resolution of the double-stranded intermediate and the subsequent replication of both ITRs (Figure 2). The smaller Rep proteins (Rep 52 and 40) also maintain these C-terminal properties. The temporal regulation of AAV gene expression is, in large part, controlled by Rep68/78 but is specific to the intracellular environment (reviewed in reference 10).
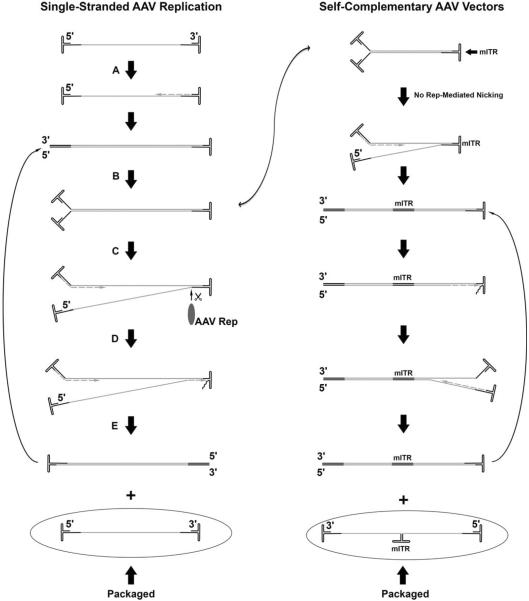
FIGURE 2.
Adeno-associated virus (AAV) genome replication is primed by the 3′ inverted terminal repeat (ITR), and primarily uses the host pol δ (A). The genome is replicated through the 5′ ITR (via strand displacement), yielding a double-stranded (dsDNA) intermediate (B). The newly generated 3′ ITR (produced when copying the 5′ ITR) then primes replication in the opposite direction (C). At the same time, the AAV Rep protein, bindings to the Rep-binding element (RBE) on the original 3′ ITR, and cuts the lower strand at the terminal resolution site (trs), generating a free 3′ end (C), which allows for the replication of the original 3′ ITR (D) and the production of a new single-stranded (ssDNA) genome as well as another dsDNA intermediate (E) which is fed back into the replication cascade. This replication mechanism results in the production of ssDNA genomes of both polarities, which are then packaged into the capsid. In the case of self-complementary (scDNA) vectors, one of the ITRs is mutanted (mITR) to remove the trs, and prevent resolution of the double-stranded intermediate. This resulting replication scheme (detailed in the right column) yields a self-complementary genome that at its center contains the mITR, which is never nicked and which serves as an intermolecular hinge. This hinge acts to form a duplex genome immediately upon uncoating and bypasses the typical need for second-strand synthesis that serves as a bottleneck for ssDNA vectors. doi:10.1128/microbiolspec.MDNA3-0052-2014.f2
Adeno-associated virus vectorology
In 1982, the entire AAV2 genome was cloned into the pBR322 plasmid (pAAV). In that work it was demonstrated that transfection of pAAV into human cells, in the presence of adenovirus, resulted in the production of AAV2 particles (4). Shortly thereafter, it was shown that the viral genes (rep, cap, and aap) were not necessary in cis for genome replication and particle production, and were effectively complemented by another plasmid without the AAV ITRs (129). The key observation was that a transgenic sequence placed between the 145-nucleotide plasmid-borne ITRs could be used for AAV “vector” production in the presence of Rep, Cap, and AAP supplied in trans (61, 62). This result suggested that AAV capsids did not have genomic packaging signal sequences required for virion production. Additionally, at the time, these data made AAV vectors among the most efficient mobile contexts for human gene delivery, and did so in the complete absence of any viral genes (61, 62). Furthermore, in the absence of the Rep proteins, site-specific integration at AAVS1 did not occur and therefore, the majority of AAV vector genomes persisted as stable episomes with low levels of “random” integration (reviewed in reference 10).
The initial demonstrations of recombinant AAV (rAAV) production, however, were complicated by the presence of the now “contaminating” adenovirus particles, which initially appeared necessary for rAAV production because they provided the necessary “helper” functions. To overcome this limitation, the necessary adenoviral genes were cloned and supplied on a plasmid, so eliminating contaminating adenovirus particles from rAAV production (62, 63). Despite over 30 years of research on AAV vector production, this “helper-free” rAAV production strategy remains the most widely used method and relies, in its most basic embodiment, on transfection with three plasmids: (i) pRepCap (provides the viral genome excluding the ITRs, (ii) pITR-sequence* (provides a variable sequence to be packaged as the transgenic genome), (iii) pAdeno-help (provides the helper functions for vector genome replication) (Figure 3); reviewed else-where (64).
ADENO-ASSOCIATED VIRUS VECTOR TRANSDUCTION AND OPTIMIZATION
Adeno-associated virus receptors
Most of our current knowledge of AAV receptor usage comes from cell culture studies that have used a combination of biochemical and genetic tools. AAVs exploit several different types of glycans as primary attachment receptors and use a broad range of cell surface receptors to mediate cellular uptake. Receptor usage varies by serotype, though heparan sulfate proteoglycan, terminal galactose (Gal), and several linkage variants of sialic acid (SA) constitute the currently identified glycan repertoire for known AAVs (Table 1). The unique combination of specific glycan and entry receptor affinity is believed to be the primary cause for the different tissue tropisms seen among the serotypes, although there is evidence for post-entry restriction in some cell/tissue types (65–68). Structural, biochemical, and genetic studies have helped to identify the glycan-binding regions for AAV1–6 (69–72) and AAV9 (73). This has enabled a wide variety of experiments that have directly used this structural information in an attempt to alter the natural trafficking patterns of AAV. As an example, a single mutation (AAV2 585E) in the AAV2 heparin-binding footprint abolishes AAV2 heparin binding and results in a different transduction profile (cardiac versus hepatic tropism) in vivo (74). As a natural extension of such results, rational design studies of AAV capsids have looked at swapping the critical motifs for primary receptor binding sites between different serotypes and in one example, demonstrated that an AAV2/AAV8 chimera (AAV2i8) displays an altered transduction profile as the result of such a swap. AAV2i8 selectively transduces cardiac and whole-body skeletal muscles with high efficiency while losing liver tropism (75). Further studies integrating the AAV9 Gal binding residues into the AAV2 capsid (AAV2G9) have found that AAV2G9 has dual receptor function and exploits Gal and heparan sulfate receptors for infection (76). Of particular interest, AAV2G9 retains a similar tropism to AAV2 but confers more rapid onset and higher liver transgene expression in mice. The strategy of domain swaps continues to be actively used and is just one tool available for generating custom tropism.
Further efforts to tailor AAV tropism and to generate AAV capsids with unique properties have exploited two broad strategies: capsid modification with targeting molecules and generation of novel AAV capsids through directed evolution. Efforts in the former have included the insertion of targeting domains such as integrin-binding motifs (77), antibody domains (78), and other targeting peptides (79–83) into the outer variable loops of AAV. In each case, there were some promising results with novel tropism or the ability to infect previously refractory cell types. Unfortunately these successes were modest in their ability to infect new cells in culture and even more so in vivo, and more often resulted in viruses that were defective for infection. Subsequent work has shown that insertional modification is highly sequence specific (79, 84), suggesting that it was difficult to predict sites and peptide candidates for successful large capsid insertions. As AAV is a small and relatively simple virus it is not surprising that large insertions into the capsid proteins are often difficult to tolerate. An alternative strategy for retargeting has used the fact that VP2 is both nonessential and able to tolerate large additional sequences at its N-terminus, which would subsequently be displayed on the capsid surface (22, 85). Although this has been a successful strategy for decorating the capsid with a myriad of proteins such as GFP, the main pitfalls of this approach have traditionally been the retention of native capsid tropism, which competes with any re-targeting molecule and low copy number of targeting molecules. This latter problem is the result of VP2 being a minor capsid protein, and while recent efforts (86, 87) have combined this VP2-based approach with de-targeted vectors with some success, the problem of low copy number remains. The upper size limit for VP2-based tags is also not clearly defined and appears to be at least partially sequence dependent.
Conversely, directed evolution as a strategy has sought to generate new capsid properties without previous mechanistic knowledge. Early directed evolution strategies relied on phage display libraries to generate peptides that would bind to a desired target and that would then be inserted into the AAV capsid (88–93). Such strategies are an extension of the capsid insertion approach previously discussed and also had to contend with the fact that successful peptide capsid insertion is highly sequence specific. Hence, subsequent attempts transitioned to random peptide insertion followed by selection to ensure that peptide inserts would bind their molecular targets in the context of the capsid (83, 94–99). Others have taken a more holistic directed evolution approach by generating libraries of novel AAV sequences either by random mutagenesis (100–102) or shuffling of capsid sequences across multiple serotypes followed by selection on either a target cell population (99, 103–107) or in vivo (108–113). The latter approach in particular has been successful in yielding capsids with conserved and functional interiors and novel tropism. Although AAV offers a compelling platform for gene therapy, the rigorous and widely varying demands of a broad range of clinical applications will ensure that the efforts to augment and tailor the natural biology of AAV will continue.
Adeno-associated virus vector trafficking/uncoating
A broad overview of recombinant AAV transduction is provided in Figure 4. This is believed to be the same process as occurs with wild-type infection, with the exception of gene transcription, which in wild-type virus is controlled by the genome and in recombinant vectors is dictated by the promoter in a given expression cassette. AAV has traditionally been thought to exploit clathrin-mediated, dynamin-dependent endocytosis (114–118). However, a growing body of work suggests that this is only one of several entry pathways that different AAVs can use with evidence implicating the caveolin pathway (119, 120), as well as the clathrin-independent carrier mediated/glycosylphosphatidylinositol-anchored-protein-enriched endosomal compartment-associated (CLIC/GEEC) pathway (121, 122). There is evidence to suggest that different AAV serotypes may be able to use different entry pathways based on their native receptor tropism, which may help to explain the post-entry restriction seen in some cell types (65–68, 123). Furthermore, although ablation of native receptor binding often results in an attenuation of infection in cell culture (AAV2 non-heparan sulfate proteoglycan-binding mutants), the same effect is not always seen in vivo, where mutants often show altered tissue tropism instead of an outright defective phenotype (74, 103, 124). AAV trafficking post-entry has been associated with a broad range of organelles from both the endosomal (88, 116, 121) and caveolin (120, 125) pathways, and appears to be influenced by a wide range of factors such as initial route of entry and initial infecting dose (126, 127). Despite the relative uncertainty over the exact trafficking route of AAV, all productive pathways share the common feature of leading to an acidified compartment. AAV trafficking through acidified endosomal compartments is required for infection. Either a pharmacological block of endosomal acidification (116, 121, 128, 129) or a bypass of the endosomal system via microinjection of AAV into the cytoplasm (3, 4, 88, 130–133), results in a failure to achieve vector transgene expression. An acidic environment has been shown to lead to structural transitions in the AAV capsid (85, 134), which activate an autocatalytic protease activity within the capsid (86, 134–136) and prime the capsid for externalization of the VP1 N-terminal domains (88, 135). The VP1 N-terminus is critical for infection and contains a phospholipase A2 domain (94, 137) along with a broad range of signaling motifs (100, 136, 137) including several nuclear localization sites (103, 104, 135, 137). Although mutation of the PLA2 domain is highly detrimental to infection and results in the perinuclear accumulation of virus, it also appears by itself to be insufficient to facilitate endosomal escape of the 3-MDa parvovirus capsid (108, 110, 112, 138, 139). It may however enable the virus to display the signaling motifs of the V1 N-terminus on the exterior of small vesicles, so enabling AAV to guide its own trafficking to the perinuclear region along the microtubule network while still contained in vesicles (114, 140, 141). Whatever the exact mechanism of endosomal escape, AAV does enter the cytoplasm where a portion of the infecting virions is subjected to phosphorylation and targeting for proteasome-mediated degradation (119, 142–144) The use of proteasome-inhibiting drugs (121, 140, 141) and surface tyrosine mutants (123, 135, 141) both result in notable increases in AAV transduction. AAV enters the nucleus as an intact capsid, most likely via active transport across the nuclear pore complex (103, 124, 141). The presence of the intact AAV capsid in the nucleus is necessary for productive infection (88, 124, 145, 146) and a carefully choreographed trafficking of the capsid to and from the nucleolus appears to influence the timing of uncoating and the success of vector-delivered transgene expression (125). There is also some evidence that genome uncoating does not involve the complete disassociation of the capsid and that the capsid may play a role in gene expression after genome uncoating (126).
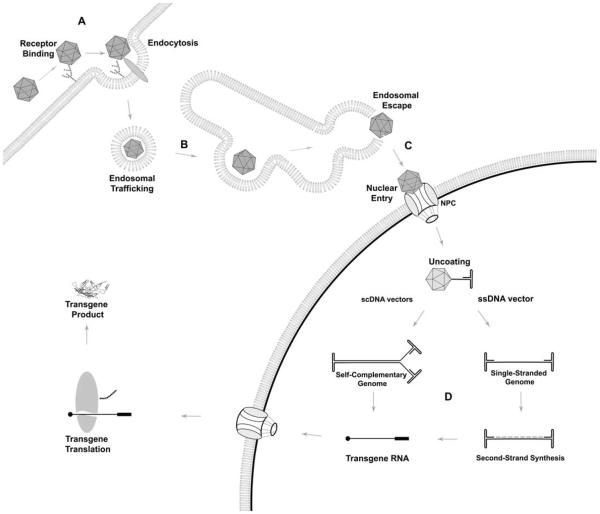
FIGURE 4.
