Abstract
Over the last decade, extraordinary progress has been made in elucidating the underlying genetic causes of gliomas. In 2008, our understanding of glioma genetics was revolutionized when mutations in isocitrate dehydrogenase 1 and 2 (IDH1/2) were identified in the vast majority of progressive gliomas and secondary glioblastomas (GBMs). IDH enzymes normally catalyze the decarboxylation of isocitrate to generate α-ketoglutarate (αKG), but recurrent mutations at Arg132 of IDH1 and Arg172 of IDH2 confer a neomorphic enzyme activity that catalyzes reduction of αKG into the putative oncometabolite D-2-hydroxyglutate (D2HG). D2HG inhibits αKG-dependent dioxygenases and is thought to create a cellular state permissive to malignant transformation by altering cellular epigenetics and blocking normal differentiation processes. Herein, we discuss the relevant literature on mechanistic studies of IDH1/2 mutations in gliomas, and we review the potential impact of IDH1/2 mutations on molecular classification and glioma therapy.
Keywords: brain tumor metabolism, D-2-hydroxyglutarate, epigenetics, glioma genetics, isocitrate dehydrogenase mutations
Normal Biochemistry of Isocitrate Dehydrogenases
Isocitrate dehydrogenase (IDH) enzymes catalyze the oxidative decarboxylation of isocitrate to form α-ketoglutarate (αKG), using NADP+ as a cofactor to generate NADPH during catalysis. IDH1 and IDH2 proteins share a high degree of sequence similarity (70% in humans) and are encoded by distinct genes (IDH1, 2q33; IDH2, 15q26). Although IDH1 and IDH2 are highly similar and catalyze identical, reversible reactions, IDH1 localizes to the cytosol and peroxisomes, while IDH2 localizes to the mitochondria. A third IDH enzyme, IDH3, catalyzes the forward decarboxylation of isocitrate to generate αKG for the tricarboxylic acid (TCA) cycle. IDH3 is an evolutionarily distinct multisubunit complex composed of 3 proteins encoded by distinct genes. In this review, we focus exclusively on the roles of IDH1 and IDH2 in cellular metabolism and glioma biology.
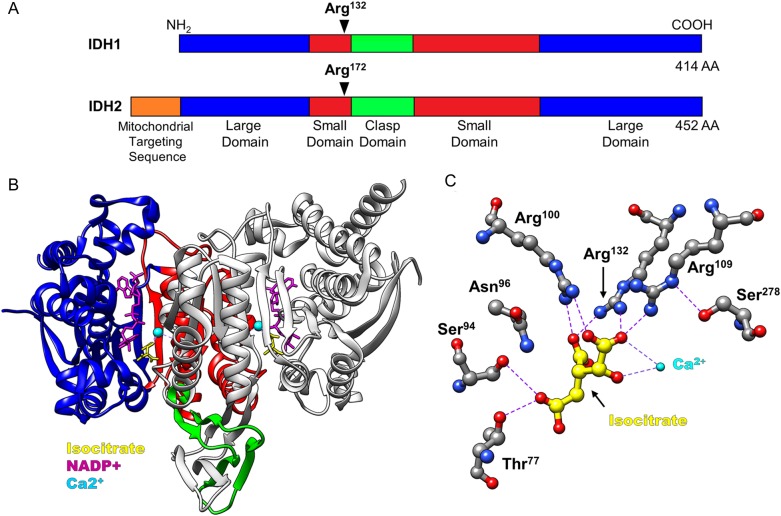
IDH1 and IDH2 are homodimeric enzymes that contain 2 active sites per dimer. Individual IDH subunits are composed of a large domain, small domain, and a clasp domain, and IDH2 contains an additional mitochondrial targeting sequence (Fig. 1A). Each active site contains binding sites for NADP(H), isocitrate, and a divalent cation1 (Fig. 1B). Catalysis proceeds by binding the NAPD+ cofactor in an inactive open conformation. This inactive conformation is characterized by a regulatory loop segment that prevents isocitrate binding to the active site by interacting with Ser94 of the IDH1 large domain. Isocitrate binding displaces the regulatory loop and is mediated by residues of both dimer subunits, including Ser94 and multiple conserved arginine residues in the active site (Fig. 1C). It is proposed that competitive binding of isocitrate to the catalytic cleft displaces the regulatory loop and induces a conformational change to a closed, catalytically active state that promotes decarboxylation of isocitrate to αKG. Interestingly, while IDH1/2 have no known allosteric regulators, bacterial IDH is inhibited by phosphorylation,2 suggesting that posttranslational modifications may modulate IDH1/2 activity.
Fig. 1.
Domain map and active site structure of IDH enzymes. (A) IDH1 and IDH2 are composed of 3 distinct domains: large domain, small domain, and clasp domain. IDH2 contains a 39 amino acid mitochondrial targeting sequence at its NH2-terminus. Conserved arginine residues at Arg132 (IDH1) and Arg172 (IDH2) are critical for catalysis. (B) Crystal structure of IDH1 homodimer, as reported in Xu et al1 (PDBID: 1T0L). Domains are color-coded as in (A), with only one subunit colored for clarity. The substrate binding pocket contains binding sites for isocitrate (yellow), a calcium ion (cyan), and NADP+ (purple). (C) Structure of critical residues in the IDH1 active site. Hydrogen bonds and hydrophilic interactions are depicted by purple-dashed lines and formed between multiple arginine residues, including Arg132, and carboxylate groups of isocitrate. IDH2 Arg172 is analogous to IDH1 Arg132 in its active site structure, and interactions with isocitrate. NADPH molecules have been removed for clarity.
IDH1 and IDH2 play important roles in a number of cellular functions, including glucose sensing, glutamine metabolism, lipogenesis, and regulation of cellular redox status. Although IDH1 and IDH2 catalyze identical reactions, these enzymes are thought to have distinct functions based primarily on their differential localizations and an altered catalytic flux between forward and reverse reactions. For example, IDHs are reported to modulate the cellular response to glucose by positively regulating insulin secretion.3,4 More recently, IDH1 has been shown to play a critical role in lipid metabolism in a variety of cellular contexts. IDH1 promotes lipogenesis during hypoxia by catalyzing the reductive carboxylation of αKG to generate acetyl-CoA for lipid synthesis.5,6 Overexpression of IDH1 in liver and adipose tissue causes obesity, fatty liver, and hyperlipidemia in transgenic mice.7 Furthermore, IDH1 regulates phospholipid metabolism in astrocytes of developing mice.8 Similar to the role of IDH1 in lipid metabolism, reverse flux of αKG through IDH2 has also been shown to promote synthesis of lipids by reductive glutamine metabolism.6,9–11 However, other studies have suggested that IDH2 may play a minor or more tissue-restricted role in reductive glutaminolysis.5,12
IDH1 is the principal source of NADPH in the human brain13 and is thought to be a major source of NADPH in other tissues.14,15 NADPH protects against oxidative damage by reducing oxidized glutathione and thioredoxin. The generation of NAPDH by IDH1, via oxidative decarboxylation of isocitrate, has been shown to protect against lipid peroxidation and oxidative DNA damage.16 IDH1/2 activity has also been shown to protect against replicative senescence by reducing oxidative DNA damage and lipid peroxidation in cell culture.17 Consistent with an antioxidant role of IDH1, other studies have shown that the NAPDH-producing activity of IDH1 protects against UVB-induced phototoxicity,18 reduces reactive oxygen species,19 and limits ischemia-reperfusion injury in the kidney.15,20 Collectively, these studies suggest that wild-type IDH1 and IDH2 play significant roles in managing the extent of oxidative stress in response to various cellular insults.
