Abstract
Background
To understand the ability of gliomas to manipulate their microenvironment, we visualized the transfer of vesicles and the effects of tumor-released extracellular RNA on the phenotype of microglia in culture and in vivo.
Methods
Extracellular vesicles (EVs) released from primary human glioblastoma (GBM) cells were isolated and microRNAs (miRNAs) were analyzed. Primary mouse microglia were exposed to GBM-EVs, and their uptake and effect on proliferation and levels of specific miRNAs, mRNAs, and proteins were analyzed. For in vivo analysis, mouse glioma cells were implanted in the brains of mice, and EV release and uptake by microglia and monocytes/macrophages were monitored by intravital 2-photon microscopy, immunohistochemistry, and fluorescence activated cell sorting analysis, as well as RNA and protein levels.
Results
Microglia avidly took up GBM-EVs, leading to increased proliferation and shifting of their cytokine profile toward immune suppression. High levels of miR-451/miR-21 in GBM-EVs were transferred to microglia with a decrease in the miR-451/miR-21 target c-Myc mRNA. In in vivo analysis, we directly visualized release of EVs from glioma cells and their uptake by microglia and monocytes/macrophages in brain. Dissociated microglia and monocytes/macrophages from tumor-bearing brains revealed increased levels of miR-21 and reduced levels of c-Myc mRNA.
Conclusions
Intravital microscopy confirms the release of EVs from gliomas and their uptake into microglia and monocytes/macrophages within the brain. Our studies also support functional effects of GBM-released EVs following uptake into microglia, associated in part with increased miRNA levels, decreased target mRNAs, and encoded proteins, presumably as a means for the tumor to manipulate its environs.
Keywords: brain tumors, exosomes, extracellular vesicles, microglia, microRNA
Glioblastoma (GBM) accounts for 12%–15% of intracranial tumors, with an incidence of 2–3 new cases per 100 000 people per year.1 The standard of care, consisting of surgical resection combined with chemotherapy and radiotherapy, provides a median survival of about 14 months from diagnosis.1 Extensive evidence indicates that cancer cells can subvert surrounding normal cells to promote tumor growth, angiogenesis, invasion, and metastases.2,3 GBM tumors rarely metastasize, but they exert influence over endogenous cell types within the brain, including microglia, macrophages, astrocytes, oligodendrocytes, neurons, and endothelial cells.2 In addition, blood monocytes enter the tumor-bearing brain and differentiate into macrophages in association with GBM.4 GBM tumor cells attract microglia and monocytes/macrophages by secreting chemokines, cytokines, and matrix proteins and stimulate their proliferation, resulting in their abundance within the tumor mass. Following interaction with tumor cells, microglia/macrophages show phenotypic activation changes that direct them toward an immunosuppressive state with increased release of cytokines, such as interleukin (IL)-10 and transforming growth factor (TGF)–β, and upregulation of arginase-1 (Arg-1) and matrix metalloprotease, which support growth and invasion associated with tumor progression.3,5,6
GBM tumors are known to mold their environment to their advantage by secretion of proteins and the display of cell surface ligands and with increasing evidence supporting transfer of instructional extracellular RNA (exRNA) and proteins/lipids contained within extracellular vesicles (EVs; including exosomes, microvesicles, and ectosomes), ribonucleoproteins (RNPs), and high-density lipoproteins (HDLs).7–9 The important role of EVs in cancer progression has been well documented.2,10 Once released, EVs can be internalized into “recipient cells,” potentially delivering genetic information to multiple cell types in the tumor microenvironment. This constitutes a new type of intercellular communication—the transfer of informative RNA between cells. Recent studies support the functional transfer of microRNA (miRNA) and other noncoding RNAs from tumor cells to normal cells.11–13
We investigated the activity of exRNA released from glioma cells and taken up by microglia and macrophages as a means by which tumor cells manipulate normal cells in their microenvironment. We monitored phenotypic changes in microglia exposed to isolated human GBM-EVs in culture, as well as the uptake of EVs and specific miRNAs and their effects on target mRNAs in an intracranial mouse glioma model. We focused on 2 miRNAs, miR-451 and miR-21, which naturally have very high levels in the EVs produced by primary GBM cells. Exposure of microglia in culture to these GBM-EVs elevated levels of these miRNAs and decreased levels of a common mRNA target encoding c-Myc. Further, utilizing a syngeneic mouse glioma model expressing red fluorescent protein in tumor cells and their EVs, and green fluorescent protein (GFP) in microglia and monocytes/macrophages, we found that infiltration of tumors by these cells was associated with their uptake of labeled tumor EVs, as visualized with multiphoton in vivo microscopy. Fluorescence activated cell sorting (FACS) of brain cells revealed increased levels of miR-21, decreased levels of c-Myc mRNA, and increases in activation-related Arg-1 mRNA. Our results are consistent with functional transfer of miRNAs from glioma cells to surrounding microglia and macrophages via EVs, as a means of modulating their phenotype, albeit EVs contain many types of RNAs and proteins which, along with the secretome of glioma cells, probably exert a combinatorial effect.
Materials and Methods
Details of methods are provided in the Supplementary material.
Cell Culture
Primary human GBM cells from 2 patients, 11/5 (GBM1) and 20/3 (GBM2),7 mouse glioma line GL261,14 mouse microglial line KW3 (J.E.K.), primary neonatal mouse microglia, and adult human microglia were cultured under standard conditions.
For stimulation experiments, EVs were isolated from two 150-mm plates of GBM1 or GBM2 cells (1–2 × 1011 EVs) and added to cultures of 0.5 × 105 microglia cells.
GBM2 cells were stably transduced using a CSCW2 lentivector (from Dr Sena-Esteves) encoding palmitoylated GFP (palmGFP).15 GL261 cells were stably transduced with lentivectors encoding firefly luciferase (Fluc), mCherry (mC), and palmtdTomato (palmdT).15
Isolation of Extracellular Vesicles
GBM/glioma cells are cultured in EV-depleted fetal bovine serum. After 48 h, conditioned media were collected and centrifuged for 10 min at 300× g, then 10 min at 2000× g, and the supernatant was filtered through a 0.8-μm filter (Millipore). EVs were pelleted by centrifugation at 100 000× g for 80 min in a Type 70 Ti Rotor (Beckman Coulter). Fractionation of EV pellets was achieved by sucrose density gradient centrifugation.
Visualization of EV Uptake in Culture and In vivo
Primary mouse and human microglia were plated on coverslips coated with poly-l-lysine and incubated with GBM2 palmGFP-EVs or GL261 palmGFP-EVs for 24 h. Cells were fixed with 4% paraformaldehyde for 10 min at room temperature. Coverslips were mounted on slides using ProLong Gold antifade reagent with 4′,6′-diamidino-2-phenylindole (DAPI) (Molecular Probes). Vesicle uptake into cells was analyzed by fluorescent microscopy using a Carl Zeiss LSM 5 Pascal laser-scanning confocal microscope. Multiphoton intravital microscopy (MP-IVM) and immunohistochemistry were used for the direct visualization of EV uptake into cells in vivo.