Adeno-associated virus (AAV) vector infectious pathway. (A) Infection is initiated by capsid binding to cell-surface receptors. In the case of AAV serotype 2 this is a two-step process involving binding to a primary glycan receptor followed by binding to a protein receptor that mediates endocytosis. Classical AAV2 internalization is believed to be a clathrin-mediated, dynamin-dependent process, although other pathways have been implicated. (B) AAV traffics through the endosomal system where it is exposed to a low-pH environment that triggers the externalization of the VP1/2 unique regions and activation of the auto-catalytic capsid protease. Exposure to the low-pH environment is necessary for productive AAV infection. (C) Escape from the endosomal compartment is mediated by the VP1 PLA2 domain, and AAV uses several nuclear localization signaling motifs to traffic to the nucleus and enter, most likely via the nuclear pore complex (NPC). (D) AAV uncoats its genome in the nucleus. In the case of single-stranded DNA vectors, second-strand synthesis must first occur before transcription, creating a bottleneck in the transduction process. Self-complementary (sc) vectors use a single mutant inverted terminal repeat (ITR) in conjunction with both + and − strands to produce a genome that is double-stranded upon uncoating so bypassing second-strand synthesis, albeit at the cost of lower packaging capacity. After a double-stranded genome is formed transcription and translation are driven by the transgene cassette and are not virus-specific. doi:10.1128/microbiolspec.MDNA3-0052-2014.f4
Adeno-associated virus vector genome fate
Single-strand DNA vector genomes are uncoated within the nucleus and form discrete foci whose abundance directly correlates to the transduction efficiency (128, 147). The generation of a DNA molecule competent for transcription is dependent upon the genome concentration within the nucleus. When vector genomes are limiting, second-strand synthesis mediated by, minimally, DNA polymerase delta, is primed by the 3′ ITR giving rise to a duplexed, yet still single-stranded, genome (Figure 2) (3, 4, 130–133, 142, 146). Additionally, it has been demonstrated that AAV vector genomes, when present in the nucleus at high concentrations, can also form double-strand molecules via opposite polarity single-strand genome annealing (134, 148–150). Regardless of the exact mechanism of second-strand generation, it is apparent that target cell replication is not necessary and that most, if not all, cell types appear to have the necessary functions to allow this aspect of AAV vector transduction. Following the generation of a duplexed DNA molecule, the vector genome primarily remains episomal and circularizes via intra- and inter-molecular ITR linkages shortly after infection (134–136, 142, 151). Using circularization-dependent AAV vectors in dividing cell cultures, it was demonstrated that RecQ helicase family members, Mre11, Nbs1, and ATM are required for efficient vector genome circularization (135, 148, 151). In nondividing skeletal muscle fibers, ATM and DNA-PKcs were necessary for genome circularization wherease Nbs1 was not (137, 151, 152). Consistent with these in vivo data, elegant work performed in the mouse liver demonstrated that Artemis and DNA-PKcs process the AAV ITRs and promote the circularization/concatamerization of vector genomes, which is also supported by other reports (62, 136, 137, 153–156). Collectively, with regards to the generation of persistent AAV vector episomes, it appears that the circularization event is probably mediated at the ITRs by different DNA repair pathways; homology directed repair in dividing cells and nonhomologous end joining (NHEJ) in terminally differentiated tissues (135, 137, 157). One of the unique features of AAV vector transduction is the remarkably long-term transgene expression from the AAV vector episome (138, 139, 158). In general, these molecules are resistant to overt transcriptional silencing and have been demonstrated to produce the transgene product for over 4 years in humans following a single injection.
It is well appreciated that the rAAV transduction elicits a DNA damage response within the host cell, which influences the overall efficiency of transduction. In general, several factors have been identified that appear to inhibit, directly or indirectly, rAAV transduction. For instance, rAAV transduction in dividing cells deficient for ATM, a key regulator of double-strand break repair, is dramatically increased, an effect attributed to enhancement of second-strand synthesis (140, 141, 159). Interestingly, several forms of DNA stress (UV, hydroxyurea, mytomycin C) also increase AAV vector transduction (142, 143, 157), although not in the absence of ATM (140, 141, 155, 156), further supporting the notion that ATM inhibits rAAV transduction at the genome level. This supposed inhibitory effect, however, is not necessarily specific to ATM, as DNA-PKcs and Rad52 are also implicated in this process, indicating the involvement of both primary DNA repair pathways, homologous recombination (HR) and NHEJ (135, 141, 155, 156). In the absence of both Ku86 and Rad52, rAAV transduction increases, and consistently both Ku86 and Rad52 have been found to bind AAV genomes following vector transduction (137, 141). In addition, Mre11, a key sensor of damaged DNA normally considered part of the HR pathway, in conjunction with Nbs1 and Rad50, also appears to inhibit AAV transduction (145, 146, 160). A fascinating study in mice demonstrated that downregulation of DNA repair proteins, in particular the Mre11–Rad50–Nbs1 (MRN) complex, via terminal differentiation coincided with increased AAV vector transduction (146, 155, 156, 160). In addition to loss of DNA repair proteins involved in HR during terminal differentiation, cells also exited the cell cycle to a quiescent state, therefore complicating the assignment of discrete roles for particular proteins in the inhibition of rAAV transduction (61, 62, 146). In particular, the role of intact cell cycle checkpoints, and the growth phase in general, also appear to affect transduction and furthermore, confound the direct implication of cell cycle regulators, such as ATM and p53, as controllers of AAV vector transduction. As mentioned above, several genotoxic stresses that also arrest the cell cycle result in increased transduction (9, 147). The mechanism of this enhancement is not completely understood but could be explained by the sequestration of repair factors that are reported to inhibit rAAV transduction away from the AAV genome (such as the MRN complex). However, this explanation, and the actual role of the cell cycle in AAV transduction, is probably not attributable to a single event. For instance, exit from the cell cycle in vivo increases transduction, whereas an earlier report demonstrated that cells in S-phase are transduced much better (200-fold increase) compared with nondividing cells (142, 146, 158). Interestingly, in wild-type diploid cultured cells, AAV transduction has been reported to induce cell cycle arrests at both the G1 and G2/M checkpoints (148–150, 161, 162). However, it has been our observation that in cells defective for particular cell cycle checkpoints, transduction increases initially, perhaps due to a quicker entry into S-phase (142, 151, 161, 162). In fact, in the absence of normal checkpoint functions, such as in p53-deficient cells or human embryonic stem cells, it has been reported that AAV vector transduction results in cell death (148, 151, 163). The reasons for the rAAV mediated cytotoxicity remain unclear and are probably cell-type-dependent; however, ITRs have been implicated as stalled replication forks and/or perhaps forming G-quadruplex/quartets, both of which would induce DNA damage cascades from human telomeres (151, 152, 164).
Although the majority of AAV vector genomes persist as episomes, as with all exogenous DNA, a low percentage will undergo illegitimate host genome integration (62, 153–156, 164–166). One major factor influencing this largely undesirable event is whether or not the cells are actively replicating/dividing (157, 167). It has been reported in dividing cell cultures that illegitimate integration of AAV vector genomes occurs at DSBs within the host genome and that efficiencies can be as high as 1% of transduced cells (158, 166, 167). Furthermore, induced DSBs, either by general genotoxic stress or by a site-specific endonuclease, resulted in the illegitimate integration of rAAV genomes with microhomologies, deletions, and insertions present at the integration junctions (159, 168–171). In contrast, an early investigation in a nondividing bronchial epithelial cell line demonstrated no AAV vector integration despite transduction in over 90% of the cells (157, 158). In addition to cellular replication, AAV vector integration prefers CpG islands and demonstrates a modest inclination towards actively transcribed regions (155, 156, 172). Consistently, a bias for vector integration was demonstrated at ribosomal DNA repeats (155, 156, 172). A large analysis of rAAV integration events in multiple murine tissues found that 30% of all integration events occurred near DNA palindromes, which are known sites of chromosome instability (137, 173). Another report found that AAV vector integration in mice occurred, in all cases, in actively transcribed genes, further demonstrating the potential risk of oncogenesis following insertional mutagenesis (160, 174). Hence, it appears that AAV vector integration is not random, and the efficiency probably depends on the species, the cell type, and the phase of the cell cycle (155, 156, 160, 174). These collective reports imply that fragile DNA sequences and or replication-induced damage of the host chromosome both facilitate the incorporation of AAV vector DNA, and probably exogenous DNA in general.
ADENO-ASSOCIATED VIRUS VECTOR APPLICATIONS
Adeno-associated virus vectors have been employed as DNA delivery vehicles for diverse applications with a primary focus on gene addition and gene editing and strategies.
Gene addition
The vectorization of AAV described above was the seminal discovery for mobile DNA applications in mammalian cells including clinical gene therapy (61, 62, 175), reviewed in references 9 and 175. Since then, the most popular use of AAV vectors has been gene addition strategies in which a gene, or more often cDNA, is placed within an expression cassette and packaged in the virion. The design and production of such vectors is straightforward when adherence to the AAV capsid packaging capacity (<5 kb) is respected. Key elements for these gene addition vectors include a promoter, cDNA or a gene, followed by a poly-adenylation sequence situated between the AAV ITRs (as depicted in Figure 2). Considering these elements, customization towards the specific application is possible (i.e. tissue-restricted promoters); however, as these sequences are not specific to AAV vectors they will not be discussed here. Regarding AAV vector-specific enhancements for gene addition applications, a pivotal advancement was spurred by the observation that transgenic cassettes less than half the size of the AAV genome (≤2.3 kb) packaged both monomer and dimer single-strand DNA species that had the capability to form a duplex vector genome through antiparallel single strand complementarity (158, 174, 176). Shortly thereafter, two reports demonstrated that these dimer species could be intentionally induced using a plasmid containing a wild-type ITR sequence, transgenic DNA (≤2.3 kb) and a truncated ITR in which the trs is deleted and therefore, is deficient for Rep-induced resolution of the typical double-stranded AAV genome intermediate (Figure 2) (161, 162, 177). Effectively, the deleted ITR functions as an intra-genomic hinge, promoting the base pairing of antiparallel DNA arms capable of duplex formation (Figure 2), and as such were termed self-complementary AAV (41, 161, 162, 178). These vectors have consistently demonstrated earlier transgene onset and in general, a more than five-fold enhancement compared with single-strand AAV vector transduction (reviewed in reference 163). In hemophilia B patients, the increased transduction efficiency by self-complementary AAV vectors has effectively allowed therapeutic correction at lower administered doses, which importantly decreases immunological complications. On the other end of the spectrum, AAV large gene (>5 kb) transduction has been extensively studied (reviewed in reference 164). These approaches all rely on host-mediated assembly of partial transgene cassettes and are currently of three general types: (i) overlapping vectors, (ii) trans-splicing vectors, and (iii) fragment AAV vectors. Regarding the most efficient strategy for AAV large gene delivery, reports have been somewhat conflicting (164–166). A recent report demonstrated that the repair mechanism of trans-splicing AAV and fragment AAV vectors for large gene transduction is inherently different (167), suggesting that the observed differences may be attributed to the specific DNA repair pathways present in different tissues (166, 167). In general, all of the collective approaches can mediate large gene transduction in vivo, albeit at a significantly decreased efficiency when compared with intact AAV vectors.
Gene editing
Another application of rAAV is as a tool for gene editing of specific sites in the human chromosome via HR. Notably, work primarily from the Russell laboratory has demonstrated that rAAV genomes are enhanced (up to 10,000-fold) for gene editing compared with other substrates, such as plasmid DNA whose efficiency of gene editing is approximately 1 in a million (168–171). In the process of rAAV-mediated gene editing, a repair sequence is used as the transgenic DNA that exhibits homologous arms to the target site with the desired modification preferably placed near the middle of the homology (158). Once inside the nucleus, the vector DNA may interact with chromosomal regions of homology, which stimulates repair via synthesis-dependent gene conversion and/or by direct strand incorporation (172). The reliance on the Rad51/54 pathway (172) suggests that this pathway is classical HR and, as such, would restrict AAV gene editing to cycling cells, which has been reported (173). Despite this academic understanding of HR, AAV gene editing has corrected a murine model of hereditary tyrosinemia type 1 (174). In this model, AAV gene-edited cells have a selective survival advantage, which is important to increase the relatively low correction efficiencies. As there appears to be a balance between HR and NHEJ, Paulk et al. elegantly demonstrated that genetic or drug inhibition of NHEJ increased AAV gene editing nearly 10-fold (174). AAV gene editing has also been reported as a potential treatment for epidermolysis bullosa simplex (175). In that work, AAV gene editing was coupled with a promoter trap clonal recovery strategy to correct the KRT14 mutation, a mutation in the basal epidermal keratin 14 protein, ex vivo in patient keratinocytes. The corrected cells were then expanded to levels relevant for skin transplant in these patients (175). These in vivo results suggest that AAV gene editing via HR occurs in the diseased liver and keratinocytes and have the ability to permanently correct disease mutations at the level of the host chromosome, especially when a selection scheme is employed (174, 176).
ADENO-ASSOCIATED VIRUS IN THE CLINIC
Adeno-associated virus is naturally replication deficient, and requires a helper virus to complete its life cycle. This fact, along with a lack of any known pathogenicity makes it an attractive candidate as a gene transfer vector for both clinical and research applications. The low immunogenic profile (177) and the ability to transencapsidate AAV2 ITRs with different serotype capsids (41, 178), in order to expand vector tissue targeting potential (179, 180), make AAV a versatile tool in the clinic. This versatility is enhanced by AAV's ability to infect both dividing and nondividing cells and to establish stable long-term gene expression (139, 181, 182), with a low risk of deleterious integration into the host genome (reviewed in reference 183). It is therefore not surprising that AAV has been in used in over one hundred clinical trials, for a broad range of conditions including systemic inherited monogenic diseases such as hemophilia (184, 185) and diseases in the central nervous system (186), eye (187–192), muscle (193), and heart (194). These trials have served to repeatedly highlight the safety, efficacy, and versatility of AAV as a clinical tool and its potential as a future widespread therapeutic platform.