Discovery of IDH Mutations
In 2008, exome-sequencing studies of glioblastoma (GBM) tumors identified recurrent missense mutations in the gene IDH1.21 Interestingly, IDH1 mutations occurred in nearly all cases of secondary GBM in the study, (ie, tumors that had progressed from WHO grade II/III gliomas) but were rare in primary GBM cases. In subsequent studies, Yan et al reported that IDH1 and IDH2 mutations occur in a mutually exclusive manner in >80% of WHO grade II/III astrocytomas, oligodendrogliomas, and oligoastrocytomas.22 Numerous studies from our laboratory and others have now shown that IDH mutations occur in the vast majority of WHO grade II/III gliomas and secondary GBMs23–30 and occur in a number of other tumors, including acute myeloid leukemia (AML),31 intrahepatic cholangiocarcinoma,32 melanoma,33,34 and cartilaginous tumors.35 Additionally, somatic mosaic mutations in IDH1 cause Ollier disease and Maffucci syndrome, conditions that are characterized by the development of multiple cartilaginous tumors.36,37 The discovery of IDH mutations in glioma and other tumors revealed an unexpected role for IDH1 and IDH2 in the genesis and progression of human malignancies and prompted a series of studies to identify the mechanisms by which mutant IDH enzymes cause cancer.
In malignant glioma, mutant IDH proteins are almost ubiquitously expressed in tumor cells, and IDH mutations precede secondary and tertiary genetic lesions, suggesting that IDH mutations are an early causative event in the genesis of this brain tumor subset.23,29,38 IDH mutations are universally missense substitutions and are almost invariably heterozygous.39–41 Remarkably, mutations in IDH1 and IDH2 occur only at specific arginine residues in the active sites of these enzymes (Table 1). For IDH1, the most common alteration is R132H (c.395G>A),22,23,26,42,43 comprising >80% of all IDH mutations.21–24,42 Other IDH1 mutations at Arg132 occur at lower frequencies, including R132S, R132C, R132G, and R132L.22,42 IDH2 mutations occur at Arg172, the analogous amino acid to IDH1 Arg132, and are most commonly an IDH2 R172K (c.515G>A) missense substitution22,42 (Table 1).
Table 1.
Frequency of specific IDH mutations in gliomas. Data are represented as the percentage of total IDH1/2 mutations in glioma patients according to Yan et al and Hartmann et al22,42
| Gene | Mutation | Amino Acid Change | Frequency (%) |
|---|---|---|---|
| IDH1 | c.395G>A | R132H | 83.5–88.9 |
| c.394C>T | R132C | 3.9–4.1 | |
| c.394C>A | R132S | 1.5–2.4 | |
| c.394C>G | R132G | 0.6–1.3 | |
| c.395G>T | R132L | 0.3–4.1 | |
| IDH2 | c.515G>A | R172K | 2.4–2.7 |
| c.515G>T | R172M | 0.8–1.8 | |
| c.514A>T | R172W | 0.0–0.7 | |
| c.514A>G | R172G | 0.0–1.2 |
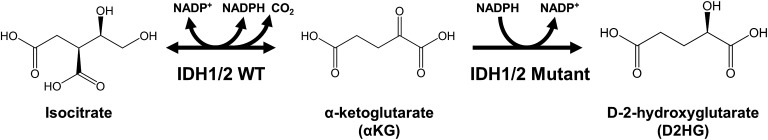
IDH1 Arg132 and IDH2 Arg172 are evolutionarily conserved residues in the active site of these enzymes and participate in isocitrate binding in the catalytic pocket (Fig. 1). Early reports suggested that IDH1/2 mutations caused a loss of normal oxidative catalytic function and dominant negative inhibition of the wild-type allele, which was supported by loss of NAPDH-producing activity and decreased affinity for isocitrate in mutant enzymes.22,44 However, in 2009, Dang et al reported the groundbreaking discovery that IDH1 mutations confer a gain-of-function neomorphic activity that reduces αKG to produce D-2-hydroxyglutarate (D2HG) in a manner that consumes NAPDH45 (Fig. 2). Subsequent studies confirmed this result and further demonstrated that leukemia-associated IDH2 mutations at Arg140, which is adjacent to IDH2 Arg172 in the active site, conferred an identical neomorphic function to produce D2HG from αKG.9,46
Fig. 2.
Neomorphic enzyme activity of mutant IDH enzymes. IDH1 and IDH2 catalyze the oxidative decarboxylation of isocitrate to generate αKG, using NADP+ as a cofactor and producing NADPH and CO2. Recurrent mutations in the active site of IDH1 and IDH2 confer a gain-of-function activity that catalyzes the conversion of αKG into D2HG in a manner that consumes NADPH.
The identification of recurrent IDH mutations in glioma and leukemia ultimately led to the discovery that IDH-mutant proteins harbor a unique gain-of-function activity to produce D2HG. These studies collectively demonstrated that IDH mutations are characteristic of a genetically and pathophysiologically distinct glioma subset.21–23,43 Accordingly, the following discussion will provide a detailed review of studies that have investigated the role of IDH mutations in glioma biology. In certain cases, experimental results from disease models other than glioma are included to highlight potential biological mechanisms underlying the role of IDH mutations in tumorigenesis.
D2HG as an Oncometabolite
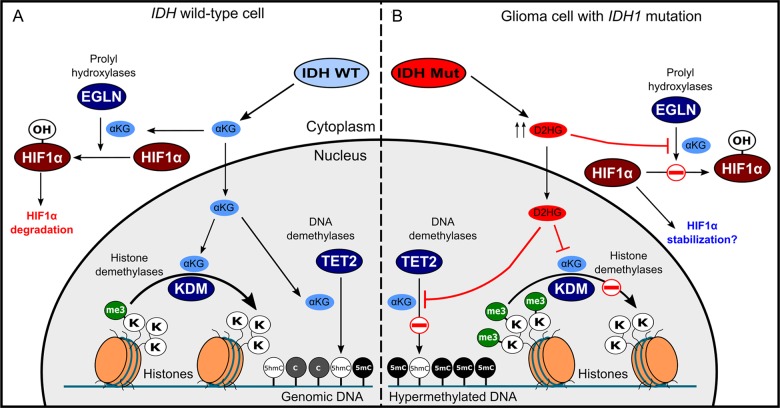
The discovery that IDH mutations cause aberrant D2HG production suggested that D2HG may play a causative role in the genesis of malignant brain tumors and led to the hypothesis that D2HG is an oncometabolite. D2HG and αKG are structurally similar metabolites and differ only by the presence of a C2 hydroxyl group in D2HG instead of the C2 carbonyl of αKG. Because of this similarity, it was hypothesized that D2HG may function as a competitive inhibitor of αKG-dependent dioxygenases, enzymes that regulate a number of important cellular processes by hydroxylating target proteins while using αKG as a cosubstrate. D2HG has been shown to inhibit several αKG-dependent dioxygenases, including histone demethylases47,48 (Fig. 3A and B). Xu et al reported that D2HG competitively inhibits the histone demethylase JHDM1A, and administration of cell-permeable D2HG is sufficient to increase histone H3K9 and H3K79 methylation in U87MG GBM cells47 and H3K9 trimethylation in HeLa cells.48 Similarly, IDH1R132H or IDH2R172K expression increases trimethylation of histone H3 at Lys9 and Lys27 in immortalized human astrocytes.49 Both D2HG and L2HG are potent inhibitors of Jumonji-domain-containing family members of histone demethylases.48 Specifically, D2HG potently inhibits the histone demethylases JMJD2A, JMJD2C, and FBXL11, with IC50 values at or below ∼100 μM for these enzymes.48
Fig. 3.