Mouse Cytokine Array and TGF-β Quantification
Primary microglia were cultured with GBM-EVs for 48 h, with Brefeldin A (1 μg/mL; Sigma-Aldrich) added during the last 8 h. Cells were lysed and cytokine expression was determined using a mouse cytokine array (R&D Systems). Membranes were scanned and analyzed for pixel intensity using ImageJ. TGF-β in GBM-EVs and microglia was measured by an enzyme-linked immunosorbent assay (ELISA) human/mouse TGF-β ELISA Ready-Set-Go kit (eBioscience).
Quantitative Real-time PCR
Total cellular and EV RNA was isolated with the miRNeasy Mini Kit (Qiagen). RNA was concentrated by ethanol precipitation. RNA yields were determined with Nanodrop (Thermo Fisher Scientific), size and quality with a 2100 Bioanalyzer (Agilent).
For miRNA analysis of GBM cells and EVs, a miRNA expression-profiling panel (1146 miRNAs; Illumina) was used. Individual miRNA expression was analyzed using TaqMan microRNA assays (Life Technologies) and normalized to U6 RNA.
For quantitative real-time PCR (qRT-PCR) of mRNAs, 50 ng RNA was reverse transcribed using Sensiscript (Qiagen) with Oligo-dT (Roche) and random nanomers (Sigma-Aldrich). Quantitative RT-PCR was performed using Power SYBR Green PCR Master Mix (Invitrogen) (primer sequences in Supplementary Table S1). Relative levels of RNA were determined using the comparative delta cycle threshold (ΔCt) method. All Ct values were normalized to glyceraldehyde 3-phosphate dehydrogenase mRNA.
Syngeneic Glioma Mouse Model
Animal experimentation was conducted under the oversight of the Massachusetts General Hospital Institution Animal Care and Use Committee. CX3CR1−GFP/GFP knock-in mice (J.E.K.)16 were bred with C57BL/6 mice (Charles River Laboratories), and the resulting heterozygous CX3CR1GFP/+ offspring were used for the intracranial injections of GL261 glioma cells.
Fluorescence Activated Cell Sorting/RNA Analysis
Eighteen days post tumor injections, the mice were anesthetized and sacrificed by transcardiac perfusion with phosphate buffered saline (PBS). Brains were removed and cells processed for FACS using a microglia isolation protocol.17 RNA was isolated from sorted cell fractions for analysis.
Immunoblot
Samples were lysed in radioimmunoprecipitation assay buffer supplemented with a protease inhibitor cocktail (Roche). Total protein concentration was measured using the Bio-Rad protein assay, and 20 μg protein was loaded onto precast 4%–12% or 10% Bis-Tris polyacrylamide gel (Invitrogen). Proteins were transferred onto polyvinylidene fluoride membranes (EMD Millipore). Membranes were blocked in 5% nonfat dry milk in Tris-buffered saline–Tween and incubated with antibodies against apoptosis-linked gene-2 interacting protein X (ALIX) (1:500; Santa Cruz: sc-53538), phosphatase and tensin homolog (PTEN) (1:200; Santa Cruz: sc-6818), migration inhibitory factor (MIF) (1:100; Santa Cruz: sc-20121), c-Myc (1:100; Santa Cruz: sc19), glyceraldehyde 3-phosphate dehydrogenase (1:2000; Millipore), TGF-β (1:500; Cell Signaling), and calcium binding protein 39 (CAB39) (1:500; Cell Signaling). Membranes were incubated with their relative horseradish peroxidase–conjugated antibody (1:5000) and imaged using a chemiluminescence detection kit (Thermo Fisher Scientific).
Viability Assay
Cell viability was determined using CellTiter-Glo Luminescent Assay (Promega). Luminescence was measured on a microtiter luminometer (Dynex Technologies).
Acetylcholinesterase Assay
Isolated EVs were resuspended in PBS, and absorbance was measured to establish a baseline. Acetylthiocholine was added to 1.25 mM in combination with 5,5′-dithiobis(2-nitrobenzoic acid) to 0.1 mM. Change in absorbance was measured at 450 nm every 5 min over 30 min using an MLX microtiter luminometer (Dynex Technologies).
Nanoparticle Tracking Analysis
Number of EVs in PBS was assayed using a Nanoparticle Tracking Analysis (NTA) Version 2.2 Build 0375 instrument (NanoSight). Particles were measured for 60 s and number of particles (30–800 nm) was determined using NTA Software 2.2.
Statistical Analysis
The unpaired 2-sample t-test was used to compare between 2 groups. One-way ANOVA, followed by Bonferroni's test, was conducted to test for significance among multiple groups, comparing all pairs of columns.
Results
GBM-derived EVs Are Internalized by Microglia
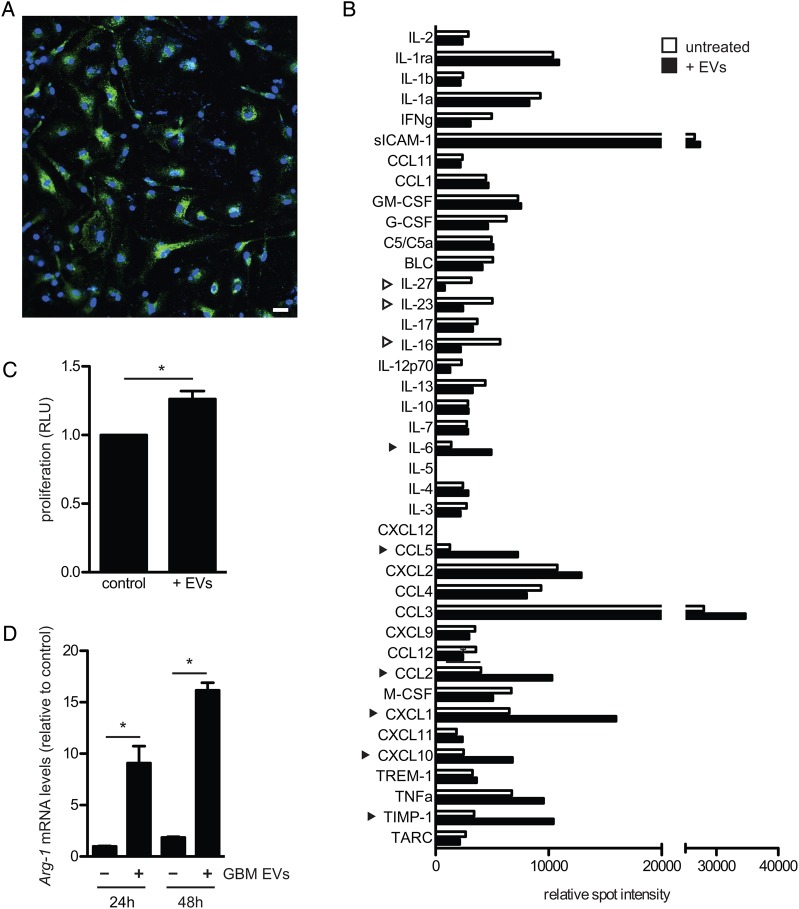
Two primary human GBM cell lines, GBM 11/5 (GBM1) and GBM 20/3 (GBM2) have been previously characterized for mRNA content in cells,7 with the most highly expressed mRNAs being consistent with the mesenchymal subtype.18 Both lines released large quantities of EVs (mg protein, Supplementary Fig. S1A, and number of particles, Supplementary Fig. S1C), the majority of which were 100–200 nm in diameter (Supplementary Fig. S1D) and contained the EV marker acetylcholinesterase (Supplementary Fig. S1B). Variations in the number of EVs released by different GBM/glioma lines have been documented.19 GBM2 cells which released more EVs than GBM1 cells were used in most experiments. Vesicle internalization by microglia was assessed by exposing primary neonatal mouse microglia to isolated EVs from GBM2 cells expressing membrane-tagged GFP (palmGFP). After 24 h, most microglia had taken up many fluorescently tagged EVs (Fig. 1A).