The eye, with its immunoprivileged status (195) and plethora of established analytical tests (196), offers an attractive target for gene therapy. One of the prominent recent successes for gene therapy has been the AAV2-based treatment of Leber's congenital amaurosis (LCA). While LCA can be caused by mutation in several genes, it consistently results in the early onset of retinal degeneration and is the most prevalent cause of childhood blindness (197). AAV-based clinical trials have focused on a subset of LCA caused by loss of function mutations in the retinal pigment epithelium-specific protein 65kDa (RPE65) gene (187–192). In a normal eye, photosensitive pigments in the retina mediate the conversion of light to electric signals. One such pigment, 11-cis-retinal is converted to all-trans-retinal when exposed to light, which initiates a chemical cascade that ultimately leads to an electrical signal that can then be interpreted by the brain. RPE65 mediates the conversion of all-trans-retinal to 11-cis-retinal, so recycling this key pigment and allowing for the visual cycle to continue. The therapy was based on the subretinal delivery of an rAAV2 vector carrying a functional copy of RPE65, which restores the conversion of all-trans-retinal to 11-cis-retinal, and would allow for the repeated signaling from photoreceptors (198). So far 30 patients have received this treatment and have demonstrated lasting improvements in visual fields, nystagmus, dark-adapted perimetry, and mobility in low light (187–192). The success of these early trials, and the gene-therapy friendly attributes of the eye have led to eight more clinical trials being initiated to treat retinal diseases with rAAV, including a phase III trial targeting LCA (http://www.abedia.com/wiley).
Although the success of LCA clinical trails is both impressive and encouraging, currently there is only one clinically approved, commercial therapeutic that is based on AAV gene delivery. Glybera® is an AAV1-based therapeutic that delivers a mutant lipoprotein lipase (LPLS447X) gene via intramuscular injection, for the treatment of lipoprotein lipase deficiency (199), an inherited disease that often leads to hyperlipidemia, recurring pancreatitis, and liver failure. The LPL S447X variant is a truncation form of LPL, which naturally occurs in 20% of the population and is associated with increased lipolytic function and an anti-atherogenic lipid profile [reviewed in (200)], so making it ideal as an LPLD therapeutic. It received European Medical Agency (EMA) approval in 2012 for use in patients with genetically confirmed lipoprotein lipase deficiency who have experienced repeated pancreatitis attacks despite dietary restrictions. In clinical trials, Glybera showed a good safety profile and demonstrated efficacy in decreasing the frequency and severity of pancreatitis (199, 201–203). These results are admirable, but continuing work on vector optimization promises to deliver even more effective and targeted AAV-based therapeutics in the future. A glimpse of the progress that has been made in the vector development arena and its direct effect on clinical trial outcomes is afforded by the hemophilia B clinical trial at St. Jude.
ADENO-ASSOCIATED VIRUS CAPSID/TRANSGENE HUMAN IMMUNE RESPONSE
Cell-mediated response
One property of AAV vectors that has been traditionally touted as an advantage has been low immunogenicity, as compared to other viral vectors such as adenovirus. The low efficiency with which AAVs transduce professional antigen-presenting cells such as macrophages and dendritic cells (67, 204, 205) was suspected as the reason behind their relatively low immunogenicity. That said, the production of cytotoxic T lymphocytes (CTLs) in response to capsid protein was documented in animal models transduced with wild-type AAV (206–208); however, such studies determined that the response was a byproduct of artificially overexpressed capsid protein and not relevant in the context of rAAV vectors. Even after initial human trials for hemophilia B revealed clearance of AAV-transduced hepatocytes by suspected anti-capsid CTLs (185), animal models once again failed to replicate the same phenomenon (209). These results highlighted the discrepancy between the inbred murine immune response and that of human patients, suggesting that better tools are necessary for characterizing anti-AAV immunity and evaluating any potential immunomodulatory strategies.
As hinted above, the first clinical trial using hepatic artery infusion of an AAV2-based clotting factor IX (F.IX) vector saw in patients who received the highest vector dose (2 × 1012 vector genomes/kg), clearance of vector-transduced hepatocytes. The same patients were able to initially produce therapeutic levels of F.IX but subsequently experienced an asymptomatic elevation of liver transaminases, accompanied by a drop in F.IX production (185). Follow-up studies revealed that the transaminitis was most likely due to AAV capsid-reactive CTL-killing of transduced hepatocytes. This initial study demonstrated therapeutic effects using AAV and showed that the relatively modest CTL activation appeared to be dose dependent, so leaving open the possibility that this issue could be addressed in the future by immuno-modulation and increased vector efficiency (lower vector dose).
A second clinical trial, used peripheral vein infusion of AAV8, F.IX-expressing vector and was able to produce significantly higher, clinically relevant levels of transgene expression while using the same vector dosing schedule as the first trial (184). As in the previous trial, the two patients receiving the highest vector doses displayed a transient increase in liver enzymes, suggesting the onset of a capsid-targeted CTL response. As this possibility was anticipated, these patients were immuno-suppressed with glucocorticoid therapy and saw a subsequent drop in liver enzymes and a continuation of therapeutic transgene production. This trial confirmed that the human anti-AAV CTL response was dose dependent and that it could potentially be managed with immunosuppressive therapy. Subsequent trials, such as the AAV1-based LPL (210) demonstrated similar dose-dependent CTL activation that appeared to be independent of route of administration.
Humoral response
As AAV is ubiquitous in nature, the majority of the human population (95% of individuals) has been exposed to AAV2, with approximately 50% of the population having capsid neutralizing antibodies (NAbs) that can inhibit rAAV transduction (42, 184, 190, 211–218). The prevalence of NAbs in children is lower, ranging from 13 to 25% (216, 219). As a result, screening for NAbs is a prerequisite before enrollment in AAV2-based clinical trials, with high pre-existing titers being a parameter for exclusion. This criterion immediately limits the broad applicability of rAAV for disease treatment until further optimization generates the ability to evade NAbs in what may likely be a patient-specific manner (patient-specific AAV capsids). Although 11 additional AAV serotypes have been explored for gene therapy purposes, minimal cross-reactivity of NAbs has been demonstrated among these types in animals (218, 220–223). However, in humans, recent studies have shown that different degrees of NAb cross-reactivity exist between AAV2 and other types (213, 214, 220, 224). Collectively, there is a lower prevalence of NAbs against AAV1, ∡5, ∡6, ∡7, and ∡8 than against AAV2, supporting the use of other natural capsids as a means to evade AAV vector neutralization by NAbs (213, 214). Additionally, studies have aimed to map NAb epitopes to the capsid surface (14) in the hope that any conserved epitopes could be mutated away, generating serologically non-reactive vectors. While those efforts are ongoing, initial results, which have mapped multiple antibody epitopes to the three-fold protrusions of the AAV capsid, have suggested that such a mutagenesis based-strategy may result in capsids that are also compromised for infection. Alternately, a decoy strategy has been put forward, using empty vector particles, which are normally present in rAAV preparations and are typically purified out (225). This strategy relied on empty capsids as antibody decoys, and was shown to provide for an increase in transduction in the face of a challenge with NAbs. Although the strategy is simple and effective it does present the obvious shortcoming of introducing even more AAV capsid protein into patients; something that has repeatedly been shown to generate a CTL response as discussed above. As AAV continues to expand into the clinic, and more work is done to characterize the interaction between the vector and host immune systems, new strategies will inevitably emerge to meet the challenge presented by pre-existing NAbs and the anti-capsid CTL response.
REFERENCES
- 1.Atchison RW, Casto BC, Hammon WM. Adenovirus-associated defective virus particles. Science. 1965;149:754–756. doi: 10.1126/science.149.3685.754. [DOI] [PubMed] [Google Scholar]
- 2.Rose JA, Berns KI, Hoggan MD, Koczot FJ. Evidence for a single-stranded adenovirus-associated virus genome: formation of a DNA density hybrid on release of viral DNA. Proc Natl Acad Sci USA. 1969;64:863–869. doi: 10.1073/pnas.64.3.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhong L, Zhou X, Li Y, Qing K, Xiao X, Samulski RJ, Srivastava A. Single-polarity recombinant adeno-associated virus 2 vector-mediated transgene expression in vitro and in vivo: mechanism of transduction. Mol Ther. 2008;16:290–295. doi: 10.1038/sj.mt.6300376. [DOI] [PubMed] [Google Scholar]
- 4.Samulski RJ, Berns KI, Tan M, Muzyczka N. Cloning of adeno-associated virus into pBR322: rescue of intact virus from the recombinant plasmid in human cells. Proc Natl Acad Sci USA. 1982;79:2077–2081. doi: 10.1073/pnas.79.6.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kotin RM, Siniscalco M, Samulski RJ, Zhu XD, Hunter L, Laughlin CA, McLaughlin S, Muzyczka N, Rocchi M, Berns KI. Site-specific integration by adeno-associated virus. Proc Natl Acad Sci USA. 1990;87:2211–2215. doi: 10.1073/pnas.87.6.2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Samulski RJ, Zhu X, Xiao X, Brook JD, Housman DE, Epstein N, Hunter LA. Targeted integration of adeno-associated virus (AAV) into human chromosome 19. EMBO J. 1991;10:3941–3950. doi: 10.1002/j.1460-2075.1991.tb04964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Linden RM, Ward P, Giraud C, Winocour E, Berns KI. Site-specific integration by adeno-associated virus. Proc Natl Acad Sci USA. 1996;93:11288–11294. doi: 10.1073/pnas.93.21.11288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sonntag F, Schmidt K, Kleinschmidt JA. A viral assembly factor promotes AAV2 capsid formation in the nucleolus. Proc Natl Acad Sci USA. 2010;107:10220–10225. doi: 10.1073/pnas.1001673107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mitchell AM, Nicolson SC, Warischalk JK, Samulski RJ. AAV's anatomy: roadmap for optimizing vectors for translational success. Curr Gene Ther. 2010;10:319–340. doi: 10.2174/156652310793180706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCarty DM, Young SM, Samulski RJ. Integration of adeno-associated virus (AAV) and recombinant AAV vectors. Annu Rev Genet. 2004;38:819–845. doi: 10.1146/annurev.genet.37.110801.143717. [DOI] [PubMed] [Google Scholar]
- 11.Young SM, Mccarty DM, Degtyareva N, Samulski RJ. Roles of adeno-associated virus Rep protein and human chromosome 19 in site-specific recombination. J Virol. 2000;74:3953–3966. doi: 10.1128/jvi.74.9.3953-3966.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kern A, Schmidt K, Leder C, Müller OJ, Wobus CE, Bettinger K, Lieth Von der CW, King JA, Kleinschmidt JA. Identification of a heparin-binding motif on adeno-associated virus type 2 capsids. J Virol. 2003;77:11072–11081. doi: 10.1128/JVI.77.20.11072-11081.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Opie SR, Warrington KH, Agbandje-McKenna M, Zolotukhin S, Muzyczka N. Identification of amino acid residues in the capsid proteins of adeno-associated virus type 2 that contribute to heparan sulfate proteoglycan binding. J Virol. 2003;77:6995–7006. doi: 10.1128/JVI.77.12.6995-7006.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gurda BL, Raupp C, Popa-Wagner R, Naumer M, Olson NH, Ng R, McKenna R, Baker TS, Kleinschmidt JA, Agbandje-McKenna M. Mapping a neutralizing epitope onto the capsid of adeno-associated virus serotype 8. J Virol. 2012;86:7739–7751. doi: 10.1128/JVI.00218-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gurda BL, Dimattia MA, Miller EB, Bennett A, McKenna R, Weichert WS, Nelson CD, Chen W-J, Muzyczka N, Olson NH, Sinkovits RS, Chiorini JA, Zolotutkhin S, Kozyreva OG, Samulski RJ, Baker TS, arrish CR, Agbandje-McKenna M. Capsid antibodies to different adeno-associated virus serotypes bind common regions. J Virol. 2013;87:9111–9124. doi: 10.1128/JVI.00622-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bleker S, Sonntag F, Kleinschmidt JA. Mutational analysis of narrow pores at the fivefold symmetry axes of adeno-associated virus type 2 capsids reveals a dual role in genome packaging and activation of phospholipase A2 activity. J Virol. 2005;79:2528–2540. doi: 10.1128/JVI.79.4.2528-2540.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kronenberg S, Böttcher B, Lieth von der CW, Bleker S, Kleinschmidt JA. A conformational change in the adeno-associated virus type 2 capsid leads to the exposure of hidden VP1 N termini. J Virol. 2005;79:5296–5303. doi: 10.1128/JVI.79.9.5296-5303.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muzyczka N. Use of adeno-associated virus as a general transduction vector for mammalian cells. Curr Top Microbiol Immunol. 1992;158:97–129. doi: 10.1007/978-3-642-75608-5_5. [DOI] [PubMed] [Google Scholar]