Cellular effects of elevated D2HG levels in glioma cells. IDH1 normally catalyzes the oxidative decarboxylation of isocitrate to generate αKG. Mutant IDH enzymes generate D2HG, which can accumulate in glioma cells to levels >100-fold compared with normal tissue. αKG functions as a cofactor for several cellular dioxygenases, including histone lysine demethylases, TET cytosine hydroxylases, and HIF prolyl hydroxylases. Excessive D2HG accumulation disrupts the normal function of αKG-dependent enzymes causing increases trimethylation of multiple histone lysine residues and decreased 5-hydoxymethlcytosine abundance as well as a concomitant increased in global 5-methylcytosine levels. Several reports also suggest that D2HG can inhibit HIF hydroxylases, preventing HIF1α degradation and increasing HIF1α-dependent transcription.
Because of the potential for D2HG to inhibit histone demethylases, recent studies have focused on the effects of IDH mutations and D2HG on cellular epigenetics and genome-wide DNA methylation. In glioma patients, IDH mutations are strongly associated with the glioma CpG island methylation phenotype (G-CIMP) in grade II/III gliomas and glioblastoma,23,26,50 and several lines of evidence now suggest that IDH mutations are sufficient to establish the G-CIMP phenotype in glioma cells. For example, Turcan et al reported that WHO grade II/III gliomas exhibit a distinct G-CIMP phenotype that can be recapitulated by expressing IDH1R132H in human astrocytes.51 A direct, mechanistic link between IDH mutations and DNA hypermethylation is provided by the observation that D2HG inhibits the activity of TET 5-methylcytosine hydroxylases.47 TETs are a family of αKG-dependent enzymes that catalyze the first step in an active DNA demethylation process that converts 5-methylcytosine to its unmethylated form through a 5-hydroxymethylcytosine (5-hmC) intermediate (Fig. 3A and B).52 Xu et al reported that expression of IDH1R132H or IDH2R172K decreases the abundance of 5-hmC in HEK293 cells by inhibiting the activity of TET1 and TET2 enzymes.47 Similarly, Sasaki et al generated a heterozygous IDH1R132H knock-in mouse model and found that IDH1R132H expression in mouse neural stem cells is sufficient to inhibit global 5-hmC levels.53 This finding is consistent with observations in glioma tissue, in which 5-hmC levels are lower in IDH-mutant gliomas relative to IDHWT gliomas.47 Interestingly, IDH mutations cause DNA hypermethylation and are mutually exclusive with TET2 loss-of-function mutations in AML.54 Collectively, these results suggest that a major mechanism by which IDH mutations contribute to tumorigenesis is the inhibition of TET enzymes and the consequent dysregulation of DNA demethylation dynamics.
Epigenetic changes induced by IDH mutations are reported to inhibit normal differentiation processes in multiple cell models, including mouse neural stem cells.49 Mutant IDH1 activity increases repressive histone methylation at promoters of astrocytic lineage markers and confers a block to differentiation in patient-derived glioma xenografts.55 Using a small-molecule inhibitor of mutant IDH1 activity, Rohle et al demonstrated that inhibition of mutant IDH1 could promote differentiation of neural stem cells and slow growth of an IDH1R132H subcutaneous xenograft in a mouse model.55 This finding is consistent with studies of IDH mutations in AML, which demonstrate that IDH2R140Q-induced epigenetic effects inhibit normal differentiation processes, and inhibition of IDH2R140Q activity induces differentiation of primary AML cells in culture.56
Studies on IDH mutations and D2HG production have led to a model in which the major oncogenic role of IDH mutations in glioma is altering DNA and histone methylation and inhibiting normal differentiation processes. Importantly, it has been reported that glioma stem-like cells with IDH mutations can be induced to differentiate by inhibiting the neomorphic activity of IDH1R132H or treating cells with the hypomethylating agent decitabine.55,57 However, in the first case, mutant IDH inhibition lowered D2HG levels in xenografts but did not reverse global DNA hypermethylation. In the second case, decitabine treatment induced differentiation while lowering DNA hypermethylation without affecting D2HG levels. Further work is therefore required to definitively identify an optimal strategy for targeting mutant IDH-dependent epigenetic effects in gliomas.
HIF1α is a hypoxia-inducible oncogenic transcription factor that regulates the expression of important modulators of tissue oxygenation and vascularization, including VEGF and EPO.58 HIF1α protein stability and transcriptional activity are controlled by proline and asparagine hydroxylation mediated by PHD2 (EGLN1) and FIH, respectively.59 HIF1α hydoxylases are αKG-dependent enzymes and may therefore be modulated by cellular D2HG levels in a competitive inhibitory manner (Fig. 3B). However, evidence supporting this hypothesis is somewhat unclear. For example, expression of an IDH1R132H transgene was initially reported to increase HIF1α protein expression due to IDH1 loss of function and decreased intracellular αKG levels.44 Expression of IDH1R132H or treatment with cell-permeable D2HG has also been shown to increase HIF1α expression.47 Similarly, inducible expression of IDH1R132H in the brains of embryonic mice causes increased HIF1α protein expression and increased steady-state levels of HIF1α-inducible genes such as VEGF and GLUT-1.53 In contrast, 2HG stereoisomers, particularly D2HG, are relatively weak inhibitors of HIF1α prolyl and asparaginyl hydroxylases in vitro, with IC50 values >1 mM.48 To further complicate the matter, D2HG has been reported to specifically increase the activity of the HIF1α hydroxylase EGLN in human astrocytes and colorectal cancer cells, leading to decreased levels of HIF1α protein.60 Further work is required to elucidate the relationship between elevated D2HG levels in glioma and the actions of HIF1α in brain tumors. It is possible that cell type and environmental factors play a dominant role in determining the extent to which D2HG accumulation can inhibit HIF hydroxylases. However, it is important to note that D2HG is known to accumulate in glioma tissue to concentrations that may exceed even the relatively high IC50 values associated with D2HG-mediated inhibition of FIH and EGLN.45
Elevated D2HG levels have also been proposed as a biomarker to guide the noninvasive detection of IDH mutations. An in-depth discussion of these methods is beyond the scope and space limitations of this article, but these techniques have been reviewed in detail elsewhere.61
Effects of IDH Mutations on Cellular Metabolism and Growth
IDH enzymes function in critical metabolic pathways in which they produce αKG and NADPH, metabolites that are required for normal macromolecule biosynthesis and redox balance. Therefore, in addition to D2HG-producing activity, mutations in IDH enzymes impair normal enzymatic function for converting isocitrate to αKG and generating NADPH. As a result, IDH mutations may have profound impacts on cellular metabolism by altering metabolic flux of αKG, depleting NAPDH, and impairing normal biosynthetic pathways that utilize IDH activity.
In their seminal study, Dang et al reported that D2HG levels reached remarkably high levels in glioma tissues harboring IDH mutations (ranging from 5–35 μmol D2HG/gram of tissue), but levels of other TCA cycle metabolites, including αKG, malate, fumarate, succinate, and isocitrate, were not significantly altered.45 This result suggested that IDH-mutant gliomas maintain normal levels of critical metabolites even in the presence of altered metabolic flux of αKG to D2HG. To investigate the effects of IDH mutations in cells, Reitman et al performed metabolomic profiling of human oligodendroglioma cells expressing IDH1R132H and IDH2R172K. In this study, mutant IDH expression caused widespread metabolic changes, including decreased levels of glutathione metabolites, alterations in free amino acids and TCA metabolite abundance, and significant reductions in N-acetyl-aspartyl-glutamate (NAAG).62 NAAG levels were also shown to be reduced in glioma tissues with IDH mutations compared with IDHWT gliomas.62 A more recent study reported that IDHR132H expression in U87 cells or human astrocytes caused similar decreases in glutamate and glutamine.63 A metabolomics study by Ohka et al showed that gliomas with IDH1 mutations had significantly decreased levels of glutamate and glutamine relative to IDHWT gliomas.64 The study also demonstrated that TCA metabolites were not reduced in the context of IDH mutations. These findings suggest that increased glutaminolysis maintains normal levels of key TCA cycle metabolites to compensate for altered flux of αKG to D2HG in IDH-mutant gliomas.