Fig. 1.
Exposure of primary mouse microglia to GBM-EVs directs them toward a tumor-associated phenotype. (A) EVs isolated from GBM2 cells expressing palmGFP were incubated for 24 h with primary mouse microglia, followed by confocal fluorescent microscopy using a 20× objective and DAPI staining. (B) Microglia were exposed for 48 h to GBM-EVs (black bars) and compared with untreated cells (white bars). Cytokines increasing over 50% are indicated by black arrowheads, those decreasing by over 50% by white arrowheads. (C) Microglia were exposed to GBM-EVs every 24 h for 5 days. Viability was measured after 7 days and compared with untreated cells (mean ± SEM, *P < .05). (D) Arg-1 mRNA was quantitated in microglia after exposure to GBM-EVs for 24 and 48 h (mean ± SEM, n = 2, *P < .05). Scale bar, 20 μm in (A).
Uptake of GBM-derived EVs Changes the Phenotype of Microglia
First, we tested whether uptake of EVs could change the cytokines produced by mouse microglia. To mimic the constant exposure of microglia to EVs in the tumor microenvironment, microglia were incubated with freshly isolated GBM-EVs every 24 h over 5 days. The relative levels of 40 different cytokines and chemokines were compared in microglia exposed to EVs versus unexposed cells using a Cytokine Antibody Array kit (R&D Systems). Densitometric analysis showed 6 cytokines that were more than 50% upregulated following GBM-EV exposure, comprising tissue inhibitor of metalloproteinase 1 (TIMP-1), chemokines CXCL10, CXCL1, CCL2, and CCL5, and the cytokine IL-6 (Fig. 1B). Of these upregulated cytokines, TIMP-1 is involved in extracellular matrix degradation,20 and the chemokines CXCL10, CXCL1, CCL2, and CCL5, and the cytokine IL-6 all induce glioma growth.3 In contrast, 3 cytokines involved in induction of immune responses were more than 50% downregulated: IL-16 (orchestrates immune response),21 IL-23 (promotes inflammation), and IL-27 (together with IL-23 enhances immunologic functions).22 Thus, exposure/uptake of GBM-EVs changes the cytokine secretion profile of microglia toward a phenotype that promotes growth and invasion of GBM cells while decreasing the immune response. In addition, we found that GBM-EVs contain high levels of TGF-β (Supplementary Fig. S2A). Exposure of mouse microglia to GBM-EVs did not increase mouse TGF-β1 mRNA (Supplementary Fig. S2B) but did increase intracellular levels of TGF-β, presumably through uptake of GBM-EVs (Supplementary Fig. S2C).
Second, we found that the presence of GBM-EVs increased the proliferation of mouse microglia by about 40% over 7 days (Fig. 1C). Further, exposure of microglia to GBM-EVs increased levels of Arg-1 (almost 10-fold) after 24 and 48 h (Fig. 1D), consistent with an activated tumor-associated phenotype.
Specific MiRNA Content in GBM Cells and EVs Compared with Microglia
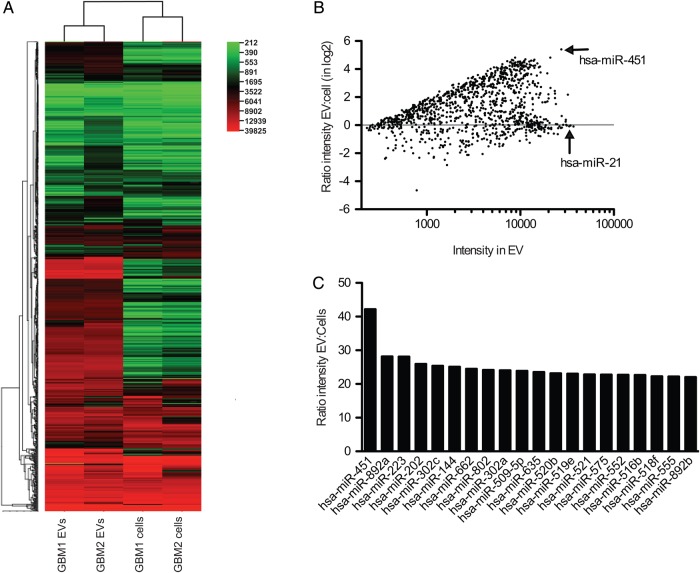
Microarray analysis of 1146 different miRNAs was performed on primary human GBM cell lines, GBM1 and GBM2, and on EVs isolated from conditioned medium from these cells (Fig. 2A). Many miRNAs were expressed at similar levels in cells and EVs, while some showed a distinct difference in levels (Fig. 2B). Two of the most abundant miRNAs in vesicles were miR-451, which was over 40-fold higher in EVs than in cells (Fig. 2C), and miR-21, a known oncomir,23 which had similarly high levels in both cells and EVs (Fig. 2B). We verified the expression levels of miR-21 and miR-451 in primary GBM1 and GBM2 cells and GBM-EVs by TaqMan assays. Relative levels of miR-21 were not significantly different between cells and EVs (Fig. 3A; Supplementary Fig. S3A), whereas miR-451 levels were 1000- to 10 000-fold higher in the vesicle fraction compared with cells (Fig. 3B; Supplementary Fig. S3B).
Fig. 2.
EVs secreted by GBM cells are enriched for many miRNAs. (A) MiRNA array analyses of 2 human primary GBM cell lines and EVs secreted by them were performed and a heat map was created using CIMminer.41 (B) Ratio of intensity in EVs compared with cells is presented in log2 scale for all 1146 miRNAs. (C) Comparison of the 20 most highly enriched miRNAs in EVs from GBM1 and GBM2 cells.