- 19.Buller RM, Rose JA. Characterization of adenovirus-associated virus-induced polypeptides in KB cells. J Virol. 1978;25:331–338. doi: 10.1128/jvi.25.1.331-338.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wistuba A, Kern A, Weger S, Grimm D, Kleinschmidt JA. Subcellular compartmentalization of adeno-associated virus type 2 assembly. J Virol. 1997;71:1341–1352. doi: 10.1128/jvi.71.2.1341-1352.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rolling F, Samulski RJ. AAV as a viral vector for human gene therapy. Generation of recombinant virus. Mol Biotechnol. 1995;3:9–15. doi: 10.1007/BF02821330. [DOI] [PubMed] [Google Scholar]
- 22.Warrington KH, Gorbatyuk OS, Harrison JK, Opie SR, Zolotukhin S, Muzyczka N. Adeno-associated virus type 2 VP2 capsid protein is nonessential and can tolerate large peptide insertions at its N terminus. J Virol. 2004;78:6595–6609. doi: 10.1128/JVI.78.12.6595-6609.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao G, Vandenberghe LH, Wilson JM. New recombinant serotypes of AAV vectors. Curr Gene Ther. 2005;5:285–297. doi: 10.2174/1566523054065057. [DOI] [PubMed] [Google Scholar]
- 24.Xie Q, Bu W, Bhatia S, Hare J, Somasundaram T, Azzi A, Chapman MS. The atomic structure of adeno-associated virus (AAV-2), a vector for human gene therapy. Proc Natl Acad Sci USA. 2002;99:10405–10410. doi: 10.1073/pnas.162250899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DiMattia M, Govindasamy L, Levy HC, Gurda-Whitaker B, Kalina A, Kohlbrenner E, Chiorini JA, McKenna R, Muzyczka N, Zolotukhin S, Agbandje-McKenna M. Production, purification, crystallization and preliminary X-ray structural studies of adeno-associated virus serotype 5. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2005;61:917–921. doi: 10.1107/S1744309105028514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xie Q, Ongley HM, Hare J, Chapman MS. Crystallization and preliminary X-ray structural studies of adeno-associated virus serotype 6. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2008;64:1074–1078. doi: 10.1107/S1744309108033290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Padron E, Bowman V, Kaludov N, Govindasamy L, Levy H, Nick P, McKenna R, Muzyczka N, Chiorini JA, Baker TS, Agbandje-McKenna M. Structure of adeno-associated virus type 4. J Virol. 2005;79:5047–5058. doi: 10.1128/JVI.79.8.5047-5058.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaludov N, Padron E, Govindasamy L, McKenna R, Chiorini JA, Agbandje-McKenna M. Production, purification and preliminary X-ray crystallographic studies of adeno-associated virus serotype 4. Virology. 2003;306:1–6. doi: 10.1016/s0042-6822(02)00037-5. [DOI] [PubMed] [Google Scholar]
- 29.Nam H-J, Lane MD, Padron E, Gurda B, McKenna R, Kohlbrenner E, Aslanidi G, Byrne B, Muzyczka N, Zolotukhin S, Agbandje-McKenna M. Structure of adeno-associated virus serotype 8, a gene therapy vector. J Virol. 2007;81:12260–12271. doi: 10.1128/JVI.01304-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lerch TF, Xie Q, Ongley HM, Hare J, Chapman MS. Twinned crystals of adeno-associated virus serotype 3b prove suitable for structural studies. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2009;65:177–183. doi: 10.1107/S1744309109000372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mitchell M, Nam H-J, Carter A, McCall A, Rence C, Bennett A, Gurda B, McKenna R, Porter M, Sakai Y, Byrne BJ, Muzyczka N, Aslanidi G, Zolotukhin S, Agbandje-McKenna M. Production, purification and preliminary X-ray crystallographic studies of adeno-associated virus serotype 9. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2009;65:715–718. doi: 10.1107/S1744309109021460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller EB, Gurda-Whitaker B, Govindasamy L, McKenna R, Zolotukhin S, Muzyczka N, Agbandje-McKenna M. Production, purification and preliminary X-ray crystallographic studies of adeno-associated virus serotype 1. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2006;62:1271–1274. doi: 10.1107/S1744309106048184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quesada O, Gurda B, Govindasamy L, McKenna R, Kohlbrenner E, Aslanidi G, Zolotukhin S, Muzyczka N, Agbandje-McKenna M. Production, purification and preliminary X-ray crystallographic studies of adeno-associated virus serotype 7. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2007;63:1073–1076. doi: 10.1107/S1744309107060289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lane MD, Nam H-J, Padron E, Gurda-Whitaker B, Kohlbrenner E, Aslanidi G, Byrne B, McKenna R, Muzyczka N, Zolotukhin S, Agbandje-McKenna M. Production, purification, crystallization and preliminary X-ray analysis of adeno-associated virus serotype 8. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2005;61:558–561. doi: 10.1107/S1744309105014132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Govindasamy L, Padron E, McKenna R, Muzyczka N, Kaludov N, Chiorini JA, Agbandje-McKenna M. Structurally mapping the diverse phenotype of adeno-associated virus serotype 4. J Virol. 2006;80:11556–11570. doi: 10.1128/JVI.01536-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walters RW, Agbandje-McKenna M, Bowman VD, Moninger TO, Olson NH, Seiler M, Chiorini JA, Baker TS, Zabner J. Structure of adeno-associated virus serotype 5. J Virol. 2004;78:3361–3371. doi: 10.1128/JVI.78.7.3361-3371.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O'Donnell J, Taylor KA, Chapman MS. Adeno-associated virus-2 and its primary cellular receptor–Cryo-EM structure of a heparin complex. Virology. 2009;385:434–443. doi: 10.1016/j.virol.2008.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Levy HC, Bowman VD, Govindasamy L, McKenna R, Nash K, Warrington K, Chen W, Muzyczka N, Yan X, Baker TS, Agbandje-McKenna M. Heparin binding induces conformational changes in Adeno-associated virus serotype 2. J Struct Biol. 2009;165:146–156. doi: 10.1016/j.jsb.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lerch TF, Xie Q, Chapman MS. The structure of adeno-associated virus serotype 3B (AAV-3B): insights into receptor binding and immune evasion. Virology. 2010;403:26–36. doi: 10.1016/j.virol.2010.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ng R, Govindasamy L, Gurda BL, McKenna R, Kozyreva OG, Samulski RJ, Parent KN, Baker TS, Agbandje-McKenna M. Structural characterization of the dual glycan binding adeno-associated virus serotype 6. J Virol. 2010;84:12945–12957. doi: 10.1128/JVI.01235-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rabinowitz JE, Bowles DE, Faust SM, Ledford JG, Cunningham SE, Samulski RJ. Cross-dressing the virion: the transcapsidation of adeno-associated virus serotypes functionally defines subgroups. J Virol. 2004;78:4421–4432. doi: 10.1128/JVI.78.9.4421-4432.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wobus CE, Hügle-Dörr B, Girod A, Petersen G, Hallek M, Kleinschmidt JA. Monoclonal antibodies against the adeno-associated virus type 2 (AAV-2) capsid: epitope mapping and identification of capsid domains involved in AAV-2-cell interaction and neutralization of AAV-2 infection. J Virol. 2000;74:9281–9293. doi: 10.1128/jvi.74.19.9281-9293.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Horowitz ED, Rahman KS, Bower BD, Dismuke DJ, Falvo MR, Griffith JD, Harvey SC, Asokan A. Biophysical and ultrastructural characterization of adeno-associated virus capsid uncoating and genome release. J Virol. 2013;87:2994–3002. doi: 10.1128/JVI.03017-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cavalier-Smith T. Palindromic base sequences and replication of eukaryote chromosome ends. Nature. 1974;250:467–470. doi: 10.1038/250467a0. [DOI] [PubMed] [Google Scholar]
- 45.Snyder RO, Im DS, Ni T, Xiao X, Samulski RJ, Muzyczka N. Features of the adeno-associated virus origin involved in substrate recognition by the viral Rep protein. J Virol. 1993;67:6096–6104. doi: 10.1128/jvi.67.10.6096-6104.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chiorini JA, Wiener SM, Owens RA, Kyöstiö SR, Kotin RM, Safer B. Sequence requirements for stable binding and function of Rep68 on the adeno-associated virus type 2 inverted terminal repeats. J Virol. 1994;68:7448–7457. doi: 10.1128/jvi.68.11.7448-7457.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mccarty DM, Pereira DJ, Zolotukhin I, Zhou X, Ryan JH, Muzyczka N. Identification of linear DNA sequences that specifically bind the adeno-associated virus Rep protein. J Virol. 1994;68:4988–4997. doi: 10.1128/jvi.68.8.4988-4997.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mccarty DM, Ryan JH, Zolotukhin S, Zhou X, Muzyczka N. Interaction of the adeno-associated virus Rep protein with a sequence within the A palindrome of the viral terminal repeat. J Virol. 1994;68:4998–5006. doi: 10.1128/jvi.68.8.4998-5006.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weitzman MD, Kyöstiö SR, Kotin RM, Owens RA. Adeno-associated virus (AAV) Rep proteins mediate complex formation between AAV DNA and its integration site in human DNA. Proc Natl Acad Sci USA. 1994;91:5808–5812. doi: 10.1073/pnas.91.13.5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Straus SE, Sebring ED, Rose JA. Concatemers of alternating plus and minus strands are intermediates in adenovirus-associated virus DNA synthesis. Proc Natl Acad Sci USA. 1976;73:742–746. doi: 10.1073/pnas.73.3.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lusby E, Bohenzky R, Berns KI. Inverted terminal repetition in adeno-associated virus DNA: independence of the orientation at either end of the genome. J Virol. 1981;37:1083–1086. doi: 10.1128/jvi.37.3.1083-1086.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Snyder RO, Im DS, Muzyczka N. Evidence for covalent attachment of the adeno-associated virus (AAV) rep protein to the ends of the AAV genome. J Virol. 1990;64:6204–6213. doi: 10.1128/jvi.64.12.6204-6213.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Im DS, Muzyczka N. Factors that bind to adeno-associated virus terminal repeats. J Virol. 1989;63:3095–3104. doi: 10.1128/jvi.63.7.3095-3104.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Im DS, Muzyczka N. The AAV origin binding protein Rep68 is an ATP-dependent site-specific endonuclease with DNA helicase activity. Cell. 1990;61:447–457. doi: 10.1016/0092-8674(90)90526-k. [DOI] [PubMed] [Google Scholar]
- 55.Owens RA, Weitzman MD, Kyöstiö SR, Carter BJ. Identification of a DNA-binding domain in the amino terminus of adeno-associated virus Rep proteins. J Virol. 1993;67:997–1005. doi: 10.1128/jvi.67.2.997-1005.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chiorini JA, Weitzman MD, Owens RA, Urcelay E, Safer B, Kotin RM. Biologically active Rep proteins of adeno-associated virus type 2 produced as fusion proteins in Escherichia coli. J Virol. 1994;68:797–804. doi: 10.1128/jvi.68.2.797-804.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brister JR, Muzyczka N. Rep-mediated nicking of the adeno-associated virus origin requires two biochemical activities, DNA helicase activity and transesterification. J Virol. 1999;73:9325–9336. doi: 10.1128/jvi.73.11.9325-9336.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wonderling RS, Kyöstiö SR, Owens RA. A maltose-binding protein/adeno-associated virus Rep68 fusion protein has DNA–RNA helicase and ATPase activities. J Virol. 1995;69:3542–3548. doi: 10.1128/jvi.69.6.3542-3548.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Snyder RO, Samulski RJ, Muzyczka N. In vitro resolution of covalently joined AAV chromosome ends. Cell. 1990;60:105–113. doi: 10.1016/0092-8674(90)90720-y. [DOI] [PubMed] [Google Scholar]