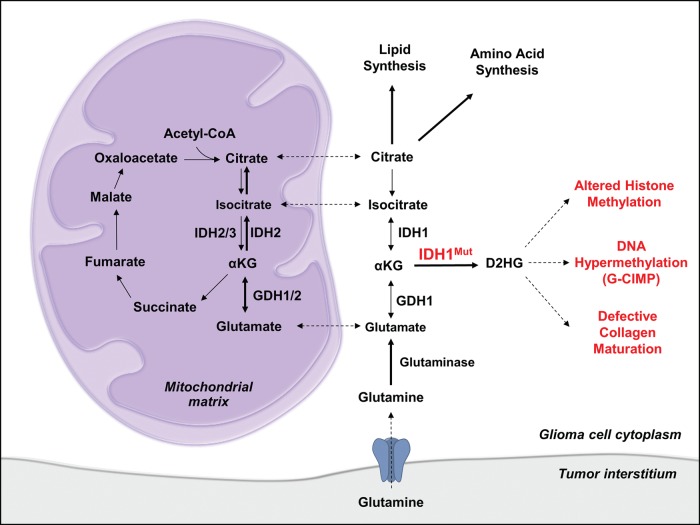
Compensatory metabolic flux, including increased glutaminolysis, is a likely mechanism by which IDH-mutant tumors maintain normal metabolite abundance in biosynthetic pathways while producing extraordinary amounts of D2HG (Fig. 4). In agreement with this hypothesis, it has been reported that glioma cells expressing IDH1R132H are sensitive to inhibition of glutaminase, a key enzyme mediating the anaplerotic flux of glutamine to αKG.65 In addition to glutaminase, glutamate dehydrogenases (GDHs), enzymes that catalyze the oxidative deamination of L-glutamate to αKG, have also been shown to be critical for maintaining normal metabolic flux in the presence of IDH1R132H expression (Fig. 4). Chen et al recently reported that mRNA expression of GDH1 and GDH2 is significantly elevated in IDH mutant GBMs relative to IDHWT GBMs.66 Additionally, they found that IDH1R132H expression in gliomagenic murine neural stem cells impaired the flux of glutamine and glucose to lipids and slowed growth of gliomas in a mouse model, suggesting that IDH1R132H-induced metabolic changes are growth-limiting in the absence of compensatory metabolic alterations. Importantly, transgenic expression of GDH2, but not GDH1, rescued glioma growth and promoted lipid synthesis in the presence of IDH1R132H expression.66 IDH1R132H lacks the reductive catalytic activity to generate citrate for lipid synthesis,67 yet IDH mutations increase the flux of glutamine to lipids, particularly under hypoxic conditions.64,66,68 It therefore seems likely that IDH1R132H-expressing tumors rely heavily on mitochondrial GDH2 and IDH2 for reductive flux of glutamine and glutamate to lipids (Fig. 4), which would explain the increased sensitivity of experimental IDH-mutant gliomas to glutaminase or GDH inhibition.65,66
Fig. 4.
Altered metabolism in gliomas with IDH1 mutations. In the presence of an IDH mutation, normal αKG flux is diverted to generate the oncometabolite D2HG, which acts as a competitive inhibitor of αKG-dependent enzymes. Elevated D2HG ultimately increases genome-wide levels of DNA methylation by inhibition TET cytosine hydroxylases, key enzymes that promote the active demethylation of 5-methylcytosine. D2HG also inhibits histone lysine demethylases and prolyl hydroxylase, thus increasing histone H3 trimethylation and disrupting normal collagen maturation. Glioma cells maintain normal levels of key metabolites in the presence of IDH mutations by increasing the relative anaplerotic flux of glutamine and glutamate into the TCA cycle. This pathway of reductive glutamine metabolism maintains levels of TCA metabolites that are critical for biosynthetic processes.
Collectively, studies on glioma metabolism have shown that IDH mutations induce widespread metabolic changes by altering critical metabolic pathways controlling macromolecule biosynthesis. Importantly, tumors harboring IDH mutations maintain normal levels of key metabolites by increasing the rate of glutaminolysis and increasing expression of enzymes involved in anaplerotic pathways, including GDH1 and GDH2 (Fig. 4). Future work should focus on the potential for therapeutically targeting compensatory metabolic pathways in IDH-mutant gliomas, including glutaminase and glutamate dehydrogenases.
Immunotherapeutic Targeting of IDH Mutations
Recent studies have begun to investigate the possibility of targeting IDH mutations by vaccination-based immunotherapy. In principle, IDH1/2 mutations are ideal tumor specific neo-antigens due to their uniform occurrence at specific codons in IDH1 and IDH2 and ubiquitous expression throughout all tumor cells. Schumacher et al recently showed that an IDH1R132H peptide vaccine is immunogenic in mice.69 Further, using MHC-humanized mice, they found that vaccination with a peptide containing IDH1R132H amino acids 123–142 elicited an MHC class II-specific antitumor response against IDH1R132H-expressing murine sarcomas. Additionally, another study recently demonstrated that immunization with an IDH1R132H-specfic peptide vaccine slowed the growth of intracranial tumors expressing an IDH1R132H transgene.70 These promising results strongly suggest that mutant IDH-targeted immunotherapies can elicit potent antitumor immune responses.
IDH1 and IDH2 Mutations as Glioma Biomarkers
In addition to their role in gliomagenesis, IDH mutations have clear value in providing more accurate diagnostic information for patients. Currently, the diagnosis of brain tumor subtype and grade is based largely on histological criteria according to guidelines established by the World Health Organization. However, accurate diagnosis and grading can be challenging due to subjectivity in the interpretation of histological features. For example, observer-specific subjectivity is most evident in discriminating between cases of astrocytomas, oligodendrogliomas, and mixed-histology oligoastrocytomas, all of which can share morphologic features. This lack of objectivity results in diagnostic inconsistencies, with one study showing concordance rates among neuropathologists as low as 52%.71 Accurate diagnosis is critical for clinical decision-making and optimizing therapy for patients as particular subtypes show increased radio and chemosensitivity, and dosages are determined by glioma type and grade.72 Genetic and molecular markers are therefore needed to supplement histological analyses to more clearly define distinct disease entities. This is perhaps best exemplified in the case of primary versus secondary GBM, which are histologically identical but have drastically different clinical courses and genetic profiles.73
In light of these diagnostic challenges and the need for clearer patient stratification, recent large-scale studies have aimed to identify genetic markers for glioma classification. These efforts have revealed clear molecular subtypes of glioma that correlate with histologic subtype and patient prognosis. In addition to IDH mutations, alterations such as 1p/19q loss of heterozygosity (LOH), EGFR amplification, MGMT promoter methylation, and mutations in TP53, ATRX, CIC, FUBP1, and the TERT promoter have contributed to identifying subtypes of glioma with distinct molecular signatures. This holds promise for more accurate diagnosis, improved clinical trial design, and tailored therapy. In this section, we will detail the diagnostic and prognostic value of IDH mutation and their roles in the molecular classification of glioma.