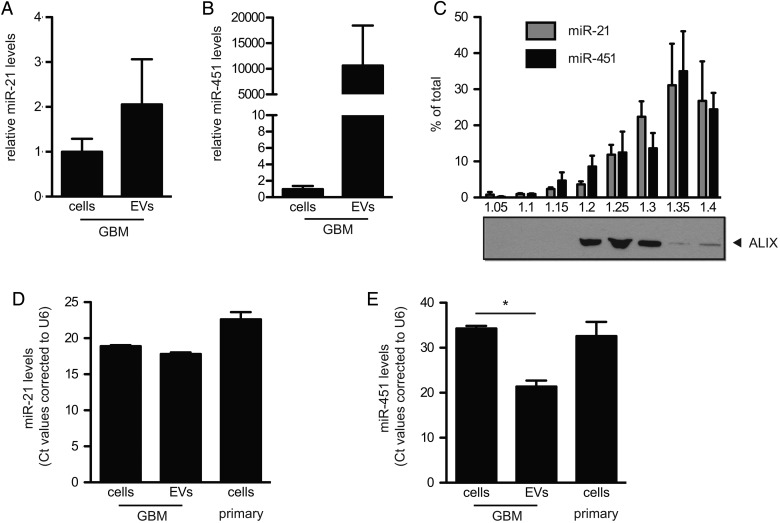
Fig. 3.
GBM cells release EVs enriched for miR-451 and miR-21 compared with microglia. (A and B) Levels of miR-21 (A) and miR-451 (B) in EVs and GBM2 cells were analyzed (mean ± SEM, n = 2). (C) Isolated GBM-EVs were separated on a sucrose density gradient. Levels of miR-21 and miR-451 in the different fractions were analyzed (mean ± SEM, n = 2). Protein levels of ALIX were analyzed by western blotting. (D and E) Levels of miR-451 (D) and miR-21 (E) in GBM cells, GBM-EVs, and primary mouse microglia were analyzed (Ct values presented, n = 2, *P < .05).
Microglia were chosen as the recipient normal brain cell based on their importance to tumor growth,3,5,6 relative ease of culture of primary cells from mouse and human sources, and the availability of transgenic mice with GFP+ microglia/macrophages.16 GBM cells are known to release abundant levels of small noncoding RNAs, such as miRNAs.24 Given the ability of EVs to carry intact miRNAs, we evaluated whether this extracellular miRNA could serve as a mechanism by which GBM cells could manipulate microglia. We hypothesized that the intercellular transfer of miRNAs from GBM cells to microglia might contribute to their phenotypic changes.
Sucrose density gradient centrifugation was used to resolve components of the GBM-EV preparation on the basis of buoyant density (Fig. 3C). Density fractions were analyzed for the presence of the exosomal marker ALIX,25 as well as for miRNA levels. ALIX was most prominent in the fractions, with densities of 1.20, 1.25, and 1.30 mg/mL (Fig. 3C), typical of classical exosomes. MiR-21 and miR-451 colocalized with the exosome marker ALIX but were also found in denser fractions which were negative for ALIX. These denser fractions might contain other types of EVs, RNPs, or HDL particles.
We also evaluated the endogenous levels of miR-451 and miR-21 in mouse primary microglia. MiR-21 and miR-451 had higher Ct levels in mouse microglia compared with GBM-EVs (Fig. 3D and E). For miR-21, the Ct difference between GBM-EVs and mouse microglia was 4.8 (equivalent to 32-fold), and for miR-451 this differential Ct was 11.2 (equivalent to about 2000-fold). Note the much higher levels of miR-21 compared with miR-451, in both GBM cells and EVs. The high levels of these miRNAs in GBM-EVs compared with the low levels in microglia was exploited to monitor functional transfer of these miRNAs from GBM cells into microglia via exRNA.
GBM-EV–Mediated Transfer of MiR-451/MiR-21 to Microglia and Decrease in c-Myc mRNA
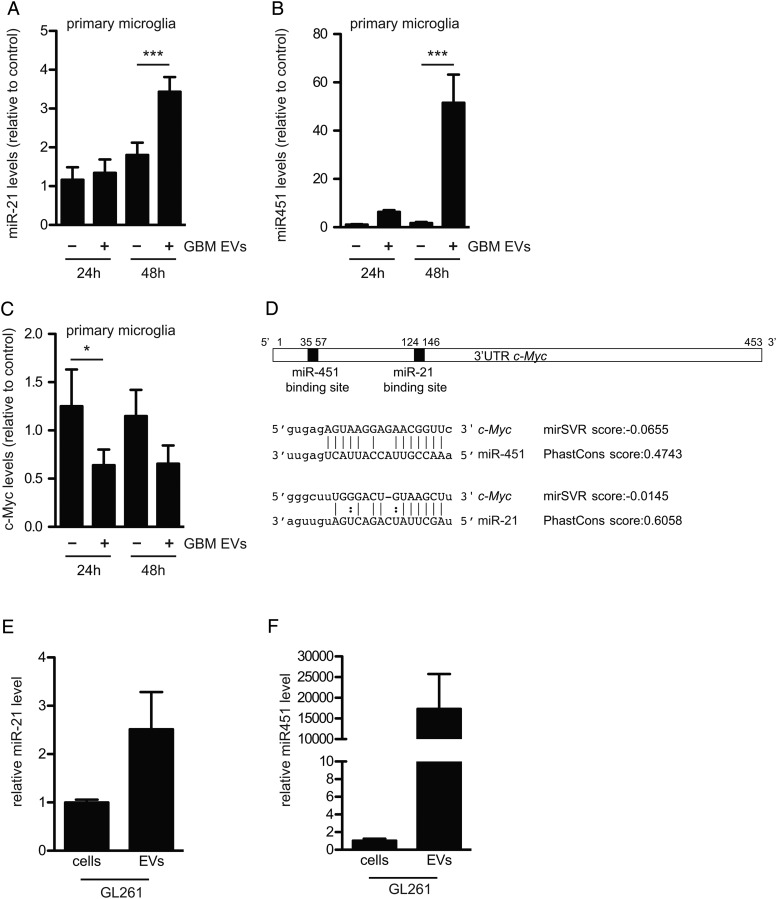
To investigate whether GBM-EVs were capable of transferring miRNAs between cells, miR-451 and miR-21 levels in mouse microglia were monitored 24 and 48 h after exposure to GBM-EVs. Initially, analysis was carried out using the murine microglial cell line KW3 with a trend seen toward higher levels of miR-21 and miR-451 after uptake of GBM-EVs (Supplementary Fig. S3C and D). To more closely mimic the in vivo situation, we tested primary mouse microglia, in which case levels of miR-21 significantly increased 1.3- and 5-fold after 24 and 48 h of exposure to GBM-EVs, respectively (Fig. 4A). Strikingly, levels of miR-451 in primary microglia increased up to 50-fold compared after 48 h exposure (Fig. 4B). In addition, human adult primary microglia exposed to labeled GBM-EVs rapidly took up the vesicles (Supplementary Fig. S6A) and displayed notably increased levels of miR-21 (1.5-fold) and miR-451 (4-fold) (Supplementary Fig. S6B and C). The GBM-EV–mediated increase in miR-451 and miR-21 levels in mouse microglia was partially blocked by heparin, which interferes with EV uptake (Supplementary Fig. S4).42 These findings support our contention that the increase in miRNA levels seen in microglia after GBM-EV exposure is due, at least in part, to uptake of EVs containing these miRNAs.