- 60.Naumer M, Sonntag F, Schmidt K, Nieto K, Panke C, Davey NE, Popa-Wagner R, Kleinschmidt JA. Properties of the adeno-associated virus assembly-activating protein. J Virol. 2012;86:13038–13048. doi: 10.1128/JVI.01675-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hermonat PL, Labow MA, Wright R, Berns KI, Muzyczka N. Genetics of adeno-associated virus: isolation and preliminary characterization of adeno-associated virus type 2 mutants. J Virol. 1984;51:329–339. doi: 10.1128/jvi.51.2.329-339.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Samulski RJ, Chang LS, Shenk T. Helper-free stocks of recombinant adeno-associated viruses: normal integration does not require viral gene expression. J Virol. 1989;63:3822–3828. doi: 10.1128/jvi.63.9.3822-3828.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ferrari FK, Xiao X, McCarty D, Samulski RJ. New developments in the generation of Ad-free, high-titer rAAV gene therapy vectors. Nat Med. 1997;3:1295–1297. doi: 10.1038/nm1197-1295. [DOI] [PubMed] [Google Scholar]
- 64.Grieger JC, Samulski RJ. Adeno-associated virus vectorology, manufacturing, and clinical applications. Meth Enzymol. 2012;507:229–254. doi: 10.1016/B978-0-12-386509-0.00012-0. [DOI] [PubMed] [Google Scholar]
- 65.Fisher KJ, Gao GP, Weitzman MD, DeMatteo R, Burda JF, Wilson JM. Transduction with recombinant adeno-associated virus for gene therapy is limited by leading-strand synthesis. J Virol. 1996;70:520–532. doi: 10.1128/jvi.70.1.520-532.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ferrari FK, Samulski T, Shenk T, Samulski RJ. Second-strand synthesis is a rate-limiting step for efficient transduction by recombinant adeno-associated virus vectors. J Virol. 1996;70:3227–3234. doi: 10.1128/jvi.70.5.3227-3234.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jooss K, Yang Y, Fisher KJ, Wilson JM. Transduction of dendritic cells by DNA viral vectors directs the immune response to transgene products in muscle fibers. J Virol. 1998;72:4212–4223. doi: 10.1128/jvi.72.5.4212-4223.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Thomas CE, Storm TA, Huang Z, Kay MA. Rapid uncoating of vector genomes is the key to efficient liver transduction with pseudotyped adeno-associated virus vectors. J Virol. 2004;78:3110–3122. doi: 10.1128/JVI.78.6.3110-3122.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wu Z, Miller E, Agbandje-McKenna M, Samulski RJ. Alpha2,3 and alpha2,6 N-linked sialic acids facilitate efficient binding and transduction by adeno-associated virus types 1 and 6. J Virol. 2006;80:9093–9103. doi: 10.1128/JVI.00895-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Summerford C, Samulski RJ. Membrane-associated heparan sulfate proteoglycan is a receptor for adeno-associated virus type 2 virions. J Virol. 1998;72:1438–1445. doi: 10.1128/jvi.72.2.1438-1445.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kaludov N, Brown KE, Walters RW, Zabner J, Chiorini JA. Adeno-associated virus serotype 4 (AAV4) and AAV5 both require sialic acid binding for hemagglutination and efficient transduction but differ in sialic acid linkage specificity. J Virol. 2001;75:6884–6893. doi: 10.1128/JVI.75.15.6884-6893.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Walters RW, Yi SM, Keshavjee S, Brown KE, Welsh MJ, Chiorini JA, Zabner J. Binding of adeno-associated virus type 5 to 2,3-linked sialic acid is required for gene transfer. J Biol Chem. 2001;276:20610–20616. doi: 10.1074/jbc.M101559200. [DOI] [PubMed] [Google Scholar]
- 73.Shen S, Bryant KD, Brown SM, Randell SH, Asokan A. Terminal N-linked galactose is the primary receptor for adeno-associated virus 9. J Biol Chem. 2011;286:13532–13540. doi: 10.1074/jbc.M110.210922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Müller OJ, Leuchs B, Pleger ST, Grimm D, Franz W-M, Katus HA, Kleinschmidt JA. Improved cardiac gene transfer by transcriptional and transductional targeting of adeno-associated viral vectors. Cardiovasc Res. 2006;70:70–78. doi: 10.1016/j.cardiores.2005.12.017. [DOI] [PubMed] [Google Scholar]
- 75.Asokan A, Conway JC, Phillips JL, Li C, Hegge J, Sinnott R, Yadav S, DiPrimio N, Nam H-J, Agbandje-McKenna M, McPhee S, Wolff J, Samulski RJ. Reengineering a receptor footprint of adeno-associated virus enables selective and systemic gene transfer to muscle. Nat Biotechnol. 2010;28:79–82. doi: 10.1038/nbt.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shen S, Horowitz ED, Troupes AN, Brown SM, Pulicherla N, Samulski RJ, Agbandje-McKenna M, Asokan A. Engraftment of a galactose receptor footprint onto adeno-associated viral capsids improves transduction efficiency. J Biol Chem. 2013;288:28814–28823. doi: 10.1074/jbc.M113.482380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Girod A, Ried M, Wobus C, Lahm H, Leike K, Kleinschmidt J, Deléage G, Hallek M. Genetic capsid modifications allow efficient re-targeting of adeno-associated virus type 2. Nat Med. 1999;5:1052–1056. doi: 10.1038/12491. [DOI] [PubMed] [Google Scholar]
- 78.Yang Q, Mamounas M, Yu G, Kennedy S, Leaker B, Merson J, Wong-Staal F, Yu M, Barber JR. Development of novel cell surface CD34-targeted recombinant adenoassociated virus vectors for gene therapy. Human Gene Ther. 1998;9:1929–1937. doi: 10.1089/hum.1998.9.13-1929. [DOI] [PubMed] [Google Scholar]
- 79.Grifman M, Trepel M, Speece P, Gilbert LB, Arap W, Pasqualini R, Weitzman MD. Incorporation of tumor-targeting peptides into recombinant adeno-associated virus capsids. Mol Ther. 2001;3:964–975. doi: 10.1006/mthe.2001.0345. [DOI] [PubMed] [Google Scholar]
- 80.Loiler SA, Conlon TJ, Song S, Tang Q, Warrington KH, Agarwal A, Kapturczak M, Li C, Ricordi C, Atkinson MA, Muzyczka N, Flotte TR. Targeting recombinant adeno-associated virus vectors to enhance gene transfer to pancreatic islets and liver. Gene Ther. 2003;10:1551–1558. doi: 10.1038/sj.gt.3302046. [DOI] [PubMed] [Google Scholar]
- 81.Rabinowitz JE, Xiao W, Samulski RJ. Insertional mutagenesis of AAV2 capsid and the production of recombinant virus. Virology. 1999;265:274–285. doi: 10.1006/viro.1999.0045. [DOI] [PubMed] [Google Scholar]
- 82.Shi W, Arnold GS, Bartlett JS. Insertional mutagenesis of the adeno-associated virus type 2 (AAV2) capsid gene and generation of AAV2 vectors targeted to alternative cell-surface receptors. Hum Gene Ther. 2001;12:1697–1711. doi: 10.1089/104303401750476212. [DOI] [PubMed] [Google Scholar]
- 83.Müller OJ, Kaul F, Weitzman MD, Pasqualini R, Arap W, Kleinschmidt JA, Trepel M. Random peptide libraries displayed on adeno-associated virus to select for targeted gene therapy vectors. Nat Biotechnol. 2003;21:1040–1046. doi: 10.1038/nbt856. [DOI] [PubMed] [Google Scholar]
- 84.Judd J, Wei F, Nguyen PQ, Tartaglia LJ, Agbandje-McKenna M, Silberg JJ, Suh J. Random insertion of mCherry into VP3 domain of adeno-associated virus yields fluorescent capsids with no loss of infectivity. Mol Ther Nucleic Acids. 2012;1:e54. doi: 10.1038/mtna.2012.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nam H-J, Gurda BL, McKenna R, Potter M, Byrne B, Salganik M, Muzyczka N, Agbandje-McKenna M. Structural studies of adeno-associated virus serotype 8 capsid transitions associated with endosomal trafficking. J Virol. 2011;85:11791–11799. doi: 10.1128/JVI.05305-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Salganik M, Venkatakrishnan B, Bennett A, Lins B, Yarbrough J, Muzyczka N, Agbandje-McKenna M, McKenna R. Evidence for pH-dependent protease activity in the adeno-associated virus capsid. J Virol. 2012;86:11877–11885. doi: 10.1128/JVI.01717-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Münch RC, Janicki H, Völker I, Rasbach A, Hallek M, Büning H, Buchholz CJ. Displaying high-affinity ligands on adeno-associated viral vectors enables tumor cell-specific and safe gene transfer. Mol Ther. 2013;21:109–118. doi: 10.1038/mt.2012.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sonntag F, Bleker S, Leuchs B, Fischer R, Kleinschmidt JA. Adeno-associated virus type 2 capsids with externalized VP1/VP2 trafficking domains are generated prior to passage through the cytoplasm and are maintained until uncoating occurs in the nucleus. J Virol. 2006;80:11040–11054. doi: 10.1128/JVI.01056-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.White AF, Mazur M, Sorscher EJ, Zinn KR, Ponnazhagan S. Genetic modification of adeno-associated viral vector type 2 capsid enhances gene transfer efficiency in polarized human airway epithelial cells. Hum Gene Ther. 2008;19:1407–1414. doi: 10.1089/hum.2008.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nicklin SA, Buening H, Dishart KL, de Alwis M, Girod A, Hacker U, Thrasher AJ, Ali RR, Hallek M, Baker AH. Efficient and selective AAV2-mediated gene transfer directed to human vascular endothelial cells. Mol Ther. 2001;4:174–181. doi: 10.1006/mthe.2001.0424. [DOI] [PubMed] [Google Scholar]
- 91.White SJ, Nicklin SA, Büning H, Brosnan MJ, Leike K, Papadakis ED, Hallek M, Baker AH. Targeted gene delivery to vascular tissue in vivo by tropism-modified adeno-associated virus vectors. Circulation. 2004;109:513–519. doi: 10.1161/01.CIR.0000109697.68832.5D. [DOI] [PubMed] [Google Scholar]
- 92.Work LM, Büning H, Hunt E, Nicklin SA, Denby L, Britton N, Leike K, Odenthal M, Drebber U, Hallek M, Baker AH. Vascular bed-targeted in vivo gene delivery using tropism-modified adeno-associated viruses. Mol Ther. 2006;13:683–693. doi: 10.1016/j.ymthe.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 93.Yu C-Y, Yuan Z, Cao Z, Wang B, Qiao C, Li J, Xiao X. A muscle-targeting peptide displayed on AAV2 improves muscle tropism on systemic delivery. Gene Ther. 2009;16:953–962. doi: 10.1038/gt.2009.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Girod A, Wobus CE, Zádori Z, Ried M, Leike K, Tijssen P, Kleinschmidt JA, Hallek M. The VP1 capsid protein of adeno-associated virus type 2 is carrying a phospholipase A2 domain required for virus infectivity. J Gen Virol. 2002;83:973–978. doi: 10.1099/0022-1317-83-5-973. [DOI] [PubMed] [Google Scholar]
- 95.Perabo L, Büning H, Kofler DM, Ried MU, Girod A, Wendtner CM, Enssle J, Hallek M. In vitro selection of viral vectors with modified tropism: the adeno-associated virus display. Mol Ther. 2003;8:151–157. doi: 10.1016/s1525-0016(03)00123-0. [DOI] [PubMed] [Google Scholar]
- 96.Waterkamp DA, Müller OJ, Ying Y, Trepel M, Kleinschmidt JA. Isolation of targeted AAV2 vectors from novel virus display libraries. J Gene Med. 2006;8:1307–1319. doi: 10.1002/jgm.967. [DOI] [PubMed] [Google Scholar]
- 97.Michelfelder S, Lee M-K, deLima-Hahn E, Wilmes T, Kaul F, Müller O, Kleinschmidt JA, Trepel M. Vectors selected from adeno-associated viral display peptide libraries for leukemia cell-targeted cytotoxic gene therapy. Exp Hematol. 2007;35:1766–1776. doi: 10.1016/j.exphem.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 98.Michelfelder S, Kohlschütter J, Skorupa A, Pfennings S, Müller O, Kleinschmidt JA, Trepel M. Successful expansion but not complete restriction of tropism of adeno-associated virus by in vivo biopanning of random virus display peptide libraries. PLoS ONE. 2009;4:e5122. doi: 10.1371/journal.pone.0005122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Grimm D, Lee JS, Wang L, Desai T, Akache B, Storm TA, Kay MA. In vitro and in vivo gene therapy vector evolution via multispecies interbreeding and retargeting of adeno-associated viruses. J Virol. 2008;82:5887–5911. doi: 10.1128/JVI.00254-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Popa-Wagner R, Porwal M, Kann M, Reuss M, Weimer M, Florin L, Kleinschmidt JA. Impact of VP1-specific protein sequence motifs on adeno-associated virus type 2 intracellular trafficking and nuclear entry. J Virol. 2012;86:9163–9174. doi: 10.1128/JVI.00282-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Maheshri N, Koerber JT, Kaspar BK, Schaffer DV. Directed evolution of adeno-associated virus yields enhanced gene delivery vectors. Nat Biotechnol. 2006;24:198–204. doi: 10.1038/nbt1182. [DOI] [PubMed] [Google Scholar]
- 102.Perabo L, Endell J, King S, Lux K, Goldnau D, Hallek M, Büning H. Combinatorial engineering of a gene therapy vector: directed evolution of adeno-associated virus. J Gene Med. 2006;8:155–162. doi: 10.1002/jgm.849. [DOI] [PubMed] [Google Scholar]
- 103.Grieger JC, Snowdy S, Samulski RJ. Separate basic region motifs within the adeno-associated virus capsid proteins are essential for infectivity and assembly. J Virol. 2006;80:5199–5210. doi: 10.1128/JVI.02723-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Johnson JS, Li C, DiPrimio N, Weinberg MS, McCown TJ, Samulski RJ. Mutagenesis of adeno-associated virus type 2 capsid protein VP1 uncovers new roles for basic amino acids in trafficking and cell-specific transduction. J Virol. 2010;84:8888–8902. doi: 10.1128/JVI.00687-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Li W, Asokan A, Wu Z, Van Dyke T, DiPrimio N, Johnson JS, Govindaswamy L, Agbandje-McKenna M, Leichtle S, Redmond DE, McCown TJ, Petermann KB, Sharpless NE, Samulski RJ. Engineering and selection of shuffled AAV genomes: a new strategy for producing targeted biological nanoparticles. Mol Ther. 2008;6:1252–1260. doi: 10.1038/mt.2008.100. [DOI] [PubMed] [Google Scholar]