Role of IDH Mutations in Glioma Classification and Prognosis
The initial genomic studies that identified IDH mutations in gliomas also revealed that patients with IDH mutations exhibit distinct disease characteristics relative to IDHWT patients. Numerous studies reported that IDH1/2 mutations are associated with a younger age of diagnosis compared with IDHWT tumors in WHO grade II/III gliomas22,24 and glioblastoma.21–24 IDH mutations are generally associated with a better prognosis, specifically among patients with glioblastoma (31 vs 15 months) and anaplastic astrocytoma (65 vs 38 months) when compared with IDHWT tumors of the same histologic types.21,22,26,74–77 Interestingly, at least one study has reported that IDH status is only associated with longer survival in the context of adjuvant temozolomide therapy in WHO grade II gliomas,78 suggesting that IDH mutation may also be indicative of enhanced chemosensitivity. Furthermore, it is now well established that decreased MGMT expression, caused by MGMT promoter methylation, results in increased sensitivity to alkylating agents such as temozolomide.79,80 GBM patients with methylated MGMT promoter have a longer median survival of 21.7 months versus 15.3 months with the addition of temozolomide treatment.80 IDH mutations are strongly associated with the glioma G-CIMP in glioblastoma and WHO grade II/III gliomas,23,81 and the majority of G-CIMP GBMs and anaplastic gliomas also have MGMT promoter methylation,26,82 although methylation of this locus also exists to a lesser extent in IDHWT gliomas. Importantly, integrated analyses of DNA methylation, copy number variation, and mutation profiles have revealed that G-CIMP occurs in nearly all IDH-mutant WHO grade II/III gliomas and can be further defined into 2 distinct G-CIMP classes (CIMP-codeleted/CIMP-A or CIMP-non-codeleted/CIMP-B) based on the co-occurrence of IDH mutations with 1p/19q LOH.83,84 The CIMP-A DNA methylation profile is associated with 1p/19q codeletion and is a defining epigenetic feature of IDH-mutant gliomas harboring TERT promoter mutations and 1p/19q LOH, which are predominantly oligodendroglioma or oligoastrocytoma tumors. On the other hand, CIMP-B is a defining epigenetic feature of gliomas with IDH mutations that lack 1p/19q LOH and do not harbor TERT promoter mutations, which are predominantly astrocytomas.84
IDH mutations are thought to be the primary initiating event in WHO grade II/III gliomas and secondary GBMs. IDH-mutant tumors invariably acquire secondary lineage-defining genetic alterations that are closely related to histological subtypes (Table 2). For example, IDH-mutant astrocytomas frequently harbor mutations in the tumor suppressor TP53,85–87 while TP53 mutations are largely absent in histologically defined oligodendrogliomas.87,88 On the other hand, IDH-mutant oligodendrogliomas frequently harbor mutations in the genes CIC/FUBP1 and exhibit LOH for chromosomal arms 1p/19q.76,89–92 Importantly, it has been shown that TP53 mutations are exclusive of CIC/FUBP1 and 1p/19q alterations in mixed-histology astrocytomas.74,88 Therefore, the co-occurrence of secondary genetic alterations with IDH mutations may provide objective molecular biomarkers that clearly delineate mixed-histology oligoastrocytomas tumors into genetically distinct oligodendroglioma or astrocytoma subsets (Table 2).
Table 2.
Genetic classification of adult gliomas
| Genetic Subtype | IDHMutTERTMut Oligodendroglioma | IDHMutTERTWT Astrocytoma | IDHWTTERTWT Astrocytoma | IDHWTTERTMut Astrocytoma |
|---|---|---|---|---|
| Major genetic alterations |
IDH mutation TERT promoter mutation 1p/19q LOH CIC mutation FUBP1 mutation |
IDH mutation TERT promoter wild-type TP53 mutation ATRX mutation |
IDH wild-type TERT promoter wild-type EGFR amplification PTEN mutation CDKN2A/B deletion |
IDH wild-type TERT promoter mutation EGFR amplification PTEN mutation CDKN2A/B deletion |
| Associated histologies/classifications | Grade II/III Oligo Grade II/III OA |
Grade II/III Astro Grade II/III OA Secondary GBM Primary GBM |
Grade III Astro Primary GBM Secondary GBM |
Grade III Astro Primary GBM Secondary GBM |
| G-CIMP status | CIMP-A | CIMP-B | Non-CIMP | Non-CIMP |
Abbreviations: Astro, astrocytoma; GBM, glioblastoma; Oligo, oligodendroglioma; OA, oligoastrocytoma; WHO, World Health Organization.
In addition to the genetic alterations mentioned above, IDH-mutant gliomas exhibit distinct mutational patterns in the telomere maintenance genes ATRX and TERT. Inactivating mutations in ATRX are common in progressive astrocytomas (diffuse, anaplastic, and secondary GBM) and lead to alternative lengthening of telomeres.76,93 Mutations in the promoter region of TERT (ie, the catalytic subunit of telomerase) are common in primary GBM and oligodendrogliomas and lead to increased TERT expression and subsequent telomerase activation.74,76,88,94 Recent studies have demonstrated that TERT promoter mutations in oligodendroglioma and oligoastroctyoma are tightly linked to 1p/19q LOH, which is a defining lesion in these tumors and is associated with a better response to PCV (procarbazine, lomustine/CCNU, vincristine) treatment.95,96 Most significantly, a large-scale study of 332 Japanese patients found that 144 of 147 participants who had IDH mutations and 1p/19q LOH also had TERT promoter mutations.84 The combination of TERT promoter mutations and IDH mutations may therefore provide an objective basis for genetically defining glioma subtypes and accurately identifying oligodendroglioma patients likely to benefit from PCV chemotherapy.
IDH-mutant tumors also exhibit distinct mRNA expression and copy-number alteration (CNA) profiles in comparison with IDHWT tumors.23,26 IDH-mutant tumors exhibit a proneural gene expression signature and lack EGFR amplifications that are characteristic of IDHWT tumors.26 Recently, a large-scale study from the German Glioma Network reported that WHO grade II/III gliomas with IDH mutations exhibited distinct CNA profiles.97 In this study, unsupervised clustering of IDH-mutant tumors revealed 3 distinct groups defined by (i) 1p/19q codeletion, (ii) 7q copy-number gain with minimal CNAs on other chromosomes, and (iii) an IDH-mutant subgroup with relatively frequent gains and losses on multiple chromosomes.97 Furthermore, stratification of patients in the study by CNA profiles yielded more distinct prognostic subgroups than stratification by histological subtypes. This finding is consistent with recent results showing that patient stratification by TERT promoter and IDH mutation status is superior to histological characterization in defining clinically distinct subgroups of malignant gliomas.74 A remaining genetic subtype of glioma is tumors that lack both TERT promoter mutations and IDH mutations. These TERTWT-IDHWT tumors consist largely of primary GBMs and anaplastic astrocytomas and harbor many of the genetic alterations seen in traditional primary GBMs (aside from TERT promoter mutation), including EGFR amplification and CDKN2A deletion.84 Importantly, WHO grade III and IV TERTWT-IDHWT gliomas exhibit a significantly worse prognosis than IDH-mutant astrocytomas of the same grade,74,98 and the median overall survival of grade IV TERTWT-IDHWT tumors is only several months longer than primary GBMs that harbor TERT promoter mutations.74,88,99
Conclusion
The discovery of IDH1 and IDH2 mutations in gliomas is a seminal example of the power of unbiased genomic analyses to elucidate novel aspects of biology. The impact of this discovery has been far-reaching and continues to expand, with recent studies focusing on diverse fields ranging from peptide-mediated immunotherapy69,70 to mutation-guided enzyme redesign.100 Moreover, while the direct therapeutic targeting of IDH-mutant enzymes in leukemia has shown promise,56,101,102 the potential benefit of this strategy in gliomas is still unclear. Future work should comprehensively characterize the effects of IDH-mutant inhibitors on patient-derived intracranial xenografts to more thoroughly examine the therapeutic potential of IDH1/2 neomorphic enzyme inhibitors in the context of brain tumors. What is now clear, however, is that IDH mutations exert profound effects on epigenetics and cellular metabolism, and these glioma-associated alterations may serve as alternative targets for glioma therapy. Furthermore, IDH1 and IDH2 mutations are defining genetic markers at the apex of an emerging molecular classification scheme for distinct glioma subsets. In this way, IDH status provides outstanding utility as an objective biomarker to complement existing histological analyses and inform accurate diagnoses.