Fig. 4.
GBM-derived EVs increase miR-21 and miR-451 levels and decrease c-Myc mRNA levels in primary mouse microglia. (A and B) Primary microglia were exposed to GBM-EVs for 24 and 48 h. Levels of miR-21 (A) and miR-451 (B) were quantitated (mean ± SEM, n = 5, ***P < .001). (C) Levels of c-Myc mRNA were measured (mean ± SEM, n = 6, *P < .05). (D) Schematic representation of 3′UTR of mouse c-Myc mRNA with miR-451 and miR-21 binding sites predicted using computational microRNA target software (http://www.microrna.org). (E and F) Levels of miR-21 (E) and miR-451 (F) in GL261 cells and EVs released from them were analyzed (mean ± SEM, n = 2).
Next, we examined the ability of the transferred miRNAs to downregulate target mRNAs. A number of targets are regulated by miR-451, including CAB39, MIF, and c-Myc.26,27 The levels of the miR-451 target mRNAs (CAB39 and MIF) and of the miR-21 target mRNA (PTEN) did not change significantly in primary mouse microglia upon exposure to GBM-EVs. Using target prediction software, we found that the 3′ untranslated region (UTR) of the c-Myc mRNA contains binding sites for both miR-451 and miR-21 (http://www.microrna.org),28 and levels of this common target mRNA (c-Myc) were significantly downregulated by about 50% when microglia were incubated with GBM-EVs (Fig. 4D and C). This increased sensitivity of c-Myc mRNA in microglia to miRNA inhibition may reflect both the low levels of this mRNA in these cells, compared with the other target mRNAs tested (Supplementary Fig. S5E), and its being a target for both miR-451 and miR-21 (Fig. 4D; Supplementary Fig. S5D). When we performed the same experiment using human microglia exposed to GBM-EVs, marked decreases in mRNA levels of c-Myc, MIF, and CAB39 were noted (Supplementary Fig. S6D–G). For mouse microglia we also evaluated the relative levels of proteins encoded in these target mRNAs by western blot analysis, with apparent decreases after GBM-EV exposure for c-Myc, PTEN, and MIF, but not CAB39 (Supplementary Fig. S5F–I). Amounts of human microglia were insufficient for western blot analysis.
In vivo Assessment of Uptake of Glioma-derived EVs by Brain Microglia and Monocytes/Macrophages
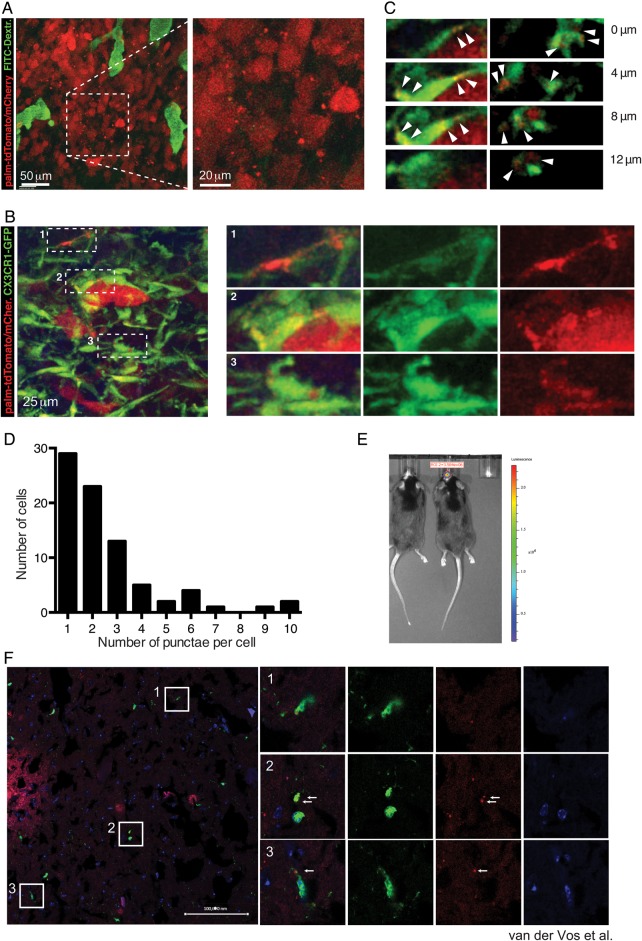
To investigate whether myeloid cells of the tumor brain stroma take up glioma-derived EVs in vivo, we used the syngeneic GL261 model.14 EVs derived from GL261 cells have elevated levels of miR-21 and miR-451 compared with these tumor cells (Fig. 4E and F), and these GL261-EVs are taken up by mouse microglia in culture (Supplementary Fig. S2D). GL261-Fluc-mC-palmtdT tumors releasing red fluorescent EVs were established in the brains of C57BL/6 wild type and C57BL/6 CX3CR1GFP/+ mice, with the latter expressing GFP in microglia and monocyte-derived macrophages.16 MP-IVM analysis of tumors was conducted using cortical brain windows, starting 1 week after tumor implantation. GL261-Fluc-mC-palmtdT tumors produced red fluorescent punctae resembling vesicles or clusters of vesicles that were located directly adjacent to as well as in the spaces between tumor cells and the stroma (Fig. 5A and B). Such vesicles were not present in tumors expressing soluble fluorescent proteins15 and were therefore likely derived from tdTomato-labeled tumor cell membranes. The apparent sizes of vesicles and vesicle clusters ranged from several micrometers down to below 1 µm. However, detection by fluorescence, on the one hand, may overestimate their actual physical size, and on the other hand, very small vesicles may escape detection by our imaging system. Therefore, visualized vesicles may represent only a fraction of EVs that are produced. As we have observed in other tumor models,15 vesicles in central areas of the tumors were mostly stationary (Supplementary Video S1), while vesicles in more peripheral areas that were densely populated by CX3CR1-GFP+ microglia and monocytes/macrophages displayed more dynamic properties (Fig. 5B; Supplementary Video S2). Importantly, our recordings revealed that most of the detectable vesicles were located either on the surface or inside of GFP+ microglia/monocyte-derived macrophages (Fig. 5C) that frequently made intimate contact with individual tumor cells (Fig. 5B, subpanel 2) but were in some cases located at some distance from labeled tumor cells. In different fields of view, the frequency of CX3CR1-GFP+ cells that contained red punctae ranged from 18% to 74%, depending on the tumor area. In most CX3CR1-GFP+ cells, 1 or 2 but in some cases up to 10 individual fluorescent punctae (single vesicles or clusters) could be identified (Fig. 5D). Rotation and stacking of GFP+ cells confirmed that many red vesicles were within cells (Supplementary Video S2).