- 106.Maguire CA, Gianni D, Meijer DH, Shaket LA, Wakimoto H, Rabkin SD, Gao G, Sena-Esteves M. Directed evolution of adeno-associated virus for glioma cell transduction. J Neurooncol. 2010;96:337–347. doi: 10.1007/s11060-009-9972-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ward P, Walsh CE. Chimeric AAV Cap sequences alter gene transduction. Virology. 2009;386:237–248. doi: 10.1016/j.virol.2009.01.012. [DOI] [PubMed] [Google Scholar]
- 108.Farr GA, Zhang L-G, Tattersall P. Parvoviral virions deploy a capsid-tethered lipolytic enzyme to breach the endosomal membrane during cell entry. Proc Natl Acad Sci USA. 2005;102:17148–17153. doi: 10.1073/pnas.0508477102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yang L, Jiang J, Drouin LM, Agbandje-McKenna M, Chen C, Qiao C, Pu D, Hu X, Wang D-Z, Li J, Xiao X. A myocardium tropic adeno-associated virus (AAV) evolved by DNA shuffling and in vivo selection. Proc Natl Acad Sci USA. 2009;106:3946–3951. doi: 10.1073/pnas.0813207106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Mani B, Baltzer C, Valle N, Almendral JM, Kempf C, Ros C. Low pH-dependent endosomal processing of the incoming parvovirus minute virus of mice virion leads to externalization of the VP1 N-terminal sequence (N-VP1), N-VP2 cleavage, and uncoating of the full-length genome. J Virol. 2006;80:1015–1024. doi: 10.1128/JVI.80.2.1015-1024.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gray SJ, Blake BL, Criswell HE, Nicolson SC, Samulski RJ, McCown TJ, Li W. Directed evolution of a novel adeno-associated virus (AAV) vector that crosses the seizure-compromised blood-brain barrier (BBB) Mol Ther. 2010;18:570–578. doi: 10.1038/mt.2009.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Cohen S, Marr AK, Garcin P, Panté N. Nuclear envelope disruption involving host caspases plays a role in the parvovirus replication cycle. J Virol. 2011;85:4863–4874. doi: 10.1128/JVI.01999-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lisowski L, Dane AP, Chu K, Zhang Y, Cunningham SC, Wilson EM, Nygaard S, Grompe M, Alexander IE, Kay MA. Selection and evaluation of clinically relevant AAV variants in a xenograft liver model. Nature. 2013;506(7488):382–366. doi: 10.1038/nature12875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Xiao P-J, Samulski RJ. Cytoplasmic trafficking, endosomal escape, and perinuclear accumulation of adeno-associated virus type 2 particles are facilitated by microtubule network. J Virol. 2012;86:10462–10473. doi: 10.1128/JVI.00935-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Duan D, Li Q, Kao AW, Yue Y, Pessin JE, Engelhardt JF. Dynamin is required for recombinant adeno-associated virus type 2 infection. J Virol. 1999;73:10371–10376. doi: 10.1128/jvi.73.12.10371-10376.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bartlett JS, Wilcher R, Samulski RJ. Infectious entry pathway of adeno-associated virus and adeno-associated virus vectors. J Virol. 2000;74:2777–2785. doi: 10.1128/jvi.74.6.2777-2785.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Uhrig S, Coutelle O, Wiehe T, Perabo L, Hallek M, Büning H. Successful target cell transduction of capsid-engineered rAAV vectors requires clathrin-dependent endocytosis. Gene Ther. 2012;19:210–218. doi: 10.1038/gt.2011.78. [DOI] [PubMed] [Google Scholar]
- 118.Liu Y, Joo K-I, Wang P. Endocytic processing of adeno-associated virus type 8 vectors for transduction of target cells. Gene Ther. 2012;20:308–317. doi: 10.1038/gt.2012.41. [DOI] [PubMed] [Google Scholar]
- 119.Zhong L, Li B, Jayandharan G, Mah CS, Govindasamy L, Agbandje-McKenna M, Herzog RW, Weigel-Van Aken KA, Hobbs JA, Zolotukhin S, Muzyczka N, Srivastava A. Tyrosine-phosphorylation of AAV2 vectors and its consequences on viral intracellular trafficking and transgene expression. Virology. 2008;381:194–202. doi: 10.1016/j.virol.2008.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bantel-Schaal U, Braspenning-Wesch I, Kartenbeck J. Adeno-associated virus type 5 exploits two different entry pathways in human embryo fibroblasts. J Gen Virol. 2009;90:317–322. doi: 10.1099/vir.0.005595-0. [DOI] [PubMed] [Google Scholar]
- 121.Douar AM, Poulard K, Stockholm D, Danos O. Intracellular trafficking of adeno-associated virus vectors: routing to the late endosomal compartment and proteasome degradation. J Virol. 2001;75:1824–1833. doi: 10.1128/JVI.75.4.1824-1833.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Nonnenmacher M, Weber T. Adeno-associated virus 2 infection requires endocytosis through the CLIC/GEEC pathway. Cell Host Microbe. 2011;10:563–576. doi: 10.1016/j.chom.2011.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zhong L, Li B, Mah CS, Govindasamy L, Agbandje-McKenna M, Cooper M, Herzog RW, Zolotukhin I, Warrington KH, Weigel-Van Aken KA, Hobbs JA, Zolotukhin S, Muzyczka N, Srivastava A. Next generation of adeno-associated virus 2 vectors: point mutations in tyrosines lead to high-efficiency transduction at lower doses. Proc Natl Acad Sci USA. 2008;105:7827–7832. doi: 10.1073/pnas.0802866105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Nicolson SC, Samulski RJ. Recombinant adeno-associated virus utilizes host cell nuclear import machinery to enter the nucleus. J Virol. 2014;88:4132–4144. doi: 10.1128/JVI.02660-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Johnson JS, Samulski RJ. Enhancement of adeno-associated virus infection by mobilizing capsids into and out of the nucleolus. J Virol. 2009;83:2632–2644. doi: 10.1128/JVI.02309-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Salganik M, Aydemir F, Nam H-J, McKenna R, Agbandje-McKenna M, Muzyczka N. Adeno-associated virus capsid proteins may play a role in transcription and second-strand synthesis of recombinant genomes. J Virol. 2014;88:1071–1079. doi: 10.1128/JVI.02093-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ding W, Zhang LN, Yeaman C, Engelhardt JF. rAAV2 traffics through both the late and the recycling endosomes in a dose-dependent fashion. Mol Ther. 2006;13:671–682. doi: 10.1016/j.ymthe.2005.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Cervelli T, Palacios JA, Zentilin L, Mano M, Schwartz RA, Weitzman MD, Giacca M. Processing of recombinant AAV genomes occurs in specific nuclear structures that overlap with foci of DNA-damage-response proteins. J Cell Sci. 2008;121:349–357. doi: 10.1242/jcs.003632. [DOI] [PubMed] [Google Scholar]
- 129.Hauck B, Zhao W, High K, Xiao W. Intracellular viral processing, not single-stranded DNA accumulation, is crucial for recombinant adeno-associated virus transduction. J Virol. 2004;78:13678–13686. doi: 10.1128/JVI.78.24.13678-13686.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Samulski RJ, Srivastava A, Berns KI, Muzyczka N. Rescue of adeno-associated virus from recombinant plasmids: gene correction within the terminal repeats of AAV. Cell. 1983;33:135–143. doi: 10.1016/0092-8674(83)90342-2. [DOI] [PubMed] [Google Scholar]
- 131.Hauswirth WW, Berns KI. Adeno-associated virus DNA replication: nonunit-length molecules. Virology. 1979;93:57–68. doi: 10.1016/0042-6822(79)90275-7. [DOI] [PubMed] [Google Scholar]
- 132.Ni TH, McDonald WF, Zolotukhin I, Melendy T, Waga S, Stillman B, Muzyczka N. Cellular proteins required for adeno-associated virus DNA replication in the absence of adenovirus coinfection. J Virol. 1998;72:2777–2787. doi: 10.1128/jvi.72.4.2777-2787.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Nash K, Chen W, McDonald WF, Zhou X, Muzyczka N. Purification of host cell enzymes involved in adeno-associated virus DNA replication. J Virol. 2007;81:5777–5787. doi: 10.1128/JVI.02651-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Nakai H, Storm TA, Kay MA. Recruitment of single-stranded recombinant adeno-associated virus vector genomes and intermolecular recombination are responsible for stable transduction of liver in vivo. J Virol. 2000;74:9451–9463. doi: 10.1128/jvi.74.20.9451-9463.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Choi VW, McCarty DM, Samulski RJ. Host cell DNA repair pathways in adeno-associated viral genome processing. J Virol. 2006;80:10346–10356. doi: 10.1128/JVI.00841-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Song S, Laipis PJ, Berns KI, Flotte TR. Effect of DNA-dependent protein kinase on the molecular fate of the rAAV2 genome in skeletal muscle. Proc Natl Acad Sci USA. 2001;98:4084–4088. doi: 10.1073/pnas.061014598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Inagaki K, Ma C, Storm TA, Kay MA, Nakai H. The role of DNA-PKcs and artemis in opening viral DNA hairpin termini in various tissues in mice. J Virol. 2007;81:11304–11321. doi: 10.1128/JVI.01225-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Duan D, Sharma P, Yang J, Yue Y, Dudus L, Zhang Y, Fisher KJ, Engelhardt JF. Circular intermediates of recombinant adeno-associated virus have defined structural characteristics responsible for long-term episomal persistence in muscle tissue. J Virol. 1998;72:8568–8577. doi: 10.1128/jvi.72.11.8568-8577.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Xiao X, Li J, Samulski RJ. Efficient long-term gene transfer into muscle tissue of immunocompetent mice by adeno-associated virus vector. J Virol. 1996;70:8098–8108. doi: 10.1128/jvi.70.11.8098-8108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Sanlioglu S, Duan D, Engelhardt JF. Two independent molecular pathways for recombinant adeno-associated virus genome conversion occur after UV-C and E4orf6 augmentation of transduction. Hum Gene Ther. 1999;10:591–602. doi: 10.1089/10430349950018661. [DOI] [PubMed] [Google Scholar]
- 141.Zentilin L, Marcello A, Giacca M. Involvement of cellular double-stranded DNA break binding proteins in processing of the recombinant adeno-associated virus genome. J Virol. 2001;75:12279–12287. doi: 10.1128/JVI.75.24.12279-12287.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Russell DW, Alexander IE, Miller AD. DNA synthesis and topoisomerase inhibitors increase transduction by adeno-associated virus vectors. Proc Natl Acad Sci USA. 1995;92:5719–5723. doi: 10.1073/pnas.92.12.5719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Miller JL, Donahue RE, Sellers SE, Samulski RJ, Young NS, Nienhuis AW. Recombinant adeno-associated virus (rAAV)-mediated expression of a human gamma-globin gene in human progenitor-derived erythroid cells. Proc Natl Acad Sci USA. 1994;91:10183–10187. doi: 10.1073/pnas.91.21.10183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Li C, He Y, Nicolson S, Hirsch M, Weinberg MS, Zhang P, Kafri T, Samulski RJ. Adeno-associated virus capsid antigen presentation is dependent on endosomal escape. J Clin Invest. 2013;123:1390–1401. doi: 10.1172/JCI66611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Schwartz RA, Palacios JA, Cassell GD, Adam S, Giacca M, Weitzman MD. The Mre11/Rad50/Nbs1 complex limits adeno-associated virus transduction and replication. J Virol. 2007;81:12936–12945. doi: 10.1128/JVI.01523-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lovric J, Mano M, Zentilin L, Eulalio A, Zacchigna S, Giacca M. Terminal differentiation of cardiac and skeletal myocytes induces permissivity to AAV transduction by relieving inhibition imposed by DNA damage response proteins. Mol Ther. 2012;20:2087–2097. doi: 10.1038/mt.2012.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Rahman SH, Bobis-Wozowicz S, Chatterjee D, Gellhaus K, Pars K, Heilbronn R, Jacobs R, Cathomen T. The nontoxic cell cycle modulator indirubin augments transduction of adeno-associated viral vectors and zinc-finger nuclease-mediated gene targeting. Hum Gene Ther. 2013;24:67–77. doi: 10.1089/hum.2012.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Raj K, Ogston P, Beard P. Virus-mediated killing of cells that lack p53 activity. Nature. 2001;412:914–917. doi: 10.1038/35091082. [DOI] [PubMed] [Google Scholar]
- 149.Winocour E, Callaham MF, Huberman E. Perturbation of the cell cycle by adeno-associated virus. Virology. 1988;167:393–399. [PubMed] [Google Scholar]
- 150.Fragkos M, Beard P. Mitotic catastrophe occurs in the absence of apoptosis in p53-null cells with a defective G1 checkpoint. PLoS ONE. 2011;6:e22946. doi: 10.1371/journal.pone.0022946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Hirsch ML, Fagan BM, Dumitru R, Bower JJ, Yadav S, Porteus MH, Pevny LH, Samulski RJ. Viral single-strand DNA induces p53-dependent apoptosis in human embryonic stem cells. PLoS ONE. 2011;6:e27520. doi: 10.1371/journal.pone.0027520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Fragkos M, Jurvansuu J, Beard P. H2AX is required for cell cycle arrest via the p53/p21 pathway. Mol Cell Biol. 2009;29:2828–2840. doi: 10.1128/MCB.01830-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Miao CH, Snyder RO, Schowalter DB, Patijn GA, Donahue B, Winther B, Kay MA. The kinetics of rAAV integration in the liver. Nat Genet. 1998;19:13–15. doi: 10.1038/ng0598-13. [DOI] [PubMed] [Google Scholar]