Funding
This work was funded by the V Foundation, the Accelerate Brain Cancer Cure Foundation, the Slomo and Cindy Silvian Foundation, the Voices Against Brain Cancer Foundation, the James S. McDonnell Foundation, American Cancer Society Research Scholar Award RSG-10–126–01-CCE, and National Cancer Institute Grants 5R01-CA140316, 1R01NS086943, and P50-CA190991. MSW is supported by the institutional training grant 5T32CA074736-15.
Conflict of interest statement. H.Y. receives royalties from Agios Pharmaceuticals and Personal Genome Diagnostics. H.Y. is a co-founder and owns stocks of Genetron Health (Beijing) Co. Ltd.
References
- 1.Xu X, Zhao J, Xu Z et al. Structures of human cytosolic NADP-dependent isocitrate dehydrogenase reveal a novel self-regulatory mechanism of activity. J Biol Chem. 2004;279(32):33946–33957. [DOI] [PubMed] [Google Scholar]
- 2.Hurley JH, Thorsness PE, Ramalingam V et al. Structure of a bacterial enzyme regulated by phosphorylation, isocitrate dehydrogenase. Proc Natl Acad Sci USA. 1989;86(22):8635–8639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Macdonald MJ, Brown LJ, Longacre MJ et al. Knockdown of both mitochondrial isocitrate dehydrogenase enzymes in pancreatic beta cells inhibits insulin secretion. Biochim Biophys Acta. 2013;1830(11):5104–5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ronnebaum SM, Ilkayeva O, Burgess SC et al. A pyruvate cycling pathway involving cytosolic NADP-dependent isocitrate dehydrogenase regulates glucose-stimulated insulin secretion. J Biol Chem. 2006;281(41):30593–30602. [DOI] [PubMed] [Google Scholar]
- 5.Metallo CM, Gameiro Pa, Bell EL et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2012;481(7381):380–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Filipp FV, Scott DA, Ronai ZA et al. Reverse TCA cycle flux through isocitrate dehydrogenases 1 and 2 is required for lipogenesis in hypoxic melanoma cells. Pigment Cell Melanoma Res. 2012;25(3):375–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koh H-J, Lee S-M, Son B-G et al. Cytosolic NADP+-dependent isocitrate dehydrogenase plays a key role in lipid metabolism. J Biol Chem. 2004;279(38):39968–39974. [DOI] [PubMed] [Google Scholar]
- 8.Bogdanovic E, Sadri A-R, Catapano M et al. IDH1 regulates phospholipid metabolism in developing astrocytes. Neurosci Lett. 2014;582:87–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ward PS, Patel J, Wise DR et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17(3):225–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Comte B, Vincent G, Bouchard B et al. Reverse flux through cardiac NADP(+)-isocitrate dehydrogenase under normoxia and ischemia. Am J Physiol Heart Circ Physiol. 2002;283(4):H1505–H1514. [DOI] [PubMed] [Google Scholar]
- 11.Wise DR, Ward PS, Shay JES et al. Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of α-ketoglutarate to citrate to support cell growth and viability. Proc Natl Acad Sci USA. 2011;108(49):19611–19616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hartong DT, Dange M, McGee TL et al. Insights from retinitis pigmentosa into the roles of isocitrate dehydrogenases in the Krebs cycle. Nat Genet. 2008;40(10):1230–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bleeker FE, Atai Na, Lamba S et al. The prognostic IDH1(R132) mutation is associated with reduced NADP+-dependent IDH activity in glioblastoma. Acta Neuropathol. 2010;119(4):487–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Winkler BS, DeSantis N, Solomon F. Multiple NADPH-producing pathways control glutathione (GSH) content in retina. Exp Eye Res. 1986;43(5):829–847. [DOI] [PubMed] [Google Scholar]
- 15.Kim J, Kim KY, Jang H-S et al. Role of cytosolic NADP+-dependent isocitrate dehydrogenase in ischemia-reperfusion injury in mouse kidney. Am J Physiol Renal Physiol. 2009;296(3):F622–F633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee SM, Koh H-J, Park D-C et al. Cytosolic NADP(+)-dependent isocitrate dehydrogenase status modulates oxidative damage to cells. Free Radic Biol Med. 2002;32(11):1185–1196. [DOI] [PubMed] [Google Scholar]
- 17.Kil IS, Huh TL, Lee YS et al. Regulation of replicative senescence by NADP+ -dependent isocitrate dehydrogenase. Free Radic Biol Med. 2006;40(1):110–119. [DOI] [PubMed] [Google Scholar]
- 18.Jo S-H, Lee S-H, Chun HS et al. Cellular defense against UVB-induced phototoxicity by cytosolic NADP(+)-dependent isocitrate dehydrogenase. Biochem Biophys Res Commun. 2002;292(2):542–549. [DOI] [PubMed] [Google Scholar]
- 19.Kim SY, Park J-W. Cellular defense against singlet oxygen-induced oxidative damage by cytosolic NADP + -dependent Isocitrate Dehydrogenase. Free Radic Res. 2003;37(3):309–316. [DOI] [PubMed] [Google Scholar]
- 20.Kim J, Kim JI, Jang H-S et al. Protective role of cytosolic NADP(+)-dependent isocitrate dehydrogenase, IDH1, in ischemic pre-conditioned kidney in mice. Free Radic Res. 2011;45(7):759–766. [DOI] [PubMed] [Google Scholar]
- 21.Parsons DW, Jones S, Zhang X et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321(5897):1807–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yan H, Parsons DWW, Jin G et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360(8):765–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lai A, Kharbanda S, Pope WB et al. Evidence for sequenced molecular evolution of IDH1 mutant glioblastoma from a distinct cell of origin. J Clin Oncol. 2011;29(34):4482–4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Balss J, Meyer J, Mueller W et al. Analysis of the IDH1 codon 132 mutation in brain tumors. Acta Neuropathol. 2008;116(6):597–602. [DOI] [PubMed] [Google Scholar]
- 25.Bleeker FE, Lamba S, Leenstra S et al. IDH1 mutations at residue p.R132 (IDH1(R132)) occur frequently in high-grade gliomas but not in other solid tumors. Hum Mutat. 2009;30(1):7–11. [DOI] [PubMed] [Google Scholar]
- 26.Brennan CW, Verhaak RGW, McKenna A et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qi S-T, Yu L, Lu Y-T et al. IDH mutations occur frequently in Chinese glioma patients and predict longer survival but not response to concomitant chemoradiotherapy in anaplastic gliomas. Oncol Rep. 2011;26(6):1479–1485. [DOI] [PubMed] [Google Scholar]
- 28.Ichimura K, Pearson DM, Kocialkowski S et al. IDH1 mutations are present in the majority of common adult gliomas but rare in primary glioblastomas. Neuro Oncol. 2009;11(4):341–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Watanabe T, Nobusawa S, Kleihues P et al. IDH1 mutations are early events in the development of astrocytomas and oligodendrogliomas. Am J Pathol. 2009;174(4):1149–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nobusawa S, Watanabe T, Kleihues P et al. IDH1 mutations as molecular signature and predictive factor of secondary glioblastomas. Clin Cancer Res. 2009;15(19):6002–6007. [DOI] [PubMed] [Google Scholar]
- 31.Paschka P, Schlenk RF, Gaidzik VI et al. IDH1 and IDH2 mutations are frequent genetic alterations in acute myeloid leukemia and confer adverse prognosis in cytogenetically normal acute myeloid leukemia with NPM1 mutation without FLT3 internal tandem duplication. J Clin Oncol. 2010;28(22):3636–3643. [DOI] [PubMed] [Google Scholar]