Fig. 5.
In vivo visualization of glioma-derived EVs and uptake by microglia/macrophages. (A) MP-IVM images from a GL261-Fluc-mC-palmtdT in a C57BL/6 mouse implanted with a brain window. Injected i.v. was 150 kDa fluorescein isothiocyanate–dextran to label the blood vasculature. (B) MP-IVM images from a GL261-Fluc-mC-palmtdT tumor in a CX3CR1GFP/+ mouse brain. Panels on the right show magnified subregions of panel on the left. (C) Individual sections highlighting intracellular localization of red punctae. (D) Frequency distribution of number of discernible red punctae per cell. (E) Tumor size of GL261-Fluc-mC-palmtdT cells implanted intracranially into CX3CR1+/GFP mice monitored by bioluminescence in vivo Fluc imaging. Cryosections were performed to visualize in vivo EV uptake. Released EV-like entities (palmtdT+) were readily observed around the tumor, as well as within GFP+ cells. Nuclei were visualized by DAPI. Arrows indicate red punctae within GFP+ cells. Bar = 100 000 nm.
In parallel experiments following direct injection of these tumor cells into the cortex of CX3CR1GFP/+ mouse brains, tumor growth was monitored by in vivo bioluminescence imaging (Fig. 5E). Mice were sacrificed after 18 days when tumors had formed and either analyzed by immunohistochemistry or used to isolate cells for FACS analyses. Immunohistochemistry of brains of tumor-bearing mice showed influx of GFP+ microglia/macrophages into the periphery of GL261-Fluc-mC-palmtdT tumors (Fig. 5F), as well as uptake of “red” tdT+ vesicles by GFP+ cells.
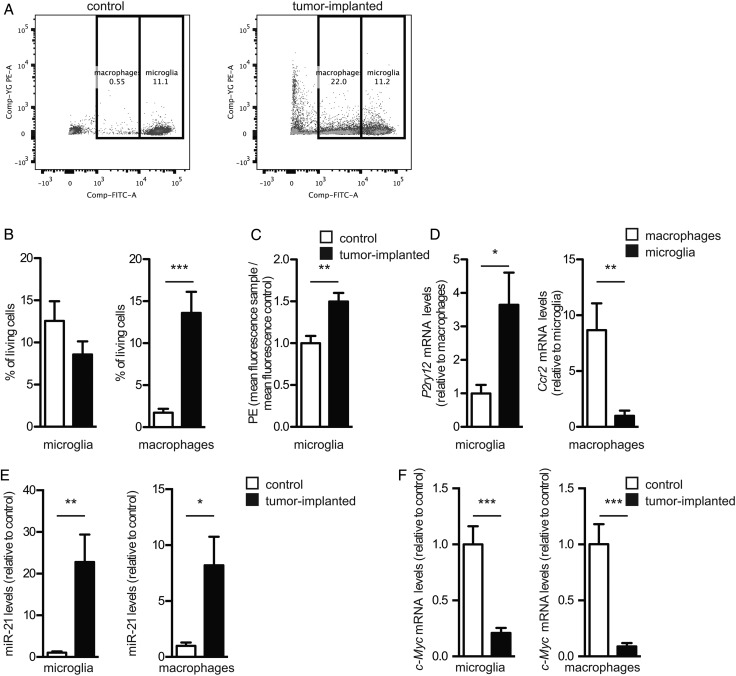
In parallel, brains of tumor-bearing and control animals were dissociated into single cells, which were resolved by FACS into fractions of tumor cells (red), microglia (high levels of GFP), and monocytes/macrophages (intermediate levels of GFP) (Fig. 6A). The numbers of monocytes/macrophages increased in the glioma-bearing brains of mice compared with control mice. While there was no significant change in the total number of microglia in the brain (Fig. 6B), there appeared to be a higher density of these cells associated with tumors (Fig. 5B). Interestingly, microglia (and to some extent macrophages) showed a significant increase in red fluorescence (phycoerythrin α [PE-A]), consistent with their uptake of mC/palm-TdTomato+ EVs released by the GBM cells in vivo (Fig. 6C). Analysis of mRNA levels in isolated cell fractions from brains showed enrichment of the mRNA for the microglia marker P2ry1229 in the microglia population and enrichment of the macrophage marker Ccr2 in the monocyte/macrophage population (Fig. 6D), confirming the validity of isolation based on GFP intensity. The levels of miR-451 and miR-21 were also measured in the isolated microglia and monocytes/macrophages. While the levels of miR-451 were below detection limits, the levels of miR-21 were 20-fold increased in microglia and 8-fold increased in monocytes/macrophages in tumor-bearing mice compared with control brains (Fig. 6E). Increased levels of miR-21 in microglia and monocytes/macrophages from brains of tumor-bearing mice correlated with a marked decrease in levels of the target c-Myc mRNA compared with controls (Fig. 6F), supporting EV-mediated functional transfer of miRNA from glioma to microglia and monocytes/macrophages in vivo.
Fig. 6.
Microglia isolated from brains of mice with glioma show increased levels of miR-21 and decreased c-Myc mRNA. (A) Representative images of FACS cells from brains of mice without and with gliomas; macrophages and microglia (GFP+, fluorescein isothiocyanate) and tumor cells (mC/palmtdTomato+, PE-A). (B) The % living FACS GFP+ cells from control brains and tumor-implanted brains were determined (mean ± SEM, ***P < .001). (C) The PE-A mean fluorescence of sorted microglia from tumor compared with control brains (**P < .01). (D) Cells sorted based on GFP+ intensity were analyzed for mRNA levels of P2ry12 and Ccr2 (*P < .05, **P < .01), (E) miR-21 levels (*P < .05, **P < .01), and (F) c-Myc levels (***P < .001). (B-F) All graphs include a total of 8 control mice and 9 mice implanted with GBM cells (mean ± SEM, n = 3).
Discussion
Tumors are known to change the phenotype of normal cells in their environs to promote tumor progression, and microglia and macrophages are key players in this process.5,6 Many interactions between gliomas and microglia/macrophages are mediated via chemokines and cytokines in the secretome. The current study supports an additional form of communication in which multipotent miRNAs are transferred from tumor cells into microglia and macrophages via EVs. Release of fluorescently labeled EVs by glioma cells and uptake by microglia/macrophages within the brain tumor environs were documented in vivo in real time using intravital microscopy. Extracellular transfer of miR-451 and miR-21 in glioma-EVs resulted in elevated levels in microglia/macrophages and associated downregulation of their target mRNAs and encoded proteins, albeit given other RNAs and proteins in these EVs, the effect may be combinatorial.