- 154.Yang CC, Xiao X, Zhu X, Ansardi DC, Epstein ND, Frey MR, Matera AG, Samulski RJ. Cellular recombination pathways and viral terminal repeat hairpin structures are sufficient for adeno-associated virus integration in vivo and in vitro. J Virol. 1997;71:9231–9247. doi: 10.1128/jvi.71.12.9231-9247.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Miller DG, Trobridge GD, Petek LM, Jacobs MA, Kaul R, Russell DW. Large-scale analysis of adeno-associated virus vector integration sites in normal human cells. J Virol. 2005;79:11434–11442. doi: 10.1128/JVI.79.17.11434-11442.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Nakai H, Wu X, Fuess S, Storm TA, Munroe D, Montini E, Burgess SM, Grompe M, Kay MA. Large-scale molecular characterization of adeno-associated virus vector integration in mouse liver. J Virol. 2005;79:3606–3614. doi: 10.1128/JVI.79.6.3606-3614.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Flotte TR, Afione SA, Zeitlin PL. Adeno-associated virus vector gene expression occurs in nondividing cells in the absence of vector DNA integration. Am J Respir Cell Mol Biol. 1994;11:517–521. doi: 10.1165/ajrcmb.11.5.7946381. [DOI] [PubMed] [Google Scholar]
- 158.Hirata RK, Russell DW. Design and packaging of adeno-associated virus gene targeting vectors. J Virol. 2000;74:4612–4620. doi: 10.1128/jvi.74.10.4612-4620.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Miller DG, Petek LM, Russell DW. Adeno-associated virus vectors integrate at chromosome breakage sites. Nat Genet. 2004;36:767–773. doi: 10.1038/ng1380. [DOI] [PubMed] [Google Scholar]
- 160.Nakai H, Montini E, Fuess S, Storm TA, Grompe M, Kay MA. AAV serotype 2 vectors preferentially integrate into active genes in mice. Nat Genet. 2003;34:297–302. doi: 10.1038/ng1179. [DOI] [PubMed] [Google Scholar]
- 161.Mccarty DM, Fu H, Monahan PE, Toulson CE, Naik P, Samulski RJ. Adeno-associated virus terminal repeat (TR) mutant generates self-complementary vectors to overcome the rate-limiting step to transduction in vivo. Gene Ther. 2003;10:2112–2118. doi: 10.1038/sj.gt.3302134. [DOI] [PubMed] [Google Scholar]
- 162.Xiao X, Xiao W, Li J, Samulski RJ. A novel 165-base-pair terminal repeat sequence is the sole cis requirement for the adeno-associated virus life cycle. J Virol. 1997;71:941–948. doi: 10.1128/jvi.71.2.941-948.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.McCarty DM. Self-complementary AAV vectors; advances and applications. Mol Ther. 2008;16:1648–1656. doi: 10.1038/mt.2008.171. [DOI] [PubMed] [Google Scholar]
- 164.Hirsch ML, Agbandje-McKenna M, Samulski RJ. Little vector, big gene transduction: fragmented genome reassembly of adeno-associated virus. Mol Ther. 2010;18:6–8. doi: 10.1038/mt.2009.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Lai Y, Zhao J, Yue Y, Wasala NB, Duan D. Partial restoration of cardiac function with ΔPDZ nNOS in aged mdx model of Duchenne cardiomyopathy. Hum Mol Genet. 2014;23:3189–3199. doi: 10.1093/hmg/ddu029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Dyka FM, Boye SL, Chiodo VA, Hauswirth WW, Boye SE. Dual adeno-associated virus vectors result in efficient in vitro and in vivo expression of an oversized gene, MYO7A. Hum Gene Ther Meth. 2014;25:166–177. doi: 10.1089/hgtb.2013.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Hirsch ML, Li C, Bellon I, Yin C, Chavala S, Pryadkina M, Richard I, Samulski RJ. Oversized AAV transductifon is mediated via a DNA-PKcs-independent, Rad51C-dependent repair pathway. Mol Ther. 2013;21:2205–2216. doi: 10.1038/mt.2013.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Russell DW, Hirata RK. Human gene targeting by viral vectors. Nat Genet. 1998;18:325–330. doi: 10.1038/ng0498-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Smithies O, Gregg RG, Boggs SS, Koralewski MA, Kucherlapati RS. Insertion of DNA sequences into the human chromosomal beta-globin locus by homologous recombination. Nature. 1985;317:230–234. doi: 10.1038/317230a0. [DOI] [PubMed] [Google Scholar]
- 170.Thomas KR, Capecchi MR. Site-directed mutagenesis by gene targeting in mouse embryo-derived stem cells. Cell. 1987;51:503–512. doi: 10.1016/0092-8674(87)90646-5. [DOI] [PubMed] [Google Scholar]
- 171.Russell DW, Hirata RK. Human gene targeting favors insertions over deletions. Hum Gene Ther. 2008;19:907–914. doi: 10.1089/hum.2008.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Vasileva A, Linden RM, Jessberger R. Homologous recombination is required for AAV-mediated gene targeting. Nucleic Acids Res. 2006;34:3345–3360. doi: 10.1093/nar/gkl455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Trobridge G, Hirata RK, Russell DW. Gene targeting by adeno-associated virus vectors is cell-cycle dependent. Hum Gene Ther. 2005;16:522–526. doi: 10.1089/hum.2005.16.522. [DOI] [PubMed] [Google Scholar]
- 174.Paulk NK, Wursthorn K, Wang Z, Finegold MJ, Kay MA, Grompe M. Adeno-associated virus gene repair corrects a mouse model of hereditary tyrosinemia in vivo. Hepatology. 2010;51:1200–1208. doi: 10.1002/hep.23481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Petek LM, Fleckman P, Miller DG. Efficient KRT14 targeting and functional characterization of transplanted human keratinocytes for the treatment of epidermolysis bullosa simplex. Mol Ther. 2010;18:1624–1632. doi: 10.1038/mt.2010.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Paulk NK, Wursthorn K, Haft A, Pelz C, Clarke G, Newell AH, Olson SB, Harding CO, Finegold MJ, Bateman RL, Witte JF, McClard R, Grompe M. In vivo selection of transplanted hepatocytes by pharmacological inhibition of fumarylacetoacetate hydrolase in wild-type mice. Mol Ther. 2012;20:1981–1987. doi: 10.1038/mt.2012.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Mueller C, Flotte TR. Clinical gene therapy using recombinant adeno-associated virus vectors. Gene Ther. 2008;15:858–863. doi: 10.1038/gt.2008.68. [DOI] [PubMed] [Google Scholar]
- 178.Rabinowitz JE, Rolling F, Li C, Conrath H, Xiao W, Xiao X, Samulski RJ. Cross-packaging of a single adeno-associated virus (AAV) type 2 vector genome into multiple AAV serotypes enables transduction with broad specificity. J Virol. 2002;76:791–801. doi: 10.1128/JVI.76.2.791-801.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Ellis BL, Hirsch ML, Barker JC, Connelly JP, Steininger RJ, Porteus MH. A survey of ex vivo/in vitro transduction efficiency of mammalian primary cells and cell lines with nine natural adeno-associated virus (AAV1-9) and one engineered adeno-associated virus serotype. Virol J. 2013;10:74. doi: 10.1186/1743-422X-10-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Zincarelli C, Soltys S, Rengo G, Rabinowitz JE. Analysis of AAV serotypes 1–9 mediated gene expression and tropism in mice after systemic injection. Mol Ther. 2008;16:1073–1080. doi: 10.1038/mt.2008.76. [DOI] [PubMed] [Google Scholar]
- 181.Podsakoff G, Wong KK, Chatterjee S. Efficient gene transfer into nondividing cells by adeno-associated virus-based vectors. J Virol. 1994;68:5656–5666. doi: 10.1128/jvi.68.9.5656-5666.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Berns KI, Pinkerton TC, Thomas GF, Hoggan MD. Detection of adeno-associated virus (AAV)-specific nucleotide sequences in DNA isolated from latently infected Detroit 6 cells. Virology. 1975;68:556–560. doi: 10.1016/0042-6822(75)90298-6. [DOI] [PubMed] [Google Scholar]
- 183.Tenenbaum L, Lehtonen E, Monahan PE. Evaluation of risks related to the use of adeno-associated virus-based vectors. Curr Gene Ther. 2003;3:545–565. doi: 10.2174/1566523034578131. [DOI] [PubMed] [Google Scholar]
- 184.Nathwani AC, Tuddenham EGD, Rangarajan S, Rosales C, McIntosh J, Linch DC, Chowdary P, Riddell A, Pie AJ, Harrington C, O'Beirne J, Smith K, Pasi J, Glader B, Rustagi P, Ng CYC, Kay MA, Zhou J, Spence Y, Morton CL, Allay J, Coleman J, Sleep S, Cunningham JM, Srivastava D, Basner-Tschakarjan E, Mingozzi F, High KA, Gray JT, Reiss UM, Nienhuis AW, Davidoff AM. Adenovirus-associated virus vector-mediated gene transfer in hemophilia B. N Engl J Med. 2011;365:2357–2365. doi: 10.1056/NEJMoa1108046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Manno CS, Pierce GF, Arruda VR, Glader B, Ragni M, Rasko JJ, Rasko J, Ozelo MC, Hoots K, Blatt P, Konkle B, Dake M, Kaye R, Razavi M, Zajko A, Zehnder J, Rustagi PK, Nakai H, Chew A, Leonard D, Wright JF, Lessard RR, Sommer JM, Tigges M, Sabatino D, Luk A, Jiang H, Mingozzi F, Couto L, Ertl HC, High KA, Kay MA. Successful transduction of liver in hemophilia by AAV-Factor IX and limitations imposed by the host immune response. Nat Med. 2006;12:342–347. doi: 10.1038/nm1358. [DOI] [PubMed] [Google Scholar]
- 186.Janson C, McPhee S, Bilaniuk L, Haselgrove J, Testaiuti M, Freese A, Wang D-J, Shera D, Hurh P, Rupin J, Saslow E, Goldfarb O, Goldberg M, Larijani G, Sharrar W, Liouterman L, Camp A, Kolodny E, Samulski J, Leone P. Clinical protocol. Gene therapy of Canavan disease: AAV-2 vector for neurosurgical delivery of aspartoacylase gene (ASPA) to the human brain. Hum Gene Ther. 2002;13:1391–1412. doi: 10.1089/104303402760128612. [DOI] [PubMed] [Google Scholar]
- 187.Testa F, Maguire AM, Rossi S, Pierce EA, Melillo P, Marshall K, Banfi S, Surace EM, Sun J, Acerra C, Wright JF, Wellman J, High KA, Auricchio A, Bennett J, Simonelli F. Three-year follow-up after unilateral subretinal delivery of adeno-associated virus in patients with Leber congenital Amaurosis type 2. Ophthalmology. 2013;120:1283–1291. doi: 10.1016/j.ophtha.2012.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Bainbridge JWB, Smith AJ, Barker SS, Robbie S, Henderson R, Balaggan K, Viswanathan A, Holder GE, Stockman A, Tyler N, Petersen-Jones S, Bhattacharya SS, Thrasher AJ, Fitzke FW, Carter BJ, Rubin GS, Moore AT, Ali RR. Effect of gene therapy on visual function in Leber's congenital amaurosis. N Engl J Med. 2008;358:2231–2239. doi: 10.1056/NEJMoa0802268. [DOI] [PubMed] [Google Scholar]
- 189.Hauswirth WW, Aleman TS, Kaushal S, Cideciyan AV, Schwartz SB, Wang L, Conlon TJ, Boye SL, Flotte TR, Byrne BJ, Jacobson SG. Treatment of Leber congenital amaurosis due to RPE65 mutations by ocular subretinal injection of adeno-associated virus gene vector: short-term results of a phase I trial. Hum Gene Ther. 2008;19:979–990. doi: 10.1089/hum.2008.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Maguire AM, High KA, Auricchio A, Wright JF, Pierce EA, Testa F, Mingozzi F, Bennicelli JL, Ying G-S, Rossi S, Fulton A, Marshall KA, Banfi S, Chung DC, Morgan JIW, Hauck B, Zelenaia O, Zhu X, Raffini L, Coppieters F, De Baere E, Shindler KS, Volpe NJ, Surace EM, Acerra C, Lyubarsky A, Redmond TM, Stone E, Sun J, McDonnell JW, Leroy BP, Simonelli F, Bennett J. Age-dependent effects of RPE65 gene therapy for Leber's congenital amaurosis: a phase 1 dose-escalation trial. Lancet. 2009;374:1597–1605. doi: 10.1016/S0140-6736(09)61836-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Maguire AM, Simonelli F, Pierce EA, Pugh EN, Mingozzi F, Bennicelli J, Banfi S, Marshall KA, Testa F, Surace EM, Rossi S, Lyubarsky A, Arruda VR, Konkle B, Stone E, Sun J, Jacobs J, Dell'Osso L, Hertle R, Ma J-X, Redmond TM, Zhu X, Hauck B, Zelenaia O, Shindler KS, Maguire MG, Wright JF, Volpe NJ, McDonnell JW, Auricchio A, High KA, Bennett J. Safety and efficacy of gene transfer for Leber's congenital amaurosis. N Engl J Med. 2008;358:2240–2248. doi: 10.1056/NEJMoa0802315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Bennett J, Ashtari M, Wellman J, Marshall KA, Cyckowski LL, Chung DC, McCague S, Pierce EA, Chen Y, Bennicelli JL, Zhu X, Ying G-S, Sun J, Wright JF, Auricchio A, Simonelli F, Shindler KS, Mingozzi F, High KA, Maguire AM. AAV2 gene therapy readministration in three adults with congenital blindness. Sci Transl Med. 2012;4:120ra15. doi: 10.1126/scitranslmed.3002865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Bowles DE, McPhee SWJ, Li C, Gray SJ, Samulski JJ, Camp AS, Li J, Wang B, Monahan PE, Rabinowitz JE, Grieger JC, Govindasamy L, Agbandje-McKenna M, Xiao X, Samulski RJ. Phase 1 gene therapy for Duchenne muscular dystrophy using a translational optimized AAV vector. Mol Ther. 2012;20:443–455. doi: 10.1038/mt.2011.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Hajjar RJ, Zsebo K, Deckelbaum L, Thompson C, Rudy J, Yaroshinsky A, Ly H, Kawase Y, Wagner K, Borow K, Jaski B, London B, Greenberg B, Pauly DF, Patten R, Starling R, Mancini D, Jessup M. Design of a phase 1/2 trial of intracoronary administration of AAV1/SERCA2a in patients with heart failure. J Card Fail. 2008;14:355–367. doi: 10.1016/j.cardfail.2008.02.005. [DOI] [PubMed] [Google Scholar]