- 32.Borger DR, Tanabe KK, Fan KC et al. Frequent mutation of isocitrate dehydrogenase (IDH)1 and IDH2 in cholangiocarcinoma identified through broad-based tumor genotyping. Oncologist. 2012;17(1):72–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lopez GY, Reitman ZJ, Solomon D et al. IDH1(R132) mutation identified in one human melanoma metastasis, but not correlated with metastases to the brain. Biochem Biophys Res Commun. 2010;398(3):585–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shibata T, Kokubu A, Miyamoto M et al. Mutant IDH1 confers an in vivo growth in a melanoma cell line with BRAF mutation. Am J Pathol. 2011;178(3):1395–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Amary MF, Bacsi K, Maggiani F et al. IDH1 and IDH2 mutations are frequent events in central chondrosarcoma and central and periosteal chondromas but not in other mesenchymal tumours. J Pathol. 2011;224(3):334–343. [DOI] [PubMed] [Google Scholar]
- 36.Pansuriya TC, van Eijk R, d'Adamo P et al. Somatic mosaic IDH1 and IDH2 mutations are associated with enchondroma and spindle cell hemangioma in Ollier disease and Maffucci syndrome. Nat Genet. 2011;43(12):1256–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Amary MF, Damato S, Halai D et al. Ollier disease and Maffucci syndrome are caused by somatic mosaic mutations of IDH1 and IDH2. Nat Genet. 2011;43(12):1262–1265. [DOI] [PubMed] [Google Scholar]
- 38.Wakimoto H, Tanaka S, Curry WT et al. Targetable signaling pathway mutations are associated with malignant phenotype in IDH-mutant gliomas. Clin Cancer Res. 2014;20(11):2898–2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reitman ZJ, Yan H. Isocitrate dehydrogenase 1 and 2 mutations in cancer: alterations at a crossroads of cellular metabolism. J Natl Cancer Inst. 2010;102(13):932–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Horbinski C. What do we know about IDH1/2 mutations so far, and how do we use it?. Acta Neuropathol. 2013;125(5):621–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cairns RA, Mak TW. Oncogenic isocitrate dehydrogenase mutations: mechanisms, models, and clinical opportunities. Cancer Discov. 2013;3(7):730–741. [DOI] [PubMed] [Google Scholar]
- 42.Hartmann C, Meyer J, Balss J et al. Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: a study of 1,010 diffuse gliomas. Acta Neuropathol. 2009;118(4):469–474. [DOI] [PubMed] [Google Scholar]
- 43.Kang MR, Kim MS, Oh JE et al. Mutational analysis of IDH1 codon 132 in glioblastomas and other common cancers. Int J Cancer. 2009;125(2):353–355. [DOI] [PubMed] [Google Scholar]
- 44.Zhao S, Lin Y, Xu W et al. Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF-1alpha. Science. 2009;324:261–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dang L, White DW, Gross S et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462(7274):739–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jin G, Reitman ZJ, Spasojevic I et al. 2-hydroxyglutarate production, but not dominant negative function, is conferred by glioma-derived NADP-dependent isocitrate dehydrogenase mutations. PLoS One. 2011;6(2):e16812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu W, Yang H, Liu Y et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19(1):17–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chowdhury R, Yeoh KK, Tian Y-M et al. The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO Rep. 2011;12(5):463–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lu C, Ward PS, Kapoor GS et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483(7390):474–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guan X, Vengoechea J, Zheng S et al. Molecular subtypes of glioblastoma are relevant to lower grade glioma. PLoS One. 2014;9(3):e91216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Turcan S, Rohle D, Goenka A et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483(7390):479–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kohli RM, Zhang Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature. 2013;502:472–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sasaki M, Knobbe CB, Itsumi M et al. D-2-hydroxyglutarate produced by mutant IDH1 perturbs collagen maturation and basement membrane function. Genes Dev. 2012;26(18):2038–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Figueroa ME, Abdel-Wahab O, Lu C et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18(6):553–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rohle D, Popovici-Muller J, Palaskas N et al. An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science. 2013;340(6132):626–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang F, Travins J, DeLaBarre B et al. Targeted inhibition of mutant IDH2 in leukemia cells induces cellular differentiation. Science. 2013;340(6132):622–626. [DOI] [PubMed] [Google Scholar]
- 57.Turcan S, Fabius AWM, Borodovsky A et al. Efficient induction of differentiation and growth inhibition in IDH1 mutant glioma cells by the DNMT Inhibitor Decitabine. Oncotarget. 2013;4(10):1729–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Manalo DJ, Rowan A, Lavoie T et al. Transcriptional regulation of vascular endothelial cell responses to hypoxia by HIF-1. Blood. 2005;105(2):659–669. [DOI] [PubMed] [Google Scholar]
- 59.Hirota K, Semenza GL. Regulation of hypoxia-inducible factor 1 by prolyl and asparaginyl hydroxylases. Biochem Biophys Res Commun. 2005;338(1):610–616. [DOI] [PubMed] [Google Scholar]
- 60.Koivunen P, Lee S, Duncan CG et al. Transformation by the (R)-enantiomer of 2-hydroxyglutarate linked to EGLN activation. Nature. 2012;483(7390):484–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Andronesi OC, Rapalino O, Gerstner E et al. Detection of oncogenic IDH1 mutations using magnetic resonance spectroscopy of 2-hydroxyglutarate. J Clin Invest. 2013;123(9):3659–3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Reitman ZJ, Jin G, Karoly ED et al. Profiling the effects of isocitrate dehydrogenase 1 and 2 mutations on the cellular metabolome. Proc Natl Acad Sci USA. 2011;108(8):3270–3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Izquierdo-Garcia JL, Viswanath P, Eriksson P et al. Metabolic reprogramming in mutant IDH1 glioma cells. PLoS One. 2015;10:e0118781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ohka F, Ito M, Ranjit M et al. Quantitative metabolome analysis profiles activation of glutaminolysis in glioma with IDH1 mutation. Tumour Biol. 2014;35(6):5911–5920. [DOI] [PubMed] [Google Scholar]
- 65.Seltzer MJ, Bennett BD, Joshi AD et al. Inhibition of glutaminase preferentially slows growth of glioma cells with mutant IDH1. Cancer Res. 2010;70(22):8981–8987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen R, Nishimura MC, Kharbanda S et al. Hominoid-specific enzyme GLUD2 promotes growth of IDH1R132H glioma. Proc Natl Acad Sci USA. 2014;111(39):14217–14222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Leonardi R, Subramanian C, Jackowski S et al. Cancer-associated isocitrate dehydrogenase mutations inactivate NADPH-dependent reductive carboxylation. J Biol Chem. 2012;287(18):14615–14620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Reitman ZJ, Duncan CG, Poteet E et al. Cancer-associated isocitrate dehydrogenase 1 (IDH1) R132H mutation and d-2-hydroxyglutarate stimulate glutamine metabolism under hypoxia. J Biol Chem. 2014;289(34):23318–23328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schumacher T, Bunse L, Pusch S et al. A vaccine targeting mutant IDH1 induces antitumour immunity. Nature. 2014;512(7514):324–327. [DOI] [PubMed] [Google Scholar]