Here we demonstrate, for the first time to our knowledge, using dynamic intravital microscopy as well as histological analyses, that EVs released from tumor cells are taken up not only by microglia in culture, but also by tumor-associated microglia and monocytes/macrophages in the glioma-bearing brain. Stacking and rotational analysis of intravital images confirmed uptake of vesicles into microglia/macrophages. Consistent with this, dissociation and FACS analysis of these cells from tumor-bearing brains demonstrated that many microglia and monocytes/macrophages have marked levels of red fluorescence, presumably reflecting the uptake of tumor EVs. FACS-isolated microglia and monocytes/macrophages from tumor-bearing animals also showed both an increase in miR-21 levels and a decrease in c-Myc mRNA. These in vivo findings are consistent with functional transfer of miRNAs from glioma cells to microglia and monocytes/macrophages via exRNA vehicles.
The role of exRNAs in communication between GBM and microglia/monocytes/macrophages was evaluated both in culture and in vivo, with these myeloid-derived cells known to infiltrate brain tumors4 and their density being proportional to glioma grade.6 Primary GBM cells shed EVs in large numbers,30 and resident brain microglia are very active in endocytosis.4 EVs derived from GBM cells were avidly taken up by microglia and monocytes/macrophages in culture and in vivo as analyzed by confocal and intravital microscopy, visualizing the internalization of EVs into recipient cells using fluorescent membrane labels.15 GBM-EV uptake correlated with changes in microglia phenotype, including increased proliferation, differences in secretion of various cytokines, and increased levels of Arg-1 mRNA associated with activation. The GBM-EV fraction contained a unique repertoire of miRNAs compared with GBM cells, with 2 miRNAs—miR-451 and miR-21—being inherently highly abundant in the EVs. We exploited the low levels of these miRNAs in microglia to demonstrate increases in cellular miR-451 and miR-21 levels conferred by exposure to GBM-EVs, consistent with, but not direct proof of, the transferred miRNAs being responsible for this increase. Exposure of mouse microglia/macrophages to GBM/glioma-EVs also decreased levels of c-Myc mRNA both in culture and in vivo, presumably through binding of elevated miR-451 and miR-21 to target sites in the 3′UTR of this mRNA. The downregulation of c-Myc mRNA in mouse microglia exposed to human GBM-EVs was unexpected, as another study reported elevated levels of this message following exposure of rat microglia to conditioned medium from rat glioma cultures.31 However, our findings of decreased c-Myc mRNA levels in microglia and monocytes/macrophages in glioma-bearing mouse brain are similar to those of another group using the same mouse tumor model.32 This suggests that secreted proteins, such as cytokines, and particulate exRNA vehicles, 2 components of the secretome, have a combinatory effect and may even convey different signals to microglia, which can have either tumor supportive or antitumor properties.33
We found a unique repertoire of miRNAs in GBM-EVs compared with GBM cells, supporting a selective loading of RNAs into EVs.12 MiR-451 was the most highly enriched miRNA in GBM-EVs, as found for a number of other cell types.12,34 Different functions have been attributed to miR-451, including tumor suppression35 and deregulation of oncogenic pathways.36,37 Thus, this miRNA may be selectively exported to enhance tumor cell growth.38 The next most abundant miRNA in GBM-EVs was miR-21, with known oncogenic properties.23 The combined increase in miR-451 and miR-21 in microglia exposed to GBM-EVs in culture presumably contributes to the decrease in their shared target c-Myc mRNA, while in vivo the decrease in c-Myc mRNA in microglia and monocytes/macrophages in the glioma-bearing brain seems to be due primarily to elevation of miR-21. We were unable to detect increased levels of miR-15b, miR-146a, and miR-223 in recipient microglia, although these were also high in GBM-EVs (data not shown). The functional activity of specific miRNAs transferred into recipient cells by EVs may depend on the type of particles/vesicles within the EV fraction with which they are associated, the mechanism of uptake of these particles/vesicles by recipient cells, levels of miRNA transferred into the cytoplasm, and levels of the endogenous target mRNAs.
In this study, downregulation of the miR-21 and miR-451 target c-Myc mRNA in microglia exposed to GBM-EVs in culture and in microglia/macrophages in glioma-bearing brains supports the functional activity of EV-transferred miRNAs, although given other factors transferred by EVs the effect could be indirect. C-Myc is a transcription factor regulating expression of many genes and has been implicated in many biological processes, including growth, energy metabolism, proliferation, differentiation, and apoptosis.39 It does not act as an on-off switch in gene activation, but rather amplifies expression of many genes.40 We hypothesize that downregulation of c-Myc expression in microglia and monocytes/macrophages might facilitate shifts in gene expression patterns that accompany the activation of these cells toward a tumor supportive phenotype.6,32
By monitoring the levels of specific miRNAs which are inherently the highest in GBM-EVs and their effects on target mRNAs following EV uptake, we have elucidated one of the mechanisms by which GBM cells can influence their microenvironment. Our results confirm that EVs released by glioma cells are taken up by microglia and monocytes/macrophages in culture and within the brain environment in vivo. MiRNAs which are high in the GBM-EV fraction appear to be transferred intact into recipient microglia and monocytes/macrophages, resulting in elevated levels of these miRNAs and downregulation of a target mRNA. This supports a means of intercellular communication via exRNA in which tumor cells can manipulate the transcriptome of normal cells, as has been described for other miRNAs and noncoding RNAs in glioma and other cancer models.11–13
Supplementary Material
Funding
This work was supported by the Dutch Scientific Organisation (NWO-Rubicon) (KvdV), U19 CA179563 (supported by the NIH Common Fund through the Office of Strategic Coordination/Office of the NIH Director [ X.O.B., A.M.K., T.R.M.]), NIH/NCI P01 CA069246 (X.O.B.), Voices Against Brain Cancer (X.O.B., C.P.L.), the Richard Floor Biorepository Fund (X.O.B.), and NIH/NCI grants R01 CA150975 and CA179563 (T.R.M.). Vectors were generated through the NIH/NINDS P30 core facility, NS045776. C.P.L. was supported by the Canadian Institute of Health Research. This work was also supported by NIH/NINDS/NIA grants R01 NS059005 and U24 AI082660 (J.E.K.).
Conflict of interest statement. J.S. is an inventor of the exosome/EV technology used in this study, which has been licensed to Exosome Diagnostics, Inc. He holds equity in and is now an employee of that company. All other authors disclose no potential conflicts of interest.