- 195.Barker SE, Broderick CA, Robbie SJ, Duran Y, Natkunarajah M, Buch P, Balaggan KS, MacLaren RE, Bainbridge JWB, Smith AJ, Ali RR. Subretinal delivery of adeno-associated virus serotype 2 results in minimal immune responses that allow repeat vector administration in immunocompetent mice. J Gene Med. 2009;11:486–497. doi: 10.1002/jgm.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Jacobson SG, Cideciyan AV, Ratnakaram R, Heon E, Schwartz SB, Roman AJ, Peden MC, Aleman TS, Boye SL, Sumaroka A, Conlon TJ, Calcedo R, Pang J-J, Erger KE, Olivares MB, Mullins CL, Swider M, Kaushal S, Feuer WJ, Iannaccone A, Fishman GA, Stone EM, Byrne BJ, Hauswirth WW. Gene therapy for leber congenital amaurosis caused by RPE65 mutations: safety and efficacy in 15 children and adults followed up to 3 years. Arch Ophthalmol. 2012;130:9–24. doi: 10.1001/archophthalmol.2011.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.McClements ME, MacLaren RE. Gene therapy for retinal disease. Transl Res. 2013;161:241–254. doi: 10.1016/j.trsl.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Cai X, Conley SM, Naash MI. RPE65: role in the visual cycle, human retinal disease, and gene therapy. Ophthalm Genet. 2009;30:57–62. doi: 10.1080/13816810802626399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Gaudet D, Méthot J, Kastelein J. Gene therapy for lipoprotein lipase deficiency. Curr Opin Lipidol. 2012;23:310–320. doi: 10.1097/MOL.0b013e3283555a7e. [DOI] [PubMed] [Google Scholar]
- 200.Ross CJD, Twisk J, Bakker AC, Miao F, Verbart D, Rip J, Godbey T, Dijkhuizen P, Hermens WTJMC, Kastelein JJP, Kuivenhoven JA, Meulenberg JM, Hayden MR. Correction of feline lipoprotein lipase deficiency with adeno-associated virus serotype 1-mediated gene transfer of the lipoprotein lipase S447X beneficial mutation. Hum Gene Ther. 2006;17:487–499. doi: 10.1089/hum.2006.17.487. [DOI] [PubMed] [Google Scholar]
- 201.Gaudet D, Méthot J, Déry S, Brisson D, Essiembre C, Tremblay G, Tremblay K, de Wal J, Twisk J, van den Bulk N, Sier-Ferreira V, van Deventer S. Efficacy and long-term safety of alipogene tiparvovec (AAV1-LPLS447X) gene therapy for lipoprotein lipase deficiency: an open-label trial. Gene Ther. 2013;20:361–369. doi: 10.1038/gt.2012.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Carpentier AC, Frisch F, Labbé SM, Gagnon R, de Wal J, Greentree S, Petry H, Twisk J, Brisson D, Gaudet D. Effect of alipogene tiparvovec (AAV1-LPL(S447X)) on postprandial chylomicron metabolism in lipoprotein lipase-deficient patients. J Clin Endocrinol Metab. 2012;97:1635–1644. doi: 10.1210/jc.2011-3002. [DOI] [PubMed] [Google Scholar]
- 203.Stroes ES, Nierman MC, Meulenberg JJ, Franssen R, Twisk J, Henny CP, Maas MM, Zwinderman AH, Ross C, Aronica E, High KA, Levi MM, Hayden MR, Kastelein JJ, Kuivenhoven JA. Intramuscular administration of AAV1-lipoprotein lipase S447X lowers triglycerides in lipoprotein lipase-deficient patients. Arterioscler Thromb Vasc Biol. 2008;28:2303–2304. doi: 10.1161/ATVBAHA.108.175620. [DOI] [PubMed] [Google Scholar]
- 204.Chuah M, VandenDriessche T. Gene therapy for hemophilia “A” and “B”: efficacy, safety and immune consequences. Verh K Acad Geneeskd Belg. 2007;69:315–334. [PubMed] [Google Scholar]
- 205.Zaiss A-K, Liu Q, Bowen GP, Wong NCW, Bartlett JS, Muruve DA. Differential activation of innate immune responses by adenovirus and adeno-associated virus vectors. J Virol. 2002;76:4580–4590. doi: 10.1128/JVI.76.9.4580-4590.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Chen J, Wu Q, Yang P, Hsu H-C, Mountz JD. Determination of specific CD4 and CD8 T cell epitopes after AAV2- and AAV8-hF.IX gene therapy. Mol Ther. 2006;13:260–269. doi: 10.1016/j.ymthe.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 207.Li C, Hirsch M, Asokan A, Zeithaml B, Ma H, Kafri T, Samulski RJ. Adeno-associated virus type 2 (AAV2) capsid-specific cytotoxic T lymphocytes eliminate only vector-transduced cells coexpressing the AAV2 capsid in vivo. J Virol. 2007;81:7540–7547. doi: 10.1128/JVI.00529-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Li H, Murphy SL, Giles-Davis W, Edmonson S, Xiang Z, Li Y, Lasaro MO, High KA, Ertl HC. Pre-existing AAV capsid-specific CD8+ T cells are unable to eliminate AAV-transduced hepatocytes. Mol Ther. 2007;15:792–800. doi: 10.1038/sj.mt.6300090. [DOI] [PubMed] [Google Scholar]
- 209.Wang Z, Allen JM, Riddell SR, Gregorevic P, Storb R, Tapscott SJ, Chamberlain JS, Kuhr CS. Immunity to adeno-associated virus-mediated gene transfer in a random-bred canine model of Duchenne muscular dystrophy. Hum Gene Ther. 2007;18:18–26. doi: 10.1089/hum.2006.093. [DOI] [PubMed] [Google Scholar]
- 210.Mingozzi F, Meulenberg JJ, Hui DJ, Basner-Tschakarjan E, Hasbrouck NC, Edmonson SA, Hutnick NA, Betts MR, Kastelein JJ, Stroes ES, High KA. AAV-1-mediated gene transfer to skeletal muscle in humans results in dose-dependent activation of capsid-specific T cells. Blood. 2009;114:2077–2086. doi: 10.1182/blood-2008-07-167510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Li C, DiPrimio N, Bowles DE, Hirsch ML, Monahan PE, Asokan A, Rabinowitz J, Agbandje-McKenna M, Samulski RJ. Single amino acid modification of adeno-associated virus capsid changes transduction and humoral immune profiles. J Virol. 2012;86:7752–7759. doi: 10.1128/JVI.00675-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Gao G-P, Alvira MR, Wang L, Calcedo R, Johnston J, Wilson JM. Novel adeno-associated viruses from rhesus monkeys as vectors for human gene therapy. Proc Natl Acad Sci USA. 2002;99:11854–11859. doi: 10.1073/pnas.182412299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Li C, Narkbunnam N, Samulski RJ, Asokan A, Hu G, Jacobson LJ, Manco-Johnson MJ, Monahan PE, Joint Outcome Study Investigators Neutralizing antibodies against adeno-associated virus examined prospectively in pediatric patients with hemophilia. Gene Ther. 2012;19:288–294. doi: 10.1038/gt.2011.90. [DOI] [PubMed] [Google Scholar]
- 214.Carlisle RC, Benjamin R, Briggs SS, Sumner-Jones S, McIntosh J, Gill D, Hyde S, Nathwani A, Subr V, Ulbrich K, Seymour LW, Fisher KD. Coating of adeno-associated virus with reactive polymers can ablate virus tropism, enable retargeting and provide resistance to neutralising antisera. J Gene Med. 2008;10:400–411. doi: 10.1002/jgm.1161. [DOI] [PubMed] [Google Scholar]
- 215.Georg-Fries B, Biederlack S, Wolf J, Hausen zur H. Analysis of proteins, helper dependence, and seroepidemiology of a new human parvovirus. Virology. 1984;134:64–71. doi: 10.1016/0042-6822(84)90272-1. [DOI] [PubMed] [Google Scholar]
- 216.Mendell JR, Campbell K, Rodino-Klapac L, Sahenk Z, Shilling C, Lewis S, Bowles D, Gray S, Li C, Galloway G, Malik V, Coley B, Clark KR, Li J, Xiao X, Samulski J, McPhee SW, Samulski RJ, Walker CM. Dystrophin immunity in Duchenne's muscular dystrophy. N Engl J Med. 2010;363:1429–1437. doi: 10.1056/NEJMoa1000228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Jacobson EM, Huber A, Tomer Y. The HLA gene complex in thyroid autoimmunity: from epidemiology to etiology. J Autoimmun. 2008;30:58–62. doi: 10.1016/j.jaut.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Taneja V, David CS. Lessons from animal models for human autoimmune diseases. Nat Immunol. 2001;2:781–784. doi: 10.1038/ni0901-781. [DOI] [PubMed] [Google Scholar]
- 219.Lee GK, Maheshri N, Kaspar B, Schaffer DV. PEG conjugation moderately protects adeno-associated viral vectors against antibody neutralization. Biotechnol Bioeng. 2005;92:24–34. doi: 10.1002/bit.20562. [DOI] [PubMed] [Google Scholar]
- 220.Erles K, Sebökovà P, Schlehofer JR. Update on the prevalence of serum antibodies (IgG and IgM) to adeno-associated virus (AAV) J Med Virol. 1999;59:406–411. doi: 10.1002/(sici)1096-9071(199911)59:3<406::aid-jmv22>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 221.Scallan CD, Jiang H, Liu T, Patarroyo-White S, Sommer JM, Zhou S, Couto LB, Pierce GF. Human immunoglobulin inhibits liver transduction by AAV vectors at low AAV2 neutralizing titers in SCID mice. Blood. 2006;107:1810–1817. doi: 10.1182/blood-2005-08-3229. [DOI] [PubMed] [Google Scholar]
- 222.Grewal IS, Flavell RA. CD40 and CD154 in cell-mediated immunity. Annu Rev Immunol. 1998;16:111–135. doi: 10.1146/annurev.immunol.16.1.111. [DOI] [PubMed] [Google Scholar]
- 223.Jiang H, Couto LB, Patarroyo-White S, Liu T, Nagy D, Vargas JA, Zhou S, Scallan CD, Sommer J, Vijay S, Mingozzi F, High KA, Pierce GF. Effects of transient immunosuppression on adenoassociated, virus-mediated, liver-directed gene transfer in rhesus macaques and implications for human gene therapy. Blood. 2006;108:3321–3328. doi: 10.1182/blood-2006-04-017913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Nathwani AC, Gray JT, McIntosh J, Ng CYC, Zhou J, Spence Y, Cochrane M, Gray E, Tuddenham EGD, Davidoff AM. Safe and efficient transduction of the liver after peripheral vein infusion of self-complementary AAV vector results in stable therapeutic expression of human FIX in nonhuman primates. Blood. 2007;109:1414–1421. doi: 10.1182/blood-2006-03-010181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Mingozzi F, Anguela XM, Pavani G, Chen Y, Davidson RJ, Hui DJ, Yazicioglu M, Elkouby L, Hinderer CJ, Faella A, Howard C, Tai A, Podsakoff GM, Zhou S, Basner-Tschakarjan E, Wright JF, High KA. Overcoming preexisting humoral immunity to AAV using capsid decoys. Sci Transl Med. 2013;5:194ra92. doi: 10.1126/scitranslmed.3005795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Chen S, Kapturczak M, Loiler SA, Zolotukhin S, Glushakova OY, Madsen KM, Samulski RJ, Hauswirth WW, Campbell-Thompson M, Berns KI, Flotte TR, Atkinson MA, Tisher CC, Agarwal A. Efficient transduction of vascular endothelial cells with recombinant adeno-associated virus serotype 1 and 5 vectors. Hum Gene Ther. 2005;16:235–247. doi: 10.1089/hum.2005.16.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Qing K, Mah C, Hansen J, Zhou S, Dwarki V, Srivastava A. Human fibroblast growth factor receptor 1 is a co-receptor for infection by adeno-associated virus 2. Nat Med. 1999;5:71–77. doi: 10.1038/4758. [DOI] [PubMed] [Google Scholar]
- 228.Kashiwakura Y, Tamayose K, Iwabuchi K, Hirai Y, Shimada T, Matsumoto K, Nakamura T, Watanabe M, Oshimi K, Daida H. Hepatocyte growth factor receptor is a coreceptor for adeno-associated virus type 2 infection. J Virol. 2005;79:609–614. doi: 10.1128/JVI.79.1.609-614.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Summerford C, Bartlett JS, Samulski RJ. AlphaVbeta5 integrin: a co-receptor for adeno-associated virus type 2 infection. Nat Med. 1999;5:78–82. doi: 10.1038/4768. [DOI] [PubMed] [Google Scholar]
- 230.Asokan A, Hamra JB, Govindasamy L, Agbandje-McKenna M, Samulski RJ. Adeno-associated virus type 2 contains an integrin alpha5beta1 binding domain essential for viral cell entry. J Virol. 2006;80:8961–8969. doi: 10.1128/JVI.00843-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Akache B, Grimm D, Pandey K, Yant SR, Xu H, Kay MA. The 37/67-kilodalton laminin receptor is a receptor for adeno-associated virus serotypes 8, 2, 3, and 9. J Virol. 2006;80:9831–9836. doi: 10.1128/JVI.00878-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Ling C, Lu Y, Kalsi JK, Jayandharan GR, Li B, Ma W, Cheng B, Gee SWY, McGoogan KE, Govindasamy L, Zhong L, Agbandje-McKenna M, Srivastava A. Human hepatocyte growth factor receptor is a cellular coreceptor for adeno-associated virus serotype 3. Hum Gene Ther. 2010;21:1741–1747. doi: 10.1089/hum.2010.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Di Pasquale G, Davidson BL, Stein CS, Martins I, Scudiero D, Monks A, Chiorini JA. Identification of PDGFR as a receptor for AAV-5 transduction. Nat Med. 2003;9:1306–1312. doi: 10.1038/nm929. [DOI] [PubMed] [Google Scholar]
- 234.Wu Z, Asokan A, Grieger JC, Govindasamy L, Agbandje-McKenna M, Samulski RJ. Single amino acid changes can influence titer, heparin binding, and tissue tropism in different adeno-associated virus serotypes. J Virol. 2006;80:11393–11397. doi: 10.1128/JVI.01288-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Weller ML, Amornphimoltham P, Schmidt M, Wilson PA, Gutkind JS, Chiorini JA. Epidermal growth factor receptor is a co-receptor for adeno-associated virus serotype 6. Nat Med. 2010;16:662–664. doi: 10.1038/nm.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Bell CL, Vandenberghe LH, Bell P, Limberis MP, Gao G-P, Van Vliet K, Agbandje-McKenna M, Wilson JM. The AAV9 receptor and its modification to improve in vivo lung gene transfer in mice. J Clin Invest. 2011;121:2427–2435. doi: 10.1172/JCI57367. [DOI] [PMC free article] [PubMed] [Google Scholar]