- 70.Pellegatta S, Valletta L, Corbetta C et al. Effective immuno-targeting of the IDH1 mutation R132H in a murine model of intracranial glioma. Acta Neuropathol Commun. 2015;3:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Coons SW, Johnson PC, Scheithauer BW et al. Improving diagnostic accuracy and interobserver concordance in the classification and grading of primary gliomas. Cancer. 1997;79:1381–1393. [DOI] [PubMed] [Google Scholar]
- 72.Van Den Bent MJ. Interobserver variation of the histopathological diagnosis in clinical trials on glioma: A clinician's perspective. Acta Neuropathol. 2010;120:297–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ohgaki H, Kleihues P. The definition of primary and secondary glioblastoma. Clin Cancer Res. 2013;19(4):764–772. [DOI] [PubMed] [Google Scholar]
- 74.Killela PJ, Pirozzi CJ, Healy P et al. Mutations in IDH1, IDH2, and in the TERT promoter define clinically distinct subgroups of adult malignant gliomas. Oncotarget. 2014;5(6):1515–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sanson M, Marie Y, Paris S et al. Isocitrate dehydrogenase 1 codon 132 mutation is an important prognostic biomarker in gliomas. J Clin Oncol. 2009;27(25):4150–4154. [DOI] [PubMed] [Google Scholar]
- 76.Jiao Y, Killela PJPJ, Reitman ZJZJ et al. Frequent ATRX, CIC, FUBP1 and IDH1 mutations refine the classification of malignant gliomas. Oncotarget. 2012;3(7):709–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Juratli Ta, Kirsch M, Geiger K et al. The prognostic value of IDH mutations and MGMT promoter status in secondary high-grade gliomas. J Neurooncol. 2012;110:325–333. [DOI] [PubMed] [Google Scholar]
- 78.Houillier C, Wang X, Kaloshi G et al. IDH1 or IDH2 mutations predict longer survival and response to temozolomide in low-grade gliomas. Neurology. 2010;75(17):1560–1566. [DOI] [PubMed] [Google Scholar]
- 79.Esteller M, Garcia-Foncillas J, Andion E et al. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med. 2000;343(19):1350–1354. [DOI] [PubMed] [Google Scholar]
- 80.Hegi ME, Diserens A-C, Gorlia T et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352(10):997–1003. [DOI] [PubMed] [Google Scholar]
- 81.Noushmehr H, Weisenberger DJ, Diefes K et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17(5):510–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Van Den Bent MJ, Erdem-Eraslan L, Idbaih A et al. MGMT-STP27 methylation status as predictive marker for response to PCV in anaplastic oligodendrogliomas and oligoastrocytomas. A report from EORTC study 26951. Clin Cancer Res. 2013;19(19):5513–5522. [DOI] [PubMed] [Google Scholar]
- 83.Wiestler B, Capper D, Sill M et al. Integrated DNA methylation and copy-number profiling identify three clinically and biologically relevant groups of anaplastic glioma. Acta Neuropathol. 2014;128(4):561–571. [DOI] [PubMed] [Google Scholar]
- 84.Suzuki H, Aoki K, Chiba K et al. Mutational landscape and clonal architecture in grade II and III gliomas. Nat Genet. 2015;0:1–14. [DOI] [PubMed] [Google Scholar]
- 85.Chung R, Whaley J, Kley N et al. TP53 gene mutations and 17p deletions in human astrocytomas. Genes Chromosom Cancer. 1991;3(I 991):323–331. [DOI] [PubMed] [Google Scholar]
- 86.Rasheed BKA, McLendon RE, Herndon JE et al. Alterations of the TP53 gene in human gliomas. Cancer Res. 1994;54:1324–1330. [PubMed] [Google Scholar]
- 87.Killela PJ, Pirozzi CJ, Reitman ZJ et al. The genetic landscape of anaplastic astrocytoma. Oncotarget. 2013;5(6):1452–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Killela PJ, Reitman ZJ, Jiao Y et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc Natl Acad Sci USA. 2013;110(15):6021–6026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cairncross JG, Ueki K, Zlatescu MC et al. Specific genetic predictors of chemotherapeutic response and survival in patients with anaplastic oligodendrogliomas. J Natl Cancer Inst. 1998;90(19):1473–1479. [DOI] [PubMed] [Google Scholar]
- 90.Labussière M, Idbaih a, Wang XW et al. All the 1p19q codeleted gliomas are mutated on IDH1 or IDH2. Neurology. 2010;74(23):1886–1890. [DOI] [PubMed] [Google Scholar]
- 91.Bettegowda C, Agrawal N, Jiao Y et al. Mutations in CIC and FUBP1 contribute to human oligodendroglioma. Science. 2011;333(6048):1453–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sahm F, Koelsche C, Meyer J et al. CIC and FUBP1 mutations in oligodendrogliomas, oligoastrocytomas and astrocytomas. Acta Neuropathol. 2012;123(6):853–860. [DOI] [PubMed] [Google Scholar]
- 93.Kannan K, Inagaki A, Silber J et al. Whole-exome sequencing identifies ATRX mutation as a key molecular determinant in lower-grade glioma. Oncotarget. 2012;3(10):1194–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Huang D, Wang Z, He X-J et al. Recurrent TERT promoter mutations identified in a large-scale study of multiple tumour types are associated with increased TERT expression and telomerase activation. Eur J Cancer. 2015;51(8):969–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cairncross G, Wang M, Shaw E et al. Phase III trial of chemoradiotherapy for anaplastic oligodendroglioma: Long-term results of RTOG 9402. J Clin Oncol. 2013;31(3):337–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Van Den Bent MJ, Brandes Aa, Taphoorn MJB et al. Adjuvant procarbazine, lomustine, and vincristine chemotherapy in newly diagnosed anaplastic oligodendroglioma: Long-term follow-up of EORTC brain tumor group study 26951. J Clin Oncol. 2013;31(3):344–350. [DOI] [PubMed] [Google Scholar]
- 97.Weller M, Weber RG, Willscher E et al. Molecular classification of diffuse cerebral WHO grade II/III gliomas using genome - and transcriptome - wide profiling improves stratification of prognostically distinct patient groups. Acta Neuropathol. 2015;129(5):679–693. [DOI] [PubMed] [Google Scholar]
- 98.Chan DT, Poon WS, Zhou L et al. TERT promoter mutations contribute to subset prognostication of lower-grade gliomas. Mod Pathol. 2014;28(2):177–186. [DOI] [PubMed] [Google Scholar]
- 99.Labussiere M, Di Stefano AL, Gleize V et al. TERT promoter mutations in gliomas, genetic associations and clinico-pathological correlations. Br J Cancer. 2014;111(10):2024–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Reitman ZJ, Choi BD, Spasojevic I et al. Enzyme redesign guided by cancer-derived IDH1 mutations. Nat Chem Biol. 2012;8(11):887–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Losman J-A, Looper RE, Koivunen P et al. (R)-2-hydroxyglutarate is sufficient to promote leukemogenesis and its effects are reversible. Science. 2013;339(6127):1621–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kernytsky A, Wang F, Hansen E et al. IDH2 mutation induced histone and DNA hypermethylation is progressively reversed by small molecule inhibition. Blood. 2014;125(2):296–304. [DOI] [PMC free article] [PubMed] [Google Scholar]