Supplementary Material
References
- 1.Johnson DR, O'Neill BP. Glioblastoma survival in the United States before and during the temozolomide era. J Neurooncol. 2012;107(2):359–364. [DOI] [PubMed] [Google Scholar]
- 2.D'Asti E, Garnier D, Lee TH et al. . Oncogenic extracellular vesicles in brain tumor progression. Front Physiol. 2012;3:294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coniglio SJ, Segall JE. Review: molecular mechanism of microglia stimulated glioblastoma invasion. Matrix Biol. 2013;32(7-8):372–380. [DOI] [PubMed] [Google Scholar]
- 4.Kushchayev SV, Kushchayeva YS, Wiener PC et al. . Monocyte-derived cells of the brain and malignant gliomas: the double face of Janus. World Neurosurg. 2014;82(6):1171–1186. [DOI] [PubMed] [Google Scholar]
- 5.Wurdinger T, Deumelandt K, van der Vliet HJ et al. . Mechanisms of intimate and long-distance cross-talk between glioma and myeloid cells: how to break a vicious cycle. Biochim Biophys Acta. 2014;1846(2):560–575. [DOI] [PubMed] [Google Scholar]
- 6.Li W, Graeber MB. The molecular profile of microglia under the influence of glioma. Neuro Oncol. 2012;14(8):958–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Skog J, Würdinger T, van Rijn S et al. . Glioblastoma microvesicles transport RNA and protein that promote tumor growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10(12):1470–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arroyo JD, Chevillet JR, Kroh EM et al. . Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A. 2011;108(12):5003–5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vickers KC, Palmisano BT, Shoucri BM et al. . MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011;13(4):423–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peinado H, Alečković M, Lavotshkin S et al. . Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med. 2012;18(6):883–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bronisz A, Wang Y, Nowicki MO et al. . Extracellular vesicles modulate the glioblastoma microenvironment via a tumor suppression signaling network directed by miR-1. Cancer Res. 2014;74(3):738–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li CC, Eaton SA, Young PE et al. . Glioma microvesicles carry selectively packaged coding and noncoding RNAs which alter gene expression in recipient cells. RNA Biol. 2013;10(8):1333–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou W, Fong MY, Min Y et al. . Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell. 2014;25(4):501–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ausman JI, Shapiro WR, Rall DP. Studies on the chemotherapy of experimental brain tumors: development of an experimental model. Cancer Res. 1970;30(9):2394–2400. [PubMed] [Google Scholar]
- 15.Lai CP, Kim EY, Mempel TR et al. . Visualization and tracking of extracellular vesicle delivery and RNA translation using multiplexed reporters. Nat Commun. 2015;6:7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jung S, Aliberti J, Graemmel P et al. . Analysis of fractalkine receptor CX(3)CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol Cell Biol. 2000;20(11):4106–4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hickman SE, Allison EK, El Khoury J. Microglial dysfunction and defective beta-amyloid clearance pathways in aging Alzheimer's disease mice. J Neurosci. 2008;28(33):8354–8360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Verhaak RG, Hoadley KA, Purdom E et al. . Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Vrij J, Maas SL, Kwappenberg KM et al. . Glioblastoma-derived extracellular vesicles modify the phenotype of monocytic cells. Int J Cancer. 2015;137(7):1630–1642. [DOI] [PubMed] [Google Scholar]
- 20.Ries C. Cytokine functions of TIMP-1. Cell Mol Life Sci. 2014;71(4):659–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Richmond J, Tuzova M, Cruikshank W et al. . Regulation of cellular processes by interleukin-16 in homeostasis and cancer. J Cell Physiol. 2014;229(2):139–147. [DOI] [PubMed] [Google Scholar]
- 22.Cocco C, Pistoia V, Airoldi I. Anti-leukemic properties of IL-12, IL-23 and IL-27: differences and similarities in the control of pediatric B acute lymphoblastic leukemia. Crit Rev Oncol Hematol. 2012;83(3):310–318. [DOI] [PubMed] [Google Scholar]
- 23.Krichevsky AM, Gabriely G. miR-21: a small multi-faceted RNA. J Cell Mol Med. 2009;13(1):39–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manterola L, Guruceaga E, Gállego Pérez-Larraya J et al. . A small noncoding RNA signature found in exosomes of GBM patient serum as a diagnostic tool. Neuro Oncol. 2014;16(4):520–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bobrie A, Krumeich S, Reyal F et al. . Rab27a supports exosome-dependent and -independent mechanisms that modify the tumor microenvironment and can promote tumor progression. Cancer Res. 2012;72(19):4920–4930. [DOI] [PubMed] [Google Scholar]
- 26.Pan X, Wang R, Wang ZX. The potential role of miR-451 in cancer diagnosis, prognosis, and therapy. Mol Cancer Ther. 2013;12(7):1153–1162. [DOI] [PubMed] [Google Scholar]
- 27.Meng F, Henson R, Wehbe-Janek H et al. . MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133(2):647–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Betel D, Koppal A, Agius P et al. . Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010;11(8):R90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hickman SE, Kingery ND, Ohsumi TK et al. . The microglial sensome revealed by direct RNA sequencing. Nat Neurosci. 2013;16(12):1896–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Balaj L, Lessard R, Dai L et al. . Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat Commun. 2011;2:180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ellert-Miklaszewska A, Dabrowski M, Lipko M et al. . Molecular definition of the pro-tumorigenic phenotype of glioma-activated microglia. Glia. 2013;61(7):1178–1190. [DOI] [PubMed] [Google Scholar]
- 32.Wei J, Gabrusiewicz K, Heimberger A. The controversial role of microglia in malignant gliomas. Clin Dev Immunol. 2013;2013:285246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gabrusiewicz K, Ellert-Miklaszewska A, Lipko M et al. . Characteristics of the alternative phenotype of microglia/macrophages and its modulation in experimental gliomas. PLoS One. 2011;6(8):e23902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guduric-Fuchs J, O'Connor A, Camp B et al. . Selective extracellular vesicle-mediated export of an overlapping set of microRNAs from multiple cell types. BMC Genomics. 2012;13:357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nan Y, Han L, Zhang A et al. . MiRNA-451 plays a role as tumor suppressor in human glioma cells. Brain Res. 2010;1359:14–21. [DOI] [PubMed] [Google Scholar]
- 36.Tian Y, Nan Y, Han L et al. . MicroRNA miR-451 downregulates the PI3K/AKT pathway through CAB39 in human glioma. Int J Oncol. 2012;40(4):1105–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Z, Luo X, Ding S et al. . MicroRNA-451 regulates p38 MAPK signaling by targeting of Ywhaz and suppresses the mesangial hypertrophy in early diabetic nephropathy. FEBS Lett. 2012;586(1):20–26. [DOI] [PubMed] [Google Scholar]
- 38.Palma J, Yaddanapudi SC, Pigati L et al. . MicroRNAs are exported from malignant cells in customized particles. Nucleic Acids Res. 2012;40(18):9125–9138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Conacci-Sorrell M, McFerrin L, Eisenman RN. An overview of MYC and its interactome. Cold Spring Harb Perspect Med. 2014;4(1):1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nie Z, Hu G, Wei G et al. . c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell. 2012;151(1):68–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weinstein JN, Myers TG, O'Connor PM et al. . An information-intensive approach to the molecular pharmacology of cancer. Science. 1997;275(5298):343–349. [DOI] [PubMed] [Google Scholar]
- 42.Atai NA, Balaj L, van Veen H et al. . Heparin blocks transfer of extracellular vesicles between donor and recipient cells. J Neurooncol. 2013;115(3):343–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.